Abstract
Genetic diversity among Borrelia burgdorferi isolates recovered from the skin of Lyme disease patients was assessed by ribosomal DNA (rDNA) spacer restriction fragment length polymorphism analysis, genomic restriction site polymorphism analysis, and plasmid content analysis. There was a significant association between the three rDNA spacer types, the six pulsed-field gel types, and plasmid content (P < 0.001). The association between distinct chromosomal and plasmid markers implies a clonal origin for each genotype.
Lyme disease, a disorder which may affect multiple organ systems, results from infection with the spirochete Borrelia burgdorferi. Between 1989 and 1997, 114,314 cases of Lyme disease were reported to the Centers for Disease Control and Prevention, making Lyme disease the most common vector-borne disease in the United States (16, 21). Early Lyme disease is manifested by a characteristic skin rash, erythema migrans (EM), and is frequently accompanied by systemic symptoms (14, 15). Molecular analysis of B. burgdorferi isolates has resulted in the differentiation of B. burgdorferi sensu lato into 10 distinct species (23). B. burgdorferi sensu stricto is the only known Lyme disease borrelia infecting patients in North America. Significant genetic heterogeneity among B. burgdorferi sensu stricto isolates in North America has been reported (1, 6, 8, 13). The type strain of B. burgdorferi sensu stricto, B31 has a large linear genome of 910,725 bp and 21 linear and circular plasmids with a total size of 610,694 bp (3, 7).
It has long been recognized that the clinical expression of B. burgdorferi infection is diverse (15). Possible explanations for this variability are differences in the genotypes of the infecting strain of B. burgdorferi. We developed a typing method based on restriction fragment length polymorphism (RFLP) in the 16S-23S ribosomal DNA (rDNA) spacer of B. burgdorferi and used it to analyze clinical isolates obtained from early Lyme disease patients (9). A highly significant association was found between the infecting RFLP type in the skin and the presence of spirochetemia or multiple EM lesions (25), suggesting that differences in the clinical presentations of Lyme disease patients may, indeed, be related to B. burgdorferi genotype. rDNA spacer analysis provides genetic typing information for a single genomic locus and interrogates a chromosomal region which is unlikely to have a direct role in pathogenesis. In order to obtain additional, broader typing information, 48 cutaneous isolates from early Lyme disease patients were analyzed by pulsed-field gel electrophoresis (PFGE).
Subjects, skin biopsy, and cultivation.
All subjects were adults with EM enrolled in a prospective study at the Lyme Disease Diagnostic Center of the Westchester Medical Center, Valhalla, N.Y. Skin biopsy specimens (2 mm) were obtained from the leading edge of primary EM lesions and cultured in BSK-II medium as described previously (19).
PFGE.
Spirochetes were harvested by centrifugation and washed twice with sterile phosphate-buffered saline. The cells were resuspended in 50 mM Tris-HCl–150 mM NaCl (pH 8.0), and an equal volume of 1.8% low-gelling-temperature agarose (SeaPlaque; FMC Corp., Rockland, Maine) was added. The cells, in agarose blocks, were lysed as described previously (5). For plasmid analysis, blocks were washed extensively with 10 mM Tris-HCl–1 mM EDTA (pH 8.0) and PFGE was performed in 0.5× Tris-borate-EDTA at 14°C on a Bio-Rad CHEF DR II electrophoresis system. One percent agarose gels were run at a constant voltage (6 V/cm) for 19 h, with a switch time of 0.7 to 2.2 s. For chromosome restriction fragment analysis, agarose blocks containing B. burgdorferi were incubated overnight at 37°C with 40 to 50 U of MluI in 0.5 ml of 50 mM Tris-HCl (pH 7.5)–10 mM MgCl2–100 mM NaCl–1 mM dithioerythritol. PFGE was performed as described above, except that a switch time of 6 to 25 s was used.
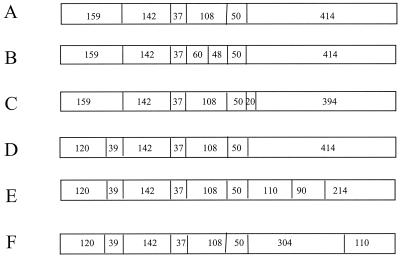
The relative proportions of the three rDNA spacer RFLP types (RSTs) among the 48 isolates (29.2, 41.7, and 29.2% for RST1, RST2, and RST3, respectively) were similar to those for the larger group of cutaneous isolates reported previously (25.1, 38.3, and 30.1% for RST1, RST2, and RST3, respectively) (10). Total genomic DNA was digested with MluI, and the resultant fragments were resolved by PFGE. The results, summarized schematically in Fig. 1, revealed six distinct pulsed-field gel (PFG) patterns. Fifty-two percent (25 of 48) displayed a pattern designated type A. An additional 16.7% (8 of 48) had the pattern designated type B, and 12.5% (6 of 48) showed a type D pattern; the remaining isolates were equally distributed between types C, E, and F. These patterns were all observed in a previous study of representative B. burgdorferi isolates by Mathiesen et al. (13) and are designated here types A to F, after their nomenclature. Of the 65 human isolates characterized in the earlier study, 54 (83%) were PFG type A, which was also the predominant PFG type (52%) among the isolates in the present investigation. Interestingly, PFG type E was observed only in ticks by Mathiesen et al. (13), whereas three PFG type E isolates were detected among the human isolates in the present analysis. Thus, all major North American B. burgdorferi sensu stricto PFG types are found in a narrowly defined geographic location (Westchester County, N.Y.) and are infectious to humans.
FIG. 1.
Schematic representation of six MluI PFG types among B. burgdorferi clinical isolates. The approximate sizes of each fragment (in kilobases) are noted.
The plasmid contents in the same isolates were also analyzed by PFGE. Since repeated culture passage is known to result in selective plasmid loss (2, 18, 20), isolates in the present study were analyzed between passages 2 and 5. Most isolates tested carried all of the plasmids previously identified in strain B31. The most distinctive difference among the isolates discernible by PFGE was the presence or absence of lp38, which was confirmed by Southern blotting with an ospD-specific probe (ospD is encoded on lp38) and by ospD-specific PCR (data not shown). Of the isolates analyzed, 21 (44%) carried a copy of lp38. Palmer et al. (17) recently reported the distribution of 12 linear plasmids in 15 B. burgdorferi sensu stricto isolates by plasmid-specific probe hybridization. These isolates were from a broad geographic distribution; the specimen sources (human, tick, etc.) were not given. They found that lp5, lp21, and lp56 were found in no more than 3 of these isolates, lp17 and lp38 were present in 10 of 15 of the isolates, and the remainder were found in at least 14 of the isolates. The results of the present study are mostly consistent with and extend those of earlier study. A difference between the two studies is the proportion of isolates which carry lp38 (67% [10 of 15] in the study of Palmer et al. [17] and 44% [21 of 27] in the present study [P = 0.12]). Since all the isolates in the present study were obtained from human EM lesions, this indicates that lp38 and its gene products are not required for infectivity.
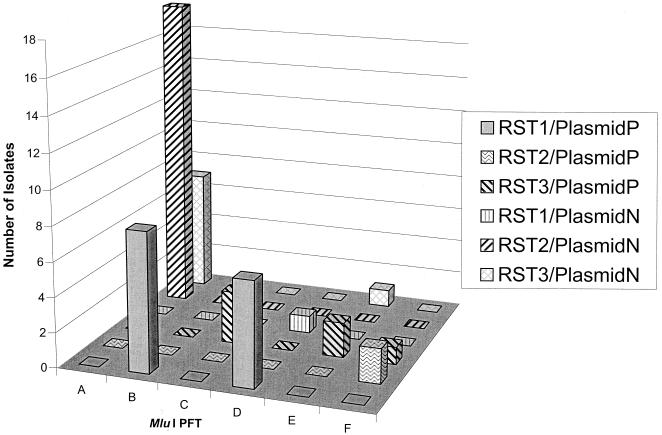
It was of interest to ascertain whether any correlation exists between the three typing methods applied to this group of isolates. For the purposes of the analysis, the plasmid type (P or N) was based on the presence or absence of lp38. Each technique scores for a specific and seemingly unrelated genotypic character: sequence heterogeneity in a ribosomal spacer, point polymorphisms in restriction sites distributed across the linear chromosome, and the presence or absence of a single linear plasmid. If these genotypic characters were independent, one would expect that all possible combinations would be observed and would occur at equal frequencies among the isolates tested. The data presented in Table 1 and Fig. 2 demonstrate that these characteristics cosegregate with each other at a highly significant rate (P < 0.001). The genotype of any individual isolate could be designated by a combination of the three separate RSTs, PFG types, and plasmid types. For example, an isolate which is RST1, is PFG type B, and contains lp38 would have the designation 1BP. Only 10 of the 36 possible genotypes were observed among the 48 isolates tested; 79% (38 of 48) were one of the four predominant genotypes (1BP, 1DP, 2AN, and 3AN) (Fig. 2).
TABLE 1.
RST, MluI PFG type, and plasmid type for 48 B. burgdorferi clinical isolates
| RST | PFG type (no. [%] of isolates)
|
Plasmid type (no. [%] of isolates)
|
||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | P | N | |
| RST1 | 0 (0) | 8 (57) | 0 (0) | 6 (43) | 0 (0) | 0 (0) | 13 (93) | 1 (7) |
| RST2 | 18 (90) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (10) | 2 (10) | 18 (90) |
| RST3 | 7 (50) | 0 (0) | 3 (21) | 0 (0) | 3 (21) | 1 (7) | 6 (43) | 8 (57) |
FIG. 2.
Association of RST, PFG type, and plasmid type. RST and plasmid types are represented as one of six possible combinations and plotted versus PFG type.
Dykhuizen et al. (4) demonstrated that there is little or no genetic exchange between chromosomal genes in B. burgdorferi and that there is little evidence for plasmid transfer between isolates; on this basis, they concluded that B. burgdorferi is clonal. In a separate study from that group, linkage between ospA and ospC genotypes was observed, even though these genes are encoded on separate plasmids (24). The data presented here are in agreement with those from the earlier studies and support the clonality of B. burgdorferi. Several groups have reported evidence for lateral exchange of plasmid-encoded genes in B. burgdorferi (11, 12, 22). The mechanisms for such gene transfer are not known, but the present data suggest that it does not occur via exchange of entire plasmids between isolates.
We previously reported an association between rDNA spacer RFLP and hematogenous dissemination of B. burgdorferi from skin (25). A limitation of that study was that sequence variation in a rDNA spacer is unlikely to be a determinant of virulence. In the present study, additional unrelated genomic loci were examined by PFGE, and a significant correlation between the results of rDNA spacer typing and those of PFGE typing was observed. Since rDNA spacer typing is a PCR-based method, it does not require cultivation and can be performed rapidly with small quantities of clinical material. The present study establishes that rDNA spacer analysis can serve as an accurate reflection of B. burgdorferi genotype.
Acknowledgments
We thank S. Casjens, R. Marconi, S. Norris, J. Purser, and B. Stevenson for providing probes or primer sequences.
This work was supported by Public Health Service grant AR41511 from the National Institutes of Health.
REFERENCES
- 1.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J-C, Assous M, Grimont P A D. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 2.Barbour A G. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C M. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete, Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 4.Dykhuizen D E, Polin D S, Dunn J J, Wilske B, Preac-Mursic V, Dattwyler R J, Luft B J. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc Natl Acad Sci USA. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferdows M S, Serwer P, Griess G A, Norris S J, Barbour A G. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1996;178:793–800. doi: 10.1128/jb.178.3.793-800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foretz M, Postic D, Baranton G. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int J Syst Bacteriol. 1997;47:11–18. doi: 10.1099/00207713-47-1-11. [DOI] [PubMed] [Google Scholar]
- 7.Fraser C M, Casjens S, Huang W, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 8.Liveris D, Gazumyan A, Schwartz I. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1995;33:589–595. doi: 10.1128/jcm.33.3.589-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, McKenna D, Nowakowski J, Nadelman R B, Wormser G P, Schwartz I. Genetic diversity of Borrelia burgdorferi in Lyme disease patients determined by culture compared to direct polymerase chain reaction on clinical specimens. J Clin Microbiol. 1999;37:565–569. doi: 10.1128/jcm.37.3.565-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liveris D, Wormser G P, Nowakowski J, Nadelman R, Bittker S, Cooper D, Varde S, Moy F H, Forseter G, Pavia C S, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 12.Marconi R T, Samuels D S, Landry R K, Garon C F. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J Bacteriol. 1994;176:4572–4582. doi: 10.1128/jb.176.15.4572-4582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathiesen D A, Oliver J H, Jr, Kolbert C P, Tullson E D, Johnson B J, Campbell G L, Mitchell P D, Reed K D, Telford III S R, Anderson J F, Lane R S, Persing D H. Genetic heterogeneity of Borrelia burgdorferi in the United States. J Infect Dis. 1997;175:98–107. doi: 10.1093/infdis/175.1.98. [DOI] [PubMed] [Google Scholar]
- 14.Nadelman R B, Nowakowski J, Forseter G, Goldberg N S, Bittker S, Cooper D, Aguero-Rosenfeld M, Wormser G P. The clinical spectrum of early Lyme borreliosis in patients with culture-confirmed erythema migrans. Am J Med. 1996;100:502–508. doi: 10.1016/s0002-9343(95)99915-9. [DOI] [PubMed] [Google Scholar]
- 15.Nadelman R B, Wormser G P. Lyme borreliosis. Lancet. 1998;352:557–565. doi: 10.1016/S0140-6736(98)01146-5. [DOI] [PubMed] [Google Scholar]
- 16.Orloski K A, Hayes E B, Campbell G L, Dennis D T. Surveillance for Lyme disease—United States, 1992–1998. Morb Mortal Wkly Rep. 2000;49(SS-3):1–11. [PubMed] [Google Scholar]
- 17.Palmer N, Fraser C, Casjens S. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J Bacteriol. 2000;182:2476–2480. doi: 10.1128/jb.182.9.2476-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz I, Wormser G P, Schwartz J J, Cooper D, Weissensee P, Gazumyan A, Zimmermann E, Goldberg N S, Bittker S, Campbell G L, Pavia C S. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J Clin Microbiol. 1992;30:3082–3088. doi: 10.1128/jcm.30.12.3082-3088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson W J, Garon C F, Schwan T G. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 21.Walker D H, Barbour A G, Oliver J H, Lane R S, Dumler J S, Dennis D T, Persing D H, Azad A F, McSweegan E. Emerging bacterial zoonotic and vector-borne diseases. Ecological and epidemiological factors. JAMA. 1996;275:463–469. [PubMed] [Google Scholar]
- 22.Wang G, van Dam A P, Dankert J. Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol Lett. 1999;177:289–296. doi: 10.1111/j.1574-6968.1999.tb13745.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, van Dam A P, Schwartz I, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin Microbiol Rev. 1999;12:633–653. doi: 10.1128/cmr.12.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang I, Dykhuizen D E, Qui W, Dunn J J, Bosler E M, Luft B J. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics. 1999;151:15–30. doi: 10.1093/genetics/151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wormser G P, Liveris D, Nowakowski J, Nadelman R B, Cavaliere L F, McKenna D, Holmgren D, Schwartz I. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis. 1999;180:720–725. doi: 10.1086/314922. [DOI] [PubMed] [Google Scholar]