Abstract
Background
Immune cell transcriptome signatures have been widely used to study the lung tumor microenvironment (TME). However, it is unclear to what extent the immune cell composition in the lung TME varies across histological and molecular subtypes (intertumor heterogeneity [inter-TH]) and within tumors (intratumor heterogeneity [ITH]) and whether ITH has any prognostic relevance.
Methods
Using RNA sequencing in 269 tumor samples from 160 lung cancer patients, we quantified the inter-TH of immune gene expression and immune cell abundance and evaluated the association of median immune cell abundance with clinical and pathological features and overall survival. In 39 tumors with 132 multiregion samples, we also analyzed the ITH of immune cell abundance in relation to overall survival using a variance-weighted multivariate Cox model not biased by the number of samples per tumor.
Results
Substantial inter-TH of 14 immune cell types was observed even within the same histological and molecular subtypes, but early stage tumors had higher lymphocyte infiltration across all tumor types. In multiregion samples, an unbiased estimate of low ITH of overall immune cell composition (hazard ratio [HR] = 0.40, 95% confidence interval [CI] = 0.21 to 0.78; P = .007), dendritic cells (HR = 0.24, 95% CI = 0.096 to 0.58; P = .002), and macrophages (HR = 0.50, 95% CI = 0.30 to 0.84; P = .009) was strongly associated with poor survival. The ITH of 3 markers, including CD163 and CD68 (macrophages) and CCL13 (dendritic cells), was enough to characterize the ITH of the corresponding immune cell abundances and its association with overall survival.
Conclusion
This study suggests that lack of immune cell diversity may facilitate tumor evasion and progression. ITH inferred from CCL13, CD163, and CD68 could be used as a prognostic tool in clinical practice.
The recent advances of lung cancer immunotherapy have fostered a growing interest in the tumor immune microenvironment (TME). To characterize the cell admixtures of immune infiltrates and their interaction with tumor cells, computational deconvolution tools are used to quantify immune cell fractions from bulk transcriptome data based on cell type–specific signatures (1–5). Using these approaches, array- and sequencing-based studies have profiled the tumor immune landscape in lung tumors from diverse ethnic and clinical and pathological groups (6-11). Some of the immune signatures were associated with clinical outcomes or response to immunotherapy (5,7,12–14). Nearly all studies analyzed a single biopsy per tumor. Two recent studies (15,16) based on multiple-regional samples per lung tumor showed substantial intratumor heterogeneity (ITH) of overall gene expression signatures (not immune-related). Moreover, using immunohistochemistry or RNA sequencing (RNA-seq) (17), the TRACERx (TRAcking non-small cell lung Cancer Evolution through therapy [Rx]) study showed ITH of the overall immune infiltration level without exploring its association with survival.
After identifying the best deconvolution approach for RNA-based immune cell detection, we performed a comprehensive analysis of intertumor heterogeneity (inter-TH) and ITH of immune gene expression and immune cell abundances in 269 tumor samples from 160 lung cancer patients and investigated their association with clinical and pathological features, including survival .
Methods
Patients and Samples
We collected fresh-frozen samples from 160 lung cancer patients from the Environment And Genetics in Lung cancer Etiology (EAGLE) study, a large population-based, case-control study conducted in Italy between 2002 and 2005 (18–20); see the Supplementary Methods (available online) for details. The study protocol was approved by the institutional review board of the US National Cancer Institute and the involved institutions in Italy. Informed consent was obtained for all subjects. In brief, 269 tumor samples and 109 paired normal samples passed quality control (Supplementary Table 1, available online) and underwent RNA-seq analysis. These samples were used to investigate inter-TH and overall immune cell abundance. After excluding low-purity samples (Supplementary Figure 1, A, available online), 132 samples from 39 multiregion tumors were used for ITH analysis.
RNA-Sequencing
RNA-seq was performed using the Illumina HiSeq platform and Illumina TruSeq Stranded Total RNA-seq protocol, generating 2x100bp paired-end reads. The FASTQ files were aligned to human reference genome GRCh37/hg19 using STAR (21) and processed by RSEM (22) for gene quantification. The resulting expression data were normalized to transcripts per million and log2 transformed.
Molecular Subtype Detection
Molecular subtypes were detected using 2 previously published nearest centroid classifiers (23,24). Log2-transformed transcripts per million data were gene median–centered. A 506-gene predictor for lung adenocarcinoma (LUAD) (23) and a 203-gene predictor for lung squamous cell carcinoma (LUSC) (24) were applied to samples of the 2 histological types, respectively. See the Supplementary Methods (available online) for details.
Quantification of Immune Cell Abundances and Tumor Purity
For tumor samples, immune cell abundances were inferred using 5 algorithms. Gene signatures from Danaher et al. (1) and Davoli et al. (2) studies were downloaded from the publications separately, and estimated immune cell abundances were determined using the average expression levels across all genes for each cell type. The toolkits CIBERSORTx (http://cibersortx.stanford.edu) (4) and TIMER (http://cistrome.org/TIMER/) (25) were used to estimate immune cell fractions. Estimation based on the Jerby-Arnon et al. study (5) used the code for cell typing from GitHub (https://github.com/livnatje/ImmuneResistance).
To evaluate the impact of tumor purity on the estimates of immune cell abundances, we inferred the tumor purity from RNA-seq data using the R package ESTIMATE (26). For tumor samples with paired normal samples, we applied paired tumor-normal (TN) estimation of immune cells based on the Danaher signatures (1). The TN estimates were derived as
where Xt and Xn are the immune cell abundances in tumor and paired normal samples, respectively, and π is the tumor purity.
Unsupervised Clustering of Abundances of Immune Cells and Immune Genes
The estimated abundances of 14 immune cells were median centered across all tumor samples and clustered by an agglomerative complete-linkage hierarchical algorithm using Euclidean distance. The same approach was then applied to the expression levels of 60 immune marker genes for clustering. The consensus clustering analysis was performed using the R package ConsensusClusterPlus (27).
Quantification of Immune Cell ITH
For each patient with k multiregion tumor samples, ITH of immune cell abundances was quantified as the average pairwise ITH or APITH (28). Specifically, for an individual immune cell type (eg, macrophage, neutrophils),
where dij is the genomic distance between a pair of samples (i, j) calculated based on the expression levels of signature genes of the immune cell type. The overall ITH summarizing all immune cell composition was quantified as
where Sij is the Euclidean distance of the relative abundances of 14 immune cells between a pair of samples (i, j).
As we discussed previously (28), the expectation of APITH does not depend on the number of tumor samples per tumor, which allows to compare ITH across groups of patients and to perform regression analysis for subjects with different numbers of tumor samples.
Immune Genewise Inter-TH and ITH
We used the metrics of genewise heterogeneity developed by Biswas et al. (15) to determine the genewise inter-TH and ITH. See the Supplementary Materials (available online) for details.
Statistical Analysis
All statistical tests were 2-sided, with a false discovery rate or P value less than .05 considered as statistically significant.
Survival analysis. We investigated the association of 1) median and 2) APITH of immune cell abundances per patient with overall survival using multivariate Cox proportional hazards models, adjusted for age, tumor stage, histological type, and smoking status. For median immune cell abundances, the overall immune cell levels were quantified as the median of the sum of all cell types. Because the variance of the APITH estimator depends on the number of samples per tumor, we used a variance-weighted Cox regression to optimize the power of detecting the association between APITH and survival (28). We used the cox.zph function (29) in the R package survival to test the proportional hazards assumption and R package survey (30) for the weighted Cox regression analysis.
Association analysis of immune cell abundances and APITH with clinical features. We tested the association of 1) median and 2) APITH of immune cell abundances per patient with stage (stage 1 vs others), tobacco smoking status (former vs current), and histological type (LUAD vs LUSC) using the Student t-test and with age at diagnosis using the Pearson correlation test. The association with smoking duration was analyzed using the ordinal regression adjusted for smoking status.
ITH analysis of single-gene markers of immune cells. We tested the association of the expression level of each signature gene with the corresponding immune cell abundance in all samples to identify single-gene markers that could characterize each immune cell type. For each selected marker, gene-level APITH was quantified as the average pairwise difference of expression across all pairs of samples from multiregion tumors and was compared with APITH of corresponding immune cells. Survival analysis was applied as described above for the APITH of immune cells.
Results
Patient Characteristics
The study patients ’ demographic characteristics are summarized in Table 1 and Supplementary Table 2 (available online). All patients were treatment naïve and had a median follow-up time of 43 months. Most patients were smokers, including 80 (50.0%) current and 73 (45.6%) former. Most tumors (98.1%) were non-small cell lung cancer, including 100 LUAD, 45 LUSC, and 5 large cell carcinomas; 3 were small cell carcinomas. Stage groups included 78 (48.8%) stage I, 41 stage II (25.6%), and 41 (25.6%) stage III-IV.
Table 1.
Demographic and clinical characteristics of patients
| Characteristic | No. of patients |
|---|---|
| Mean age at first diagnosis (range), y | 65.6 (38.8-79.7) |
| Sex | |
| Male | 134 |
| Female | 26 |
| Pathological subtype | |
| Adenocarcinoma | 100 |
| Squamous/Epidermoid carcinoma | 45 |
| Large cell carcinoma | 5 |
| Mixed type | 5 |
| Other non-small cells | 2 |
| Small cell carcinoma | 3 |
| Tumor stage | |
| IA | 40 |
| IB | 38 |
| IIA | 23 |
| IIB | 18 |
| IIIA | 35 |
| IIIB | 2 |
| IV | 4 |
| Chemotherapy | |
| Yes | 12 |
| No | 146 |
| Distant metastasis | |
| Yes | 84 |
| No | 74 |
| Death from any cause | 102 |
| Smoking status | |
| Never | 5 |
| Former | 73 |
| Current | 80 |
| Unknown | 2 |
| Cigarettes per day | |
| Mean (SD) | 22.5 (10.7) |
| ≤10 | 22 |
| >10 to ≤20 | 72 |
| >20 to ≤30 | 31 |
| >30 | 24 |
| Cigarette smoking duration, y | |
| Mean (SD) | 43.5 (10.6) |
| ≤30 | 13 |
| >30 to ≤40 | 47 |
| >40 to ≤50 | 48 |
| >50 | 41 |
Deconvolution of Immune Cells From RNA-Seq Data
To characterize the immune cell landscape in lung cancer, we used the tumor RNA-seq data and the immune signatures from 5 published studies (1–4,31) to infer the immune cell components based on gene expression. The abundances of B cells and CD8 T cells were highly consistent across all 5 methods, whereas the abundances of regulatory T cells, natural killer (NK) cells, and macrophages showed consistency across only 3 methods, by Danaher et al. (1), Davoli, et al. (2) and Jerby-Arnon et al. (5) (Supplementary Figure 2, available online). Among them, the Danaher et al. (1) method required the minimum set of genes to define immune cell signatures compared with the other 2 approaches. In addition, the TRACERx study benchmarked computational deconvolution tools with pathology tumor-infiltrating lymphocyte estimates and showed optimal concordance with the CD8 cell abundances inferred using the Danaher approach (17). Therefore, we used the Danaher approach and characterized 14 immune cell types using 60 immune genes in our study.
All immune cells were statistically significantly correlated between tumor-only estimates and estimates based on paired tumor and TN samples (P < .05; Supplementary Figure 1, available online). For CD45, CD8 T cells, exhausted CD8, overall T cells, macrophages, and dendritic cells (DC), the Pearson correlation was approximately 0.9; for all other cell types, the correlation was greater than 0.65 (Supplementary Figure 1, available online). We therefore used tumor-only estimates as a surrogate of TN estimates to characterize the relative immune cell abundances. Similarly, APITH (28) of all immune cells had a statistically significant correlation between tumor-only and TN estimates (Supplementary Figure 3, available online), after excluding low purity samples (Supplementary Methods, available online). RNA integrity scores of all tumor samples were 6 or more, and no association between APITH of any immune cells and APITH of RNA integrity was observed.
Characterization of Inter-TH and ITH of Immune Cells in Lung Cancer
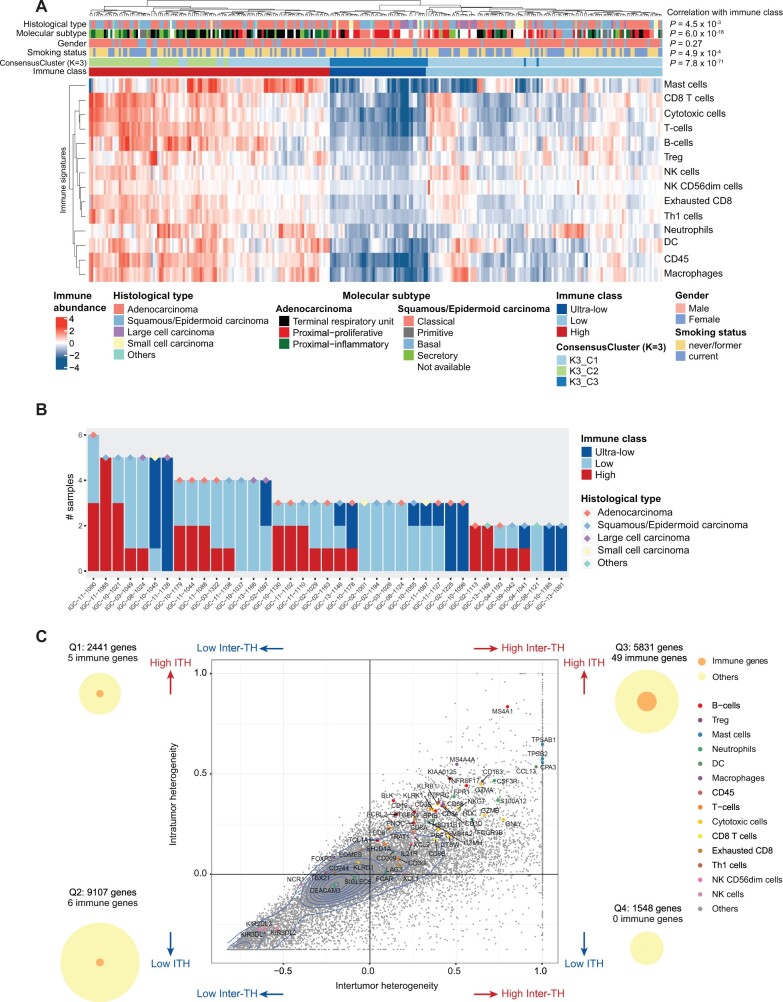
Unsupervised hierarchical clustering of the 14 immune cell types across 269 tumor samples revealed 2 clusters with high (113 samples) and low (156 samples) levels of immune infiltration (Figure 1, A), consistent with the TRACERx study (17). In the immune-low cluster, a subset of 45 samples had low levels of all immune cells and was defined as the ultra-low subgroup. Substantial inter-TH was observed across and within non-small cell lung cancer histological types (Supplementary Table 3, available online), but small cell and large cell carcinomas mostly had low or ultra-low immune gene clusters. Previous studies (23,24) had defined molecular subtypes by gene expression within histological types. Using this classification, we found that all molecular subtypes contained immune-high and -low tumors (Supplementary Table 3, available online). Notably, the ultra-low clusters were dominated by the LUAD proximal-proliferative subtype and LUSC classical subtype (Figure 1, A). Within the 39 multiregion tumors, 3 (7.7%) had immune-high samples exclusively, 17 (43.6%) had immune-low samples exclusively, and 19 (48.7%) had both immune-high and -low samples, including 3 (7.7%) with both immune-high and ultra-low samples (Figure 1, B). Clustering of the 60 immune marker genes revealed similar patterns, with 227 of 269 (84.4%) samples classified within the same immune classes as the 14 immune cell abundances, and 15 (28.5%) tumors had both immune-high and -low samples (Supplementary Table 1, available online). Consensus clustering analysis confirmed the presence of 3 robust clusters that accounted for 211 of 269 (78.4%) of all samples (Figure 1, A).
Figure 1.
Summary of inter-tumor heterogeneity (inter-TH) and intra-tumor heterogeneity (ITH) of EAGLE-lung samples. A) Unsupervised clustering of RNA-based abundance of 14 immune cells across 269 tumor samples from 160 tumors. Histological types, molecular subtypes, sex, smoking status, consensus clusters (cluster count K = 3), and immune classes are indicated by different colors of the column sidebars. The P values are based on the χ2 test of the immune classes and clinical features. B) Summary of immune classes of samples from the same subjects. 39 subjects with at least 2 samples per subject that passed the purity filtering were analyzed. Histological types of subjects are indicated by different colors of the diamond symbols. C) Inter- and intratumor mRNA heterogeneity quadrant of 18 927 expressed genes in 132 samples from 39 multiregion tumors. Each dot represents a gene. Genes of immune signatures are indicated by colors. 2D density contour is indicated by blue lines. The plot is split into 4 quadrants by average inter-TH and ITH, defined as Q1 to Q4, respectively (ie, x- and y-axes indicate mean values of ITH and inter-TH scores, respectively). The numbers of all genes and immune genes are indicated by circles on the left and right sides of the figure. All statistical tests were 2-sided. DC = dendritic cells; NK = natural killer; Treg = regulatory T cells.
Next, we investigated inter-TH and ITH of immune marker genes (n = 60) from the Danaher signatures relative to inter-TH and ITH of all expressed genes (n = 18 927). The inter-TH of all expressed genes across 39 multiregion tumors was highly consistent with the inter-TH across all tumors (R = 0.969, P < .001). Therefore, we evaluated ITH and inter-TH in the multiregion tumors (Figure 1, C). We investigated the distributions of the overall genes and immune genes in the quadrants. Specifically, 5 (8.3%) immune genes had low inter-TH and high ITH (defined as quadrant Q1) ; 6 (10.0%) immune genes had low inter-TH and low ITH (Q2); 49 (81.7%) immune genes had high inter-TH and high ITH (Q3); and no immune gene had high inter-TH and low ITH (Q4). Thus, immune genes were statistically significantly enriched in Q3 (P < .001, Fisher exact test).
Association of Immune Cell Abundance With Clinical and Pathological Features
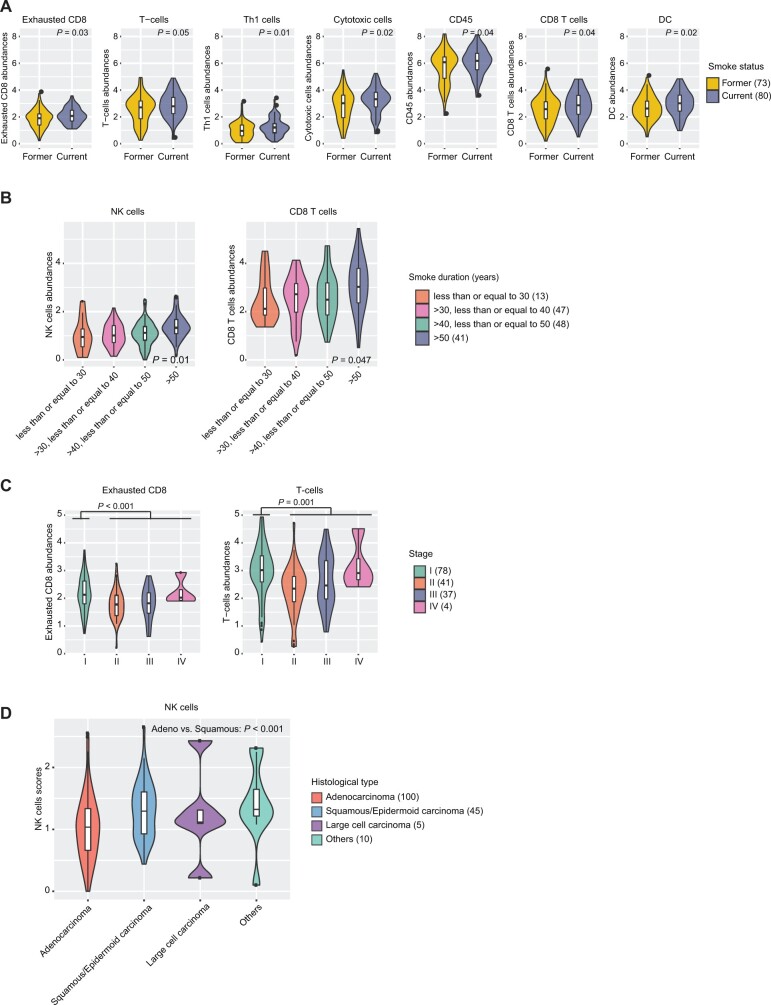
We first tested the association of individual immune cell abundances with clinical and pathological features (Supplementary Table 4, available online). The median abundance per tumor of most immune cells was suggestively enriched in current smokers (Figure 2, A; nominal P < .05). Further, NK and CD8 T-cell abundances showed suggestive associations with tobacco smoking duration adjusted for smoking status (Figure 2, B; nominal P = .01 and 0.047, ordinal regression). Several immune cells had suggestively higher abundance in stage I tumors (nominal P < .05), among which the results in exhausted CD8 and T cells were statistically significant (Figure 2, C; adjusted P = .02 for both), with similar results in LUAD and LUSC. No statistically significant association was observed between histological type or age at diagnosis and immune cell abundances except for a higher NK cell abundance in LUSC vs LUAD (Figure 2, D). Similarly, no statistically significant association was identified between APITH of immune cell abundances and clinical or pathological features (data not shown).
Figure 2.
Association of immune cell abundances with clinical and pathological features. A) Comparison of 8 immune cell abundances in former (n = 73) and current smokers (n = 80). The immune cell abundances were determined by the average expression level across all marker genes for each immune cell type and were plotted in the same scale across all figures in the panel. The nominal P value is based on a 2-sided Student t-test. B) Distribution of the natural kill (NK) cell and CD8 T-cell abundances in patients with different smoke duration periods. P values are based on ordinal regression adjusted for smoking status. C) Comparison of 5 immune cell abundance in subjects with different tumor stages. P values are based on a 2-sided Student t-test comparing the stage I patients vs stage II-IV patients. D) Comparison of NK cell abundance in different histological types. The P value is based on a 2-sided Student t-test of adenocarcinoma (n = 100) and squamous and/or epidermoid carcinoma (n = 45). DC = dendritic cell.
Association of Immune Cell Abundance and ITH of Immune Cells With Lung Cancer Survival
To evaluate the prognostic relevance of ITH and inter-TH of immune cell abundances, we first tested the association of median immune cell abundances per tumor with overall survival in 160 tumors (Supplementary Table 5, available online). We observed that higher abundances of CD8 T cells and T cells were associated with longer overall survival, but the associations were not statistically significant after correcting for multiple comparisons.
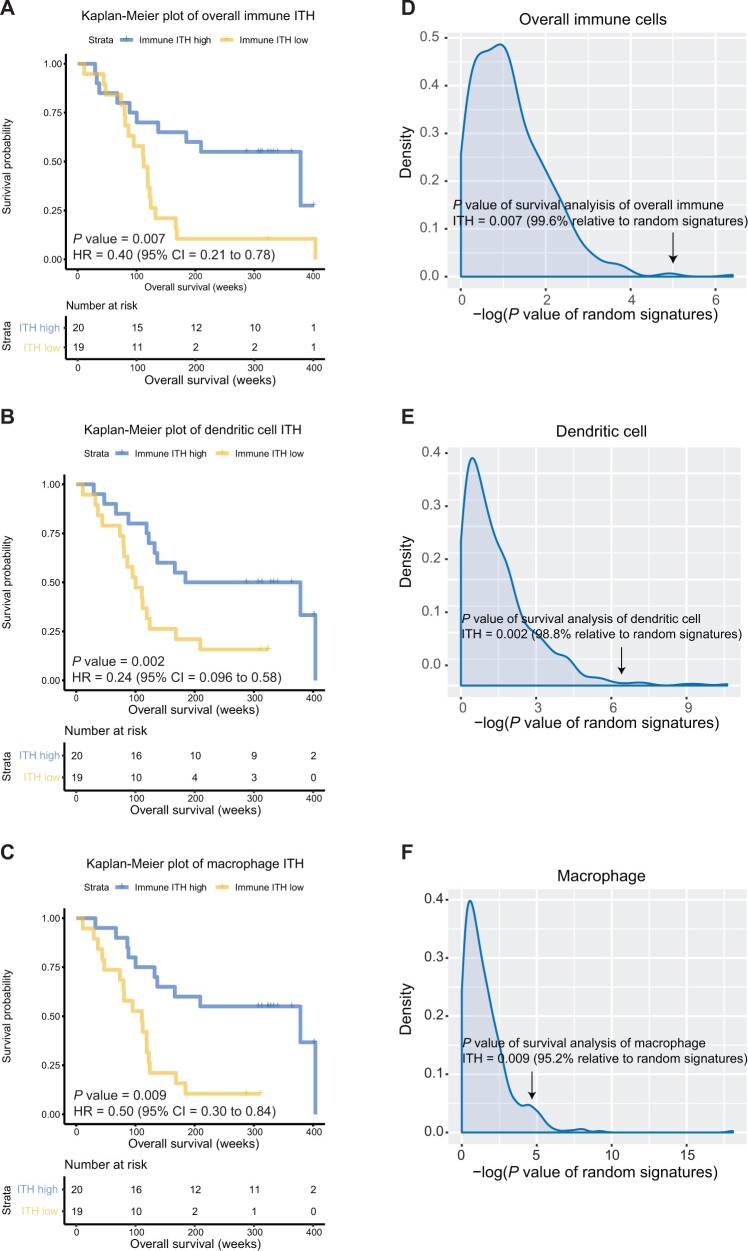
We then evaluated the association of APITH of individual immune cells and overall immune cell composition with overall survival (Figure 3; Supplementary Table 6 and Figure 4, available online). Low APITH of overall immune cell composition was statistically significantly associated with poor survival (hazard ratio [HR] = 0.40, 95% confidence interval [CI] = 0.21 to 0.78; P = .007). Similarly, among individual immune cells, low APITH of DC and macrophage abundances was statistically significantly associated with poor survival (DC: HR = 0.24, 95% CI = 0.096 to 0.58; P = .002; macrophage: HR = 0.50, 95% CI = 0.30 to 0.84; P = .009). The results remained statistically significant after adjusting for chemotherapy or radiation therapy (Supplementary Table 7, available online). We next included the median abundance of corresponding immune cells as a covariate of the proportional-hazards model and confirmed the statistically significant association of the overall APITH of immune cells and APITH of DC and macrophages with survival (Supplementary Table 6, available online). All survival analyses for APITH of immune cells remained statistically significant after correction for multiple comparisons.
Figure 3.
Association between overall survival and intratumor heterogeneity (ITH) of immune cells in subjects with multiregion tumor samples. A-C) Kaplan-Meier curves for overall survival stratified by the median APITH of (A) overall immune cells, (B) dendritic cells, and (C) macrophages. P values and hazard ratios indicated are based on the multivariate model. D-F) Distribution of log10 (P values) of the multivariate Cox proportional-hazards model for 500 random signatures of identical sizes of (D) overall immune cells, (E) dendritic cells, and (F) macrophages. P values of corresponding immune ITH are indicated by arrows. All statistical tests were 2-sided. CI = confidence interval; HR = hazard ratio.
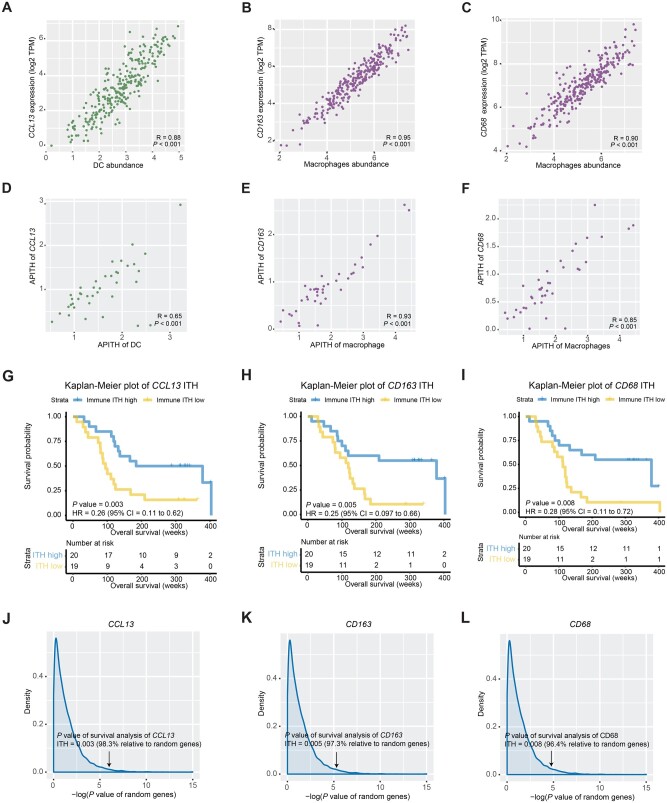
Figure 4.
Association between overall survival and intratumor heterogeneity (ITH) of single immune gene markers. A-C) Scatter plots of the gene expression levels of immune markers and the corresponding immune cell abundances for (A) CCL13 (DC marker), (B) CD163 (macrophage marker), and (C) CD68 (macrophage marker). D-F) Scatter plots of the single gene-based APITH and APITH of the corresponding immune cells for (D) CCL13, (E) CD163, and (F) CD68. G-I) Kaplan-Meier curves for overall survival stratified by the median APITH of (G) CCL13, (H) CD163, and (I) CD68. P values and hazard ratios indicated are based on the multivariate model. J-L) Distribution of log10 (P values) of the multivariate Cox proportional hazards model for all expressed genes (n = 18 927). P values of (J) CCL13, (K) CD163, and (L) CD68 are indicated by arrows. CI = confidence interval; DC = dendritic cell; HR = hazard ratio; TPM = transcripts per million .
A previous study showed that a substantial proportion of randomly picked genes could be associated with clinical outcomes possibly because of the confounding effect of the proliferation-related signals (32). To verify whether our results based on immune gene signatures were superior to those based on random genes, we generated 500 random signatures of sizes identical to each immune cell signature and then computed APITH of individual random signatures corresponding to each immune cell and the overall random signatures. We tested the prognostic value of the APITH of random signatures using the weighted Cox proportional hazards model. We confirmed that the associations of overall APITH with survival were statistically significantly stronger than the random signatures (Figure 3, D; P = .004). The APITH of DC and macrophages was also statistically significantly more associated with survival than the random signatures (Figure 3, E and F; DC: P = .01; macrophages: P = .048).
Next, we explored whether a few immune markers could capture the expression of all genes in DC and macrophages and investigated their association with overall survival. Across all tumor samples, immune markers CCL13 and CD163 had the highest correlation with DC and macrophage abundances, respectively (Figure 4, A and B; CCL13: R = 0.88; CD163: R = 0.95). In addition, 3 macrophage markers—CD68, CD84, and MS4A4A—were also highly correlated with the macrophage abundances (Figure 4, C; CD68: R = 0.90; CD84: R = 0.93, MS4A4A: R = 0.94). We then selected 2 markers with the best correlation, CCL13 and CD163, as well as CD68, which is routinely used as a histochemical marker for pan-macrophages, and analyzed the APITH of individual genes in 39 multiregion tumors. The single gene-based APITH of 3 markers was highly correlated with APITH of the corresponding immune cells (Figure 4, D-F). We further investigated the prognostic capability of single gene-based APITH. Lower APITH of CCL13, CD163, and CD68 was statistically significantly associated with worse survival using multivariate models (Figure 4, G-I). Compared with APITH of random genes, the prognostic capability of APITH of CCL13, CD163, and CD68 remained statistically significant (Figure 4, J-L; CCL13: P = .02; CD163: P = .03; CD68: P = .04).
Discussion
In this study, we characterized immune gene expression patterns and immune cell abundances using RNA-seq in 160 tumors of different histological types, including 39 multiregion tumors. We demonstrated extensive inter-TH and ITH of 14 immune cell types inferred from the expression of 60 immune-related genes. The overall immune infiltration level varied substantially across tumors even within histological and molecularly defined (33,34) subtypes, highlighting the importance of investigating the TME role in defining these subtypes. However, some patterns were consistent across all tumors, including higher immune cell abundances in current smokers and lower lymphocytic infiltration in advanced stages. High CD8 T cells in tumors were also associated with better survival, as previously observed (35–37). These results confirm the important role for lymphocytes in the TME in tumor progression and prognosis.
At the gene level, 80% of immune markers were high inter-TH and ITH genes. The results confirmed a previous observation that overall immune infiltration levels exhibit ITH (17) and additionally revealed ITH of the levels of individual immune genes and immune cells. Importantly, we found that low ITH of overall immune cells and ITH of individual cells, such as DC and macrophages, were statistically significantly associated with poor overall survival, suggesting that lack of immune cell diversity may facilitate tumor evasion and progression. For this analysis, we implemented the APITH and used a variance-weighted proportional hazards model so, differently from previous ITH studies of T-cell density or clonality (38), our results were not biased by the number of samples per tumor. Additionally, we performed simulation analyses to demonstrate the superior performance of immune gene ITH compared with random genes.
Finally, we demonstrated that 3 single-gene markers, including the DC marker CCL13 and macrophage markers CD163 and CD68, were able to recapitulate the overall relative abundances and ITH of the corresponding cell types. The ITH of these markers showed prognostic capability as the ITH from the polygenic signatures. In the clinic, the evaluation of only these 3 genes by immunohistochemistry or gene expression in 2 or more samples could aid in prognosis assessment.
The major limitation of this study is the lack of any publicly available cohort we could use for validation. To overcome this limitation, we validated our results using simulation analyses. We confirmed strong statistically significant results with large effect size of the immune ITH on mortality risk. This study mostly included smokers of European descent, and only 5 of 160 (3.1%) were lung cancer in never-smokers (LCINS). Previous studies showed an upregulation of immune-related genes in LCINS of East Asian descent (9,39), suggesting a potentially different role of the TME in these tumors. Therefore, a larger study focusing on LCINS is needed to verify the prognostic effects of immune cell diversity.
In conclusion, our analysis revealed substantial immune cell ITH and inter-TH in lung tumors, with strong prognostic relevance of specific immune cells and immune genes.
Funding
This work was supported by the Intramural Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH.
Notes
Role of the funder: The funder had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of this manuscript; or the decision to submit the manuscript.
Disclosures: The authors declare no competing interests.
Author contributions: MTL conceived the study. ACP, DC, NEC oversaw the surgical sampling design, sample and data collection, and field activities. WZ performed the bioinformatics and association analyses. JS and BZ helped with the statistical analyses. TZ and DW helped with the bioinformatics analyses. AH conducted the laboratory analyses. WZ, JS, and MTL discussed the results and wrote the manuscript. All authors reviewed the manuscript.
Acknowledgements: This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We are grateful to the patients and families who contributed to this study and the many investigators who are involved in the EAGLE study (https://eagle.cancer.gov/).
Data Availability
The RNA-seq data underlying this article are available in SRA and can be accessed through dbGaP with the accession number phs002346.v1.p1.
Supplementary Material
References
- 1. Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355(6322):eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jerby-Arnon L, Shah P, Cuoco MS, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984–997.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faruki H, Mayhew GM, Serody JS, et al. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12(6):943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seo JS, Kim A, Shin JY, et al. Comprehensive analysis of the tumor immune micro-environment in non-small cell lung cancer for efficacy of checkpoint inhibitor. Sci Rep. 2018;8(1):14576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thorsson V, Gibbs DL, Brown SD, et al. ; for the Cancer Genome Atlas Research Network. The immune landscape of cancer. Immunity. 2019;51(2):411–412. [DOI] [PubMed] [Google Scholar]
- 9. Wang C, Yin R, Dai J, et al. Whole-genome sequencing reveals genomic signatures associated with the inflammatory microenvironments in Chinese NSCLC patients. Nat Commun. 2018;9(1):2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang XC, Wang J, Shao GG, et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10(1):1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li B, Cui Y, Diehn M, et al. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017;3(11):1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Givechian KB, Garner C, Benz S, et al. An immunogenic NSCLC microenvironment is associated with favorable survival in lung adenocarcinoma. Oncotarget. 2019;10(19):1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mony JT, Schuchert MJ.. Prognostic implications of heterogeneity in intra-tumoral immune composition for recurrence in early stage lung cancer. Front Immunol. 2018;9:2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki K, Kachala SS, Kadota K, et al. Prognostic immune markers in non-small cell lung cancer. Clin Cancer Res. 2011;17(16):5247–5256. [DOI] [PubMed] [Google Scholar]
- 15. Biswas D, Birkbak NJ, Rosenthal R, et al. ; for the TRACERx Consortium. A clonal expression biomarker associates with lung cancer mortality. Nat Med. 2019;25(10):1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee WC, Diao L, Wang J, et al. Multiregion gene expression profiling reveals heterogeneity in molecular subtypes and immunotherapy response signatures in lung cancer. Mod Pathol. 2018;31(6):947–955. [DOI] [PubMed] [Google Scholar]
- 17. Rosenthal R, Cadieux EL, Salgado R, et al. ; for the TRACERx consortium. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Consonni D, Pierobon M, Gail MH, et al. Lung cancer prognosis before and after recurrence in a population-based setting. J Natl Cancer Inst. 2015;107(6):djv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Landi MT, Consonni D, Rotunno M, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi J, Hua X, Zhu B, et al. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li B, Dewey CN.. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilkerson MD, Yin X, Walter V, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7(5):e36530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilkerson MD, Yin X, Hoadley KA, et al. Lung squamous cell carcinoma mRNA expression subtypes are reproducible, clinically important, and correspond to normal cell types. Clin Cancer Res. 2010;16(19):4864–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilkerson MD, Hayes DN.. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hua X, Zhao W, Pesatori AC, et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat Commun. 2020;11(1):2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grambsch PM, Therneau TM.. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):515–526. [Google Scholar]
- 30. Binder DA. Fitting cox proportional hazards models from survey data. Biometrika. 1992;79(1):139–147. [Google Scholar]
- 31. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. ; for the TRACERx Consortium. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. [DOI] [PubMed] [Google Scholar]
- 32. Venet D, Dumont JE, Detours V.. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7(10):e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Al-Shibli KI, Donnem T, Al-Saad S, et al. Prognostic effect of epithelial and stromal lymphocyte infiltration in non-small cell lung cancer. Clin Cancer Res. 2008;14(16):5220–5227. [DOI] [PubMed] [Google Scholar]
- 36. Chen B, Li H, Liu C, et al. Prognostic value of the common tumour-infiltrating lymphocyte subtypes for patients with non-small cell lung cancer: a meta-analysis. PLoS One. 2020;15(11):e0242173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim SH, Go SI, Song DH, et al. Prognostic impact of CD8 and programmed death-ligand 1 expression in patients with resectable non-small cell lung cancer. Br J Cancer. 2019;120(5):547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reuben A, Gittelman R, Gao J, et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 2017;7(10):1088–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J, Yang H, Teo ASM, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet. 2020;52(2):177–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data underlying this article are available in SRA and can be accessed through dbGaP with the accession number phs002346.v1.p1.