Summary
It is unknown whether transient transgenerational epigenetic responses to environmental challenges affect the process of evolution, which typically unfolds over many generations. Here, we show that in C. elegans, inherited small RNAs control genetic variation by regulating the crucial decision of whether to self-fertilize or outcross. We found that under stressful temperatures, younger hermaphrodites secrete a male-attracting pheromone. Attractiveness transmits transgenerationally to unstressed progeny via heritable small RNAs and the Argonaute Heritable RNAi Deficient-1 (HRDE-1). We identified an endogenous small interfering RNA pathway, enriched in endo-siRNAs that target sperm genes, that transgenerationally regulates sexual attraction, male prevalence, and outcrossing rates. Multigenerational mating competition experiments and mathematical simulations revealed that over generations, animals that inherit attractiveness mate more and their alleles spread in the population. We propose that the sperm serves as a “stress-sensor” that, via small RNA inheritance, promotes outcrossing in challenging environments when increasing genetic variation is advantageous.
Keywords: small RNA inheritance, transgenerational inheritance, epigenetics, non-Mendelian inheritance, genetic variation, C. elegans, sexual attractiveness, sex pheromones, germ granules, evolution, germline
Graphical abstract
Highlights
-
•
Growth at 25°C increases sexual attractiveness of C. elegans hermaphrodites
-
•
Increased attractiveness transmits transgenerationally via HRDE-1 and small RNAs
-
•
Attractiveness is associated with sperm defects and small RNAs targeting sperm genes
-
•
Stress-induced transient epigenetic inheritance enhances mating and can affect evolution
It is unclear whether transient heritable epigenetic responses can have a long-lasting impact on evolution. Toker et al. describe a small-RNA-based heritable response to environmental stress that makes worm progeny more sexually attractive for multiple generations, leading to more mating events and higher allele prevalence in the population.
Introduction
Hermaphrodites of the androdioecious nematode species Caenorhabditis elegans can either self-reproduce or outcross. Outcrossing comes at a great cost, as it requires finding a partner, exposes the animals to physical damage, increases predation risk, and, importantly, dilutes each parent’s genetic contribution (Otto, 2009). However, theory predicts that an increased tendency for outcrossing under stress would evolve in many cases and could facilitate adaptation (Hadany and Otto, 2009; Ram and Hadany, 2016). Accordingly, it was shown that C. elegans outcross more when exposed to stressful conditions (Morran et al., 2009, 2011) and that the genetic variability gained following outcrossing is essential for adaptation to changing environments (Parrish et al., 2016). Recent studies revealed that when aging C. elegans hermaphrodites run out of self-made sperm and lose the ability to self-reproduce, they secrete a volatile sex pheromone that elevates their chances of mating with males (Leighton et al., 2014; Wan et al., 2019). The secretion of this pheromone is triggered when there is no longer contact between the depleted sperm and the oocytes in the gonads (Leighton et al., 2014). The exact chemical nature of the pheromone is unknown; however, it was shown that males detect it using the G-protein-coupled receptor SRD-1 in the bilateral AWA sensory neurons (Wan et al., 2019).
The inheritance of experiences has been a controversial topic for centuries (Chen et al., 2016; Jablonka and Lamb, 2008). Nevertheless, when C. elegans are stressed, they can generate physiological responses that carry on transgenerationally (Ewe et al., 2020; Kaletsky et al., 2020; Klosin et al., 2017; Moore et al., 2019; Ni et al., 2016; Rechavi et al., 2011, 2014; Schott et al., 2014). Some of the effects were shown to be transmitted independently of DNA changes by heritable small RNAs that can be re-synthesized and amplified in the progeny by RNA-dependent RNA polymerases (Rechavi et al., 2011; Sapetschnig et al., 2015). Dedicated machinery regulates the transmission and maintenance of these effects (Buckley et al., 2012; Houri-Ze’evi et al., 2016; Houri-Zeevi et al., 2020; Lev et al., 2017; Perales et al., 2018; Rechavi and Lev, 2017; Shukla et al., 2020; Spracklin et al., 2017; Xu et al., 2018), and disruption of small RNA inheritance results in damages to the germ cells that accumulate over generations and can lead to full sterility (Buckley et al., 2012; Lev et al., 2017; Sakaguchi et al., 2014; Simon et al., 2014; Spracklin et al., 2017). The germline, therefore, both serves as a rheostat that controls the secretion of male-attracting pheromones and reflects the effects of environmental responses on the pools of heritable small RNAs. Hence, we hypothesized that the secretion of the male-attracting pheromone might be induced by environmental stress and memorized by heritable small RNAs.
Results
Environmental stress induces transgenerational premature attraction via heritable small RNAs
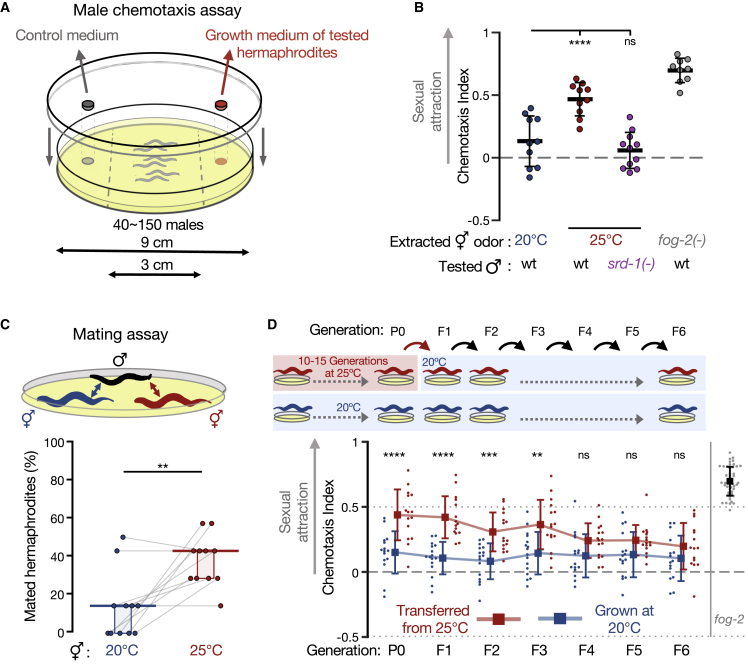
To test our hypothesis, we first performed chemotaxis experiments to measure the male attraction to odors extracted from the growth medium of 1-day-old hermaphrodites exposed to different environmental stresses (Figure 1A). Under standard growth conditions, wild-type hermaphrodites do not secrete the male-attracting pheromone at this young age. As a positive control, we used odors extracted from fog-2(q71) adult hermaphrodites that do not produce sperm and are, therefore, constitutively attractive (Leighton et al., 2014; Schedl and Kimble, 1988; Wan et al., 2019).
Figure 1.
Environmental stress induces transgenerational premature attractiveness
(A) Scheme of the volatile chemotaxis assay with males.
(B) Male chemotaxis experiments are shown. Growth temperatures of the hermaphrodites used for odor extraction and males used for chemotaxis appear below the panel. Each dot represents one biological replicate (chemotaxis plate) with 37–152 males. Bars: mean ± SD; results are from 3 independent experiments. One-way ANOVA and Dunnett’s correction for multiple comparisons to the 20°C condition were used.
(C) Proportions (%) of hermaphrodites mated during mating choice experiments are shown. Young hermaphrodites previously cultivated at the indicated temperatures interacted with wild-type males for exactly 1 h. Each gray line represents one biological replicate; results were collected over three independent experiments. Horizontal bar: median. Boxes: interquartile range (IQR). Two-tailed Wilcoxon matched-pairs signed rank test. See negative (non-attracting) and positive (fog-2 mutants) control groups in Figure S1C.
(D) Male chemotaxis experiments with the odors extracted from hermaphrodites grown at 25°C for 10–15 generations (P0) and transferred back to 20°C for 6 generations (red). Hermaphrodites continuously grown at 20°C were used as control (blue). Each dot represents one biological replicate (chemotaxis plate) with 38–144 males. Bars: mean ± SD; results were from 4 independent experiments. Two-way ANOVA and Sidak’s correction for multiple comparisons for every generation were used. (B–D) ∗∗∗∗p < 10−4, ∗∗∗p < 0.001, ∗∗p < 0.01, ns p > 0.05. See also Figure S1.
We examined a few stressful conditions: short-term incubation at 25°C, short-term starvation at L1 larval stage, and long-term starvation at the dormant dauer stage. None of these conditions induced premature attractiveness (Figure S1A). Previous studies have shown that temperature-induced gene expression changes accumulate over generations (Klosin et al., 2017; Manage et al., 2020). When we continuously cultivated the animals for 10–15 generations at 25°C, which is mildly stressful but within the standard temperature range for lab cultures, we discovered that the adult hermaphrodites were prematurely attractive (Figure 1B). Attractiveness seemed to accumulate over generations among hermaphrodites grown at 25°C for 1, 4, 8, and 12 generations (Figure S1B). Attraction required the activity of the pheromone receptor SRD-1 in the males (Figure 1B), indicating that this premature attractiveness is detected by the same sensory pathway that mediates attraction toward aged or spermless hermaphrodites (Wan et al., 2019). To examine whether premature attractiveness also translates to an increase in mating, we performed mating choice assays in which males carrying fluorescently labeled sperm had to choose between synchronized control (grown at 20°C) and prematurely attractive hermaphrodites (grown at 25°C). In the physical presence of both mating partners, the males copulated more with the hermaphrodites grown at 25°C (43% versus 14% mated hermaphrodites, p = 0.008 ; Figure 1C and Figure S1C for controls). The increase in insemination rates was srd-1-independent under these co-culture conditions (Figure S1C), suggesting that distinct locally acting cues, in addition to the secreted volatile odor, could further stimulate the males or the stressed hermaphrodites to mate (Aprison and Ruvinsky, 2015; Fagan et al., 2018; Kleemann and Basolo, 2007; Morsci et al., 2011; Srinivasan et al., 2008; Susoy et al., 2021). For example, a subset of soluble ascarosides were shown in the past to trigger male attraction without requiring functional srd-1 (Fagan et al., 2018).
Strikingly, we found that premature attractiveness was maintained transgenerationally for three generations among the descendants transferred to standard growth conditions (20°C) (Figure 1D), before it petered out. How is premature attractiveness inherited across generations? We hypothesized that this process could be mediated by changes in heritable small RNAs because germline small RNAs are modulated in response to high temperatures (Manage et al., 2020; Ni et al., 2016) and regulate key germline functions (Conine et al., 2010; Gu et al., 2009; Kamminga et al., 2012; Phillips et al., 2015). The inheritance of many germline endogenous small interfering RNAs (endo-siRNAs) (Buckley et al., 2012; Ni et al., 2016; Sapetschnig et al., 2015) and the transgenerational inheritance of different small RNA-mediated phenotypes (Lev et al., 2019; Luteijn et al., 2012; Posner et al., 2019; Rechavi et al., 2014; Shirayama et al., 2012) require the Argonaute HRDE-1 (Heritable RNAi Deficient-1), which induces the silencing of the cognate mRNA targets of its bound siRNAs.
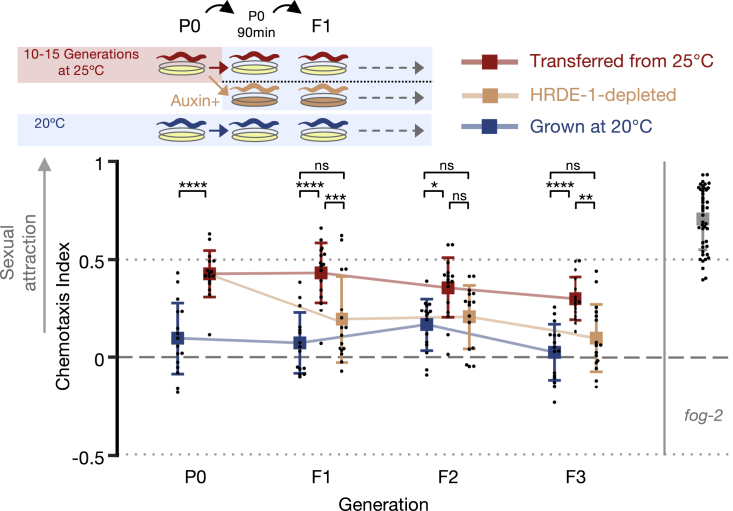
Because hrde-1 mutants are sterile when cultivated at 25°C (Buckley et al., 2012; Spracklin et al., 2017), we used CRISPR-Cas9 to engineer strains permitting conditional HRDE-1 depletion via the auxin-inducible degradation (AID) system (Nishimura et al., 2009; Zhang et al., 2015) (see STAR Methods). This approach allowed the depletion of the HRDE-1 protein down to undetectable levels within 1 h of the exposure to auxin (Figure S2A). We validated that the auxin-dependent degradation of HRDE-1 indeed abolishes the transgenerational RNAi inheritance by examining its effect on the heritable silencing of a germline-expressed gfp transgene (Figure S2B). Next, we extracted odors from aid::hrde-1 hermaphrodites grown at 25°C for 10–15 generations and from their HRDE-1-depleted descendants cultivated at 20°C on auxin-containing plates (see scheme in Figure 2). These experiments revealed that the heat- (25°C) triggered inheritance of premature attractiveness depends on HRDE-1 and, therefore, on the transgenerational transmission of the silencing small RNAs (Figure 2; see Figure S2C for additional controls). The histone-3-lysine-9 histone-methyltransferase SET-25 was previously shown to participate in certain transgenerational temperature-induced effects (Klosin et al., 2017), but our results indicate that it is not required for the inheritance of premature attractiveness (Figure S2D). Together, these results demonstrate that exposure to environmental stress induces enhanced sexual attractiveness in young hermaphrodites, which is transmitted transgenerationally via the small RNA inheritance machinery.
Figure 2.
Inheritance of premature attractiveness depends on HRDE-1
Odors were extracted from 25°C-grown hrde-1(pig-6[aid::ha::hrde-1]);sun-1p::tir-1 worms cultivated with (beige) or without auxin (red). The experiment was performed with additional control groups (see Figure S2C) accounted for in the presented statistical analysis. To ensure HRDE-1 depletion, P0 hermaphrodites were transferred to an intermediate auxin (or control) plate for 90 min at 20°C before being re-transferred to a new auxin plate where they laid the F1. Each dot represents one biological replicate (chemotaxis plate) with 38–138 males. Bars: mean ± SD; results were from 4 independent experiments. Two-way ANOVA and Tukey’s correction for multiple comparisons were used. ∗∗∗∗p < 10−4, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05. See also Figure S2.
MEG-3/4-associated small RNAs regulate heritable sexual attraction
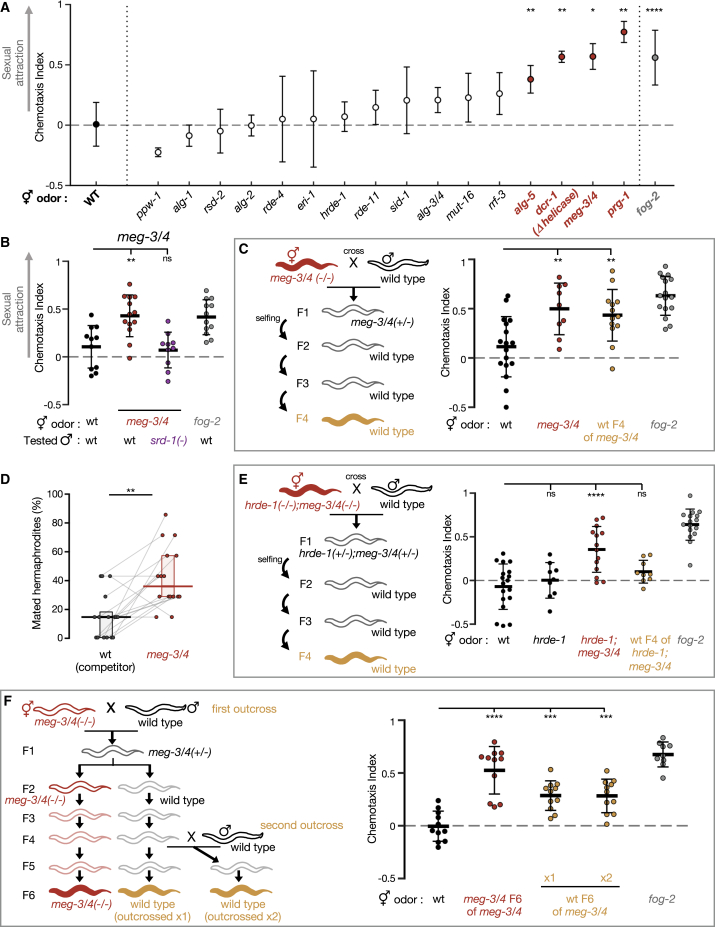
To dissect the small RNA pathways involved in heritable premature attractiveness, we assayed males for chemotaxis toward the odorants extracted from 1-day-old hermaphrodites carrying lesions in genes required in different small RNA pathways (Figure 3A; Table S1). We tested 16 small RNA mutant strains and discovered that the meg-3(tm4259);meg-4(ax2026) double mutants are prematurely attractive to males in an SRD-1-dependent manner and, moreover, that they transmit the premature attractiveness transgenerationally to their wild-type descendants (Figures 3B and 3C). Three other strains, prg-1(n4357), dcr-1(mg375), and alg-5(tm1163), were found to be prematurely attractive; however, they did not transmit the attractiveness transgenerationally (Figures 3A, S3A, and S3B). In addition to secreting the attractive odors, hermaphrodites from these four strains mated more with males in mating choice assays (Figures 3D and S3C).
Figure 3.
Small RNAs associated with MEG-3/4 regulate heritable sexual attraction
(A) Premature attractiveness screening of mutants impaired in genes acting in different small RNA pathways. Odors were extracted from mutated (x axis) young hermaphrodites and tested for the attraction of wild-type males. Dots and bars: Mean ± SD. Kruskal-Wallis test with Dunn’s correction for multiple comparisons to wild type was used. Full information appears in Table S1.
(B) Male chemotaxis experiments are shown. The genotypes of hermaphrodites used for odor extraction and those of tested males appear below panel. Results from one of the meg-3/4 experiments shown here are also represented in Figure 3A.
(C) Male chemotaxis experiments testing for the odor extracted from young wild-type descendants of meg-3/4 (homozygote double mutants for >80 generations).
(D) Proportions (%) of mated hermaphrodites during 1-h-long mating choice experiments; genotypes are indicated below panels. Gray lines represent biological replicates; results were collected over 4 independent experiments. Horizontal bar: median. Boxes: IQR. Two-tailed Wilcoxon matched-pairs signed rank test.
(E) Male chemotaxis experiments testing for the odor extracted from the wild-type descendants of hrde-1;meg-3/4 triple mutants. Data for the hrde-1 group were collected separately and were incorporated in the panel for the convenience of visualization.
(F) Male chemotaxis experiments testing for the odors extracted from F6 wild-type descendants of meg-3/4 double mutants outcrossed either once (P0) or twice (P0 and F4) with wild-type males. meg-3/4 double mutants were homozygote for >80 generations. (C, E, and F) Tested odors are color-coded according to schemes, and all hermaphrodites contained an integrated single-copy mex-5p::gfp transgene. (B, C, E, and F) Each dot represents one biological replicate (chemotaxis plate) with 30–190 males. Bars: mean ± SD. One-way ANOVA, Dunnett’s correction for multiple comparisons with wild type. All conditions were tested across at least 3 independent experiments. ∗∗∗∗p < 10−4, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05. See also Tables S1 and S2; Figures S3–S5.
Similar to the transgenerational effect of long-term cultivation at 25°C, we found that the inheritance of premature attractiveness from meg-3/4 mutants to wild-type progeny depended on the Argonaute HRDE-1 (Figure 3E). This experiment also revealed that functional hrde-1 is required specifically for the inheritance, but not for mere induction, of premature attractiveness (see hrde-1;meg-3/4 triple mutants in Figure 3E), in line with the fact that hrde-1 functions in the transgenerational transmission of RNAi and not in RNAi per se (Ashe et al., 2012; Buckley et al., 2012; Luteijn et al., 2012). Moreover, the gene sets displaying altered small RNA levels both in the progeny of meg-3/4 mutants and in animals grown at 25°C for multiple generations (Lev et al., 2019; Manage et al., 2020) were enriched with known targets of HRDE-1-bound small RNAs (Figure S4A). HRDE-1-bound small RNAs are characterized by them not being poly-uridylated (in contrast to other small RNA species; de Albuquerque et al., 2015; van Wolfswinkel et al., 2009). Accordingly, we found these gene sets to be enriched with the gene targets of small RNAs that were not poly-uridylated in these experiments (Figure S4B; see STAR Methods).
The heritable attractiveness of meg-3/4 was particularly potent and lasted for six generations (Figure 3F). This result resonates with the previous findings showing that meg-3/4 mutants exhibit a transgenerational disruption in the expression patterns of small RNAs pools in the germline (Dodson and Kennedy, 2019; Lev et al., 2019; Ouyang et al., 2019). These misregulated heritable small RNAs lead to an extraordinarily strong inheritance of RNAi defects, which can be detected in their wild-type descendants even after >10 generations (Dodson and Kennedy, 2019; Lev et al., 2019). The disruption of germ granules could, in theory, cause phenotypes in the progeny by affecting the maternally deposited mRNAs. However, the findings that heritable premature attractiveness lasts in the wild-type progeny of meg-3/4 for multiple generations and relies on functional HRDE-1 together support a model wherein it is the changes in heritable small RNAs that mediate the transgenerational inheritance of premature attractiveness.
Interestingly, all the RNA processing proteins that we found to affect premature attractiveness are located in the germ granules (Batista et al., 2008; Beshore et al., 2011; Brown et al., 2017; Wang and Reinke, 2008; Wang et al., 2014), which are cytoplasmic condensates found in the germline of a great number of organisms and made of RNA and proteins. meg-3/4 double mutants display severe defects in germ granules (Folkmann et al., 2021; Wang et al., 2014). Moreover, in accordance with a previous study (Putnam et al., 2019), we found that exposure to heat reduced the size of the P granules in the adult germline (Figures S4C and S4D). As previously shown (Dodson and Kennedy, 2019; Lev et al., 2019; Ouyang et al., 2019), transiently disrupting the germ granules’ stability can have major heritable consequences on the germline gene expression, as it dramatically alters the pool of heritable small RNAs (see more below). Thus, the small-RNA-mediated heritable attractiveness of the worms subjected to 25°C may have been derived from the effect of elevated temperatures on the stability of germ granules and on the processing of germline small RNAs therein.
Premature attractiveness is associated with the misregulation of sperm genes and sperm defects
How do heritable small RNAs control attraction? Numerous endo-siRNAs, emanating from throughout the genome, were misregulated in the worms grown at 25°C for multiple generations, in meg-3/4 mutants and in the wild-type descendants of meg-3/4 mutants (Dodson and Kennedy, 2019; Lev et al., 2019; Manage et al., 2020; Ni et al., 2016; Ouyang et al., 2019). We used genetic mutants and RNAi to disrupt the targets of the top endo-siRNA candidates overexpressed in the attractive wild-type progeny of meg-3/4 mutants (Figure S5; Table S2). With the exception of gem-4 mutants that displayed a partial effect (chemotaxis index = 0.36, p = 0.002 and 0.26, p = 0.15 for the alleles dx77 and ok878, respectively), the disruption of these targets was not sufficient to induce premature attractiveness (Figure S5). GEM-4 (Gon-2 Extragenic Modifier 4) encodes a copine family protein (Creutz et al., 1998; Nakayama et al., 1998) (binding cytoplasmic Ca2+-dependent phosphatidylserine) expressed in both the gonad and the spermatheca and was implicated in the gonad development (Church and Lambie, 2003). We found that gem-4 is targeted by the small RNAs in meg-3/4 mutants and their wild-type progeny and that the gem-4 mRNA expression is downregulated in meg-3/4 mutants (Figure S5).
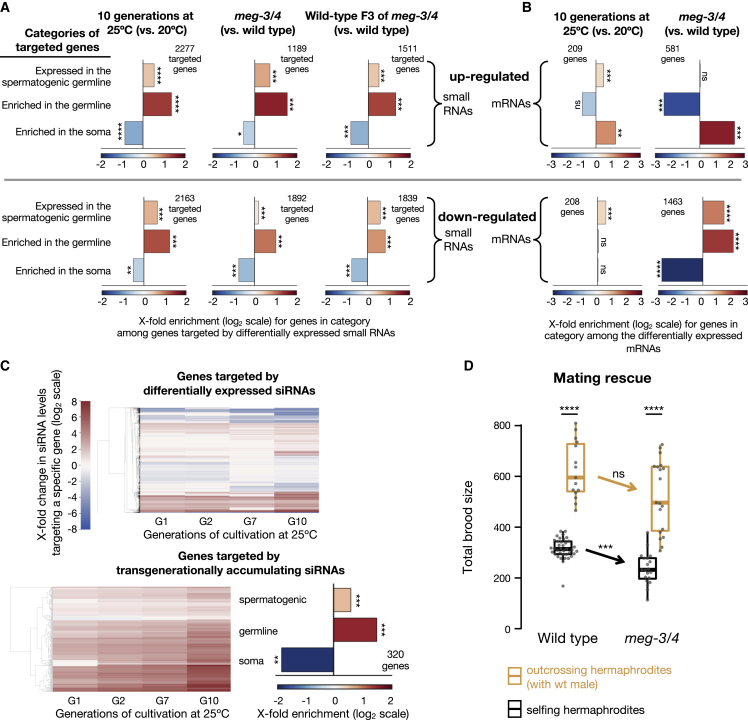
Further analyses revealed a global enrichment for sperm-expressed genes among the mRNAs and the targets of endo-siRNAs that are differentially expressed under conditions that trigger premature attractiveness (Figures 4A and 4B). A subset of 320 protein-coding genes were targeted by small RNAs that transgenerationally accumulate in worms cultivated at 25°C, and these genes were also enriched for sperm-expressed genes (Figure 4C).
Figure 4.
Prematurely attractive hermaphrodites exhibit the misregulation of sperm genes and sperm defects
(A and B) Enrichment analysis of small RNAs (A) and mRNAs (B) upregulated (top panels) and downregulated (bottom panels) in worms upon cultivation at 25°C for 10 generations (Manage et al., 2020) in meg-3/4 mutants (Lev et al., 2019; Ouyang et al., 2019) or in F3 wild-type progeny (two generations of homozygosity) of meg-3/4 mutants (Lev et al., 2019). All small RNAs targeting a specific protein-coding gene were grouped together into one value that was used to assess whether the total amount of small RNAs targeting the gene was different (analyzed with DESeq2, adjusted p value < 0.1). The number of differentially targeted (small RNAs [A]) or expressed (mRNA, [B]) protein-coding genes for each condition appears next to the relevant panel. Shown are fold enrichment results (observed/expected, log2 scale) for genes known to be expressed in the spermatogenic germline (Ortiz et al., 2014), for genes enriched in the whole germline (Serizay et al., 2020) and for genes enriched in the soma (Serizay et al., 2020), in the lists of genes differentially targeted by small RNAs or with significantly different mRNA levels. Adjusted p values were obtained using a randomization bootstrapping test and corrected for multiple comparisons using the Benjamini-Hochberg step-up procedure.
(C) Clustering of siRNA expression in worms grown at 25°C for 1, 2, 7, and 10 generations. Data were generated by re-analysis of the available dataset by Manage et al., 2020. (Top) Hierarchical clustering analysis of 5,223 genes targeted by differentially expressed siRNAs in worms grown at 25°C for 10 generations compared with control. (Bottom) A subset of 320 protein-coding genes targeted by transgenerationally accumulating siRNAs at 25°C. siRNAs were considered “transgenerationally accumulating” if they displayed significantly higher differential expression levels in each time point compared with the previous one and were significantly upregulated at the 10th generation. Color bar indicates the log2 fold changes in siRNA expression in both panels (top and bottom, compared with worms grown at 20°C, adj. p < 0.1). Non-significant fold-change values (adj. p > 0.1) appear in white. (Right) Enrichment analysis for the subset of 320 protein-coding genes.
(D) Total brood size (y axis) of wild-type and meg-3/4 hermaphrodites reproducing via selfing (black) or via outcrossing (yellow). Insemination by wild-type males similarly increased the brood size of wild-type and meg-3/4 hermaphrodites, indicating that meg-3/4 oocytes can be fertilized when provided with wild-type sperm. Data were collected over 3 independent experiments. Dots represent values for individual hermaphrodites. Boxplot (Tukey’s style): median and IQR, whiskers extend to the most extreme value within 1.5 × IQR from the 25th or 75th percentile. Welch’s ANOVA, Games-Howell post-hoc correction for multiple comparisons. ∗∗∗∗p < 10−4, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05. See also Table S2; Figures S5 and S6.
Wild-type animals grown at 25°C for 10 generations were previously shown to have sperm defects (Argon and Ward, 1980; Conine et al., 2010; Manage et al., 2020). We found that both meg-3/4 mutants and their wild-type descendants exhibit a reduction in selfing-derived brood size (−22% and −13%, respectively; Figure S6A). In mating rescue experiments (Figure 4D), the brood size of meg-3/4 mutants robustly increased following mating with wild-type males (2.2×-fold, p < 10−4), i.e., when meg-3/4 hermaphrodites were provided with a functional sperm. These results imply that the reduction in brood size in selfing meg-3/4 hermaphrodites is at least in part due to the defects in the self-produced sperm. The three other small RNA mutants that we found to be prematurely attractive exhibited sperm defects such as abnormal sperm pseudopod formation (Pavelec et al., 2009; Wang and Reinke, 2008) or shorter spermatogenesis (Brown et al., 2017). Collectively, these findings suggest that the disruption of germ granules and mistargeting of sperm genes by small RNAs are associated with mild sperm defects that could trigger premature attractiveness. Indeed, as detailed above, severe defects in sperm are known to trigger the secretion of the male-attracting volatile pheromone (Leighton et al., 2014; Wan et al., 2019).
Premature attractiveness is associated with an increase in male prevalence
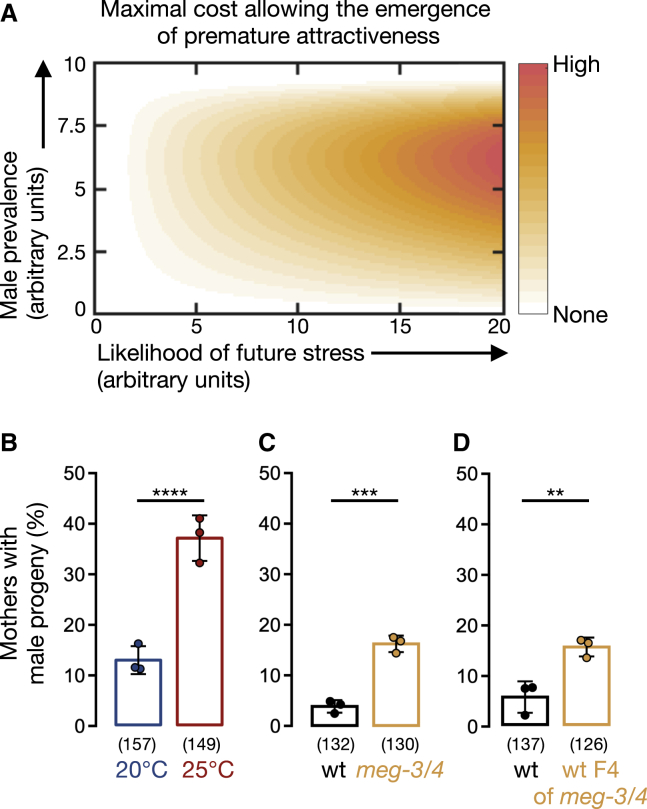
Secreting a male-attracting pheromone could be costly (due to the costs of the production of the pheromone itself or the consequences of the mating it facilitates) and may, thus, negatively impact the fitness of the hermaphrodites that have the cheaper option of reproducing via selfing (Maures et al., 2014; Shi and Murphy, 2014; Shi et al., 2017). We used a theoretical population genetics approach to computationally examine under which conditions a transient increase in premature male attraction would spread in the population or, in other words, under which environmental conditions it would be adaptive to acquire the ability to transgenerationally transmit premature attractiveness (see STAR Methods for details). The model constitutes an “invasion analysis,” assessing the evolutionary trajectory of a rare allele for premature attraction in a population that does not display premature attraction and deriving the conditions that allow the increase in the frequency of this allele in the population (Otto and Day, 2011). The model was based on new empirical data that we collected (Figure S6B and chemotaxis results).
For the secretion of the male-attracting pheromone to be adaptive, our model predicted two requirements: (1) the environmental stress should persist for multiple generations before triggering premature attractiveness, and (2) there should be a mild increase in the number of males (Figure 5A). The first condition is in line with our abovementioned observation that long-term, but not short-term, cultivation at 25°C induces premature attraction (Figures 1A, S1A, and S1B). Interestingly, a recent study has found that C. remanei nematodes cope relatively better in elevated temperatures only if the temperatures in previous generations were stably high or were gradually increasing in a predictable manner (Lind et al., 2020). In accordance with the second prediction of the model, a high incidence of males (Him phenotype) was documented in worms grown at elevated temperatures (Figure 5B; Zhang et al., 2011). The Him phenotype also accompanies the transgenerational gradual loss of fertility (Mrt Phenotype) of C. elegans wild isolates and mutants impaired in small RNA and chromatin modification pathways when cultivated at high temperatures (Frézal et al., 2018; reviewed in Rechavi and Lev, 2017). We discovered that the selfing meg-3/4 hermaphrodites and their wild-type descendants also have more male progeny than wild-type animals (Figures 5C and 5D).
Figure 5.
Premature attractiveness is associated with an increase in male prevalence
(A) Model for maximal cost (color-coded) that allows the evolution of premature attractiveness as a function of male prevalence in the population (y axis) and likelihood of stress in future generations (x axis). Model derivation and parameters are found in STAR Methods.
(B–D) Proportion (%) of hermaphrodites with males in their selfing-derived progeny. Tested conditions are indicated below data. Animals in (B) were wild-type N2 worms, while in (C) and (D), all animals bear an integrated single-copy mex-5p::gfp transgene in their genetic background. Dots represent results from independent experiments. Bars: mean ± SD. Numbers below bars: total n of tested hermaphrodites. Fisher’s exact test, ∗∗∗∗p < 10−4, ∗∗∗p < 0.001, ∗∗p < 0.01. See also Figure S6.
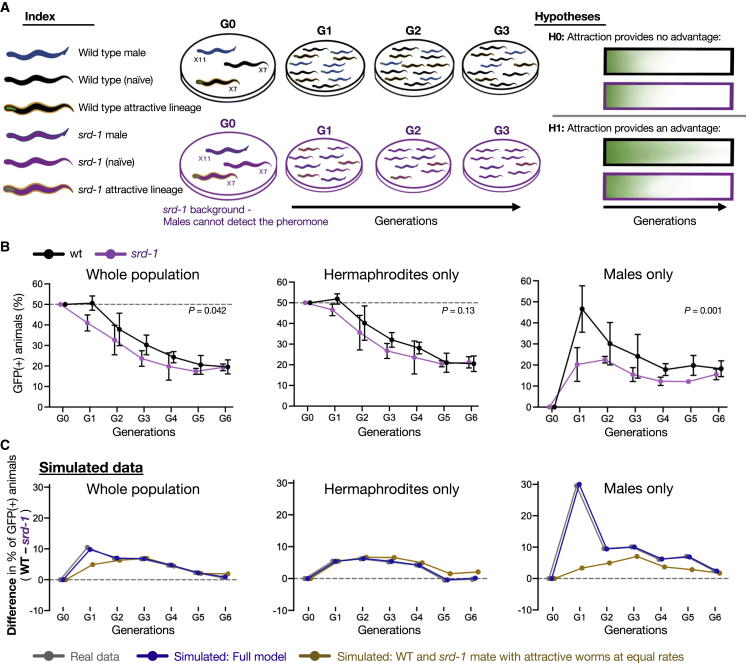
Transgenerational premature attractiveness can affect the genetic structure of the population
To examine whether heritable small-RNA-mediated premature attractiveness can impact the genetic structure of a population, we conducted multigenerational competition experiments (scheme in Figure 6A and STAR Methods). These experiments were conducted with genetically identical (wild-type) worms that differed in their epigenetic heritage (F3 wild-type descendants of meg-3/4 versus naive wild type). We took advantage of the fact that premature attractiveness is strongly transmitted to the wild-type descendants of meg-3/4 mutants and let these two groups of genetically identical worms compete for 7 generations over the same resources (food and males). To determine whether the effects of the epigenetic heritage on the population are due to the male-attracting pheromone, we ran the experiments side-by-side using both wild types and srd-1(−) mutants that cannot sense the secreted pheromone.
Figure 6.
Transgenerational premature attractiveness can affect the genetic structure of the population
(A) Scheme depicting the multigenerational competition experiments. In these experiments, the GFP-tagged wild-type animals originating from meg-3/4(−) ancestry (“attractive lineage”) competed against the naive GFP-negative wild-type animals (non-attractive lineage). Large populations were imaged and analyzed in every generation. Experiments were performed side by side using two genetic backgrounds: wild-type (black) and srd-1 mutants (purple). Attractive worms in the G0 generation were F3 wild-type descendants of meg-3/4 (or meg-3/4;srd-1) hermaphrodites.
(B) Epigenetically attractive lineages mate more frequently and are more prevalent in populations when males sense the secreted male-attracting pheromone. Proportion (%) of GFP-positive animals (y axis) over generations (x axis) in whole populations (left) or sexual subgroups (center and right) are shown. Because outcrossing progeny contains ∼50% males versus only ∼0.1% in selfing-derived progeny, males that are present in the populations likely derive from outcrossing events. Experiments with wild-type (black) and srd-1(−) (purple) animals were performed side by side in three biological replicates. Dots and bars: mean ± SD. n per group/replicate/generation = 368±93 (range: 184–576). Indicated p values were calculated using the generalized linear mixed model (GLMM).
(C) The difference (wild type – srd-1) in the proportions (%) of GFP-positive animals (y axis) over generations (x axis). Shown are the experimental results (gray, as depicted in [B]), the simulated results (dark blue), and the simulated results where the underlying assumption that wild type and srd-1(−) mate differentially with attractive hermaphrodites is relaxed (brown). See STAR Methods and Figure S6 for full details about the models and additional tested assumptions. See also Table S3; Figure S6.
As described earlier, the “attractive” wild-type descendants of meg-3/4 produce a ∼15% lower brood size compared with that of the naive wild-type worms (Figure S6A) and are, thus, expected to decrease in frequency across generations. Remarkably, we found that the ability of the males to sense the male-attracting odor increased the relative frequency of the “attractive lineage” (compare srd-1(+) and srd-1(−) populations; Figure 6B; full results in Table S3). Moreover, the prevalence of the males derived from the “attractive lineage” was substantially higher in the population with functional SRD-1, indicating an increase in mating events involving the “attractive lineage” across multiple generations. We also conducted in silico simulations of population dynamics incorporating the empirical data that we collected on the differences in male attraction, brood size, and lineage fitness (see Figure S6C and STAR Methods for more details). These analyses corroborated that the increase in male prevalence of the “attractive lineage” (i.e., mating) is largely explained by srd-1-dependent premature attraction (Figures 6C and S6C). Together, these results imply that the detection of the male-attracting pheromone transgenerationally increases the mating frequency of prematurely attractive hermaphrodites and the spread of the alleles derived from the heritably attractive lineages in the population.
Discussion
Our findings reveal that in response to mildly stressful temperatures, C. elegans hermaphrodites elevate their attractiveness and the attractiveness of their stress-naive descendants for multiple generations. Mechanistically, premature attractiveness is associated with sperm defects and with disturbances in germ granules and is transgenerationally inherited to ensuing generations via heritable small RNAs and the germline Argonaute HRDE-1. Because increased premature attractiveness also elevates the frequency of mating events, we propose that transient small-RNA-based responses to environmental challenges could impact genetic variation and could thus potentially shape the course of evolution. This phenomenon may constitute a novel manifestation of stress-induced evolvability, analogous to stress-induced mutagenesis, stress-triggered mobile element transposition, horizontal gene transfer, and others (Galhardo et al., 2007; Ram and Hadany, 2014; Slotkin and Martienssen, 2007; Thomas and Nielsen, 2005). Yet, the enhanced mating driven by premature attractiveness differs from these examples because it is induced by an inherited entity. Thus, the heritable small RNAs can affect genetic material by altering sexual behavior even when the original environmental stimulus is no longer present.
In addition to the potential increase in genetic variability, we speculate that the inherited enhanced attractiveness and mating may facilitate the elimination of the accumulated detrimental epigenetic information that could harm the animal’s reproductive fitness. Indeed, many mutants that are defective in small RNA inheritance exhibit a Mrt phenotype and become gradually sterile if forced to self-fertilize in the lab (reviewed in Rechavi and Lev, 2017).
A previous study showed that to avoid pathogenic bacteria, worms monitor their health and notice when they become sick (they learn to avoid bacterial food that inactivates vital cellular processes). This strategy is very useful because it allows the worms to detect the action of numerous toxins, even when they do not have specific receptors to identify each and every one of the toxic molecules (Melo and Ruvkun, 2012). Similarly, our results raise the intriguing possibility that the small-RNA-mediated regulation of sperm functions may act as a stress rheostat that induces enhanced male attraction whenever the worms are stressed. Interestingly, such a mechanism would not be dependent on specific sperm gene targets (as those could vary between stress conditions) and could instead respond to the accumulative effect of even partial silencing of various sperm-related genes. High temperatures impact sperm function in a wide range of species including mammals. Future studies may show whether such a generalized mechanism is also conserved. In C. elegans hermaphrodites, elevated temperatures that induce sperm defects also trigger a heritable increase in sexual attraction, which could be adaptive under environmental stress. Deepening our understanding of the biological strategies that help to better cope with high temperatures is critical for the preservation of biodiversity amid global warming.
Limitations of the study
There could be many genes that affect sperm health and activate premature attractiveness due to their transgenerational silencing by heritable small RNAs. At present, we were not able to determine whether enhanced attractiveness is driven by the regulation of a group of genes or by a single gene targeted by heritable small RNAs. The large number of candidates based on differentially expressed small RNAs and mRNAs (Table S2; Figure S5), the probable additive/synergistic actions of individual affected genes, and the sterility phenotype of knockout mutations impairing with sperm function make it currently impractical for exhaustive genetic examinations. A better understanding of the chemical nature of the secreted pheromone, the enzymatic pathway, and the inter-tissue communication involved in its synthesis (Leighton et al., 2014) may narrow the list of promising targets for subtle genetic manipulations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-β-actin | Sigma-Aldrich | Cat#: A5441; RRID: AB_476744 |
| Mouse monoclonal anti-HA | Biolegend | Cat#: 901501; RRID: AB_2565006 |
| Donkey polyclonal anti-mouse IgG conjugated with Horseradish peroxidase. | Jackson ImmunoResearch | Cat#: 715-035-150; RRID: AB_2340770 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium Azide | Sigma-Aldrich | Cat#: S2002 |
| MitoTracker Red CMXRos | Invitrogen | Cat#: M7512 |
| indole-3-acetic acid (IAA, natural auxin) | Alfa Aesar | Cat#: A10556 |
| Levamisol hydrochloride | Sigma-Aldrich | Cat#: L0380000 |
| cOmplete™, Mini Protease Inhibitor Cocktail | Roche | Cat#: 11836153001 |
| NuPAGE™ LDS Sample Buffer | Invitrogen | Cat#: NP0007 |
| 7.5% Mini-PROTEAN® TGX™ Precast Protein Gels | Bio-Rad | Cat#: 4561023 |
| Trans-Blot Turbo Mini 0.2 μm Nitrocellulose Transfer | Bio-Rad | Cat#: 1704158 |
| SuperSignal™ WestPico PLUS Chemiluminescent Substrate | Thermo Scientific | Cat#: 34580 |
| Critical Commercial Assays | ||
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat#: 23227 |
| Deposited Data | ||
| Original Code | This manuscript | https://doi.org/10.5281/zenodo.5651221 |
| Sequencing dataset | Lev et al., 2019 | GSE128112 |
| Sequencing dataset | Manage et al., 2020 | GSE138220 |
| Sequencing dataset | Ouyang et al., 2019 | GSE134638 |
| Original Western Blot images | This manuscript | https://doi.org/10.17632/sgvpkjnmr4.1 |
| Experimental Models: Organisms/Strains | ||
| Wild type | CGC | N2 |
| fog-2(q71) V | CGC | JK574 |
| srd-1(eh1) II | CGC | CB5414 |
| bqSi577[myo-2p::GFP + unc-119(+)] IV | This manuscript | BFF53 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II; unc-119(ed3) III | Eric Miska lab | SX1263 |
| unc-119(ed3) III; ieSi38[sun-1p::TIR1::mRuby::sun-1 3’UTR + Cbr-unc-119(+)] IV | CGC | CA1199 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II; hrde-1(pig6[aid::ha::hrde-1]) III | This manuscript | BFF68 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II; hrde-1(pig6[aid::ha::hrde-1]) III; ieSi38[sun-1p::TIR1::mRuby::sun-1 3′UTR + Cbr-unc-119(+)] IV | This manuscript | BFF69 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II; hrde-1(tm1200) III | Rechavi lab | BFF48 |
| set-25(n5021) III | CGC | MT17463 |
| ppw-1(pk1425) I | CGC | NL3511 |
| alg-1(gk214) X. | CGC | VC446 |
| rsd-2(pk3307) IV | CGC | NL3307 |
| alg-2(ok304) II | CGC | WM53 |
| rde-4(ne299) III | Rechavi lab | BFF12 |
| eri-1(mg366) IV | CGC | GR1373 |
| hrde-1(tm1200) III | CGC | YY538 |
| rde-11(hj37) IV | CGC | VS27 |
| sid-1(qt9) V | CGC | HC196 |
| alg-4(ok1041) III; alg-3(tm1155) IV | CGC | WM300 |
| mut-16(pk710) I | CGC | NL1810 |
| rrf-3(pk1426) II | CGC | NL2099 |
| alg-5(tm1163) I | CGC | WM159 |
| dcr-1(mg375) III | CGC | YY470 |
| meg-3(tm4259);meg-4(ax2026) X | CGC | JH3225 |
| prg-1(n4357) I | CGC | SX922 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II;meg-3(tm4259);meg-4(ax2026) X | Rechavi lab | BFF40 |
| mjIs134 [mex-5p::gfp::h2b::tbb-2_3’UTR] II;hrde-1(tm1200) III;meg-3(tm4259);meg-4(ax2026) X | Rechavi lab | BFF49 |
| wago-4(tor117[wago-4::gfp::flag]) II; pgl-1(gg547[pgl-1::3xflag::tagRFP]) IV | Rechavi lab | BFF44 |
| bqSi577[myo-2p::GFP + unc-119(+)] IV;meg-3(tm4259);meg-4(ax2026) X | This manuscript | BFF70 |
| srd-1(eh1) II; bqSi577 [myo-2p::GFP + unc-119(+)] IV; meg-3(tm4259) X; meg-4(ax2026) X | This manuscript | BFF57 |
| gem-4(dx77) IV | CGC | EJ808 |
| gem-4(ok878) IV | CGC | RB974 |
| Oligonucleotides | ||
| N’ hrde-1 crRNA: 5’-CAUAAUUUUGUCGAGCAAGU-3’ | IDT | N/A |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | N/A |
| FASTQC | Andrews, 2010 | N/A |
| Cutadapt | Martin, 2011 | N/A |
| Shortstack | Axtell, 2013 | N/A |
| HTSeq count | Langmead and Salzberg, 2012 | N/A |
| R DESeq2 | Love et al., 2014 | N/A |
| RNAlysis | github.com/GuyTeichman/RNAlysis | N/A |
| Seaborn | Waskom et al., 2020 | N/A |
| Wormachine | Hakim et al., 2018 | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Oded Rechavi (odedrechavi@gmail.com).
Materials availability
C. elegans strains generated in this study will be deposited to the Caenorhabditis Genetics Center (CGC).
Experimental model and subject details
Culture of worms and genetic crosses
All strains used in this study are listed in key resources table. Worms were cultured using standard procedures in NGM plates, fed with Escherichia coli (OP50 strain) and kept at 20°C unless noted otherwise. For the genetic crosses depicted in Figures 3, S3, and S6, 3–4 L4 hermaphrodites were placed with 10-15 males of the indicated genotypes in plates previously seeded with one drop of OP50. P0 Mated hermaphrodites were transferred to fresh plates without males after 16-24 hours of interaction. F1 hermaphrodites were separated to fresh plates as L4s to prevent mating by F1 males. Genotyping of F1s and F2s was performed by PCR.
Method details
Extraction of Worm-Conditioned Media (WCM)
WCM (containing the odors secreted by the hermaphrodites) was produced as previously described (Leighton et al., 2014) with minor modifications. To obtain large synchronized populations of hermaphrodites on the first day of adulthood, 12-20 gravid adults were transferred to fresh seeded plates and were allowed to lay eggs for 2-3 hours before removal. This procedure was performed in triplicates. 72-80 hours later (or 60-64h for animals grown at 25°C due to faster development) the adults were rinsed off the plates and washed 4 times with M9 buffer in 1.7ml tubes to remove bacteria. In every round of wash, worms were allowed to settle at the bottom of the tube for 90-120 seconds, then we removed the supernatant, applied 1ml of fresh M9 and gently mixed the tube via finger-tapping. After the last removal of the supernatant, worms were transferred to a non-seeded NGM plate with no cover using a Pasteur pipette. Once the buffer fully evaporated, 75 worms were transferred by picking into the inverted cap of a standard 1.7ml tube containing 75μl of M9. Caps were then sealed with parafilm and maintained for 24 hours at 20°C. Following incubation, the full contents of the caps were transferred into 1.7ml tubes and rested on ice for 3 minutes to let the worms and eggs settle at the bottom of the tube. 50μl of supernatant with no worms/eggs were then transferred into a new tube and stored at -20°C. WCMs from wild-type young hermaphrodites grown at 20°C were collected side-by-side with WCMs from biological groups of interest and used as negative control in subsequent male chemotaxis experiments.
25%-30% of meg-3/4 adults are sterile and noticeably bear empty uteri (Wang et al., 2014), thus special care was taken to select only fertile worms containing visible embryos for WCM production.
In some cases, we used an alternative synchronization protocol in which L4 hermaphrodites were picked to new plates 24 hours before beginning of WCM incubation. These cases included the obligatory-outcrossing strain fog-2, strains with pronounced developmental delays (alg-1, prg-1), or animals deriving from genetic crosses and whose parents required PCR-genotyping. Control groups were collected similarly side-by-side and WCM extraction was performed normally.
Male chemotaxis experiments
Male chemotaxis assays were inspired from Leighton et al. (2014) with a few modifications. To obtain large populations of wild-type or srd-1(-) males, ten mating plates were prepared, that included twelve L4 hermaphrodites and twenty young males and a small lawn of food. 24h later, mated hermaphrodites were transferred to fresh seeded plates without males. 3 days later (one day before chemotaxis experiment), 1600-2000 young males were separated from hermaphrodites by picking and transferred to fresh seeded plates for 24 hours. This isolation was meant to reduce the impact of recent mating on the chemotaxis performance. Chemotaxis plates (90mm diameter, 14 ml, 2% agar, 5 mM KP04, 1mM CaCl2 and 1mM MgS04) were poured on the day of male picking, let dry with no cover for 30 minutes, then with cover overnight at room temperature. On the day of experiment, remaining excess of humidity on the plates’ cover was removed with a Kimwipe. 1μl of 1M sodium azide was applied in the agar on each far side of the petri dish. Right after application of sodium azide on plates, WCMs were taken out of -20°C storage to thaw at room temperature, and males were rinsed from plates and washed four times in M9 (no spinning, just gravity). Once washes were completed, ∼60-120 males were pipetted to the center of a chemotaxis plate. While the males were still swimming in the excess M9, 10μl of WCM were pipetted to the bottom side of the lid (facing the dish), above the sodium azide spot, and 10μl of M9 above the opposite spot (see scheme Figure 1A). Excess of M9 in the plate was then gently dried out of the males’ area using a kimwipe, and the plate was covered with the WCM-applied lid. Plates were sealed with parafilm and incubated overnight in the dark at 20°C. At the conclusion of the assay, males on each side of plates were quantified, and all males within the 3cm central “buffer” zone were ignored. Chemotaxis index was calculated as (#males at WCM−#males at M9)/(#males at WCM+#males at M9). All chemotaxis experiments included a negative control (plates testing WCM extracted from young wild types) and a positive control (plates testing WCM from fog-2 worms). Each experiment typically included 3-5 tested groups (controls included) and 3-4 biological replicates (chemotaxis plates) per group. Independent chemotaxis experiments were performed on different days and tested independently-produced WCMs. All biological groups of WCMs were tested across at least three independent experiments, with the exception of some of the screened mutants shown in Figure 3A that did not trigger male-attraction.
Exposure to environmental stress
L1 starvation
Synchronized gravid adults were bleached with standard hypochlorite treatment to obtain embryos and to remove traces of food. Embryos were transferred to NGM plates with no food, while a portion was transferred to OP50-seeded plates (fed controls). After 6 days of starvation, OP50 was added to the starved plates. WCM from starved and fed animals were collected side-by-side on the first day of adulthood (synchronized by picking of L4s 24h prior to WCM production). Due to differences in developmental pace, the WCM of fed worms were extracted from the F1 progeny of the “bleached” embryos.
Dauer induction
Dauer induction was conducted as previously described (Webster et al., 2018). Wild-type gravid adults were bleached with standard hypochlorite treatment. Embryos were resuspended in 5ml of S-Complete buffer containing 1mg/ml HB101 E. coli, at a concentration of 5 embryos/μl (i.e. 25,000 embryos in total). Worms were cultured in a 25mL Erlenmeyer flask at 20°C and 180 rpm for 40-45 days. Worms were eventually rescued upon transfer to NGM plates seeded with OP50. WCM from post-dauer animals was produced on the first day of adulthood. Control fed worms were cultivated in similar conditions after bleach except a higher HB101 concentration (38mg/ml) and lower embryo concentration (1 embryo/μl). The control worms were grown in liquid for 48 hours (they are L3 larvae by this time), and transferred to seeded NGM plates at the same time as post-dauer worms.
Cultivation at 25°C
For all data displayed in Figure 1, wild-type worms had been cultivated at 25°C for 10-15 generations before initiation of the experiment. WCM production started 60h-64h after synchronized egg-laying and the 24h-incubation was performed at 20°C, similar to all other WCMs tested. WCM from wild-type 20°C controls were produced side-by-side, using worms that were synchronized by egg-laying 8h-12h prior to the 25°C synchronization (due to differences in developmental time). For the experiment depicted in Figure S1B, G1 is the first generation grown at 25°C, and collection of WCM was performed similarly to the above. For transgenerational experiments (Figures 1D and 2), the depicted P0 generation is the last generation cultivated at 25°C. P0 young adults were transferred to 20°C for 2-3 hours, to lay the synchronized F1 embryos. From this moment on, the F1s and the consecutive generations continuously grew at 20°C.
Mating choice experiments
In those experiments, eleven males with fluorescent sperm interacted for exactly one hour on a small patch of food with seven “test” hermaphrodites and seven “competitor” hermaphrodites, and the mated hermaphrodites were subsequently scored based on fluorescence in their spermathecae.
One day before mating assays, young males (larvae with visible male morphology) were transferred into NGM plates containing 2μM MitoTracker Red CMXRos (Invitrogen, M7512) and previously seeded with OP50. On the same day, L4 hermaphrodites from the strains to be tested were picked into fresh plates, to isolate synchronized populations. For all experiments, the “competitor” hermaphrodites belonged to the pseudo-wt BFF53 strain that bears a myo-2p::gfp single-copy integrated transgene. After overnight incubation, males were rinsed off the MitoTracker plates and washed four times in M9, then recovered for 90 minutes on regular NGM plates with OP50. Seven myo-2p::gfp hermaphrodites (“competitor”) and seven hermaphrodites of the strain of interest (“test”) were then transferred to the center of a mating 60mm NGM plate (seeded with 50μl of OP50 at the center of the plate, 24h prior to experiment). Eleven Mitotracker-stained males were added to the mating plate already containing the fourteen hermaphrodites. All 25 worms interacted for exactly one hour, at the end of which all males were removed. The 14 remaining hermaphrodites were mounted on a microscope slide with a 2% agarose pad, at least 1h after removal of males, to allow enough time for mating-derived sperm to reach the spermatheca (Kubagawa et al., 2006). Hermaphrodites were imaged with x10 magnification using a BX63 Olympus microscope (Tel Aviv University) or a TiE Nikon microscope (Bio-Imaging Resource Center, Rockefeller University). Images were used to quantify the events of successful mating in each group of hermaphrodites, based on the presence of MitoTracker-positive sperm in the spermathecae. “Competitor” and “test” worms were distinguished based on GFP fluorescence in the pharynx. Groups were typically tested in biological quadruplicates, and side-by-side with plates testing for wild-type and fog-2 controls on the same day. Independent experiments were performed on separate dates, and all biological groups were tested across at least three independent experiments. In two instances (one plate in Figure 1C and one plate in Figure S1C, bottom right panel), one hermaphrodite from the competitor group was lost during slide preparation. In these two cases, the depicted proportion value was calculated out of a total of 6 instead of 7.
Generation of aid::hrde-1 strains
We used a previously described CRISPR strategy with hybrid dsDNA donors (Dokshin et al., 2018) to tag the N’ terminus of the hrde-1 gene with aid::ha. crRNA sequence: CAUAAUUUUGUCGAGCAAGU. The crRNA and aid::ha DNA sequence (PCR amplified to generate the repair templates) were synthesized by IDT. The ribonucleoprotein mix was injected as described (Dokshin et al., 2018), into animals of the SX1263 strain bearing a germline-expressed mex-5p::gfp transgene. The obtained engineered strain (BFF68) was then crossed with the CA1199 strain (Zhang et al., 2015) bearing a sun-1p::tir-1 transgene. The resulting strain (BFF69) was used for conditional HRDE-1 depletion experiments. Experiments to validate the auxin-dependent depletion of HRDE-1 are depicted in Figures S2A and S2B.
Conditional hrde-1 depletion experiments
Auxin treatment was performed as previously described (Zhang et al., 2015) using the natural auxin indole-3-acetic acid (IAA) purchased from Alfa Aesar (#A10566). Auxin-containing plates were prepared by adding auxin in ethanol (1mM final concentration) to freshly-autoclaved NGM medium cooled to 50°C, just prior to the pouring of the plates. Concentrated (5x) OP50 was used on auxin-containing plates due to slower growth of bacteria on auxin.
Transgenerational RNAi experiments
These experiments were performed to test BFF69 animals for auxin-dependent disruption of heritable RNAi, the classic phenotype of hrde-1(-) worms (Buckley et al., 2012). HT115 E. coli bacteria that transcribe anti-gfp dsRNA (or empty-vector controls) were grown overnight in LB supplemented with 25 μg/ml Carbenicillin. Bacterial cultures were then seeded onto NGM plates containing IPTG (1mM), Carbenicillin (25 μg/ml) and auxin (1mM). Bacteria were seeded on auxin+RNAi plates 3-4 days before addition of worms since bacteria grow slower on auxin-containing plates. L4-stage larvae of the BFF69 (or control strains, all bearing a mex-5p::gfp transgene) were transferred to auxin+RNAi plates overnight. The next morning, the adults (10-12 per plate) were allowed to lay eggs for 2-3 hours on fresh auxin+RNAi plates for synchronization of the P0 generation. Three days later, P0 adult worms were transferred to auxin-containing NGM plates with OP50 (no RNAi) and were allowed to lay eggs for 2-3 hours, for synchronization of the F1 generation. Synchronizations proceeded similarly every three days on auxin-containing plates seeded with OP50 until the F3 generation. On the day of synchronization, adults were collected for imaging on microscope slides with 2% agarose pads and a drop of M9 with 2mM levamisole. Images were acquired using a TiE Nikon microscope equipped with a Andor Neo sCMOS camera (Bio-Imaging Resource Center, Rockefeller University) with x10 magnification. Images were used to quantify the GFP silencing in the germline. Analysis of GFP fluorescence was done using the Fiji software (Schindelin et al., 2012), by manually defining in each worm the area of the proximal oocyte and three background regions. CTCF values were calculated in this manner: Integrated density values of the oocyte – (area of measured oocyte ∗ mean fluorescence of background regions). We normalized the CTCF value to the average CTCF value obtained from photographs of control animals of the same genotype, generation and age which were fed on empty-vector control plates.
Western blot
200-300 1-Day adult hermaphrodites were washed 4 times with M9 buffer, then resuspended in 100μl of ice-cold RIPA buffer supplemented with protease inhibitors (cOmplete™,11836153001, Roche) and homogenized using a Dounce homogenizer. Following 3 minutes centrifugation at 850g, supernatants were collected to a new eppendorf tube and the protein concentration was determined with BCA (ThermoFisher, 23227). Samples were diluted to equally contain 10μg total protein, denatured in LDS buffer (Invitrogen™, NP0007) and incubated at 70°C for 5 minutes, prior to loading reduced with 50mM DTT and run on a 7.5% polyacrylamide gel (TGX #4561023, Bio-Rad) in Tris-glycine-SDS running buffer. Proteins were transferred onto a 0.2-μm nitrocellulose membrane (#1704158, Bio-Rad) using the TGX turbo protocol (Trans blot turbo, #1704150, Bio-Rad). Following Ponceau staining and membrane annotation, the membrane was cut at ∼75 kDa to parallel analysis of the AID::HA-tagged HRDE-1 protein (∼130 kDa, upper half of membrane) and actin protein (∼42 kDa, lower half of membrane). Membranes were blocked with 5% milk in TBST for 30 minutes and incubated overnight (4°C) with primary antibody (anti-β-actin [Sigma-Aldrich, A5441], anti-HA [Biolegend, 901501]). After 3 washes with TBST, membranes were incubated with Horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature (Jackson ImmunoResearch, 715-035-150), and visualized using ECL substrate (Thermo Fisher, 34580) in an Amersham Imager 600 (GE).
Transgenerational premature attractiveness experiments
BFF69 worms were cultivated at 25°C for 10-15 generations, and the last generation of worms grown at 25°C were considered the P0 generation. Once they reached the first day of adulthood, P0 hermaphrodites were transferred to 20°C on auxin-containing NGM plates (or control NGM plates) for 90 minutes, then transferred again to new auxin/control plates for 2-3 hours to lay the synchronized F1 embryos. The 90 minutes pre-incubation was performed to assure that all the F1 progeny derive from HRDE-1-depleted P0 mothers. For the rest of the experiment, all worms continuously grew at 20°C on auxin-containing/control plates, and WCM was extracted from young adults on every generation until the F3. WCM production and extraction was performed as described above and did not involve exposure to auxin. WCM from all conditions tested were collected side by side and appear in Figure S2C. The control BFF68 strain (that does not express the TIR-1 enzyme needed for auxin-dependent degradation of AID-tagged proteins) was exposed to auxin already from the P0 generation (25°C) and in following generations (at 20°C).
Imaging of germ granules in the adult germline
BFF44 hermaphrodites were synchronized by picking young L4s 20h (20°C) or 16h (25°C for ten generations) prior to imaging. Young adults (with only one line of embryos in their uterus) were mounted on a microscope slide with a 3% agarose pad. Animals were imaged at their cultivation temperature thanks to a controlled closed chamber. Images were acquired at x100 magnification using a Nikon Ti-2 eclipse microscope equipped with a 100X CFI Plan-Apo 1.45 NA objective (Nikon, Tokyo, Japan), a CSU-W1 spinning-disk confocal head (Yokogawa Corporation, Tokyo, Japan), a DPSS-Laser (Gataca, France) and a Prime95B sCMOS camera (Photometrics, Tucson, AZ), controlled by the MetaMorph software (Molecular Devices, Sunnyvale, CA). We photographed the germline of young adult worms using ∼80 z stacks per worm with three channels: Bright field, 488nm and 561nm. Image analysis was performed with the Fiji software (Schindelin et al., 2012) using the 3D object counter plugin (Bolte and Cordelières, 2006).
Analysis of RNA-Seq data
We used previously published datasets for meg-3/4(-) mRNAs (Ouyang et al., 2019), meg-3/4(-) germline small RNAs (Lev et al., 2019), and germline small RNAs from F3 wild-type progeny of meg-3/4 (second generation of homozygosity) (Lev et al., 2019). We performed re-analysis of recently published small RNA and mRNA expression data extracted from worms grown at 25°C (Manage et al., 2020), Accession number GSE138220. The quality of the fastq files were assessed with FastQC (Andrews, 2010). Adapters were cut using Cutadapt (Martin, 2011) based on the adapter sequences taken from the original paper. Reads that were not cut or that were either less than 15 bp long (small RNA) or 17 bp (mRNA) long were removed. We also applied a quality threshold of 30, similar to the original publication. We used the following parameters:
For small RNAs: “cutadapt -q 30 -m 15 --discard-untrimmed -a TGGAATTCTCGGGTGCCAAGG”.
For mRNAs: “cutadapt -q 30 -m 17 --nextseq-trim=20 --max-n 2 -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCA”
Next, small RNA reads were mapped to the C. elegans genome (WS235) using Shortstack (Axtell, 2013) allowing no mismatches; mRNA reads were aligned using Bowtie2 (Langmead and Salzberg, 2012). The mapped reads were counted using HTseq_count (Langmead and Salzberg, 2012) using a.gff feature file retrieved from wormbase.org (version WBcel235). The following parameters were used:
For small RNAs: “HTSeq.scripts.count–stranded=reverse–mode=intersection-nonempty input.sam Genes.gff.”
For mRNAs: “HTSeq.scripts.count --mode=intersection-nonempty input.sam Genes.gtf”
Differential expression was analysed using DESeq2 (Love et al., 2014). P-adjusted value <0.1 was regarded as statistically significant.
Poly-U analysis
We examined poly-uridylation of small RNAs by adapting an approach previously described in (Lev et al., 2019). We conducted three additional iterations of alignment of the small RNA libraries to the C. elegans genome (using Shortstack (Axtell, 2013)), requiring a perfect match with no mismatches. In each iteration we trimmed an additional T nucleotide from the 3’ of the small RNA read and re-aligned the small RNA reads. We then processed the alignment.sam files and discarded reads that were already aligned in previous alignment iterations. Next, we used HTseq (Langmead and Salzberg, 2012) to count the antisense aligned reads. Then via a custom code we calculated the fraction of poly-uridylated reads by dividing the number of reads with untemplated 3’ Ts out of the total of aligned reads. For the analysis of the mean proportions of uridylated small RNAs among all small RNAs targeting specific genes, we considered only protein coding genes that had at least 1 RPM of small RNAs targeting them. Next, we set a threshold of < 10% poly-uridylation to define which genes were targeted by non-poly-uridylated small RNAs (as we found this threshold to efficiently distinguish known targets of HRDE-1-bound small RNAs). Finally, we defined a list of genes that were consistently not poly-uridylated across all repeats of a given tested dataset and used it for further gene-set enrichment analysis. For this gene enrichment analysis, the background list of genes was restricted to gene targets of small RNAs that appeared in the tested experiment (at least 1 RPM), to accommodate the bias in focusing on genes that had poly-U data.
Gene enrichment analysis
To test for enrichment for different gene list we used the “enrichment” function of the RNAlysis package (https://github.com/GuyTeichman/RNAlysis). The calculated fold enrichment values denote the ratio between the observed representation of a specific gene set to the expected one based on all genes (in this study we focused on all protein coding genes). Adjusted P values were obtained using a randomization test (10,000 random genesets), with the formula P = (successes + 1)/(repeats + 1). The P values were corrected for multiple comparisons using the Benjamini–Hochberg step-up procedure. For more information see RNAlysis documentation. We performed the analysis on 3 gene sets: (A) 752 germline-enriched genes (Serizay et al., 2020), (B) 672 soma-enriched genes (Serizay et al., 2020), (C) 6936 genes expressed in the spermatogenic germline (Ortiz et al., 2014). For this list we also applied a threshold of at least 10 RPKM.
Hierarchical clustering of siRNA expression
Hierarchical clustering was done using the clustermap function of the Seaborn (Waskom et al., 2020) python package. Log2 fold change values in siRNA expression were taken from the DESeq2 output files. Default parameters were used: the distance metric was euclidic and the linkage method was average.
Quantification of brood size experiments
Wild-type and meg-3/4 L3 larvae were transferred individually to seeded plates (Day 0). After 24h (Day 1), each adult hermaphrodite was transferred to a new seeded plate (50μl drop of OP50) containing 4 wild-type males, which were removed after 2 hours of interaction to avoid detrimental effects on hermaphrodite fitness (Booth et al., 2019; Shi et al., 2019). The hermaphrodites were transferred to new plates every 24h for 8 days. For each plate the progeny were quantified 3 days after transfer of the hermaphrodite. Hermaphrodites with numerous males in their progeny were considered mated, and non-mated hermaphrodites were used as (self-reproducing) controls. Results in Figure 4D were collected in three independent experiments and included 2-16 hermaphrodites/group/replicate. For the data collected for the mathematical model (Figure S6B), similar experiments were performed using wild-type hermaphrodites and BFF53 males bearing a myo-2p::gfp integrated single-copy transgene. Quantified progeny were scored for their sex and for their genetic background (selfing/cross progeny) based on gfp expression using a fluorescent dissection microscope. Data from hermaphrodites that died before Day 6 of adulthood were excluded from the analysis. The experiment was conducted independently twice (R1 nmated = 15, R1 nunmated = 24, R2 nmated = 11, R2 nunmated = 23). For the experiment depicted in Figure S6A, experiments were run similarly to the above but did not involve any interaction with males. The experiment was conducted independently three times, and included 6-13 hermaphrodites/group/replicate.
Mathematical model
We constructed a theoretical population genetics model to examine the range of conditions in which premature attraction would be favored by natural selection. Code available at: https://doi.org/10.5281/zenodo.5651221.
Model outline
For simplicity, we analysed a discrete-time model and assumed very large populations (so that demographic stochasticity and random genetic drift can be neglected) at carrying capacity, nonoverlapping generations, and single locus haploid inheritance. We performed an invasion analysis, exploring the success of a rare allele for premature attraction in a population of worms that do not display premature attraction. We derived the conditions for increase from rarity of said allele.
Model parameters
Full descriptions of the abbreviated parameters used in the formulas of the mathematical model appear in Table S4.
Mating function
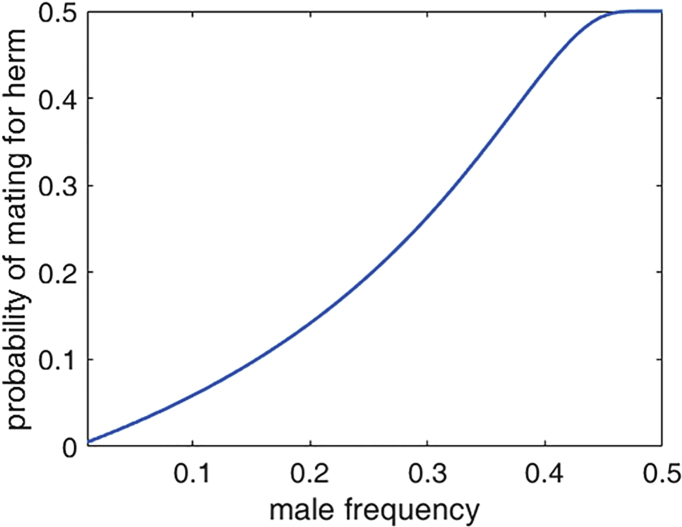
We define , level of attraction, measured in arbitrary units between 0 (= no attraction) and 1 (=max possible attraction), corresponding to chemotaxis values. We adapted a function from (Kuno, 1978), to describe the probability of mating for an hermaphrodite as a function of the frequency of males in the population and the level of attraction the hermaphrodites exude .
Shape of the mating function, parameters:
In the results presented in the manuscript, the values tested were where , as we assumed in the invasion analysis that a rare variant will initially arise with a low level of attraction. For the rest of the population, where , as we assumed the general population does not exhibit premature attraction.
Offspring types
We account for the possible offspring types that may exhibit the trait of premature attraction. We assume that males can carry and transmit the premature attraction trait to their offspring, but it does not affect their own phenotype.
Parameters and probabilities representing offspring types appear in Table S5.
We adapted the different brood size numbers from the empirical data we gathered (Figure S6B), as well as the relative fitness for every type of offspring. When there is no stress, we assume for simplicity that all types of offspring have the same fitness (thus, ). When there is stress, we assume that offspring born from outcrossing have an advantage over offspring born from selfing, and that males have an advantage over hermaphrodites.
Mean offspring fitness
Population mean mating probability:
Total number of individuals in offspring generation ( = number of hermaphrodites in the current generation):
Mean offspring fitness without stress:
Mean offspring fitness under stress:
Mean fitness of individuals exhibiting premature attraction, in the following generation (without stress):
Mean fitness of individuals exhibiting premature attraction, in the following generation (under stress):
Evolution of premature attraction
We derive the frequency of individuals with premature attraction in the next generation, , given that in the current generation it is given by . We separate the derivations into two scenarios, with stress and without stress.
Without stress: Under stress:
Where is the cost of premature attraction, is the level of premature attraction and is the likelihood of future stress.
We are interested in the case of a rare variant of type in a population of individuals of type . We want to find the condition for its increase from rarity, meaning . To do this, we find , the critical for which . is the maximal value for which , presented in Figure 5A.
Male incidence in selfing-derived progeny
Early L4 worms were transferred individually into 42-54 NGM wells (in standard 6-wells plates) seeded with 1-2 drops of OP50. 4 days later (or 3.5 for worms grown at 25°C), when the progeny reached adulthood but food was still visible, the wells were examined for the presence of males using a dissection microscope. Each well was scanned for exactly 75 seconds and scored as positive if at least one male was detected during this time.
For experiments testing meg-3/4 mutants (BFF40) and control wild types (SX1263), L4s were picked for synchronization, and young adults were individually transferred into the wells one day later to start the incubation period. This allowed us to include only fertile meg-3/4 adults in the experiment.
Multigeneration competition experiment
The “attractive” wild-type worms were myo-2p::gfp;meg-3/4(+/+) F3 progeny of myo-2p::gfp;meg-3/4(-/-) hermaphrodites outcrossed with myo-2p::gfp males. Seven “attractive” hermaphrodites (GFP+) and seven naïve wild-type hermaphrodites (GFP-negative) interacted 1h with eleven wild-type males (GFP-negative) on a mating plate seeded with a dried 50μl drop of OP50. All worms were synchronized by picking L4s into a separate plate 16h prior to the 1h interaction. Side-by-side with the wild-type experiment, the same procedure was performed in a srd-1(eh1) background in which all worms involved (including for the ancestral crosses) carried the mutation. Both groups were tested in biological triplicates (a total of 6 groups). After 1h of interaction, all fourteen hermaphrodites (but not the males) were transferred together to fresh seeded plates (one per replicate), and were considered the P0 generation. Step A: After 40h, the worms were rinsed out of the plates and allowed to settle for 2 minutes in a 1.7ml tube. Hundreds of F1 larvae were transferred into a new tube by pipetting the supernatant (by this time, the P0 adults reached the bottom of the tube). For each group, F1 larvae were transferred into seeded plates (∼300-400 larvae per plate, in technical triplicates, 18 plates in total). Step B: After 56h, all F1s (now young adults) were collected, washed twice in M9, and transferred to fresh seeded plates in technical triplicates. This step permitted more interaction time for the adults without consuming all the food. At this occasion, 25 young males (GFP-negative, either wt or srd-1 according to the group) were artificially added to each plate, representing 6%-8% of the cultured population. Step C: After 40h of incubation, all worms were rinsed out of plate using M9, F1 worms were kept for imaging (see below) and F2 larvae were separated like in Step A to form the next generation. Steps A-C repeated until the imaging of the F6 generation.
Imaging
Collected adult worms (Step C of the above) were washed 8-10 times in M9 to eliminate the remaining food and larvae, with special care not to wash out the males that precipitate more slowly. After the last round of washes, worms were left in ∼100μl and paralyzed in Sodium Azide (25-50mM final concentration). Worms were then transferred to imaging plates (60mm diameter, 8ml, 2% agarose, 0.3% NaCl, 5 mM KPO4, 1 mM CaCl2, 1 mM MgSO4) and physically separated from each other using a platinum-wired pick. Separation of the worms is important for allowing the Wormachine software to correctly process each worm individually (Hakim et al., 2018). Images were acquired with x4 magnification using a TiE Nikon microscope equipped with a Andor Neo sCMOS camera (Bio-Imaging Resource Center, Rockefeller University). High throughput analysis of images was performed using the Wormachine software (Hakim et al., 2018). Animals were binarily scored for GFP expression and for sex.
Computer simulations of competition experiments
We developed a computer simulation to reproduce the results of the multigenerational competition experiment under various settings (using Python version 3.7.9). The simulation receives as input the initial (G0) number of worms of each type: 7 gfp(+/+) hermaphrodites (attractive lineage), 7 gfp(-/-) hermaphrodites (naïve), 11 gfp(-/-) males. In every generation, the offspring distribution is calculated according to the following parameters: mating rates for hermaphrodites of the attractive lineage [gfp(+/+) or gfp(+/-)] or naïve lineage [gfp(-/-)], relative fitness (brood size) of attractive worms [i.e. gfp(+/+) or gfp(+/-)], brood sizes and the distribution of selfing-derived and cross-derived offspring. The amount of offspring for each genetic background combination is calculated. Then, 300 worms are sampled at random to form the next generation, and so forth.
Simulation parameters (derived from empirical results):
-
•
Brood size (of cross-derived offspring only) for mated hermaphrodites: 450.
-
•
Brood size (of selfing-derived offspring only) for mated hermaphrodites: 100.
-
•
Brood size for unmated hermaphrodites: 330.
-
•
Frequency of spontaneous males: 0.001.
We used manual parameter optimization to fit the unknown variables: mating rates and fitness of the green worms, such that the resulting dynamics aligns with the experimental data most accurately. Additionally, we tested different scenarios relaxing various underlying assumptions of the experiment. Full code available at: https://doi.org/10.5281/zenodo.5651221.
Tested scenarios (see Figure S6C):
-
1.
Full model predicting the real data
-
2.
Fitness of the attractive lineage (gfp+) remains constant throughout the generations.
-
3.
Attractive worms have the same mating rates in srd-1 and wild-type experiments.
-
4.
Attractive and naïve worms have the same mating rates.
Statistical analyses
Statistical analyses and generation of graphs were performed using Prism, R v4.0.0 or MATLAB R2019b. Statistical details of experiments appear in figure legends. For the chemotaxis experiments, we applied the d’Agostino-Pearson omnibus test (α=0.05) to assess the normality of the values’ distribution in each tested group. In cases where assumption of normality was rejected or the test could not apply (n<8), we used the a-parametric Kruskal-Wallis test with Dunn’s correction for multiple comparisons to control. Otherwise, we used one-way ANOVA with Dunnett’s correction for multiple comparisons to control. For transgenerational experiments involving only two conditions, we used two-way ANOVA with Sidak’s correction for multiple comparisons. For the transgenerational HRDE-1-depletion experiment (involving five conditions), we used two-way ANOVA with Tukey post-hoc correction for multiple comparisons. For the mating choice assays, we used the Wilcoxon matched-pairs signed rank test (two-tailed). For brood size and mating rescue experiments, and because the assumption of equal variance was rejected (Brown-Forsythe test with p < 0.05), we used Welch’s ANOVA with Games-Howell post-hoc correction for multiple comparisons. For spontaneous male incidence experiments, we used Fisher’s exact test. For the multigenerational competition test, we fit a generalized linear mixed model (GLMM) with generation, genetic background (wt vs. srd-1) and their interaction as fixed effects, using the lme4 v1.1-21 package in R. Replicate populations were included as a random effect.
Acknowledgments
We thank the members of the Rechavi Lab, the Shaham Lab, and the Hadany Lab for helpful discussions. We would like to thank Daniel Leighton, Paul Sternberg, Gillian Stanfield, and Ehud Cohen for their experimental advice and Annie Mais for her help with data analysis. We thank Anat Nitzan, Ronen Zaidel-Bar, and the Rockefeller University Bio-Imaging Resource Center for their expertise and assistance with imaging experiments. We thank Guy Teichman for his assistance with coding and bioinformatic analysis. Special thanks to Ofir Cahana for contributing to the illustration of the graphical abstract. Some strains were provided by the Caenorhabditis Genetics Center which is funded by the N.I.H. Office of Research Infrastructure Programs (P40 OD010440), and we thank Eric Miska for providing the SX1263 strain. This research was supported by the NIH grant R35NS105094 (S.S.), the ISF 2064/18 (L.H.), the Minerva Stiftung grant #713740 (O.R. and L.H.), the Adelis Foundation grant #0604916191 (O.R.), the DFG grant SCHU 2494/10-1 (O.R.), and the ERC grants #335624 and #819151 (O.R.). O.R. is grateful to funding from the Eric and Wendy Schmidt Fund for Strategic Innovation (Polymath Award #0140001000).
Author contributions
I.A.T., I.L., Y.M., and O.R. conceived the project and designed the experiments. I.A.T., I.L., and Y.M. performed the experiments and analyzed the data with the assistance of Y.G., D.F., and H.G. Y.G. and L.H. designed and implemented the mathematical model and computer simulations. L.H.-Z. and O.A. generated the aid::ha::hrde-1 strains. H.D. and S.A. helped with the validation of the aid::ha::hrde-1 conditional degradation. S.S. provided guidance and resources. I.A.T., I.L., Y.M., and O.R. wrote the manuscript with input from all the authors.
Declaration of interests
The authors declare no competing interests.
Published: February 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2022.01.005.
Contributor Information
Itai Antoine Toker, Email: itaitoker@gmail.com.
Itamar Lev, Email: itamai.et@gmail.com.
Yael Mor, Email: yael.moryosef@gmail.com.
Oded Rechavi, Email: odedrechavi@gmail.com.
Supplemental information
The analyses for mRNA changes (Ouyang et al., 2019) and small RNA changes (Lev et al., 2019) in meg-3/4 mutants as well as small RNA changes in the wild-type F3 progeny (two generations of homozygosity) of meg-3/4 mutants (Lev et al., 2019) are shown. Missing adjusted p values are “NaN” values generated according to the DESeq2 algorithm
GFP-positive animals are myo-2p::gfp;meg-3/4(+,+) descendants of myo-2p::gfp;meg-3/4(-/-) hermaphrodites outcrossed with myo-2p::gfp males and represent the “attractive lineage”. The same experiment was performed side by side in srd-1(+,+) (“wt”) and srd-1(-/-) backgrounds.
Data and code availability
-
•
This paper analyses existing, publicly available data. Accession numbers for the datasets are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online] Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Aprison E.Z., Ruvinsky I. Sex pheromones of C. elegans males prime the female reproductive system and ameliorate the effects of heat stress. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argon Y., Ward S. Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics. 1980;96:413–433. doi: 10.1093/genetics/96.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C., Doebley A.L., Goldstein L.D., Lehrbach N.J., Le Pen J., et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J., Ruby J.G., Claycomb J.M., Chiang R., Fahlgren N., Kasschau K.D., Chaves D.A., Gu W., Vasale J.J., Duan S., et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshore E.L., McEwen T.J., Jud M.C., Marshall J.K., Schisa J.A., Bennett K.L. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev. Biol. 2011;350:370–381. doi: 10.1016/j.ydbio.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Booth L.N., Maures T.J., Yeo R.W., Tantilert C., Brunet A. Self-sperm induce resistance to the detrimental effects of sexual encounters with males in hermaphroditic nematodes. Elife. 2019;8 doi: 10.7554/eLife.46418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.C., Svendsen J.M., Tucci R.M., Montgomery B.E., Montgomery T.A. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 2017;45:9093–9107. doi: 10.1093/nar/gkx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H., Kimble J., Fire A., Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Yan W., Duan E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 2016;17:733–743. doi: 10.1038/nrg.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D.L., Lambie E.J. The promotion of gonadal cell divisions by the Caenorhabditis elegans TRPM cation channel GON-2 is antagonized by GEM-4 copine. Genetics. 2003;165:563–574. doi: 10.1093/genetics/165.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C.C., Batista P.J., Gu W., Claycomb J.M., Chaves D.A., Shirayama M., Mello C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2010;107:3588–3593. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz C.E., Tomsig J.L., Snyder S.L., Gautier M.C., Skouri F., Beisson J., Cohen J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 1998;273:1393–1402. doi: 10.1074/jbc.273.3.1393. [DOI] [PubMed] [Google Scholar]
- de Albuquerque B.F.M., Placentino M., Ketting R.F. Maternal piRNAs are essential for germline development following de novo establishment of endo-siRNAs in Caenorhabditis elegans. Dev. Cell. 2015;34:448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Dodson A.E., Kennedy S. Germ granules coordinate RNA-based epigenetic inheritance pathways. Dev. Cell. 2019;50:704–715.e4. doi: 10.1016/j.devcel.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin G.A., Ghanta K.S., Piscopo K.M., Mello C.C. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 2018;210:781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewe C.K., Torres Cleuren Y.N., Flowers S.E., Alok G., Snell R.G., Rothman J.H. Natural cryptic variation in epigenetic modulation of an embryonic gene regulatory network. Proc. Natl. Acad. Sci. USA. 2020;117:13637–13646. doi: 10.1073/pnas.1920343117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan K.A., Luo J., Lagoy R.C., Schroeder F.C., Albrecht D.R., Portman D.S. A single-neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Curr. Biol. 2018;28:902–914.e5. doi: 10.1016/j.cub.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann A.W., Putnam A., Lee C.F., Seydoux G. Regulation of biomolecular condensates by interfacial protein clusters. Science. 2021;373:1218–1224. doi: 10.1126/science.abg7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frézal L., Demoinet E., Braendle C., Miska E., Félix M.A. Natural genetic variation in a multigenerational phenotype in C. elegans. Curr. Biol. 2018;28:2588–2596.e8. doi: 10.1016/j.cub.2018.05.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo R.S., Hastings P.J., Rosenberg S.M. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J., et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadany L., Otto S.P. Condition-dependent sex and the rate of adaptation. Am. Nat. 2009;174(Supplement 1) doi: 10.1086/599086. S71–S78. [DOI] [PubMed] [Google Scholar]
- Hakim A., Mor Y., Toker I.A., Levine A., Neuhof M., Markovitz Y., Rechavi O. WorMachine: machine learning-based phenotypic analysis tool for worms. BMC Biol. 2018;16:8. doi: 10.1186/s12915-017-0477-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houri-Ze’evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y., Awad L., Degani L., Alon U., Rechavi O. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Houri-Zeevi L., Korem Kohanim Y., Antonova O., Rechavi O. Three rules explain transgenerational small RNA inheritance in C. elegans. Cell. 2020;182:1186–1197.e12. doi: 10.1016/j.cell.2020.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E., Lamb M.J. Soft inheritance: challenging the modern synthesis. Genet. Mol. Biol. 2008;31:389–395. [Google Scholar]
- Kaletsky R., Moore R.S., Vrla G.D., Parsons L.R., Gitai Z., Murphy C.T. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature. 2020;586:445–451. doi: 10.1038/s41586-020-2699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga L.M., van Wolfswinkel J.C., Luteijn M.J., Kaaij L.J., Bagijn M.P., Sapetschnig A., Miska E.A., Berezikov E., Ketting R.F. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann G.A., Basolo A.L. Facultative decrease in mating resistance in hermaphroditic Caenorhabditis elegans with self-sperm depletion. Anim. Behav. 2007;74:1339–1347. [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- Kubagawa H.M., Watts J.L., Corrigan C., Edmonds J.W., Sztul E., Browse J., Miller M.A. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat. Cell Biol. 2006;8:1143–1148. doi: 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- Kuno E. Simple mathematical models to describe the rate of mating in insect populations. Res. Popul. Ecol. 1978;20:50–60. [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton D.H., Choe A., Wu S.Y., Sternberg P.W. Communication between oocytes and somatic cells regulates volatile pheromone production in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2014;111:17905–17910. doi: 10.1073/pnas.1420439111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev I., Seroussi U., Gingold H., Bril R., Anava S., Rechavi O. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- Lev I., Toker I.A., Mor Y., Nitzan A., Weintraub G., Antonova O., Bhonkar O., Ben Shushan I., Seroussi U., Claycomb J.M., et al. Germ granules govern small RNA inheritance. Curr. Biol. 2019;29:2880–2891.e4. doi: 10.1016/j.cub.2019.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind M.I., Zwoinska M.K., Andersson J., Carlsson H., Krieg T., Larva T., Maklakov A.A. Environmental variation mediates the evolution of anticipatory parental effects. Evol. Lett. 2020;4:371–381. doi: 10.1002/evl3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn M.J., Van Bergeijk P., Kaaij L.J., Almeida M.V., Roovers E.F., Berezikov E., Ketting R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–3430. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manage K.I., Rogers A.K., Wallis D.C., Uebel C.J., Anderson D.C., Nguyen D.A.H., Arca K., Brown K.C., Cordeiro Rodrigues R.J.C., de Albuquerque B.F., et al. A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. Elife. 2020;9 doi: 10.7554/eLife.56731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- Maures T.J., Booth L.N., Benayoun B.A., Izrayelit Y., Schroeder F.C., Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J.A., Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.S., Kaletsky R., Murphy C.T. Piwi/PRG-1 Argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell. 2019;177:1827–1841.e12. doi: 10.1016/j.cell.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L.T., Parmenter M.D., Phillips P.C. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009;462:350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L.T., Schmidt O.G., Gelarden I.A., Parrish R.C., Lively C.M., Lively C.M. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsci N.S., Haas L.A., Barr M.M. Sperm status regulates sexual attraction in Caenorhabditis elegans. Genetics. 2011;189:1341–1346. doi: 10.1534/genetics.111.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Yaoi T., Yasui M., Kuwajima G. N-copine: a novel two C2-domain-containing protein with neuronal activity-regulated expression. FEBS Lett. 1998;428:80–84. doi: 10.1016/s0014-5793(98)00497-9. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Kalinava N., Chen E., Huang A., Trinh T., Gu S.G. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016;9:3. doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Fukagawa T., Takisawa H., Kakimoto T., Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Ortiz M.A., Noble D., Sorokin E.P., Kimble J. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 2014;4:1765–1772. doi: 10.1534/g3.114.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P. The evolutionary enigma of sex. Am. Nat. 2009;174(Supplement 1):S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- Otto S.P., Day T. Princeton University Press; 2011. A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution. [Google Scholar]
- Ouyang J.P.T., Folkmann A., Bernard L., Lee C.Y., Seroussi U., Charlesworth A.G., Claycomb J.M., Seydoux G. P granules protect RNA interference genes from silencing by piRNAs. Dev. Cell. 2019;50:716–728.e6. doi: 10.1016/j.devcel.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish R.C., Penley M.J., Morran L.T. The integral role of genetic variation in the evolution of outcrossing in the Caenorhabditis elegans-Serratia marcescens host-parasite system. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelec D.M., Lachowiec J., Duchaine T.F., Smith H.E., Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R., Pagano D., Wan G., Fields B.D., Saltzman A.L., Kennedy S.G. Transgenerational epigenetic inheritance is negatively regulated by the HERI-1 chromodomain protein. Genetics. 2018;210:1287–1299. doi: 10.1534/genetics.118.301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C.M.M., Brown K.C.C., Montgomery B.E.E., Ruvkun G., Montgomery T.A.A. piRNAs and piRNA-dependent siRNAs protect conserved and essential C. elegans genes from misrouting into the RNAi pathway. Dev. Cell. 2015;34:457–465. doi: 10.1016/j.devcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner R., Toker I.A., Antonova O., Star E., Anava S., Azmon E., Hendricks M., Bracha S., Gingold H., Rechavi O. Neuronal small RNAs control behavior transgenerationally. Cell. 2019;177:1814–1826.e15. doi: 10.1016/j.cell.2019.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam A., Cassani M., Smith J., Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019;26:220–226. doi: 10.1038/s41594-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram Y., Hadany L. Stress-induced mutagenesis and complex adaptation. Proc. Biol. Sci. 2014;281:20141025. doi: 10.1098/rspb.2014.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram Y., Hadany L. Condition-dependent sex: who does it, when and why? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150539. doi: 10.1098/rstb.2015.0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze’evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 2017;27:R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- Rechavi O., Minevich G., Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Sarkies P., Simon M., Doebley A.L., Goldstein L.D., Hedges A., Ikegami K., Alvares S.M., Yang L., LaRocque J.R., et al. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA. 2014;111:E4323–E4331. doi: 10.1073/pnas.1406131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A., Sarkies P., Lehrbach N.J., Miska E.A. Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D., Yanai I., Hunter C.P. Natural RNA interference directs a heritable response to the environment. Sci. Rep. 2014;4:7387. doi: 10.1038/srep07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizay J., Dong Y., Jänes J., Chesney M., Cerrato C., Ahringer J. Distinctive regulatory architectures of germline-active and somatic genes in C. elegans. Genome Res. 2020;30:1752–1765. doi: 10.1101/gr.265934.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Booth L.N., Murphy C.T. Insulin-like peptides and the mTOR-TFEB pathway protect Caenorhabditis elegans hermaphrodites from mating-induced death. Elife. 2019;8 doi: 10.7554/eLife.46413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Murphy C.T. Mating induces shrinking and death in Caenorhabditis mothers. Science. 2014;343:536–540. doi: 10.1126/science.1242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Runnels A.M., Murphy C.T. Mating and male pheromone kill Caenorhabditis males through distinct mechanisms. Elife. 2017;6 doi: 10.7554/eLife.23493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H.C., Gu W., Ishidate T., Conte D., Mello C.C. PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Yan J., Pagano D.J., Dodson A.E., Fei Y., Gorham J., Seidman J.G., Wickens M., Kennedy S. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature. 2020;582:283–288. doi: 10.1038/s41586-020-2323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A.L., Goldstein L.D., Mitchell J., Sakaguchi A., Miska E.A., Ahmed S. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 2014;7:762–773. doi: 10.1016/j.celrep.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Spracklin G., Fields B., Wan G., Becker D., Wallig A., Shukla A., Kennedy S. The RNAi inheritance machinery of Caenorhabditis elegans. Genetics. 2017;206:1403–1416. doi: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Kaplan F., Ajredini R., Zachariah C., Alborn H.T., Teal P.E.A.A., Malik R.U., Edison A.S., Sternberg P.W., Schroeder F.C. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susoy V., Hung W., Witvliet D., Whitener J.E., Wu M., Park C.F., Graham B.J., Zhen M., Venkatachalam V., Samuel A.D.T. Natural sensory context drives diverse brain-wide activity during C. elegans mating. Cell. 2021;184:5122–5137.e17. doi: 10.1016/j.cell.2021.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Wan X., Zhou Y., Chan C.M., Yang H., Yeung C., Chow K.L. SRD-1 in AWA neurons is the receptor for female volatile sex pheromones in C. elegans males. EMBO Rep. 2019;20 doi: 10.15252/embr.201846288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3 doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom, M., Gelbart, M., Botvinnik, O., Ostblom, J., Hobson, P., Lukauskas, S., Gemperline, D.C., Augspurger, T., Halchenko, Y., Warmenhoven, J., et al. (2020). mwaskom/seaborn: v0.11.1 (December 2020).
- Webster A.K., Jordan J.M., Hibshman J.D., Chitrakar R., Baugh L.R. Transgenerational effects of extended dauer diapause on starvation survival and gene expression plasticity in Caenorhabditis elegans. Genetics. 2018;210:263–274. doi: 10.1534/genetics.118.301250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Feng X., Chen X., Weng C., Yan Q., Xu T., Hong M., Guang S. A cytoplasmic Argonaute protein promotes the inheritance of RNAi. Cell Rep. 2018;23:2482–2494. doi: 10.1016/j.celrep.2018.04.072. [DOI] [PubMed] [Google Scholar]
- Zhang C., Montgomery T.A., Gabel H.W., Fischer S.E.J., Phillips C.M., Fahlgren N., Sullivan C.M., Carrington J.C., Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:1201–1208. doi: 10.1073/pnas.1018695108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ward J.D., Cheng Z., Dernburg A.F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The analyses for mRNA changes (Ouyang et al., 2019) and small RNA changes (Lev et al., 2019) in meg-3/4 mutants as well as small RNA changes in the wild-type F3 progeny (two generations of homozygosity) of meg-3/4 mutants (Lev et al., 2019) are shown. Missing adjusted p values are “NaN” values generated according to the DESeq2 algorithm
GFP-positive animals are myo-2p::gfp;meg-3/4(+,+) descendants of myo-2p::gfp;meg-3/4(-/-) hermaphrodites outcrossed with myo-2p::gfp males and represent the “attractive lineage”. The same experiment was performed side by side in srd-1(+,+) (“wt”) and srd-1(-/-) backgrounds.
Data Availability Statement
-
•
This paper analyses existing, publicly available data. Accession numbers for the datasets are listed in the key resources table. Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.