Abstract
The diversity of herbivorous insects is attributed to their propensity to specialize on toxic plants. In an evolutionary twist, toxins betray the identity of their bearers when herbivores coopt them as cues for host-plant finding, but the evolutionary mechanisms underlying this phenomenon are poorly understood. We focused on Scaptomyza flava, an herbivorous drosophilid specialized on isothiocyanate (ITC)-producing (Brassicales) plants, and identified Or67b paralogs that were triplicated as mustard-specific herbivory evolved. Using in vivo heterologous systems for the expression of olfactory receptors, we found that S. flava Or67bs, but not the homologs from microbe-feeding relatives, responded selectively to ITCs, each paralog detecting different ITC subsets. Consistent with this, S. flava was attracted to ITCs, as was Drosophila melanogaster expressing S. flava Or67b3 in the homologous Or67b olfactory circuit. ITCs were likely coopted as olfactory attractants through gene duplication and functional specialization (neofunctionalization and subfunctionalization) in S. flava, a recently derived herbivore.
Keywords: Drosophila melanogaster, Scaptomyza flava, herbivory, evolution, olfaction, isothiocyanate, chemoreceptor, SSR, olfactory receptor, wasabi, Brassicales, Or67b, gene duplication, neofunctionalization, subfunctionalization, specialization, olfactory specialization
Introduction
Many plant molecules used in food, agriculture, and medicine first evolved as defenses against natural enemies (Fraenkel 1959). Among the most familiar are reactive electrophiles that produce pain when eaten, including diallyl disulfide and thiosulfinates in Alliaceae (e.g., garlic), α, β-unsaturated aldehydes in Lauraceae (e.g., cinnamon), and isothiocyanates (ITCs) in mustards (Brassicaceae) and relatives in the Brassicales (e.g., arugula, radish, and wasabi). These electrophiles activate the “wasabi taste receptor” TrpA1, which is conserved in flies and humans (Kang et al. 2010). Although ITCs are potent natural insecticides aversive to most insects, including Drosophila melanogaster (Lichtenstein et al. 1962), some insect species are specialized on ITC-bearing Brassicales. This insect–plant interaction has been important in advancing the field of coevolution (Berenbaum and Zangerl 2008).
Brassicales specialists from many insect orders (e.g., Diptera; Eckenrode and Arn 1972, Heteroptera; Pivnick et al. 1991, Hemiptera; Demirel and Cranshaw 2006, and Lepidoptera; Pivnick et al. 1994) can be trapped with ITC baits in crop fields, revealing an evolutionary twist of fate in which ancestrally aversive electrophiles are coopted as olfactory cues for host-plant finding (Kergunteuil et al. 2012). Identifying the mechanisms that allow insects to exploit plants containing toxic chemicals can help us to understand the evolution of herbivorous species, 90% of which are specialized on a limited set of host plants (Bernays and Graham 1988).
Scaptomyza represents a compelling genus to investigate how specialist herbivores evolved to coopt plant defenses as host finding cues. Phylogenetically nested within Drosophila, Scaptomyza is a sister group to the Hawaiian Drosophila radiation (Lapoint et al. 2013), and contains many herbivorous species with varying degrees of specialization on Brassicales and Caryophyllaceae (Gloss et al. 2019) (fig. 1A). Adult females create feeding punctures on the undersides of living leaves using sclerotized dentate ovipositors (Pelaez et al. 2020), and larvae mine the mesophyll of leaf tissue (Whiteman et al. 2011), an unusual drosophilid life history. Scaptomyzaflava specializes on Brassicales (Mitchell-Olds 2001) and like humans and D. melanogaster, uses the mercapturic pathway to detoxify ITCs (Gloss et al. 2014). Scaptomyzaflava was first reported from Arabidopsis thaliana in North America (Chittenden 1902). Thus, genomic and genetic tools of both Arabidopsis and Drosophila can be utilized to dissect both sides of the plant-herbivore equation. Herbivory evolved ca. 10–15 Ma in Scaptomyza (fig. 1A) and so it provides an useful context to understand the evolutionary and functional mechanisms underlying chemosensory specialization, because the major herbivorous insect radiations (e.g., Lepidoptera; Kawahara et al. 2019, Phytophaga; McKenna et al. 2019) are far more ancient in origin. Scaptomyzaflava lost three olfactory receptor (Or) genes encoding canonical yeast-associated receptors compared with microbe-feeding Drosophila (Goldman-Huertas et al. 2015). However, the loss of function does not explain how herbivorous Scaptomyza species evolved attraction to Brassicales plants.
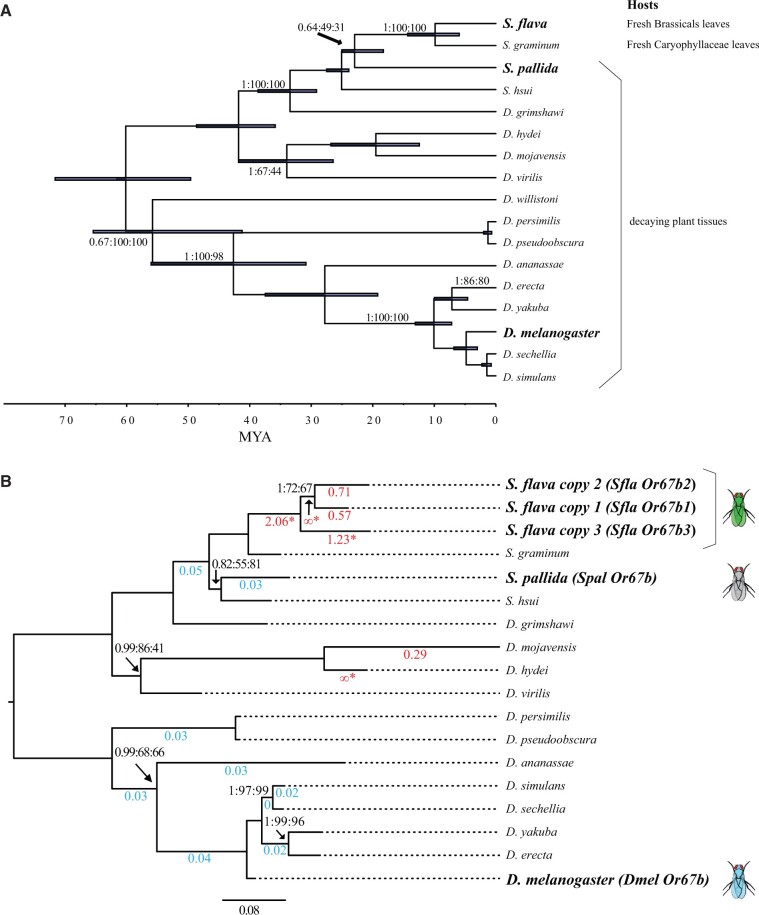
Fig. 1.
Phylogeny of Scaptomyza and the orthologs of the odorant receptor Or67b (A) Time-calibrated Bayesian chronogram of Scaptomyza and Drosophila spp. inferred from nine protein coding and two ribosomal genes. Bars indicate 95% highest posterior density (HPD) age estimates in millions of years ago (Ma). Tree topology labeled as follows: Posterior Probability (PP):ML BS: Parsimony BS on branches. Scale bar proportional to Ma. (B) ML phylogeny reconstructed based on the coding sequence of Or67b orthologs from 12 Drosophila species, S. pallida, S. hsui, S. graminum, and S. flava. Source species for the Or67b coding sequences crossed into the empty neuron systems in subsequent figures are indicated. Support values are only indicated for bifurcations with less than 100% support in any analysis, labeled as in (A). Branches with significant support (FDR P-value < 0.05) for dN/dS values different from the background rate are indicated with colored branch labels (blue where the foreground rate is less than the background, and red/enlarged fonts where dN/dS is greater than the background). Only S. flava and D. mojavensis branches have significantly elevated dN/dS according to branch model tests. Scaptomyza flava, S. pallida, and D. melanogaster labeled identically to (A). Scale bar units are substitutions per site.
Gain of function through gene duplication and subsequent divergence plays an important role in evolutionary innovation and is often associated with trophic transitions (Ohno 1970). Gene and whole genome duplications in Brassicales in the last 90 My resulted in the evolution of glucosinolates, the precursors of ITC compounds (Edger et al. 2015). Reciprocally, in Brassicales-specialist pierid butterflies, enzymes that divert hydrolysis of aliphatic glucosinolates away from ITC production to less toxic nitriles evolved in tandem, underpinning their diversification, and highlighting the importance of gene family evolution in this coevolutionary arms race (Wheat et al. 2007). Although our understanding of detoxification in plant-herbivore systems has grown rapidly over the past decade (Heckel 2018), the evolutionary mechanisms underlying the chemosensory basis of host plant orientation and finding are less clear. Here we investigated how the evolution of olfactory receptor genes in the S. flava lineage contributes to attraction to host-plant volatiles, including ITCs.
We took a genes-first approach to study olfactory host-plant specialization in S. flava. Three chemoreceptor protein families (olfactory receptors -Ors-, ionotropic receptors -Irs-, and nociceptive receptors -Trps-) are candidates for mediating responses to host-specific volatile electrophiles such as ITCs (Joseph and Carlson 2015). In particular, insect Ors are collectively sensitive to a variety of odorants important for feeding, reproduction, and survival, including aversive chemicals (e.g., Stensmyr et al. 2012), pheromones (e.g., Butenandt 1959; Sakurai et al. 2004; Kurtovic et al. 2007), and host-, oviposition-, and food-related attractive compounds (Dekker et al. 2006; Dweck et al. 2013; Linz et al. 2013). Accordingly, we scanned the genome sequence of S. flava to identify rapidly evolving Ors and found a lineage-specific triplication of the olfactory receptor gene Or67b. We named these paralogs Or67b1, Or67b2, and Or67b3. The coding sequences of each S. flava paralog exhibit signatures of positive selection. In contrast, Or67b is present as a single copy under strong evolutionary constraint across the Drosophilidae (Robertson et al. 2003; Guo and Kim 2007; Goldman-Huertas et al. 2015), including the microbe-feeding species Scaptomyzapallida and D. melanogaster. Our in vivo functional characterizations of all Or67b proteins from S. flava, S. pallida, and D. melanogaster show that the three S. flava Or67b paralogs, but not the Or67b orthologs from the microbe-feeding relatives, responded selectively and sensitively to volatile ITCs. Among these species, only S. flava is attracted to single ITC compounds. Sfla Or67b3 can confer odor-oriented responses toward ITCs when this Or is expressed in the D. melanogaster homologous Or67b olfactory circuit. In summary, our results suggest a relatively simple sensory mechanism by which mustard-specialist herbivores evolved olfactory attraction toward ITCs, via gene duplication followed by neofunctionalization and subfunctionalization of a specific clade of otherwise conserved Ors.
Results
Phylogenetic Analysis and Antennal Transcriptome Identify S. flava Or67b Paralogs as Candidates Mediating the Detection of ITCs
To search for candidate ITC-detecting chemosensory receptors in S. flava, we conducted a phylogenetic analysis of the Or protein sequences of S. flava and four other Drosophila species to identify Scaptomyza- and S. flava-specific gene losses and gains (supplementary fig. 1, Supplementary Material online), as well as those that were rapidly evolving at the protein level, regardless of duplication history. The Or topology was largely congruent with previous Drosophila Or gene trees (Goldman-Huertas et al. 2015), except for some deeper nodes with low bootstrap support. We fit foreground/background branch models, as implemented in PAML, to detect S. flava Ors whose coding sequences were evolving under different selection regimes compared with the Drosophila background (Yang 2007). Out of 75 S. flava branches tested, 7 branch models, related to Or63a, Or67b, and Or98a in D. melanogaster, inferred a foreground rate larger than one, consistent with a high rate of fixation of nonsynonymous mutations during positive selection. Albeit the known functions and expression patterns of individual Ors in Drosophila species cannot be directly extrapolated to those of Scaptomyza spp homologs, we noted that Or63a is only expressed in D. melanogaster larvae (Hoare et al. 2011), and the Or98a-like genes found in Scaptomyza have no D. melanogaster homologs and have not been characterized functionally. In contrast, Or67b modulates oviposition behavior in adult D. melanogaster (Chin et al. 2018), and is implicated in selection of oviposition sites in D. mojavensis (Crowley-Gall et al. 2016), making it a good candidate for the olfactory adaptation of Scaptomyza to a novel host niche. After expanding the representation of Or67b orthologs in a phylogenetic analysis, and conducting branch tests in PAML on all single branches on the Or67b phylogeny, we found significantly elevated dN/dS exclusively in S. flava paralogs and D. mojavensis Or67b (fig. 1B). Furthermore, synteny analysis confirmed that homologous regions upstream and downstream of the two nonsyntenic Or67b copies in S. flava are present in both S. pallida and D. melanogaster. Thus, the absence of these paralogs in S. pallida and D. melanogaster is likely not a genome assembly or sequencing artifact but rather, an actual absence (supplementary fig. 2A and B and files 1 and 2, Supplementary Material online).
Additionally, we analyzed the antennal transcriptomes of S. flava and S. pallida. This confirmed expression of each annotated Or67b gene copy in the two species. Relative expression of Or67b paralogs was higher in S. flava antennae than in S. pallida in all replicates compared with other Ors. This was not the case for Or98a (supplementary fig. 3 and file 3, Supplementary Material online), which further supports our choice of Or67b as a candidate for functional studies.
In summary, Or67b is uniquely triplicated in S. flava among the species analyzed, the coding sequences of these paralogs show signatures of positive selection, and the three paralogs are expressed at higher levels in S. flava antennae compared with the orthologous copy in the outgroup.
Scaptomyza flava Or67bs Respond Specifically to Mustard Plant Odors
Next, we investigated whether Sfla Or67bs acquired novel ligand-binding sensitivity toward odorants characteristic of mustard host plants. To study the odor-response profile of Or67b across the three species, we heterologously expressed their Or67b proteins in D. melanogaster antennal trichoid (at1) or antennal basiconic 3A (ab3A) olfactory sensory neurons (OSNs) lacking endogenous Ors (Dobritsa et al. 2003; Kurtovic et al. 2007). We then recorded electrophysiological responses to an array of mustard and nonmustard plant volatile organic compounds (VOCs), other odors, and odorants known to be activators of D. melanogaster Or67b (figs. 2 and 3 and supplementary fig. 3, Supplementary Material online). We first expressed each Or67b transgene in ab3A OSNs because Or67b is expressed in basiconic sensilla in D. melanogaster. Ab3A neurons expressing Sfla Or67b1, Sfla Or67b3, Spal Or67b, and Dmel Or67b showed spontaneous bursts of action potentials, as described for other Ors expressed in these neurons (Dobritsa et al. 2003), whereas neurons expressing Sfla Or67b2 showed spontaneous activity only when we expressed this protein in at1 OSNs (fig. 2A and supplementary fig. 5A, Supplementary Material online). Thus, we used the “at1 empty neuron” system for expression of Sfla Or67b2, and the “ab3A empty neuron” system for all other Or67b proteins. Note that comparisons of odor-evoked response intensity between OSNs expressing Sfla Or67b2 and those of OSNs expressing the other Or67b proteins should be interpreted cautiously (but see supplementary fig. 5, Supplementary Material online), as the response magnitude is smaller for Ors expressed in the at1 empty neuron (e.g., Syed et al. 2010).
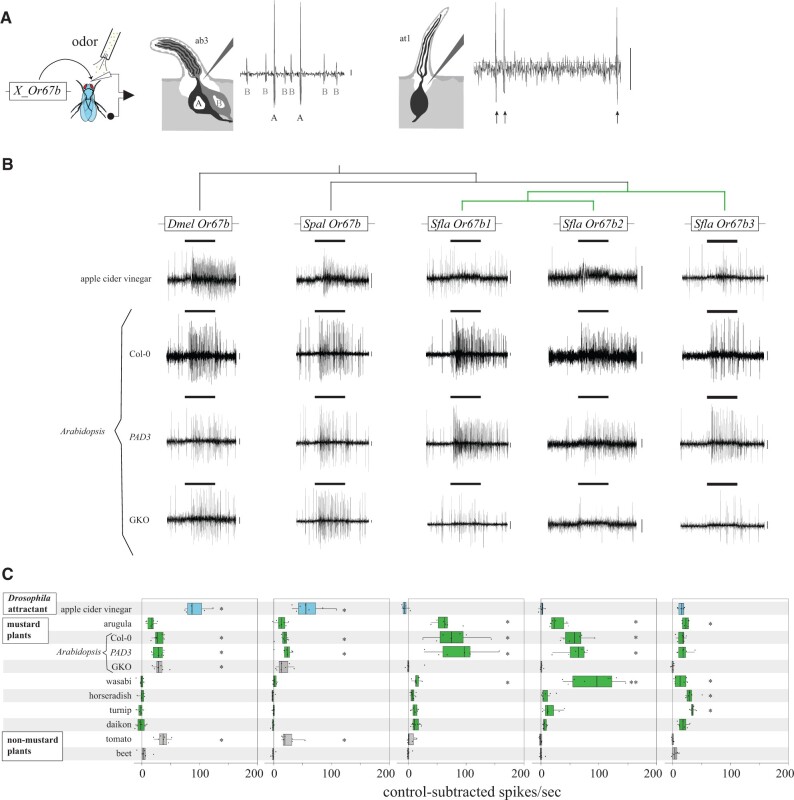
Fig. 2.
Responses of homologous Or67bs from D. melanogaster, S. pallida, and S. flava expressed in the D. melanogaster empty neuron systems to stimulation with natural odor blends. (A) Schematic representation of the single sensillum recording (SSR) using two “empty neuron systems”. Or67b proteins (X_Or67b, where X refers to the fly species) were expressed in a D. melanogaster mutant that lacks its endogenous Or22a in ab3A (antennal basiconic 3A) OSNs (Hallem and Carlson 2006; left), or Or67d in at1 (antennal trichoid 1) OSNs (Kurtovic et al. 2007; right). Note that the at1 empty neuron system was used only for expression of Sfla Or67b2, as this Or was not functional in the ab3A empty neuron system. Although comparison between the responses of this Or with other Or67b proteins should be interpreted cautiously (Syed et al. 2010), Or tuning (but not response intensity) is independent from the empty neuron system expressed for Or expression (supplementary fig. 5, Supplementary Material online). The antennal basiconic sensilla houses the A (which expresses one of the Or67b proteins) and the native intact B neuron; the A neuron has larger amplitude spikes than the B neuron, allowing discrimination of spikes originating from either neuron. The antennal trichoid sensilla houses a single OSN expressing Sfla Or67b2; arrows indicate spikes (see also supplementary fig. 4, Supplementary Material online). Calibration bars (vertical lines to the right of traces): 10 mV throughout all figures unless noted. (B) Representative electrophysiological recordings obtained from the targeted sensilla of flies expressing Or67b in OSNs in response to stimulation with apple cider vinegar, crushed rosette leaves of wild-type Col-0 Arabidopsis thaliana, and phytoalexin deficient 3 (PAD3) and aliphatic and indolic Glucosinolate Knock Out (GKO; myb28 myb29 cyp79b2 cyp79b3) A. thaliana mutants. Although all three A. thaliana genotypes have the same genetic background (Glazebrook and Ausubel 1994; Beekwilder et al. 2008), PAD3 plants are a more appropriate control for this study than GKO or Col-0, because PAD3 is deficient (as is GKO) in the production of camalexin but still produces normal levels of aliphatic or indolic glucosinolates. The bars above records indicate the onset and duration (1 s) of the stimulation throughout all figures unless otherwise noted. (C) Responses (net number of spikes/second, control subtracted, n = 6–9 obtained from 2 to 4 animals) evoked by stimulation with apple cider vinegar, odors from homogenized mustard leaves (arugula and A. thaliana), grated mustard root odors (wasabi, horseradish, turnip, and daikon), homogenized nonmustard leaf odors (tomato), and grated nonmustard root odors (beet, control). The outer edges of the horizontal bars represent the 25% and 75% quartiles, the vertical line inside the bars represents the median, and the whiskers represent 10% and 90% quartiles; each dot represents an individual response. Blue, gray, and green rectangles, respectively, show responses by stimulation with the Drosophila melanogaster attractant apple cider vinegar, non-ITC-bearing plants, and ITC-bearing plants. Asterisks indicate significant differences between the control-subtracted net number of spikes and a threshold value for responsiveness (10 spikes/second), as explained in Materials and Methods (one-sample signed rank tests; *P < 0.05, **P < 0.01). Neurons expressing Dmel Or67b and Spal Or67b, but not those expressing any of the S. flava Or67b paralogs, responded to stimulation with apple cider vinegar. Conversely, only neurons expressing S. flava Or67b paralogs responded to arugula odors (which bear ITCs). None of the S. flava paralogs responded to stimulation with the nonhost (non-ITC bearing) tomato. Dmel Or67b responded to all A. thaliana genotypes, whereas Sfla Or67b1-2 responded only to ITC-bearing A. thaliana, indicating that the presence of ITCs within plants is necessary to evoke responses from these two S. flava paralogs. Stimulation with wasabi root odors evoked responses from all Sfla Or67b paralogs but not from the Dmel or the Spal paralogs.
Fig. 3.
Responses of homologs Or67bs from D. melanogaster, S. pallida, and S. flava expressed in the D. melanogaster empty neuron systems to stimulation with single odorants. Experiments were conducted and analyzed as in figure 2. As before, the at1 empty neuron system was only used for expressing Sfla Or67b2 (see supplementary fig. 5, Supplementary Material online for responses to ITC from at1 OSNs expressing S. pallida and S. flava proteins). (A) Representative electrophysiological recordings obtained from the targeted sensilla of flies expressing Or67b genes in the empty neuron systems in response to stimulation with acetophenone and BITC at 1:100 vol/vol. (B) Responses evoked by stimulation with single odorants (tested at 1:100 vol/vol) categorized as follows: Dmel Or67b activators (Database of Odor Responses; Münch and Galizia 2016; blue), GLVs (gray), ITCs (green), benzyl thiocyanate (yellow), nitrile (pink), and TrpA1 activators (purple). OSNs expressing any of the Sfla Or67b paralogs respond strongly and selectively to ITCs (see also supplementary fig. 5, Supplementary Material online); these OSNs do not respond to benzyl thiocyanate stimulation (yellow bars), indicating that the ITC functional group (–N = C=S) is necessary for evoking responses from the S. flava paralogs. Spal Or67b and Dmel Or67b have similar odor-response profiles, responding mostly to stimulation with D. melanogaster activators and GLVs (*P < 0.05, **P < 0.01, one-sample signed rank tests performed as explained in fig. 2). Most odorants were diluted in mineral oil but some were diluted in other solvents as needed (see Materials and Methods). Responses to the appropriate solvent control were subtracted from odorant-evoked responses. (C) Tuning curves for each Or67b, showing the distribution of median responses to the 42 odorants tested (color coded as in A). The odorants are displayed along the horizontal axis according to the net responses they elicit from each Or. The odorants (numbers) eliciting the strongest responses for each Or are located at the center of the distribution and weaker activators are distributed along the edges (Hallem and Carlson 2006). Note that the strongest responses (center of the distribution) from Dmel Or67b and Spal Or67b are evoked by D. melanogaster activators and GLVs (blue and gray bars), whereas the strongest responses from all Sfla Or67b paralogs are evoked by ITCs (green bars). The tuning breadth of each Or is quantified by the kurtosis value (k) of the distribution (Willmore and Tolhurst 2001), with higher values indicating narrower odor-response profiles. The chemical structure of the top seven Sfla Or67b3 activators, as well as AITC, citronellal, acetophenone, cis-3-hexenyl butyrate, and benzyl thiocyanate (BTC) are shown in bottom.
To test which odors specifically activate Sfla Or67b paralogs, we measured the responses of all Or67b copies to stimulation with apple cider vinegar, a potent D. melanogaster attractant (Faucher et al. 2013), crushed arugula leaves (Eruca vesicaria, a mustard), and crushed tomato leaves (Solanum lycopersicum), a nonmustard plant that releases large quantities of VOCs (Schauer et al. 2004). OSNs expressing Dmel Or67b and Spal Or67b, but not those expressing any of the Sfla Or67b paralogs, responded strongly to stimulation with apple cider vinegar (one-sample signed rank tests, P < 0.05; fig. 2), indicating that the Sfla Or67b paralogs may have lost the ancestral odorant sensitivity. VOCs from the nonhost tomato activated both Dmel Or67b and Spal Or67b (P < 0.05), but not any of the Sfla Or67b paralogs (P > 0.05; fig. 2C). In contrast, all Sfla Or67b proteins responded to arugula leaf volatiles (P < 0.05), suggesting that Sfla Or67bs respond specifically to VOCs characteristic of the hosts S. flava attacks.
In contrast to tomato plants, Brassicales plants, including arugula, produce glucosinolates, some of which are hydrolyzed into ITCs upon tissue damage (Fahey et al. 2001). To test if responses of Sfla Or67bs to mustard leaf odors are mediated by ITCs and/or by other mustard VOCs, we used grated leaves of wild-type 4–6 weeks old A. thaliana (Col-0) plants and two loss of function A. thaliana mutants in the Col-0 background. One of these mutants (myb28 myb29 cyp79b2 cyp79b3; Glucosinolate Knock Out) is deficient in the production of ITC precursors derived from aliphatic and indolic glucosinolates as well as camalexin (Glazebrook and Ausubel 1994; Beekwilder et al. 2008), and therefore does not release ITCs upon wounding (Zhao et al. 2002; Schuhegger et al. 2006; Sønderby et al. 2007; Beekwilder et al. 2008), whereas the control mutant line phytoalexin deficient 3 (PAD3) produces these glucosinolates but not camalexin (Glazebrook and Ausubel 1994; Schuhegger et al. 2006).
Stimulation with wild-type leaves of Col-0 and PAD3, both of which produce ITCs, produced responses from OSNs expressing Sfla Or67b1, b2, and b3 (one-sample signed rank tests, P < 0.05), whereas stimulation with GKO leaves did not (P > 0.05; fig. 2C). Furthermore, the responses of all S. flava Or67b paralogs to VOCs from Col-0 and PAD3 were different from the responses to VOCs from GKO plants (Kruskal–Wallis ANOVA followed by Tukey tests, P < 0.05). Thus, ITCs, but not other A. thaliana leaf volatiles in our experiments, likely activated the three S. flava Or67b copies. In contrast, OSNs expressing Dmel Or67b or Spal Or67b showed similar responses to all three A. thaliana genotypes (fig. 2C, Kruskal–Wallis ANOVAs, P > 0.05), in agreement with the reported responses of this Or to green leaf volatiles (GLVs) in D. melanogaster (Münch and Galizia 2016). Because GKO plants differed from both Col-0 and PAD3 plants only in their capacity to produce aliphatic and indolic glucosinolates and their hydrolysis products, including ITCs (Glazebrook and Ausubel 1994; Beekwilder et al. 2008), these results demonstrate that Sfla Or67b paralogs are selectively activated by these signature Brassicales VOCs.
Because mustard plant roots release a variety of VOCs (including ITCs) but not GLVs (Ishii et al. 1989; Sultana et al. 2003; Xue et al. 2020), we also used these sources as stimuli to further explore natural, ITC-rich volatile blends on Or67b responses. We prepared root homogenates of four mustard plant species, including wasabi (Eutrema japonicum; Sultana et al. 2003), horseradish (Armoracia rusticana; Sultana et al. 2003), turnip (Brassica rapa; Xue et al. 2020), daikon (Raphanus sativus; Ishii et al. 1989), and beet (Beta vulgaris; Richardson 2013), a nonmustard root (control) vegetable species. OSNs expressing D. melanogaster or S. pallida Or67b paralogs did not respond to any root stimuli (one-sample signed rank tests, P > 0.05, fig. 2C), whereas OSNs expressing any of the three S. flava Or67b paralogs responded to wasabi root volatiles (P < 0.05 in all three cases). Finally, Sfla Or67b3 additionally responded to horseradish and turnip (P < 0.05 in both cases). OSNs expressing Sfla Or67b3 showed small (but significant) responses to mustard plant (vegetative or root) VOCs (<40 spikes/second, median). Given the species diversity within Brassicales, it is possible that the natural activator of Sfla Or67b3 was not included in this study.
Scaptomyza flava Or67b Paralogs Have Different ITC Selectivity
We next compared the odor-tuning profiles of Or67b copies from all species using a panel of 42 individual odorants (all tested at 1:100 vol/vol). This included ITCs, nitriles, GLVs, and odorants that activate Dmel Or67b, including ketones, esters, and alcohols (Münch and Galizia 2016; fig. 3). Testing this varied array of VOCs covered a diverse range of known secondary plant chemicals and other appetitive odors (Allmann et al. 2013; Liu et al. 2020).
We first verified the previously reported Dmel Or67b odor-response profile (Münch and Galizia 2016). Most of the odorants categorized as Dmel activators and GLVs evoked significant responses (13/17 odorants, one-sample signed rank tests, P < 0.05; fig. 3). Spal Or67b responded to a smaller number of odorants within those categories (7/17 odorants; P < 0.05). Strikingly, only one of the compounds that elicited responses from DmelOr67b and Spal Or67b (trans-2-hexen-1-ol) evoked responses from Sfla Or67b1 (median = 22 net spikes/second; P < 0.05), but none of the 17 odorants categorized as Dmel activators and GLVs evoked responses from Sfla Or67b2 or Sfla Or67b3 (fig. 3; P > 0.05). Although various ITCs evoked responses from each of the S. flava paralogs (6–7/10 ITCs depending on the paralog; P < 0.05), none evoked responses from Dmel Or67b or Spal Or67b (P > 0.05; fig. 3A and B). Tuning curves (fig. 3C) of all Or67b proteins show that Sfla Or67bs have response profiles distinct from Dmel Or67b and Spal Or67b, whereas non-ITC compounds evoked the strongest responses (acetophenone and cis-3-hexenyl-butyrate, respectively, center of the distribution, yellow bars; fig. 3C). All Sfla Or67b paralogs had the strongest responses to ITC compounds (center of the distribution, green bars; fig. 3C). Sfla Or67b1 had the narrowest odorant-receptive range, responding most strongly to a smaller subset of ITC compounds tested, indicated by the high kurtosis value and sharp peak of the tuning curve. In contrast, Sfla Or67b3 had the widest odorant-receptive range, responding similarly to many ITCs, indicated by the low kurtosis and broad peak of the tuning curve (fig. 3C). In sum, Dmel Or67b and Spal Or67b do not respond to ITCs and have similar odor-response profiles, whereas each Sfla Or67b paralog is differentially responsive to ITCs found in diverse mustard plant species (Fahey et al. 2001).
We confirmed that the differences in ITC selectivity across Or67b proteins were not an artifact resulting from the use of two different empty neurons. We expressed Spal Or67b, Sfla Or67b1, and Sfla Or67b3 in at1 empty OSNs and compared their responses to acetophenone and ITCs (which serve to discriminate Or67bs, fig. 3) with those of ab3A OSNs expressing each of these proteins. In both cases, Spal Or67b responded only to acetophenone, whereas Sfla Or67b1 and Sla Or67b3 selectively responded to ITCs (fig. 3 and supplementary fig. 5, Supplementary Material online). Although the response intensity was lower for at1 OSNs than for ab3A OSNs (supplementary fig. 5B, Supplementary Material online), the response profiles of Sfla paralogs were similar for both OSN types. For example, BITC activated all paralogs but SBITC only activated Or67b3 (fig. 3 and supplementary fig. 5, Supplementary Material online).
We next tested whether the presence of the ITC functional group (–N = C=S) is necessary for evoking responses from OSNs expressing Sfla proteins. We stimulated OSNs with two linkage isomers, BITC (which bears the ITC functional group) and benzyl thiocyanate BTC (which bears the thiocyanate functional group, –S≡C–N; fig. 3B and C). Stimulation with BITC evoked robust responses in OSNs expressing any of the three paralogs (one-sample signed rank tests, P < 0.05), whereas stimulation with BTC had no effect (P > 0.05; fig. 3B and C). This differential activation pattern is therefore likely due to the ITC functional group, as these compounds are not only linkage isomers but also have similar volatilities.
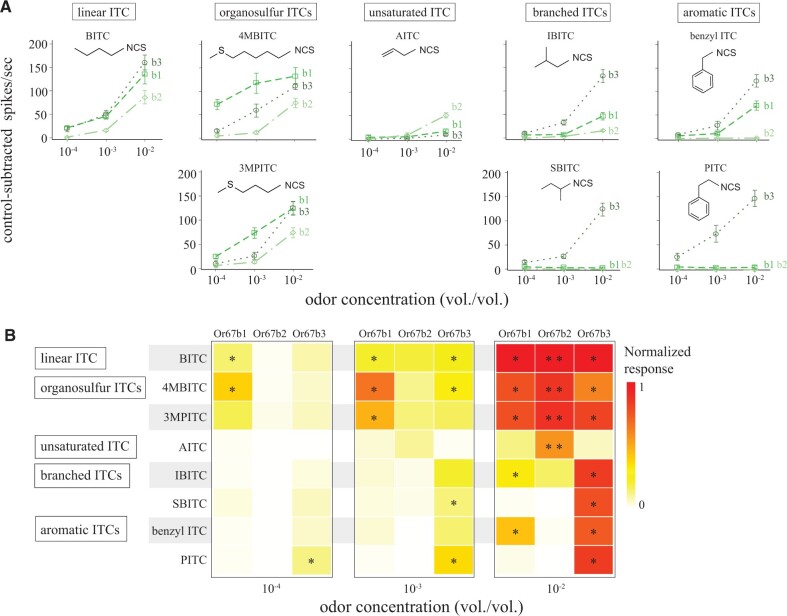
We also stimulated OSNs expressing Sfla paralogs with serial dilutions of eight ITCs that evoked the strongest responses at 1:100 vol/vol (fig. 3B) to further asses paralog ITC selectivity, which may allow detection and discrimination of different mustard plant accessions or species. Because the magnitude of the responses may be reduced when an Or is expressed in at1 OSNs (vs. responses when expressed in ab3A OSNs, supplementary fig. 5, Supplementary Material online, Syed et al. 2010), we included both net and normalized median responses (fig. 4A and B). In general, OSN responses increased with increasing odorant concentration (fig. 4A and B). All paralogs have similar dose-responses to BITC stimulation, whereas other ITCs elicited Or-specific dose-responses (fig. 4). For example, Sfla Or67b1 responded sensitively to BITC and 4MBIT down to 10−4 vol/vol, and Sfla Or67b3 was the only paralog that responded to PITC at all concentrations (fig. 4B). We also used principal component analysis to analyze the responses of OSNs expressing each of the Or67b paralogs in odor space. Most ITC responses clustered in an odorant-concentration-dependent manner: responses separated best at 1:100 vol/vol, except for BITC, 4MBITC, and 3MPITC, which separated best at 1:1,000 vol/vol (supplementary fig. 6 and file 4, Supplementary Material online).
Fig. 4.
Sfla Or67b1-3 have distinct ITC selectivity. (A) Dose responses of OSNs expressing Sfla Or67b1, Sfla Or67b2, and Sfla Or67b3 (abbreviated as b1, b2, and b3) to stimulation with increasing concentrations (vol/vol) of eight different ITCs (categorized according to molecular structure, top boxes; odorant abbreviations are as in fig. 3). As before, the at1 empty neuron system was used for expressing Sfla Or67b2 (see also supplementary fig. 5, Supplementary Material online). Data represent the control-subtracted net number of spikes (average ± SE; n = 6–8, obtained from 2 to 3 animals). (B) Heatmap of dose-responses (median, color coded) from the three Sfla paralog normalized (to allow comparisons across paralogs) by each paralog’s median response to 1:100 vol/vol of BITC (the strongest ITC activator across all paralogs). Asterisks indicate significant differences as explained in Materials and Methods (one-sample signed rank tests; * P < 0.05; ** P < 0.01). The strongest responses were evoked by the highest ITC concentration, with many compounds evoking responses from all paralogs, particularly in the case of Sfla Or67b1 and Or67b3. The number of stimuli that evoked responses decreased with decreasing odorant concentration. Overall, three ITCs evoked responses from two paralogs down to 10−4 vol/vol. See supplementary figure 6, Supplementary Material online for analysis of responses in odor space.
A Population of S. flava Antennal OSNs Are Responsive to BITC
Because Sfla Or67b paralogs responded sensitively to diverse ITCs when expressed in the heterologous empty neuron systems of D. melanogaster (fig. 4), we next investigated whether ITC-sensitive Ors are present in the antennae of S. flava by recording responses of OSNs in antennal basiconic-like (n = 36) and trichoid-like sensilla (n = 36) to stimulation with BITC 1:1,000 vol/vol (supplementary fig. 7, Supplementary Material online). We used BITC at this low, biologically relevant concentration (Jacobsson et al. 2004; Rohloff and Bones 2005; Gruber et al. 2009), because it evoked responses from all S. flava paralogs (fig. 4B; middle panel). Nearly one-fifth (19%) of recorded OSNs in basiconic-like and in trichoid-like sensilla showed medium to strong responses to stimulation with BITC (median: 130 and 166 net spikes/second, range: 74–252 and 65–200 net spikes/second, respectively, in each sensilla type; supplementary fig. 7A and B, Supplementary Material online). These ITC-sensitive OSNs are located proximally in basiconic-like sensilla, and distally in trichoid-like sensilla (supplementary fig. 7C, Supplementary Material online). The finding that S. flava has BITC-sensitive OSNs in at least two different morphological types of antennal olfactory sensilla is consistent with our finding that Sfla Or67b2 is functional only when expressed in the at1 empty neuron system, whereas Sfla Or67b1 and Sfla Or67b3 are functional in both at1 and ab3 basiconic sensilla of D. melanogaster (figs. 2 and 3 and supplementary fig. 5, Supplementary Material online). Future work is needed to confirm both expression of Or67b paralogs in S. flava ITC-sensitive sensilla and necessity of that expression for ITC detection.
Scaptomyza flava Is Attracted to Mustard Plant Odors and Volatile ITCs
The Or67b paralogs of S. flava—but not those of its nonherbivore relatives—responded selectively to ITCs. Thus, we hypothesized that S. flava evolved attraction to these odorants, like most other members of the mustard-feeding guild of herbivorous insects. We addressed this using a dual-choice olfactory assay (based on Reisenman et al. 2013; supplementary fig. 8, Supplementary Material online) in which flies are allowed to choose between an odor-laden and an odorless arm of a “y-maze” olfactometer (e.g., Auer et al. 2020). We first tested whether flies are differentially attracted to mustard and nonmustard leaf VOCs under our experimental conditions.
In the y-maze, S. flava was attracted to arugula and A. thalianaPAD3 leaf VOCs (two-tailed binomial tests, P < 0.05), whereas S. pallida was not (P > 0.05; fig. 5 and supplementary fig. 9, Supplementary Material online). Leaf VOCs from the nonhost plant tomato did not attract or repel S. flava (P > 0.05), but attracted S. pallida (P < 0.01; fig. 5), which has been reported from rotting tomato (Maca 1972). Scaptomyzaflava tested with leaf VOCs from A. thaliana PAD3 and ITC-lacking GKO mutants similarly preferred VOCs from either of these genotypes over clean air (supplementary fig. 9, Supplementary Material online). This is not surprising because GKO plants release many VOCs other than ITCs that may mediate long-distance olfactory attraction (Glazebrook and Ausubel 1994; Zhao et al. 2002; Sønderby et al. 2007; Beekwilder et al. 2008). As expected (Faucher et al. 2013), D. melanogaster was strongly attracted to apple cider vinegar odors (P < 0.05; fig. 5). Both S. flava and S. pallida, in contrast, distributed at random between the apple cider vinegar odor-laden and the odorless arm of the maze (P > 0.05; fig. 5). In summary, S. flava is specifically attracted to mustard plant odors, but not to nonhost plant odors or those that attract D. melanogaster, including acetic acid and ester-, carbonyl-, and hydroxyl-containing compounds (Aurand et al. 1966; Cousin et al. 2017).
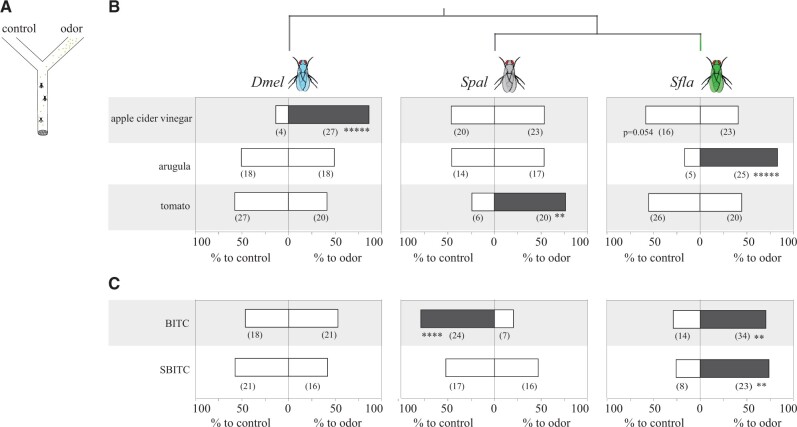
Fig. 5.
Olfactory behavioral responses of S. flava and its microbe-feeding relatives S. pallida and D. melanogaster to ecologically relevant odors and ITCs. (A) Schematic representation of the dual choice y-maze used to test the olfactory responses of flies (see details in supplementary fig. 8, Supplementary Material online). One arm of the maze offered constant odor/odorant airflow (apple cider vinegar, arugula, tomato, or single ITC compounds at 1:100 vol/vol), whereas the control arm offered a constant odorless airflow (controls: water for odor sources, and mineral oil for single odorants). In each test, a group of nonstarved flies (n = 3–4) was released at the base of the maze and allowed to choose between the two arms of the maze. Each test (maximum duration = 5 min) ended when the first insect (out of all released) made a choice. (B) Olfactory behavioral responses of D. melanogaster, S. pallida, and S. flava to apple cider vinegar odors and VOCs from leaves of arugula and tomato plants. Data represent the percentage of tests in which animals choose the odorous or odorless arms of the maze; numbers between parentheses indicate the number of tests with choices for one or the other arm. For each fly species and odor/odorant compound, data were analyzed using two-tailed Binomial tests (**P < 0.01, ****P < 0.001, *****P < 0.0001; gray-shaded bars serve to visualize that the proportion of tests with flies orienting toward that arm of the y-maze is significantly different from random). Scaptomyza flava was attracted to mustard (arugula) VOCs but not to nonmustard (tomato) VOCs. Scaptomyza flava tended to avoid apple cider vinegar odors although differences were not statistically significant (P = 0.054). Drosophila melanogaster was strongly attracted to apple cider vinegar but not to arugula or tomato leaf VOCs. Scaptomyza pallida was only attracted to tomato leaf VOCs. See supplementary figure 9, Supplementary Material online for responses to A. thaliana volatiles. (C) Olfactory behavioral responses of flies from the three species to single ITC compounds (20 µl of a 1:100 vol/vol dilution in mineral oil loaded in filter paper); the control arm had 20 µl of mineral oil loaded in filter paper. Assays were conducted as described in (B). Scaptomyza flava was strongly attracted to both ITC compounds tested, S. pallida was strongly repelled by BITC, and D. melanogaster was indifferent to either ITC.
We next investigated whether ITCs alone can mediate olfactory attraction in S. flava. We chose BITC and SBITC because these compounds evoked distinct odor responses from Sfla Or67b paralogs (fig. 4; BITC strongly activates all S. flava paralogs, whereas SBITC activates only Sfla Or67b3). Scaptomyzaflava was attracted to both BITC and SBITC (P < 0.05; fig. 5B). Interestingly, S. pallida strongly avoided BITC (P < 0.005; fig. 5), which must occur via an Or67b independent olfactory pathway because Spal Or67b does not respond to any of the ITCs tested, even at high concentrations (fig. 3). We also conducted dual-choice trapping experiments with D. melanogaster widely used for microbe-feeding Drosophila spp. (e.g., Prieto-Godino et al. 2017; supplementary fig. 10A, Supplementary Material online). As previously reported (e.g., Auer et al. 2020), flies were captured in traps baited with apple cider vinegar over solvent control (Wilcoxon-matched pairs tests, P < 0.005, n = 9; supplementary fig. 10A, Supplementary Material online), whereas flies distributed at random between the control and the BITC-baited test traps (P > 0.9, n = 24; supplementary fig. 10A, Supplementary Material online). Thus, BITC does not attract D. melanogaster in a short-distance (trap) olfactory assay either. We could not use this trap assay with S. flava, because neither trap captured flies, likely due to the particular biology of this fly species, females of which only attack and oviposit on the underside of leaves. It is possible that the use of intact leaves as an oviposition substrate selected for differences in search behavior. Overall, single ITCs compounds that evoke strong responses from Sfla Or67b paralogs expressed in D. melanogaster and from S. flava antennal OSNs (figs. 3 and 4; supplementary fig. 7, Supplementary Material online) can mediate olfactory orientation in S. flava, but do not attract (and can even repulse) its microbe-feeding relatives.
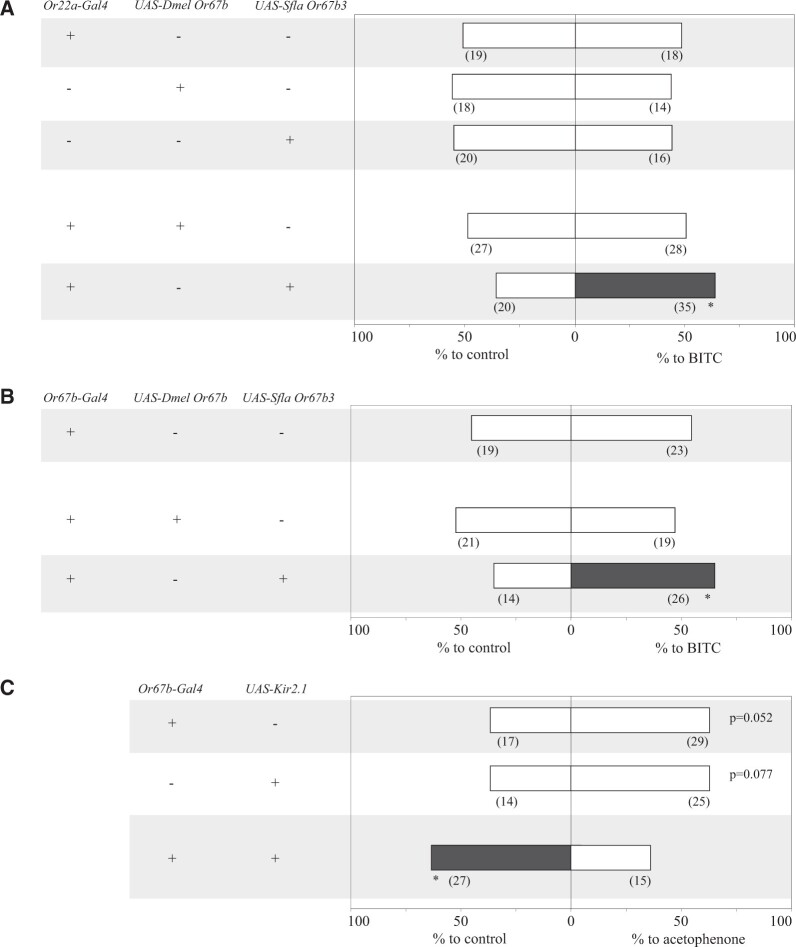
Expression of Sfla Or67b in the Homologous Olfactory Circuit of D. melanogaster Confers Behavioral Responses to ITCs
Because Sfla Or67b paralogs selectively respond to ITCs, we tested if ectopic expression of these receptors in the homologous olfactory circuit of the microbe-feeder species is sufficient to mediate behavioral responses to these compounds. To test this, we first used the Or22a olfactory circuit because it mediates detection of ecologically relevant chemicals in diverse drosophilids (Dekker et al. 2006; Linz et al. 2013; Mansourian et al. 2018). We focused on Sfla Or67b3, as this paralog has the broadest ITC receptive range (figs. 3 and 4). As before, we used a dual-choice olfactometer and tested flies in which the expression of Sfla Or67b3 or Dmel Or67b was under the control of Gal4 in the ab3A neuron. We used BITC at 1:1,000 vol/vol because Sfla Or67b3 responds reliably to this odorant concentration (fig. 4).
Although genetic control lines carrying Gal4 or UAS transgenes showed no preference for either arm of the maze (one-sample signed rank tests, P > 0.05, fig. 6A), flies expressing Sfla Or67b3, but not Dmel Or67b, preferred the BITC-bearing arm (P < 0.05, fig. 6A). Importantly, the genetic parental control flies showed normal odor responses toward apple cider vinegar, a D. melanogaster attractant, in the dual-choice trap assay (supplementary fig. 10B, Supplementary Material online). We next tested whether Sfla Or67b3 can confer behavioral responses to BITC when expressed in D. melanogaster Or67b-OSNs. In this experiment, flies did not lack expression of the endogenous Or67b. Gal4 control flies and flies expressing Dmel Or67b under the control of Gal4 distributed at random between the two arms of the y-maze (P > 0.05, fig. 6B), whereas flies expressing Sfla Or67b3 preferred the ITC-bearing arm of the maze (P < 0.05; fig. 6B, gray bar). The attraction to BITC in these transgenic flies could be due to selective activation of OSNs expressing Sfla Or67b3, alone or concurrent with activation of other unknown attractive olfactory pathways by BITC moieties other than the ITC functional group.
Fig. 6.
Ectopic expression of Sfla Or67b3 in Or22a OSNs or Or67b OSNs conferred behavioral responses to BITC in D. melanogaster. The behavioral responses of D. melanogaster flies expressing different Or67b transgenes were tested in dual-choice assays (odorant vs. mineral oil) and experiments were conducted and analyzed as in figure 5 but flies were starved 24 h previous to testing. (A) Responses of flies expressing Dmel Or67b or Sfla Or67b3 in Or22a OSNs lacking its cognate Or and tested with BITC 1:1,000 vol/vol versus mineral oil. The two parental control lines (first two groups) and flies expressing Dmel Or67b (third group) were not attracted or repelled by BITC (binomial tests, P > 0.05 in all cases), whereas flies expressing Sfla Or67b3 were attracted to the odorant (*P < 0.05). Importantly, parental genetic control lines retain olfactory attraction toward apple cider vinegar, a potent D. melanogaster olfactory attractant and activator (supplementary fig. 10, Supplementary Material online). These results show that ITCs can evoke olfactory behavioral responses when Sfla Or67b3 is expressed in the D. melanogaster Or22a olfactory pathway, an important circuit for detection of host odorants in many drosophilid species (Dekker et al. 2006; Linz et al. 2013; Mansourian et al. 2018). (B) Same as (A), but flies expressed Dmel Or67b or Sfla Or67b3 in Or67b OSNs. Note that flies have the endogenous Or67b expressed in OSNs, in addition to the transgene. As in (A), only flies carrying the Sfla Or 67b3 transgene were attracted to BITC (*P < 0.05, third group). (C) Responses of D. melanogaster expressing a silencer of synaptic activity (Kir2.1) in Or67b OSNs (third group), along with the responses of the two parental genetic control lines (first and second group) in tests with acetophenone 1:50,000 vol/vol, a strong Dmel Or67b activator (Münch and Galizia 2016; fig. 3). Genetic control flies showed a trend for attraction (0.05 < P < 0.08) to low concentrations of acetophenone, whereas flies with Or67b OSNs silenced lost attraction and were instead repelled by the odorant. These behaviors mirrored those of wild-type D. melanogaster flies tested with low and high concentrations of acetophenone, respectively (supplementary fig. S11, Supplementary Material online; Strutz et al. 2014). In (A–C), transgenes are indicated to the left; gray-shaded bars serve to visualize that the proportion of tests with flies orienting toward that arm of the y-maze is significantly different (P < 0.05) from random.
Or67b-mediated host attraction has been proposed for other drosophilid species besides S. flava, including the distantly related D. mojavensis (Crowley-Gall et al. 2016). However, even in D. melanogaster, it remains unclear if the Or67b circuit mediates olfactory attraction to host odors. Addressing this question could facilitate interpretation of the results of our gain of function experiments in which Sfla Or67b3 was expressed in the native Dmel Or67b circuit (fig. 6B). Our electrophysiological recordings showed that Dmel Or67b was activated by esters, alcohol, and ketones (fig. 3), which are released from fruits (Maldonado-Celis et al. 2019). Of these chemicals, we used acetophenone, because this compound is a naturally occurring odorant released from guava pulp (Idstein et al. 1985), which strongly activates Dmel Or67b (fig. 3; Münch and Galizia 2016). Importantly, acetophenone attracts D. melanogaster at low concentrations, but repels these flies at high concentrations (Strutz et al. 2014). We then tested if Dmel Or67b mediates olfactory attraction to host odors by silencing Dmel Or67b OSNs using UAS-Kir2.1, an inwardly rectifying potassium channel that suppresses neuronal activity (Baines et al. 2001) under the control of Or67b-Gal4. Thus, we expect these flies to no longer be attracted to low concentrations of acetophenone.
Our experimental conditions faithfully reproduced previously reported results: wild-type D. melanogaster (Canton-S) was attracted and repelled by low and high concentrations of acetophenone, respectively (supplementary fig. 11, Supplementary Material online). Flies with Or67b OSNs silenced were repelled by low concentrations of acetophenone (fig. 6C; P < 0.05), whereas Gal4 and UAS genetic controls tended to be attracted to the same concentrations of this odorant compound (63% in both cases; binomial tests, P = 0.052 and 0.077; fig. 6C). Altogether, these results suggest that the D. melanogaster Or67b olfactory circuit is necessary for mediating attraction towards low concentrations of the Or67b cognate ligand acetophenone.
Thus, as was proposed for D. mojavensis (Crowley-Gall et al. 2016), the Or67b circuit is a reasonable candidate for mediating olfactory attraction to host odors in drosophilids, including S. flava. Importantly, it remains to be investigated whether S. flava Or67b is necessary for host finding, which can be tested by developing knockout mutants in the future. Finally, it is possible that the Dmelanogaster Or67b circuit does not reflect the ancestral character state for S. flava or D. mojavensis, given that these lineages may have split over 60 Ma (fig. 1A).
Discussion
Most herbivorous insect species specialize on a narrow range of host plant species that each synthesize similar secondary chemicals (Jaenike 1990). Although these toxins serve to defend plants against herbivory, ancestrally aversive molecules are coopted as attractants by herbivores specializing on a new host plant lineage (Fraenkel 1959). We investigated the evolution of attraction to toxic host-plants using S. flava as a model. A candidate Or lineage (Or67b) was triplicated in a recent ancestor of S. flava and experienced rapid protein evolution, resulting in three divergent paralogs (Sfla Or67b1-b3; fig. 1) expressed in the antennae (supplementary fig. 3, Supplementary Material online). Each Sfla Or67b paralog specifically responded to stimulation with mustard-plant odors and volatile ITCs in heterologous expression systems (figs. 2 and 3, and supplementary fig. 5, Supplementary Material online). In addition, each paralog responded to a specific subset of ITCs (fig. 4). In contrast, S. pallida and D. melanogaster Or67b orthologs did not respond to ITCs but showed strong responses to stimulation with apple cider vinegar and a broad range of aldehydes, alcohols, and ketones (figs. 2and 3 and supplementary fig. 5, Supplementary Material online), consistent with their microbe-feeding niche (Galizia and Sachse 2010). Recordings from S. flava antennal sensilla revealed OSNs sensitive to ITCs (supplementary fig. 7, Supplementary Material online). We found that S. flava, but not S. pallida or D. melanogaster, was attracted to volatile ITC compounds (fig. 5). Ectopic expression of S. flava Or67b3 in the D. melanogaster homologous Or67b olfactory circuit conferred odor-oriented behavioral responses to ITCs (fig. 6B). Finally, suppression of activity in Or67b OSNs in D. melanogaster decreased preference to its Or cognate ligand (fig. 6C). These results suggest that the ancestral Or67b olfactory circuit likely mediates olfactory attraction. Thus, gene duplication followed by specialization is a mechanism by which specialist herbivores evolve Ors that may confer olfactory attraction toward ancestrally aversive chemical compounds.
Evolutionary Path of Olfactory Receptor Specialization
The S. flava Or67b triplication reported here raises several molecular evolutionary questions, including how the expansion contributed to the generation of novel gene functions. In this regard, different scenarios have been proposed (Ohno 1970; Heidel-Fischer et al. 2019): 1) neofunctionalization, when one of the duplicated genes (paralogs) acquires a new function after accumulating de novo mutations, whereas the other copy retains ancestral function; 2) subfunctionalization sensu stricto, where mutations accumulate in both copies leading to partitioning of ancestral function; and 3) specialization, when subfunctionalization and neofunctionalization evolve simultaneously, yielding gene copies that are functionally different from one another and the ancestral copy (He and Zhang 2005; Assis and Bachtrog 2013).
Rapidly evolving Slfa Or67b paralogs strongly and selectively respond to ITCs, whereas the more conserved orthologous proteins from both a close and a more distant microbe-feeding relative did not respond to ITCs at all (figs. 3 and 4 and supplementary fig. 5, Supplementary Material online). Sfla Or67b paralogs thus evolved entirely new ligand-binding affinities (to ITCS) in the S. flava lineage at least since divergence from the common ancestor with S. pallida Or67b. This is consistent with a neofunctionalization event that predates the duplications of Sfla Or67b. Further, each Sfla Or67b paralog selectively responds to different subsets of ITCs across a range of odorant concentrations, which comports with subfunctionalization. We therefore conclude that our data are most consistent with a specialization scenario to explain the evolution and function of Sfla Or67b paralogs.
An Olfactory Receptor Sensitive to a New Niche-Specific Chemical Class of Odorant Compounds
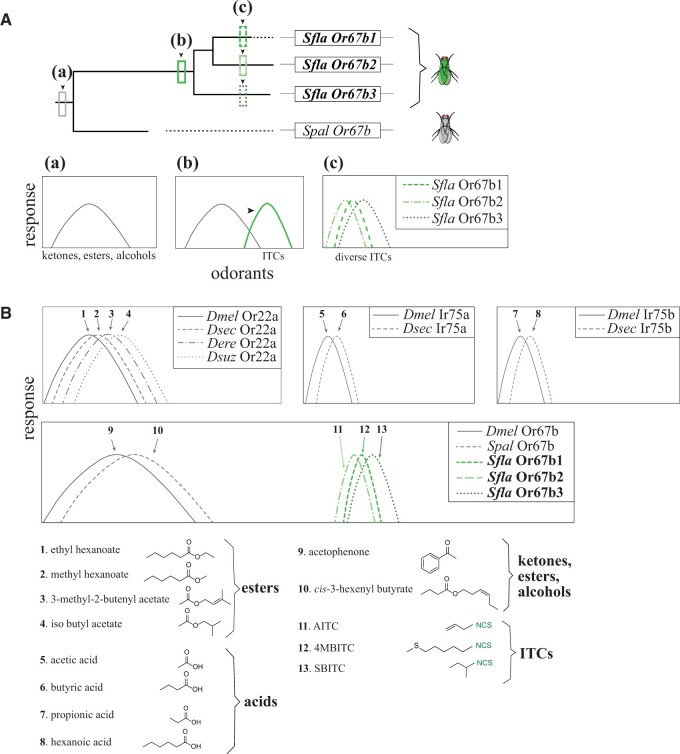
Across Drosophilidae, several orthologous chemoreceptors respond in a species-specific manner to ecologically relevant ligands, suggesting that adaptive evolution has shaped their ligand-binding capabilities. Or22a is an example of a hot spot for sensory evolution in Drosophila because shifts in ligand binding are associated with different odor-preference behaviors (Mansourian et al. 2018; Auer et al. 2020). Or22a and other highly variable receptors such as Ir75a/b have typically evolved new specificities toward different odorants within one chemical class (Prieto-Godino et al. 2017; fig. 7). However, this is not the case for Or67b, which responds to alcohols, aldehydes and ketones in nonherbivorous D. melanogaster and S. pallida, but not in S. flava, where the paralogs responds to volatile ITCs, an entirely different chemical class (figs. 3 and 4). By comparing the responses of BITC and its linkage isomer BTC, we found that the presence of the ITC functional group (–N = C=S) is key for activation of these olfactory receptors, highlighting a functional shift in ligand-binding specificity (fig. 3B and C). The striking difference in odor selectivity among Or67b orthologs may result from the evolution of herbivory in Scaptomyza or subsequent specialization on mustard plants.
Fig. 7.
A model for the evolution of Or67b and comparison with the known evolution of other Or and Ir orthologs in D. melanogaster and specialist drosophilid species. (A) Model for the evolution of Scaptomyza Or67b. The evolution of this Or begins with a shift in the ligand specificity of an ancestral Or67b (a) tuned to Dmel activators, GLVs, and ITCs (neofunctionalization, b). Subsequent gene triplication of Sfla Or67b gave rise to two additional paralogous Or67b genes (Sfla Or67b1, Sfla Or67b2, and Sfla Or67b3; c), each of the paralogs has different but overlapping ITC odorant-receptive ranges (figs. 3 and 4). (B) Evolution of drosophilid orthologs with known ligand specificities (Or22a, Ir75a, Ir75b, top; and Or67b, bottom). The Or22a protein orthologs from D. melanogaster (Dmel Or22a), D. sechellia (Dsec Or22a), D. erecta (Der Or22a), and D. suzukii (Dsuz Or22a) are all strongly activated by species-specific host-derived esters (compounds 1–4; top left; Dekker et al. 2006; Linz et al. 2013; Mansourian et al. 2018; Keesey et al. 2019). The Ir75a protein orthologs from D. melanogaster (Dmel Ir75a), and D. sechellia (Dsec Ir75a) are strongly activated respectively by the acid compounds 5 and 6 (Prieto-Godino et al. 2017). Similarly, the Ir75b protein orthologs from D. melanogaster (Dmel Ir75b) and D. sechellia (Dsec Ir75b) are respectively activated by the acids 7 and 8 (top right; Prieto-Godino et al. 2016). Dmel Or67b and Spal Or67b are strongly activated respectively by acetophenone and cis-3-hexenyl butyrate (compounds 9 and 10), whereas Sfla Or67b copies are activated by ITCs only (bottom, paralog-specific activation by compounds 11–13, abbreviations as in fig. 3). Note that Or22a, Ir75a, and Ir75b orthologs are all divergent but activated by ligands belonging to a single chemical class (whether esters or acids). On the other hand, the ligands of orthologs Or67b from Dmel and Spal are responsive to a variety of chemical classes which include alcohols, aldehydes, and ketones, whereas Sfla Or67b orthologs are responsive to ITCs, an entirely different compound chemical class. Notably, copies of Or22a, Or42b, and Or85d have been evolutionarily lost in S. flava. These Ors are important in mediating attraction to canonical microbe (especially yeast)-associated odors (Goldman-Huertas et al. 2015).
An Olfactory Receptor Likely Mediating Attraction to Mustard Host Plants
Through stimulation with VOCs from leaves of A. thaliana genotypes with and without ITCs, S. flava has at least two different olfactory pathways for mustard-plant attraction: ITC dependent and ITC independent (fig. 5 and supplementary fig. 9, Supplementary Material online). This is consistent with the fact that plant VOCs are complex bouquets of diverse, widely distributed and lineage-specific molecules that are variably capable of releasing attraction behaviors (Bruce and Pickett 2011). Our results do not preclude that other chemoreceptors and OSNs also contribute to mediate olfactory attraction to mustard plant VOCs and ITC compounds in S. flava. The necessity of Or67bs for orientation toward ITCs requires generating loss of function mutants and testing whether flies can still orient toward these odorants. Because S. flava females lay eggs in the leaf mesophyll and cannot be reared on available media, generation of Or67b mutants using CRISPR-Cas9 is not yet feasible. Newer technologies such as ReMOT Control may enable such experiments (Chaverra-Rodriguez et al. 2018). It also remains to be explored if evolution also sculpted neural wiring to modulate responses to ITC compounds (Auer et al. 2020; Zhao and McBride 2020).
ITC taste detection in vertebrates and insects leads to aversive behaviors mediated by the contact chemoreceptor TrpA1 (Hinman et al. 2006; Macpherson et al. 2007; Kang et al. 2010). However, volatile ITCs are widely used to trap pests of Brassicales (Kergunteuil et al. 2012) and the antennae of some of these insects, including S. flava (Goldman-Huertas et al. 2015; supplementary fig. 7, Supplementary Material online), respond to volatile ITCs (Renwick et al. 2006; Liu et al. 2020; Piersanti et al. 2020). Similarly, two Ors in the mustard specialist diamondback moth Plutella xylostella respond selectively to ITCs (Liu et al. 2020), including 3MPITC and PITC, which strongly activate S. flava Or67b paralogs. Thus, these ITCs are likely important for host orientation in distantly related mustard specialists.
Facing highly inducible host plant defenses in vegetative tissues is a distinct challenge for leaf-mining Scaptomyza, compared with other herbivorous drosophilids like D. sechellia and D. suzukii, which feed on ripe fruit. Although glucosinolates and jasmonate-inducible defenses reduce growth rates of S. flava larvae (Whiteman et al. 2011, 2012), these flies have evolved highly efficient glutathione S-transferases through gene duplication events (Gloss et al. 2014), some of which detoxify ITCs more efficiently in vitro than those from any known animal (Gloss et al. 2019). Yet, here we show that volatile ITCs at biologically relevant concentrations are actually attractive to S. flava adult females. The attraction to ITCs by S. flava may in part be explained by the Or67b triplication, which is only known from Brassicales-specialist Scaptomyza species (Kim et al. 2021) and not S. graminum, which attacks leaves of Caryophyllaceae (fig. 1A).
Although paradoxical, these results are entirely consistent with Fraenkel’s (1959) overarching hypothesis that plant toxins evolve to dissuade and punish herbivores, but specialists eventually overcome and then coopt the toxins as faithful signposts of their host plants. Our study adds to the growing body of evidence that gene duplication and subsequent evolution have played an important role in coevolution (sensu lato) between Brassicales plants and the diverse guild of herbivorous insects that use them as hosts. More generally, chemosensory specialization can result from relatively simple genetic modifications in the peripheral nervous system that changes olfactory receptor tuning, contributing to major niche shifts in insects.
Materials and Methods
Time-Calibrated Molecular Phylogeny of Scaptomyza and Drosophila spp
A time-calibrated species tree was inferred using the loci: 16S, COI, COII, ND2, 28S, Cad-r, Gpdh1, Gstd1, Marf, l(2)tid (also known as Alg3 or Nltdi), and AdhR from 13 species of Drosophila and four Scaptomyza spp. Scaptomyza sequences were accessed from the genomes in Gloss et al. (2019) and Kim et al. (2021) using tblastn searches for protein coding genes and blastn searches for the ribosomal RNA genes. The Adh Related gene appears to be deleted in S. flava and was coded as missing data. Two uniform priors on the age of Scaptomyza and the age of the combined virilis-repleta radiation, Hawaiian Drosophila and Scaptomyza clade were set as in Katoh et al. (2017). Protein coding genes were portioned into first and second combined and third position partitions, except COI and Gpdh1, which were divided into first, second, and third partitions. The partitioning scheme was chosen based on Partition Finder 2.1.1 (Lanfear et al. 2017) and nucleotide substitution models chosen with IQ-Tree (Nguyen et al. 2015). Model parameters, posterior probabilities, accession numbers, and genome coordinates can be found in supplementary file 5, Supplementary Material online. Five independent runs of BEAST v2.6.2 (Bouckaert et al. 2019) each with 25 million generations were run logging after every 2,500th generation to infer the chronogram with 10% burn-in. Phylogenies were also inferred in RAxML v8.2.12 (Stamatakis 2014) with the GTR+GAMMA+I model and 1,000 rapid bootstraps and with parsimony in PAUP* 4.0a (Swofford 2002) with TBR branch swapping and 1,000 bootstraps. Phylogeny parameters, sequence accession numbers, and likelihood scores available in supplementary file 5, Supplementary Material online.
Molecular Phylogeny of Drosophilid Or67b Genes
Or67b coding sequences (CDS) from D. grimshawi, D. mojavensis, D. virilis D. sechellia, D. simulans, D. erecta, D. yakuba, D. pseudoobscura, D. persimilis, D. ananassae, and D. melanogaster (builds dgri r1.3, dmoj r1.3, dvir r1.07, dsec r1.3, dsim r1.4, dere r1.3, dyak r1.3, dpse r3.2, dper r1.3, dana r1.3, and dmel r6.28, respectively) were downloaded from Flybase (www.flybase.org, last accessed January 3, 2022; Thurmond et al. 2019) and D. hydei from GenBank (accession number XM_023314350.2). The S. pallida DNA sequence was obtained through PCR and Sanger sequencing as described below. Scaptomyzaflava DNA sequences were previously published (Goldman-Huertas et al. 2015). Two more Scaptomyza sequences were obtained from Gloss et al. (2019) and Kim et al. (2020) including the nonleaf-mining species S. hsui (subgenus Hemiscaptomyza and a microbe feeder) and S. graminum (subgenus Scaptomyza and a leaf miner on Caryophyllaceae). DNA sequences were aligned, partitioned by codon position, models fitted to all three partitions, and chosen according to AICc (GTR+I + G) in IQ-Tree (Nguyen et al. 2015). Trees were inferred using RAxML (v8.2.10) with the GTRGAMMA+I model and 1,000 rapid bootstraps, and MrBayes (v3.2.6) setting Nst to 6, nucmodel to 4by4, rates to Invgamma, number of generations to 125,000, burnin equal to 20% of generations, heating to 0.2, number of chains to 4, runs to 2, and priors set to default setting (Ronquist et al. 2012). An additional parsimony analysis was performed in Paup 4.0 (Swofford 2002) with TBR branchswapping and 1,000 bootstraps. Model parameters, accession numbers, and likelihood scores are available in supplementary file 5, Supplementary Material online.
Analysis of Molecular Evolutionary Rates
CDS of homologs of every Or gene in S. flava found in the 12 Drosophila genome builds were aligned to S. flava Or CDS. Homology was assessed according to inclusion in well-supported clades in the Or translation phylogeny from the 12 species; S. flava sequences were previously published (Goldman-Huertas et al. 2015). Sequences were aligned in MAFFT (v7.017; Katoh 2002) and adjusted manually to preserve codon alignments. Or98a-like genes found in subgenus Drosophila species were split into three separate clades, as were a group of Or83c paralogs not found in D. melanogaster, and a group of Or85a-like genes. All examined sequences of Or46a contain two alternatively spliced exons, so this gene was analyzed with all gene exon sequences in a single alignment. Or69a, however, contains alternatively spliced exons only in species within the subgenus Sophophora, and therefore these alternative splice forms were analyzed as separate taxa. Phylogenies were generated for every alignment using PhyML (Guindon et al. 2010) with the GTR+G substitution models. If >70% bootstrap support was found for a topology contrary to the known species topology, or if the Or homology group contained duplicates, these trees were used in PAML analyses instead of the species tree.
Our next goal was to identify Or genes experiencing rapid rates of protein evolution potentially consistent with episodic positive selection. Branch models of sequence evolution were fit using PAML 4.9h (Yang 2007). A foreground/background branch model was fit for every S. flava tip branch and every ancestral branch in a Scaptomyza-specific Or gene duplication clade, and compared in a likelihood ratio test to a null model with one dN/dS rate (ratio of nonsynonymous to synonymous substitution rate) for every unique phylogeny (75 tests in total). After focusing on Or67b and selecting this as a candidate (as explained above), patterns of molecular evolution among the drosophilid Or67b homologs were explored using the expanded Or67b CDS phylogeny above. Foreground/background branch models were fit for every branch in the Or67b phylogeny and the S. flava Or67b paralogs clade with likely ratio tests performed as before (34 tests total, supplementary file 6, Supplementary Material online). P-values were adjusted for multiple comparisons using the false-discovery rate (FDR) method (Benjamini and Hochberg 1995). Branch test results and model parameters are included in supplementary file 6, Supplementary Material online.
Fly Husbandry and Lines
D. melanogaster (wild-type Canton-S and transgenic lines) were reared using cornmeal media, yeast, and agar medium prepared by UC-Berkeley core facilities. Isofemale lines of S. pallida (collected in Berkeley, California, US) were maintained on Nutri-Fly medium (Genesee Scientific). S. flava (collected in New Hampshire, US) were maintained on fresh A. thaliana Col-0 plants and 10% honey water solution. All flies were cultured at 23 °C and 60% relative humidity under a 12-h light/12-h dark cycle. S. flava and S. pallida were ca. 7–10 days old at the time of experiments; D. melanogaster (wild-type or transgenic) were ca. 3–10 days old at the time of the experiments.
Flies used for heterologous expression of Ors were of the following genotypes: for ab3A “empty neuron” flies, Or22abGal4::3xP3-DsRed was used. This “M2-MD” line was generated by a CRISPR-Cas9-mediated deletion of Or22a/b and a knock-in of Gal4 and DsRed (Gross et al. 2000) by homology directed repair (HDR); in these flies Gal4 is not functional. Therefore, Or22abGal4::3xP3-DsRed; Or22a-Gal4/UAS-Or67b flies were used for experiments. For at1 “empty neuron” flies, Or67dGal4 (Kurtovic et al. 2007) was used. The Or67b-Gal4 fly line (BDSC# 9996) was crossed with UAS-Or67b lines or UAS-Kir2.1 (Baines et al. 2001) for behavioral assays.
Scaptomyza Or67b Gene Cloning, UAS Line Generation
The UAS-Or67b transgene lines were constructed as follows: RNA was extracted from 20 to 25 days postemergence adults of both sexes from laboratory-reared S. pallida (collected from the White Mountains, NM, USA) and S. flava (collected from near Portsmouth, NH, USA). RNA was extracted using Trizol (Thermo-Fisher, Waltham, MA, USA) and precipitated with isopropanol. Extracted RNA was treated with DNaseI, cDNA was generated using qScript cDNA Supermix (Quantabio, Beverly, MA, USA). PCR conditions and primers are detailed in Lapoint et al. (2013). EcoRI and KpnI cut sites were introduced using restriction enzyme cut-site primers (supplementary file 7, Supplementary Material online) with 10 ng/µl diluted plasmids (as template) with 3% DMSO vol/vol. The pUAST attB plasmid (Bischof et al. 2007) and the four S. pallida and S. flava Or67b PCR amplicons with RE flanking sites were individually double digested with KpnI and EcoRI high-fidelity enzyme in cut smart buffer for 3 h, according to the manufacturer’s protocol (NEB). Cut fragments were gel-purified using the Qiaquick Gel Cleanup Kit (Qiagen) and ligated in a 1:3 vector: insert molar ratio using T4 ligase (Promega). Ligations were transformed into JM109 cells. Some cells were preserved as glycerol stocks and a portion were used for injection into the y1 w67c23; P{CaryP}attP2 D. melanogaster line for generating Sfla Or67b1, Sfla Or67b2, Sfla Or67b3, Spal Or67b, or into the y1 w67c23; P{CaryP}attP40 for Sfla Or67b2 (BestGene Inc., Houston, TX, USA). Transformants were selected from individually injected flies with compound eye color rescue phenotypes.
Single Sensillum Recordings
Adult female flies that were fed and 3–15 days old were prepared for single sensillum recordings (SSR) as previously described (Hallem and Carlson 2004). We recorded the responses of 6–10 sensilla obtained from 2–4 individuals for each experiment/odorant. Signals were amplified 100× (A-M systems, Differential AC Amplifier model 1700), digitized using a 16-bit analog-digital converter, filtered (low cut-off: 300 Hz, high cut-off: 500 Hz), and analyzed off-line using WinEDR (v3.9.1; University of Strathclyde, Glasgow). A tube delivering a constant flow of charcoal-filtered air (16 ml/min, using a flowmeter; Gilmont Instruments, USA) was placed near the fly’s head, and the tip of the stimulation pipette (50 ml) was inserted into the constant air stream. The stimulation pipette contained a piece of filter paper loaded with 20 µl of odorant solution or the solvent control. One second pulse of clean air was delivered to the stimulus pipette using a membrane pump operated by a Stimulus Controller CS 55 (Syntech, Germany). Ab3 sensilla were identified by using diagnostic odorants (Gonzalez et al. 2016; all odorants were obtained from Sigma-Aldrich, US, purity > 95%): ethyl hexanoate (CAS # 123-66-0), ethyl acetate (CAS # 141-78-6), and 2-heptanone (CAS # 110-43-0; supplementary fig. 4, Supplementary Material online). We were able to distinguish at1 sensilla from all other at sensilla because at1 is the only antennal trichoid sensilla known to house a single OSN (Gonzalez et al. 2016).
The following odor sources were used (purchased from Berkeley Bowl in Berkeley, CA, USA, 20 µl of material were loaded on filter paper unless noted): grated roots of the four Brassicaceae species; W. japonica (wasabi), A. rusticana (horseradish), B. rapa (turnip), R. sativus (daikon), and the Amaranthaceae species B. vulgaris (beet). Approximately 10 g of roots were grated immediately before experiments. We also used pure apple cider vinegar as a stimulus (40 µl, O Organics, USA). Brassicaceae species E. vesicaria (arugula), A. thaliana, and S. lycopersicum (tomato) were grown from seeds at 23 °C and 60% relative humidity under a 12-h light: 12-h dark cycle, and leaves from 3 to 8 weeks old plants were used for odor stimulation. The following A. thaliana genotypes were used: wild-type (Col-0), aliphatic and indolic Glucosinolate Knock Out (GKO) mutant in myb28 myb29 cyp79b2 cyp79b3, which has no detectable aliphatic and indolic glucosinolates or camalexin (Zhao et al. 2002; Sønderby et al. 2007; Beekwilder et al. 2008), and camalexin-deficient phytoalexin deficient 3 (PAD3) mutants that have wild-type levels of aliphatic and indolic glucosinolates but no camalexin. Therefore, PAD3 plants are more appropriate controls for comparisons with GKO plants than Col-0 plants for our study (Schuhegger et al. 2006). Three-four leaves were excised from plants and homogenized with a grater immediately before tests; the homogenate was replaced every 30 min since OSN responses were stable at least during this time window. All synthetic odorants were diluted in mineral oil (1:100 vol/vol) unless otherwise noted. The following odorants (all from Sigma-Aldrich, US, purity > 95%) were diluted in dimethyl sulfoxide (DMSO): mandelonitrile (CAS # 532-28-5), iodoacetamide (CAS # 144-48-9), N-methylmaleimide (CAS # 930-88-1), N-hydroxysuccinimide (CAS # 6066-82-6), and benzyl thiocyanate (CAS # 3012-37-1). 11-cis vaccenyl acetate (CAS # 6186-98-7) and 4MBITC (CAS # 4430-36-8) were diluted in ethanol. All chemicals used in this study are listed in supplementary file 8, Supplementary Material online.
The “net number of spikes” was obtained by counting the number of spikes originating from the OSN of interest during a 1-s window which started 0.2 s after the onset of stimulation, and subtracting from this count the background spiking activity (obtained by counting the number of spikes in a 1-s window prior to the onset of the odor stimulation). In all figures unless otherwise noted, the net response to each odor or odorant stimulation was subtracted from the median net response to the solvent control used. Some odorants were dissolved in solvents other than mineral oil and so the respective control was used (supplementary file 9, Supplementary Material online).
In SSR experiments, it is common to observe unspecific, slight changes in spiking activity which are not considered biologically meaningful, including small responses to solvent controls. It is common to find reports, including investigations using the empty neuron system of D. melanogaster (Hallem and Carlson 2006), in which net spiking responses smaller than 10–25 spikes/second are not considered true odor-evoked responses (e.g., Stensmyr et al. 2003; Olsson et al. 2006). Therefore, we asked whether the net responses of a given Or to a given stimulus are statistically significant (P < 0.05) using one-sample signed rank tests under the following null and alternative hypotheses: Ho: net number of spikes >−10 and <10, and Ha: net number of spikes <−10 or >10. The responses of a given paralog to various stimuli were also compared using Kruskal–Wallis ANOVAs (followed by posthoc tests if significant). Although results were considered significant only if P < 0.05, we indicated cases in which P-values were slightly larger (0.05 < P < 0.08). We report these values because “a result that does not meet the P < 0.05 threshold should not be considered meaningless when in fact provides…at least preliminary evidence that requires further attention” (Makin and de Xivry 2019), especially with small sample sizes due to the nature of the experiment, as in this report.
Tuning curves and kurtosis values were generated and calculated in Microsoft Excel (2016). Similarly, a matrix of median responses (control subtracted) was produced and used for PCA in R statistical software. For generation of the heatmap in R version 1.4.1717 (fig. 4B), the median responses of OR-odorant pairs were normalized to the maximum median response (which was evoked by BITC at 1:100 vol/vol) for each Or.
Y-Maze Binary Choice Assay
The olfactory responses of mated, fed adult female D. melanogaster, S. pallida, and S. flava were tested using a dual-choice “Y-shaped” olfactometer (Reisenman et al. 2013; supplementary fig. 8, Supplementary Material online). The “Y piece” of the olfactometer was a propylene connector, and the arms of the “Y” were each connected to a 1-ml syringe containing a piece of filter paper (6 × 50 mm) loaded with the odor or control stimuli. The odorous stimulus was loaded on a piece of filter paper and inserted into the 1-ml syringe immediately prior to each test; control syringes had a piece of filter paper loaded with the mineral oil solvent (for odorant solutions) or water (in tests apple cider vinegar). The odorants (20 µl of 1:100 or 1:1,000 vol/vol mineral oil solution, depending on the experiment) used in experiments were BITC (CAS # 592-82-5) and SBITC (CAS # 15585-98-5), and acetophenone (CAS # 98-86-2) at various concentrations. We also used apple cider vinegar (40 µl, O Organics, USA). For tests of host orientation, leaves from four to 6 weeks old E. sativa, A. thaliana, or S. lycopersicum plants, grown in an insect and insecticide/pesticide free chamber or greenhouse, were excised and broken just before tests and placed in 5 ml syringes connected to the Y-maze; control syringes had two pieces of wet tissue paper. In all cases, the Y-mazes, tubing, and syringes were washed with 70% ethanol and allowed to air-dry before reuse.
Flies were starved 24 h for these experiments. Flies were anesthetized under CO2 for 2 h (in the case of D. melanogaster and S. pallida) or ca. 20 h before tests (in the case of S. flava), and placed in groups of 3–4 individuals in open-top and mesh-bottom cylindrical release containers (20 mm long × 10 mm diameter) constructed using silicon tubing. The open tops of the containers were capped with tissue paper soaked in distilled water (in the case of D. melanogaster and S. pallida) or with a piece of tissue soaked with 10% vol/vol aqueous honey solution (in the case of S. flava). Before tests, each release tube was placed on ice for 45–60 s to slow down insect activity; the cap was then removed and the open top of the tube was slid into the open end of the Y-maze. Only the first choice (and the time of the choice) was recorded among each group of 3–4 flies per trial. A choice was considered as such only if the fly walked at least 10 mm into one of the arms, orienting upwind. Each test lasted a maximum of 5 min. Experiments were conducted during the 2–5 h of the insects’ photophase at 24 °C under white light (Feit electric, 100 W; in the case of S. pallida and D. melanogaster). Green light (Sunlite green, 100 W) was used in the case of experiments with S. flava, as our observations suggest that this species is more active under this condition. The test was discarded if two or more insects chose different arms of the maze within a 5-s window of each other. Results from a test session with a given species/fly line were discarded if insects did not make a choice in at least 50% of tests. This happened in less than 5–10% of experimental sessions except for S. pallida, which often had low activity levels. Insects from the same cohort were tested in the same day with different stimuli as much as possible. In the case of experiments using transgenic flies, we conducted experiments with the progeny of at least 4–5 independent crosses; genetic control and test flies were tested in parallel as much as possible. Tests with each combination of fly line or species and stimulus were conducted in at least five different days with different progeny. The position of the odor and the odorless arm was switched every 1–2 tests to control for positional asymmetries; the mazes and odor sources were changed and replaced for clean/new ones every 4 tests or 10 min, whichever occurred first.
For each odor/odorant, species, and fly line, the number of tests in which an insect made a choice for one or the other arm of the Y-maze (with the criteria described above) were tested against a 50% expected random distribution using two-tailed binomial tests (Zar 1999). Results were considered statistically significant if P < 0.05. We established a priori a minimum sample size (n ≥ 30), and data collection for each experimental series/species/condition ended when significance was achieved, or when n = 55, regardless of the outcome. Scaptomyzapallida was unusually inactive and so sample sizes were sometimes lower but averaged n = 28 across experimental series. This criterion was adopted because binomial tests require an untenably large sample size to evince small deviations from an expected proportion (e.g., n ≥ 78 to detect a 10% deviation from the expected 50% random choice with P < 0.05). In cases where n < 55, we conducted a similar number of tests for control and experimental lines to ensure that results were comparable. Similarly, we also noted behavioral results when 0.05 < P < 0.08, because such outcomes indicate that significance at P < 0.05 could likely be achieved by increasing sample size (Makin and de Xivry 2019), although increasing sample size is often difficult for this type of behavioral experiment. For example, a 60% choice with n = 55 yields P = 0.08, and sample size needs to be ≥74 for achieving P < 0.05. Statistical power was in most significant cases >0.8; exceptions include cases where deviations from the expected distribution were small (e.g., a significant 10% deviation requires n = 188 to achieve 0.8 statistical power; supplementary file 10, Supplementary Material online).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Drs Chauda Sebastian, Dennis Mathew, John Carlson, Barry J. Dickson, and Bloomington Drosophila Stock Center (NIH P40OD018537) for sharing M2-MD, UAS-Dmel Or67b, and Or67dGal4, and to Drs Johannes Bischof and Konrad Basler for donation of the pUASTattB plasmid. C.E.R. thanks Dr Kristin Scott for support and encouragement. T.M. thanks Dr Makoto Hiroi for advice on SSR experiments. We thank members of the Whiteman and Scott Laboratories for discussions and comments on the manuscript. T.M. was supported by the Uehara Memorial Foundation (award number 201931028); N.K.W. by the National Institute of General Medical Sciences of the National Institutes of Health (award number R35GM119816); B.G.-H. by the National Science Foundation (award number IOS 1755188 and DEB 1601355).
References
- Allmann S, Späthe A, Bisch-Knaden S, Kallenbach M, Reinecke A, Sachse S, Baldwin IT, Hansson BS.. 2013. Feeding-induced rearrangement of green leaf volatiles reduces moth oviposition. Elife 2:e00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R, Bachtrog D.. 2013. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A. 110(43):17409–17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Khallaf MA, Silbering AF, Zappia G, Ellis K, Álvarez-Ocaña R, Roman Arguello J, Hansson BS, Gregory SX, Caron SJC, et al. 2020. Olfactory receptor and circuit evolution promote host specialization. Nature 579(7799):402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurand LW, Singleton JA, Bell TA, Etchells JL.. 1966. Volatile components in the vapors of natural and distilled vinegars. J Food Sci. 31(2):172–177. [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M.. 2001. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 21(5):1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J, Van Leeuwen W, Van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, et al. 2008. The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS One 3(4):e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B Methodol. 57:289–300. [Google Scholar]
- Berenbaum MR, Zangerl AR.. 2008. Facing the future of plant-insect interaction research: le retour à la “raison d’être.” Plant Physiol. 146(3):804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernays E, Graham M.. 1988. On the evolution of host specificity in phytophagous arthropods. Ecology 69(4):886–892. [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K.. 2007. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc Natl Acad Sci U S A. 104(9):3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4):e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce TJA, Pickett JA.. 2011. Perception of plant volatile blends by herbivorous insects–finding the right mix. Phytochemistry 72(13):1605–1611. [DOI] [PubMed] [Google Scholar]
- Butenandt A. 1959. Über den Sexual-Lockstoff des Seidenspinners Bombyx mori: Reindarstellung und Konstitution. Verlag d. Zeitschr. f. Naturforschung. Available from: https://play.google.com/store/books/details?id=j-uNPgAACAAJ. Accessed January 3, 2022.
- Chaverra-Rodriguez D, Macias VM, Hughes GL, Pujhari S, Suzuki Y, Peterson DR, Kim D, McKeand S, Rasgon JL.. 2018. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun. 9:3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin SG, Maguire SE, Huoviala P, Jefferis GSXE, Potter CJ.. 2018. Olfactory neurons and brain centers directing oviposition decisions in drosophila. Cell Rep. 24(6):1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden HM. 1902. The American fur trade of the far west: a history of the pioneer trading posts and early fur companies of the Missouri valley and the Rocky Mountains and the overland commerce with Santa Fe. F.P. Harper. Available from: https://play.google.com/store/books/details?id=EkFEAQAAMAAJ. Accessed January 3, 2022.
- Cousin FJ, Le Guellec R, Schlusselhuber M, Dalmasso M, Laplace J-M, Cretenet M.. 2017. Microorganisms in fermented apple beverages: current knowledge and future directions. Microorganisms 5(3):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley-Gall A, Date P, Han C, Rhodes N, Andolfatto P, Layne JE, Rollmann SM.. 2016. Population differences in olfaction accompany host shift in Drosophila mojavensis. Proc Biol Sci. 283(1837):20161562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS.. 2006. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 16(1):101–109. [DOI] [PubMed] [Google Scholar]
- ΡDemirel N, Cranshaw W.. 2006. Relative attraction of color traps and plant extracts to the false chinch bugNysius raphanus and its parasitoid, Phasia occidentis, on brassica crops in Colorado. Phytoparasitica 34(2):197–203. [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR.. 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37(5):827–841. [DOI] [PubMed] [Google Scholar]
- Dweck HKM, Ebrahim SAM, Kromann S, Bown D, Hillbur Y, Sachse S, Hansson BS, Stensmyr MC.. 2013. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr Biol. 23(24):2472–2480. [DOI] [PubMed] [Google Scholar]
- Eckenrode CJ, Arn H.. 1972. Trapping cabbage maggots with plant bait and allyl isothiocyanate. J Econ Entomol. 65(5):1343–1345. [DOI] [PubMed] [Google Scholar]
- Edger PP, Heidel-Fischer HM, Bekaert M, Rota J, Glöckner G, Platts AE, Heckel DG, Der JP, Wafula EK, Tang M, et al. 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 112(27):8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P.. 2001. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56:5–51. [DOI] [PubMed] [Google Scholar]
- Faucher CP, Hilker M, de Bruyne M.. 2013. Interactions of carbon dioxide and food odours in Drosophila: olfactory hedonics and sensory neuron properties. PLoS One. 8(2):e56361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel G. 1959. The raison d’etre of secondary plant substances. Science 129:1466–1470. [DOI] [PubMed] [Google Scholar]
- Galizia CG, Sachse S. 2010. Odor Coding in Insects. In: Menini A, editor. The Neurobiology of Olfaction. Chapter 2. Boca Raton (FL): CRC Press/Taylor & Francis. [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM.. 1994. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci U S A. 91(19):8955–8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss AD, Nelson Dittrich AC, Lapoint RT, Goldman-Huertas B, Verster KI, Pelaez JL, Nelson ADL, Aguilar J, Armstrong E, Charboneau JLM, et al. 2019. Evolution of herbivory remodels a Drosophila genome. bioRxiv767160. Available from: https://www.biorxiv.org/content/10.1101/767160v1.abstract. Accessed January 3, 2022.
- Gloss AD, Vassão DG, Hailey AL, Nelson Dittrich AC, Schramm K, Reichelt M, Rast TJ, Weichsel A, Cravens MG, Gershenzon J, et al. 2014. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the drosophilidae. Mol Biol Evol. 31(9):2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Huertas B, Mitchell RF, Lapoint RT, Faucher CP, Hildebrand JG, Whiteman NK.. 2015. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc Natl Acad Sci U S A. 112(10):3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Witzgall P, Walker WB.. 2016. Protocol for heterologous expression of insect odourant receptors in Drosophila. Front Ecol Evol. 4:189. [Google Scholar]
- Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY.. 2000. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 97(22):11990–11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber MY, Xu N, Grenkow L, Li X, Onyilagha J, Soroka JJ, Westcott ND, Hegedus DD.. 2009. Responses of the crucifer flea beetle to Brassica volatiles in an olfactometer. Environ Entomol. 38(5):1467–1479. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Guo S, Kim J.. 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol. 24(5):1198–1207. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR.. 2004. The odor coding system of Drosophila. Trends Genet. 20(9):453–459. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR.. 2006. Coding of odors by a receptor repertoire. Cell 125(1):143–160. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J.. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169(2):1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel DG. 2018. Insect detoxification and sequestration strategies. Annu Plant Rev. 77–114. Available from: https://onlinelibrary.wiley.com/doi/10.1002/9781119312994.apr0507. Accessed January 3, 2022. [Google Scholar]
- Heidel-Fischer HM, Kirsch R, Reichelt M, Ahn S-J, Wielsch N, Baxter SW, Heckel DG, Vogel H, Kroymann J.. 2019. An insect counteradaptation against host plant defenses evolved through concerted neofunctionalization. Mol Biol Evol. 36(5):930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman A, Chuang H-H, Bautista DM, Julius D.. 2006. TRP channel activation by reversible covalent modification. Proc Natl Acad Sci U S A. 103(51):19564–19568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare DJ, Humble J, Jin D, Gilding N, Petersen R, Cobb M, McCrohan C.. 2011. Modeling peripheral olfactory coding in Drosophila larvae. PLoS One. 6(8):e22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idstein H, Bauer C, Schreier P.. 1985. Flüchtige Säuren in Tropenfrüchten: cherimoya (Annona cherimolia, Mill.), Guava (Psidium guajava, L.), Mango (Mangifera indica, L., var. Alphonso), Papaya (Carica papaya, L.) [Volatile acids in tropical fruits: cherimoya (Annona cherimolia, Mill.), guava (psidium guajava, L.), mango (Mangifera indica, L., var. Alphonso), papaya (Carica papaya, L.)]. Z Lebensm Unters Forsch. 180(5):394–397. [DOI] [PubMed] [Google Scholar]
- Ishii G, Saijo R, Mizutani J.. 1989. A quantitative determination of 4-methylthio-3-butenyl glucosinolate in daikon (Raphanus sativus L.) roots by gas liquid chromatography. Engei Gakkai Zasshi. 58(2):339–344. [Google Scholar]
- Jacobsson A, Nielsen T, Sjöholm I.. 2004. Influence of temperature, modified atmosphere packaging, and heat treatment on aroma compounds in broccoli. J Agric Food Chem. 52(6):1607–1614. [DOI] [PubMed] [Google Scholar]
- Jaenike J. 1990. Host specialization in phytophagous insects. Annu Rev Ecol Syst. 21(1):243–273. [Google Scholar]
- Joseph RM, Carlson JR.. 2015. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genetics. 31(12):683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, Garrity PA.. 2010. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464(7288):597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-I, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Izumitani HF, Yamashita S, Watada M.. 2017. Multiple origins of Hawaiian drosophilids: phylogeography of Scaptomyza hardy (Diptera: Drosophilidae). Entomol Sci. 20(1):33–44. [Google Scholar]
- Kawahara AY, Plotkin D, Espeland M, Meusemann K, Toussaint EFA, Donath A, Gimnich F, Frandsen PB, Zwick A, dos Reis M, et al. 2019. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc Natl Acad Sci U S A. 116(45):22657–22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesey I, Zhang J, Depetris-Chauvin A, Obiero GF, Knaden M, Hansson BS.. 2019. Evolution of a pest: towards the complete neuroethology of Drosophila suzukii and the subgenus Sophophora. bioRxiv [Internet]. Available from: https://pure.mpg.de/pubman/faces/ViewItemOverviewPage.jsp?itemId=item_3150127. [DOI] [PMC free article] [PubMed]
- Kergunteuil A, Dugravot S, Mortreuil A, Le Ralec A, Cortesero AM.. 2012. Selecting volatiles to protect brassicaceous crops against the cabbage root fly, Delia radicum. Entomol Exp Appl. 144(1):69–77. [Google Scholar]
- Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D’Agostino ER, Pelaez J, et al. 2021. Highly contiguous assemblies of 101 drosophilid genomes. Elife 10:e66405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ.. 2007. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446(7135):542–546. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B.. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. [DOI] [PubMed] [Google Scholar]
- Lapoint RT, O’Grady PM, Whiteman NK.. 2013. Diversification and dispersal of the Hawaiian Drosophilidae: the evolution of Scaptomyza. Mol Phylogenet Evol. 69(1):95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein EP, Strong FM, Morgan DG.. 1962. Naturally occurring insecticides, identification of 2-Phenylethylisothiocyanate as an insecticide occurring naturally in the edible part of turnips. J Agric Food Chem. 10(1):30–33. [Google Scholar]
- Linz J, Baschwitz A, Strutz A, Dweck HKM, Sachse S, Hansson BS, Stensmyr MC.. 2013. Host plant-driven sensory specialization in Drosophila erecta. Proc. Biol. Sci. 280:20130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-L, Zhang J, Yan Q, Miao C-L, Han W-K, Hou W, Yang K, Hansson BS, Peng Y-C, Guo J-M, et al. 2020. The molecular basis of host selection in a crucifer-specialized moth. Curr Biol. 30(22):4476–4482.e5. [DOI] [PubMed] [Google Scholar]
- Maca J. 1972. Czechoslovak species of the genus Scaptomyza Hardy (Diptera, Drosophilidae) and their bionomics. Acta Entomol Bohemoslov. 69:119–132. [Google Scholar]
- Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, Patapoutian A.. 2007. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature 445(7127):541–545. [DOI] [PubMed] [Google Scholar]
- Makin TR, de Xivry J-JO.. 2019. Science forum: ten common statistical mistakes to watch out for when writing or reviewing a manuscript. Elife 8:e48175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S, Enjin A, Jirle EV, Ramesh V, Rehermann G, Becher PG, Pool JE, Stensmyr MC.. 2018. Wild African Drosophila melanogaster are seasonal specialists on marula fruit. Curr Biol. 28(24):3960–3968.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna DD, Shin S, Ahrens D, Balke M, Beza-Beza C, Clarke DJ, Donath A, Escalona HE, Friedrich F, Letsch H, et al. 2019. The evolution and genomic basis of beetle diversity. Proc Natl Acad Sci U S A. 116(49):24729–24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Celis ME, Yahia EM, Bedoya R, Landázuri P, Loango N, Aguillón J, Restrepo B, Guerrero Ospina JC.. 2019. Chemical composition of mango (Mangifera indica L.) fruit: nutritional and phytochemical compounds. Front Plant Sci. 10:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T. 2001. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol Evol. 16(12):693–700. [Google Scholar]
- Münch D, Galizia CG.. 2016. DoOR 2.0–comprehensive mapping of Drosophila melanogaster odorant responses. Sci Rep. 6:21841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. The enormous diversity in genome sizes of fish as a reflection of natureˈs extensive experiments with gene duplication. Trans Am Fish Soc. 99(1):120–130. [Google Scholar]
- Olsson SB, Linn CE Jr, Roelofs WL.. 2006. The chemosensory basis for behavioral divergence involved in sympatric host shifts. I. Characterizing olfactory receptor neuron classes responding to key host volatiles. J Comp Physiol A. 192(3):279–288. [DOI] [PubMed] [Google Scholar]
- Pelaez JN, Gloss AD, Ray JF, Whiteman NK.. 2020. Evolution and genetic basis of the plant-penetrating ovipositor, a key adaptation in the transition to herbivory within the Drosophilidae. Cold Spring Harbor Lab. 2020.05.07.083253. Available from: https://www.biorxiv.org/content/10.1101/2020.05.07.083253v1.abstract. Accessed January 3, 2022. [Google Scholar]
- Piersanti S, Rebora M, Ederli L, Pasqualini S, Salerno G.. 2020. Role of chemical cues in cabbage stink bug host plant selection. J Insect Physiol. 120:103994. [DOI] [PubMed] [Google Scholar]
- Pivnick KA, Jarvis BJ, Slater GP.. 1994. Identification of olfactory cues used in host-plant finding by diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). J Chem Ecol. 20(7):1407–1427. [DOI] [PubMed] [Google Scholar]
- Pivnick KA, Reed DW, Millar JG, Underhill EW.. 1991. Attraction of northern false chinch bug Nysius niger (Heteroptera: Lygaeidae) to mustard oils. J Chem Ecol. 17(5):931–941. [DOI] [PubMed] [Google Scholar]
- Prieto-Godino LL, Rytz R, Bargeton B, Abuin L, Roman Arguello J, Peraro MD, Benton R.. 2016. Olfactory receptor pseudo-pseudogenes. Nature 539(7627):93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, D, Peraro M, Benton R.. 2017. Evolution of acid-sensing olfactory circuits in drosophilids. Neuron 93(3):661–676.e6. [DOI] [PubMed] [Google Scholar]
- Reisenman CE, Lee Y, Gregory T, Guerenstein PG.. 2013. Effects of starvation on the olfactory responses of the blood-sucking bug Rhodnius prolixus. J Insect Physiol. 59(7):717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick JAA, Haribal M, Gouinguené S, Städler E.. 2006. Isothiocyanates stimulating oviposition by the diamondback moth, Plutella xylostella. J Chem Ecol. 32(4):755–766. [DOI] [PubMed] [Google Scholar]
- Richardson B. 2013. Identification and characterization of potent odorants in selected beet root (Beta vulgaris) products. Available from: https://www.ideals.illinois.edu/handle/2142/45435. Accessed January 3, 2022.
- Robertson HM, Warr CG, Carlson JR.. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100(Suppl 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohloff J, Bones AM.. 2005. Volatile profiling of Arabidopsis thaliana - putative olfactory compounds in plant communication. Phytochemistry 66(16):1941–1955. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T.. 2004. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci U S A. 101(47):16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Zamir D, Fernie AR.. 2004. Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J Exp Bot. 56(410):297–307. [DOI] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E.. 2006. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141(4):1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ.. 2007. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One. 2(12):e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensmyr MC, Dweck HKM, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, et al. 2012. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151(6):1345–1357. [DOI] [PubMed] [Google Scholar]
- Stensmyr MC, Giordano E, Balloi A, Angioy A-M, Hansson BS.. 2003. Novel natural ligands for Drosophila olfactory receptor neurones. J Exp Biol. 206(4):715–724. [DOI] [PubMed] [Google Scholar]
- Strutz A, Soelter J, Baschwitz A, Farhan A, Grabe V, Rybak J, Knaden M, Schmuker M, Hansson BS, Sachse S.. 2014. Decoding odor quality and intensity in the Drosophila brain. Elife 3:e04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana T, Savage GP, McNeil DL, Porter NG, Clark B.. 2003. Comparison of flavour compounds in wasabi and horseradish. J Food Agric Environ. 1:117–121. [Google Scholar]
- Swofford DL. 2002. PAUP: phylogenetic analysis using parsimony, version 4.0 b10.
- Syed TH, Famiglietti JS, Chambers DP, Willis JK, Hilburn K.. 2010. Satellite-based global-ocean mass balance estimates of interannual variability and emerging trends in continental freshwater discharge. Proc Natl Acad Sci U S A. 107(42):17916–17921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Vogel H, Wittstock U, Braby MF, Underwood D, Mitchell-Olds T.. 2007. The genetic basis of a plant–insect coevolutionary key innovation. Proc Natl Acad Sci U S A. 104(51):20427–20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Gloss AD, Sackton TB, Groen SC, Humphrey PT, Lapoint RT, Sønderby IE, Halkier BA, Kocks C, Ausubel FM, et al. 2012. Genes involved in the evolution of herbivory by a leaf-mining, Drosophilid fly. Genome Biol Evol. 4(9):900–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Groen SC, Chevasco D, Bear A, Beckwith N, Gregory TR, Denoux C, Mammarella N, Ausubel FM, Pierce NE.. 2011. Mining the plant–herbivore interface with a leafmining Drosophila of Arabidopsis. Mol Ecol. 20(5):995–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore B, Tolhurst DJ.. 2001. Characterizing the sparseness of neural codes. Network 12(3):255–270. [PubMed] [Google Scholar]
- Xue Y-L, Han H-T, Liu C-J, Gao Q, Li J-H, Zhang J-H, Li D-J, Liu C-Q.. 2020. Multivariate analyses of the volatile components in fresh and dried turnip (Brassica rapa L.) chips via HS-SPME-GC-MS. J Food Sci Technol. 57(9):3390–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zar JH. 1999. Biostatistical analysis. New Jersey: Prentice Hall. Available from: https://play.google.com/store/books/details?id=zLUxhpbgxKYC. Accessed January 3, 2022. [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL.. 2002. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16(23):3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, McBride CS.. 2020. Correction to: evolution of olfactory circuits in insects. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 206(4):663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.