ABSTRACT
Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 is an RNA-binding posttranscriptional regulator. We recently applied an affinity-purified anti-ORF57 antibody to conduct ORF57 cross-linking immunoprecipitation (CLIP) in combination with RNA-sequencing (CLIP-seq) and analyzed the genome-wide host RNA transcripts in association with ORF57 in BCBL-1 cells with lytic KSHV infection. Mapping of the CLIP RNA reads to the human genome (GRCh37) revealed that most of the ORF57-associated RNA reads were from rRNAs. The remaining RNA reads mapped to several classes of host noncoding and protein-coding mRNAs. We found that ORF57 binds and regulates expression of a subset of host long noncoding RNAs (lncRNAs), including LINC00324, LINC00355, and LINC00839, which are involved in cell growth. ORF57 binds small nucleolar RNAs (snoRNAs) responsible for 18S and 28S rRNA modifications but does not interact with fibrillarin or NOP58. We validated ORF57 interactions with 67 snoRNAs by ORF57 RNA immunoprecipitation (RIP)-snoRNA array assays. Most of the identified ORF57 rRNA binding sites (BS) overlap the sites binding snoRNAs. We confirmed ORF57-snoRA71B RNA interaction in BCBL-1 cells by ORF57 RIP and Northern blot analyses using a 32P-labeled oligonucleotide probe from the 18S rRNA region complementary to snoRA71B. Using RNA oligonucleotides from the rRNA regions that ORF57 binds for oligonucleotide pulldown-Western blot assays, we selectively verified ORF57 interactions with 5.8S and 18S rRNAs. Polysome profiling revealed that ORF57 associates with both monosomes and polysomes and that its association with polysomes increases PABPC1 binding to polysomes but prevents Ago2 association with polysomes. Our data indicate a functional correlation with ORF57 binding and suppression of Ago2 activities for ORF57 promotion of gene expression.
IMPORTANCE As an RNA-binding protein, KSHV ORF57 regulates RNA splicing, stability, and translation and inhibits host innate immunity by blocking the formation of RNA granules in virus-infected cells. In this study, ORF57 was found to interact with many host noncoding RNAs, including lncRNAs, snoRNAs, and rRNAs, to carry out additional unknown functions. ORF57 binds a group of lncRNAs via the RNA motifs identified by ORF57 CLIP-seq to regulate their expression. ORF57 associates with snoRNAs independently of fibrillarin and NOP58 proteins and with rRNA in the regions that commonly bind snoRNAs. Knockdown of fibrillarin expression decreases the expression of snoRNAs and CDK4 but does not affect viral gene expression. More importantly, we found that ORF57 binds translationally active polysomes and enhances PABPC1 but prevents Ago2 association with polysomes. Data provide compelling evidence on how ORF57 in KSHV-infected cells might regulate protein synthesis by blocking Ago2’s hostile activities on translation.
KEYWORDS: Ago2, ORF57, Kaposi’s sarcoma-associated herpesvirus, lincRNA, snoRNA, polysomes, rRNA
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV-8), is a DNA tumor virus of lymphotropic gammaherpesvirus subfamily (1, 2). KSHV is the causative agent for the development of Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman’s disease (MCD) (3–5). KSHV displays two phases of infection: a short lytic phase and a persistent latent infection (6). During the latent infection, a few viral proteins are expressed to maintain the KSHV genome in the infected cells. However, the lytic KSHV infection can be reactivated from the cells with latent KSHV infection under a hypoxia or redox condition or by chemical induction. Expression of a viral replication and transcription activator (RTA; ORF50) during reactivation induces the cascaded expression of viral lytic genes (7, 8), including KSHV ORF57, for viral DNA replication and efficient virus production (9–11).
KSHV ORF57, also called mRNA transcript accumulation (MTA), comprises 455 amino acid residues and has been characterized as an RNA-binding protein (RBP) for posttranscriptional regulation. ORF57 is present in the cytoplasm of infected cells but localizes primarily in the nucleus, where it is enriched in the nuclear speckles containing pre-mRNAs and spliceosomal components (12, 13). Structurally, KSHV ORF57 protein has two domains, a disordered N-terminal domain (NTD) and a highly structured α-helix-rich C-terminal domain (CTD). The ORF57 NTD bears three nuclear localization signals (NLSs) and phosphorylation sites and is required for various interactions with target RNAs and cellular cofactors (14–16). The ORF57 CTD functions mainly for ORF57 dimerization and stability (14–16).
As a posttranscriptional regulator, ORF57 regulates RNA stability, splicing, and translation (10–12, 14, 17). ORF57 stabilizes viral polyadenylated nuclear (PAN) RNA by binding to an MTA-responsive element (MRE) in the PAN RNA 5′ end via interaction with PABPC1 [poly(A)-binding protein cytoplasmic 1] (18, 19). ORF57 promotes the splicing of K8 pre-mRNA by interacting with the splicing factor SRSF3 (serine/arginine-rich splicing factor 3 or SRp20), small nuclear RNAs (snRNAs), and other factors (13, 20). ORF57 enhances viral interleukin-6 (vIL-6) translation by preventing microRNA (miRNA)-mediated vIL-6 RNA degradation and translation inhibition (21, 22). ORF57 interacts with protein kinase R (PKR) and PACT to block phosphorylation of eIF2α (α subunit of the eukaryotic translation initiation factor) and stress granule formation, and interacts with Ago2, GW182, and RNA helicase A to inhibit P-body formation and host innate immune responses during lytic KSHV infection, thus promoting robust KSHV replication and virion production (23, 24).
The ribosome is a macromolecular machine comprised of ∼80 proteins and four species of rRNA (5S, 5.8S, 18S, and 28S rRNA) (25, 26). Ribosome biogenesis is regulated by transcription, processing, and modification of rRNAs and, finally, assembly of these components. The rRNAs are extensively chemically modified during their transcription and subsequent maturation in the nucleolus, nucleus, and cytoplasm (26). Chemical modifications of rRNA pseudouridylation (Ψ) and 2′-O-methylation (2′-Ome) are guided, respectively, by box H/ACA small nucleolar RNAs (snoRNAs) and box C/D snoRNAs (26). These two groups of snoRNAs are also involved in various physiological and pathological cellular processes (27), including pre-mRNA splicing (28, 29) and development of cancers (30–32).
Widespread involvement of snoRNAs in virus-host interactions has been reported with regard to host cell susceptibilities to virus infection or virus-induced cell death (33, 34). Knockdown of expression of individual snoRNAs in host cells has shown their roles in both DNA and RNA virus infections (33, 35). In addition to snoRNAs, virus infections and viral proteins that regulate rRNA biogenesis have been well documented (36–39). Epstein-Barr virus (EBV) and its relative rhesus lymphocryptovirus encode a viral snoRNA, v-snoRNA1, a C/D box-like snoRNA (40). Human cytomegalovirus (HCMV) (41) and herpes simplex virus 1 (HSV-1) (42, 43) inhibit or stimulate rRNA gene transcription or modulate pre-rRNA maturation by interaction of viral proteins with rRNAs. KSHV infection stimulates 45S rRNA gene transcription and its level of pseudouridylation in endothelial cells (44). KSHV lytic infection also alters nuclear and nucleolar architectures in iSLK-Bac16 cells (45).
To identify host RNA targets of ORF57, we first applied UV cross-linking and immunoprecipitation (CLIP) with an anti-ORF57-specific antibody to detect ORF57-associated RNAs in BCBL-1 cells with lytic KSHV infection. This approach had successfully detected ORF57-associated ORF59 RNA (12). Subsequently, we applied this technique in combination with cloning and Sanger sequencing to identify KSHV RNA targets of ORF57. We showed that ORF57 in lytic KSHV infection is associated with more than 11 different viral RNA transcripts, including PAN RNA, vIL-6, and ORF56 (19, 21, 22, 46). In the present study, we combined ORF57 CLIP with high-throughput RNA sequencing (CLIP-seq) (47) to analyze genome-wide host RNA targets of ORF57 in BCBL-1 cells with lytic KSHV infection. We discovered ORF57 association with various classes of host RNAs during KSHV lytic infection. Further analysis showed ORF57 binding to not only host coding RNAs but also numerous noncoding RNAs, including rRNAs, snoRNAs, and long noncoding RNAs (lncRNAs). We demonstrated that ORF57 associates with snoRNAs independently of fibrillarin and NOP58 proteins and with rRNA in the regions that commonly bind snoRNAs. More importantly, we found that ORF57 association with PABPC1 and translationally active polysomes prevents Ago2 association with polysomes, indicating a role for ORF57 in regulation of protein translation.
RESULTS
Identification of genome-wide host RNA targets of KSHV ORF57 by CLIP-seq.
We and others previously showed that ORF57 functions as an RNA processing regulator of KSHV gene expression (12, 13, 21, 46, 48–52). However, host RNA targets of ORF57 have not been described carefully owing to use of a low-quality anti-ORF57 antibody in high-throughput transcriptome sequencing (RNA-seq) studies (50, 53). We therefore performed ORF57 CLIP-seq (47) with our affinity-purified, highly specific anti-ORF57 antibody to identify the entire repertoire of host RNAs associated with ORF57 during KSHV lytic infection (Fig. 1A). As diagrammed in Fig. 1A, BCBL-1 cells with KSHV lytic infection were UV irradiated to covalently cross-link RNA-protein complexes in the cells. The cell lysates were then subjected to anti-ORF57 immunoprecipitation. The ORF57-RNA complexes pulled down by anti-ORF57 antibody were treated with RNase A/T1 mixture to remove the RNA regions without the binding of ORF57 or ORF57-associated proteins, followed by dephosphorylation with recombinant shrimp alkaline phosphatase (rSAP) and proteinase K digestion to remove ORF57 protein from the remaining RNA fragments. The resultant ORF57-bound RNA fragments protected from RNase digestion were extracted, ligated with a 3′ linker, reverse transcribed, circulated, and then amplified for library preparation and RNA-seq.
FIG 1.
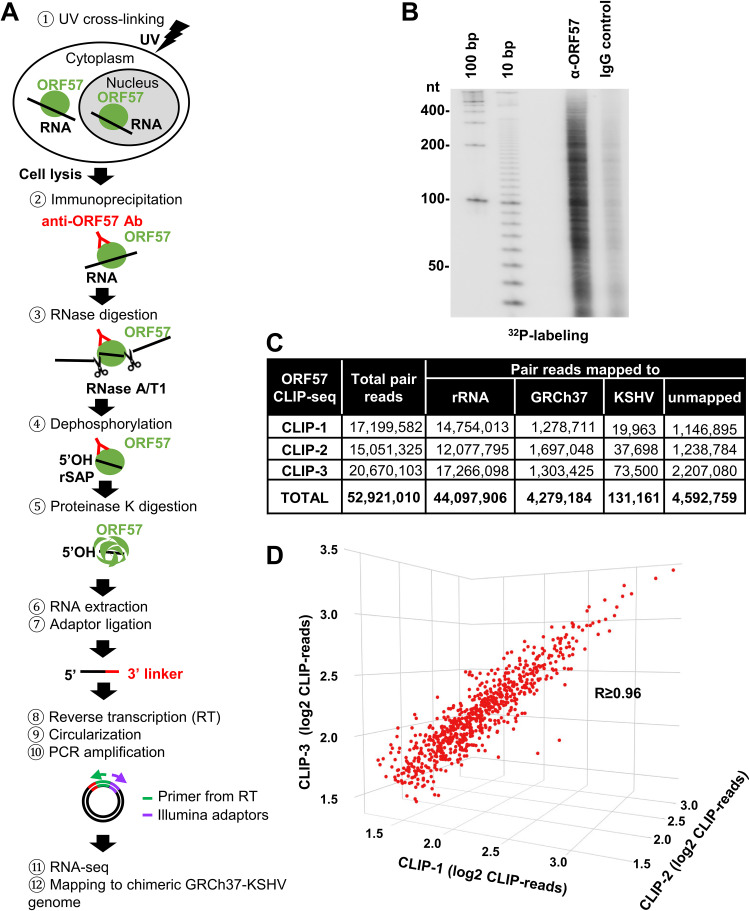
Identification of ORF57-associated RNAs in BCBL-1 cells using CLIP-seq. (A) Schematic illustration of ORF57 CLIP-seq procedure. BCBL-1 cells treated with valproic acid were UV irradiated and ORF57-RNA complexes were immunoprecipitated with an affinity-purified, highly specific anti-ORF57 antibody. The unbound RNA regions were digested by RNase A and T1, followed by dephosphorylation of ORF57-bound RNA with recombinant shrimp alkaline phosphatase (rSAP). Proteinase K was used to digest ORF57 to release the RNA from the complexes for RNA extraction. The obtained RNA fragments were then ligated with a 3′ linker, reverse transcribed, circularized, and amplified by PCR to generate RNA-seq libraries for high-throughput sequencing. The obtained sequence reads were then mapped to the chimeric GRCh37-KSHV genome. (B) Autoradiogram of 32P-labeled RNA cross-linked to ORF57 and identified by anti-ORF57 CLIP, with nonspecific IgG serving as a negative control. (C) After being mapped to rRNA, the paired reads from three separate ORF57-CLIP-seq repeats (CLIP-1, CLIP-2, and CLIP-3) were mapped to the chimeric GRCh37-KHSV genome. (D) Correlation of the mapped reads from three separate ORF57 CLIP-seq experiments in a 3D scatterplot. Each dot represents a single gene, with coordinates showing a number of reads in each sample mapped to that gene obtained using RSEM software (RNA-Seq by Expectation Maximization). The R value (Pearson’s correlation efficient) is ≥0.96.
To check the anti-ORF57 CLIP quality, a small fraction of the extracted RNA fragments was end labeled with 32P, resolved in a denaturing urea-PAGE gel, and autoradiographed. The results showed the enriched RNA fragments in various sizes from anti-ORF57 CLIP over the control IgG CLIP, a negative control (Fig. 1B). We conducted three independent anti-ORF57 CLIP experiments (CLIP-1, CLIP-2, and CLIP-3) and constructed three separate RNA-seq cDNA libraries for sequencing by using an Illumina HiSeq 2500 platform. In total, the three ORF57 CLIP-seq experiments generated 52,921,010 paired reads with high quality (NCBI Gene Expression Omnibus [GEO] accession number GSE179726). Using STAR aligner, we aligned 44,097,906 paired reads (83.3%) to rRNA (GenBank accession number U13369; human ribosomal DNA complete repeating unit), 4,279,184 paired reads (8.1%) to the other parts of the human genome, and 131,161 paired reads (0.3%) to the KSHV genome (GenBank accession number U75698.1) (Fig. 1C). By Pearson's correlation efficient analysis, we found that the three sets of CLIP-seq data were very consistent with an R value of ≥0.96, indicating the high quality of anti-ORF57 CLIPs and cDNA library construction and sequencing (Fig. 1D).
ORF57 interacts with both host protein-coding and noncoding RNA transcripts by binding to a specific RNA region.
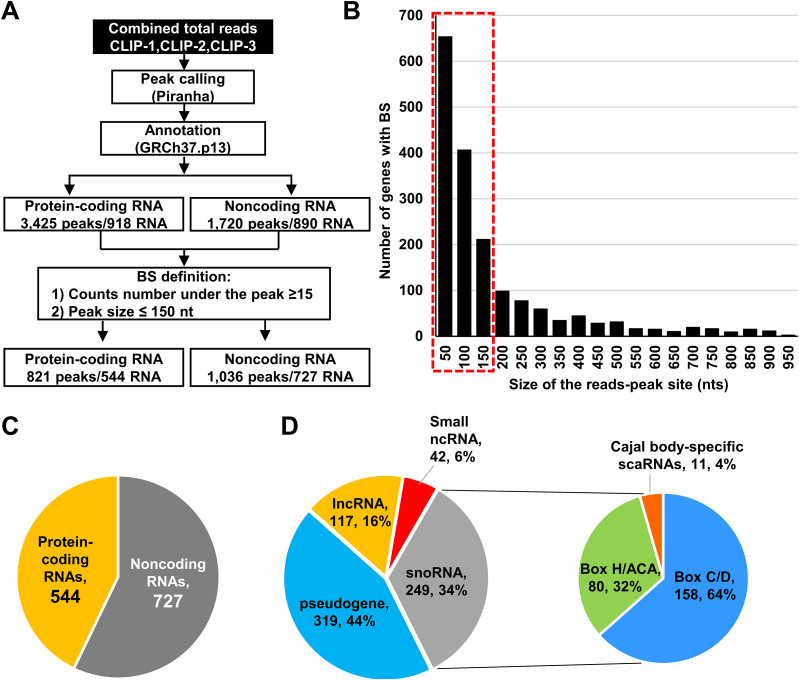
By mapping all reads to the human genome using STAR aligner in combination with Piranha software for peak calling (54), we were able to determine the regions (peaks) significantly enriched with RNA strand-specific sequence reads over a background model and thus identify the regions as the ORF57 binding sites (BS) from the CLIP-seq data. By annotation of all obtained ORF57 peaks against the annotated genes in the human genome, we identified a total of 5,145 ORF57 BS, of which 3,425 were mapped to the RNAs from 918 protein-coding genes and 1,720 to the RNAs from 890 noncoding genes (Fig. 2A). The determined peaks were in a broad range of size, from most prevalent small peaks at the minimal size of 50 nucleotides (nt) to fewer common peaks larger than 950 nt. We next focused on the peaks in the size less than or equal to 150 nt supported by 15 or more reads as a more defined ORF57 BS for further analyses and validation (Fig. 2B). With these criteria, we identified 821 BS mapped to 544 protein coding genes and 1,036 peaks to 727 noncoding genes (Fig. 2A and C and Table S1 in the supplemental material). The higher number of ORF57 BS than mapped RNAs was caused by multiple ORF57 BS found in a particular set of RNAs. In the rest of this report, we focus only on the ORF57 interactions with noncoding RNA transcripts; the coding gene transcripts associated with ORF57 will be reported elsewhere.
FIG 2.
ORF57 binds both host protein-coding and noncoding RNAs. (A) Workflow used to identify host RNAs as ORF57 targets. Reads from three ORF57 CLIP-seq experiments were combined to increase the probability of read peak identification. Peak-calling was performed by using Piranha software. Obtained peaks were annotated to the human genome GRCh37. Peaks of ≤15 reads were excluded from further consideration. (B) Size distribution and frequency of the peaks detected by Piranha software. The red dashed line indicates a threshold (≤150 nt) used to define an ORF57 binding site (BS). (C) Pie chart showing noncoding and protein-coding RNAs containing the ORF57 BS. (D) Distribution of the ORF57 BS in various species of noncoding RNAs. The pie plot shows the number and percentage of ORF57 RNA targets in each category.
ORF57 is associated with several classes of host noncoding RNAs with distinct functionality.
Among 727 ncRNAs containing ORF57 BS (Fig. 2C), 319 (44%) were pseudogene transcripts, 249 (34%) were snoRNAs, 117 (16%) were lncRNAs (lincRNAs, antisense RNAs, and intronic RNAs), and 42 (6%) were other small ncRNAs, including small nuclear RNAs (snRNAs), miRNAs, and misc_RNAs (miscellaneous small RNAs), such as RNY (Ro-associated Y RNAs) and vault RNAs (vtRNAs) (Fig. 2D, left pie). Among 249 snoRNAs bound to ORF57, 158 (64%) were C/D box snoRNAs, 80 (32%) were H/ACA box snoRNAs, and 11 (4%) were small Cajal body-associated RNAs (scaRNAs) (Fig. 2D, right pie, and Table S1).
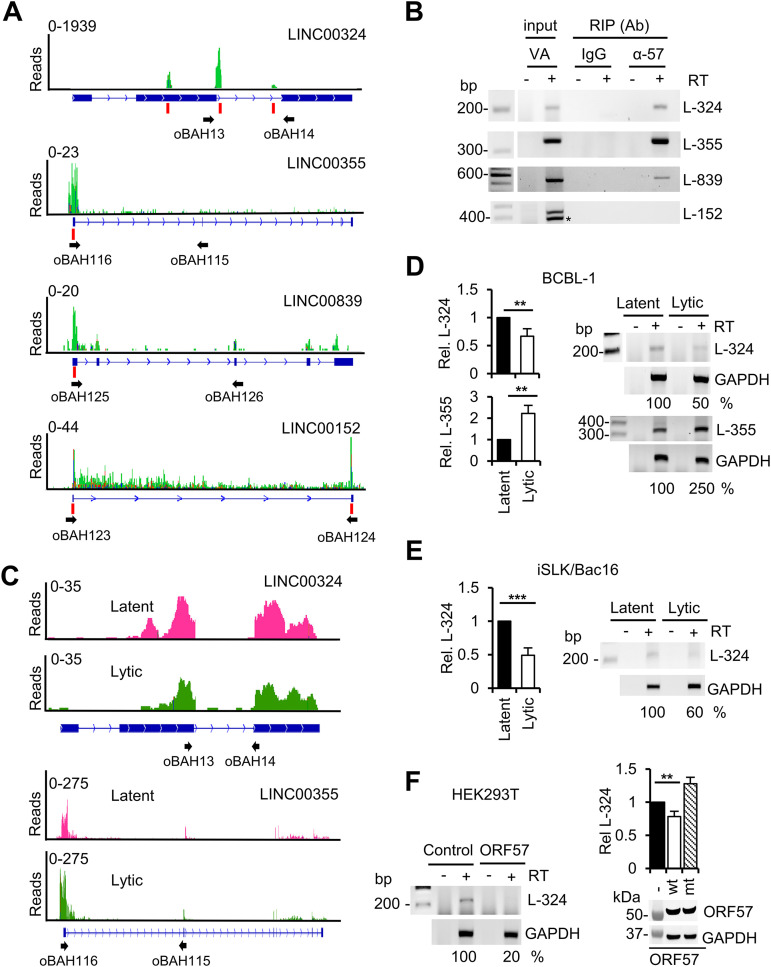
Pseudogene transcription and function remain largely unknown, although some pseudogenes may have regulatory effects (55, 56); lncRNAs have been deliberated in recent years to be involved in both nuclear and cytoplasmic functions (57–60). Thus, by anti-ORF57 RIP and reverse transcription-PCR (RT-PCR), we selectively verified in BCBL-1 cells under KSHV lytic infection the three of four lncRNAs which are associated with cell growth and contain 1 to 3 well-defined ORF57 BS (Fig. 3A and B). LINC00355 expressed from an intergenic region of chromosome 13 promotes cell proliferation and tumor progression (61–63). LINC00839 is derived from an intergenic region of chromosome 10, and LINC00324 (C17orf44) is expressed from an intergenic region of chromosome 17. They have been shown to regulate cell proliferation, invasion and migration, and possibly tumorigenesis (64–68). LINC00152 (C2orf59) is highly expressed in gastric and colorectal cancer tissues and myeloma cells from the chromosome 2p11.2 region and promotes cell cycle progression and cell proliferation (69, 70). We identified three ORF57 BS with up to 1,900 reads at the second exon and intron of LINC00324 and a single ORF57 BS with a depth of 20 to 23 reads at the 5′ end of LINC00355 and LINC00839 (Fig. 3A). Although LINC00152 also showed two ORF57 BS with up to 30 reads, we found that a notable proportion of the mapped reads had color-coded mutations and were unable to verify LINC00152 association with ORF57 in the cells by anti-ORF57 RIPs and RT-PCR (Fig. 3A and B). RNA-seq (Fig. 3C), RT-quantitative PCR (RT-qPCR), and RT-PCR (Fig. 3D) analyses of total cellular RNA extracted from BCBL-1 cells showed a relative decrease (∼2-fold) of LINC00324 but an increase (∼2.5-fold) of LINC00355 RNA expression in KSHV lytic infection compared with latent infection, whereas LINC00839 showed only minimal, not significant, change between the two infection phases (data not shown). Subsequently, we verified by RT-qPCR and RT-PCR the decreased expression of LINC00324 during lytic KSHV infection in iSLK/Bac16 cells (Fig. 3E). This decreased expression of LINC00324 expression in both BCBL-1 and iSLK/Bac16 cells with viral lytic infection was specific to ORF57. We found that ectopic wild-type (wt) ORF57, but not its dysfunctional mutant (mt ORF57), in HEK293T cells decreases the expression of LINC00324 (Fig. 3F).
FIG 3.
Association of ORF57 with noncoding RNAs and its possible roles in regulation of their expression. (A) ORF57 in BCBL-1 cells with lytic KSHV infection is associated with the selected host long intergenic noncoding RNAs (lincRNAs). IGV visualization and distribution of ORF57 CLIP-seq reads in green along with selected lincRNAs: LINC00324 (L-324), LINC00355 (L-355), and LINC00839 (L-839). The read peaks identified by Piranha software are marked with vertical red lines. Numbers in the upper left corners represent CLIP-seq read coverage depths. The names and locations of the oligonucleotide primers used in RT-PCR detections (B) are shown below each lincRNA diagram. (B) Validation of ORF57 association with the selected three host LINCRNAs in BCBL-1 cells with viral lytic infection. The RT-PCR in the absence (-) or presence (+) of reverse transcriptase (RT) was carried out using RNA isolated from ORF57 RIPs. IgG served as a negative RIP control. Total RNA from BCBL-1 cells induced with valproic acid (VA; 1 mM) for 24 h was used as an input control. *, a nonspecific band identified by Sanger sequencing. (C and D) lincRNAs associated with ORF57 are differentially expressed during viral lytic infection. RNA-seq read coverage maps by the integrative genomics viewer (IGV) (C) show the expression of LINC00324 and LINC00355 in BCBL-1 cells under viral lytic (green) over latent (pink) infection. Numbers in the upper left corners represent read coverage depths. Representative validation of differential expression of the selected host lincRNAs L-324 and L-355 was performed by TaqMan RT-qPCR (D, bar graphs on the left) and semiquantitative RT-PCR (D, gel images on the right) with the primer pair shown below each lincRNA diagram (C). RT-PCR in the absence or presence of reverse transcriptase was carried out using RNA isolated from BCBL-1 cells with viral latent or lytic infection induced by 24 h of VA treatment. (E) Differential expression of L-324 was also confirmed by TaqMan RT-qPCR (bar graphs on the left) and semiquantitative RT-PCR (gel images on the right) of total RNA isolated from iSLK/Bac16 cells with latent or lytic infection induced by 48 h of Dox and butyrate treatment as described for panel D. (F) Ectopic wild-type (wt) ORF57, but not its dysfunctional mutant (mt), in HEK293T cells regulates the expression of L-324. Total RNA was extracted from HEK293T cells 24 h after Flag-tagged ORF57 plasmid transfection for RT-PCR (gel images on the left) and RT-qPCR (bar graphs on the right) detection of L-324 as described for panels D and E. Western blotting with total cell lysate and anti-Flag antibody was used to detect both wt and mt ORF57 protein (below the bar graph). GAPDH served as a protein sample loading control. Densitometry analysis was performed for RT-PCR by normalizing to each internal GAPDH RNA loading control, with the RNA band intensity in latent or vector control as 100%. All gel images are one representative of three biological replicates, and statistical analysis for RT-qPCR was performed by two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001.
KSHV ORF57 is associated with snoRNAs in BCBL-1 cells with viral lytic infection.
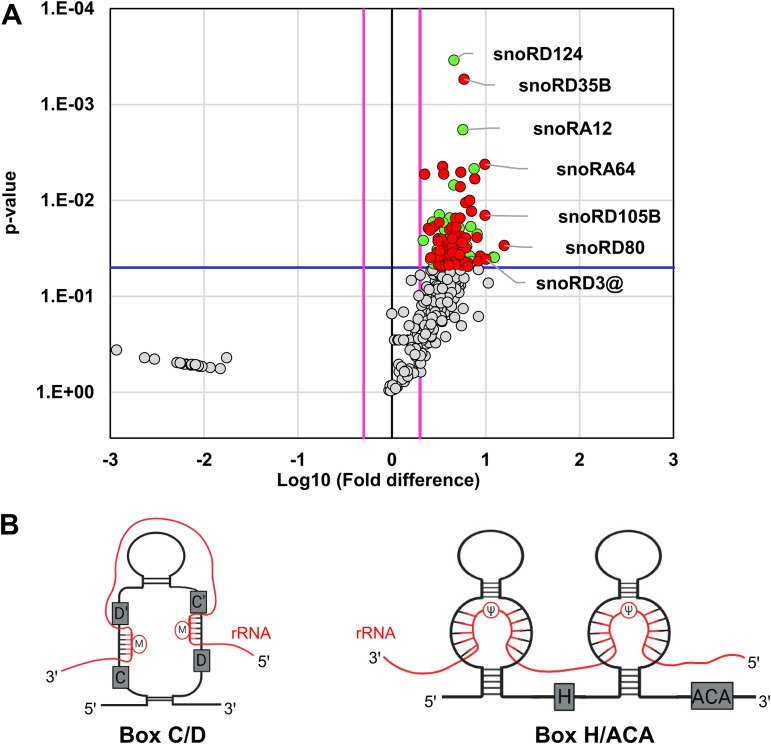
Given that snoRNAs were the second largest group of the ncRNA species bound to ORF57 in ORF57 CLIP-seq, we applied the nrStar human snoRNA PCR array to verify the ORF57-bound snoRNAs in BCBL-1 cells with KSHV lytic infection. This array contains a panel of 359 snoRNAs and 7 snRNAs, of which 207 and 4 were identified, respectively, by ORF57 CLIP-seq. To obtain the ORF57-associated snoRNAs and snRNAs, we performed anti-ORF57 RIP, with nonspecific IgG as a negative RIP control. The extracted RNAs from the RIP pulldowns after proteinase K digestion were directly used for the snoRNA array analysis. With this approach, we found a total of 97 snoRNAs and 4 snRNAs significantly enriched in the ORF57 RIP over the IgG RIP control (Fig. 4A, green and red dots) with a fold change (FC) of ≥2 (P value ≤ 0.05, Student’s t test). This included 67 (27.3%) of 249 snoRNAs identified by ORF57 CLIP-seq (Fig. 4A, red dots), of which snoRD80, snoRD3@ (including all five human snoRD3A-D) (34), and snoRD105B showed 15-fold, 10-fold, and 9-fold enrichments, respectively, in the anti-ORF57 RIP over the IgG control (Table S2). The ORF57 RIP-snoRNA array also identified additional 30 snoRNAs in association with ORF57, which were missed in our ORF57 CLIP-seq (Fig. 4A, green dots). The omission of RNase digestion in isolation of ORF57-associated RNAs in the RIP and a higher input of radioimmunoprecipitated RNAs in the snoRNA array might contribute to this detection discrepancy from the CLIP-seq in addition to the different filtering parameters being applied to the calculation in two different methods.
FIG 4.
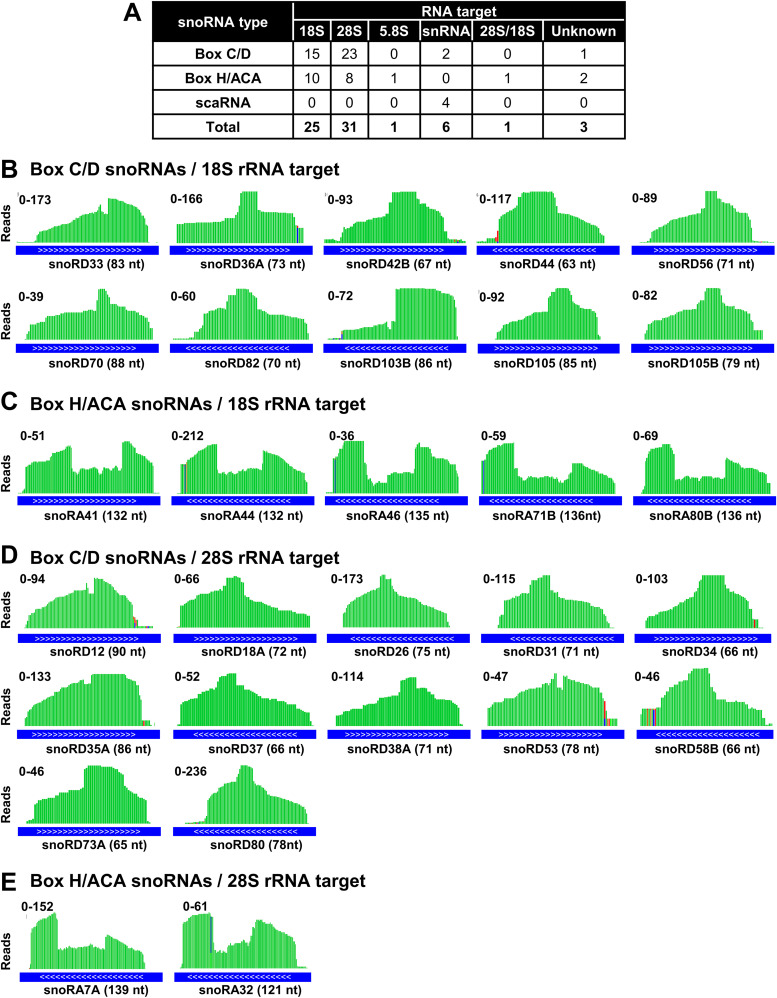
Validation of ORF57 association with snoRNAs by ORF57 RIP-snoRNA array analysis. (A) Volcano plot showing the snoRNAs enriched in ORF57 RIP over IgG RIP from snoRNA array analyses based on three independent experiments. The threshold shown by the blue line indicates statistically significant change (P ≤ 0.05, Student’s t test), and the pink lines indicate at least 2-fold change. Highlighted in green are snoRNAs significantly enriched in ORF57 RIP-snoRNA array analyses. Highlighted in red are snoRNAs significantly enriched in both ORF57 CLIP-seq and ORF57 RIP-snoRNA array analyses. (B) Structural features of box C/D and H/ACA snoRNAs and their association with rRNA.
The ORF57 RIP-snoRNA array also identified U1 (RNU1), U3 (RNU3), U4 (RNU4), and U6 (RNU6) snRNA in interaction with ORF57, with U1 and U3 being the strongest ORF57 binders (FC = ∼12) (Table S2). These data were consistent with our previous report (13).
Two functional classes of snoRNAs are associated with KSHV ORF57.
snoRNAs are mostly nucleolus, nonpolyadenylated ncRNAs and are classified by highly conserved “box” sequences into either box C/D- or box H/ACA-containing snoRNAs (Fig. 4B). The snoRNA with an additional CAB box are called scaRNAs (71). The box C/D snoRNAs mediating 2′-O-methylation (2′-Ome) (Fig. 4B, left diagram) of the target RNAs represent the largest group, 1,391 reported members, followed by the box H/ACA snoRNAs, with 651 members to mediate pseudouridylation (Fig. 4B, right diagram) of target RNAs (snoRNA Atlas [http://snoatlas.bioinf.uni-leipzig.de/]) (72). The scaRNAs involved in snRNA modifications are the smallest group, with 32 members. A primary role of box C/D and H/ACA snoRNAs is serving as a molecular “guide” for posttranscriptional chemical modifications of other host RNAs, primarily rRNAs, tRNAs, and snRNAs (25, 73, 74). To uncover a possible connection of snoRNAs in ORF57 binding and snoRNA-mediated RNA modifications, we classified the ORF57-bound snoRNAs based on their target RNAs. As expected, we found that a majority of the box C/D and H/ACA snoRNAs (Fig. 4B) associated with ORF57 target rRNAs.
As summarized in the snOPY database (http://snoopy.med.miyazaki-u.ac.jp/), the posttranscriptional modifications in 28S rRNA are guided by 134 snoRNAs (77 C/D and 57 H/ACA), in 18S rRNA by 116 snoRNAs (55 C/D and 61 H/ACA), and in 5.8S rRNA by 5 snoRNAs (3 C/D and 2 H/ACA). Among 67 snoRNAs identified both by ORF57 CLIP-seq and by nrStar snoRNA array that targets rRNA, snRNAs, and others, we found that 31 target 28S rRNA (23 C/D, and 8 H/ACA), 25 target 18S rRNA (15 C/D and 10 H/ACA), and 1 H/ACA snoRNA targets 5.8S rRNA (Fig. 5A). As predicted, more C/D snoRNAs than H/ACA snoRNAs were proportionally associated with ORF57. IGV visualizations of individual snoRNA reads distribution from ORF57 CLIP-seq are selectively shown in Fig. 5B to E. We also observed a characteristic binding pattern across all snoRNAs associated with ORF57, similar to the previous study in which photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP) was performed with the snoRNP core proteins fibrillarin, NOP58, and dyskerin (75). These data indicate that ORF57 interacts predominantly with the snoRNAs which are associated with rRNA modifications.
FIG 5.
ORF57-associated snoRNAs target rRNAs. (A) A table showing the type and number of predicted RNA targets from snoRNAs associated with ORF57. snoRNA type and target RNA were obtained from the snOPY database http://snoopy.med.miyazaki-u.ac.jp/. (B and C) IGV visualization of read distribution of selective box C/D (B) and H/ACA (C) snoRNAs guiding 18S rRNA modifications which bind to ORF57. The snoRNAs shown were identified by ORF57 CLIP-seq with a P value of ≤1 × 10−15 and number of reads of ≥70 and also validated by ORF57 RIP-snoRNA array analyses. ENSEMBL transcript annotation tracks (blue lines) shown at the bottom indicate the orientation (white arrows 5′ to 3′) and size in nucleotides of each snoRNA. Values on the y axis represent the scale of ORF57 CLIP-seq read counts. (D and E) IGV visualization of read distribution of selective box C/D (D) and H/ACA (E) snoRNAs guiding 28S rRNA modification which bind to ORF57. See other details described for panels B and C.
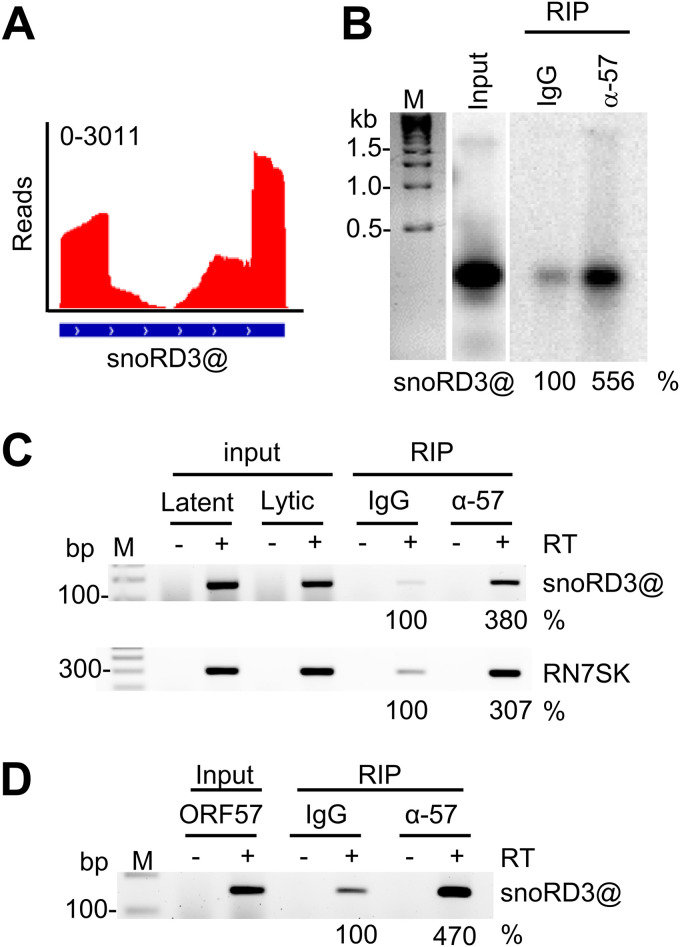
According to the snOPY database, 3 snoRNAs (snoRA16, snoRA53, and snoRD3@) associated with ORF57 in our study have no annotated RNA targets. snoRD3A, 3B-1, 3B-2, -3C, and -3D, derived from a snoRD3@ (previously called U3 snoRNA) cluster region, are highly homologous and were abundant in ORF57 CLIP-seq (Fig. 6A). The snoRD3@ transcribed by RNA polymerase II functions as a scaffold during early ribosome assembly and can be processed to produce miRNA miR-U3 (76). To validate the ORF57-snoRD3@ interaction, we extracted total RNA from BCBL-1 cells with viral lytic infection and examined anti-ORF57 immunoprecipitated RNAs by Northern blotting using an antisense oligonucleotide probe specific to all members of snoRD3@. As shown in Fig. 6B, Northern blotting confirmed the specific ORF57-snoRD3@ binding. This ORF57-snoRD3@ interaction in BCBL-1 cells with lytic KSHV infection could be verified by anti-ORF57 RIP and RT-PCR (Fig. 6C), where ORF57 was found to specifically interact with transcription elongation RNA regulator RN7SK (77) as identified by ORF57 CLIP-seq (Table S1) (Fig. 6C). The specific ORF57-snoRD3@ interaction was further determined in HEK293T cells by ectopic expression of ORF57 by anti-ORF57 RIP and RT-PCR (Fig. 6D), thus excluding possible involvement of other viral proteins in the identified ORF57-snoRD3@ interaction during KSHV lytic infection.
FIG 6.
ORF57 binds the snoRD3@ with unknown RNA targets. (A and B) Distribution of ORF57 CLIP-seq reads mapped to snoRD3@ (A) and Northern blot verification of snoRD3@ interaction with ORF57 in BCBL-1 cells by anti-ORF57 antibody RIP and then by 32P-labeled antisense DNA oligonucleotide probe oBAH135, specific to all members of snoRD3@ (B). IgG served as a RIP control. (C) RT-PCR of ORF57 immunoprecipitated RNAs from BCBL-1 cells with VA-induced KSHV lytic infection in the absence or presence of reverse transcriptase. IgG served as a negative RIP control. Total RNA extracted from the cells with latent or lytic KSHV infection was examined for relative levels of snoRD3@ and RN7SK RNAs, and the RNA from the cells with KSHV lytic infection also served as a lytic RNA input control. (D) RT-PCR in the absence or presence of reverse transcriptase using ORF57 immunoprecipitated RNAs from HEK293T cells with ectopic ORF57 expression. IgG served as a negative RIP control. Total RNA from HEK293T cells 24 h after ORF57 plasmid transfection was used as an input control. The following primer pairs were used: oBAH198 and oBAH199 for detection of snoRD3@ and oBAH192 and oBAH193 for detection of RN7SK. Northern blot or RT-PCR band intensity representing the immunoprecipitated RNA was calculated after normalized to the corresponding input, with IgG control as 100%.
Relationship of ORF57 in snoRNA binding and expression with snoRNA-binding proteins NOP58 and fibrillarin.
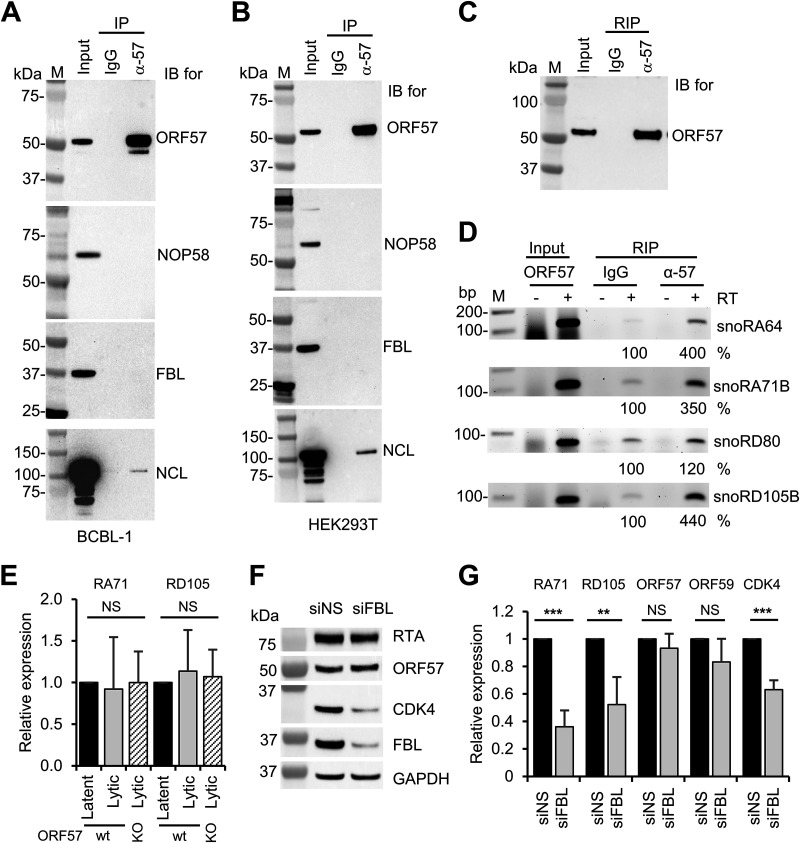
To determine whether the snoRNAs associate with ORF57 indirectly through ORF57 interactions with snoRNA-associated proteins such as NOP58 and fibrillarin, we performed ORF57 coimmunoprecipitation (co-IP) experiments using the proteins extracts from BCBL-1 cells with lytic KSHV infection or HEK293T cells with ectopic expression of ORF57. By Western blotting of the proteins in anti-ORF57 co-IP pulldowns, we demonstrated the interaction of ORF57 with nucleolin, a protein previously proved to be an ORF57-interacting protein (16), but did not find interaction of ORF57 with fibrillarin or NOP58 in BCBL-1 or HEK293T cells (Fig. 7A and B), indicating that ORF57 interacts with the snoRNAs independently of these two snoRNA-binding proteins. However, we showed by anti-ORF57 RIP, as expected, that ectopic ORF57 in HEK293T cells was capable of interacting with snoRA64, snoRA71B, snoRD80, and snoRD105B as examined. The ORF57 interactions with snoRNAs appeared not to affect snoRNA expression. In a comparison of the expression levels of snoRA71B and snoRD105B in iSLK/Bac16 cells with lytic KSHV infection, there was no difference in expression levels of these two snoRNAs from the cells containing a wt KSHV genome and the cells with an ORF57-null genome (Fig. 7E). Interestingly, knockdown of fibrillarin expression in iSLK/Bac16 cells with lytic KSHV infection was found to reduce the expression of snoRA71B, snoRD105B, and surprisingly CDK4 RNA and protein but did not affect the expression of viral ORF57 and ORF59 RNAs or of ORF57 and RTA proteins (Fig. 7F and G). These data suggest that in contrast to host RNAs, viral RNA and protein expression in the infected cells is relatively resistant to snoRNA-mediated regulation.
FIG 7.
Relationship of ORF57 in snoRNA binding and expression with snoRNA-binding proteins fibrillarin and NOP58. (A) ORF57 does not interact with fibrillarin or NOP58 in BCBL-1 cells with KSHV lytic infection. Total cell extract from BCBL-1 cells under viral lytic infection was immunoprecipitated with an anti-ORF57 antibody, and the ORF57 complexes in the IP pulldowns were resolved by SDS-PAGE for detection of ORF57, fibrillarin (FBL), and NOP58 by Western blotting with the corresponding antibodies. Nucleolin (NCL) served as a positive control of cointeraction with ORF57. (B) Total cell extract from HEK293T cells transfected with an ORF57 expressing vector (pVM7) was also used for the co-IP and Western blotting as performed for panel A. (C and D) Ectopic ORF57 in HEK293T cells interacts with selective snoRNAs. (C) Representative Western blot of immunoprecipitated ORF57 from HEK293T cells expressing ORF57 by mouse monoclonal anti-ORF57 antibody. (D) RT-PCR for detection of snoRA64, snoRA71B, snoRD80, and snoRD105B in the absence or presence of reverse transcriptase (RT) using ORF57 immunoprecipitated RNAs from HEK293T cells with ectopic ORF57 expression for 24 h. IgG served as a negative RIP control. Total RNA from HEK293T cells expressing ORF57 served as an input control. See Table 1 for primer pair details for individual snoRNA detection. RT-PCR band intensity representing the immunoprecipitated RNA was calculated after normalization to the corresponding input, with IgG control as 100%. (E) ORF57 expression in iSLK/Bac16 cells does not affect the expression of snoRA71B (RA71) or snoRD105B (RD105). Total RNA isolated from iSLK/Bac16 cells bearing a wild-type KSHV genome or an ORF57-null genome (ORF57 knockout [KO]) (11) in viral latent or lytic infection induced by Dox and sodium butyrate for 48 h was used for RT-qPCR to detect the expression of representative snoRA71B and snoRD105B. GAPDH RNA served as an internal control to normalize the quantification of each snoRNA level. (F and G) Knockdown of fibrillarin expression in iSLK/Bac16 cells carrying a wt KSHV genome led to reduce the expression of snoRA71B and snoRD105B and CDK4 but not the expression of viral ORF57, ORF59, or RTA. The iSLK/Bac16 cells were first treated with a siRNA specific for fibrillarin (siFBL) or a nontargeting control siRNA (siNS) for 36 h and then induced for lytic KSHV infection for 12 h by Dox and sodium butyrate treatment before total RNA and protein extraction. Total protein lysates were blotted for viral RTA and ORF57 and host CDK4, FBL, and GAPDH by the individual corresponding antibodies. GAPDH protein served as a sample loading control (F). Total cell RNA was examined by RT-qPCR for the expression of snoRA71B, snoRD105B, viral ORF57 and ORF59, and host CDK4 (G). The bar graphs in panels E and G show means ± SD from three separate experiments, with latent RNA (E) and siNS RNA (G) as 1. Statistical analysis for RT-qPCR in panels E and G was performed by two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001. NS, no significance.
ORF57 associates with host rRNAs mainly through the regions interacting with the ORF57-bound snoRNAs.
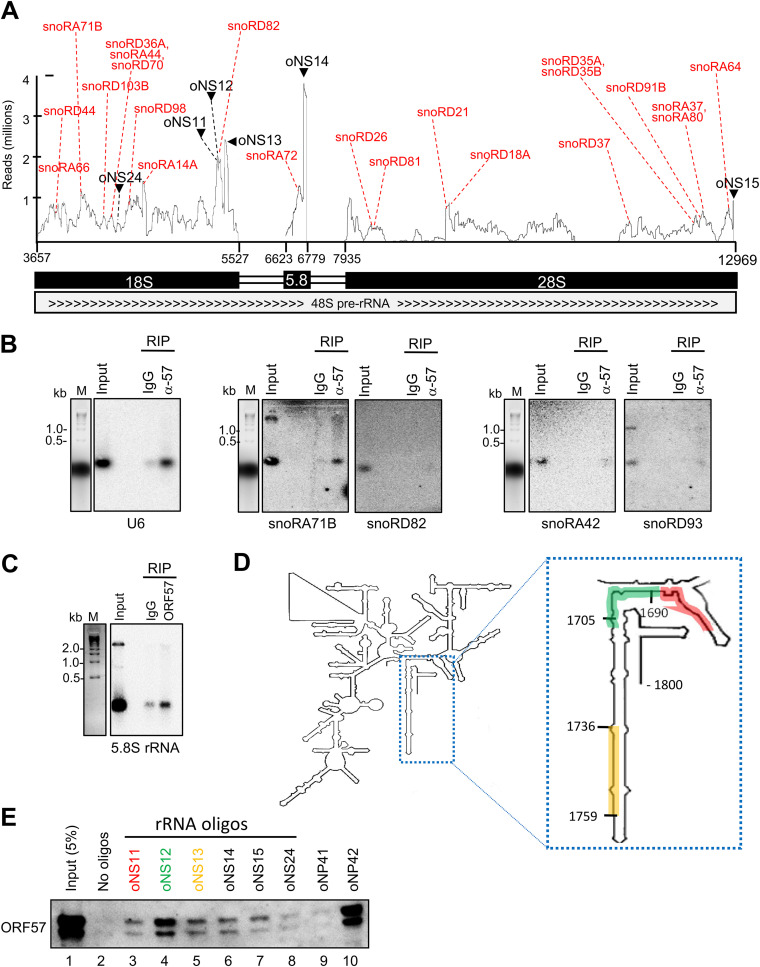
Considering that ORF57 preferentially binds the snoRNAs that mediate rRNA modifications (Fig. 5) and ORF57 CLIP-seq also identified a high number of rRNA reads (Fig. 1C), we investigated the possible connection of ORF57-snoRNAs to rRNAs. We visualized the rRNA read distribution along the entire 18S, 5.8S, and 28S rRNAs by IGV and then overlaid the rRNA read peaks with the rRNA regions that bind snoRNAs. As showed in Fig. 8A, we found that the majority of the rRNA read peaks identified by ORF57 CLIP-seq were the regions that bind snoRNAs, suggesting that these rRNA regions with the bound snoRNAs were co-associated with ORF57 and thus both were protected from RNase digestion during anti-ORF57 CLIP. Some selected snoRNA-binding sites are shown in Fig. 8A, including snoRA66, snoRD44, snoRA71B, snoRD103B, snoRD36A, snoRA44, snoRD70, snoRD98, snoRA14A, snoRD82, snoRA72, snoRD26, snoRD21, snoRD81, snoRD18A, snoRD37, snoRD35A and -35B, snoRD91B, snoRA80, snoRA37, and snoRA64. All of these snoRNAs were detected by both ORF57 CLIP-seq and ORF57 RIP snoRNA array.
FIG 8.
ORF57 association with host rRNA and snoRNAs in BCBL-1 cells with lytic infection. (A) Distribution of the ORF57 CLIP-seq reads mapped to 18S, 5.8S, and 28S rRNAs from three combined experiments. Red dotted lines indicate the read peak sites where ORF57 and snoRNA co-bind. Black arrows and dotted lines indicate the synthetic biotinylated RNA oligonucleotides from the indicated peaks used for oligonucleotide pulldown of ORF57 and Western blot assays. (B) Northern blot verification of the enriched snoRNAs in ORF57 RIP over control IgG RIP. U6 served as a positive control in interacting with ORF57 in BCBL-1 cells (13). Both snoRA42 and snoRD93 not binding to ORF57 both in the ORF57 CLIP-seq and in ORF57 RIP-snoRNA array analyses served as two negative snoRNA controls. Northern blotting was performed with a 32P-labeled oligonucleotide antisense to the region of individual snoRNAs binding to rRNA. (C) Verification of ORF57 binding to 5.8S rRNA in the ORF57 RIP over the control IgG RIP by Northern blotting using a 32P-labeled oligonucleotide antisense to the 5.8S rRNA region overlapping snoRA72 binding. (D and E) Verification of ORF57 binding to rRNA by RNA oligonucleotide pulldown-Western blot assays. Biotinylated RNA oligonucleotides shown in panel A and colored oligonucleotides oNS11 to -13, whose positions on 18S rRNA are shown in panel D, were mixed with total protein lysate from BCBL-1 cells with 24 h of lytic KSHV infection and pulled down by avidin-coated beads. The amount of ORF57 protein associated with each RNA oligonucleotide in the pulldown was detected by an anti-ORF57 antibody. RNA oligonucleotides oNP41 and oNP42, derived from KSHV vIL-6 (22), served, respectively, as negative and positive controls for ORF57-RNA interaction. A region from 18S rRNA (oNS24) not enriched in ORF57 CLIP-seq also served as an additional negative control.
Subsequently, we performed Northern blot analyses to confirm these snoRNAs pulled down from BCBL-1 cells with lytic KSHV infection by ORF57 RIP using a 32P-labeled antisense oligonucleotide to individual snoRNAs as a probe. snoRA71B and snoRD82 were selected for Northern blotting because they were detected by both ORF57 CLIP-seq and snoRNA array, whereas snoRA42 and snoRD93 served as two negative controls because they were not associated with ORF57 in our study. Similar to the binding of ORF57 to U6 (13), we found a specific interaction of ORF57 with snoRA71B, but not with snoRD82 (Fig. 8B). As expected, we did not find ORF57 binding to snoRA42 and snoRD93 (Fig. 8B). By using an antisense oligonucleotide probe to the snoRA72 binding site in 5.8S rRNA, we also demonstrated by Northern blotting ORF57 binding to this small rRNA in BCBL-1 cells (Fig. 8C). The snoRA71 family has four members, snoRA71A to -D, of which all share the identical complementary sequence to 18S rRNA to guide the same modification. Thus, using this complementary sequence as a 32P-labeled probe would allow us to detect all three snoRA71 members (snoRA71A, -B, and -D) identified by ORF57 CLIP-seq (Table S1). The negative detection of snoRD82 by Northern blotting indicates that ORF57 binds to this region of 18S rRNA presumably free of snoRD82 in addition to its low abundance in the cells (Table S1).
Next, we performed RNA oligonucleotide pulldown assays using biotinylated RNA oligonucleotides derived from three rRNA read peaks, one each from 18S, 5.8S (oNS14), and 28S (oNS15) rRNAs, to validate ORF57-rRNA interaction and three oligonucleotides (oNS11, -12, and -13) running over the entire 18S peak region binding to snoRD82. We used a region from 18S rRNA with low ORF57 CLIP-seq read coverage as a negative control (oNS24), together with previously described KSHV vIL6 RNA oligonucleotides (oNP41, a negative control, and oNP42, an ORF57-binding positive control) with well-defined features of binding to ORF57 (22). The RNA oligonucleotides were mixed with total cell extracts from BCBL-1 cells with lytic KSHV infection, and the relative amount of ORF57 protein in the oligonucleotide pulldowns was measured by Western blotting. As shown in Fig. 8D and E, we observed ORF57 binding to various RNA oligonucleotides, with the strongest binding of ORF57 to the RNA oligonucleotide oNS12 from 18S rRNA and the RNA oligonucleotide oNP42 from vIL-6 but only weak binding to oNS24 and oNP41, as expected. These data further suggest that ORF57 binding to an rRNA motif containing a snoRNA binding site could have been either in complex with a snoRNA or free of the snoRNA in the cells when the CLIP-seq was performed.
ORF57 associates with polysomes along with PABPC1 and prevents Ago2 from association with polysomes.
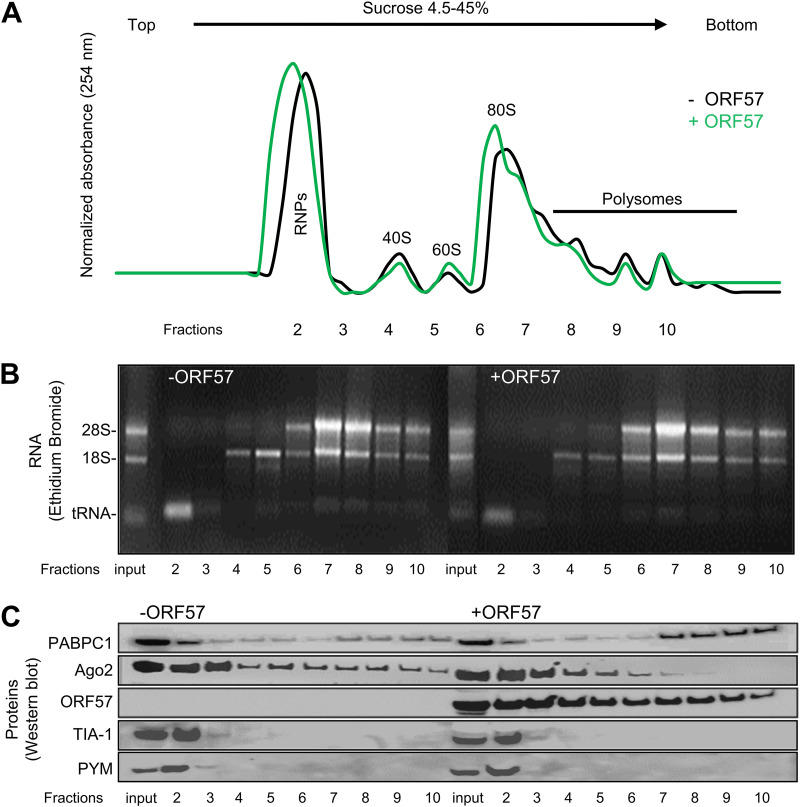
Findings of ORF57 association with rRNAs and interaction with ribosomal proteins L3 and L4 in our previous study (16) prompted us to further investigate the possible association of ORF57 with translating ribosomes. We conducted polysome profiling analyses for HEK293T cells with or without ectopic ORF57 expression and examined the translation-related proteins associated with translating ribosomes in the presence or absence of ORF57. To do so, we obtained HEK293T cell cytoplasmic fractions 24 h after transfection of ORF57 and analyzed for the polysome profiling across a 4.5% to 45% sucrose density gradient.
As shown in Fig. 9A, the A254 absorption spectrum of the obtained fractions showed a distinguishable profile of peaks representing free ribonucleoproteins (RNPs), monosomal 40S, 60S, and 80S, and polysomal complexes in the presence or absence of ORF57. Ethidium bromide staining of total RNA isolated from each fraction further confirmed the presence of monosomal forms in the lighter, low-density fractions as defined by 18S rRNA and polysomal complexes in the heavy density (up to 45%) fractions as defined by the presence of both 28S and 18S rRNAs (Fig. 9B). When the total protein from each fraction was analyzed by Western blotting, we discovered the dramatic difference of polysomal RNA-binding protein composition from the absence to the presence of ORF57 (Fig. 9C).
FIG 9.
ORF57 association with polysomes enhances PABPC1 binding to, but prevents Ago2 association with, polysomes by polysome profiling of HEK293T cytoplasmic fractions. HEK293T cells were transfected with or without an ORF57-expressing vector (pVM7). At 24 h posttransfection, cytoplasmic fractions (after nucleus removal) were analyzed for polysome profiling across a 4.5% to 45% sucrose density gradient. The A254 absorption spectrum of the fractions was automatically collected using a fractionator attached to the spectrophotometer to show a distinct pattern of peaks of free RNPs, monosomal 40S, 60S, and 80S, and polysomal complexes (A). The input and collected sample fractions (2 to 10) were used for RNA extraction and rRNA detection by 1.5% agarose gel electrophoresis using ethidium bromide staining (B). Total protein from each fraction was immunoblotted for detection of PABPC1, Ago2, ORF57, TIA-1, and PYM by the corresponding antibodies (C). Panels show one representative of three independent experiments. RNPs, ribonucleoproteins.
PABPC1 [poly(A)-binding protein cytoplasmic 1] is a cytoplasmic protein involved in mRNA translation initiation and stability (78). Ago2 is a core component of the RNA-induced silencing complex (RISC) responsible for small interfering RNA (siRNA)- and miRNA-mediated translational suppression and degradation of targeted mRNAs (79, 80). We found that ORF57, despite having no effect on PABPC1, Ago2, and two other translational regulators, TIA-1 (81) and PYM (82), associated with 40S ribosomes in fractions 2 to 4, enhanced PABPC1, but simultaneously prevented Ago2 association with the translationally active polysomes (Fig. 9C, compare lanes 7 to 10 in the absence and presence of ORF57). These data are consistent with our previous findings that ORF57 promotes RNA translation and stability by blocking miRNA-mediated Ago2 binding to mRNA (21, 22).
DISCUSSION
As an RNA-binding protein responsible for the posttranscriptional regulation of viral RNA transcripts during viral lytic infection, KSHV ORF57 in BCBL-1 cells in this study was found to interact with host noncoding RNAs, including lncRNAs, snoRNAs, and rRNAs, to execute additional novel functions. Importantly, we found that ORF57 is associated with translationally active polysomes and, together with PABPC1, prevents Ago2 from association with polysomes, by which ORF57 facilitates its role in promoting protein translation.
Seeking specific RBP-interacting RNAs is vital for our understanding of RBP functions in various cell types and tissues, but it has been very challenging in the context of physiological and diseased conditions. Various CLIP assays using a specific antibody were developed to identify RNA targets of each RBP and its specific binding site(s) on the targeted RNAs under cell culture conditions (83–85). By using these techniques, we and others previously demonstrated some of viral and host RNAs specifically interacting with ORF57 in KSHV-infected cells (18, 19, 21, 22, 47, 50, 53). Although most CLIP-seq studies focus on the protein-coding RNAs, we found that the noncoding RNA reads in our ORF57 CLIP-seq represented a large proportion of ORF57-associated RNAs. RBP binding to noncoding RNAs with known or unknown functions, such as rRNAs, snRNAs, and lncRNAs, was also reported for more than 70% of RBPs in enhanced CLIP (eCLIP)-seq or other CLIP-seq techniques (86, 87). In many studies, rRNA reads were often considered contaminants and discarded, and thus, the biological significance of these interactions was often ignored.
Most of the known host and viral lncRNAs have been characterized as regulatory RNAs, which function as chromatin modifiers or posttranscriptional regulators by their specific sequence motifs to interact with DNA, RNA, or proteins (60, 88). Functional lncRNA-protein interactions have been implied in diverse diseases, including virus infection and cancer development (89–91). For example, KSHV lncRNA PAN RNA interacts with viral and host proteins, including viral ORF57 and LANA and host PABPC1 and demethylases UTX and JMJD3, to regulate viral lytic reactivation and infection (18, 19, 92–97). On the other hand, host lncRNA GAS5 interacts with the hepatitis C virus (HCV) NS3 protein to repress HCV replication (98). Hepatitis B virus (HBV) HBx protein binds directly to lncRNA DLEU2 to modulate HBV replication (99). In this study, we identified ORF57 interaction with 117 host lncRNAs by ORF57 CLIP-seq and selectively verified ORF57 interactions with LINC00324, LINC00355, and LINC00839 by RIP during lytic KSHV infection, all of which are involved in cell proliferation, migration, and invasion (61, 62, 64, 100, 101). Furthermore, we demonstrated that these ORF57-lncRNA interactions during KSHV lytic infection decrease the expression of LINC00324 but increase the expression of LINC00355. Although lncRNAs are differentially expressed from different cell types and tissues and not all types of cells express equal amounts of certain lncRNAs, the altered lncRNA expression in our study appeared to be specific only for wt ORF57 and not for a dysfunctional ORF57 mutant. Since both LINC00324 and LINC00355 are involved in promotion of cell proliferation, migration, and invasion (61, 62, 64, 100, 101), the ORF57-induced decrease of host LINC00324 and increase of LINC00355 expression might facilitate KSHV carcinogenesis and make viral ORF57 possibly play a tumorigenic role during KSHV infection.
snoRNAs are a growing family of noncoding RNAs highly concentrated in the nucleoli and mainly involved in rRNA biogenesis by guiding rRNA base modifications via formation of a short duplex with rRNA (26, 102). The last step of rRNA maturation occurs in the cytoplasm (103). Recent studies showed that snoRNAs in the nucleus and the cytoplasm are also involved in regulation of host gene expression, RNA splicing and 3′ end processing, the phosphatidylinositol 3-kinase (PI3K)-Akt pathway, production of snoRNA-derived small RNAs, and interaction with Dicer (27, 31, 76, 104, 105). Many snoRNAs were also reported being commonly associated with protein kinase R (PKR) (87). Over the years, we have found that KSHV ORF57 interacts with host splicing factors, snRNAs, and PABPC1 to regulate RNA splicing and stability (14). Findings of ORF57 interactions with a large number of snoRNAs independently of canonical snoRNPs NOP58 and fibrillarin in this study suggest additional ORF57 regulation of host cell functions probably through the interacting snoRNAs. In this study, despite that these ORF57-snoRNA interactions do not affect the expression of snoRNAs, knockdown of fibrillarin expression in iSLK/Bac16 cells with KSHV lytic infection reduced the production of snoRNAs and the levels of host CDK4 RNA and protein, an important protein kinase for cell cycle control, but showed no effect on viral ORF57, ORF59, or RTA expression. Data suggest that these viral RNAs are more tolerable than host CDK4 RNA for snoRNA-mediated regulation and the ORF57-snoRNA interaction might contribute partially to the host shutdown during lytic KSHV infection. Although the majority of snoRNAs are derived from the intron region of protein-coding and noncoding transcripts, the snoRD3@ cluster has its own promoter and thus is produced uniquely by its own transcripts. Functionally, the snoRD3@ serves as a scaffold for early ribosome assembly and was recently discovered as a source of miRNA miR-U3 to target SNX27 mRNA (76). Demonstration of ORF57 association with snoRD3@ implies a potential role for ORF57 in the described snoRD3@ functions. Together, these findings provide a new path to investigate how other individual snoRNAs might be involved in DNA and RNA virus infections (33, 35).
Herpesvirus infections, including HSV-1 (43), HCMV (41), and KSHV (44), dysregulate rRNA biogenesis, but the mechanism of this dysregulation remains elusive. We found that KSHV ORF57 in BCBL-1 cells with viral lytic infection and in HEK293T cells with ectopic expression is highly associated with rRNAs. We demonstrated ORF57 association with rRNAs by ORF57 CLIP-seq, rRNA oligonucleotide pulldown assays, ORF57 RIP, and polysome profiling. Interestingly, we found that many ORF57-binding sites on rRNAs overlap the rRNA sites where individual snoRNAs bind during rRNA modifications (103, 106). Various modified CLIP-seq technologies have been applied to study RBP-associated RNA-RNA interactions (107–109). By Northern blotting using an oligonucleotide probe with the rRNA sequence complementary to snoRA71B, we were able to detect ORF57-bound snoRA71B after ORF57 RIP, suggesting that this rRNA region with bound snoRNA71B could be co-associated with ORF57 and thus both were protected from RNase digestion during anti-ORF57 CLIPs. However, we were unable to detect the snoRD82 by Northern blotting with the same experimental approach (Fig. 8B), despite the fact that the same rRNA region-derived RNA oligonucleotide was found to strongly bind ORF57 (Fig. 8D and E). Although the Northern blot data could have resulted from the abundances of individual snoRNAs, these data may imply that the rRNA-snoRNA interactions are dynamic and that in many cases, these snoRNA binding sites on an rRNA region might be free from snoRNA binding, particularly after completion of rRNA modification. The finding of ORF57 association with snoRA71B is interesting because snoRA71 interacts with endogenous PKR (87) and ORF57 interaction with PKR prevents PKR activation (24).
KSHV ORF57 interacts with the RRM motifs of PABPC1 to regulate KSHV PAN RNA expression (18, 19). ORF57 also interacts with the double-stranded RNA (dsRNA)-binding domain of PKR to block the formation of stress granules involved in translational suppression (24) and with the N domain of Ago2 (23), a major component of RISC by which siRNA and miRNA could mediate mRNA degradation and translation inhibition (79, 80), to inhibit the formation of RNA P-bodies (processing bodies) (23) and enhance the production of IL-6 during KSHV infection (21, 22). Consistently, our polysome profiling revealed that ORF57 associates with ribosomes from the beginning of ribosome assembly to the late, translationally active polysome stage. ORF57 association with 40S ribosomal subunits was expected because one of the ORF57 functions is regulating translational initiation by interaction with PYM (82) and eIF4E (24) to recruit the preinitiation complex. We found PYM, TIA-1, Ago2, and PABPC1 association with 40S ribosome subunits in the absence of ORF57, but we did not see the related PYM to accompany with ORF57 (Fig. 9C) during ribosomal assembly as suggested previously (82). Strikingly, our polysome profiling studies found that the association of ORF57 with polysomes promotes recruitment of PABPC1, but not Ago2, to the polysomes. Together, these observations provide compelling evidence that ORF57 plays a functional role in protein translation by preventing Ago2 from association with the translationally active polysomes.
MATERIALS AND METHODS
Cells.
Renal carcinoma cell line-derived iSLK cells carrying a KSHV Bac16 genome (iSLK/Bac16 wt cells) (110) or containing an ORF57-null KSHV genome from CRISPR-Cas9 knockout (iSLK/Bac16 clone 2G1 cells) (11) and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose and HEPES (12430054; Thermo Fisher Scientific, Waltham, MA). Primary effusion lymphoma BCBL-1 cells carrying the KSHV genome were cultivated in RPMI 1640 medium (1879020; Thermo Fisher Scientific). All media were supplemented with 10% HyClone fetal bovine serum (SH30070.03; Cytiva, Marlborough, MA) and 1× penicillin-streptomycin-glutamine (Thermo Fisher Scientific). The iSLK/Bac16 cells were grown under selection with the addition of 150 μg/mL of hygromycin B, 1 μg/mL of puromycin, and 250 μg/mL of G418. KSHV lytic infection in BCBL-1 cells was induced by adding valproic acid (VA; P4543; Sigma-Aldrich, St. Louis, MO) at a final concentration of 1 mM for 24 h. The lytic infection in iSLK/Bac16 cells was induced by adding 1 mM sodium butyrate (NaBu) (B5887; Millipore Sigma) and 1 μg/mL of doxycycline (Dox; NC0424034; Fisher Scientific) for 12 or 48 h.
CLIP-seq.
ORF57 CLIP was performed in three biological replicates as described previously (47). Briefly, 6 × 107 VA-induced BCBL-1 cells were washed twice with cold 1× phosphate-buffered saline (PBS) and exposed to UV light at 480 mJ/cm2 at 254 nm. After UV cross-linking, the cells were lysed on ice in 1× radioimmunoprecipitation assay (RIPA) buffer (BP-115; Boston Bioproducts, Ashland, MA) (50 mM Tris base/Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40 substitute) supplemented with protease inhibitors (complete mini-EDTA-free protease inhibitor cocktail, 469315900; Millipore Sigma, Burlington, MA) for 30 min, followed by a brief sonication. The obtained cell lysates were cleared by 10 min of centrifugation at 20,000 × g and 4°C. Protein A-agarose beads (16-266; Millipore Sigma) were washed with 1× immunoprecipitation (IP) buffer (50 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% NP-40) before coating with an affinity-purified, highly specific anti-ORF57 rabbit polyclonal antibody (used in all experiments described in this report) (12, 22) or control rabbit nonspecific IgG (02-6102; Thermo Fisher Scientific). To further eliminate nonspecific interactions, the cell lysates were precleaned three times with nonspecific (control) rabbit IgG-coated beads and then used in immunoprecipitation with the anti-ORF57 or control rabbit IgG antibody-coated beads overnight at 4°C. After IP, the beads were washed 3 times with 1× IP buffer, and RNAs in the pulldowns were digested with an RNase A/T1 mix (EN0551; Thermo Fisher Scientific) for 5 s, followed by dephosphorylation with recombinant shrimp alkaline phosphatase (rSAP; M0371S; New England BioLabs, Ipswich, MA). Finally, proteinase K (71049; Millipore Sigma) was added to remove the proteins from RNA. The obtained RNA was purified with phenol-chloroform extraction, ethanol precipitated, ligated to a 3′ RNA linker, and reverse transcribed. The obtained cDNA was circularized and used as a template for library construction using PCR amplification, followed by size selection of PCR fragments at 200 to 350 nt. High-throughput sequencing (HITS) was performed by a standard stranded RNA-seq protocol on the Illumina HiSeq 2500 platform with a 2 × 50-bp paired-end (PE) modality.
Raw data processing and read counting per transcript.
The following pipeline was applied for each ORF57 CLIP-seq replicate. First, the raw reads were trimmed by Cutadapt (version 1.18) (111). Next, the trimmed reads were mapped to the rRNA (GenBank accession number U13369) and to the chimeric human genome GRCh37/KSHV (GenBank accession number U75698.1) by running STAR version 2.7.6a. Then bam files were converted to bedGraph with the bedtools (112) for easier IGV visualization. Transcript strand-specific feature counts for the human genome were obtained using the RSEM (RNA-Seq by Expectation Maximization) software package (version 1.3.0) (113). Plotly was used to perform a three-dimensional (3D) scatterplot. Pearson's coefficient analysis was performed with the statistical software Prism 6 (GraphPad).
Identification of ORF57-binding sites in host gene transcripts.
A peak caller Piranha software (version 4.0.164) (54) (https://github.com/smithlabcode/piranha) was used to identify regions with a statistically significant read enrichment in ORF57 CLIP-seq. Since we observed high reproducibility among ORF57 CLIP-seq replicates, we combined reads from all three ORF57-CLIPs to ensure higher read depth. The combined library was first trimmed by Cutadapt (version 1.18) (111) and then mapped to the human genome sequence (GRCh37) by STAR aligner (version 2.7.6a) (114). Next, we used the first mode of Piranha operation, which uses a single regular distribution that fits the input data, with each region having assigned a P value based on this distribution. Piranha called peak boundaries were truncated to a width of 50 nt (bin). The parameters used for peak calling were “-aligner bed -dist_type 0 40 -landerwaterman 0.01 -subpeaks 0.5 -bedgraph -readahead_window.” The list of peaks was annotated by the intersection with the GRCh37.p13 GTF file (https://www.gencodegenes.org/human/release_19.html) using the “intersectBed” command of the bedtools package (version 2.29.2) (112). The peaks were annotated by gene identifier (ID) and name.
Anti-ORF57 RNA RIP, RT-PCR, snoRNA array, and Northern blot analyses.
To confirm ORF57 association with selected host transcripts, we performed RIP using the specific anti-ORF57 antibody described above, with nonspecific IgG serving as a negative RIP control. The first two steps in ORF57 RIP are the same as described for the ORF57 CLIP-seq (Fig. 1A). However, RNase A/T1 digestion was skipped to recover the full-length RNAs for the downstream RT-PCR, snoRNA array, and Northern blot assays. RNA from the RIP or input total RNAs from BCBL-1, iSLK/Bac16, and HEK293T cells were extracted by TRIpure reagent (11667157001; Millipore Sigma).
ORF57 RIP RT-PCR was performed with 2 × 107 of VA-induced BCBL-1 cells or HEK293T cells expressing ORF57. RNA extracted was DNase treated with a TURBO DNA-free kit (AM1907; Thermo Fisher Scientific). The cDNA synthesis from RIP and control total RNA was carried out with a SuperScript first-strand synthesis system kit (11904018; Thermo Fisher Scientific) in the absence or presence of reverse transcriptase, followed by PCR using AmpliTaq DNA polymerase (N8080160; Thermo Fisher Scientific). The following specific primer pairs were used to amplify the selected ORF57 targets: oBAH13 and oBAH14 for LINC00324, oBAH115 and oBAH116 for LINC00355, oBAH125 and oBAH126 for LINC00839, oBAH123 and oBAH124 for LINC00152, oBAH198 and oBAH199 for snoRD3@ (for snoRD3A, B-1, -B2, -3C, and -3D), oBAH200 and oBAH201 for snoRA64, oBAH202 and oBAH219 for snoRA71B (also for snoRA71D), oBAH204 and oBAH205 for snoRD105B, oBAH195 and oBAH206 for snoRD80, and oBAH192 and oBAH193 for RN7SK (see Table 1 for details).
TABLE 1.
List of DNA and RNA oligonucleotides used in this studya
F, forward; R, reverse; B, biotinylated; @, family members; r, ribonucleotide; d, deoxyribonucleotide; P, TaqMan probe for RT-qPCR; FAM, 6-carboxyfluorescein.
ORF57 RIP snoRNA RT-qPCR array (Arraystar Inc., Rockville, MD) was performed with 3 × 107 of VA-induced BCBL-1 cells. The RIP-obtained RNAs and input control total RNA from BCBL-1 cells were converted to cDNA using an rtStar first-strand cDNA synthesis kit (AS-FS-001; Arraystar Inc.). The snoRNA array (quantification by RT-qPCR) was performed according to the manufacturer’s protocols using the nrStar human snoRNA PCR array (AS-NR-003-1; Arraystar Inc.). The 5S rRNA was used as an internal positive control. The 2−ΔΔCT method was used to determine fold change (FC) of enriched individual snoRNAs in ORF57 RIP versus control IgG RIP. The statistical significance was determined by Student’s t test based on three independent experiments.
For Northern blot analysis, ORF57 RIP was performed using 2 × 107 of VA-induced BCBL-1 cells. The RIP-obtained RNA was separated in a 3% agarose gel containing formaldehyde and transferred to a nylon membrane. RNA transcripts snoRD3@, snoRA71B, snoRD82, snoRA42, snoRD93, and snRNA U6 were detected by individual 32P-labeled antisense DNA oligonucleotide probes (see Table 1 for details).
RT-PCR and RT-qPCR.
Total RNA was prepared with TRIpure reagent (11667157001; Millipore Sigma) from 2 × 106 of BCBL-1 cells and 1 × 106 of iSLK/Bac16 cells with latent or lytic KSHV infection. HEK293T cells (1 × 106) were transfected for 24 h with 2 μg of wt (pVM7) ORF57, mt (pVM89) ORF57 (20), or empty vector pFLAG-CMV5.1.
For RT-PCR, total RNA (2 μg) was reverse transcribed using random hexamers in the absence or presence of murine leukemia virus (MuLV) reverse transcriptase (4311235; Thermo Fisher Scientific) at 42°C for 1 h, followed by PCR amplification using AmpliTaq DNA polymerase (N8080160; Thermo Fisher Scientific) with the primer pairs described above for each lincRNA.
For RT-qPCR, total RNA (1 μg) treated with DNase for quantification of snoRNAs, but no DNase treatment for quantification of the rest of the gene transcripts, was reverse transcribed using the SuperScript first-strand synthesis system kit (11904018; Thermo Fisher Scientific). For RT-qPCR, TaqMan assays from Thermo Fisher Scientific with probe-spanning exons were used to perform real-time PCR quantification of the selected lncRNAs: Hs00543397_m1 for LINC00324, Hs04939567_m1 for LINC00355, Hs03309498_sH for snoRA71B, Hs03301054_s1 for snoRD105B, and Hs02758991_g1 for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). TaqMan IDT PrimeTime custom-designed probes were used for specific detection and quantification for expression of ORF57 and ORF59 (46) (see Table 1 for details) and IDT TaqMan probe Hs.PT.58.38531977 for CDK4.
RNA-protein pulldown assays.
Biotinylated RNA oligonucleotides were used to pull down RNA-binding proteins (22, 115). Briefly, VA-induced BCBL-1 cells were washed twice with 1× PBS and lysed on ice in 1× RIPA buffer containing protease inhibitors. After brief sonication, the cell extract was cleared by 15 min of centrifugation at 12,000 × g and 4°C, and the supernatant was used for pulldown assays with one of the following biotinylated RNA oligonucleotides (Integrated DNA Technologies [IDT], Coralville, IA): oNS15 derived from 28S rRNA; oNS11, oNS12, oNS13, oNS24 derived from 18S rRNA; and oNS14 derived from 5.8S rRNA (see Table 1 for details). Previously reported oNP41 and oNP42 (22) derived from viral interleukin-6 (vIL-6) RNA were used as negative and positive controls, respectively (see Table 1 for details). Individual RNA oligonucleotides (800 pM) were immobilized on NeutrAvidin beads (29201; Thermo Fisher Scientific) in 300 μL of 1× Tris-buffered saline (TBS) at 4°C for 2 h. After two washes in 1× TBS, the beads coated with a biotinylated RNA oligonucleotide were incubated with 100 μL of cell extract from ∼2 × 106 VA-induced BCBL-1 cells. The mixture with a final volume of 400 μL was incubated overnight at 4°C on a rotating mixer. After the incubation, the beads were washed three times with 1× TBS, and proteins in the pulldown were eluted in 40 μL of 2× SDS protein sample buffer supplemented with 5% (vol/vol) 2-mercaptoethanol (2-ME). Proteins were analyzed by Western blotting using an anti-ORF57 antibody.
Co-IP and Western blotting.
For co-IP assays, we used 1 × 107 of VA-induced BCBL-1 cells or HEK293T cells transfected with an ORF57-expressing vector (pVM7, 8 μg) using LipoD293 transfection reagent (SL100668; SignaGen Laboratories, Frederick, MD). The cells were washed with 1× PBS and resuspended in 500 μL of ice-cold 1× RIPA buffer. Precleaned protein lysates were mixed with 100 μL (50% slurry) of protein A beads (16-125; Millipore Sigma), coated by either rabbit anti-ORF57 antibody or rabbit nonspecific IgG (Thermo Fisher Scientific) in 1 mL of 1× IP buffer. The mixture was incubated overnight at 4°C, followed by extensive washing with IP buffer. The immunoprecipitated complexes were eluted with 2× SDS protein sample buffer supplemented with 10% (vol/vol) 2-ME and were analyzed by Western blotting to detect individual cellular or viral proteins with antibodies against NOP58 (14409-1-AP; Proteintech, Rosemont, IL), fibrillarin (FBL, 2639; Cell Signaling, Danvers, MA), nucleolin (NCL, Ab22758; Abcam, Cambridge, MA), and ORF57 proteins.
siRNA-mediated knockdown of fibrillarin expression and Western blot analysis.
iSLK/Bac16 cells were used to knock down fibrillarin expression by SMARTpool human siRNAs targeting fibrillarin (L-011269-00-0005; Horizon, Lafayette, CO) or nontargeting control siRNA (D-001810-10; Horizon) at 40 nM using a LipoJet in vitro transfection kit (version II, SL100468; SignaGen Laboratories). After 36 h of transfection, the cells were induced for 12 h with butyrate plus doxycycline for lytic KSHV infection before total protein or RNA sample preparation for the corresponding experiments.
Protein lysates were obtained by lysing the cells directly with 1× SDS protein sample buffer supplemented with 5% (vol/vol) 2-ME and were analyzed by Western blotting to detect individual cellular or viral proteins with antibodies against CDK4 (2906; Cell Signaling), GAPDH (97166; Cell Signaling), fibrillarin (2639; Cell Signaling), RTA (24), and viral ORF57 proteins.
Polysome profiling.
HEK293T cells (5 × 106) were transfected with an ORF57-expressing vector (pVM7, 2.5 μg/dish) using LipoD293 transfection reagent (SignaGen Laboratories). Cells without ORF57 were used as a negative control. A total of 3 samples were prepared for each group. Twenty-four hours after the transfection, the culture medium was replaced with a fresh medium containing 100 μg/mL of cycloheximide for 10 min at 37°C. The cells were harvested on ice in 5 mL of cold PBS containing 100 μg/mL of cycloheximide. After short centrifugation, the cell pellet was resuspended in 750 μL of polysome buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.25 mM MgCl2, 5 U/mL of RNase inhibitor, complete mini-EDTA-free protease inhibitor cocktail, 1 mM dithiothreitol [DTT], 1% NP-40). After 15 min of incubation on ice, the lysates were clarified by 20 min of centrifugation at 12,000 × g and 4°C. The concentration of each sample was adjusted based on A260. An equal amount of each sample was loaded on the top of a 4.5% to 45% continuous sucrose density gradient and spun for 3 h at 39,000 rpm in an SW-41 rotor at 4°C. An automated fractionator was used to collect 1 mL per fraction. The fraction’s A254 absorbance was recorded using the attached spectrophotometer to identify the monosomes (40S, 60S, and 80S) and polysomes in the individual fractions. The total protein and RNA samples were prepared from the input and individual fractions by lysis in 2× SDS/2-ME protein sample buffer and by RNA extraction with TRIpure reagent. The protein samples were analyzed by Western blotting to detect cellular PABPC1 (ab21060; Abcam), TIA-1 (sc-1751; Santa Cruz), PYM (WIBG, sc-169812; Santa Cruz, Dallas, TX), Ago2 (07-590; Millipore Sigma), and viral ORF57. The total RNA isolated from fractions 2 to 10 was subjected to electrophoresis under denaturing conditions in a formaldehyde-containing 1.5% agarose gel. The distribution of 28S and 18S rRNAs in individual fractions was visualized by ethidium bromide.
RNA-seq analysis of BCBL-1 cells with latent and lytic KSHV infection.
Total RNA was extracted from BCBL-1 cells with latent or VA-induced lytic KSHV infection using miRNeasy (217004; Qiagen, Hilden, Germany). RNA samples were evaluated using Fragment Analyzer and NanoDrop. Library preparation and sequencing were done by ACGT Inc. (Germantown, MD). Indexed cDNA sequencing libraries were prepared from the RNA samples using a True-Seq stranded RNA sample preparation kit (Illumina, Inc.) and bar-coded with individual tags according to the manufacturer’s recommendations. Indexed libraries were prepared as equimolar pools and run on HiSeq 4000 (2 × 150 paired-end runs) in order to generate 40 million paired-end reads per sample. All RNA-seq data were aligned to human genome GRCh37 using STAR aligner v2.5.2b (114). Differential expression was analyzed by Limma-voom (116).
Data availability.
The ORF57 CLIP-seq data have been deposited in NCBI’s GEO database under accession number GSE179726. The BCBL-1 cell RNA-seq data are accessible in NCBI’s GEO database under accession number GSE179727.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH (ZIASC010357 to Z.M.Z.).
We thank Alan Hinnebusch of NICHD, NIH, for his advice on polysome profiling experiments.
Footnotes
Supplemental material is available online only.
Contributor Information
Zhi-Ming Zheng, Email: zhengt@exchange.nih.gov.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266:1865–1869. 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci USA 93:14862–14867. 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L. 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86:1276–1280. 10.1182/blood.V86.4.1276.bloodjournal8641276. [DOI] [PubMed] [Google Scholar]
- 4.Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708–2714. 10.1182/blood.V86.7.2708.2708. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan RJ, Pantanowitz L, Casper C, Stebbing J, Dezube BJ. 2008. HIV/AIDS: epidemiology, pathophysiology, and treatment of Kaposi sarcoma-associated herpesvirus disease: Kaposi sarcoma, primary effusion lymphoma, and multicentric Castleman disease. Clin Infect Dis 47:1209–1215. 10.1086/592298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan L, Majerciak V, Zheng ZM, Lan K. 2019. Towards better understanding of KSHV life cycle: from transcription and posttranscriptional regulations to pathogenesis. Virol Sin 34:135–161. 10.1007/s12250-019-00114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukac DM, Renne R, Kirshner JR, Ganem D. 1998. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304–312. 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 8.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. 1998. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA 95:10866–10871. 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukac DM, Kirshner JR, Ganem D. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol 73:9348–9361. 10.1128/JVI.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Z, Swaminathan S. 2006. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J Virol 80:5251–5260. 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.BeltCappellino A, Majerciak V, Lobanov A, Lack J, Cam M, Zheng ZM. 2019. CRISPR/Cas9-mediated knockout and in situ inversion of the ORF57 gene from all copies of the Kaposi’s sarcoma-associated herpesvirus genome in BCBL-1 cells. J Virol 93:e00628-19. 10.1128/JVI.00628-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem 281:28365–28378. 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- 13.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. 2008. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol 82:2792–2801. 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majerciak V, Zheng ZM. 2015. KSHV ORF57, a protein of many faces. Viruses 7:604–633. 10.3390/v7020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan F, Gao ZQ, Majerciak V, Bai L, Hu ML, Lin XX, Zheng ZM, Dong YH, Lan K. 2018. The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function. PLoS Pathog 14:e1007232. 10.1371/journal.ppat.1007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majerciak V, Pripuzova N, Chan C, Temkin N, Specht SI, Zheng ZM. 2015. Stability of structured Kaposi’s sarcoma-associated herpesvirus ORF57 protein is regulated by protein phosphorylation and homodimerization. J Virol 89:3256–3274. 10.1128/JVI.03721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura K, Ueda K, Guwanan E, Sakakibara S, Do E, Osaki E, Yada K, Okuno T, Yamanishi K. 2004. A posttranscriptional regulator of Kaposi’s sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. Virology 325:364–378. 10.1016/j.virol.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 18.Massimelli MJ, Kang JG, Majerciak V, Le SY, Liewehr DJ, Steinberg SM, Zheng ZM. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int J Biol Sci 7:1145–1160. 10.7150/ijbs.7.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massimelli MJ, Majerciak V, Kruhlak M, Zheng ZM. 2013. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J Virol 87:243–256. 10.1128/JVI.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majerciak V, Lu M, Li X, Zheng ZM. 2014. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 20:1747–1758. 10.1261/rna.045500.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang JG, Majerciak V, Uldrick TS, Wang X, Kruhlak M, Yarchoan R, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J Pathol 225:378–389. 10.1002/path.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol 85:2620–2630. 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma NR, Majerciak V, Kruhlak MJ, Yu L, Kang JG, Yang A, Gu S, Fritzler MJ, Zheng ZM. 2019. KSHV RNA-binding protein ORF57 inhibits P-body formation to promote viral multiplication by interaction with Ago2 and GW182. Nucleic Acids Res 47:9368–9385. 10.1093/nar/gkz683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma NR, Majerciak V, Kruhlak MJ, Zheng ZM. 2017. KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog 13:e1006677. 10.1371/journal.ppat.1006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojha S, Malla S, Lyons SM. 2020. snoRNPs: functions in ribosome biogenesis. Biomolecules 10:783. 10.3390/biom10050783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. 2017. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14:1138–1152. 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bratkovic T, Bozic J, Rogelj B. 2020. Functional diversity of small nucleolar RNAs. Nucleic Acids Res 48:1627–1651. 10.1093/nar/gkz1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, Nevo Y, Eyras E, Sperling R, Stamm S. 2016. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci USA 113:E1625–E1634. 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. 2010. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19:1153–1164. 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong J, Li Y, Liu CJ, Xiang Y, Li C, Ye Y, Zhang Z, Hawke DH, Park PK, Diao L, Putkey JA, Yang L, Guo AY, Lin C, Han L. 2017. A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep 21:1968–1981. 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 31.Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. 2019. Small nucleolar RNAs: insight into their function in cancer. Front Oncol 9:587. 10.3389/fonc.2019.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abel Y, Rederstorff M. 2019. SnoRNAs and the emerging class of sdRNAs: multifaceted players in oncogenesis. Biochimie 164:17–21. 10.1016/j.biochi.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Murray JL, Sheng J, Rubin DH. 2014. A role for H/ACA and C/D small nucleolar RNAs in viral replication. Mol Biotechnol 56:429–437. 10.1007/s12033-013-9730-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamm S, Lodmell JS. 2019. C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks. Noncoding RNA Res 4:46–53. 10.1016/j.ncrna.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian X, Xu C, Wu B, Tang H, Zhao P, Qi Z. 2020. SNORD126 promotes hepatitis C virus infection by upregulating claudin-1 via activation of PI3K-AKT signaling pathway. Front Microbiol 11:565590. 10.3389/fmicb.2020.565590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Learned RM, Smale ST, Haltiner MM, Tjian R. 1983. Regulation of human ribosomal RNA transcription. Proc Natl Acad Sci USA 80:3558–3562. 10.1073/pnas.80.12.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponti D, Troiano M, Bellenchi GC, Battaglia PA, Gigliani F. 2008. The HIV Tat protein affects processing of ribosomal RNA precursor. BMC Cell Biol 9:32. 10.1186/1471-2121-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizuno CM, Guyomar C, Roux S, Lavigne R, Rodriguez-Valera F, Sullivan MB, Gillet R, Forterre P, Krupovic M. 2019. Numerous cultivated and uncultivated viruses encode ribosomal proteins. Nat Commun 10:752. 10.1038/s41467-019-08672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong HJ, Zhang R, Kuang Y, Wang XJ. 2021. Selective regulation in ribosome biogenesis and protein production for efficient viral translation. Arch Microbiol 203:1021–1032. 10.1007/s00203-020-02094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Huttenhofer A. 2009. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog 5:e1000547. 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianco C, Mohr I. 2019. Ribosome biogenesis restricts innate immune responses to virus infection and DNA. Elife 8:e49551. 10.7554/eLife.49551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madjar JJ, Simonin D, Massé T, Diaz JJ. 1997. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J Gen Virol 78:435–443. 10.1099/0022-1317-78-2-435. [DOI] [PubMed] [Google Scholar]
- 43.Belin S, Kindbeiter K, Hacot S, Albaret MA, Roca-Martinez JX, Therizols G, Grosso O, Diaz JJ. 2010. Uncoupling ribosome biogenesis regulation from RNA polymerase I activity during herpes simplex virus type 1 infection. RNA 16:131–140. 10.1261/rna.1935610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadagopan S, Sharma-Walia N, Veettil MV, Bottero V, Levine R, Vart RJ, Chandran B. 2009. Kaposi’s sarcoma-associated herpesvirus upregulates angiogenin during infection of human dermal microvascular endothelial cells, which induces 45S rRNA synthesis, antiapoptosis, cell proliferation, migration, and angiogenesis. J Virol 83:3342–3364. 10.1128/JVI.02052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atari N, Rajan KS, Chikne V, Cohen-Chalamish S, Orbaum O, Jacob A, Kalt I, Michaeli S, Sarid R. 2020. Lytic reactivation of the Kaposi’s sarcoma-associated herpesvirus (KSHV) is accompanied by major nucleolar alterations. bioRxiv 10.1101/2020.05.15.097808. [DOI] [PMC free article] [PubMed]
- 46.Massimelli MJ, Majerciak V, Kang JG, Liewehr DJ, Steinberg SM, Zheng ZM. 2015. Multiple regions of Kaposi’s sarcoma-associated herpesvirus ORF59 RNA are required for its expression mediated by viral ORF57 and cellular RBM15. Viruses 7:496–510. 10.3390/v7020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Y, Liu P, Majerciak V, Zhu J, Zheng ZM. 2016. CLIP-seq to identify KSHV ORF57-binding RNA in host B cells. Curr Protoc Microbiol 41:1E.11.1–1E.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi’s sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J Virol 81:9990–9998. 10.1128/JVI.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li DJ, Verma D, Swaminathan S. 2012. Binding of cellular export factor REF/Aly by Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 protein is not required for efficient KSHV lytic replication. J Virol 86:9866–9874. 10.1128/JVI.01190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verma D, Li DJ, Krueger B, Renne R, Swaminathan S. 2015. Identification of the physiological gene targets of the essential lytic replicative Kaposi’s sarcoma-associated herpesvirus ORF57 protein. J Virol 89:1688–1702. 10.1128/JVI.02663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Majerciak V, Uranishi H, Kruhlak M, Pilkington GR, Massimelli MJ, Bear J, Pavlakis GN, Felber BK, Zheng ZM. 2011. Kaposi’s sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J Virol 85:1528–1540. 10.1128/JVI.01709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J Virol 80:11968–11981. 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sei E, Wang T, Hunter OV, Xie Y, Conrad NK. 2015. HITS-CLIP analysis uncovers a link between the Kaposi’s sarcoma-associated herpesvirus ORF57 protein and host pre-mRNA metabolism. PLoS Pathog 11:e1004652. 10.1371/journal.ppat.1004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. 2012. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28:3013–3020. 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. 2011. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA 17:792–798. 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milligan MJ, Lipovich L. 2014. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet 5:476. 10.3389/fgene.2014.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Q, Hao Q, Prasanth KV. 2018. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 34:142–157. 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guh CY, Hsieh YH, Chu HP. 2020. Functions and properties of nuclear lncRNAs—from systematically mapping the interactomes of lncRNAs. J Biomed Sci 27:44. 10.1186/s12929-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rashid F, Shah A, Shan G. 2016. Long non-coding RNAs in the cytoplasm. Genomics Proteomics Bioinformatics 14:73–80. 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. 2018. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA 9:e1471. 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu S, Sun Z, Tang L, Chen L. 2020. LINC00355 promotes tumor progression in HNSCC by hindering microRNA-195-mediated suppression of HOXA10 expression. Mol Ther Nucleic Acids 19:61–71. 10.1016/j.omtn.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Zhao W, Jin Y, Wu P, Yang J, Chen Y, Yang Q, Huo X, Li J, De W, Chen J, Yang F. 2020. LINC00355 induces gastric cancer proliferation and invasion through promoting ubiquitination of P53. Cell Death Discov 6:99. 10.1038/s41420-020-00332-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Liang Y, Rong X, Luo Y, Li P, Han Q, Wei L, Wang E. 2020. A novel long non-coding RNA LINC00355 promotes proliferation of lung adenocarcinoma cells by down-regulating miR-195 and up-regulating the expression of CCNE1. Cell Signal 66:109462. 10.1016/j.cellsig.2019.109462. [DOI] [PubMed] [Google Scholar]
- 64.Chen Q, Shen H, Zhu X, Liu Y, Yang H, Chen H, Xiong S, Chi H, Xu W. 2020. A nuclear lncRNA Linc00839 as a Myc target to promote breast cancer chemoresistance via PI3K/AKT signaling pathway. Cancer Sci 111:3279–3291. 10.1111/cas.14555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Guo H, Ma L, Chen X, Chen G. 2020. Long noncoding RNA LINC00839 promotes the malignant progression of osteosarcoma by competitively binding to microRNA-454-3p and consequently increasing c-Met expression. Cancer Manag Res 12:8975–8987. 10.2147/CMAR.S269774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Zou Z, Ma T, He X, Zhou J, Ma H, Xie M, Liu Y, Lu D, Di S, Zhang Z. 2018. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis 9:717. 10.1038/s41419-018-0758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J, Dai C, Yu X, Yin XB, Zhou F. 2020. Long noncoding RNA LINC00324 exerts protumorigenic effects on liver cancer stem cells by upregulating fas ligand via PU box binding protein. FASEB J 34:5800–5817. 10.1096/fj.201902705RR. [DOI] [PubMed] [Google Scholar]
- 68.Xu J, Li Z, Su Q, Zhao J, Ma J. 2020. Suppression of long noncoding RNA LINC00324 restricts cell proliferation and invasion of papillary thyroid carcinoma through downregulation of TRIM29 via upregulating microRNA-195-5p. Aging (Albany NY) 12:26000–26011. 10.18632/aging.202219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D, Huang Z. 2017. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis 6:395. 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Binder S, Zipfel I, Friedrich M, Riedel D, Ende S, Kampf C, Wiedemann K, Buschmann T, Puppel SH, Reiche K, Stadler PF, Horn F. 2020. Master and servant: LINC00152—a STAT3-induced long noncoding RNA regulates STAT3 in a positive feedback in human multiple myeloma. BMC Med Genomics 13:22. 10.1186/s12920-020-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kufel J, Grzechnik P. 2019. Small nucleolar RNAs tell a different tale. Trends Genet 35:104–117. 10.1016/j.tig.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 72.Bouchard-Bourelle P, Desjardins-Henri C, Mathurin-St-Pierre D, Deschamps-Francoeur G, Fafard-Couture E, Garant JM, Elela SA, Scott MS. 2020. snoDB: an interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res 48:D220–D225. 10.1093/nar/gkz884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dupuis-Sandoval F, Poirier M, Scott MS. 2015. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6:381–397. 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bratkovic T, Rogelj B. 2011. Biology and applications of small nucleolar RNAs. Cell Mol Life Sci 68:3843–3851. 10.1007/s00018-011-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. 2013. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol 14:R45. 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lemus-Diaz N, Ferreira RR, Bohnsack KE, Gruber J, Bohnsack MT. 2020. The human box C/D snoRNA U3 is a miRNA source and miR-U3 regulates expression of sortin nexin 27. Nucleic Acids Res 48:8074–8089. 10.1093/nar/gkaa549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.C Quaresma AJ, Bugai A, Barboric M. 2016. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res 44:7527–7539. 10.1093/nar/gkw585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goss DJ, Kleiman FE. 2013. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4:167–179. 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197. 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 80.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441. 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 81.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J 19:4154–4163. 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. 2010. Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J 29:1851–1864. 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302:1212–1215. 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 84.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456:464–469. 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141. 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Nostrand EL, Pratt GA, Yee BA, Wheeler EC, Blue SM, Mueller J, Park SS, Garcia KE, Gelboin-Burkhart C, Nguyen TB, Rabano I, Stanton R, Sundararaman B, Wang R, Fu XD, Graveley BR, Yeo GW. 2020. Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins. Genome Biol 21:90. 10.1186/s13059-020-01982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Youssef OA, Safran SA, Nakamura T, Nix DA, Hotamisligil GS, Bass BL. 2015. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci USA 112:5023–5028. 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17:47–62. 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 89.Schmitt AM, Chang HY. 2016. Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463. 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu W, Ding C. 2017. Roles of lncRNAs in viral infections. Front Cell Infect Microbiol 7:205. 10.3389/fcimb.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shin TJ, Lee KH, Cho JY. 2020. Epigenetic mechanisms of lncRNAs binding to protein in carcinogenesis. Cancers (Basel) 12:2925. 10.3390/cancers12102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossetto CC, Pari G. 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8:e1002680. 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rossetto CC, Pari GS. 2011. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol 85:13290–13297. 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rossetto CC, Tarrant-Elorza M, Verma S, Purushothaman P, Pari GS. 2013. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J Virol 87:5540–5553. 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Withers JB, Li ES, Vallery TK, Yario TA, Steitz JA. 2018. Two herpesviral noncoding PAN RNAs are functionally homologous but do not associate with common chromatin loci. PLoS Pathog 14:e1007389. 10.1371/journal.ppat.1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campbell M, Kim KY, Chang PC, Huerta S, Shevchenko B, Wang DH, Izumiya C, Kung HJ, Izumiya Y. 2014. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol 88:1843–1848. 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Campbell M, Izumiya Y. 2020. PAN RNA: transcriptional exhaust from a viral engine. J Biomed Sci 27:41. 10.1186/s12929-020-00637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian X, Xu C, Zhao P, Qi Z. 2016. Long non-coding RNA GAS5 inhibited hepatitis C virus replication by binding viral NS3 protein. Virology 492:155–165. 10.1016/j.virol.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 99.Salerno D, Chiodo L, Alfano V, Floriot O, Cottone G, Paturel A, Pallocca M, Plissonnier ML, Jeddari S, Belloni L, Zeisel M, Levrero M, Guerrieri F. 2020. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut 69:2016–2024. 10.1136/gutjnl-2019-319637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen M, Zhang M, Xie L, Wu S, Zhong Y. 2021. LINC00324 facilitates cell proliferation through competing for miR2145p in immature ovarian teratocarcinoma. Int J Mol Med 47:397–407. 10.3892/ijmm.2020.4800. [DOI] [PubMed] [Google Scholar]
- 101.Dong Y, Wan G, Yan P, Qian C, Li F, Peng G. 2020. Long noncoding RNA LINC00324 promotes retinoblastoma progression by acting as a competing endogenous RNA for microRNA-769-5p, thereby increasing STAT3 expression. Aging (Albany NY) 12:7729–7746. 10.18632/aging.103075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bachellerie JP, Cavaille J, Huttenhofer A. 2002. The expanding snoRNA world. Biochimie 84:775–790. 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 103.Pelletier J, Thomas G, Volarevic S. 2018. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18:51–63. 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 104.Huang C, Shi J, Guo Y, Huang W, Huang S, Ming S, Wu X, Zhang R, Ding J, Zhao W, Jia J, Huang X, Xiang AP, Shi Y, Yao C. 2017. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res 45:8647–8660. 10.1093/nar/gkx651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rybak-Wolf A, Jens M, Murakawa Y, Herzog M, Landthaler M, Rajewsky N. 2014. A variety of dicer substrates in human and C. elegans. Cell 159:1153–1167. 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 106.Lafontaine DL. 2015. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22:11–19. 10.1038/nsmb.2939. [DOI] [PubMed] [Google Scholar]
- 107.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. 2014. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell 54:1042–1054. 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gumienny R, Jedlinski DJ, Schmidt A, Gypas F, Martin G, Vina-Vilaseca A, Zavolan M. 2017. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Nucleic Acids Res 45:2341–2353. 10.1093/nar/gkw1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, Kim VN. 2018. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol Cell 71:1051–1063.e6. 10.1016/j.molcel.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 110.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 112.Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47:11.12.1–11.12.34. 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. 2009. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol 83:167–180. 10.1128/JVI.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1<br>. Download jvi.01782-21-s0001.xlsx, XLSX file, 0.07 MB (76.8KB, xlsx)
Table S2<br>. Download jvi.01782-21-s0002.xlsx, XLSX file, 0.02 MB (18KB, xlsx)
Data Availability Statement
The ORF57 CLIP-seq data have been deposited in NCBI’s GEO database under accession number GSE179726. The BCBL-1 cell RNA-seq data are accessible in NCBI’s GEO database under accession number GSE179727.