Abstract
Objectives
To investigate the acquired resistome in 18 colistin-resistant Escherichia coli isolated from different poultry farms in Lebanon, analyse Inc plasmids associated with mcr and assess potential transmission to humans.
Methods
A total of 18 E. coli were recovered from poultry faeces collected from different poultry farms in Lebanon. Broth microdilution (BMD) assay was performed to determine the antimicrobial resistance profiles. WGS was used to identify the genetic determinants behind the resistance in these isolates.
Results
BMD results showed that all of the 18 isolates were colistin resistant. Furthermore, resistance to trimethoprim/sulfamethoxazole was the most recorded among the isolates and only one isolate was resistant to cefepime. Sequencing results showed that the isolates were distributed into seven different STs and that the most abundant was ST1140. The number of antimicrobial resistant determinants ranged from 4 to 21 among the 18 isolates, with tet(A) and floR being the most frequent. Moreover, a total of 15 different plasmid replicon types were identified. The mcr-1 gene was shown to be predominantly located on IncX4 plasmids. Additionally, two isolates harboured the IncI2-type self-conjugative plasmid.
Conclusions
The findings show that mcr and other important resistance determinants occur in MDR E. coli isolated poultry farms in Lebanon. The occurrence of mcr on mobile plasmids and the zoonotic potential and clinical relevance of some strains highlight a risk of transmission to humans.
Introduction
The emergence of MDR bacteria has resulted in the use of colistin (polymyxin E) as a last resort for treatment of severe Gram-negative bacterial infections.1 This induced a reliance on this antibiotic and the likely selection of colistin-resistant strains.2 Initially, colistin resistance was believed to be solely caused by chromosomal mutations. However, the discovery of the plasmid-borne mobile colistin resistance gene, mcr-1, in 2015 in China indicated that resistance can also be transmitted laterally. Today, mcr is spreading globally and colistin use in animal farming is thought to be a major driver in this dissemination.3 The mcr-1 gene has been reported in approximately all food-producing animal species in 57 countries distributed over five continents.4 Therefore, this study aimed to investigate the acquired resistome, Inc plasmid types, and STs of 18 colistin-resistant Escherichia coli isolates from different poultry farms in Lebanon.
Methods
Bacterial isolates
The faecal samples were suspended in sterile buffered peptone water and homogenized gently by pipetting. An aliquot (100 μL) from each suspension was spread onto the selective medium RAPID’E. coli 2 agar (Bio-Rad, USA), which was supplemented with 4 mg/L colistin (Sigma–Aldrich, USA). The isolates (n = 90) that exhibited a typical E. coli phenotype on the agar (violet to pink colour) were further purified and their identity was confirmed by PCR targeting a species-specific fragment of the 16S rRNA gene.5 A total of 18 E. coli isolates were included in this study. Ten (56%) and eight (44%) E. coli isolates were recovered between 2017 and 2018 from broiler faeces collected from poultry farms located in the north and south of Lebanon, respectively.6 These animals were housed in three major farms in Lebanon (two farms in the north and one in the south).
Broth microdilution (BMD) assay
BMD was performed using 19 different antibiotics. Serial dilution ranged from 1024 mg/L to 1 mg/L. The plate was incubated at 37°C for 18 h and the experiments were run in duplicate for each antibiotic. The results were interpreted according to the CLSI M100 guidelines.7
Transformation
Plasmids were extracted using the QIAGEN® Plasmid Mini Kit (QIAGEN, USA) and transformed into recipient chemically competent E. coli JM109 cells using the heat-shock method.8,9 Competent E. coli (50 μL) were mixed with 10 μL of the extracted plasmid and incubated on ice for 30 min. In a separate reaction, 10 μL deionized water (no plasmids added) was used as a control. The mixtures were heat shocked at 42°C for 60 s in a water bath and incubated on ice for 2 min. Sterile LB broth (940 μL) was added, and the mixtures were placed in a shaking incubator (180 rpm) for 90 min at 37°C. The mixtures were centrifuged for 2 min at 14 000 rpm, and the pellet was resuspended in 0.1 mL LB broth, which was spread onto RAPID’E. coli 2 agar plates supplemented with 2 mg/L of colistin. The plates were incubated at 37°C for 18–24 h. Putative transformants were harvested and analysed for the acquisition of mcr-1 using PCR and colistin resistance using the BMD assay as described above.6
WGS and bioinformatics
Genomic DNA was extracted from overnight cultures using the QIAamp DNA mini kit (QIAGEN, USA) according to manufacturer's instructions. The quality and yield of the extracted DNA were checked on the nanodrop and qubit and then prepared for sequencing using the Nextera XT DNA library preparation kit (Illumina, UK). Sequencing was performed on the MiSeq instrument using the 2 × 300 paired-end protocol. Generated reads were assembled into contigs using shovill 1.0.9 (https://github.com/tseemann/shovill) with the default options. MLST were inferred from assembled genomes with the mlst-v2.18.1 software, while antimicrobial resistance genes were identified using Abricate 0.9.8 (https://github.com/tseemann/abricate). Chromosomal mutations associated with antimicrobial resistance were detected using gene_finder (https://github.com/phe-bioinformatics/gene_finder). Plasmids were determined using PlasmidFinder on CGE (https://cge.cbs.dtu.dk/services/PlasmidFinder/). To confirm the complete structure of the mcr-1-bearing IncX4- and IncI2-type plasmids, DNA from isolates ECOL79 and ECOL80 were subjected to low coverage sequencing on the MinION sequencer using the nanopore ligation sequencing kit according to the manufacturer’s instructions. Hybrid assembly of short and long reads were performed using Unicycler 0.4.8.10 Plasmid contigs from assembled genomes using the Illumina reads were compared using BLASTn and BRIG.11
Results
Antimicrobial susceptibility testing
BMD results showed that all of the 18 (100%) E. coli isolates were resistant to colistin with MICs ranging between 4 and 8 mg/L. In total, 16 (89%) isolates were resistant to trimethoprim/sulfamethoxazole, 15 (83%) to tetracycline, 14 to (78%) ciprofloxacin and levofloxacin, 13 (72%) to tigecycline, 12 (67%) to gentamicin, 11 (61%) to cefuroxime, 10 (56%) to fosfomycin, 9 (50%) to azithromycin and ceftolozane/tazobactam, 8 (44%) to ceftazidime and 7 (39%) to aztreonam, and only 1 (6%) isolate showed resistance to cefepime. None of the isolates was resistant to piperacillin/tazobactam, amikacin, ertapenem, meropenem or imipenem (Table 1 and Table S1, available as Supplementary data at JAC-AMR Online). Moreover, all the isolates were MDR (Table S1).
Table 1.
Antimicrobial susceptibility testing results of the 18 E. coli isolates
| Antimicrobial family | Antibiotic | No. resistant (%) | MIC range | MIC50 | MIC90 |
|---|---|---|---|---|---|
| Cephalosporins | Cefuroxime | 11 (61%) | 2 to 1024 | 64 | 1024 |
| Ceftazidime | 8 (44%) | ≤ 1 to 64 | ≤ 1 | 32 | |
| Cefepime | 1 (6%) | ≤ 1 to 32 | ≤ 1 | 8 | |
| Carbapenems | Meropenem | 0 (0%) | ≤ 1 | ≤ 1 | ≤ 1 |
| Imipenem | 0 (0%) | ≤ 1 to 2 | ≤ 1 | ≤ 1 | |
| Ertapenem | 0 (0%) | ≤ 1 | ≤ 1 | ≤ 1 | |
| Monobactams | Aztreonam | 7 (39%) | ≤ 1 to 64 | 8 | 32 |
| Fluoroquinolones | Ciprofloxacin | 14 (78%) | ≤ 1 to 32 | 16 | 32 |
| Levofloxacin | 14 (78%) | ≤ 1 to 32 | 8 | 32 | |
| Aminoglycosides | Gentamicin | 12 (67%) | ≤ 1 to 512 | 64 | 512 |
| Amikacin | 0 (0%) | ≤ 1 to 4 | ≤ 1 | 4 | |
| Tetracyclines | Tetracycline | 15 (83%) | ≤ 1 to 256 | 128 | 256 |
| Tigecycline | 13 (72%) | 0.25 to 2 | 0.5 | 1 | |
| Polymyxins | Colistin | 18 (100%) | 4 to 8 | 4 | 8 |
| Macrolides | Azithromycin | 9 (50%) | ≤ 0.125 to > 128 | 16 | > 128 |
| Fosfomycin | Fosfomycin | 10 (56%) | 2 to > 1024 | 1024 | > 1024 |
| Sulphonamides | Trimethoprim/sulfamethoxazole | 16 (89%) | ≤ 0.25 to > 256 | > 256 | > 256 |
| β-lactam + β-lactamase inhibitor | Piperacillin/tazobactam | 0 (0%) | ≤ 1 to 16 | 8 | 16 |
| Ceftolozane/tazobactam | 9 (50%) | ≤ 1 to 8 | 2 | 8 |
MIC50, MIC value at which 50% of the 18 E. coli isolates were inhibited; MIC90, MIC value at which 90% of the 18 E. coli isolates were inhibited.
Transmissibility of the mcr-1-carrying plasmids
Plasmids extracted from the mcr-1-positive E. coli were successfully transformed into recipient E. coli cells, which was evident by the formation of transformant colonies on the RAPID’E. coli 2 agar plates supplemented with colistin. The transformants were found positive for mcr-1 using PCR analysis. Additionally, the tested transformants were colistin resistant (MIC > 2 mg/L).
WGS
Genome sequence analyses showed that the 18 E. coli isolate were distributed into seven different STs, which included ST1140 (n = 7), ST226 (n = 3), ST2705 (n = 3), ST162 (n = 2), ST2936 (n = 1), ST3288 (n = 1) and ST6448 (n = 1). The E. coli isolates recovered from poultry faeces from the south belonged to ST1140 (n = 5), ST162 (n = 2) and ST6448 (n = 1). Furthermore, E. coli isolates obtained from poultry faeces from farms in the north belonged to ST2705 (n = 3), ST226 (n = 3), ST1140 (n = 2), ST2936 (n = 1) and ST3288 (n = 1). The number of antimicrobial resistance determinants ranged from 4 to 21 among the 18 isolates. The tet(A) and floR genes were the most frequent, and they were detected in 15 (83%) out of the 18 E. coli isolates. Moreover, several β-lactamase genes were detected, including: blaTEM-1 (n = 12) and blaTEM-122 (n = 5) as penicillinases, blaCTX-M-3 (n = 4) and blaCTX-M-55 (n = 1) as ESBLs, and blaCMY-2 (n = 7) as plasmid-borne cephalosporinase. Furthermore, resistance to fluoroquinolones was either due to two mutations in the gyrA gene (S83L and D87N) and one in the parC gene (S80I) (n = 13) or one mutation in the gyrA gene (S83L) (n = 1). In addition, 12 out of the 18 E. coli harboured the macrolide resistance genes lnu(F) and mph(A). Finally, eight different aminoglycosides resistance genes were detected (Table S2).
A total of 15 different plasmid replicon types were detected in the isolates; distributed as follows: IncX4 (n = 16), IncFII(pCoo) (n = 10), IncI2(Delta) (n = 7), IncHI2 (n = 6), IncHI2A (n = 6), IncFIB(K) (n = 6), IncFIB(AP001918) (n = 5), IncFIA (n = 4), IncI1-I(Alpha) (n = 4), IncFII (n = 4), IncFIC(FII) (n = 3), p0111 (n = 3), IncFII(29) (n = 2), IncI2 (n = 1) and IncFIB(pLF82-PhagePlasmid) (n = 1) (Table S2).
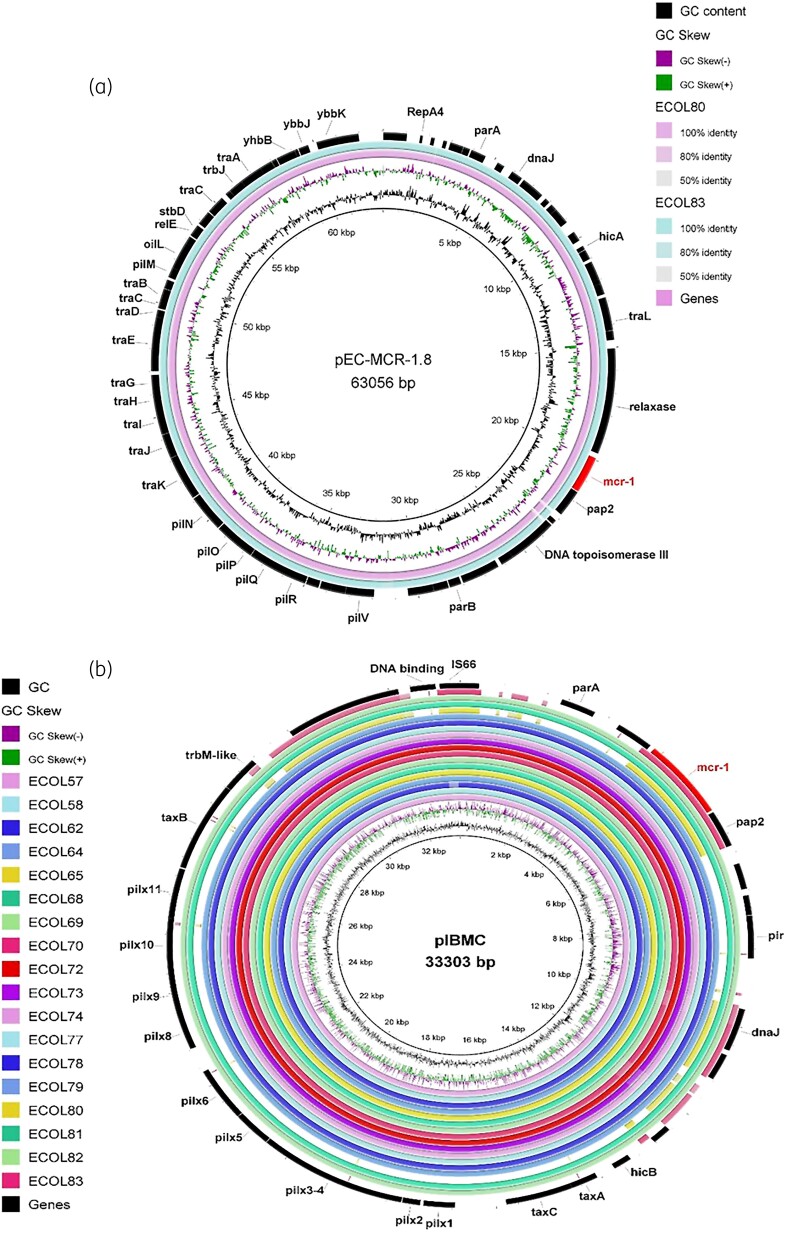
The mcr-1.1 gene in the majority of sequenced isolates (16/18, 88.9%) was located on a 33.3 kb circular contig sharing high similarity with IncX4-type plasmids harbouring mcr variants (coverage 97%–100%, identity 99%–100%) that were previously reported in various Enterobacterales species, including Salmonella enterica, E. coli and Klebsiella pneumoniae (Figure 1b).12 The draft genome of Isolate 80 located mcr-1 on a 64.7 kb circular contig that was most similar to the IncI2-type self-conjugative plasmid bearing mcr-1.8 (92% coverage with 99.46% identity, accession number: KY792081.2). Sequence comparison confirmed that Isolate 83 had the same IncI2-type mcr-1-harbouring plasmid (Figure 1a). Moreover, no other resistance genes were detected on both plasmids.13
Figure 1.
Circular graphical map showing BLAST comparisons of the mcr-1 carrying plasmids identified in (a) Isolates 80 and 83 against IncI2-type plasmid pEC-MCR1.8 (accession number: KY792081) and (b) the remaining isolates against IncX4-type plasmid pIBMC_mcr1 (accession number: MF449287.1).The mcr-1 gene location is shown in the figure with all the other genes. The colour intensity in each ring represents the BLASTn match identity as shown in the legend of (a).
Discussion
In 2017, an E. coli isolate belonging to ST2705 was recovered from a stool sample of a patient admitted to Hôtel-Dieu de France hospital in Beirut.14 This matches with the STs of three of the E. coli in this study. Moreover, in the University Hospital of Larissa-Central Greece an E. coli isolate belonging to ST1140 was detected in a patient,15 and this is the most abundant ST in this study. These findings show that MDR strains harbouring mcr with zoonotic potential occur in poultry farming in Lebanon. Urgent action is needed to control the dissemination of these determinants and strains on poultry farms in order to control the emergence of pandrug resistant bacteria that can affect both humans and animals in a One Health approach.
Supplementary Material
Funding
This work was funded by the Medical Practice Plan at the American University of Beirut Medical Center.
Transparency declarations
None to declare.
Supplementary data
Tables S1 and S2 are available as Supplementary data at JAC-AMR Online.
References
- 1. Ahmed ZS, Elshafiee EA, Khalefa HSet al. Evidence of colistin resistance genes (mcr-1 and mcr-2) in wild birds and its public health implication in Egypt. Antimicrob Resist Infect Control 2019; 8: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Islam S, Urmi UL, Rana Met al. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci Rep 2020; 10: 17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y-Y, Wang Y, Walsh TRet al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 4. Nagy Á, Székelyhidi R, Hanczné Lakatos Eet al. Review on the occurrence of the mcr-1 gene causing colistin resistance in cow’s milk and dairy products. Heliyon 2021; 7: e06800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kassem II, Mann D, Li Set al. Draft genome sequences and resistome analysis of multidrug-resistant mcr-1-harbouring Escherichia coli isolated from pre-harvest poultry in Lebanon. J Glob Antimicrob Resist 2021; 25: 114–6. [DOI] [PubMed] [Google Scholar]
- 6. Hmede Z, Kassem II. The colistin resistance gene mcr-1 is prevalent in commensal Escherichia coli isolated from preharvest poultry in Lebanon. Antimicrob Agents Chemother 2018; 62: e01304–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100. 2020. [Google Scholar]
- 8. Sulaiman AAA, Kassem II. First report on the detection of the plasmid-borne colistin resistance gene mcr-1 in multi-drug resistant E. coli isolated from domestic and sewer waters in Syrian refugee camps in Lebanon. Travel Med Infect Dis 2019; 30: 117–20. [DOI] [PubMed] [Google Scholar]
- 9. Hmede Z, Sulaiman AAA, Jaafar Het al. Emergence of plasmid-borne colistin resistance gene mcr-1 in multidrug-resistant Escherichia coli isolated from irrigation water in Lebanon. Int J Antimicrob Agents 2019; 54: 102–04. [DOI] [PubMed] [Google Scholar]
- 10. Wick RR, Judd LM, Gorrie CLet al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alikhan N-F, Petty NK, Ben Zakour NLet al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011; 12: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun J, Fang L-X, Wu Zet al. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep 2017; 7: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li R, Xie M, Zhang Jet al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 2017; 72: 393–401. [DOI] [PubMed] [Google Scholar]
- 14. Al-Mir H, Osman M, Azar Net al. Emergence of clinical mcr-1-positive Escherichia coli in Lebanon. J Glob Antimicrob Resist 2019; 19: 83–4. [DOI] [PubMed] [Google Scholar]
- 15. Mavroidi A, Miriagou V, Liakopoulos Aet al. Ciprofloxacin-resistant Escherichia coli in Central Greece: mechanisms of resistance and molecular identification. BMC Infect Dis 2012; 12: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.