Abstract
Background
Approximately 20% of lung adenocarcinoma (LUAD) is negative for the lineage-specific oncogene Thyroid transcription factor 1 (TTF-1) and exhibits worse clinical outcome with a low frequency of actionable genomic alterations. To identify molecular features associated with TTF-1–negative LUAD, we compared the transcriptomic and proteomic profiles of LUAD cell lines. SRGN , a chondroitin sulfate proteoglycan Serglycin, was identified as a markedly overexpressed gene in TTF-1–negative LUAD. We therefore investigated the roles and regulation of SRGN in TTF-1–negative LUAD.
Methods
Proteomic and metabolomic analyses of 41 LUAD cell lines were done using mass spectrometry. The function of SRGN was investigated in 3 TTF-1–negative and 4 TTF-1–positive LUAD cell lines and in a syngeneic mouse model (n = 5 to 8 mice per group). Expression of SRGN was evaluated in 94 and 105 surgically resected LUAD tumor specimens using immunohistochemistry. All statistical tests were 2-sided.
Results
SRGN was markedly overexpressed at mRNA and protein levels in TTF-1–negative LUAD cell lines (P < .001 for both mRNA and protein levels). Expression of SRGN in LUAD tumor tissue was associated with poor outcome (hazard ratio = 4.22, 95% confidence interval = 1.12 to 15.86, likelihood ratio test, P = .03), and with higher expression of Programmed cell death 1 ligand 1 (PD-L1) in tumor cells and higher infiltration of Programmed cell death protein 1–positive lymphocytes. SRGN regulated expression of PD-L1 as well as proinflammatory cytokines, including Interleukin-6, Interleukin-8, and C-X-C motif chemokine 1 in LUAD cell lines; increased migratory and invasive properties of LUAD cells and fibroblasts; and enhanced angiogenesis. SRGN was induced by DNA demethylation resulting from Nicotinamide N-methyltransferase–mediated impairment of methionine metabolism.
Conclusions
Our findings suggest that SRGN plays a pivotal role in tumor–stromal interaction and reprogramming into an aggressive and immunosuppressive tumor microenvironment in TTF-1–negative LUAD.
Thyroid transcription factor-1 (TTF-1), also known as NKX2-1, is a homeodomain transcription factor required for lung morphogenesis and epithelial cell differentiation (1). TTF-1 is amplified in 10%-15% of lung adenocarcinoma (LUAD) and plays a crucial role as a lineage-survival oncogene in LUAD (2-5). On the other hand, TTF-1 also possesses tumor-suppressive functions such as inhibition of metastasis and tumorigenesis induced by oncogenic Kirsten rat sarcoma viral oncogene (KRAS) as well as regulation of epithelial differentiation status (6,7). TTF-1–negative LUAD, accounting for approximately 20% of LUAD (8–10), is associated with worse clinical outcomes (9–14) and exhibits a low frequency of actionable genomic alterations, including Epidermal growth factor receptor (EGFR) mutation (9,12,15). Because loss of Ttf-1 in murine lung tumors is not sufficient to induce metastasis (16), the mechanisms underlying acquisition of aggressive properties of TTF-1–negative LUAD are not well understood.
In this study, we initially compared the transcriptomic and proteomic profiles of 41 LUAD cell lines to identify molecular features associated with TTF-1–negative LUAD. Serglycin (SRGN) was identified as a markedly overexpressed gene in TTF-1–negative LUAD cell lines. SRGN is a chondroitin sulfate proteoglycan expressed in most hematopoietic cells and plays an intricate role in inflammation by interacting with many inflammatory mediators, including proteases, chemokines, cytokines, and growth factors (17,18). SRGN is also expressed in several types of epithelial cancers with aggressive phenotypes (19–23). We thus further investigated the functional relevance and regulation of SRGN in TTF-1–negative LUAD.
Methods
Detailed experimental procedures are provided in the Supplementary Methods (available online).
Tissue Samples
All human tissue samples were obtained following institutional review board approval and informed consent. Set 1 comprised 94 surgically resected LUAD tumor specimens collected at The University of Texas MD Anderson Cancer. Set 2 comprised 105 surgically resected LUAD tumor specimens collected at the Nagoya University Hospital. An additional set of tumor tissues from 17 SRGN-positive and 11 SRGN-negative surgically resected LUAD, collected at the Aichi Cancer Center Hospital, was used for the elastic Verhoeff-Van Gieson staining and CD31 immunohistochemical staining.
Mouse Models
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at MD Anderson and Aichi Cancer Center. Details are found in the Supplementary Methods (available online).
Statistical Analysis
Categorical data were compared using Fisher’s exact test or a χ2 test. Continuous variables were compared using the Mann-Whitney U test or an unpaired t test. Spearman correlation was used to assess the correlation between 2 continuous variables.
Analyses were carried out using Prism 7 software (GraphPad) and the R computing language. A 2-sided statistical significance level of .05 was used for all statistical analyses. Details on analysis of gene expression data, survival analysis, and cluster analysis are provided in the Supplementary Methods (available online).
Results
Comparison of Molecular Profiling of TTF-1–Positive and –Negative LUAD Cell Lines
Based on TTF-1 protein (Figure 1, A) and mRNA expression levels (GSE32036 and GSE63882), we classified 19 LUAD cell lines as TTF-1–positive and 22 as TTF-1–negative (Supplementary Table 1, available online). TTF-1–negative expression status was associated with lower frequency of EGFR mutation, Liver kinase B1 (LKB1) inactivation, and more aggressive migratory and invasive properties (Figure 1, B;Supplementary Table 1, available online) (24).
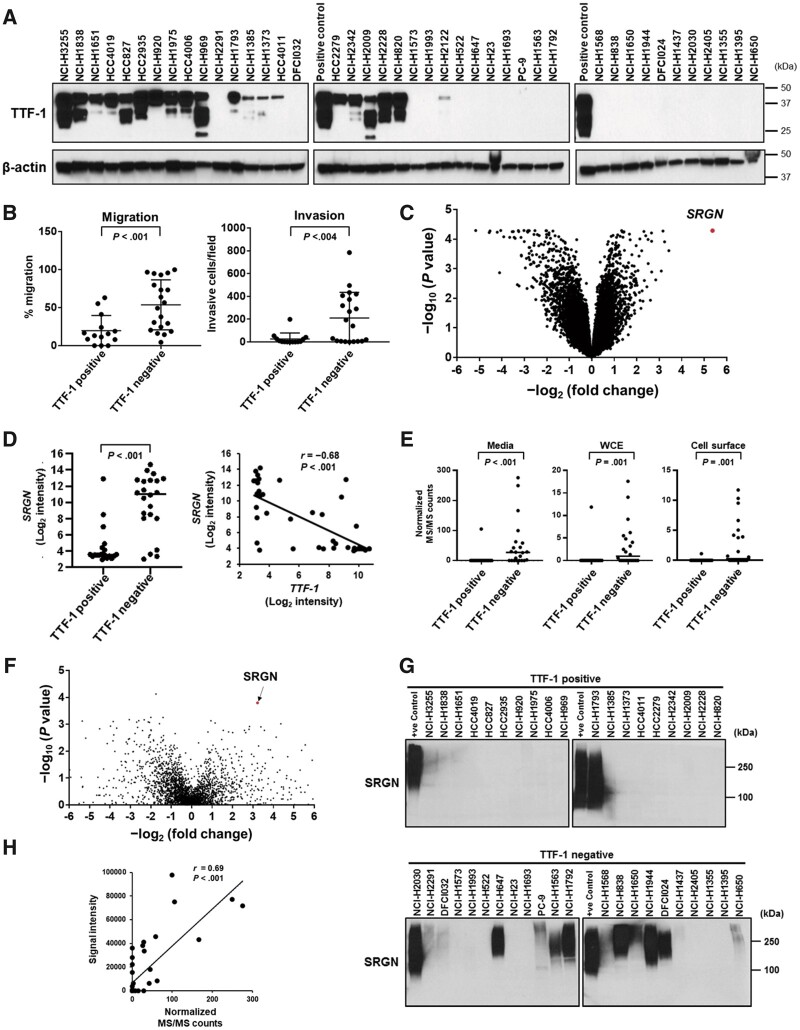
Figure 1.
Expression of Serglycin (SRGN) in lung adenocarcinoma (LUAD) cell lines. A) Immunoblot analysis of Thyroid transcription factor 1 (TTF-1) in LUAD cell lines. H3255 cell lysates were used as a TTF-1–positive control. β-Actin served as a loading control. B) Cell migration and invasion in TTF-1–positive and –negative LUAD cell lines. Data are not available for 5 TTF-1–positive and 2 –negative LUAD cell lines (24). C) Volcano plot of genes differently expressed between TTF-1–positive and –negative LUAD cell lines. The x-axis is log2 fold-change in gene expression between TTF-1–negative and –positive LUAD cell lines, and the y-axis is log10P values adjusted for false discovery rate (FDR) based on the Benjamini and Hochberg method (Supplementary Table 2, available online). D) (Left) mRNA expression of SRGN in TTF-1–positive and –negative LUAD cell lines. (Right) Correlation between TTF-1 and SRGN mRNA expression in LUAD cell lines. E) SRGN protein expression levels in 3 cellular compartments, including conditioned media, whole-cell extracts (WCE), and cell surface proteins, from TTF-1–positive and –negative LUAD cell lines based on normalized tandem mass spectrometry (MS/MS) spectral counts. F) Volcano plot of secreted proteins (normalized MS/MS counts >1; n = 2809) differently expressed between TTF-1–positive and –negative LUAD cell lines. The x-axis is log2 fold-change in normalized MS/MS counts between TTF-1–negative and –positive LUAD cell lines, and the y-axis is log10P values calculated by the Mann-Whitney U test. G) Immunoblot analysis of SRGN expression in the conditioned media from TTF-1–positive and –negative LUAD cell lines. Conditioned media from H2030 cells were used as SRGN-positive controls. H) Correlation of SRGN protein expression quantified by immunoblotting and mass spectrometry. Signal intensity of SRGN in immunoblotting was measured by ImageJ. In B, D (left) and E, horizontal lines indicate mean values and SD, and P values were calculated using Mann-Whitney U test. In D (right) and H, correlation coefficients and P values were calculated using Spearman correlation test. All statistical tests were 2-sided.
To identify molecular features associated with TTF-1–negative LUAD, we compared the gene expression profiles of TTF-1–positive and –negative LUAD cell lines. SRGN, encoding a chondroitin sulfate proteoglycan Serglycin, was the most statistically significantly overexpressed gene in TTF-1–negative cell lines compared with TTF-1–positive cell lines (average log2 intensity [SD] = 9.90 [3.61] for TTF-1–negative and 4.52 [2.46] for TTF-1–positive cell lines, respectively; P < .001, significance analysis of microarrays) and showed a strong inverse correlation with TTF-1 gene expression (Spearman correlation coefficient r = −0.68 and P < .001) (Figure 1, C and D; Supplementary Table 2, available online). Proteomic profiling of 3 cellular compartments (conditioned media, whole-cell extracts, and cell surface proteins) of 41 LUAD cell lines (25) revealed that SRGN was secreted into the extracellular space and markedly increased in TTF-1–negative cell lines (average normalized tandem mass spectrometry counts [SD] in the conditioned media = 51.79 [79.22] for TTF-1–negative and 5.55 [24.17] for TTF-1–positive cell lines, respectively; P < .001, Mann-Whitney U test; Figure 1, E and F). SRGN protein expression in the conditioned media of LUAD cell lines was further confirmed by immunoblotting (Figure 1, G and H).
SRGN Expression and Prognosis in LUAD
We next examined SRGN and TTF-1 protein expression using a tissue microarray of 94 LUAD tumors (set 1; Table 1). SRGN expression was inversely associated with TTF-1 expression (Figure 2, A and B). SRGN-positive LUAD exhibited worse disease-free survival and worse overall survival (OS) (Figure 2, C). In addition, SRGN expression was a statistically independent predictor of OS (multivariable Cox regression analysis, hazard ratio = 4.22, 95% confidence interval = 1.12 to 15.86, P = .03) (Table 2), suggesting SRGN as a potential prognostic biomarker for LUAD. Association of SRGN expression with TTF-1 expression and OS was validated in an independent set consisting of 105 surgically resected Japanese LUAD patients (set 2; Table 1; Figure 2, D and E). Although overall SRGN positivity rate was higher in set 2 (49 of 105 LUAD; 46.7%) compared with set 1 (9 of 94 LUAD; 9.6%), the SRGN positivity rate in set 2 was consistent with the previous study in which SRGN was positive in tumor cells of 39 (48.1%) of 81 Taiwanese non-small cell lung cancer patients (19), suggesting potential molecular diversity between Caucasian and Asian LUAD patients (26–29).
Table 1.
Clinical characteristics in the 2 tissue sample sets
| Clinical characteristics | Set 1 | Set 2 |
|---|---|---|
| No. (%) | No. (%) | |
| Total | 94 | 105 |
| Sex | ||
| Male | 47 (50.0) | 68 (64.8) |
| Female | 47 (50.0) | 37 (35.2) |
| Age | ||
| >65 y | 39 (41.5) | 80 (76.2) |
| ≤65 y | 55 (58.5) | 25 (23.8) |
| Smoking status | ||
| Smoker (current and former) | 82 (87.2) | 67 (63.8) |
| Nonsmoker | 12 (12.8) | 38 (36.2) |
| Stage | ||
| I | 56 (59.6) | 61 (58.1) |
| II | 21 (22.3) | 22 (21.0) |
| III | 17 (18.1) | 17 (16.2) |
| IV | 0 (0.0) | 5 (4.8) |
| Mutationa | ||
| KRAS | 28 (30.1) | — |
| EGFR | 14 (15.1) | 40 (40.8) |
| Wild type | 51 (54.8) | 58 (59.2) |
| TTF-1 immunostaining | ||
| Positive | 62 (66.0) | 70 (66.7) |
| Negative | 32 (34.0) | 35 (33.3) |
Mutations were not investigated in 1 sample in set 1 and 7 samples in set 2. KRAS mutation was not analyzed in set 2.
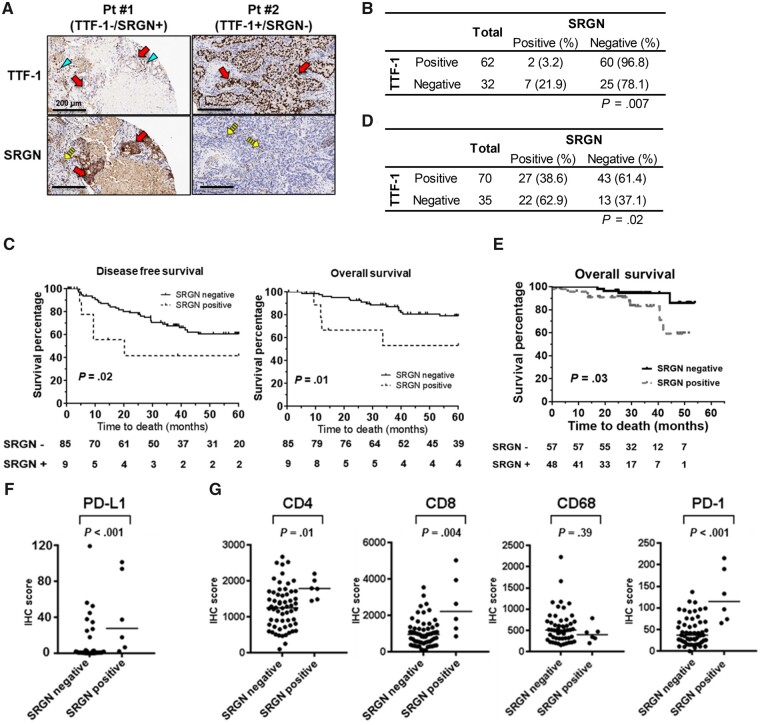
Figure 2.
Association of Serglycin (SRGN) expression and survival of lung adenocarcinoma (LUAD) patients. A) Representative images of immunohistochemical analysis of SRGN and Thyroid transcription factor 1 (TTF-1) in set 1. Arrows indicate tumor cells, arrowheads indicate alveolar type II epithelial cells with nuclear TTF-1 expression, and dashed arrows indicate SRGN expression in inflammatory cells of the tumor stroma. Scale bars represent 200 µm. B) SRGN and TTF-1 expression in set 1 (n = 94). P values were calculated by Fisher’s exact test. C) Kaplan-Meier survival curves for disease-free survival (left) and overall survival (right) in set 1, stratified according to protein expression of SRGN (SRGN-negative, n = 85; SRGN-positive, n = 9). P values were calculated using Gehan-Breslow-Wilcoxon test. D) SRGN and TTF-1 expression in set 2. P values were calculated by Fisher’s exact test. E) Kaplan-Meier survival curves for overall survival in set 2, stratified according to protein expression of SRGN (SRGN-negative, n = 56; SRGN-positive, n = 49). P values were calculated using Gehan-Breslow-Wilcoxon test. F) Programmed cell death 1 ligand 1 (PD-L1) expression in tumor cells and (G) CD4, CD8, CD68, and Programmed cell death protein 1 (PD-1) expression in tumor-infiltrating lymphocytes in the SRGN-positive (n = 6) and SRGN-negative (n = 60) LUAD in set 1. Horizontal lines indicate median values, and P values were calculated using Mann-Whitney U test. All statistical tests were 2-sided.
Table 2.
Univariate and multivariable analysis of overall survival in set 1
| Univariate analysis |
Multivariable analysis |
|||
|---|---|---|---|---|
| Variable | HR (95% CI) | P a | HR (95% CI) | P a |
| Sex | ||||
| Female | 1.00 (Reference) | 1.00 (Reference) | ||
| Male | 1.28 (0.52 to 3.16) | .59 | 0.73 (0.26 to 2.01) | .54 |
| Age | ||||
| ≤65 years | 1.00 (Reference) | 1.00 (Reference) | ||
| >65 years | 0.62 (0.23 to 1.63) | .33 | 0.66 (0.23 to 1.84) | .43 |
| Smoking status | ||||
| Never smoker | 1.00 (Reference) | 1.00 (Reference) | ||
| Former smoker | 0.98 (0.25 to 3.55) | .93 | 0.44 (0.11 to 1.83) | .26 |
| Current smoker | 0.96 (0.25 to 3.63) | .95 | 0.23 (0.05 to 1.11) | .07 |
| Stage | ||||
| I | 1.00 (Reference) | 1.00 (Reference) | ||
| II | 5.41 (1.63 to 17.93) | .006 | 6.06 (1.71 to 21.46) | .005 |
| III | 9.04 (2.64 to 31.03) | <.001 | 16.57 (4.01 to 68.31) | <.001 |
| TTF-1 expression | ||||
| Positive | 1.00 (Reference) | 1.00 (Reference) | ||
| Negative | 2.75 (1.11 to 6.76) | .03 | 2.51 (0.90 to 7.00) | .08 |
| SRGN expression | ||||
| Negative | 1.00 (Reference) | 1.00 (Reference) | ||
| Positive | 3.32 (1.10 to 10.02) | .03 | 4.22 (1.12 to 15.86) | .03 |
P values were calculated by likelihood ratio test. All tests were 2-sided. CI = confidence interval; HR = hazard ratio.
Interestingly, assessment of immune profiles of LUAD tumors in set 1 revealed higher expression of Programmed cell death 1 ligand 1 (PD-L1) in tumor cells and higher infiltration of Programmed cell death protein 1 (PD-1)–positive T lymphocytes in SRGN-positive tumors (Figure 2, F and G), suggesting an immunosuppressive tumor microenvironment in SRGN-positive LUAD.
Analysis of SRGN Function in Vitro and in Vivo
To elucidate the impact of cancer cell–derived SRGN on the tumor microenvironment, we compared the secretome of SRGN-positive (n = 14) and SRGN-negative (n = 24) LUAD cell lines (Supplementary Table 3, available online). Several cytokines, including C-X-C motif chemokine 1 (CXCL1), Interleukin-6 (IL-6), and Interleukin-8 (IL-8), were highly abundant in the secretome of SRGN-positive LUAD cell lines compared with SRGN-negative LUAD cell lines (Supplementary Table 4, available online). Knockdown of SRGN decreased both mRNA and protein levels of CXCL1, IL-6, and IL-8 as well as PD-L1 gene expression (Figure 3, A and B; Supplementary Figure 1, A and B, available online). Knockdown of SRGN did not affect cell growth but reduced cell migration and invasion (Figure 3, C and D; Supplementary Figure 1, C, available online), as recently reported (19).
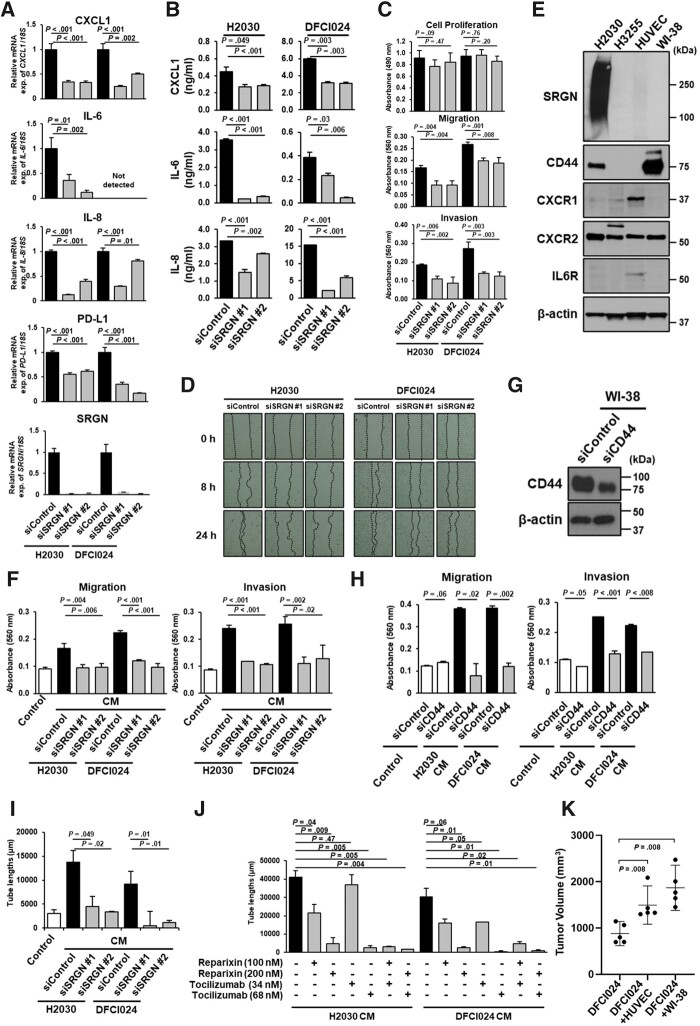
Figure 3.
The regulatory role of Serglycin (SRGN) on expression of proinflammatory cytokines and cancer cell-stromal cell interaction via SRGN in Thyroid transcription factor 1 (TTF-1)–negative lung adenocarcinoma (LUAD). A) mRNA expression levels of C-X-C motif chemokine 1 (CXCL1), Interleukin-6 (IL-6), Interleukin-8 (IL-8), Programmed cell death 1 ligand 1 (PD-L1), and SRGN in H2030 and DFCI024 cell lines with SRGN knockdown. B) Levels of CXCL1, IL-6, and IL-8 in the conditioned media from H2030 and DFCI024 cell lines with SRGN knockdown. C) Cell proliferation, migration, invasion, and D) scratch assays in H2030 and DFCI024 cells with SRGN knockdown. E) Immunoblot analysis of SRGN, CD44, C-X-C chemokine receptor 1 (CXCR1), C-X-C chemokine receptor 2 (CXCR2), and IL-6 receptor (IL6R) expression in umbilical vein endothelial cells (HUVEC) and WI-38 cell lines. Conditioned media and cell lysates of H2030 and H3255 were used as positive and negative controls for SRGN and CD44, respectively. β-Actin served as a loading control. F) Migration and invasion assays in WI-38 cells, cultured in the conditioned media from H2030 and DFCI024 cells treated with negative control siRNA or SRGN siRNA. G) Immunoblot analysis of CD44 expression in WI-38 cell lines treated with negative control siRNA or CD44 siRNA. H) Migration and invasion assay in WI-38 cells with CD44 knockdown, cultured in the conditioned media from H2030 and DFCI024 cells. I) Tube formation assay of HUVEC cells cultured in the conditioned media from H2030 and DFCI024 cells treated with negative control siRNA or SRGN siRNA. J) Tube formation assay of HUVEC cells with CXCR1/2 inhibitor reparixin and/or IL6R inhibitor tocilizumab, cultured in the conditioned media from H2030 and DFCI024 cells. K) Tumor volumes in nude mice 4 weeks after subcutaneous injection with DFCI024 cells alone, with WI-38 cells or with HUVEC cells (n = 5 per group). Before coinjection, WI-38 and HUVEC cells were cultured for 96 hours with conditioned media from DFCI024 cells. Horizontal lines indicate mean and SD. P values were calculated using Mann-Whitney U test. In A-C, F, H-J, experiments have been performed with at least 3 independent biological repeats. Columns indicate the average of triplicate samples from a representative experiment, and bars indicate SD. P values were calculated using unpaired t test, compared with siControl (A-C, F, H, and I) and with control (black) (J). CM = conditioned media. All statistical tests were 2-sided.
To further determine SRGN-mediated interactions of cancer cells with cells in the tumor microenvironment, we cultured fibroblast cell line WI-38 and umbilical vein endothelial cells (HUVECs) using conditioned media from SRGN-positive LUAD cell lines with or without SRGN knockdown. SRGN was not expressed in WI-38 cells but weakly expressed in HUVEC cells (Figure 3, E). Conditioned media from SRGN-positive LUAD cell lines, but not conditioned media from these same cell lines after SRGN knockdown, increased migratory and invasive properties of WI-38 cells (Figure 3, F;Supplementary Figure 1, D, available online), indicative of activation of fibroblasts. In addition, knockdown of CD44, a known receptor of SRGN, in WI-38 cells suppressed migration and invasion induced by cancer cell–derived SRGN (Figure 3, G and H), suggesting a critical role of SRGN/CD44 axis in fibroblast activation. Whereas SRGN was weakly expressed in HUVEC cells (Figure 3, E), conditioned media from SRGN-positive LUAD cell lines, except for H1793 cells, markedly promoted tube formation of HUVEC cells (Figure 3, I;Supplementary Figure 1, E, available online; data not shown for H1793). H1793 cells are the only TTF-1 and SRGN-positive cell line (Supplementary Table 3, available online) with low expression levels of IL-6, IL-8, and CXCL1 (Supplementary Figure 1, F, available online). Despite lack of CD44 expression, IL-6 receptor (IL6R), C-X-C chemokine receptor 1 (CXCR1), and C-X-C chemokine receptor 2 (CXCR2), receptors of IL-6, IL-8, and CXCL1, were expressed in HUVEC cells (Figure 3, E). Both IL6R inhibitor tocilizumab and CXCR1/2 inhibitor reparixin reduced tube formation induced by cancer cell–derived SRGN in a dose-dependent fashion and showed additive effects when combined (Figure 3, J), suggesting that these SRGN-regulated cytokines play an important role in promoting blood vessel remodeling in the tumor microenvironment. Coinjection of DFCI024 cells with WI-38 cells or HUVEC cells activated by conditioned media from DFCI024 cells markedly promoted growth of subcutaneous DFCI024 xenografts in nude mice (Figure 3, K), indicating relevance of reciprocal interaction of LUAD cells with their microenvironment in tumor progression. On the other hand, ex vivo experiments revealed that cancer cell–derived SRGN did not induce PD-1 expression in activated T cells (data not shown). These findings suggest that other immunosuppressive factors in the tumor microenvironment, such as IL-10 and Transforming growth factor beta, may induce expression of PD-1 in tumor-infiltrating lymphocytes.
We next examined the effect of SRGN on the tumor microenvironment in vivo using an immunocompetent syngeneic tumor mouse model with orthotopic transplantation of 393P mouse LUAD cell line in which either Ttf-1 or Srgn was not expressed (Supplementary Figure 2, A, available online). Tail vein injection of parental 393P or a clone overexpressing mouse Srgn revealed that Srgn had increased tumor burden as well as the number and size of tumors in mouse lungs (Figure 4, A-E). Although Srgn also promoted growth of subcutaneously transplanted 393P tumors (Supplementary Figure 2, B, available online), spontaneous metastasis was not observed by 5 weeks, suggesting that interaction between lung microenvironment and tumor cells may be critical in development of metastasis. We found increased fibrosis in Srgn-overexpressing tumors (Figure 4, F;Supplementary Figure 2, C, available online), suggesting increased production and remodeling of the extracellular matrix as the consequence of fibroblast activation. Blood vessels were larger in Srgn-overexpressing tumors (Figure 4, G;Supplementary Figure 2, C and D, available online), suggesting the occurrence of blood vessel remodeling. Remodeling of the microenvironment was also observed in SRGN-positive human LUAD tumors (Figure 4, H–J; Supplementary Figure 2, E, available online).
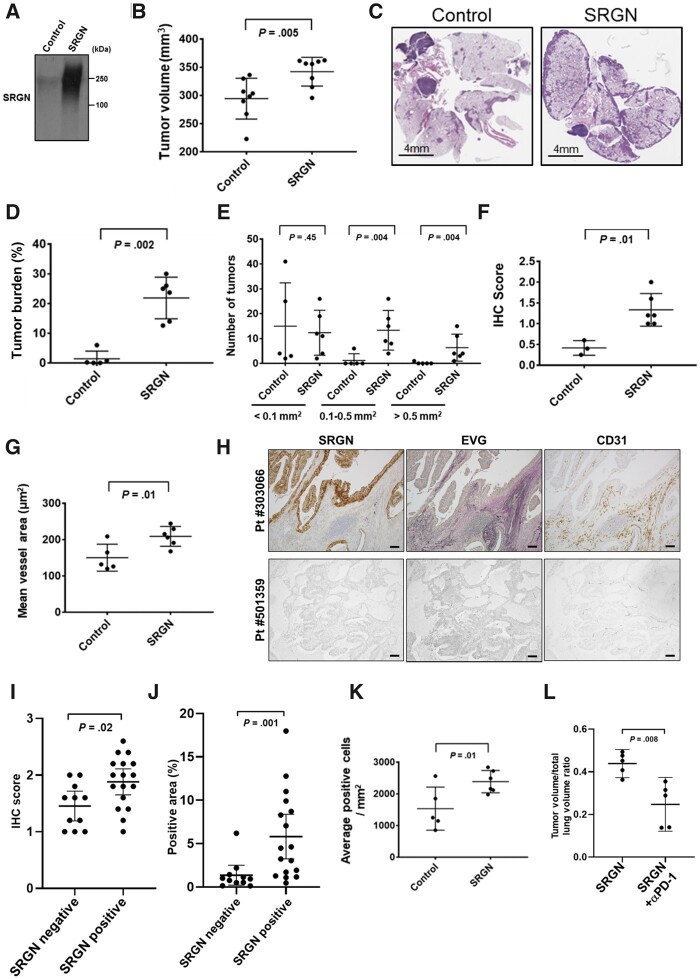
Figure 4.
The contribution of Serglycin (Srgn) to tumor progression and reprogramming of tumor microenvironment in vivo. A) Immunoblot analysis of mouse Srgn using Myc-tag antibody in a control clone and a 393P-derived clone stably expressing mouse Srgn. B) Tumor volumes in mice 2 weeks after tail vein injection of Srgn-overexpressing 393P and control 393P cells (n = 8 per group). Tumor volumes were determined using the Seg3D tool. C) Representative hematoxylin and eosin (H&E) staining of lung sections from mice injected with Srgn-overexpressing 393P clone and control cells. Scale bars represent 4 mm. D) Tumor burden and E) number of tumors were assessed in 6 mice with SRGN-overexpressing tumors and 5 mice with control tumors. Tumor burden was defined as the percentage of area of tumors in H&E staining of lung sections and was quantified using Aperio ImageScope software. F) Tumor fibrosis was assessed by Masson’s trichrome staining in 6 mice with SRGN-overexpressing tumors and 3 mice with control tumors. (G) CD31-positive vessel area was measured using Aperio ImageScope software in 6 mice with SRGN-overexpressing tumors and 5 mice with control tumors. H) Representative images of SRGN and CD31 immunohistochemical staining and Elastica van Gieson (EVG) staining in SRGN-positive (#303066) and -negative (#501359) LUAD tumors. Scale bars represent 100 µm. I) Tumor fibrosis assessed by EVG staining and (J) CD31-positive vessel area in 17 SRGN-positive and 11 negative LUAD tumors. CD31-positive vessel area was measured using Hybrid Cell Count software. K) Number of PD-1–positive T lymphocytes in 6 mice with SRGN-overexpressing tumors and 5 mice with control tumors. L) Tumor volumes in mice 4 weeks after tail vein injection of Srgn-overexpressing 393P cells with or without intraperitoneal administration of anti–PD-1 antibodies (n = 5 per group). Tumor volumes were measured using Image J software. In B, D-G, I-L, horizontal lines indicate mean and SD. P values were calculated using Mann-Whitney U test. All statistical tests were 2-sided.
Consistent with the findings in the human LUAD tissue microarray (set 1), the number of PD-1–positive T lymphocytes was greater in Srgn-overexpressing tumors compared with control tumors, whereas no tumor cells or immune cells were positive for PD-L1 staining in this mouse model (Figure 4, K;Supplementary Figure 2, C, available online). However, PD-1 blockade inhibited growth of Srgn-overexpressing tumors (Figure 4, L;Supplementary Figure 2, F, available online), suggesting the occurrence of immunosuppressive tumor microenvironment in Srgn-overexpressing tumors. The numbers of CD4+ or CD8+ T lymphocytes and macrophages were not different between Srgn-overexpressing tumors and control tumors (Supplementary Figure 2, B and C, available online).
Regulatory Mechanism of SRGN Gene Expression
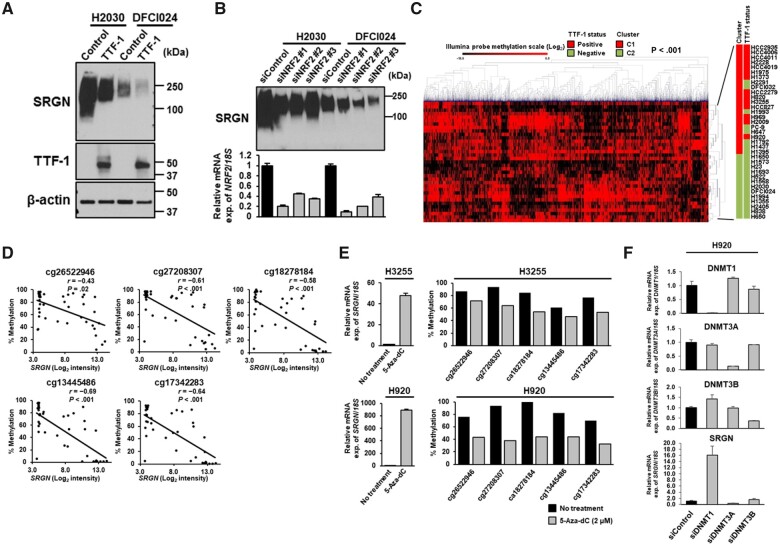
Next, we investigated regulation of SRGN expression in LUAD. Ingenuity Pathway Analysis (http://www.ingenuity.com/) for genes increased in SRGN-positive cell lines compared with SRGN-negative cell lines (Supplementary Tables 3 and 5, available online) identified Nuclear factor erythroid 2-related factor 2 (NRF2)-mediated oxidative stress response as the most statistically significantly associated pathway (−log[P value] = 11.0, Fisher’s exact test) (Supplementary Figure 3, A, available online). Kelch-like ECH-associated protein 1 (KEAP1) mutation and subsequent activation of NRF2, a master transcription factor of the antioxidant response, have been associated with TTF-1–negative LUAD (13). Interestingly, SRGN was highly overexpressed in LUAD cell lines with KEAP1 mutation (Supplementary Figure 3, B, available online). TTF-1 overexpression or NRF2 knockdown decreased SRGN expression at both mRNA and protein levels as well as the activity of luciferase reporter containing the 1.8-kb SRGN promoter region (Figure 5, A and B; Supplementary Figure 3, C and D, available online). On the other hand, TTF-1 and/or KEAP1 knockdown did not induce SRGN expression in TTF-1–positive cell lines (Supplementary Figure 3, E, available online), suggesting the occurrence of additional regulatory mechanisms for suppressing SRGN expression in TTF-1–positive LUAD. Neither overexpression nor knockdown of SRGN altered TTF-1 mRNA expression (Supplementary Figure 3, F, available online).
Figure 5.
Regulation of Serglycin (SRGN) gene expression by DNA methylation. Immunoblot analyses of SRGN expression in the conditioned media from H2030 and DFCI024 cell lines A) after TTF-1 overexpression and B) treated with negative control siRNA or NRF2 siRNA. Experiments have been performed with at least 3 independent biological repeats. C) Hierarchical clustering analysis of the top 15% methylated CpG sites (n = 4137) of an Illumina Infinium HumanMethylation27 BeadChip microarray in 35 LUAD cell lines. P values were calculated using Fisher’s exact test. C1 and C2, clusters 1 and 2. D) Correlation of SRGN mRNA expression and the methylation levels of the 5 CpG sites in the SRGN promoter region in 41 LUAD cell lines. Correlation coefficients and P values were calculated using Spearman correlation test. To adjust P values for multiple comparisons, the Bonferroni correction was conducted. E) Quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of SRGN mRNA expression levels (left) and the methylation levels of the 5 CpG sites in the SRGN promoter region (right) after treatment with 5-Aza-dC at 2 µM in H3255 and H920 cell lines. F) Quantitative RT-PCR analysis of DNA methyltransferase 1 (DNMT1), DNMT3A, DNMT3B, and SRGN mRNA expression levels in H920 cells treated with negative control siRNA or siRNA against DNMT1, DNMT3A, or DNMT3B. In E and F, experiments have been performed with at least 3 independent biological repeats. mRNA expression was presented as the average and SD of triplicate samples from a representative experiment. All statistical tests were 2-sided.
We hypothesized that DNA methylation may regulate SRGN gene expression in LUAD. Given that analyses of global DNA methylation in tumor tissues have identified distinct subtypes of LUAD (27), we first examined the association of genome-wide DNA methylation profiles with cell line characteristics using our previous DNA methylation array data of 35 LUAD cell lines (GSE63940) (24). Interestingly, unsupervised hierarchical clustering analysis revealed that DNA methylation patterns of the most variable probes formed 2 distinctive clusters that were most statistically significantly associated with TTF-1 expression status (P < .001, Fisher’s exact test) (Figure 5, C). Methylation levels of 4 of 5 CpG sites in the SRGN promoter region were statistically significantly and inversely correlated with SRGN mRNA expression levels (r = −0.61, P < .001 for cg27208307; r = −0.58, P < .001 for cg18278184; r = −0.69, P < .001 for cg13445486; and r = −0.64, P < .001 for cg17342283, Spearman correlation; Figure 5, D;Supplementary Figure 3, G and H, available online). Treatment with DNA methylation inhibitor 5-aza-2'-deoxycytidine (5-Aza-dC) markedly increased SRGN mRNA expression levels and decreased methylation levels of the 5 CpG sites in the SRGN promoter region (Figure 5, E). Whereas knockdown of DNA methyltransferases indicated a crucial role of DNA Methyltransferase 1 (DNMT1) in silencing SRGN gene expression (Figure 5, F), gene expression levels of DNA methyltransferases were not associated with TTF-1 expression status or SRGN gene expression levels (Supplementary Figure 3, I and J, available online), suggesting that additional factors may determine DNA methylation status in the promoter region of the SRGN gene.
Analysis of Methionine Metabolism in Regulating SRGN Gene Expression
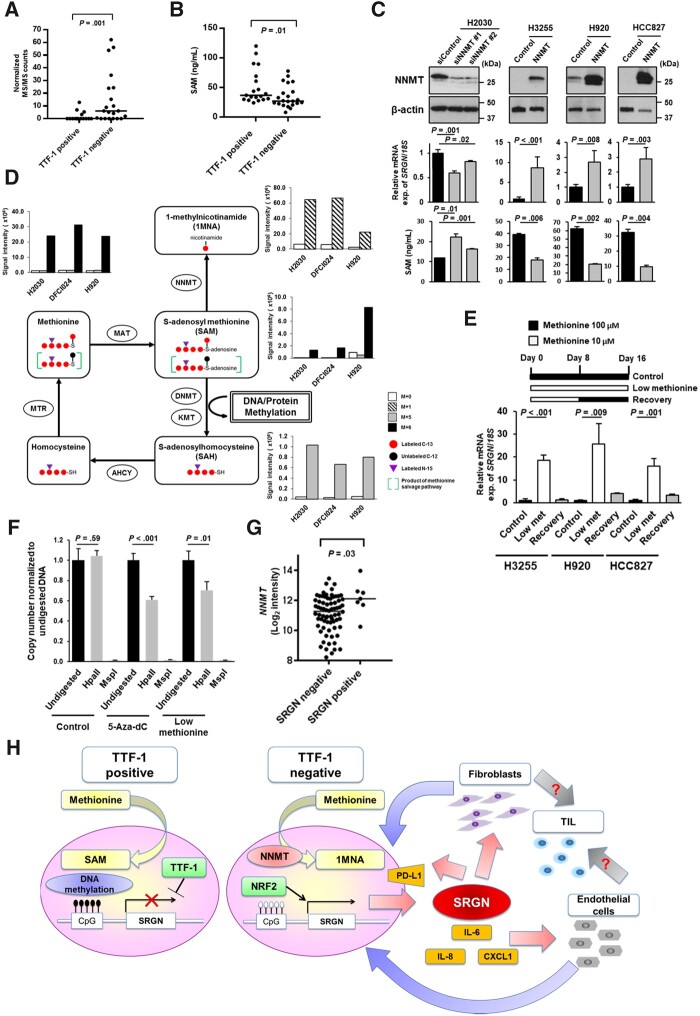
We found that Nicotinamide N-methyltransferase (NNMT) was overexpressed in the whole-cell extracts of TTF-1–negative LUAD cell lines (Figure 6, A). NNMT is a cytosolic enzyme that can impair the methylation potential by consuming S-adenosyl methionine (SAM), the primary methyl group donor for DNA methylation in the methionine cycle (30). In line with the findings, intracellular SAM levels were decreased in TTF-1–negative LUAD cell lines (Figure 6, B). As opposed to previous studies (31,32), SAM levels were decreased in LUAD cell lines with LKB1 inactivation (Supplementary Figure 3, K, available online), possibly due to different expression of SAM-consuming enzymes among LUAD cell lines, because SAM is one of the most widely used enzyme substrates in diverse biological processes. Knockdown of NNMT decreased SRGN mRNA expression levels and increased SAM levels, whereas overexpression of NNMT increased SRGN mRNA expression levels and decreased SAM levels (Figure 6, C), suggesting that NNMT may determine the availability of SAM for DNA methylation and SRGN gene expression levels.
Figure 6.
Induction of Serglycin (SRGN) expression by impaired methionine metabolism in Thyroid transcription factor 1 (TTF-1)–negative lung adenocarcinoma (LUAD). A) Nicotinamide N-methyltransferase (NNMT) protein expression levels in the whole-cell extracts (WCE) of TTF-1–positive and –negative LUAD cell lines based on normalized mass spectrometry (MS/MS) spectral counts. B) S-adenosyl methionine (SAM) levels in TTF-1–positive and –negative LUAD cell lines. SAM levels were normalized according to total protein concentration. C) SRGN mRNA expression levels and SAM levels after NNMT knockdown in H2030 cells or overexpression in H3255, H920, and HCC827 cells. D) Schema of the methionine cycle and methionine metabolic flux in LUAD cell lines 8 hours after exposure to 13C- and 15N-labeled methionine. E) SRGN mRNA expression levels in H3255, H920, and HCC827 cell lines cultured with 100 µM methionine (control), 10 µM methionine (low methionine), and 10 µM methionine for 8 days followed by 100 µM methionine for 8 days (recovery). F) Quantitative polymerase chain reaction (PCR) analysis of DNA methylation in the SRGN promoter region after treatment with 5-Aza-dC or low methionine. At the Chr10: 70847430 (hg19) in the promoter region of the SRGN gene where methylation was presented in H920 cells, the methylation-sensitive enzyme HpaII did not digest the DNA at this site, whereas methylation-insensitive enzyme MspI did digest DNA at this site. Quantitative PCR was conducted after DNA was digested by HpaII or MspI. Both 5-Aza-dC and low methionine (10 µM) treatment statistically significantly decreased DNA methylation at this site in H920 cells. All results were obtained from 3 repetitive experiments with 3 replicates. G) mRNA expression levels of NNMT in the SRGN-positive (N = 7) and SRGN–negative (N = 72) LUAD tumors in the tissue microarray for whom NNMT gene expression data are available. H) A schematic model showing that NNMT-induced perturbation of methionine metabolism induces SRGN-mediated reprogramming of tumor microenvironment in TTF-1–negative LUAD. Overexpression of NNMT consumes the SAM pool available for DNA methylation in TTF-1–negative LUAD, and impaired methionine metabolism results in induction of SRGN gene expression through loss of DNA methylation in the promoter region of the SRGN gene and transcriptional activation by Nuclear factor erythroid 2-related factor 2 (NRF2). Cancer cell-derived SRGN regulates expression of Programmed cell death 1 ligand 1 (PD-L1) and pro-inflammatory cytokines including Interleukin-6 (IL-6), Interleukin-8 (IL-8), and C-X-C motif chemokine 1 (CXCL1) in cancer cells, and reprograms tumor microenvironment via interaction with fibroblasts and endothelial cells, leading to enhanced aggressiveness and immune suppression in TTF-1–negative LUAD. In A, B, and G, horizontal lines indicate median values, and P values were calculated using Mann-Whitney U test. In C, E, and F, experiments have been performed with at least 3 independent biological repeats. Columns indicate the average of triplicate samples from a representative experiment, and bars indicate SD. P values were calculated using unpaired t test compared with control (C and E) and with undigested control (F). All statistical tests were 2-sided.
Analysis of stable-isotope tracer studies after exposure to 13C- and 15N-labeled methionine confirmed sequestration of the methyl group from SAM to 1-methylnicotinamide, a product of NNMT, in TTF-1–negative LUAD cell lines (Figure 6, D). Recycling of methionine occurs via the remethylation of homocysteine by methionine synthase and can be assessed through enrichment of the M + 5 isotopologue of methionine or SAM. Our tracer analysis also indicated that enrichment at the M + 5 position for methionine and SAM was very low in all LUAD cell lines (Figure 6, D), suggesting limited methionine recycling, possibly due to constitutive activation of transsulfuration pathway to maintain the cellular cysteine pool (33) and greater dependence on exogenous supply of methionine or protein catabolism. We then examined the effects of methionine deficiency on SRGN gene expression. Compared with the standard cell culture conditions with 100 µM methionine, SRGN gene expression levels were increased at 10 µM methionine with decreasing DNA methylation levels in the SRGN promoter region, and the increased SRGN gene expression returned to close to the baseline expression levels on subsequent addition of methionine (Figure 6, E and F). However, neither overexpression of NNMT nor methionine deficiency increased protein levels of proinflammatory cytokines in the conditioned media of TTF-1–positive LUAD cell lines (Supplementary Figure 3, L and M, available online), suggesting cell lineage–specific regulation of cytokine expression in LUAD cell lines. Expression levels of NNMT mRNA were higher in SRGN-positive tumors compared with SRGN-negative tumors in set 1 (Figure 6, G), further supporting the notion that SRGN is epigenetically dysregulated through NNMT-induced perturbation of methionine metabolism in TTF-1–negative LUAD.
Discussion
We have demonstrated that SRGN contributes to the aggressiveness of TTF-1–negative LUAD (Figure 6, H). Cancer cell–derived SRGN regulated expression of PD-L1 and pro-inflammatory cytokines, including CXCL1, IL-6, and IL-8. These play important and intricate roles in tumor-stromal interaction and cause reprogramming into an aggressive and immunosuppressive tumor microenvironment, which contributes to poor outcomes in TTF-1–negative LUAD.
SRGN expression was transcriptionally activated by NRF2 and epigenetically induced through NNMT-induced perturbation of methionine metabolism in TTF-1–negative LUAD (Figure 6H). Given that NRF2 activity may be enhanced in a nongenomic fashion such as through decreased expression of TTF-1–regulated RNA-binding motif protein 47 (34) or epigenetic silencing of KEAP1 (35), SRGN may be an important epigenetic driver (36) in TTF-1–negative LUAD without actionable driver mutations. In addition, Guo et al. (19) demonstrated that SRGN/CD44 axis promotes stemness via Nanog induction, leading to increased chemo and anoikis resistance and poor survival in non-small cell lung cancer. Because LUAD with NRF2 activation also exhibits a stem-like phenotype (37) and worse prognosis (38–40), NRF2 may in part promote aggressiveness of TTF-1–negative LUAD through induction of SRGN. On the other hand, aberrant KEAP1-NRF2 pathway activation has been associated with decreased T-cell infiltration (41) and resistance to immune checkpoint blockade in LUAD (40,42). Conversely, we observed higher PD-L1 expression in tumor cells and higher infiltration of PD-1–positive T lymphocytes in SRGN-positive tumors, suggesting the occurrence of immune-inflamed tumor microenvironment, which shows better response to immune checkpoint blockade (43). We indeed demonstrated inhibitory effects of PD-1 blockade on growth of Srgn-overexpressing tumors in a syngeneic mouse model, suggesting the potential of SRGN as a predictive biomarker for immune checkpoint blockade, which is indicated for LUAD patients without actionable driver mutations, either as monotherapy or in combination with chemotherapy (44).
Although SRGN plays an essential role in immune cells, SRGN deficiency in mice does not affect longevity or fertility (17,18,45), suggesting the promising potential of SRGN as a therapeutic target. Despite the lack of SRGN inhibitors, which currently is a potential limitation of the study to hamper immediate application to therapy, therapeutic strategies targeting a SRGN receptor CD44 (46) are currently under investigation (47). Whereas specific NRF2 inhibitors are not currently available (48), based on preclinical data demonstrating glutamine dependence (49,50), clinical trials of the glutaminase inhibitor CB-839 are ongoing in KEAP1/NRF2-mutated, metastatic, nonsquamous non-small cell lung cancer. In addition, a small-molecule NNMT inhibitor has recently shown promise in targeting stomal NNMT in an ovarian cancer mouse model (51). Given critical roles of SRGN-mediated reprogramming of the microenvironment in tumor progression, targeting other cellular and noncellular components in the tumor microenvironment, such as fibroblasts and extracellular matrix, and endothelial cells and vasculature (52,53), may represent a promising therapeutic approach. A recent study has demonstrated that a subset of cancer-associated fibroblasts with CD10 and GPR77 expression promotes tumor formation and chemoresistance by sustaining cancer stemness in lung and breast cancer (54). Because SRGN induces stem-like properties in lung cancer cells (19), targeting the CD10+GPR77+ cancer-associated fibroblast subset with a neutralizing anti-GPR77 antibody (54) as well as targeting cancer stemness (55,56) could be also an effective therapeutic strategy against SRGN-positive LUAD.
In conclusion, transcriptomic and proteomic characterization of LUAD cell lines identified SRGN as a key molecule in reprogramming the tumor microenvironment in TTF-1–negative LUAD, suggesting the potential of SRGN as a therapeutic target and a biomarker for predicting clinical outcome as well as response to immune checkpoint blockade.
Funding
This work was supported in part by the National Cancer Institute (NCI) Early Detection Research Network, the LUNGevity Foundation, the Canary Foundation, the U.S. Department of Defense (W81XWH-09-LCRP-CTRA and W81XWH-15-1-0127), the NIH/NCI (U01 CA186150, P30CA016672 [the Characterized Cell Line Core, the Functional Genomics Core, the DNA methylation Analysis Core, the Research Histology Core Laboratory, and Small Animal Imaging Facility], P50CA70907 [Lung Specialized Program of Research Excellence], and 1S10OD012304-01 [Proteomics and Metabolomics Facility]), the Cancer Prevention & Research Institute of Texas (Multi-Investigator Research Award RP160668 and Core Facility Grant RP130397 [Proteomics and Metabolomics Facility]), the MD Anderson Moon Shot Proteomics Platform, the MD Anderson Moon Shots Program, MD Anderson start-up funds, the Grants-in-Aid for Young Scientists (grant number: 18K15949) from the Japan Society for the Promotion of Science, Takeda Science Foundation, and the Daiko Foundation. KAD was supported in part by a Cancer Center Support Grant NCI Grant P30 CA016672, NIH grants UL1TR003167, 5R01GM122775, and the Moon Shots funding at MD Anderson Cancer Center.
Notes
Role of the funder: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors declare no competing interests to disclose.
Author contributions: Conceptualization, SH and AT; Data curation, JFF, HW, HK, and JBD; Formal analysis, CAB, MN, FS, VB, and KAD; Investigation, IT, DD, MCT, HM, LMS, STC, JFF, NU, GP, RJ, ERP, YM, BM, MC, DD, JPC, PLL, JF, CB, EJO, JRC, TK, and SK; Methodology, IT, DD, MCT, HM, and AT; Resources, TH, TF, YY, WH, KK, KY, TFC, YH, AFG, IIW, SH and AT; Writing—Original Draft, IT, DD, MCT, HM, SH, and AT; Writing—review & editing, All authors.
Acknowledgements: We thank Dr Jonathan Kurie for providing mouse 393P and 344SQ cells; Genentech for providing tocilizumab; and the Department of Scientific Publications at MD Anderson Cancer Center for editorial assistance.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
References
- 1. Maeda Y, Dave V, Whitsett JA.. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87(1):219–244. [DOI] [PubMed] [Google Scholar]
- 2. Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kendall J, Liu Q, Bakleh A, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA. 2007;104(42):16663–16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwei KA, Kim YH, Girard L, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27(25):3635–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67(13):6007–6011. [DOI] [PubMed] [Google Scholar]
- 6. Yamaguchi T, Hosono Y, Yanagisawa K, et al. NKX2-1/TTF-1: an enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression. Cancer Cell. 2013;23(6):718–723. [DOI] [PubMed] [Google Scholar]
- 7. Cheung WK, Nguyen DX.. Lineage factors and differentiation states in lung cancer progression. Oncogene. 2015;34(47):5771–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yatabe Y, Mitsudomi T, Takahashi T.. TTF-1 expression in pulmonary adenocarcinomas. Am J Surg Pathol. 2002;26(6):767–773. [DOI] [PubMed] [Google Scholar]
- 9. Tang X, Kadara H, Behrens C, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17(8):2434–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anagnostou VK, Syrigos KN, Bepler G, et al. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol. 2009;27(2):271–278. [DOI] [PubMed] [Google Scholar]
- 11. Qian HH, Xu TS, Cai XQ, et al. Prognostic value of TTF-1 expression in patients with non-small cell lung cancer: a meta-analysis. Clin Chim Acta. 2015;451(Pt B):208–214. [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y, Wang R, Li Y, et al. Negative thyroid transcription factor 1 expression defines an unfavorable subgroup of lung adenocarcinomas. J Thorac Oncol. 2015;10(10):1444–1450. [DOI] [PubMed] [Google Scholar]
- 13. Cardnell RJ, Behrens C, Diao L, et al. An integrated molecular analysis of lung adenocarcinomas identifies potential therapeutic targets among TTF1-negative tumors, including DNA repair proteins and Nrf2. Clin Cancer Res. 2015;21(15):3480–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solis LM, Behrens C, Raso MG, et al. Histologic patterns and molecular characteristics of lung adenocarcinoma associated with clinical outcome. Cancer. 2012;118(11):2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yatabe Y, Kosaka T, Takahashi T, et al. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol. 2005;29(5):633–639. [DOI] [PubMed] [Google Scholar]
- 16. Snyder EL, Watanabe H, Magendantz M, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell. 2013;50(2):185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korpetinou A, Skandalis SS, Labropoulou VT, et al. Serglycin: at the crossroad of inflammation and malignancy. Front Oncol. 2014;3:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kolset SO, Pejler G.. Serglycin: a structural and functional chameleon with wide impact on immune cells. J Immunol. 2011;187(10):4927–4933. [DOI] [PubMed] [Google Scholar]
- 19. Guo JY, Hsu HS, Tyan SW, et al. Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene. 2017;36(17):2457–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Korpetinou A, Skandalis SS, Moustakas A, et al. Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PLoS One. 2013;8(10):e78157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He L, Zhou X, Qu C, et al. Serglycin (SRGN) overexpression predicts poor prognosis in hepatocellular carcinoma patients. Med Oncol. 2013;30(4):707. [DOI] [PubMed] [Google Scholar]
- 22. Li XJ, Ong CK, Cao Y, et al. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 2011;71(8):3162–3172. [DOI] [PubMed] [Google Scholar]
- 23. Roy A, Attarha S, Weishaupt H, et al. Serglycin as a potential biomarker for glioma: association of serglycin expression, extent of mast cell recruitment and glioblastoma progression. Oncotarget. 2017;8(15):24815–24827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schliekelman MJ, Taguchi A, Zhu J, et al. Molecular portraits of epithelial, mesenchymal, and hybrid states in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015;75(9):1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Celiktas M, Tanaka I, Tripathi SC, et al. Role of CPS1 in cell growth, metabolism and prognosis in LKB1-inactivated lung adenocarcinoma. J Natl Cancer Inst. 2017;109(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang C, Yin R, Dai J, et al. Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat Commun. 2018;9(1):2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu JY, Zhang C, Wang X, et al. Integrative proteomic characterization of human lung adenocarcinoma. Cell. 2020;182(1):245–261. e17. [DOI] [PubMed] [Google Scholar]
- 29. Gillette MA, Satpathy S, Cao S, et al. ; Clinical Proteomic Tumor Analysis Consortium. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182(1):200–225. e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ulanovskaya OA, Zuhl AM, Cravatt BF.. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9(5):300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kottakis F, Nicolay BN, Roumane A, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539(7629):390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kitajima S, Ivanova E, Guo S, et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discov. 2019;9(1):34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu J, Berisa M, Schworer S, et al. Transsulfuration activity can support cell growth upon extracellular cysteine limitation. Cell Metab. 2019;30(5):865–876.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sakurai T, Isogaya K, Sakai S, et al. RNA-binding motif protein 47 inhibits Nrf2 activity to suppress tumor growth in lung adenocarcinoma. Oncogene. 2016;35(38):5000–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rojo de la Vega M, Chapman E, Zhang DD.. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34(1):21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okazaki K, Anzawa H, Liu Z, et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non-small cell lung cancers. Nat Commun. 2020;11(1):5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goeman F, De Nicola F, Scalera S, et al. Mutations in the KEAP1-NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J Thorac Oncol. 2019;14(11):1924–1934. [DOI] [PubMed] [Google Scholar]
- 39. Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16(14):3743–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Singh A, Daemen A, Nickles D, et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin Cancer Res. 2021;27(3):877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411): aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24(2):334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Havel JJ, Chowell D, Chan TA.. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19(3):133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herbst RS, Morgensztern D, Boshoff C.. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. [DOI] [PubMed] [Google Scholar]
- 45. Abrink M, Grujic M, Pejler G.. Serglycin is essential for maturation of mast cell secretory granule. J Biol Chem. 2004;279(39):40897–40905. [DOI] [PubMed] [Google Scholar]
- 46. Toyama-Sorimachi N, Sorimachi H, Tobita Y, et al. A novel ligand for CD44 is serglycin, a hematopoietic cell lineage-specific proteoglycan. Possible involvement in lymphoid cell adherence and activation. J Biol Chem. 1995;270(13):7437–7444. [DOI] [PubMed] [Google Scholar]
- 47. Chen C, Zhao S, Karnad A, et al. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol. 2018;11(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cuadrado A, Rojo AI, Wells G, et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18(4):295–317. [DOI] [PubMed] [Google Scholar]
- 49. Romero R, Sayin VI, Davidson SM, et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med. 2017;23(11):1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galan-Cobo A, Sitthideatphaiboon P, Qu X, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79(13):3251–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eckert MA, Coscia F, Chryplewicz A, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569(7758):723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valkenburg KC, de Groot AE, Pienta KJ.. Targeting the tumour stroma to improve cancer therapy. Nat Rev Clin Oncol. 2018;15(6):366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fukumura D, Kloepper J, Amoozgar Z, et al. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat Rev Clin Oncol. 2018;15(5):325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Su S, Chen J, Yao H, et al. CD10(+)GPR77(+) Cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172(4):841–856.e16. [DOI] [PubMed] [Google Scholar]
- 55. Saygin C, Matei D, Majeti R, et al. Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell. 2019;24(1):25–40. [DOI] [PubMed] [Google Scholar]
- 56. Clara JA, Monge C, Yang Y, et al. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol. 2020;17(4):204–232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.