ABSTRACT
Adeno-associated virus (AAV) is extensively used as a viral vector to deliver therapeutic genes during human gene therapy. A high-affinity cellular receptor (AAVR) for most serotypes was recently identified; however, its biological function as a gene product remains unclear. In this study, we used AAVR knockdown cell models to show that AAVR depletion significantly attenuated cells to activate unfolded protein response (UPR) pathways when exposed to the endoplasmic reticulum (ER) stress inducer, tunicamycin. By analyzing three major UPR pathways, we found that ATF6 signaling was most affected in an AAVR-dependent fashion, distinct from CHOP and XBP1 branches. AAVR capacity in UPR regulation required the full native AAVR protein, and AAV2 capsid binding to the receptor altered ATF6 dynamics. Conversely, the transduction efficiency of AAV2 was associated with changes in ATF6 signaling in host cells following treatment with different small molecules. Thus, AAVR served as an inhibitory molecule to repress UPR responses via a specificity for ATF6 signaling, and the AAV2 infection route involved the release from AAVR-mediated ATF6 repression, thereby facilitating viral intracellular trafficking and transduction.
IMPORTANCE The native function of the AAVR as an ER-Golgi localized protein is largely unknown. We showed that AAVR acted as a functional molecule to regulate UPR signaling under induced ER stress. AAVR inhibited the activation of the transcription factor, ATF6, whereas receptor binding to AAV2 released the suppression effects. This finding has expanded our understanding of AAV infection biology in terms of the physiological properties of AAVR in host cells. Importantly, our research provides a possible strategy which may improve the efficiency of AAV-mediated gene delivery during gene therapy.
KEYWORDS: AAV receptor, UPR, ATF6 signal, AAV transduction, AAV receptor
INTRODUCTION
Adeno-associated viruses (AAVs) belong to the Parvoviridae family in the Dependovirus genus. Twelve major human AAV serotypes (AAV 1 to 12) have been identified and exhibit broad expression differences in tissue tropisms and different transduction efficiencies (1). The absence of a discernible pathology post-host infection has facilitated AAVs and other modifiable recombinant variants as ideal vectors for gene therapy against some human diseases (2). AAVs have been successfully used in several clinical trials to treat hereditary and neurological diseases and, most recently, were approved for the treatment of genetic metabolic diseases, such as lipoprotein lipase deficiency (3–6).
Various host factors facilitate AAV entry during infection. Heparan sulfate proteoglycan mediates AAV2 attachment in most permissive cells (7). Human fibroblast growth factor receptor-1 (FGFR1) and hepatocyte growth factor receptor serve as putative coreceptors to promote viral infection (8, 9). However, FGFR1 or MET knockout only displays minor effects on AAV2 infection, thereby failing to explain tropism differences in different tissue types (10). Recently, a genome-wide insertional mutagenesis screen in haploid human cells identified an adeno-associated virus receptor (AAVR or KIAA0319L) as a high-affinity receptor for multiple AAV serotypes. This receptor identification greatly expanded our understanding of AAV transduction biology and may improve the application of diverse AAV serotypes to different biological techniques (10, 11). Nonetheless, an important question has remained; what is the fundamental cell biological role of AAVR as a gene product? Even if the sole function of AAVR was to mediate AAV cell entry, how does AAVR function in intracellular viral infection pathways, and what are its effects on AAV transduction efficiency?
Currently, studies exploring the KIAA0319L gene and the AAVR protein are limited. From animal and transgenic models, the protein/gene was linked to dyslexia and was potentially implicated in neuronal migration and axon guidance (12). AAVR contains five polycystic kidney disease (PKD) domains (PKD 1 to 5) in the N-terminal extracellular portion which are linked to a single-pass membrane-spanning helix (10). These PKD domains are widely distributed and conserved structural elements, but without well-defined signatures. The first member of the PKD protein family to be identified was polycystin-1 (PDK1), which is implicated in polycystic kidney disease. PDK1 is a large cell-surface glycoprotein involved in protein-protein and protein-carbohydrate adhesive interactions (13, 14). PKD domains are found in many other proteins, usually in the extracellular domains of transmembrane proteins, including AAVR (10). However, it is unclear how exactly these PKD domains mediate interactions between themselves and other PKD-containing proteins.
AAVR is a common and critical viral entry receptor that mediates the infection of many AAV serotypes and variants (10, 11). It was suggested that the viral capsid of different AAV serotypes interacted with the receptor via distinctive PKD domains (11, 15–17). Recently, Pillay et al. reported that AAVR was associated with trans-Golgi network 46 (TGN46), a cis-medial marker (giantin) and colocalized with a marker. By chasing a pool of labeled AAVR in 4°C precooled cells, the authors showed that AAVR rapidly migrated from the plasma membrane to the Golgi, immediately after the temperature rose to initiate endocytosis (10). These data suggested that AAVR shared a behavior profile with several trafficking proteins and possibly had functions other than binding AAV at the cell surface.
In our study, we showed that the widely expressed native AAVR acted as an inhibitory factor to pacify endoplasmic reticulum (ER) stress, with selectivity primarily toward the ATF6-mediated unfolded protein response (UPR) pathway. The binding and interaction of AAVR with AAV2 virions deprived the suppressive function of AAVR on ER stress induced by chemical reagents. These results provided valuable insights on the physiological functions of AAVR. They also exemplified the UPR pathway as a novel mechanism in AAV infections and intracellular trafficking. These findings not only explain the effects of several agents that modulate AAV transduction efficiency, but also provide a discovery platform for modifiable host targets to improve AAV infection efficiency.
RESULTS
AAVR depletion enhances ER stress responses induced by tunicamycin (TM).
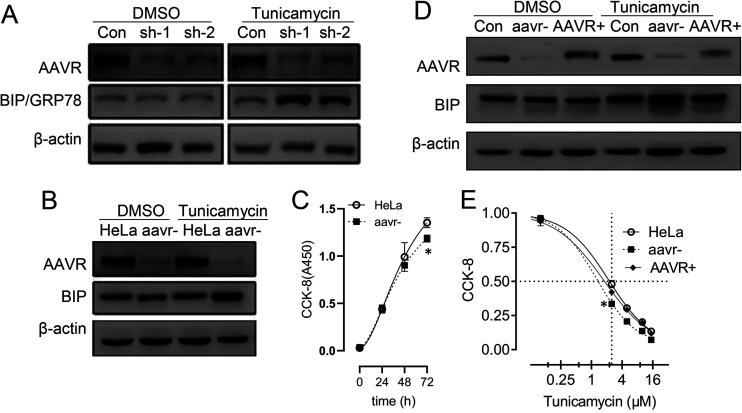
In addition to the plasma membrane, AAVR also localizes to ER-Golgi compartments (10). Also, AAVR knockout appears to cripple cell responses to environmental stresses. Several reagents, including teniposide (18), bortezomib (MG132) (19), and chloroquine (20, 21), are known to alter AAV infection and transduction and disrupt ER stress pathways (22), therefore, we explored whether AAVR was involved in ER stress response regulation using these reagents. We used TM, which is a typical ER stress inducer and affects viral uptake or trafficking, to treat host cells. In HeLa cells transfected with plasmids carrying short hairpin RNA (shRNA) sequences targeting AAVR, AAVR expression was significantly reduced (Fig. 1A). Also, the BIP molecule, as an indicator ER of stress activation, was detected by Western blotting. Thus, AAVR knockdown had significantly increased BIP/GRP78 levels following TM induction. To verify these findings, we generated a stable AAVR knockdown strain (aavr–) using lentiviral infection procedures. Similar results were identified when compared with the native HeLa parental line (Fig. 1B). Cell growth assays showed that AAVR downregulation exerted inhibitory effects at 72 h and should have no significant influence on the examination of ER stress markers at 24 or 48 h (Fig. 1C). To further exclude the possibility of nonspecific effects from TM treatment or shRNA transfection, we transfected aavr– cells with an AAVR overexpression plasmid to perform rescue experiments. AAVR recovery in aavr– cells significantly suppressed induced BIP protein expression from TM treatments (Fig. 1D). We also compared TM dose responses between aavr– and AAV+ cells at 72 h and observed that AAVR rescue recovered cell viability due to AAV knockdown (Fig. 1E). These data suggested a functional involvement of AAVR in regulating ER stress responses.
FIG 1.
AAVR depletion enhances tunicamycin (TM)-induced endoplasmic reticulum (ER) stress responses in HeLa cells. (A) HeLa cells were transfected with short hairpin RNAs (shRNAs) sh-1 and sh-2 targeting AAVR for 24 h. Western blotting for BIP/GRP78 was performed with or without 2.5 μmol/L TM for 6 h. (B) An AAVR knockdown cell line (aavr–) was prepared from shRNA lentivirus infection and compared with parental HeLa cells on TM treatment in BIP expression. (C) A CCK-8 time course assay comparing aavr– and HeLa cell growth. (D) Transfection of an AAVR-expressing plasmid into aavr– cells (AAVR+) and Western blotting for BIP with or without TM. (E) 48-h dose response curves of TM treatments by CCK-8 assay. Data are expressed as the mean ± standard deviation (n = 3). Statistical significance was determined using Student’s t test. *, P < 0.05; **, P < 0.01.
AAVR knockdown enhances ATF6 signaling in the UPR induced by TM.
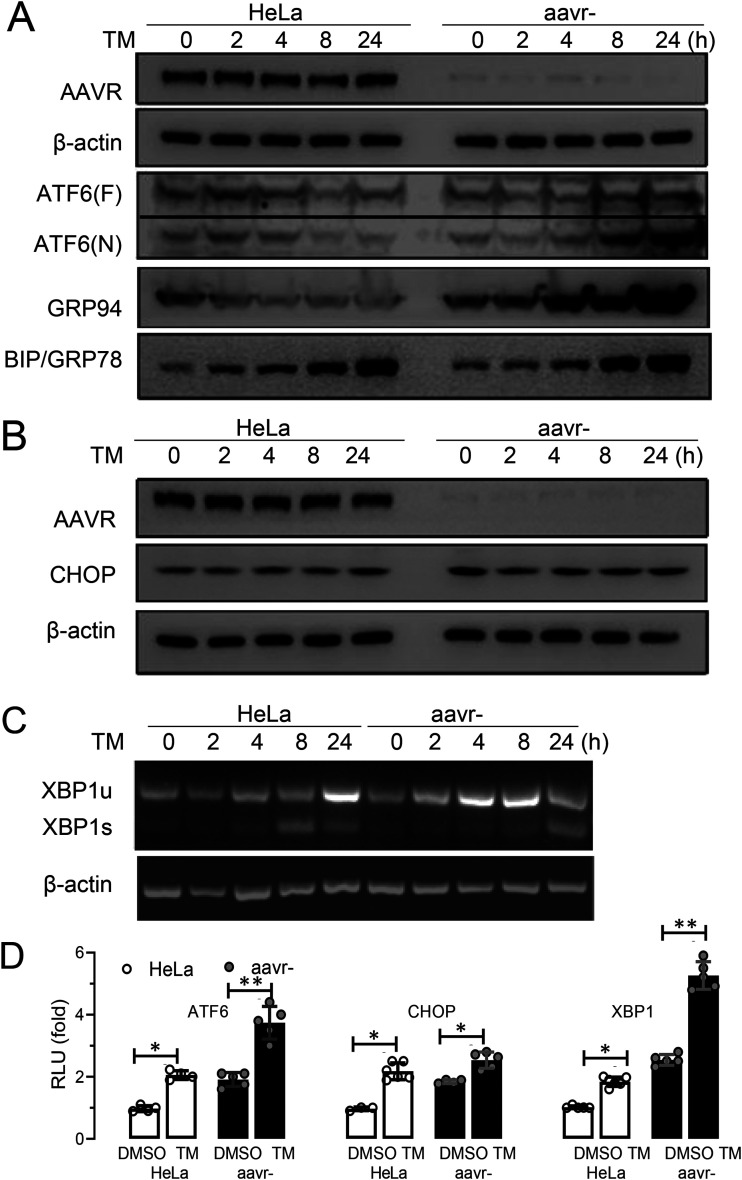
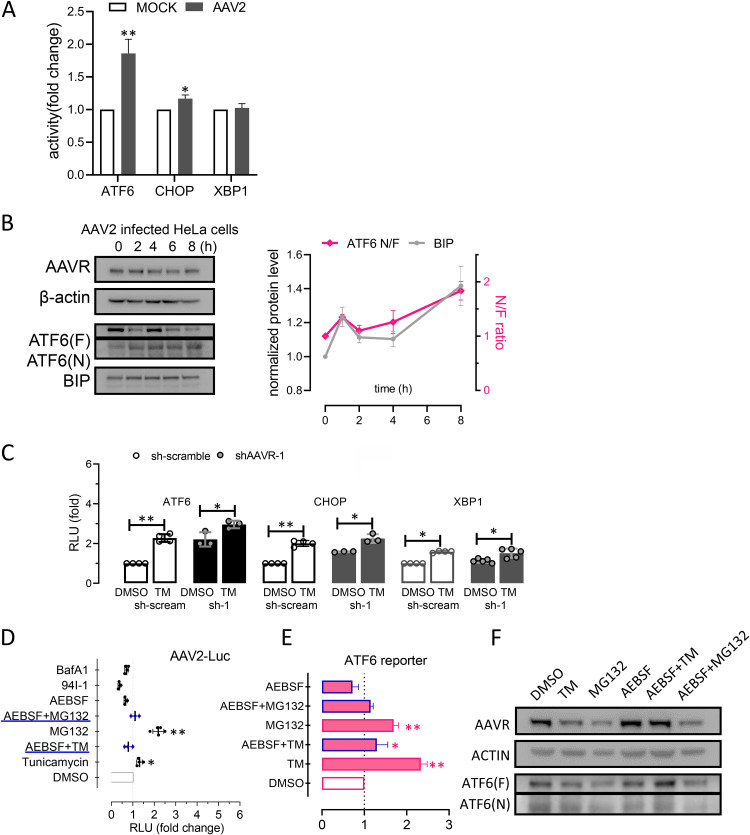
ER stress is induced by various assaults and elicits complex downstream signaling. The UPR is a common cause of ER stress and occurs when protein synthesis overwhelms protein folding capacity in the ER or improperly folded proteins accumulate and block normal ER homeostasis (23). ER stress linked to the BIP sensor could be activated by three different pathways from UPR signals, represented by key molecules of ATF6, PERK, and IRE1α, respectively. When dissociated from BIP at the Golgi, ATF6 is cleaved and becomes an active transcription factor. PERK phosphorylates the initiation factor 2α (eIF2) and produces CHOP, which activates UPR target genes. Then, IRE1α induces the mRNA cleavage of X box-binding protein 1 (XBP1) to translate spliced XBP1 protein (XBP1s), which is a key regulator of ER folding (24, 25). To study AAVR regulatory roles in UPR processes, we investigated changes in cleaved ATF6, CHOP, and XBP1s levels using Western blotting and/or reverse transcriptase PCR (RT-PCR). As shown (Fig. 2A), cleaved ATF6 N-terminal protein levels increased following TM induction for 8 h in aavr– cells. Also, GRP94 expression levels (an ATF6 regulated UPR gene) were significantly increased. Although CHOP expression levels were also significantly increased, it appeared that activation of PERK signaling was less dependent on AAVR abundance (Fig. 2B). The fold changes in XBP1s mRNA levels were not significantly different between conditions, despite TM treatment increasing XBP1 expression in both aavr– and native HeLa cells (Fig. 2C). To quantitatively evaluate the activation of different UPR signal pathways, we generated luciferase-based reporters for ATF6, CHOP, and XBP1s. Reporter assay data, with or without TM, are summarized (Fig. 2D) and indicated that ATF6 was the primary signal molecule that conveyed AAVR functions of UPR regulation during ER stress responses following TM treatment.
FIG 2.
AAVR knockdown enhances ATF6 signaling in tunicamycin (TM)-induced stress on the unfolded protein response (UPR) in HeLa cells. (A) Western blots for ATF6 activation on both expression of full-length ATF(F) and cleaved ATF6(N) induced by TM for 6 h in aavr– and parental HeLa cells. Levels of BIP and GRP94 upstream and downstream of ATF6 signaling were detected. (B) CHOP expression in aav– and HeLa cells with or without TM stress. (C) XBP1s and XBP1u mRNA levels during IRE1 pathway activation by semiquantitative PCR. (D) Luciferase reporter assays showing UPR pathway activation. HeLa and aavr– cells were transfected with 0.2 μg firefly luciferase reporter and 0.04 μg pTK-Renilla plasmid for 24 h with/without 2.5 μM TM for 6 h. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
AAVR mutants attenuate the inhibiting role of ATF6 signaling during UPR progress.
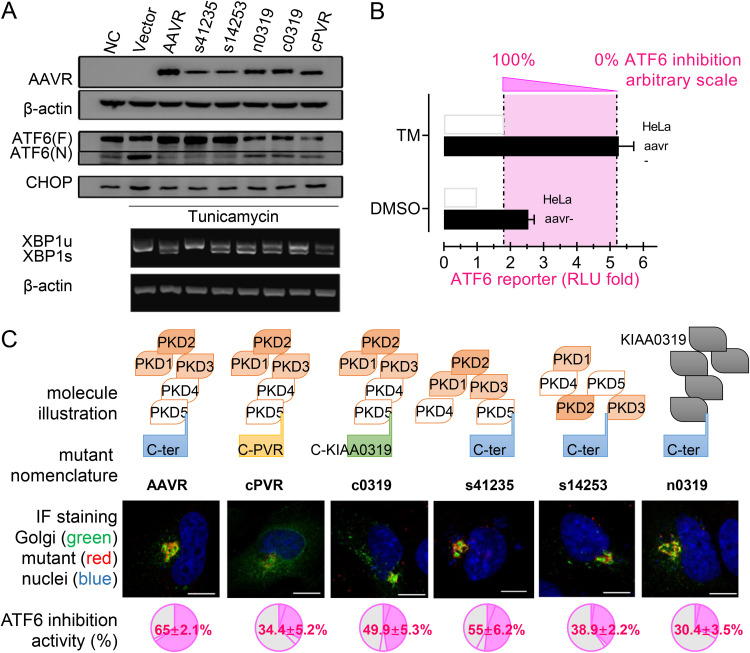
Different AAVR segments have been assigned distinct functions related to viral binding, subcellular localization, etc. For example, the AAVR C-terminal tail is required for AAVR migration from the plasma membrane to the TGN. Therefore, we dissected which AAVR segment was required to regulate ATF6 activities. Accordingly (Fig. 3A), five AAVR mutants were constructed (structures are shown in Fig. 3C) to test their influence on UPR signaling. Western blotting showed that the rescued expression of AAVR in aavr– cells significantly reduced the levels of cleaved N-terminal ATF6 as increased by TM treatment, and all the mutants in both the N-terminal and C-tail of AAVR attenuated its inhibiting role in ATF6 signaling (Fig. 3A). These results indicated that mutations in the N-terminal and C-tail of AAVR attenuated its inhibitory effects on CHOP and XBP1 signal activation (Fig. 3A). To quantitatively measure ATF6 activity inhibition, we generated indices from normalized reporter assays comparing TM responses under specific conditions (Fig. 3B). The data are shown in Fig. 3C.
FIG 3.
The effect of AAVR and mutants on inhibition of ATF6-mediated unfolded protein response (UPR) following tunicamycin stress. (A) Transfection of various AAVR mutants into aavr– cells and UPR assessment via ATF6, CHOP, and XBP1 activation (Western blots). (B) Illustration of defining a quantitative measurement on AAVR inhibition of ATF6 signaling by luciferase assays. (C) Intracellular localization of transfected AAVR mutants and their impact on inhibiting ATF6 activation. Scale bar = 10 μm. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
From immunofluorescence assays, mutant AAVR with a C-terminal replacement altered its intracellular distribution as previously reported, including cPVR (the C terminus of AAVR replaced by the C terminus of poliovirus receptor) and c0319 (the C terminus of AAVR replaced by the C terminus of KIAA0319 protein, a homologous protein of AAVR) as shown in Fig. 3C. Wild-type AAVR and its mutants at the N terminus, including shuffled PKD domains (s41235 and s14253) and a replacement mutant (n0319, the AAVR N terminus, was replaced by the N terminus of KIAA0319 protein), were localized to the TGN and colocalized with TGN46 (Fig. 3C). Both cPVR and c0319 attenuated the inhibitory role of AAVR toward ATF6 transactivation, from 65.0% to 34.4% and 49.9%, respectively, indicating the importance of the AAVR C-tail on ATF6 inhibition. Other N-terminal AAVR mutants impaired the inhibition of ATF6 activation to different extents, including s41235, s14253, and n0319 (Fig. 3C). When the five PKD domains were replaced by the KIAA0319 sequence (n0319), a low inhibitory measurement was observed at 30.4%. These results suggested that complete AAVR activity required the full protein sequence. N-terminal ectopic and C-tail intravehicular portions were equally important for inhibiting UPR; however, the interface for AAV binding (PKD 1 to 3) had to be intact. However, whether AAVR directly or indirectly interacts with cleaved or uncleaved ATF6 forms requires future investigation.
ATF6 signaling activation elicited by AAV2 infection influences AAV transduction in HeLa cells.
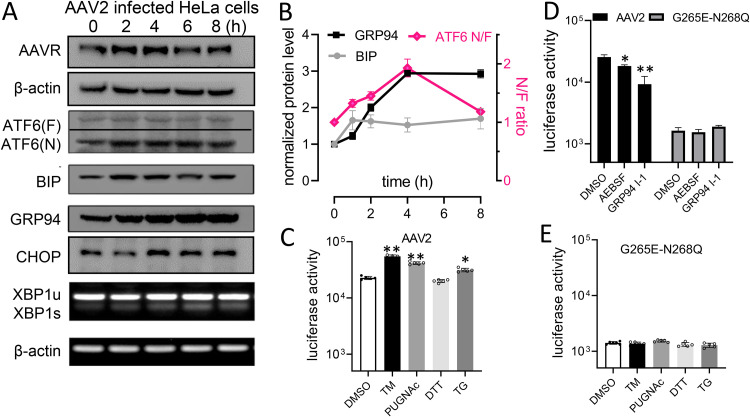
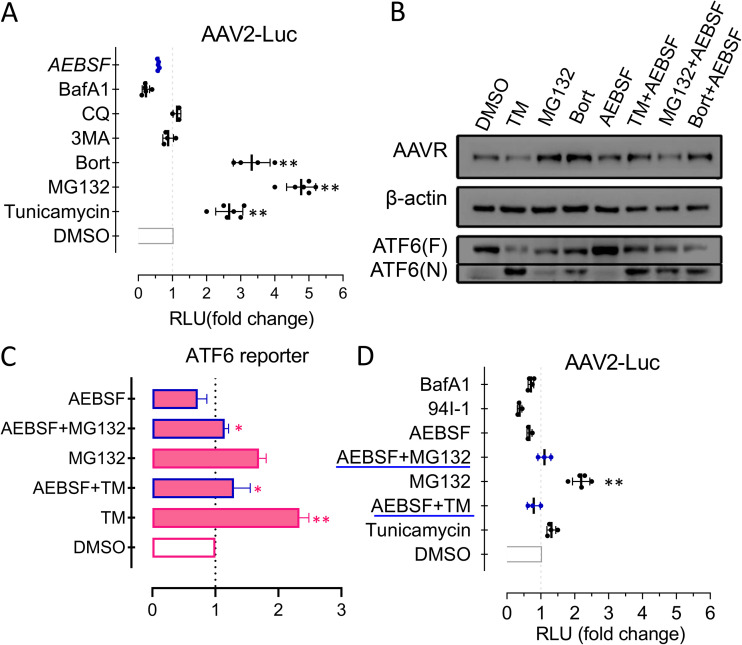
To explore the impact of AAVR in UPR signal modulation during AAV infection, we compared wild-type AAV2 and an AAV2 G265E-N268Q mutant (lacking receptor binding) in HeLa cells, with or without different chemical treatments. Cleaved ATF6 appeared during a 2- to 8-h infection time course and was accompanied by increased BIP and GRP94 protein levels (Fig. 4A). No significant changes were identified in CHOP protein or XBP1s mRNA levels (Fig. 4A). These results suggested that the UPR-related activation of ATF6 signaling was dependent on AAVR binding to its ligand and could be a physiological requirement during AAV2 infection. Our semiquantitative assays (Fig. 4B) also appeared to support this theory. Following treatment with various ER stress inducers (Fig. 4C), including TM, PUGNAc, TG, etc., we noted that AAV2-induced ATF6 activation was enhanced, whereas in AAV2 G265E-N268Q cells, it was not (Fig. 4E). However, AEBSF (blocks ATF6 cleavage) significantly inhibited AAV2 transduction. Similar changes were observed when GRP94 inhibitior-1, a GRP94-selective inhibitor (26), was used (Fig. 4D). Thus, mutant AAV2 G265E-N268Q failed to respond to GRP94 inhibitor-1 treatment in reporter activities for the quantification of transduction efficiency. These results suggested that AAV2 exploited the UPR, as provoked by ATF6 activation signaling, rather than the CHOP and XBP1 pathways. Also, modulating ATF6 and other downstream molecules could facilitate a natural AAV infection route.
FIG 4.
AAV2 infection alters ATF6-mediated unfolded protein response (UPR) signaling and activates transduction in HeLa cells. (A) Western blots showing ATF6, CHOP, and XBP1 expression during AAV2 infection time courses. (B) Quantification of data from panel A by densitometry (n = 3). (C) Comparison of AAV2 with a nonreceptor binding mutant, G265D-N268Q, for transduction in HeLa cells following treatments of various reagents of UPR modulation. (D) Comparison of infection of AAV2 and the G265D-N268Q mutant following inhibition of ATF6 or downstream GRP94. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
Alterations in ATF6 signaling influence AAV2 transduction.
From previous studies, particular agents (i.e., proteasome inhibitors) potently promoted AAV transduction efficiency (21). However, these mechanistic investigations did not identify correlations with virus degradation. We speculated if drug-induced UPR effects could enhance AAV infection. HeLa cell treatment with TM, MG132, and Bort significantly enhanced AAV transduction (Fig. 5A). Using these treatments, ATF6 activation levels appeared to be correlated with cleaved ATF6 fragments seen using Western blotting (Fig. 5B). In addition, 3-MA and BAFA1, which mainly trigger ER stress by inhibiting autophagy without a requirement for ATF6 signaling (data not shown), appeared less effective in promoting AAV infection (Fig. 5A). To further evaluate the effects of AEBSF on blocking ATF6 cleavage, ATF6 luciferase reporter assays (Fig. 5C) were conducted to examine AAV2 transduction efficiency (Fig. 5D) in different combinations. We identified a significant correlation between AAV transgene expression and ATF6 activity, and also, AEBSF inhibited AAV transduction under broad conditions. These data demonstrated that the effects on activation of ATF6 signal could indeed influence the AAV infection efficiency of AAV.
FIG 5.
The effects of ATF6-mediated unfolded protein response following treatment with different reagents to modulate AAV2 transduction. (A) AAV2 infection of a luciferase reporter in HeLa cells treated with reagents to assess AAV transduction. (B) Western blots showing ATF6 activation comparing AEBSF treatments (with/without) to inhibit ATF6 cleavage. (C) Reporter assays for ATF6 activation. (D) AAV2 transduction efficiency in HeLa cells. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
Activation of ATF6-mediated UPR promotes AAV2 transduction in primary hepatocytes.
We next investigated the effects of ATF6 activation via AAV2 infection on AAV transduction in primary monkey hepatocytes. From our luciferase reporter assays, ATF6 was significantly activated in cells after AAV2 infection for 6 h (Fig. 6A). No significant differences were identified in CHOP and XBP1 activation levels (Fig. 6A). Thus, AAV2 induced UPR primarily through ATF6 activation and was dependent on AAVR. As shown (Fig. 6B), cleaved ATF6 N-terminal protein levels increased after 1 h when induced by AAV2 infection, together with increased BIP levels. Under AAVR knockdown conditions (Fig. 6C), TM-induced ATF6 signals were enhanced similarly to aavr– HeLa cells. Luciferase reporter assay data for XBP1 and CHOP were also altered following TM treatment; therefore, we theorized these may have been compensatory effects due to AAVR depletion in primary cells. For hepatocytes subjected to chemical treatment, AEBSF inhibition of ATF6 downstream was found to reduce AAV transduction under all experimental conditions, and the correlation in AAV transgene expression (Fig. 6D) and ATF6 activity (Fig. 6E) was maintained. These data verified our previous finding in HeLa cells and indicated that ATF6 signal activation involved AAV infection, again suggesting that the transduction efficiency of AAV2 could be altered by modulating ATF6 and downstream signals in primary hepatocytes.
FIG 6.
Activation of ATF6-mediated unfolded protein response and AAV2 transduction in monkey primary hepatocytes. (A) Reporter assays for ATF6, CHOP, and XBP1 activation in monkey primary hepatocytes following AAV2 infection for 6 h. (B) Western blot and densitometric quantification of ATF6, BIP, and GRP94 during AAV2 infection time courses. (C) Luciferase reporter assays showing ATF6, CHOP, and XBP1 activation. Monkey primary hepatocytes were transfected with AAVR-targeted short hairpin RNA (shRNA) or a scrambled control(sh-scream) for 24 h. (D) Reporter assays showing ATF6 activation in monkey primary hepatocytes treated with reagents. (E) AAV2 infection of a luciferase reporter in monkey primary hepatocytes. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
ATF6 activation is involved in AAV5 transduction in HeLa cells.
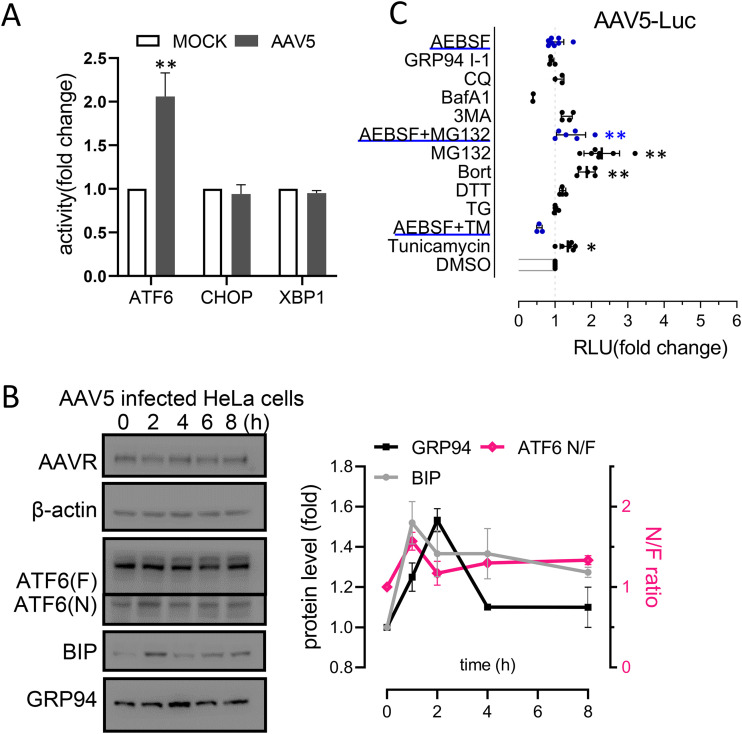
As AAVR binds the AAV5 capsid, we evaluated ATF6 signaling during AAV5 infection in HeLa cells. Luciferase reporter assays showed a significant increase in ATF6 activation at 6 h postinfection (Fig. 7A). Western blotting indicated that ATF6 N-terminal fragment levels were also increased, correlating with increased BIP and GRP94 expression levels (Fig. 7B). Various chemical treatments altered AAV5-mediated transgenic expression (Fig. 7C), which correlated well with ATF6 activity as previously shown in HeLa cells (Fig. 5C). These results suggested that ATF6 signal activation, which was dependent on AAVR, also facilitated the transduction efficiency of AAV5.
FIG 7.
Activation of ATF6-mediated unfolded protein response facilitates AAV5 transduction in HeLa cells. (A) Reporter assays showing ATF6, CHOP, and XBP1 activation in HeLa cells at 6 h post-AAV5 infection. (B) Western blot and densitometric quantification of ATF6, BIP, GRP94, and CHOP during AAV5 infection time courses. (C) AAV5 transduction in HeLa cells treated with various reagents that modulate AAV transduction. Data are presented as the mean ± standard deviation (n ≥ 3). Statistical significance was determined using the unpaired Student’s t test. *, P < 0.05; **, P < 0.01.
DISCUSSION
The ER contains many chaperone proteins that facilitate the proper conformational folding and modification (if required) of newly synthesized peptides. ER dysfunction leads to the accumulation of misfolded peptides or, occasionally, a massive influx of exogenous proteins which trigger UPR-mediated ER homeostasis and the restoration of normal cell functions (27). UPR failure to respond to ER stress endangers cell survival; therefore; this pathway is key for cell homeostasis, and also viral infection (23). Earlier studies of AAVR functions, although few in number, suggested that limited AAVR expression damaged cell robustness in terms of adapting to stress environments. Since AAVR predominantly localized in the ER/Golgi (10), and ER stress with activation of the UPR can be a very frequent cause of cell stresses (23), it was plausible that AAVR could have roles in ER function and UPR regulation. As ER stress regulation involves multiple pathways, with possible redundancy and cross talk, the technical dissection of these processes was highly challenging. However, we not only detected up- and downstream factors in major UPR pathways by Western blotting, but we also used luciferase-based reporter quantitative assays in host cell transfections. Our approach greatly facilitated a greater understanding of the terminal and PKD domains of the AAVR protein. We postulated that, as a gene of long history and a protein of localization, AAVR could serve as a native regulator (or a participant) of ER homeostatic maintenance. AAV infection could utilize such a function of AAVR and be partially used for transduction activation. This could elucidate why different AAV serotypes use a common receptor and behave similarly during uncoating and transduction processes after cell internalization.
In cell biology, viral infection is closely related to UPR processes. Herpes simplex virus, cytomegalovirus, hepatitis C virus, and others induce ER stress and activate UPR signaling pathways (28–31). However, the UPR pathway appears to be required to establish productive infection of many viruses, such as Japanese encephalitis virus, Tula virus, and Zika virus (32–35). The interplay of ER stress signals with viral entry is more frequently observed for coated viruses, such as influenza virus and rotavirus, which differentially cross-activate UPR mechanisms (36–38). As a primary choice for gene therapy vectors, whether AAV infection alters UPR signals in host cells remains poorly characterized, as most target tissues tolerate AAV infection even at high doses (multiplicity of infection/cell measures), where changes in intracellular trafficking profiles were observed (39). Balakrishnan et al. observed IRE1α and PERK signal activation at 12 to 48 h after AAV2 infection (22). We observed that UPR alterations occurred much quicker during AAV infection. In our studies, the induction of the ATF6 signaling pathway occurred 1 to 8 h after the infection of AAV2 (Fig. 4). AAVR is a high-affinity receptor for multiple AAV serotypes; it mediates postattachment events for viral entry and rapidly undergoes endocytosis from the plasma membrane to the TGN (40, 41). This allows the virus to mimic UPR conditions with the AAVR in the loaded state and release its inhibition on ATF6 signal, which tricks host cells into activating the expression of downstream genes to facilitate AAV transduction. This suggests that the tissue tropism of different AAV serotypes not only depends on the abundance of receptors or coreceptors, but is also determined by ATF6 activation toward tissue-specific AAV-facilitating genes. Thus, profiling ATF6 activity or analyzing the expression of downstream genes could sufficiently and dynamically approximate AAV infection behaviors in both in vivo and in vitro applications. The underlining molecular mechanism needs to be further studied.
The ER is a highly dynamic organelle, with complex functions significantly influenced by multiple parameters, both inside the cell and the local microenvironment. In addition, complex cross talk during ER stress signaling modules also leads to the instability of endoplasmic reticulum signal and the difficulty of accurate quantification (23, 42). Moreover, experimental transfection procedures can induce transient disruption of cell measurements, especially when performing chemical stimulation studies or investigating cell models. These issues contributed to study limitations and complexity. For example, our data were often equivocal in terms of what was usually expected from, e.g., reporter assays or mutant analyses. Our data showed the N-terminal ectopic and C-tail intravehicle portions were important for inhibiting UPR to different extents (Fig. 3). Using our data, we were unable to definitively assign precise functional contribution percentages for each PKD domain. Thus, multiple AAVR protein segments appeared to contribute to ATF6 signal modulation, as many chaperone proteins, especially PKD family members, do not contain kinase-like domains for specific activities. Maximum AAVR activity appears to require an intact protein structure, similar to AAV2, which mainly interfaces with PKD2, whereas AAV5 contacts the PKD1 domain of AAVR. Other serotypes (including AAV1 and AAV8) require a combination of PKD1 and PKD2 for efficient viral transduction, but both AAV1 and AAV8 bind only to PKD2 in virus overlay assays (11, 15, 16). AAVR is a multiserotype receptor for AAVs and is the first example of a single receptor responsible for multiple viral strains. Although divergent interactions exist between multiple virus strains and AAVR, the PKD 1 to 3 domains appear to be important and necessary. Coincidentally, PKD 1 to 3 were required for the inhibiting activity of AAVR in ATF6 signal (Fig. 3), suggesting an intracellular function for PKD 1 to 3 as well as an interface for AAV binding. The mechanism of AAVR inhibiting UPR, such as the interaction with ER chaperones or specific kinase of ER stress signal, requires investigation in future studies.
An unresolved issue was the role of AAVR in ER stress regulation, which also affected non-AAVR-dependent transduction. Also, how do variations in AAVR binding affinities translate to altered ATF6 activation? The strength of the UPR was provoked differentially by the distinct AAV serotypes during their intracellular trafficking (21, 39), which could be due to the different binding properties between AAV and AAVR and the different suppression effect released from the binding. Among the 13 distinct AAV serotypes, AAV2 is traditionally used in broad tissue tropisms (3). However, recent research demonstrated that AAV1 was more efficient in transducing muscle than AAV2 (43). It was speculated that this higher transduction efficiency was due to AAV1’s remarkably high AAVR binding affinity (13.9 nM) compared with that of AAV2 (57 nM) (15, 16). AAV5 is distinct among the parvoviruses, as it was originally isolated from a patient sample and has unique sequences and biochemical features (44, 45). AAV2 exhibited 6- to 32-fold higher transduction efficiencies than AAV5 in COS, HEK293, HeLa, IB3, and MCF7 cells, but both viruses exhibited poor transduction efficiencies in NIH 3T3, SKBR3, and T-47D cell lines (44). These features may be related to the binding of virus particles to the AAVR or the inhibitory function of AAVR toward ATF6 signaling, which may regulate infectious pathways and efficiency.
We used a spectrum of small molecule reagents to either induce ER stress via UPR or modulate AAV transduction. In fact, a significant fraction of these reagents displayed both functions. The regulation of ER stress by proteasome inhibitors in cellular models was shown to influence AAV transduction (21). Remarkably, blocking the ATF6 pathway by inhibiting ATF6 hydrolysis or activating GRP94 significantly reduced the infection efficiency of AAV (Fig. 4). Correspondingly, promoting ATF6 signaling by proteasome inhibitors (TM, MG132, and Bort) significantly increased the infection efficiency of AAV (Fig. 5). This finding suggests that proteasome inhibitors benefit AAV transduction through the regulation of activation of ATF6 signaling instead of preventing the protein degradation associated with ER stress. Notably, the chemical stimulation inhibiting the activation of ATF6 signaling suppressed AAV infection efficiency (Fig. 5D), suggesting a novel and effective strategy to regulate AAV transduction. We demonstrated that effects of suppressing ATF6 signal on attenuating AAV transduction were consistent with different chemical treatments. However, it remains to be seen if enhanced AAV infection in ATF6 mutant models can be achieved. These observations provide new insights on the search for compounds which could facilitate improved AAV trafficking and transduction in human gene therapy applications.
Deciphering the exact role of ATF6 on AAV transduction required further comprehensive and in-depth mechanistic explorations. Due to the limitation of the present study, several important questions remained to be unanswered, including finding the interactive protein partner that is directly in contact with AAVR and its specific domains and identifying the key participant involved in ATF6 upstream signal molecules and specific downstream regulated genes. More elegant or advanced technology may be of great help to address the unsolved issues, such as applications of AAVR knockout (or humanized knock-in mutant) transgenic mouse models, establishing single cell clones with different AAVR-KD levels, and conducting high-throughput screening experiments involving immunoprecipitation mass spectrum and sequencing technologies.
Conclusions.
AAVR participated in the regulation of UPR processes and relieving ER stress, specifically by suppressing ATF6 activation during different chemical stimulation assays. The binding of AAV capsids to AAVR during infection released the inhibition of AAVR on ATF6 cleavage for activation. ATF6 pathway and downstream signaling suppression appeared to reduce AAV transduction efficiency. These observations provide invaluable cell biological insights on the physiological role of AAVR, transduction activation, and intracellular trafficking during AAV infection.
MATERIALS AND METHODS
Tissue culture and cell treatments.
The human embryonic kidney cell line HEK293T and the human cervical carcinoma cell line HeLa were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Cryopreserved male cynomolgus monkey (Macaca fascicularis) hepatocytes were purchased from Milestone Biotechnologies (Shanghai, China). A stable AAVR knockdown HeLa cell line was generated as described previously (15). Briefly, shRNA sequences targeting AAVR were designed and cloned into the pHS-GFP-2A-Puro plasmid. These shRNA sequences are shRNA-1, 5′ AGATGTCTGCCAATAATATCC3′, and shRNA-2, 5′GAACTGAGGTGACACAATAGC3′. Cells treated with a scram-sequence were used as controls. HeLa cells were seeded in 12-well plates and transfected with 1 μg shRNA plasmid using 3 μL polyethylenimine (PEI) transfection reagent (Invitrogen) (1 mg mL−1) in 50 μL Dulbecco modified Eagle medium (DMEM; Gibco) per well. The medium was changed after 4 h, and cells were incubated for another 48 h before immunoblotting. Stably transfected cells were selected with 1.5 μg/mL puromycin (Sigma-Aldrich) over 2 weeks until AAVR-silenced strains were generated. HeLa parental and knocked down cells were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco) at 37°C in 5% CO2.
Monkey hepatocytes were seeded in seeding medium in precoated 24-well plates at 6 × 104 cells/well. After 6 h, the medium was changed to culture medium according to the manufacturer’s instructions (Milestone Biotechnologies). TM (HY-A0098; MCE), thapsigargin (HY-13433; MCE), dithiothreitol (HY-15917; MCE), 3-MA (189490; Merck), chloroquine (HY-17589A; MCE), MG132 (C2211; Merck), bortezomib (Bort) (HY-10227; MCE), melatonin (HY-B0075; MCE), AEBSF hydrochloride (AEBSF) (HY-12821; MCE), GRP94 inhibitor-1 (HY-112910; MCE), and PUGNAc (A7229; Merck) were used to evaluate AAVR biological functions.
Vector preparation and transfection.
AAVR cDNA was cloned into the pcDNA3.1(+)-Myc tag vector to generate the Myc tagged-AAVR expression construct, as described previously (15). Constructs of c0319 (containing the N-terminal 1 to 982 amino acids [aa] of AAVR and C-tail 1,006 to 1072 aa of KIAA0319), cPVR (containing the N-terminal 1 to 982 aa of AAVR and C-tail 344 to 417 aa of poliovirus receptor), and n0139 (containing the N-terminal 1 to 1,005 aa of KIAA0319 and C-tail 983 to 1,049 aa of AAVR) were PCR amplified and subcloned into pcDNA3.1/CT-Myc as Myc fusion proteins. PKD 1 to 5 domain sequences were amplified and subcloned into pcDNA3.1 to generate the AAVR mutants s41235 and s14253 using NEBuilder HiFi DNA assembly master mix (E2621S; New England Biolabs, UK). Cells were transfected with expression plasmids or shRNA constructs using PEI as described. At 48 h posttransfection, Western blotting was performed to assess protein levels from corresponding genes.
pGL3-ATF6-Luc, pGL3-CHOP-Luc, or pGL3-XBP1-Luc reporters were used in HeLa cells to determine the transactivation of ATF6, CHOP, and XBP1 induced by ER stress inducers. pGL3-ATF6-Luc was purchased from Yeasen (11533ES03; Yeasen, China) and contained the ATF6 binding sequence, 5′CTCGAGACAGGTGCTGACGTGGCATTC3′. pGL3-CHOP-Luc and pGL3-XBP1-Luc were constructed based on previous reports (46–48). Briefly, the XBP-1 promoter (GenBank accession no. NG_012266, −330 to +129 region; these are nucleotide positions relative to the transcription start site) and the CHOP promoter (GenBank accession no. NG_027674, −870 to +17) were amplified from HeLa genomic DNA and cloned into the KpnI-BglI restriction sites of pGL3 (Promega), which contained firefly luciferase coding sequence but lacked eukaryotic promoter or enhancer elements.
Virus production and purification.
The packaging and purification of AAV2 or AAV5 was described previously (15). Briefly, a triple-plasmid transfection (see below) was performed to generate recombinant AAV2 or AAV5. HEK293T cells were cultured in 150-mm plates and at 80% confluence; they were transfected with a pHelper plasmid (12 μg), pRC5/pRC2/pRC2-G265E-N268Q plasmids (10 μg), and pAV plasmid (6 μg). The pRC2-G265E-N268Q plasmid was generously provided by Z. Lou (15) and contained two mutated AAVR-AAV2 compound binding sites. At 72 h posttransfection or when cytopathic effects were observed, cells were centrifuged at 1,000 × g for 30 min at 4°C, and the pellet was resuspended in buffer containing 10 mM Tris-HCl (pH 8.0). This suspension was subjected to four freeze-thaw cycles in dry ice/ethanol and a 37°C water bath. Cell debris was sonicated and digested in DNase I (200 units in 1.5 mL 1% sodium deoxycholate) plus 0.05% trypsin for 1 h at 37°C. Following centrifugation at 10,000 × g for 10 min at 4°C, the supernatant was collected as an AAV crude lysate.
This crude lysate was diluted in 10 mM Tris-HCl (pH 8.0) to a final volume of 4 mL and bottom-loaded in a discontinuous gradient of CsCl (1.25 g/cm3) and CsCl (1.50 g/cm3) in an 8.9-mL ultracentrifuge tube (3621623; OptiSeal). After ultracentrifugation at 200,000 × g for 8 h at 4°C, 1000-μL fractions were collected. The titer-abundant fractions were combined and desalted using a 3-kDa cutoff ultrafiltration kit (4 mL; Millipore). Fraction titers were determined by quantitative PCR as previously described and stored at −80°C.
Cell viability.
We seeded 5 × 103 cells/well in 96-well plates and cultured the cells for 1 to 3 days. Cell viability was determined using a cell counting kit (KeyGen Biotech, China) according to the manufacturer’s protocols. After processing, plates were scanned at 450 nm in a spectrophotometer (BioTek). Each final data point generated an average from six replicates. Experiments were independently repeated three times.
Western blotting.
Western blotting was performed as previously described (15). Briefly, cell lysates were separated using 10% SDS-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes. Probing antibodies were against the following antigens: KIAA0319 (ab105385; Abcam), Myc (2276; Cell Signaling Technology [CST]), Phospho-eIF2α (Ser51) (3398; CST), eIF2α (D7D3) (5324; CST), CHOP (60304; Proteintech), GRP78/BIP (11587; Proteintech), GRP94 (14700; Proteintech), β-actin (66009; Proteintech), and ATF-6α (sc-166659; Santa Cruz).
Immunofluorescence.
HeLa cells, pretransfected with AAVR and AAVR mutant vectors, were seeded onto glass coverslips in 24-well plates at 5.0 × 104 per well. Cells were fixed, permeabilized, blocked, and finally washed in phosphate-buffered saline. Then, cells were probed with an antimyc marker and a trans-Golgi marker, TGN46 (Novus Biologicals). After incubation with an Alexa Fluor 488 (Life Technologies) and Alexa Fluor 594 secondary antibody (Life Technologies), cells were nuclear-stained with Hoechst 33258 (Sigma-Aldrich). Finally, cells were visualized using a confocal microscope (TCS SP8; Leica Microsystems, Germany). Control samples, without primary antibody, were prepared to assess nonspecific noise levels.
Dual-luciferase reporter assay.
Cells were preseeded in 48-well plates for 24 h and transfected with pGL3-ATF6-Luc, pGL3-CHOP-Luc, or pGL3-XBP1-Luc plasmids. Then, cells were assayed for luciferase activity using the Dual-Glo Luciferase assay system (Promega) according to the manufacturer’s instructions. Firefly luciferase activities were normalized to Renilla luciferase activities. Data from at least three independent experiments were used for analysis.
RNA extraction and RT-PCR.
Total RNA was isolated using TRIzol (Life Technologies, Carlsbad, CA), and the HiScript II Q reverse transcriptase (RT) kit (Vazyme, Nanjing, China) was used for reverse transcription. PCR assays were conducted to amplify XBP1u and XBP1s. The XBP1u primers were forward 5′-CTTTTGCTAGAAAATCAGCTTTTACGAG-3′ and reverse 5′- AGAGGTGCACGTAGTCTGAGTGC-3′. The XBP1s primers were forward 5′-TTGCTGAAGAGGAGGCGGAAGCC-3′ and reverse 5′- CCTGCACCTGCTGCGGACT-3′. β-actin was used as a reference gene (forward 5′- CATGTACGTTGCTATCCAGGC-3′ and reverse 5′- CTCCTTAATGTCACGCACGAT-3′).
AAV infections and luciferase reporter assays.
Recombinant AAV2 and AAV5 vectors expressing luciferase expression cassettes were used to evaluate viral transduction efficiency. At least 12 h prior to infection, HeLa cells or monkey hepatocytes were seeded in 24-well plates at 80% confluence (approximately 5 × 105 cells per well). Purified AAV5-Luc and AAV2-Luc, in either wild-type or binding ability-lose mutant AAV2-Luc (AAV2 G265E-N268Q), at 2,000 vg/cell were used to infect cells over 24 h. Luciferase activities were then measured as described. Results from at least three independent experiments were normalized to infected cell numbers and plotted as the mean ± standard error.
Statistical analysis.
Analysis of variance (ANOVA) was used to determine statistical significance in multiple groups. Student’s t test was used to compare cell functions between paired groups. A P value of <0.05 indicated statistical significance. Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA) was used to plot the data.
ACKNOWLEDGMENTS
We thank Z. Lou and R. Zhang for providing plasmids.
This work was supported by the National Natural Science Foundation of China (no. 82073215).
W.D. and S.C. conceived the project and designed the experiments. M.C., Q.Z., J.W., and Y.S. performed the experiments. W.D., S.C., and M.C. analyzed the data. W.D., S.C., and M.C. wrote the manuscript. All authors discussed the experiments and read and approved the final manuscript.
Contributor Information
Shan Cheng, Email: chengs@ccmu.edu.cn.
Wei Ding, Email: weiding@ccmu.edu.cn.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA 99:11854–11859. 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, Goodspeed K, Gray SJ, Kay CN, Boye SL, Boye SE, George LA, Salabarria S, Corti M, Byrne BJ, Tremblay JP. 2021. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther 29:464–488. 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hacker UT, Bentler M, Kaniowska D, Morgan M, Büning H. 2020. Towards clinical implementation of adeno-associated virus (AAV) vectors for cancer gene therapy: current status and future perspectives. Cancers 12:1889. 10.3390/cancers12071889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruntman AM, Flotte TR. 2018. The rapidly evolving state of gene therapy. FASEB J 32:1733–1740. 10.1096/fj.201700982R. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2017. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol 91:e00710-17. 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 72:1438–1445. 10.1128/JVI.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. 2005. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol 79:609–614. 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med 5:71–77. 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 10.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, Deng X, Wosen JE, Davulcu O, Yan Z, Engelhardt JF, Brown KE, Chapman MS, Qiu J, Carette JE. 2017. Adeno-associated virus (AAV) serotypes have distinctive interactions with domains of the cellular AAV receptor. J Virol 91:e00391-17. 10.1128/JVI.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platt MP, Adler WT, Mehlhorn AJ, Johnson GC, Wright KA, Choi RT, Tsang WH, Poon MW, Yeung SY, Waye MM, Galaburda AM, Rosen GD. 2013. Embryonic disruption of the candidate dyslexia susceptibility gene homolog Kiaa0319-like results in neuronal migration disorders. Neuroscience 248:585–593. 10.1016/j.neuroscience.2013.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibraghimov-Beskrovnaya O, Bukanov NO, Donohue LC, Dackowski WR, Klinger KW, Landes GM. 2000. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Hum Mol Genet 9:1641–1649. 10.1093/hmg/9.11.1641. [DOI] [PubMed] [Google Scholar]
- 14.Bycroft M, Bateman A, Clarke J, Hamill SJ, Sandford R, Thomas RL, Chothia C. 1999. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J 18:297–305. 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Cao L, Cui M, Sun Z, Hu M, Zhang R, Stuart W, Zhao X, Yang Z, Li X, Sun Y, Li S, Ding W, Lou Z, Rao Z. 2019. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat Microbiol 4:675–682. 10.1038/s41564-018-0356-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Xu G, Cao L, Sun Z, He Y, Cui M, Sun Y, Li S, Li H, Qin L, Hu M, Yuan Z, Rao Z, Ding W, Rao Z, Lou Z. 2019. Divergent engagements between adeno-associated viruses with their cellular receptor AAVR. Nat Commun 10:3760. 10.1038/s41467-019-11668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer NL, Hu G, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich DS, Trzynka A, David L, Stagg SM, Chapman MS. 2019. Structure of the gene therapy vector, adeno-associated virus with its cell receptor, AAVR. Elife 8:e44707. 10.7554/eLife.44707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakashima T, Miura M, Hara M. 2000. Tetrocarcin A inhibits mitochondrial functions of Bcl-2 and suppresses its anti-apoptotic activity. Cancer Res 60:1229–1235. [PubMed] [Google Scholar]
- 19.Fribley A, Wang CY. 2006. Proteasome inhibitor induces apoptosis through induction of endoplasmic reticulum stress. Cancer Biol Ther 5:745–748. 10.4161/cbt.5.7.2971. [DOI] [PubMed] [Google Scholar]
- 20.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14:1435–1455. 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolson SC, Li C, Hirsch ML, Setola V, Samulski RJ. 2016. Identification and validation of small molecules that enhance recombinant adeno-associated virus transduction following high-throughput screens. J Virol 90:7019–7031. 10.1128/JVI.02953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakrishnan B, Sen D, Hareendran S, Roshini V, David S, Srivastava A, Jayandharan GR. 2013. Activation of the cellular unfolded protein response by recombinant adeno-associated virus vectors. PLoS One 8:e53845. 10.1371/journal.pone.0053845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hetz C, Zhang K, Kaufman RJ. 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21:421–438. 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. 2013. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol 5:a013169. 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, Luís A, McCarthy N, Montibeller L, More S, Papaioannou A, Püschel F, Sassano ML, Skoko J, Agostinis P, de Belleroche J, Eriksson LA, Fulda S, Gorman AM, Healy S, Kozlov A, Muñoz-Pinedo C, Rehm M, Chevet E, Samali A. 2019. Endoplasmic reticulum stress signalling: from basic mechanisms to clinical applications. FEBS J 286:241–278. 10.1111/febs.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang F, Guo AP, Xu JC, You QD, Xu XL. 2018. Discovery of a potent Grp94 selective inhibitor with anti-inflammatory efficacy in a mouse model of ulcerative colitis. J Med Chem 61:9513–9533. 10.1021/acs.jmedchem.8b00800. [DOI] [PubMed] [Google Scholar]
- 27.Senft D, Ronai ZA. 2015. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci 40:141–148. 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinte F, van Anken E, Tirosh B, Brune W. 2020. Repression of viral gene expression and replication by the unfolded protein response effector XBP1u. Elife 9:e51804. 10.7554/eLife.51804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Kang R, Huang H, Xi X, Wang B, Wang J, Zhao Z. 2014. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway-mediated MAP1LC3B and ATG12 expression. Autophagy 10:766–784. 10.4161/auto.27954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinte F, Müller J, Brune W. 2021. Viral mediated tethering to SEL1L facilitates endoplasmic reticulum-associated degradation of IRE1. J Virol 95:e01990-20. 10.1128/JVI.01990-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sir D, Chen W-L, Choi J, Wakita T, Yen TSB, Ou J-HJ. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061. 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya S, Sen U, Vrati S. 2014. Regulated IRE1-dependent decay pathway is activated during Japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J Gen Virol 95:71–79. 10.1099/vir.0.057265-0. [DOI] [PubMed] [Google Scholar]
- 33.Su HL, Liao CL, Lin YL. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol 76:4162–4171. 10.1128/jvi.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y, Lin Q, Huo Z, Chen C, Zhou S, Ma X, Gao H, Lin Y, Li X, He J, Zhang P, Liu C. 2020. Inositol-requiring enzyme 1α promotes Zika virus infection through regulation of stearoyl coenzyme A desaturase 1-mediated lipid metabolism. J Virol 94:e01229-20. 10.1128/JVI.01229-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XD, Lankinen H, Putkuri N, Vapalahti O, Vaheri A. 2005. Tula hantavirus triggers pro-apoptotic signals of ER stress in Vero E6 cells. Virology 333:180–189. 10.1016/j.virol.2005.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao D, Xue R, Zhang M, Lu C, Ma T, Ren C, Zhang T, Yang J, Teng Q, Li X, Li Z, Liu Q. 2021. N-linked glycosylation plays an important role in budding of neuraminidase protein and virulence of influenza viruses. J Virol 95:e02042-20. 10.1128/JVI.02042-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trujillo-Alonso V, Maruri-Avidal L, Arias CF, López S. 2011. Rotavirus infection induces the unfolded protein response of the cell and controls it through the nonstructural protein NSP3. J Virol 85:12594–12604. 10.1128/JVI.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duarte M, Vende P, Charpilienne A, Gratia M, Laroche C, Poncet D. 2019. Rotavirus infection alters splicing of the stress-related transcription factor XBP1. J Virol 93:e01739-18. 10.1128/JVI.01739-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JS, Gentzsch M, Zhang L, Ribeiro CM, Kantor B, Kafri T, Pickles RJ, Samulski RJ. 2011. AAV exploits subcellular stress associated with inflammation, endoplasmic reticulum expansion, and misfolded proteins in models of cystic fibrosis. PLoS Pathog 7:e1002053. 10.1371/journal.ppat.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madigan VJ, Berry GE, Tyson TO, Nardone-White D, Ark J, Elmore ZC, Murlidharan G, Vincent HA, Asokan A. 2020. The Golgi calcium ATPase pump plays an essential role in adeno-associated virus trafficking and transduction. J Virol 94:e01604-20. 10.1128/JVI.01604-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry GE, Asokan A. 2016. Cellular transduction mechanisms of adeno-associated viral vectors. Curr Opin Virol 21:54–60. 10.1016/j.coviro.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Teuber V, Albert-Gasco H, Auyeung VC, Papa FR, Mallucci GR, Hetz C. 2019. Small molecules to improve ER proteostasis in disease. Trends Pharmacol Sci 40:684–695. 10.1016/j.tips.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Xiao W, Chirmule N, Berta SC, McCullough B, Gao G, Wilson JM. 1999. Gene therapy vectors based on adeno-associated virus type 1. J Virol 73:3994–4003. 10.1128/JVI.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiorini JA, Kim F, Yang L, Kotin RM. 1999. Cloning and characterization of adeno-associated virus type 5. J Virol 73:1309–1319. 10.1128/JVI.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callaway HM, Feng KH, Lee DW, Allison AB, Pinard M, McKenna R, Agbandje-McKenna M, Hafenstein S, Parrish CR. 2017. Parvovirus capsid structures required for infection: mutations controlling receptor recognition and protease cleavages. J Virol 91:e01871-16. 10.1128/JVI.01871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20:6755–6767. 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park JS, Luethy JD, Wang MG, Fargnoli J, Fornace AJ Jr, McBride OW, Holbrook NJ. 1992. Isolation, characterization and chromosomal localization of the human GADD153 gene. Gene 116:259–267. 10.1016/0378-1119(92)90523-R. [DOI] [PubMed] [Google Scholar]
- 48.Ponath PD, Fass D, Liou HC, Glimcher LH, Strominger JL. 1993. The regulatory gene, hXBP-1, and its target, HLA-DRA, utilize both common and distinct regulatory elements and protein complexes. J Biol Chem 268:17074–17082. 10.1016/S0021-9258(19)85304-8. [DOI] [PubMed] [Google Scholar]