Abstract
Background
Many factors impact survival in patients with glioblastoma, including age, Karnofsky Performance Status, postoperative chemoradiation, IDH1/2 mutation status, MGMT promoter methylation status, and extent of resection. High-throughput next-generation sequencing is a widely available diagnostic tool, but the independent impact of tumors harboring specific mutant genes on survival and the efficacy of extent of resection are not clear.
Methods
We utilized a widely available diagnostic platform (FoundationOne CDx) to perform high-throughput next-generation sequencing on 185 patients with newly diagnosed glioblastoma in our tertiary care center. We performed multivariate analysis to control for clinical parameters with known impact on survival to elucidate the independent prognostic value of prevalent mutant genes and the independent impact of gross total resection.
Results
When controlling for factors with known prognostic significance including IDH1/2 mutation and after multiple comparisons analysis, CDKN2B and EGFR mutations were associated with reduced overall survival while PTEN mutation was associated with improved overall survival. Gross total resection, compared to other extent of resection, was associated with improved overall survival in patients with tumors harboring mutations in CDKN2A, CDKN2B, EGFR, PTEN, TERT promoter, and TP53. All patients possessed at least one of these 6 mutant genes.
Conclusions
This study verifies the independent prognostic value of several mutant genes in glioblastoma. Six commonly found mutant genes were associated with improved survival when gross total resection was achieved. Thus, even when accounting for known predictors of survival and multiple mutant gene comparisons, extent of resection continues to be strongly associated with survival.
Keywords: DNA, glioblastoma, high-throughput nucleotide analysis, multivariate analysis, retrospective studies, sequence analysis
Key Points.
Complete resection of tumors with any of 6 common mutant genes improves survival.
Multivariate and multiple comparisons analyses are critical in sequencing studies.
Importance of the Study.
This study underscores the importance of utilizing multivariate analysis while taking into account multiple comparisons when determining the clinical significance of a large panel of genetic mutations in glioblastoma. First, our findings verify the prognostic value of several previously reported mutant genes with increased scientific rigor. Second, we find that gross total resection is beneficial for patients with tumors harboring any of 6 common mutant genes. A rigorous analytical approach is important to control for the heterogeneous clinical and genetic profiles of patients with glioblastomas, which is critical for patient selection in translational neuro-oncology research.
Glioblastoma is the most common primary malignant central nervous system tumor in adults. Median survival for patients with newly diagnosed glioblastoma is approximately 20 months, and 5-year survival remains poor at 5%.1–3 Prior to the era of clinical genetic sequencing, several factors had been known to be associated with prolonged overall survival (OS) including higher initial Karnofsky Performance Status (KPS), younger age, extent of standard-of-care adjuvant chemoradiation, and higher extent of tumor resection (EOR).4–6 Knowledge of these factors has paved the way for establishing the standard of care for newly diagnosed glioblastoma—maximal safe resection followed by radiotherapy and concurrent temozolomide.
Modern clinical genetic testing in glioblastoma has led to further improvements in the classification of glioblastoma into several molecular subgroups, which were found to have prognostic and potentially therapeutic significance.7 Two molecular features with particularly significant clinical implications are the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) gene mutations and O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation. IDH1/2 mutations are found in nearly 80% of secondary glioblastoma (also referred to as IDH-mutant glioblastoma) and in approximately 5% of de novo glioblastoma and are associated with better prognosis.8,9 The DNA repair gene MGMT can be epigenetically silenced through promoter methylation, which renders tumors more susceptible to alkylating agents such as temozolomide. MGMT promoter methylation is more common in IDH1/2-mutant glioblastoma and is associated with a better prognosis in patients receiving temozolomide.10 Glioblastoma has been shown to have a highly heterogeneous genetic profile beyond the IDH1/2 mutation and MGMT promoter methylation, but with few exceptions, surprisingly little is known about the prognosis of patients with tumors harboring other genetic mutations. Even among the few reported prognostic mutant genes, the literature contains conflicting findings.11–13 The prognostic significance of EGFRvIII is controversial14,15; for example, PTEN mutation has been associated with poor survival in glioma patients.16CDKN2A and CDKN2B deletions, but not TERT mutation or EGFR amplification, have been recently reported to be an independent prognostic marker for IDH1/2-wildtype glioblastoma.17 Few studies investigating the prognostic value of particular gene alterations have taken into account factors that are known to impact survival. One study investigated the prognostic significance of TERT promoter mutation.18 The authors demonstrated in a univariate analysis that age, KPS, EOR, MGMT promoter methylation status, and postoperative chemoradiation were prognostic for survival. In a subsequent multivariate analysis, the authors found that TERT promoter mutation was an independent predictor of poor prognosis. In their exploratory analysis, the authors found that TERT promoter mutation may be prognostic only if subtotal resection (STR) was achieved and if patients did not undergo adjuvant chemotherapy, leading to the conclusion that patients with TERT promoter mutations may require aggressive treatment.
Knowledge of the relationship between particular genetic events in a tumor and EOR have implications for how aggressively tumors should be resected. For example, in a study of 282 patients with newly diagnosed anaplastic astrocytomas or glioblastomas stratified by IDH1 mutation status, patients with tumors harboring IDH1 mutations had an additional survival benefit if further resection of the nonenhancing disease (supramaximal resection) in addition to the enhancing disease was achieved.19 More recently, a survival benefit was demonstrated in younger patients with even IDH1-wildtype glioblastomas after supramaximal resection.20 In theory, however, more discrete knowledge of genetic subtypes within the main subgroups of IDH1-wildtype and -mutant glioblastomas could influence the impact of EOR, and other practices, and ultimately affect patient survival. Therefore, there is an urgent need to study the independent prognostic significance of EOR on the known genetic subtypes of glioblastoma by controlling for factors with a known impact on survival. Moreover, while it is known that gross total resection (GTR) positively impacts survival, it is not known to what degree this is true when stratifying by individual mutant genes.
With the knowledge of factors with known prognostic significance and the availability of high-throughput next-generation sequencing (NGS), there is an opportunity to study the independent prognostic significance of multiple known mutant genes in glioblastoma. In this retrospective cohort study, we used age, KPS, extent of adjuvant chemoradiation, MGMT promoter methylation status, EOR, and IDH1/2 mutation status as covariates while accounting for multiple gene comparisons to investigate this hypothesis. We also explored the independent impact of GTR on survival in tumors with the most prevalent mutant genes. Ultimately, genetic tumor classification can be utilized as a molecular diagnostic tool or a prognostic indicator of OS, further leading to the development and characterization of new treatment strategies.
Material and Methods
Patient Selection
The study was conducted with the approval of the institutional review board. Patients aged 18 and older with newly diagnosed IDH1/2-wildtype and -mutant glioblastoma (WHO 2016 criteria), who were treated with surgery at our tertiary center and had samples sent to Foundation Medicine between 2015 and 2020, were retrospectively reviewed. All pathology slides were reviewed for confirmation of diagnosis of glioblastoma by a board-certified neuropathologist (S.D.). Exclusion criteria included patients treated at another facility whose adjuvant chemoradiation regimen was unknown, secondary glioblastoma (prior low-grade or anaplastic astrocytoma), secondary debulking or ablative surgery after recurrence, patients lost to follow-up after surgery, post-resection MRI performed greater than 72 h after surgery, and patients who died within 60 days after surgery to exclude confounding variables affecting survival such as sepsis and acute cardiopulmonary events. To determine MGMT promoter methylation status, DNA was isolated from formalin-fixed, paraffin-embedded tumor tissue specimens. Molecular analysis of the MGMT gene was performed by methylation-specific PCR and detected on ABI 7900 (Labcorp Center for Molecular Biology and Pathology). The MGMT and beta-actin copy numbers were used to calculate the ratio of the MGMT/beta-actin ×1000. This assay detects the target gene at 45–50 copies per reaction. Methylation score ≥2.00 was considered positive for promoter methylation.
Next-Generation Sequencing
NGS was exclusively performed using the FDA-approved, commercially available FoundationOne CDx test (F1CDx Foundation Medicine), which is a Clinical Laboratory Improvement Amendments (CLIA)-certified in vitro diagnostic test that uses targeted high-throughput hybridization-based capture technology to detect substitutions, insertions, deletion, and copy number alterations in 324 genes and select gene rearrangements using DNA isolated from formalin-fixed paraffin-embedded tumor tissue specimens (https://info.foundationmedicine.com/hubfs/FMI%20Labels/FoundationOne_CDx_Label_Technical_Info.pdf). This sequencing platform has been used routinely at our institution since 2015 as an additional tool to further characterize the molecular changes within central nervous system neoplasms.
Treatment and Outcome
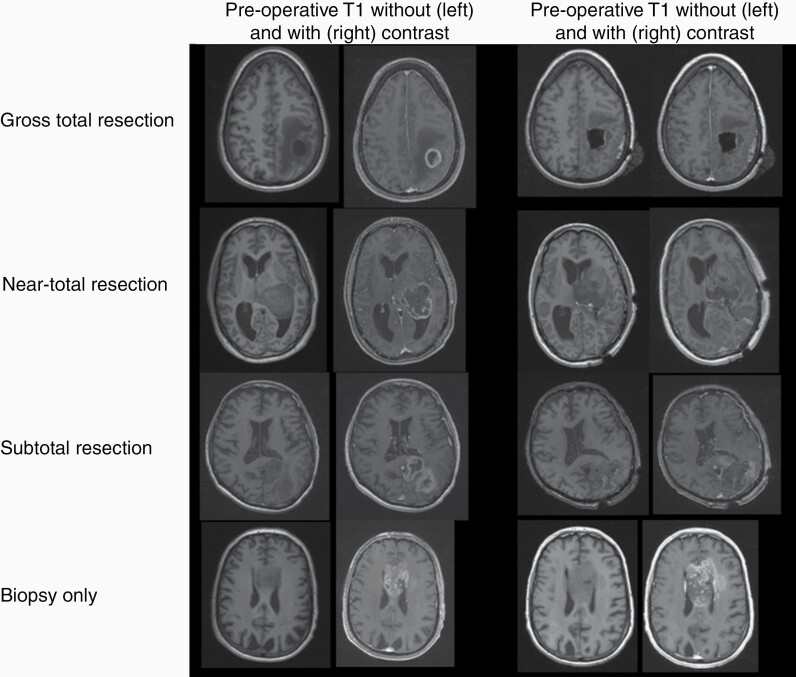
Clinical parameters collected included age, EOR, progression-free survival (PFS; defined as the time between the date of surgery and the date of the MRI read as concerning for recurrence), OS (defined as the time between the date of surgery and the date of hospice or death), baseline KPS as determined by first postoperative oncology visit, MGMT promoter status, and the extent to which patients received adjuvant chemoradiation (full, partial, or none). All patients underwent standard of practice postoperative 3T brain MRI (including a slice thickness 1.0 mm MPRAGE sequence) without and with gadolinium contrast within 72 h of surgery. EOR for all cases was independently assessed by a subspecialty board-certified neuroradiologist (A.R.C.) with greater than 7 years of experience based on RANO criteria.21,22 The assessments were compared to the original neuroradiology report for concordance. Discrepancies between central review and neuroradiology reports were reconciled by direct review of images by another author (P.H.Y.) and resulted in an agreement with a central review. We defined postoperative MRI GTR as 100% complete resection of enhancing tissue, near-total resection (NTR) as resection of between approximately 80–99% of enhancing tissue (less than 5 cm3 residual contrast-enhancing tissue volume), and STR as resection of less than 80% (greater than 5 cm3 residual contrast-enhancing tissue volume) (Figure 1). Any areas of intrinsic T1 hyperintensity representing postoperative blood products were distinguished and excluded from true enhancing tissue assessment. Biopsy only was also included and was defined as a needle biopsy without tumor debulking via craniotomy.
Figure 1.
Extent of resection as depicted by T1-weighted sequences without and with gadolinium contrast on preoperative and postoperative magnetic resonance imaging.
Statistics
Kaplan–Meier (KM) analysis was conducted to generate survival curves for PFS and OS of parameters with previously known impact on survival as a validation of our patient cohort (Figure 2). The log-rank test was used to examine the statistical significance of the differences observed between the groups. A prognostic study was performed using a multivariate Cox proportional hazards model to compute hazard ratios (HRs) and 95% confidence intervals of the most frequently mutated tumor genes while controlling for covariates with known impact on survival. A multivariate analysis was performed to investigate the impact on survival of GTR versus all other EOR in patients with tumors that harbored the most frequently mutated genes. The Benjamini–Hochberg procedure controlling false discovery rate (FDR) at a 5% level was used to adjust the survival analysis. The 2-tailed t test with P < .05 was considered statistically significant. All analyses were performed within Statistical Analysis System (SAS, version 9.4; SAS Institute Inc.).
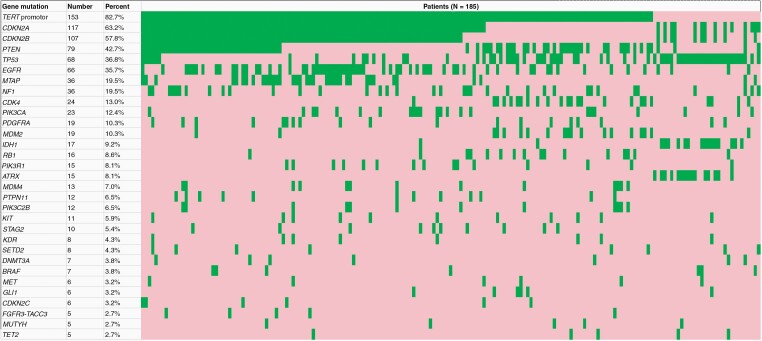
Figure 2.
Waterfall plot depicting the most commonly mutated genes in our cohort, sorted by frequency. The most commonly mutated genes included TERT promoter (82.7%), CDKN2A (63.2%), CDKN2B (57.8%), PTEN (42.7%), TP53 (36.8%), and EGFR (35.7%). IDH1 mutation frequency was 9.2%. Mutated genes are depicted in green and non-mutated genes are depicted in red.
Exploratory Validation Analysis
As an exploratory validation, we collected the clinical and radiological data as above and performed an identical analysis of glioblastoma patients who underwent surgery between 2011 and 2015 at our institution. All specimens underwent previously validated NGS testing through an institutional platform.23–25 We used the bootstrapping technique on the FoundationOne data set to perform a power analysis (Supplementary Methods, Supplementary Table 1).
Results
Patient Characteristics
About 269 patients were screened and 185 patients (108 male, 58.4%) with a median age of 62.2 years (range 23.3–84.6 years) met criteria for further analysis. About 87.0% of patients (N = 161) had a postoperative KPS of 70 or higher. 9.7% of patients (N = 18) had IDH1 or IDH2 mutation and 42.4% (N = 75 out of 177 with completed results) had MGMT promoter methylation. 27.0% (N = 50) patients underwent GTR, 10.3% (N = 19) underwent NTR, 35.1% (N = 65) underwent STR, and 27.6% (N = 51) underwent biopsy only. 95.1% (N = 176) of all patients underwent some adjuvant chemoradiation, of which 79.0% (N = 139 out of 176) underwent full adjuvant chemoradiation (Table 1).
Table 1.
Demographic, Clinical, and Radiological Data for the Patient Cohort
| Patient Data | Number | Percent |
|---|---|---|
| Demographics | ||
| Patients | 185 | 100.0% |
| Male | 108 | 58.4% |
| Female | 77 | 41.6% |
| Mean age at diagnosis (range) | 59.5 (23.3–84.6) | |
| Median age at diagnosis | 62.2 | |
| Karnofsky Performance Status | ||
| <70 | 24 | 13.0% |
| ≥70 | 161 | 87.0% |
| Tumor features | ||
| Primary glioblastoma | 185 | 100.0% |
| IDH mutation (IDH1 or IDH2) | 18 | 9.7% |
| MGMT promoter methylationa | 75 | 42.4% |
| Extent of resection | ||
| Gross total resection | 50 | 27.0% |
| Near-total resection | 19 | 10.3% |
| Subtotal resection | 65 | 35.1% |
| Biopsy | 51 | 27.6% |
| Adjuvant treatment | ||
| No adjuvant treatment | 9 | 4.9% |
| Adjuvant treatment | 176 | 95.1% |
| Full chemoradiation | 139 | 75.1% |
| Partial chemoradiation | 37 | 20.0% |
aCensored, 177 with results.
A waterfall plot demonstrated the mutant genes found in our patient cohort sorted by mutation frequency (Figure 2). TERT promoter mutation (82.7% mutation frequency, N = 153), CDKN2A mutation (63.2%, N = 117), CDKN2B mutation (57.8%, N = 107), PTEN mutation (42.7%, N = 79), TP53 mutation (36.8%, N = 68), and EGFR mutation (35.7%, N = 66) were among the most frequently mutated genes. IDH1 mutation was found in 9.2% (N = 17) of patients.
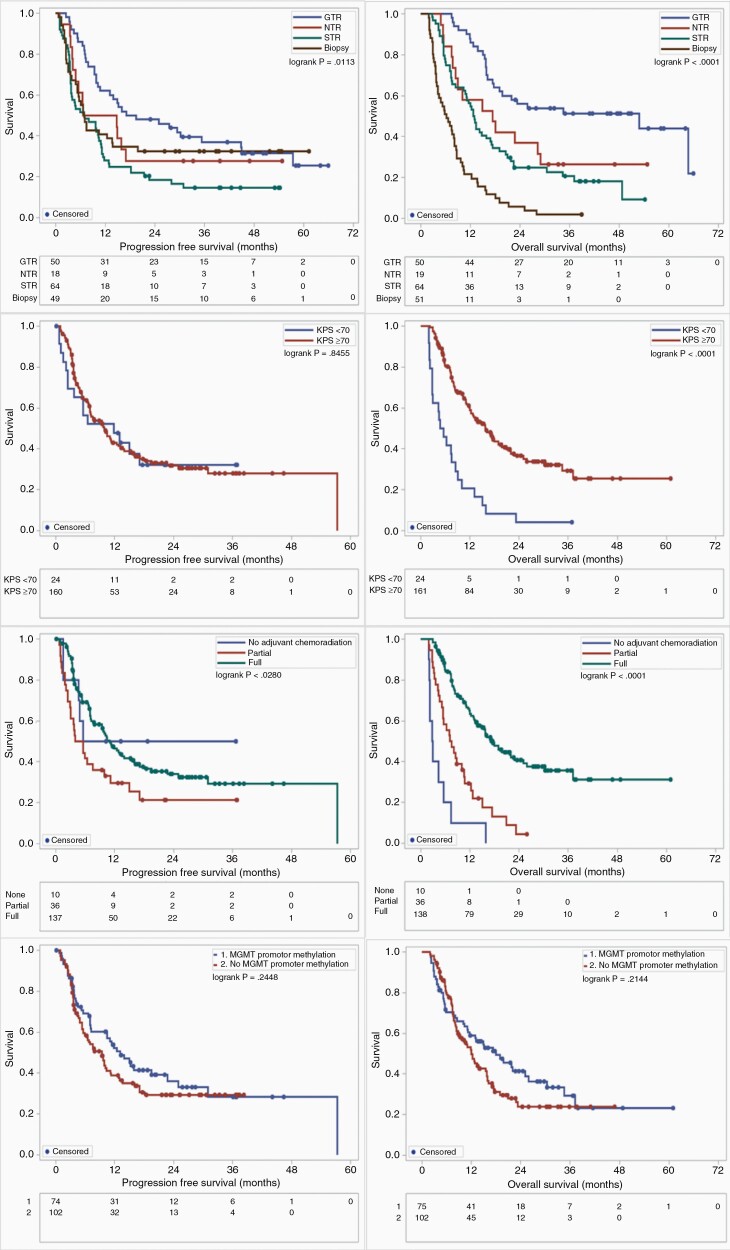
KM plots of factors with known impact on survival demonstrated that EOR was significantly associated with PFS (P = .0113) and OS (P < .0001). KPS ≥70 was associated with improved OS (P < .0001), extent of adjuvant chemoradiation was associated with improved OS (P < .0001), and IDH1 or IDH2 mutation was associated with improved OS (P < .0001) (Figure 3).
Figure 3.
Overall survival and progression-free survival delineated by factors with known impact on prognosis. Greater extent of resection (gross total resection [GTR], near-total resection [NTR], subtotal resection [STR], and biopsy), Karnofsky Performance Status (KPS) ≥70, and extent of adjuvant chemoradiation were associated with improved overall survival. O6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation was not associated with survival.
Independent Prognostic Value of Gene Mutations
We performed a prognostic study using a subset of 10 genes with prognostic significance reported in the literature (Table 2). These were TERT promoter, CDKN2A, CDKN2B, PTEN, TP53, EGFR, NF1, PIK3CA, PDGFRA, and IDH1/2. To determine the independent prognostic value of these mutant genes compared to wildtype, we performed the analysis using covariates with known prognostic significance on survival: age, KPS, extent of adjuvant chemoradiation, MGMT promoter methylation status, and EOR without and with IDH1/2 as a covariate.
Table 2.
Independent Prognostic Value of a Subset of Commonly Mutated Genes Using Multivariate Analysis and Multiple Comparisons (N = 185)
| Progression-Free Survival | Overall Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene mutation | P | FDR-adjusted P value | HR | 95% CI | P value | FDR-adjusted P value | HR | 95% CI |
| A | ||||||||
| CDKN2A | .0950 | .3837 | 1.40 | 0.94–2.07 | .0215 | .0430 | 1.57 | 1.07–2.32 |
| CDKN2B | .1960 | .3847 | 1.28 | 0.88–1.85 | .0039 | .0120 | 1.74 | 1.19–2.53 |
| EGFR | .1648 | .3847 | 1.30 | 0.90–1.88 | .0010 | .0100 | 1.85 | 1.28–2.68 |
| NF1 | .2656 | .3847 | 1.29 | 0.82–2.03 | .4850 | .5389 | 0.85 | 0.53–1.35 |
| PDGFRA | .9788 | .9788 | 1.01 | 0.54–1.88 | .7427 | .7427 | 1.10 | 0.61–1.99 |
| PIK3CA | .5273 | .6292 | 1.18 | 0.70–2.00 | .3403 | .4254 | 1.28 | 0.77–2.12 |
| PTEN | .5663 | .6292 | 0.90 | 0.62–1.29 | .0295 | .0492 | 0.66 | 0.46–0.96 |
| TERT promoter | .1151 | .3837 | 1.63 | 0.89–2.99 | .0048 | .0120 | 2.49 | 1.32–4.70 |
| TP53 | .2693 | .3847 | 0.80 | 0.54–1.19 | .2097 | .2996 | 0.77 | 0.52–1.16 |
| IDH1/2 | .0171 | .1710 | 0.33 | 0.14–0.82 | .0044 | .0120 | 0.12 | 0.03–0.52 |
| Covariates: age, KPS, adjuvant chemoradiation, MGMT promoter methylation, EOR | ||||||||
| B | ||||||||
| CDKN2A | .1307 | .6304 | 1.35 | 0.91–2.00 | .0320 | .0720 | 1.53 | 1.04–2.26 |
| CDKN2B | .2154 | .6304 | 1.26 | 0.87–1.83 | .0028 | .0185 | 1.78 | 1.22–2.61 |
| EGFR | .3467 | .6304 | 1.20 | 0.82–1.73 | .0062 | .0186 | 1.67 | 1.16–2.42 |
| NF1 | .3502 | .6304 | 1.24 | 0.79–1.95 | .3586 | .5379 | 0.80 | 0.51–1.28 |
| PDGFRA | .9915 | .9915 | 1.00 | 0.54–1.87 | .7030 | .7030 | 1.12 | 0.62–2.02 |
| PIK3CA | .5504 | .8033 | 1.17 | 0.69–1.99 | .4359 | .5604 | 1.23 | 0.73–2.04 |
| PTEN | .2441 | .6304 | 0.80 | 0.56–1.16 | .0041 | .0185 | 0.58 | 0.40–0.84 |
| TERT promoter | .6248 | .8033 | 1.18 | 0.62–2.25 | .1234 | .2221 | 1.67 | 0.87–3.22 |
| TP53 | .7233 | .8137 | 0.93 | 0.62–1.40 | .6888 | .7030 | 0.92 | 0.62–1.37 |
| Covariates: age, KPS, adjuvant chemoradiation, MGMT promoter methylation, EOR, IDH1/2 mutation |
Bolded values indicate P < .05.
In the FDR-adjusted OS analysis, without IDH1/2 mutation as a covariate, CDKN2A mutation (P = .0430, HR 1.57), CDKN2B mutation (P = .0120, HR = 1.74), EGFR mutation (P = .0100, HR = 1.85), and TERT promoter mutation (P = .0120, HR = 2.49) were associated with decreased OS while PTEN mutation (P = .0492, HR = 0.66) and IDH1/2 mutation (P = .0120, HR = 0.12) were associated with improved OS (Table 2, A). When adding IDH1/2 mutation as a covariate to the FDR-adjusted analysis, CDKN2B mutation (P = .0185, HR = 1.78) and EGFR mutation (P = .0186, HR = 1.67) were associated with decreased OS while PTEN mutation (P = .0185, HR = 0.58) was associated with improved OS (Table 2, B). CDKN2A mutation was not associated with OS in the FDR-adjusted analysis with addition of IDH1/2 mutation as a covariate.
In the FDR-unadjusted PFS analysis, IDH1/2 mutation (P = .0171, HR = 0.33) was associated with improved PFS, but not after FDR adjustment.
Subset OS analysis of the 167 patients with IDH1/2-wildtype glioblastomas showed similar findings to the above analysis with IDH1/2 mutation as a covariate (Supplementary Table 2). In the FDR-adjusted analysis, CDKN2B mutation (P = .0171, HR = 1.81) and EGFR mutation (P = .0231, HR = 1.65) were associated with decreased OS while PTEN mutation (P = .0171, HR = 0.58) was associated with improved OS. CDKN2A mutation was not significantly associated with OS in the FDR-adjusted analysis.
Independent Prognostic Value of GTR
Next, we sought to determine the independent prognostic significance of GTR compared to all other EOR among the 10 genes previously described using age, KPS, extent of adjuvant chemoradiation, and MGMT promoter methylation status without and with IDH1/2 as covariates. In the FDR-adjusted analysis without IDH1/2 mutation as a covariate, GTR was associated with improved OS among patients with tumors harboring CDKN2A mutation (P = .0178, HR = 0.49), CDKN2B mutation (P = .0110, HR = 0.45), EGFR mutation (P = .0110, HR = 0.35), TERT promoter mutation (P = .0020, HR = 0.40), and TP53 mutation (P = .0110, HR = 0.26) (Table 3, A). GTR was associated with improved OS in patients harboring PTEN mutation on the FDR-unadjusted analysis (P = .0303, HR = 0.43), but this was not significant after FDR adjustment.
Table 3.
Independent Prognostic Value of Gross Total Resection Versus Other Extent of Resection Among Patients With Specific Gene Mutations, Using Multivariate Analysis and Multiple Comparisons
| Progression-Free Survival | Overall Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene mutation | P | FDR-adjusted P value | HR | 95% CI | P value | FDR-adjusted P value | HR | 95% CI |
| A | ||||||||
| CDKN2A | .8471 | .9387 | 0.95 | 0.57–1.58 | .0089 | .0178 | 0.49 | 0.28–0.83 |
| CDKN2B | .9387 | .9387 | 0.98 | 0.58–1.65 | .0041 | .0110 | 0.45 | 0.26–0.78 |
| EGFR | .4060 | .9387 | 0.76 | 0.40–1.45 | .0044 | .0110 | 0.35 | 0.17–0.72 |
| NF1 | .8088 | .9387 | 1.18 | 0.31–4.57 | .7254 | .8060 | 0.76 | 0.16–3.52 |
| PDGFRA | .0441 | .2675 | 12.98 | 1.07–157.37 | .2833 | .3541 | 3.15 | 0.39–25.59 |
| PIK3CA | .5117 | .9387 | 0.63 | 0.16–2.52 | .2000 | .2857 | 0.37 | 0.08–1.69 |
| PTEN | .7663 | .9387 | 1.11 | 0.57–2.14 | .0303 | .0505 | 0.43 | 0.20–0.92 |
| TERT promoter | .5856 | .9387 | 0.88 | 0.56–1.38 | .0002 | .0020 | 0.40 | 0.25–0.65 |
| TP53 | .2544 | .848 | 0.63 | 0.28–1.4 | .0034 | .0110 | 0.26 | 0.10–0.64 |
| IDH1/2 | .0535 | .2675 | 28.76 | 0.95–869.79 | .9998 | .9998 | 0.0 | 0-. |
| Covariates: age, KPS, adjuvant chemoradiation, MGMT promoter methylation | ||||||||
| B | ||||||||
| CDKN2A | .8471 | .9387 | 0.95 | 0.57–1.58 | .0224 | .0403 | 0.53 | 0.31–0.91 |
| CDKN2B | .9387 | .9387 | 0.98 | 0.58–1.65 | .0089 | .0267 | 0.48 | 0.28–0.83 |
| EGFR | .4060 | .9387 | 0.76 | 0.4–1.45 | .0044 | .0198 | 0.35 | 0.17–0.72 |
| NF1 | .8088 | .9387 | 1.18 | 0.31–4.57 | .7254 | .7254 | 0.76 | 0.16–3.52 |
| PDGFRA | .0441 | .3969 | 12.98 | 1.07–157.37 | .3039 | .3907 | 2.99 | 0.37–24.03 |
| PIK3CA | .5117 | .9387 | 0.63 | 0.16–2.52 | .4283 | .4818 | 0.56 | 0.13–2.37 |
| PTEN | .7663 | .9387 | 1.11 | 0.57–2.14 | .0303 | .0455 | 0.43 | 0.20–0.92 |
| TERT promoter | .5856 | .9387 | 0.88 | 0.56–1.38 | .0005 | .0045 | 0.42 | 0.26–0.69 |
| TP53 | .2544 | .9387 | 0.63 | 0.28–1.40 | .0135 | .0304 | 0.29 | 0.11–0.78 |
| Covariates: age, KPS, adjuvant chemoradiation, MGMT promoter methylation, IDH1/2 mutation |
Bolded values indicate P < .05.
In the FDR-adjusted analysis adding IDH1/2 mutation as a covariate, GTR was associated with improved OS in patients harboring CDKN2A mutation (P = .0403, HR = 0.53), CDKN2B mutation (P = .0267, HR = 0.48), EGFR mutation (P = .0198, HR = 0.35), PTEN mutation (P = .0455, HR = 0.43), TERT promoter mutation (P = .0045, HR = 0.42), and TP53 mutation (P = .0304, HR = 0.29) (Table 3, B).
In the FDR-unadjusted PFS analysis, GTR in patients harboring PDGFRA mutation (P = .0441, HR = 12.98) was associated with reduced PFS, but not after FDR-adjustment.
Patients with IDH1/2-wildtype glioblastomas harboring CDKN2B, EGFR, and TERT promoter mutations who underwent GTR were associated with improved OS when compared to other EOR in the FDR-adjusted analysis (Supplementary Table 3). CDKN2A mutation and PTEN mutation were significantly associated with improved OS in the FDR-unadjusted analysis but were no longer significant in the FDR-adjusted analysis.
Exploratory Validation Analysis
In an attempt to validate our findings on the impact of GTR on tumor with specific mutant genes, we examined a separate institutional patient cohort (N = 108) with NGS data derived from an alternative verified platform. Mutations were categorized by varying levels of clinical significance (Supplementary Methods, Supplementary Figure 1A). Among the most frequently tested genes, the most common mutant genes were TERT promoter (80.6%), PIK3CA (40.0%), EGFR (38.9%), TP53 (38.9%), PTEN (31.5%), and NF1 (20.0%) (Supplementary Figure 1B). KM plots demonstrated findings similar to those found in the literature, with improved OS with greater EOR (P = .0025), KPS (P = .0001), and extent of standard adjuvant chemoradiation (P < .0001), and improved PFS with adjuvant chemoradiation (P < .0001) and MGMT promoter methylation (P = .0084) (Supplementary Figure 2). Out of the 10 genes examined in FoundationOne analysis, HR could be calculated for only 5 genes in the validation data set (EGFR [N = 42, 108 tested], NF1 [N = 11, 85 tested], PTEN [N = 34, 108 tested], TERT promoter [N = 58, 72 tested], and TP53 [N = 42, 108 tested]) because of insufficient sample size or testing of the other 5 genes (CDKN2A [N = 4], CDKN2B [N = 0], PDGFRA [N = 3], PIK3CA [N = 16, 40 tested], and IDH1 [N = 8]).
We first ascertained if this validation cohort was sufficiently powered to detect the associations observed in our initial data set. In the bootstrapping analysis, to achieve a power of >0.80 and P < .05, the minimum sample sizes for each gene mutation for OS ranged from 100 to >1000 (Supplementary Table 1). In total, however, our institutional validation data set contained much fewer than 100 patients per mutation. Nevertheless, we thought it was still potentially informative to pursue an analysis of this validation cohort. We found that PTEN mutation was associated with improved OS if GTR was achieved without and with IDH1/2 mutation as a covariate (P = .0473); however, this only trended toward significance in the FDR-adjusted analysis. EGFR, NF1, TERT promoter, and TP53 mutations did not reach statistical significance in the unadjusted and adjusted analysis without IDH1/2 mutation as a covariate (Supplementary Table 4A). We also performed a similar analysis with IDH1/2 mutation (although IDH2 mutation was not found in this data set) as a covariate, which showed that no single mutant gene reached statistical significance in the FDR-adjusted analysis (Supplementary Table 4B). Similarly, the results were not significant among the IDH1/2-wildtype group (N = 100) in the FDR-adjusted analysis (Supplementary Table 4C). Overall, in this exploratory validation analysis, although no single gene mutation was significantly associated with prognosis in the setting of GTR, there was a trend with HR < 1.0 for each of the 5 genes tested (EGFR, NF1, PTEN, TERT promoter, and TP53).
Discussion
Prognostic studies are important to elucidate potential fundamental differences in glioblastoma with different molecular features. For example, IDH1/2-wildtype and -mutant glioblastomas have markedly different prognoses, and the WHO 2016 criteria used IDH1/2 mutation status as a criterion when classifying glioblastomas.26 Several studies in the past decade have investigated the association of patient survival with mutations in genes such as IDH1/2, TP53, PTEN, EGFR, and TERT promoter, and more recent data show that CDKN2A/B deletions are associated with decreased survival when controlling for prognostic factors.17,27,28 For example, Shinojima et al.14 found that EGFRvIII overexpression in the setting of EGFR amplification was an independent marker for poor prognosis in a multivariate study. However, this study, like many others, did not take into account other significant covariates that may impact survival such as the effects of EOR and adjuvant chemoradiation, the latter of which was found to have significant implications for improved survival in more recent studies. As more information is discovered about factors with known impact on prognosis, more recent studies have begun to utilize clinical data for multivariate analyses of the prognostic significance of specific genetic mutations; however, none performed multivariate analysis while accounting for multiple gene comparisons.17,29 To our knowledge, this is the first study to investigate both the prognosis of tumors and the impact of EOR on glioblastoma patient outcomes using a wide spectrum of genetic events while controlling for major factors with known impact on survival. Importantly, our assessment of EOR was externally validated according to standardized criteria.21,22
EOR in glioblastoma has been an area of intensive study for the past 2 decades, but few studies have investigated the correlation between molecular findings of glioblastoma, EOR, and their independent impact on prognosis. For example, Felsberg et al.30 found that MGMT promoter hypermethylation and near-complete resection were independently associated with better prognosis in patients treated with resection followed by chemoradiation. A review of the current available literature suggests that GTR increases the likelihood of 1- and 2-year survival.31 Recent evidence also suggests that complete resection of the T1 contrast-enhancing portion in addition to a portion of the surrounding T2 hyperintensity is associated with a survival benefit over resection of the T1 contrast-enhancing portion alone in glioblastoma and that maximizing EOR may confer an additional survival advantage in patients with tumors harboring certain mutant genes including IDH1.20,32
We found that, after controlling for the effects of age, KPS, adjuvant chemoradiation, MGMT promoter methylation, EOR, and IDH1/2 mutation status and accounting for multiple gene comparisons, CDKN2B and EGFR mutations were independently associated with reduced OS while PTEN mutation was associated with improved OS. These results validate some of the conclusions from prior studies and may provide clarification for specific mutant genes whose prognostic value is controversial.7,15–18 For example, the prognostic value of PTEN mutation has been reported to be associated with worse outcomes, in large clinical data sets,33,34 or to have no clear association with outcome in other studies with multivariate analyses.35 These studies, however, did not all control for prognostic factors such as MGMT methylation status, EOR, and IDH1/2 mutation status. In addition, large studies that combine clinical and genetic data from different institutions often do not properly take into account EOR, either because the data are not available or because the definition may be nonstandardized across institutions. But our controlled results, using 6 known prognostic factors including centrally reviewed neuroradiology for EOR and FDR-adjustment, suggest that PTEN mutation was independently associated with improved survival. These findings illustrate that rigorous multivariate analysis and multiple comparisons adjustment should be performed when evaluating the effect of individual gene mutations to determine their prognostic value.
Using this approach, we also asked whether glioblastomas harboring certain genetic events were associated with survival if GTR was achieved. We found that among patients with mutations in CDKN2A, CDKN2B, EGFR, PTEN, TERT promoter, and TP53, GTR was associated with improved OS, compared to other EOR when accounting for known predictors of survival including IDH1/2 mutation status. Moreover, in the IDH1/2-wildtype cohort, CDKN2B and EGFR mutations were associated with worse outcomes, but GTR still resulted in a survival benefit over other EOR.
In this study, the impact of GTR was found to be associated with 6 out of the 10 mutant genes analyzed, and this remained significant in the FDR-adjusted analysis. Since all patients in our cohort possessed at least one of these mutant genes, the result of improved survival when GTR is achieved in patients harboring these mutations is in fact a reflection of the previously known observation that GTR is beneficial to survival. Despite the common knowledge that GTR is beneficial for OS in glioblastoma patients, an intriguing hypothesis raised by our findings is that patients with tumors that do not harbor one of these 6 common mutations may not have derived benefit from GTR. However, this hypothesis remains to be verified. These findings suggest that, given the prevalence of these 6 gene mutations, following the diagnosis of glioblastoma and knowledge of IDH1/2 mutation status, GTR (or maximum safe resection) should be attempted on all patients regardless of results from sequencing studies. The identification of druggable targets is still in its early stages, and the findings in this study suggest that patients, even those in low-resource settings, are not necessarily at a disadvantage if they do not undergo sequencing.
Although limited somewhat by sample size, this study highlights an overall approach to evaluating the contribution of genetic mutations and, in the future, other potential omic variables (such as transcriptomics, metabolomics, and proteomics) to prognosis and response to specific treatments, including surgical resection, radiation therapy, and medical therapies. This analytical framework is important for controlling for inherently heterogeneous patient populations and tumor characteristics and may have greater implications on translational studies of glioblastoma such as in clinical trial design.
Modern massively parallel DNA sequencing techniques provide a degree of granularity to glioblastoma research previously not possible by standard histopathological diagnostic methods. Unique mutations detected by NGS, such as using the FoundationOne CDx platform in this study, allow for a greater understanding of the spectrum of abnormal protein isoforms with varying degrees of clinical significance. For example, IDH1 R132H is the most common IDH1 mutation, but our study revealed multiple other isoforms with known and unknown clinical significance, such as R132C and R132G, which are undetectable by standard antibodies to IDH1.
There are several limitations to this study. This study was retrospective, performed at a single center, and is limited in power. In addition, other genes, due to their relatively low mutation frequency with insignificant P values in our analysis, may have an impact on clinical outcomes, but this cannot be determined given the sample size in our analysis. EOR was determined by qualitative assessment of enhancement on post-resection MRI by neuroradiology instead of by volumetric quantification of EOR. We performed a validation analysis on a separate group of 108 patients who underwent institutional NGS, and although we found that no single gene mutation was prognostic after FDR adjustment, there was a trend toward improved survival if GTR was achieved in patients harboring one of 5 common gene mutations. However, this exploratory analysis was underpowered. The findings in the current study warrant further validation using a consistent diagnostic platform and statistical analyses in a larger, multi-institutional cohort.
Conclusion
In a single-center retrospective study of the genomic landscape of 185 glioblastoma patients, multivariate analysis controlling for 6 covariates with known impact on survival while accounting for multiple gene comparisons revealed that CDKN2B, EGFR, and PTEN mutations were independently associated with survival. The survival benefit of GTR was specifically seen in patients harboring mutations in CDKN2A, CDKN2B, EGFR, PTEN, TERT promoter, or TP53, which represented the entirety of the patient cohort. Therefore, the benefit of GTR seen globally in glioblastoma patients may be the result of improved survival among patients harboring any of these 6 commonly found mutant genes. These findings and our methodological approach may help to clarify results from other sequencing studies and guide future clinical and translational investigations on this topic.
Supplementary Material
Funding
This work was supported by the National Institutes of Health grants R01 NS094670 (to A.H.K.), R01 NS106612 (to A.H.K.), the Christopher Davidson and Knight Family Fund (to A.H.K. and G.P.D.), and the Duesenberg Research Fund (to A.H.K.).
Conflict of interest statement. A.H.K. has research grants from Monteris Medical, Stryker, and Collagen Matrix, which are not directly relevant to this publication. G.P.D. is co-founder of Immunovalent, which is not directly relevant to this publication. S.R. is a senior director of pathology at Foundation Medicine Inc.
Authorship statement. P.H.Y. and A.H.K. conceived of the presented idea. P.H.Y. performed data collection, created figures and tables, and wrote the manuscript with feedback from A.H.K., G.P.D., S.M., J.W.H., E.D., and S.D. H-C.L., S.R., and S.D. provided genetics information. Y.T. and J.L. performed statistical analyses. A.R.C. performed neuroradiology central review and manuscript editing. M.P. assisted with data collection.
References
- 1. Lieberman F. Glioblastoma update: molecular biology, diagnosis, treatment, response assessment, and translational clinical trials. F1000Res. 2017;6:1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Weller M, Butowski N, Tran DD, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanai N, Polley M-Y, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. [DOI] [PubMed] [Google Scholar]
- 5. Grabowski MM, Recinos PF, Nowacki AS, et al. Residual tumor volume versus extent of resection: predictors of survival after surgery for glioblastoma. J Neurosurg. 2014;121(5):1115–1123. [DOI] [PubMed] [Google Scholar]
- 6. Zinn PO, Colen RR, Kasper EM, Burkhardt J-K. Extent of resection and radiotherapy in GBM: A 1973 to 2007 surveillance, epidemiology and end results analysis of 21,783 patients. Int J Oncol. 2013;42(3):929–934. [DOI] [PubMed] [Google Scholar]
- 7. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brat DJ, Aldape K, Colman H, et al. cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 11. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahlokozera T, Vellimana AK, Li T, et al. Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro Oncol. 2017;20(4):472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shinojima N, Tada K, Shiraishi S, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 15. Montano N, Cenci T, Martini M, et al. Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia (New York, NY). 2011;13(12):1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Han F, Hu R, Yang H, et al. PTEN gene mutations correlate to poor prognosis in glioma patients: a meta-analysis. Onco Targets Ther. 2016;9:3485–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma S, Rudra S, Campian JL, et al. Prognostic impact of CDKN2A/B deletion, TERT mutation, and EGFR amplification on histological and molecular IDH-wildtype glioblastoma. Neuro Oncol Adv 2020;2(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon M, Hosen I, Gousias K, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015;17(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glenn CA, Baker CM, Conner AK, et al. An examination of the role of supramaximal resection of temporal lobe glioblastoma multiforme. World Neurosurg. 2018;114:e747–e755. [DOI] [PubMed] [Google Scholar]
- 20. Molinaro AM, Hervey-Jumper S, Morshed RA, et al. Association of maximal extent of resection of contrast-enhanced and non–contrast-enhanced tumor with survival within molecular subgroups of patients with newly diagnosed glioblastoma. JAMA Oncol. 2020;6(4):495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vogelbaum MA, Jost S, Aghi MK, et al. Application of novel response/progression measures for surgically delivered therapies for gliomas: Response Assessment in Neuro-Oncology (RANO) Working Group. Neurosurgery. 2012;70(1):234–243; discussion 243. [DOI] [PubMed] [Google Scholar]
- 22. Karschnia P, Vogelbaum MA, van den Bent M, et al. Evidence-based recommendations on categories for extent of resection in diffuse glioma. Eur J Cancer. 2021;149:23–33. [DOI] [PubMed] [Google Scholar]
- 23. Cottrell CE, Al-Kateb H, Bredemeyer AJ, et al. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16(1):89–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carter JH, McNulty SN, Cimino PJ, et al. Targeted next-generation sequencing in molecular subtyping of lower-grade diffuse gliomas: application of the World Health Organization’s 2016 revised criteria for central nervous system tumors. J Mol Diagn. 2017;19(2):328–337. [DOI] [PubMed] [Google Scholar]
- 25. McNulty SN, Cottrell CE, Vigh-Conrad KA, et al. Beyond sequence variation: assessment of copy number variation in adult glioblastoma through targeted tumor somatic profiling. Hum Pathol. 2019;86:170–181. [DOI] [PubMed] [Google Scholar]
- 26. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 27. Shiraishi S, Tada K, Nakamura H, et al. Influence of p53 mutations on prognosis of patients with glioblastoma. Cancer. 2002;95(2):249–257. [DOI] [PubMed] [Google Scholar]
- 28. Li Q-J, Cai J-Q, Liu C-Y. Evolving molecular genetics of glioblastoma. Chin Med J (Engl). 2016;129(4):464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salvati M, Bruzzaniti P, Relucenti M, et al. Retrospective and randomized analysis of influence and correlation of clinical and molecular prognostic factors in a mono-operative series of 122 patients with glioblastoma treated with STR or GTR. Brain Sci. 2020;10(2):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Felsberg J, Rapp M, Loeser S, et al. Prognostic significance of molecular markers and extent of resection in primary glioblastoma patients. Clin Cancer Res. 2009;15(21):6683–6693. [DOI] [PubMed] [Google Scholar]
- 31. Brown TJ, Brennan MC, Li M, et al. Association of the extent of resection with survival in glioblastoma: a systematic review and meta-analysis. JAMA Oncol. 2016;2(11):1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li YM, Suki D, Hess K, Sawaya R. The influence of maximum safe resection of glioblastoma on survival in 1229 patients: can we do better than gross-total resection? J Neurosurg. 2016;124(4):977–988. [DOI] [PubMed] [Google Scholar]
- 33. Bäcklund LM, Nilsson BR, Goike HM, et al. Short postoperative survival for glioblastoma patients with a dysfunctional Rb1 pathway in combination with no wild-type PTEN. Clin Cancer Res. 2003;9(11):4151–4158. [PubMed] [Google Scholar]
- 34. Xu J, Li Z, Wang J, Chen H, Fang J-Y. Combined PTEN mutation and protein expression associate with overall and disease-free survival of glioblastoma patients. Transl Oncol. 2014;7(2):196–205.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carico C, Nuño M, Mukherjee D, et al. Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era. PLoS One. 2012;7(3):e33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.