Abstract
Background
Tumors with homologous recombination deficiency (HRD) show high sensitivity to platinum salts and poly(ADP-ribose) polymerase–inhibitors in several malignancies. In colorectal cancer (CRC), the role of HRD alterations is mostly unknown.
Methods
Next-generation sequencing, whole transcriptome sequencing, and whole exome sequencing were conducted using CRC samples submitted to a commercial Clinical Laboratory Improvement Amendments certified laboratory. Tumors with pathogenic and/or presumed pathogenic mutations in 33 genes involved in the homologous recombination pathway were considered HRD, the others were homologous recombination proficient (HRP). Furthermore, tumor samples from patients enrolled in the phase III TRIBE2 study comparing upfront FOLFOXIRI+bevacizumab vs FOLFOX+bevacizumab were analyzed with next-generation sequencing. The analyses were separately conducted in microsatellite stable or proficient mismatch repair (MSS/pMMR) and microsatellite instable-high or deficient mismatch repair (MSI-H/dMMR) groups. All statistical tests were 2-sided.
Results
Of 9321 CRC tumors, 1270 (13.6%) and 8051 (86.4%) were HRD and HRP, respectively. HRD tumors were more frequent among MSI-H/dMMR than MSS/pMMR tumors (73.4% vs 9.5%; P < .001; q < 0.001). In MSS/pMMR group, HRD tumors were more frequently tumor mutational burden high (8.1% vs 2.2%; P < .001; q < 0.001) and PD-L1 positive (5.0% vs 2.4%; P < .001; q = 0.001), enriched in all immune cell and fibroblast populations and genomic loss of heterozygosity-high (16.2% vs 9.5%; P = .03). In the TRIBE2 study, patients with MSS/pMMR and HRD tumors (10.7%) showed longer overall survival compared with MSS/pMMR and HRP tumors (40.2 vs 23.8 months; hazard ratio [HR] = 0.66, 95% confidence interval [CI] = 0.45 to 0.98; P = .04). Consistent results were reported in the multivariable model (HR = 0.67, 95% CI = 0.45 to 1.02; P = .07). No interaction effect was evident between homologous recombination groups and treatment arm.
Conclusions
HRD tumors are a distinctive subgroup of MSS/pMMR CRCs with specific molecular and prognostic characteristics. The potential efficacy of agents targeting the homologous recombination system and immune checkpoint inhibitors in this subgroup is worthy of clinical investigation.
BRCA1 and BRCA2 proteins are involved in DNA repair through the homologous recombination system. In tumors bearing alterations in BRCA1 and BRCA2 genes, this system is largely ineffective and fails to repair DNA double-strand breaks (1). Moreover, genomic alterations other than BRCA1 and 2 mutations may cause homologous recombination deficiency (HRD) (2-4). The consequence of these alterations is that tumor cells either progress to programmed cell death or attempt to repair the resultant DNA lesions through the nonhomologous end-joining pathway, including poly(ADP-ribose) polymerase (PARP) proteins (5,6).
From a clinical point of view, this translates into high sensitivity to platinum salts that are able to induce covalent cross-links within the DNA double helix, frequently resulting in double-strand breaks, and to PARP-inhibitors that lead to a synthetic lethal response by blocking the nonhomologous end-joining pathway (7–10). Recently, the use of PARP inhibitors has entered clinical practice for the treatment of ovarian, breast, and more recently, pancreatic and prostate cancers, carrying deleterious BRCA1/2 mutations or HRD (11–14).
In colorectal cancer (CRC), the role of homologous recombination alterations is still widely unknown, and few data about their clinical impact are available (15,16). However, recent studies reported on the emerging role of germline pathogenic variants of BRCA1, ATM, and PALB2 as risk factors for CRC, particularly for early onset CRC, and showed that germline or somatic alterations in homologous recombination genes are carried by up to 15% of CRCs (17–20).
Drawing from these considerations, we performed a large-scale molecular study using a comprehensive tumor profiling platform to address genomic and transcriptomic alterations in key homologous recombination pathways and their association with common clinical and molecular features in CRC patients. In addition, a more extensive clinical, molecular, and prognostic characterization with regard to these genomic alterations was conducted among metastatic CRC patients enrolled in the randomized TRIBE2 trial of first-line chemotherapy plus bevacizumab (bev) (21). Considering the high enrichment of homologous recombination–related gene mutations in microsatellite instability high or mismatch repair deficient (MSI-H/dMMR) tumors (22), the analyses were separately conducted in microsatellite stable or mismatch repair proficient (MSS/pMMR) and MSI-H/dMMR groups.
Methods
Study Population
A total of 9321 formalin-fixed paraffin-embedded (FFPE) tumor samples from CRC patients were analyzed for molecular profiles by a commercial Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory (Caris Life Sciences, Phoenix, AZ). Next-generation sequencing (NGS) and whole transcriptome sequencing (WTS) data were available for 9321 and 1529 tumors, respectively.
In addition, 296 tumor samples derived from patients enrolled in the phase III TRIBE2 study (21) were analyzed with NGS. TRIBE2 randomly assigned 679 unresectable previously untreated mCRC patients to receive 5-Fluorouracil, Levofolinic acid and Oxaliplatin (FOLFOX)+bevacizumab (bev) followed by 5-Fluorouracil, Levofolinic acid and Irinotecan (FOLFIRI) +bev after disease progression or 5-Fluorouracil, Levofolinic acid, Oxaliplatin and Irinotecan (FOLFOXIRI) +bev followed by the reintroduction of the same agents after disease progression; all treatments were administrated for up to 8 cycles followed by 5-fluorouracil plus bev maintenance until disease progression, unacceptable adverse events, or consent withdrawal. For this subgroup, survival data were available as well as clinical and molecular analysis.
Definition of HRD and Homologous Recombination Proficient (HRP) Groups
In this study, we focused on 33 genes within homologous recombination pathways (4) that were included in the Caris MI TumorSeek Panel (23). These genes were categorized as involving core homologous recombination machinery (“core”) or a closely related process such as DNA damage signaling or replication-associated break repair (“related”) (4). The detailed list of genes is depicted in Supplementary Table 1 (available online). The samples with 1 or more pathogenic or presumed pathogenic mutations, categorized according to the American College of Medical Genetics and Genomics standards, in any of the 33 homologous recombination–related genes were categorized in the HRD group and the remaining samples in the HRP.
Genome, Transcriptome, and Immunohistochemistry Analyses
NGS using a custom-designed panel enriching 592 gene targets (Caris MI TumorSeek panel) and WTS were conducted using DNA and RNA isolated from FFPE samples, respectively. The consensus molecular subtype classifier was developed using RNA sequencing data collected from the WTS platform, as previously described (24).
Microsatellite (MS) and mismatch repair system (MMR) status was assessed with a combination method using immunohistochemistry, fragment analysis, and NGS, with resulting status defined as either MSI-H/dMMR or MSS/pMMR, as previously described (25,26).
Tumor mutational burden (TMB) was measured by counting all nonsynonymous missense, nonsense, inframe insertion and/or deletion, and frameshift mutations found per tumor that had not been previously described as germline alterations in dbSNP151, Genome Aggregation Database, or benign variants identified by Caris geneticists. The threshold adopted for the definition of TMB-high (TMB-H) was at least 10 mutations per megabase based on the KEYNOTE-158 trial showing higher clinical activity with pembrolizumab in patients with a TMB of at least 10 mutations per megabase across several tumor types than patients with a TMB less than 10 mutations per megabase (27). Caris Life Sciences is a participant in the Friends of Cancer Research TMB Harmonization Project (28).
PD-L1 expression was tested via immunohistochemistry using SP142 antibody (Spring Biosciences, Pleasanton, California, USA). The staining intensity on the tumor cell membrane was assessed on a semiquantitative scale: 0 for no staining, 1+ for weak staining, 2+ for moderate staining, and 3+ for strong staining. Tumors exhibiting more than 5% of tumor cells stained as 2+ or 3+ were considered PD-L1 positive.
In an independent cohort of 1120 CRC samples profiled by whole exome sequencing (WES), the functional impact of homologous recombination alterations was measured by evaluation of genomic loss of heterozygosity (LOH), defined as the percentage of examined genomic segments (max 552) with an average SNP variant frequency skewed at least 15% from the expected heterozygous frequency (50%). Tumors with a LOH of at least 16% were regarded as LOH-high, and tumors with a LOH less than 16% as LOH-low (29,30).
Ethics Approval and Consent to Participate
All patients provided written informed consent to study procedures before enrollment. TRIBE2 studies were conducted in accordance with the Declaration of Helsinki. Approval for TRIBE2 protocol was obtained from local ethics committees of participating sites.
Statistical Analysis
The χ2 test, Fisher exact test, and Mann-Whitney test were used, when appropriate, to compare clinical and molecular characteristics between HRD and HRP groups. The Microenvironment Cell Population-counter [MCP-counter (31)] was used for the quantification of the abundance of immune and stromal cell populations using WTS data. The median gene expression levels were compared between each subgroup, and the fold change was calculated. Patients with any missing data were not included in the analysis. To adjust P values for multiple hypothesis testing, the q values were calculated using the Benjamini-Hochberg method.
For TRIBE2 patients, progression-free survival (PFS), defined as the time from randomization to the evidence of disease progression or death, whichever occurred first, and overall survival (OS), defined as the time from randomization to death, were determined according to the Kaplan-Meier estimates method. Survival curves were compared using the log-rank test and hazard ratios (HR) with 95% confidence interval (CI) were estimated by Cox proportional hazards model. The impact of clinical and molecular prognostic variables on PFS and OS was first assessed in univariate analyses. Statistically significantly prognostic covariates (P < .10) were included in a multivariable Cox proportional hazard model. Hazard proportionality was assumed and verified using the goodness-of-fit χ2 test. Subgroup analyses to assess the benefit of FOLFOXIRI+Bev vs FOLFOX+Bev based on homologous recombination status in terms of PFS and OS were carried out using interaction test. The data cut-off for patients enrolled in the TRIBE2 study was December 28, 2020. All statistical analyses were conducted with SPSS v23 (IBM SPSS Statistics, Armonk, NY, USA) and JMP 13.2.1 (SAS Institute Inc, Cary, North Carolina, USA), and all tests were 2-sided at a statistical significance level set to a P value less than .05.
Results
Overall Study Population
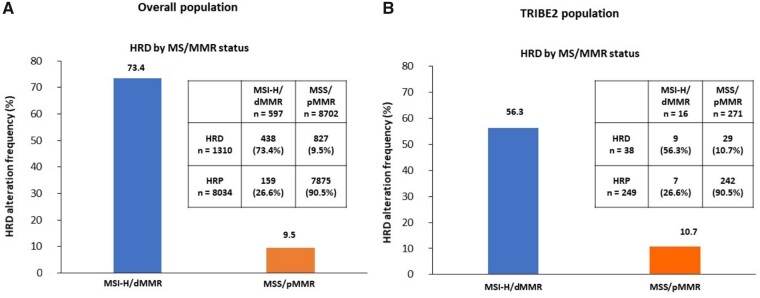
Of 9321 patients included in the platform, 1270 (13.6%) and 8051 (86.4%) were HRD and HRP, respectively. Overall, 597 (6.4%) and 8702 (93.4%) were MSI-H/dMMR and MSS/pMMR, respectively, and in 22 tumors MS status was not available. MSI-H/dMMR patients were enriched in HRD alteration compared with MSS/pMMR (73.4% vs 9.5%; P < .001; q < 0.001) (Figure 1, A).
Figure 1.
Frequency of HRD tumors in MSI-H/dMMR and MSS/pMMR cohorts. A) The overall population and (B) TRIBE2 population are shown. dMMR = deficient mismatch repair; HRD = homologous recombination deficiency; HRP = homologous recombination proficient; MMR = mismatch repair; MS = microsatellite; MSI-H = microsatellite instability-high; MSS = microsatellite stable; pMMR = proficient mismatch repair.
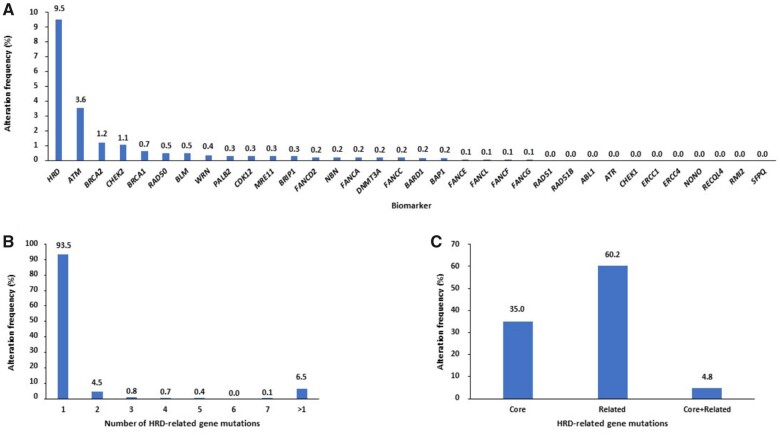
In the MSS/pMMR cohort, among 33 homologous recombination–related genes, ATM (3.6%), BRCA2 (1.2%), CHEK2 (1.1%), and BRCA1 (0.7%) were the most commonly altered (Figure 2, A). Among 827 HRD tumors, 289 (35.0%) and 498 (60.2%) showed mutations of genes involved in the core and in the related homologous recombination system, respectively. In addition, 54 (6.5%) samples showed more than 1 homologous recombination alteration, and 40 (4.8%) had mutations in both core and related homologous recombination machinery (Figure 2, B and C). In the MSI-H/dMMR cohort, among 33 homologous recombination–related genes, BRCA2 (23.8%), ATM (17.8%), NBN (12.7%), and CDK12 (10.1%) were the most commonly altered (Supplementary Figure 1, A, available online). As expected, most of MSI-H/dMMR tumors had more than 1 homologous recombination alteration (55.3%), and 29.1% of cases showed mutations in both core and related homologous recombination machinery (Supplementary Figure 1, B and C, available online).
Figure 2.
(A) Frequency of homologous recombination–related gene alterations and (B) of tumors according to number of mutated homologous recombination–related genes, stratified (C) between core and related homologous recombination systems in MSS/pMMR cohort of the overall population. HRD = homologous recombination deficiency; MSS = microsatellite stable; pMMR = proficient mismatch repair.
Patient characteristics are summarized in Table 1. In the MSS/pMMR group (n = 8702), HRD tumors (n = 827) were more frequently PD-L1 positive (5.0% vs 2.4%; P = .001; q = 0.001) and TMB-high (8.1% vs 2.2%; P < .001; q < 0.001) and had a higher median TMB (5 vs 4 mut/Mb; P < .001; q < 0.001). Similar data were reported comparing HRP tumors with subgroups of HRD tumors defined according to the presence of alterations in the core and/or related homologous recombination machinery (Supplementary Table 2, available online) or according to the specific altered genes, with the exception of ATM-mutated tumors (Supplementary Table 3, available online).
Table 1.
Patient characteristics in the overall population
| Characteristics | Overall population | MSS/pMMR population (n = 8702) |
MSI-H/dMMR population (n = 597) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HRD | HRP | P | q a | HRD | HRP | P | q a | ||
| Total, No. (%) | 9321 | 827 (9.5) | 7875 (90.5) | 438 (73.4) | 159 (26.6) | ||||
| Median age (range), y | 60 (14-90) | 60 (20-90) | 60 (14-90) | .28b | 0.32b | 67 (16-90) | 65 (24-90) | .27b | 0.32b |
| Sex, No. (%) | |||||||||
| Male | 5011 (53.8) | 431 (52.1) | 4299 (54.6) | .17c | 0.25c | 194 (44.3) | 79 (49.7) | .24c | 0.32c |
| Female | 4310 (46.2) | 396 (47.9) | 3576 (45.4) | 244 (55.7) | 80 (50.3) | ||||
| Primary tumor location, No. | 6752 | 602 | 5678 | 346 | 113 | ||||
| Left and rectum, No. (%) | 4455 (66.0) | 394 (65.4) | 3932 (69.2) | .06c | 0.09c | 84 (24.3) | 36 (31.9) | .11c | 0.16c |
| Right, No. (%) | 2297 (34.0) | 208 (34.6) | 1746 (30.8) | 262 (75.7) | 77 (68.1) | ||||
| NA, No. | 2569 | 225 | 2197 | 92 | 46 | ||||
| RAS mutational status, No. | 9316 | 827 | 7870 | 438 | 159 | ||||
| Mut, No. (%) | 4888 (52.5) | 461 (55.7) | 4245 (53.9) | .32c | 0.35c | 122 (27.9) | 48 (30.2) | .58c | 0.58c |
| WT, No. (%) | 4428 (47.5) | 366 (44.3) | 3625 (46.1) | 316 (72.1) | 111 (69.8) | ||||
| NA, No. | 5 | — | 5 | — | — | ||||
| BRAF mutational status, No. | 9312 | 825 | 7870 | 437 | 158 | ||||
| Mut, No. (%) | 846 (9.1) | 66 (8.0) | 532 (6.8) | .18c | 0.25c | 186 (42.6) | 59 (37.3) | .25c | 0.33c |
| WT, No. (%) | 8466 (90.9) | 759 (92.0) | 7338 (93.8) | 251 (57.4) | 99 (62.7) | ||||
| NA, No. | 9 | 2 | 5 | 1 | 1 | ||||
| MS/MMR status, No. | 9299 | — | — | — | — | ||||
| MSI-H/dMMR, No. (%) | 597 (6.4) | — | — | — | — | — | — | — | — |
| MSS/pMMR, No. (%) | 8702 (93.6) | — | — | — | — | ||||
| NA, No. | 22 | — | — | — | — | ||||
| TMB-high (≥10 mut/Mb), No. | 5163 | 479 | 4359 | 249 | 75 | ||||
| Yes, No. (%) | 453 (8.8) | 39 (8.1) | 97 (2.2) | <.001c | <0.001c | 248 (99.6) | 69 (92.0) | <.001c | 0.001c |
| No, No. (%) | 4710 (91.2) | 440 (91.9) | 4262 (97.8) | 1 (0.4) | 6 (8.0) | ||||
| NA, No. | 4158 | 348 | 3516 | 189 | 84 | ||||
| TMB (mut/Mb), No. | 5163 | 479 | 4359 | 249 | 75 | ||||
| Median (range) | 8 (1-513) | 5 (0-446) | 4 (0-115) | <.001b | <0.001b | 37 (4-476) | 27 (5-56) | <.001b | <0.001b |
| PD-L1, No. | 8934 | 786 | 7562 | 416 | 155 | ||||
| Yes, No. (%) | 331 (3.7) | 39 (5.0) | 178 (2.4) | <.001c | <0.001c | 79 (19.0) | 35 (22.6) | .34c | 0.36c |
| No, No. (%) | 8603 (96.3) | 747 (95.0) | 7384 (97.6) | 337 (81.0) | 120 (77.4) | ||||
| NA, No. | 387 | 41 | 313 | 22 | 4 | ||||
| CMS subtypes, No. | 1529 | 135 | 1301 | 77 | 16 | ||||
| CMS1, No. (%) | 201 (13.1) | 17 (12.6) | 122 (9.4) | .28c | 0.34c | 51 (66.2) | 11 (68.7) | .30c | 0.35c |
| CMS2, No. (%) | 449 (29.4) | 33 (24.4) | 415 (31.9) | 0 (0.0) | 1 (6.3) | ||||
| CMS3, No. (%) | 165 (10.8) | 15 (11.1) | 138 (10.6) | 10 (13.0) | 2 (12.5) | ||||
| CMS4, No. (%) | 714 (46.7) | 70 (51.9) | 626 (48.1) | 16 (20.8) | 2 (12.5) | ||||
| NA, No. | 7792 | 692 | 6574 | 361 | 143 | ||||
q is the Benjamini-Hochberg adjusted P value. All statistical tests were 2-sided. CMS = consensus molecular subtype; dMMR = deficient mismatch repair; HRD = homologous recombination deficiency; HRP = homologous recombination proficient; Mb = megabase; MMR = mismatch repair; MS = microsatellite; MSI-H = microsatellite instability high; MSS = microsatellite stable; Mut = mutation; NA = not available; pMMR = proficient mismatch repair; TMB = tumor mutational burden; WT = wild-type; — = unavailable data for subclassification and statistical analysis of clinical characteristics.
P value was calculated using a 2-sided Mann-Whitney test.
P value was calculated using a 2-sided χ2 test.
Similarly to MSS/pMMR tumors, also in the MSI-H/dMMR group (n = 597), HRD tumors (n = 438) were more frequently TMB-high (99.6% vs 92.0%; P < .001; q = 0.001) and had a higher median TMB (37 vs 27 mut/Mb; P < .001; q < 0.001) (Table 1).
MCP-counter analysis showed that HRD tumors had numerically higher levels of all immune cell and fibroblast populations than HRP irrespective of MS/MMR status (Supplementary Table 4 and Supplementary Figure 2, A and C, available online). Similarly, gene expression levels of all immune checkpoints were higher in HRD than in HRP tumors. However, in the MSI-H/dMMR subgroup, LAG3 and CTLA4 expression was slightly decreased (fold change = 0.9, not statistically significant) in HRD compared with HRP tumors (Supplementary Table 4 and Supplementary Figure 2, B and C, available online).
A total of 1194 samples were available for the homologous recombination–related gene expression analysis. The expression of homologous recombination–related genes was statistically significantly different between MSI-H/dMMR and MSS/pMMR groups. However, no substantial differences were reported between HRD and HRP groups in terms of homologous recombination–related genes expression levels in both MSS/pMMR and MSI-H/dMMR populations (Supplementary Figure 3, available online).
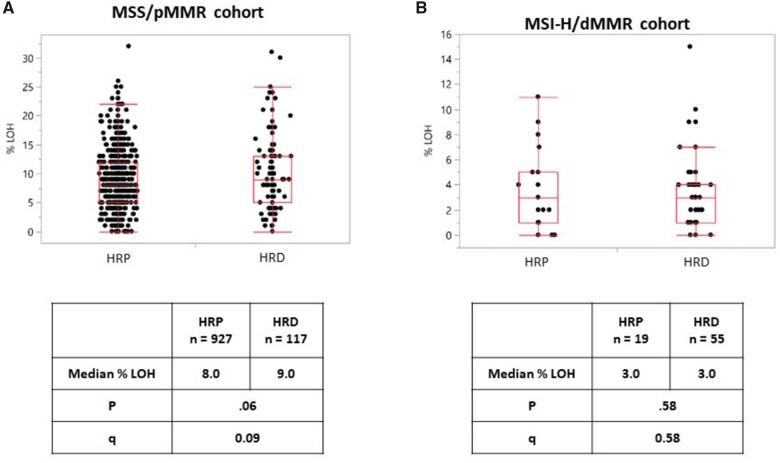
In the independent cohort of 1120 CRC samples profiled by WES, 172 (15.4%) and 948 (84.6%) were HRD and HRP, respectively. Interestingly, in the MSS group (n = 1044, 93.2%), HRD tumors (n = 117; 11.2%) were more frequently LOH-high (16.2% vs 9.5%; P = .03; q = 0.07) and showed a trend for a higher median LOH (9.0% vs 8.0%; P = .06; q = 0.09). On the other hand, in MSI-high group (n = 74; 6.6%), no difference was shown between homologous recombination subgroups in terms of median LOH (P = .58; q = 0.58), and none of these tumors was LOH-high (Table 2 and Figure 3). Among the 33 homologous recombination–related genes, only BRCA1 mutations were more frequent in LOH-high compared to LOH-low tumors in the MSS/pMMR group (26.3% vs 2.0%; P = .001; q = 0.04) (Supplementary Figure 4, available online).
Table 2.
Association between homologous recombination status and LOH
| Characteristics | Overall population | MSS/pMMR population (n = 1044) |
MSI-H/dMMR population (n = 74) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HRD | HRP | P | q a | HRD | HRP | P | q a | ||
| Total, No. (%) | 1120 | 117 (11.2) | 927 (88.8) | 55 (74.3) | 19 (25.7) | ||||
| LOH-high (≥16%), No. | 1120 | 117 | 927 | 55 | 19 | ||||
| Yes, No. (%) | 108 (9.6) | 19 (16.2) | 88 (9.5) | .03c | 0.07c | 0 (0) | 0 (0) | — | — |
| No, No. (%) | 1012 (90.4) | 98 (83.8) | 839 (90.5) | 55 (100) | 19 (100) | ||||
| LOH, No. | 1120 | 117 | 927 | 55 | 19 | ||||
| Median (range), % | 8.0 (0-34) | 9.0 (0-31) | 8.0 (0-32) | .06b | 0.09b | 3 (0-15) | 3 (0-11) | .58b | 0.58b |
q is the Benjamini-Hochberg adjusted P value. All statistical tests were 2-sided. dMMR = deficient mismatch repair; HRD = homologous recombination deficiency; HRP = homologous recombination proficient; LOH = loss of heterozygosity; MSI-H = microsatellite instability high; MSS = microsatellite stable; pMMR = proficient mismatch repair.
P value was calculated using a 2-sided Mann-Whitney test.
P value was calculated using a 2-sided χ2 test.
Figure 3.
Frequency of loss of heterozygosity according to homologous recombination status. Results for (A) MSS/pMMR population and (B) MSI-H/dMMR population are shown. LOH percentage were compared with the Mann-Whitney test and a 2-sided P value calculated; q values were calculated using the Benjamini-Hochberg method. dMMR = deficient mismatch repair; HRD = homologous recombination deficiency; HRP = homologous recombination proficient; LOH = loss of heterozygosity; MSI-H= microsatellite instability-high; MSS = microsatellite stable; pMMR = proficient mismatch repair.
TRIBE2 Study Population
Of 679 patients enrolled in the TRIBE2 study, 296 cases with available NGS assessment were included in this analysis. Baseline characteristics of patients in the NGS cohort as compared with those of patients in the intention-to-treat population are summarized in the Supplementary Table 5 (available online). Forty-one (13.9%) and 255 (86.1%) patients were HRD and HRP, respectively (Supplementary Figure 5). Overall, 16 (5.6%) and 271 (94.4%) were MSI-H/dMMR and MSS/pMMR, respectively, and in 9 tumors, MS status was not available. MSI-H/dMMR tumors were enriched in HRD alteration compared with MSS/pMMR (56.3% vs 10.7%; P < .001; q < 0.001) (Figure 1, B).
In the MSS/pMMR group, ATM (6.3%), BRCA2 (2.2%), and BARD1 (1.1%) were the most commonly altered homologous recombination–related genes (Supplementary Figure 6, A, available online). In the MSI-H/dMMR group, ATM (18.8%), BRCA2 (18.8%), RAD50 (12.5%), and WNR (12.5%) were the most commonly altered homologous recombination–related genes (Supplementary Figure 6, B, available online).
The distribution of homologous recombination alterations in the HRD group is depicted in Supplementary Figure 7 (available online). Among 41 HRD tumors, 11 (26.8%) and 24 (58.5%) showed mutations of genes involved in the core and in the related homologous recombination system, respectively. In addition, 9 (21.9%) samples showed more than 1 homologous recombination alteration, and 6 (14.6%) had mutations in both core and related homologous recombination machinery. Of 13 tumors, 7 (53.8%) with MSI-H/dMMR or POLE mutation or TMB-high had more than 1 homologous recombination–related alteration.
No substantial differences in baseline characteristics were evident among HRD and HRP tumors in both MSS/pMMR and MSI-H/dMMR groups (Supplementary Table 6, available online). However, in the MSS/pMMR subgroup, HRD tumors had a higher median TMB (6 vs 5 mut/Mb; P = .008; q = 0.01) and were more frequently TMB-high (13.6% vs 3.8%; P = .08; q = 0.11). Interestingly, among MSS/pMMR patients, 2 HRD tumors with a TMB of more than 150 mut/Mb showed mutations in the POLE proofreading domain.
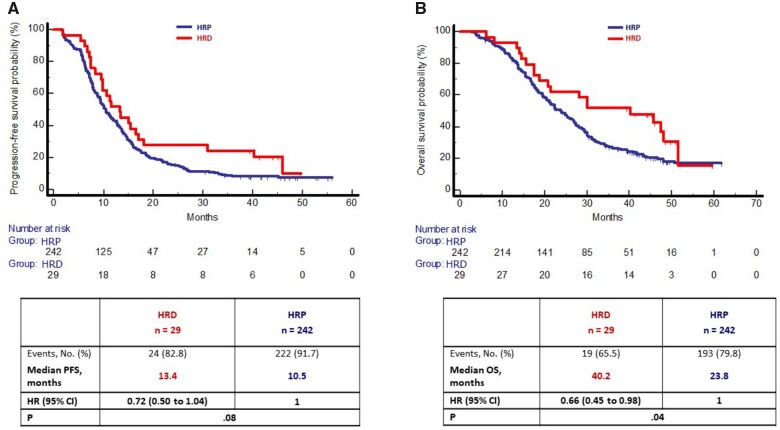
In the MSS/pMMR population, at a median follow-up of 47.7 months, progression and death events occurred in 246 (90.7%) and 212 (78.2%) patients, respectively. Median PFS was 13.4 months in the HRD group and 10.5 months in the HRP group (HR = 0.72, 95% CI = 0.50 to 1.04; P = .08) (Figure 4, A), and median OS was 40.2 and 23.8 months, respectively (HR = 0.66, 95% CI = 0.45 to 0.98; P = .04) (Figure 4, B). In the multivariable model, the trend of better prognosis for patients bearing HRD tumors was confirmed in terms of both PFS (HR = 0.67, 95% CI = 0.45 to 1.02; P = .07) and OS (HR = 0.64, 95% CI = 0.39 to 1.03; P = .07) (Supplementary Table 7, available online). No interaction effect was observed between homologous recombination groups and treatment arm in terms of both PFS (Pinteraction = 0.23) (Supplementary Figure 8, A, available online) and OS (Pinteraction = 0.67) (Supplementary Figure 8, B, available online). However, among patients in the HRP group, those treated with upfront FOLFOXIRI+Bev reported statistically significantly longer PFS (12.7 vs 9.4 months; HR = 0.63, 95% CI = 0.48 to 0.82; P < .001) and OS (27.4 vs 22.0 months; HR = 0.75, 95% CI = 0.56 to 0.99; P = .045). On the other hand, no difference was observed among patients with HRD tumors in terms of PFS (P = .87) and OS (P = .87).
Figure 4.
Kaplan-Meier curves of (A) progression-free survival and (B) overall survival according to HR status in MSS/pMMR cohort of the TRIBE2 population. Survival curves were compared, and P values determined using a 2-sided log-rank test. Hazard ratios with 95% confidence interval were estimated by Cox proportional hazards model. CI = confidence interval; HR = hazard ratio; HRD: homologous repair deficiency; HRP = homologous recombination proficient; MSS = microsatellite stable; OS = overall survival; pMMR = proficient mismatch repair; PFS = progression-free survival.
Discussion
Homologous recombination–related alterations are novel targets for platinum compounds and PARP inhibitors across different cancer types initially including breast and ovarian tumors and currently prostate and pancreatic cancers with HRD (11–14). In CRC, the role of these alterations is poorly investigated, and only a few data about their clinical impact are available (15,16). In a recent paper by Arai et al. (22), alterations in DNA damage response pathways are described by adopting a different selection of genes to define HRD and HRP. In the present analysis, we mainly focused on the MSS/pMMR subgroup of mCRC to limit the confounding effect related to the higher frequency of HRD alterations in the MSI-H/dMMR subgroup and to disclose the potential therapeutic implications of HRD alterations. We also evaluated the HRD phenotype by means of LOH analysis, and because of the availability of a well-annotated series of patients, we explored the prognostic impact of HRD and the potential predictive implications with regard to the intensification of the upfront chemotherapy backbone.
Using a large dataset of more than 9300 FFPE colorectal cancers, we were able to reliably characterize CRCs bearing alterations in the homologous recombination pathways. Considering that HRD tumors were strongly enriched in the MSI-H/dMMR subgroup, likely as a consequence of the underlying DNA repair defect, we separately analyzed these 2 populations to better identify characteristics of HRD tumors. HRD tumors accounted for approximately 10% of MSS/pMMR CRCs, were more frequently TMB-high and PD-L1 positive, and showed an immune-enriched microenvironment. These results were supported by preclinical studies showing that deficient damage response systems increase immunogenicity and, by the evidence of higher neoantigen loads, tumor-infiltrating lymphocyte, TMB, and PD-L1 expression in tumors with a HRD phenotype (32–36). These findings may have important therapeutic implications. Indeed, CRCs bearing homologous recombination pathway alterations could be sensitive not only to platinum compounds and PARP inhibitors but also to immune checkpoint inhibitors or to combinations of these agents (ie, anti-PD1/PD-L1 in combinations to oxaliplatin-based chemotherapy or PARP inhibitors). Recently, a phase I and II study showed promising antitumor activity and safety with the combination of the PARP inhibitor olaparib and the anti–PD-L1 durvalumab in patients with BRCA1/2-mutated metastatic breast cancer (37). However, specific studies are needed in mCRC with homologous recombination–related alterations to assess the efficacy and safety of agents targeting homologous recombination system such as PARP, ATR, ATM, WEE1, and CHK1/2 inhibitors (38,39) and immune checkpoint inhibitors, or their combinations.
Because only tumor tissues were available, we could not determine whether mutations affecting homologous recombination–related genes were somatic or germline or mono- or bi-allelic. However, a previous pancancer analysis revealed that bi-allelic pathogenic alterations in homologous recombination system–related genes were more often associated with the genomic features of HRD than monoallelic mutations (4). To this regard, we assessed the association between homologous recombination alterations and genomic LOH, a signature of HRD, in an independent cohort of 1120 CRC samples profiled by WES. Actually, only HRD tumors belonging to the MSS/pMMR group were enriched in LOH-high. On the other hand, no LOH-high tumor was found in the MSI-H/dMMR group, irrespectively of homologous recombination status. As a consequence, homologous recombination alterations in MSI-H/dMMR tumors may be regarded as passenger mutations, and agents targeting the homologous recombination system may be ineffective. Conversely, in MSS/pMMR tumors, homologous recombination alterations are more likely to be drivers and therefore targetable. The low incidence of LOH-high in HRD tumors of the MSS/pMMR population (16.2%) could suggest that most alterations of homologous recombination–related genes are monoallelic or passenger also in this subgroup. At the same time, even if other genomic signatures of HRD (ie, Telomeric Allelic Imbalance, Large-scale State Transitions and Signature 3) (40–43) were not assessed in our study, genomic features associated with homologous recombination repair diagnostic assays do not entirely capture colorectal cancer tumors with HRD pathway and susceptible to PARP inhibition (15). In a previous single-arm phase II study, the PARP inhibitor olaparib showed no activity in chemorefractory mCRC patients both in MSS/pMMR and MSI-H/dMMR groups (44). Based on present data, only patients with HRD MSS/pMMR tumors should be included in future trials assessing the role of agents targeting the homologous recombination system.
Although information about the stage of disease and survival outcome of patients included in the overall population were lacking, these data were available for patients included in the TRIBE2 study (21), whose samples were analyzed with the same molecular profiling assay. In this homogeneous cohort of mCRC patients, we were able to confirm the enrichment of HRD among MSI-H/dMMR tumors, an incidence of homologous recombination–related alterations around 10% in the MSS/pMMR group, and the association with high TMB among MSS/pMMR tumors.
Moreover, as already reported in ovarian cancer (45,46), HRD mCRC patients had a better prognosis in patients with MSS/pMMR tumors with a positive trend confirmed in the multivariable model. No formal interaction effect was observed between homologous recombination status and treatment arm, but the benefit from the addition of irinotecan to FOLFOX+Bev seems quite limited in HRD tumors. This may be explained by the higher sensitivity to oxaliplatin of these tumors (15) where the added value of the intensification of the chemotherapy backbone may be less relevant.
Unfortunately, a unanimously accepted gene set for the definition of HRD is currently lacking, and the published lists of genes are heterogeneous (4,20,45,47,48). Moving from the widest list of genes published by Riaz et al. including 102 genes (4), we focused on the 33 genes assessed by the Caris panel (23). As a consequence, we acknowledge that the incidence of homologous recombination–related alterations might be even higher than reported in our study.
In conclusion, HRD tumors are a distinctive subgroup of MSS/pMMR CRCs with specific molecular and prognostic characteristics. The potential efficacy of novel treatments, including agents targeting the homologous recombination system and immune checkpoint inhibitors, in this subgroup is worthy of clinical investigation. Further efforts are needed to standardize genomic or functional tests to define HRD and to determine its predictive value.
Funding
The study was supported by GONO (Gruppo Oncologico del Nord Ovest) and ARCO Foundations, and by Bando Salute Regione Toscana “In bilico project” (no grant number applicable).
Notes
Role of the funders: The funders had no role in the writing of this article or the decision to submit it for publication.
Disclosures: AE: Caris employee. JZ: Caris employee. JX: Caris employee. PS: Caris employee. MO: Caris employee with equity. JA: Caris employee. DS: Caris employee. DR: travel, accommodations and expenses—Takeda. SL: consulting or advisory role—Amgen, Lilly, Merck Serono, Astra Zeneca, Incyte, Daiichi-Sankyo, Bristol-Myers Squibb and Servier Speakers’ Bureau—Bristol-Myers Squibb, Lilly, Merck Serono, Roche, Pierre-Fabre, GSK, Amgen and Servier Research funding—Amgen, Bayer, Roche, Lilly, Astra Zeneca, Bristol-Myers Squibb and Merck Serono. FP: honoraria/speaker’s bureau from Roche, Amgen, Merck Serono, Lilly, Sanofi, Bayer, Servier; research grants from BMS and AstraZeneca. GT: advisory boards from Lilly, Amgen, Novartis. AF: honoraria—Amgen, Lilly, Merck, Roche and Servier Consulting or advisory role—Amgen, Bayer, Bristol-Myers Squibb, Lilly, Merck, Roche and Servier Research funding—Amgen (Inst), Bayer (Inst), Merck (Inst), MSD (Inst), Roche (Inst), Sanofi (Inst) and Servier (Inst) Travel, accommodations and expenses—Amgen, Bayer, Merck, Roche and Servier. MK: Caris employee, Consultant of Merck CC: honoraria—Amgen, Bayer, Merck, Roche and Servier Consulting or advisory role—Amgen, Bayer, MSD, Roche Speakers’ Bureau—Servier Research funding—Bayer, Merck, Servier Travel, accommodations and expenses—Roche and Servier. All other authors have declared no conflicts of interest.
Author contributions: Conceptualization: RM, CC, AE, MK, HA, HJL. Methodology: RM, CC, AE, MK, HA, HJL. Resources: RM, DR, AE, JX, PS, MO, JA, DS. Supervision: RM, AE, JX, PS, MO, JA, DS. Data curation: RM, CC, AE, MK. Formal analysis: RM, DR, AE, JX, PS, MO, JA, DS. Writing—original draft: RM, CC. Writing—review and editing: all authors.
Acknowledgements: The authors are grateful to Caris Life Science for molecular analyses, GONO and ARCO Foundations, to all participating patients and their families, and to the GONO investigators from the participating Italian centres.
Supplementary Material
Data Availability
Datasets supporting the results of this work are available to editors, referees, and readers promptly upon request.
References
- 1. Roy R, Chun J, Powell SN.. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12(1):68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pilié PG, Tang C, Mills GB, Yap TA.. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16(2):81–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lord CJ, Ashworth A.. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi:10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- 4. Riaz N, Blecua P, Lim RS, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8(1):857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. [DOI] [PubMed] [Google Scholar]
- 6. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumors with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. [DOI] [PubMed] [Google Scholar]
- 7. Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123–134. [DOI] [PubMed] [Google Scholar]
- 8. Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28(15):2512–2519. [DOI] [PubMed] [Google Scholar]
- 9. Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–244. [DOI] [PubMed] [Google Scholar]
- 10. Kummar S, Kinders R, Gutierrez ME, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27(16):2705–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mirza MR, Coleman RL, González-Martín A, et al. The forefront of ovarian cancer therapy: update on PARP inhibitors. Ann Oncol. 2020;31(9):1148–1159. [DOI] [PubMed] [Google Scholar]
- 12. Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robson M, Im S-A, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. [DOI] [PubMed] [Google Scholar]
- 14. de Bono J, Mateo J, Fizazi K, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382(22):2091–2102. [DOI] [PubMed] [Google Scholar]
- 15. Arena S, Corti G, Durinikova E, et al. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin Cancer Res. 2020;26(6):1372–1384. [DOI] [PubMed] [Google Scholar]
- 16. Reilly NM, Novara L, Di Nicolantonio F, Bardelli A.. Exploiting DNA repair defects in colorectal cancer. Mol Oncol. 2019;13(4):681–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oh M, McBride A, Yun S, et al. BRCA1 and BRCA2 gene mutations and colorectal cancer risk: systematic review and meta-analysis. J Natl Cancer Inst. 2018;110(11):1178–1189. [DOI] [PubMed] [Google Scholar]
- 18. Mauri G, Arena S, Siena S, Bardelli A, Sartore-Bianchi A.. The DNA damage response pathway as a land of therapeutic opportunities for colorectal cancer. Ann Oncol. 2020;31(9):1135–1147. [DOI] [PubMed] [Google Scholar]
- 19. Knijnenburg TA, Wang L, Zimmermann MT, et al. ; for the Cancer Genome Atlas Research Network. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. J Clin Oncol Precis Oncol. 2018;PO.17.00286:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cremolini C, Antoniotti C, Rossini D, et al. Upfront FOLFOXIRI plus bevacizumab and reintroduction after progression versus mFOLFOX6 plus bevacizumab followed by FOLFIRI plus bevacizumab in the treatment of patients with metastatic colorectal cancer (TRIBE2): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2020;21(4):497–507. [DOI] [PubMed] [Google Scholar]
- 22. Arai H, Elliott A, Xiu J, et al. The landscape of alterations in DNA damage response pathways in colorectal cancer. Clin Cancer Res. 2021;27(11):3234–3242. [DOI] [PubMed] [Google Scholar]
- 23.Caris Molecular Intelligence; Caris Life Sciences. June 29, 2020. carismolecularintelligence.com/wp-content/uploads/2018/10/MI-Tumor-Seek-Profile-Menu.pdf. Accessed January 24, 2021.
- 24. Abraham JP, Magee D, Cremolini C, et al. Clinical validation of a machine-learning-derived signature predictive of outcomes from first-line oxaliplatin-based chemotherapy in advanced colorectal cancer. Clin Cancer Res. 2021;27(4):1174–1183. [DOI] [PubMed] [Google Scholar]
- 25. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J.. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7(3):746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salem ME, Puccini A, Grothey A, et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD-L1 expression in a large patient cohort of gastrointestinal cancers. Mol Cancer Res. 2018;16(5):805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marabelle A, Le DT, Ascierto PA, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2020;38(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merino DM, McShane LM, Fabrizio D, et al. ; for the TMB Harmonization Consortium. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer. 2020;8(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coleman RL, Swisher EM, Oza AM, et al. Refinement of prespecified cutoff for genomic loss of heterozygosity (LOH) in ARIEL2 part 1: a phase II study of rucaparib in patients (pts) with high grade ovarian carcinoma (HGOC). J Clin Oncol. 2016;34(suppl 15):5540–5540. [Google Scholar]
- 30. Coleman RL, Brady MF, Herzog TJ, et al. Bevacizumab and paclitaxel-carboplatin chemotherapy and secondary cytoreduction in recurrent, platinum-sensitive ovarian cancer (NRG Oncology/Gynecologic Oncology Group study GOG-0213): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18(6):779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Becht E, de Reyniès A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22(16):4057–4066. [DOI] [PubMed] [Google Scholar]
- 32. Sen T, Rodriguez BL, Chen L, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9(5):646–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parkes EE, Walker SM, Taggart LE, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109(1):djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teo MY, Bambury RM, Zabor EC, et al. DNA damage response and repair gene alterations are associated with improved survival in patients with platinum-treated advanced urothelial carcinoma. Clin Cancer Res. 2017;23(14):3610–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chae YK, Anker JF, Oh MS, et al. Mutations in DNA repair genes are associated with increased neoantigen burden and a distinct immunophenotype in lung squamous cell carcinoma. Sci Rep. 2019;9(1):3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park S, Lee H, Lee B, et al. DNA damage response and repair pathway alteration and its association with tumor mutation burden and platinum-based chemotherapy in SCLC. J Thorac Oncol. 2019;14(9):1640–1650. [DOI] [PubMed] [Google Scholar]
- 37. Domchek SM, Postel-Vinay S, Im S-A, et al. Olaparib and durvalumab in patients with germline BRCA-mutated metastatic breast cancer (MEDIOLA): an open-label, multicentre, phase 1/2, basket study. Lancet Oncol. 2020;21(9):1155–1164. [DOI] [PubMed] [Google Scholar]
- 38. Blackford AN, Jackson SP.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017;66(6):801–817. [DOI] [PubMed] [Google Scholar]
- 39. Forment JV, O’Connor MJ.. Targeting the replication stress response in cancer. Pharmacol Ther. 2018;188:155–167. [DOI] [PubMed] [Google Scholar]
- 40. Birkbak NJ, Wang ZC, Kim J-Y, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Popova T, Manié E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–5462. [DOI] [PubMed] [Google Scholar]
- 42. Telli ML, Timms KM, Reid J, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22(15):3764–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. ; for the ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leichman L, Groshen S, O’Neil BH, et al. Phase II study of olaparib (AZD-2281) after standard systemic therapies for disseminated colorectal cancer. Oncologist. 2016;21(2):172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Konstantinopoulos PA, Spentzos D, Karlan BY, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28(22):3555–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park W, Chen J, Chou JF, et al. Genomic methods identify homologous recombination deficiency in pancreas adenocarcinoma and optimize treatment selection. Clin Cancer Res. 2020;26(13):3239–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets supporting the results of this work are available to editors, referees, and readers promptly upon request.