ABSTRACT
Rift Valley fever virus (RVFV) belongs to the order Bunyavirales and is the type species of genus Phlebovirus, which accounts for over 50% of family Phenuiviridae species. RVFV is mosquito-borne and causes severe diseases in both humans and livestock, and consists of three segments (S, M, L) in the genome. The L segment encodes an RNA-dependent RNA polymerase (RdRp, L protein) that is responsible for facilitating the replication and transcription of the virus. It is essential for the virus and has multiple drug targets. Here, we established an expression system and purification procedures for full-length L protein, which is composed of an endonuclease domain, RdRp domain, and cap-binding domain. A cryo-EM L protein structure was reported at 3.6 Å resolution. In this first L protein structure of genus Phlebovirus, the priming loop of RVFV L protein is distinctly different from those of other L proteins and undergoes large movements related to its replication role. Structural and biochemical analyses indicate that a single template can induce initiation of RNA synthesis, which is notably enhanced by 5′ viral RNA. These findings help advance our understanding of the mechanism of RNA synthesis and provide an important basis for developing antiviral inhibitors.
IMPORTANCE The zoonosis RVF virus (RVFV) is one of the most serious arbovirus threats to both human and animal health. RNA-dependent RNA polymerase (RdRp) is a multifunctional enzyme catalyzing genome replication as well as viral transcription, so the RdRp is essential for studying the virus and has multiple drug targets. In our study, we report the structure of RVFV L protein at 3.6 Å resolution by cryo-EM. This is the first L protein structure of genus Phlebovirus. Strikingly, a single template can initiate RNA replication. The structure and assays provide a comprehensive and in-depth understanding of the catalytic and substrate recognition mechanism of RdRp.
KEYWORDS: Rift Valley fever virus, RNA-dependent RNA polymerase, structure, RNA synthesis, Cryo-EM
INTRODUCTION
The order Bunyavirales, negative-sense, single-stranded RNA virus, is divided into 12 families (Arenaviridae, Cruliviridae, Fimoviridae, Hantaviridae, Leishbuviridae, Mypoviridae, Nairoviridae, Peribunyaviridae, Phasmaviridae, Phenuiviridae, Tospoviridae, and Wupedeviridae) (https://talk.ictvonline.org/taxonomy/), including over 300 identified species (1). The order Bunyavirales can infect plants, animals, and humans, posing a serious threat to agriculture, animal husbandry, human life, and property. For example, Tomato spotted wilt virus can infect more than 1,000 plant species, Hantaan orthohantavirus and Lassa mammarenavirus cause severe hemorrhagic fevers with a mortality rate of up to 50%, Crimean-Congo hemorrhagic fever virus also causes severe hemorrhagic fever in humans with mortality rates of up to 30% (2, 3). Rift Valley fever virus (RVFV) belongs to the order Bunyavirales, family Phenuiviridae, genus Phlebovirus (https://talk.ictvonline.org/taxonomy/), and is one of the most prominent and well-known bunyaviruses (4). The genus Phlebovirus includes 60 species and accounts for more than half of the family Phenuiviridae species (117 species). RVFV, the type species of this genus, is a zoonosis, leading to severe infections in animals and humans and significant economic losses.
RVFV was first isolated in Kenya in 1931 by Daubney et al. through inoculating lambs with the serum from a moribund sheep (5). Rift Valley fever (RVF) is widespread in Africa, and multiple outbreaks have been recorded in Africa and the Arabian Peninsula in the 21st century. Recent outbreaks in a French overseas department and some seropositive and imported cases in Turkey, Libya, and China have raised concerns that RVF could extend to other parts of the world, which poses a serious threat to global health and economics (6–11). RVFV is transmitted among ruminants by bites from mosquitos mainly of the Aedes and Culex genera, or by direct contact with body fluids of viremic animals (12). RVFV is classified as a category A high priority pathogen by the National Institute of Allergy and Infectious Diseases and virus handling requires high- or maximum-containment facilities, such as a biosafety level 3 enhanced (BSL-3E) or BSL-4 laboratory (13). RVFV leads to severe disease in livestock, with up fatality rates of up to 70% in young animals and as high as 20%–30% in adult ruminants. In addition, infected pregnant animals undergo spontaneous abortion at an alarmingly high rate, in the range of 40–100% (14). After being infected with RVFV, humans may experience generalized fatigue, low-grade fever, headache, photophobia, and joint pains. Some patients infected with RVFV will develop encephalitis, retinopathy, or disseminated intravascular coagulation leading to hemorrhage and even death, with a case fatality rate of approximately 20% (11). The cross-border movement of goods and animals means that there is now a threat of RVFV dissemination to Asia and Europe (15).
RVFV has a single-stranded, tripartite RNA genome, composed of L, M, and S segments. The large negative-sense L RNA segment encodes a single-polypeptide L protein, and the negative-sense M RNA segment encodes the glycoprotein precursor. The S segment adopts an ambisense strategy for expressing NSs and nucleoprotein (16). The RVFV viral genome is wrapped by the nucleoprotein and the conserved 3′ and 5′ ends of the genomic RNA (vRNA) are bound to L protein, resembling the influenza virus (17, 18). The L protein is a multifunctional enzyme that catalyzes genome replication and viral gene transcription, which are initiated de novo by prime-and-realign and cap-snatching mechanisms, respectively (19).
To date, precautions to avoid new RVFV outbreaks mostly rely on weather monitoring, and no specific drugs or licensed vaccines are available to treat or prevent infection (20). Although there have been reports about the structures of Bunyavirus LACV and SFTSV L proteins (21–23), no RNA-dependent RNA polymerase (RdRp) structures of genus Phlebovirus have been reported. To understand the role of L protein in the viral life cycle and identify a feasible drug target, we solved an RVFV polymerase structure using cryogenic electron microscopy (cryo-EM) at 3.6 Å resolution. This first RdRp structure of the genus Phlebovirus guided the elucidation of the mechanism of viral RNA synthesis and transcription. Using biochemical assays we verified the endonuclease activity and polymerase initiation activity of the RVFV L protein. The RVFV L protein endonuclease activity has a preference toward MnCl2 and has polymerase initiation activity in the presence of only template RNA or both template and allosteric activator RNA.
RESULTS
Enzymatic properties of RVFV L protein in vitro.
After extensive screening, we successfully constructed and expressed the full-length L protein using the Pichia pastoris expression system. Full-length L protein existed as a monomer in solution (Fig. 1A and B). In vitro endonuclease activity experiments revealed that the L protein, but not the endonuclease active site mutant (D111A) (lane 2) (24), had strong endonuclease activity in the presence of MnCl2 (Fig. 1C, lane 1). Using 30 nt polyA as an RNA substrate, the L protein had ion-concentration dependent endonuclease activity in the presence of MnCl2 (Fig. 1D, lanes 6, 7, and 8) (25), but no activity in the presence of CaCl2 (Fig. 1D, lanes 9, 10, and 11) (23, 26). To understand the polymerase activity, we tested the ability of the purified RVFV L protein to incorporate fluorescently labeled nucleotides in vitro. To prevent RNA degradation during the experiment, we used the D111A endonuclease active site mutant and tested its activity. We added fluorescein-12-UTP into the assay system as the substrate, and monitored the RNA synthesis products by electrophoresis on 20% (wt/vol) polyacrylamide gels containing 7 M urea. The D111A/D1133A negative-control mutant, in which the conserved Asp1133 in the motif C of the polymerase domain was mutated to alanine, lacked polymerization activity (Fig. 1E, lane 12). Interestingly, clear RNA synthesis products were produced by the D111A mutant (Fig. 1E, lanes13, 14).
FIG 1.
In vitro enzymatic activities of RVFV L protein. (A) SDS–PAGE profile of Rift Valley fever virus (RVFV) L protein. (B) Size-exclusion chromatogram of RVFV L protein. Absorbance curves for the sample at wavelengths of 260 nm and 280 nm. (C) Endonuclease assay with high concentration protein. 0.1 μΜ RVFV L protein was incubated at 30°C for 1 h with 0.45 μM fluorescently labeled 30 nt PolyA RNA substrate and 25 μM MnCl2. The band in the bottom of the gel was the degradation product. (D) Endonuclease assay. RVFV L protein (0.06 μΜ) was incubated with 0.45 μM fluorescently labeled 30 nt PolyA RNA substrate at 30°C for 40 min in the presence of 10, 25, or 50 μM MnCl2 or CaCl2. Reactions without protein, in the presence of 50 μM EDTA or the known endonuclease-specific inhibitor 2,4-dioxo-4-phenylbutanoic acid (DPBA), were negative controls in the presence of 25 μM MnCl2. All inputs are shown as red arrows in panels C, D, and E. (E) In vitro replication initiation assay. The endonuclease and polymerase inactivation site double mutant (D111A/D1133A) was added in the left lane as a negative control. Mutant (D111A) was added in the middle and right lanes. These three lanes were incubated with the conserved 5′ 20 nt of the M segment (5′ M: 5′-ACACAAAGACGGUGCAUUAA-3′) or/and a 20 nt template RNA named RNA* (5′-UGUGUUUCUGGCCACGUUGA-3′). Nucleotide incorporation assay was detected by fluorescence (fluorescein-12-UTP). (F) Effect of metal ions (Mn2+ or Mg2+) and reducing agent (DTT or TCEP (Tris[2-carboxyethyl]phosphine)) on nucleotide incorporation assay by endonuclease active site mutant (D111A). The assay was performed with 5 mM MnCl2/MgCl2 and 2.5 mM DTT/TCEP for 40 min at 30°C. (G) The schematic diagram of the template and product in the polymerase experiment, “T” represents the template, and ‘’P2-P4’ represents the dinucleotide product to the tetranucleotide product, respectively.
Examination of the synthesized products revealed that most were dinucleotides, and several oligonucleotide bands were also observed (Fig. 1F and G). The low yield of long products suggests that the fluorophore-labeled UTP may impair subsequent synthesis after the formation of the dinucleotide. Nevertheless, the observation of dinucleotide as the primary product species indicates that RVFV L is capable of de novo initiation, thus allowing us to use this assay in characterizing fundamental features of the polymerase. We further tested how Mg2+ and Mn2+ metal ions affected RVFV L protein polymerase initiation activity in the presence of different reducing agents (DTT or TCEP) (27). Urea PAGE analysis showed that the type of reducing agent did not significantly affect RVFV L protein polymerase initiation activity. However, RVFV L protein polymerase initiation activity was stronger in the presence of MnCl2 than in the presence of MgCl2 (Fig. 1F). Interestingly, the RVFV L protein was also capable of initiating RNA synthesis in the presence of only one template RNA (Fig. 1E, lane 13). Therefore, the RVFV L protein can initiate de novo RNA synthesis in the absence of 5′ vRNA. This activity is notably enhanced by the addition of 5′ vRNA. This is different from the mode of most bunyaviruses that need both 5′ RNA and 3′ RNA for successful initiation of RNA synthesis. To explore the mechanism of RVFV L protein property we decided to reveal the RVFV L protein structure.
Overall RVFV L protein structure.
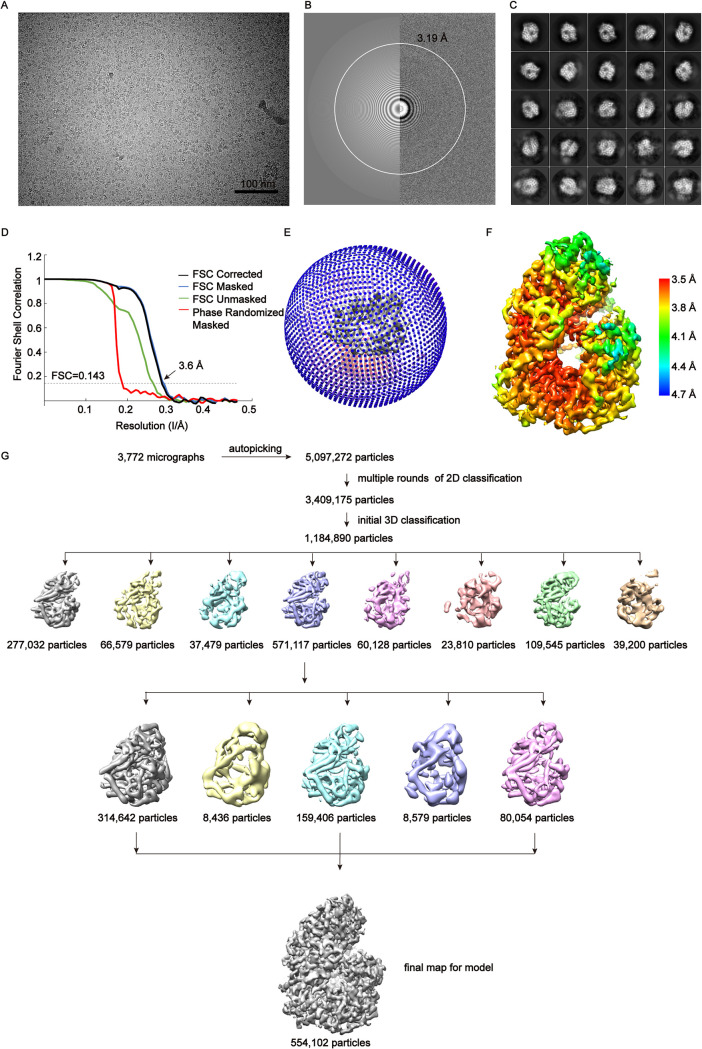
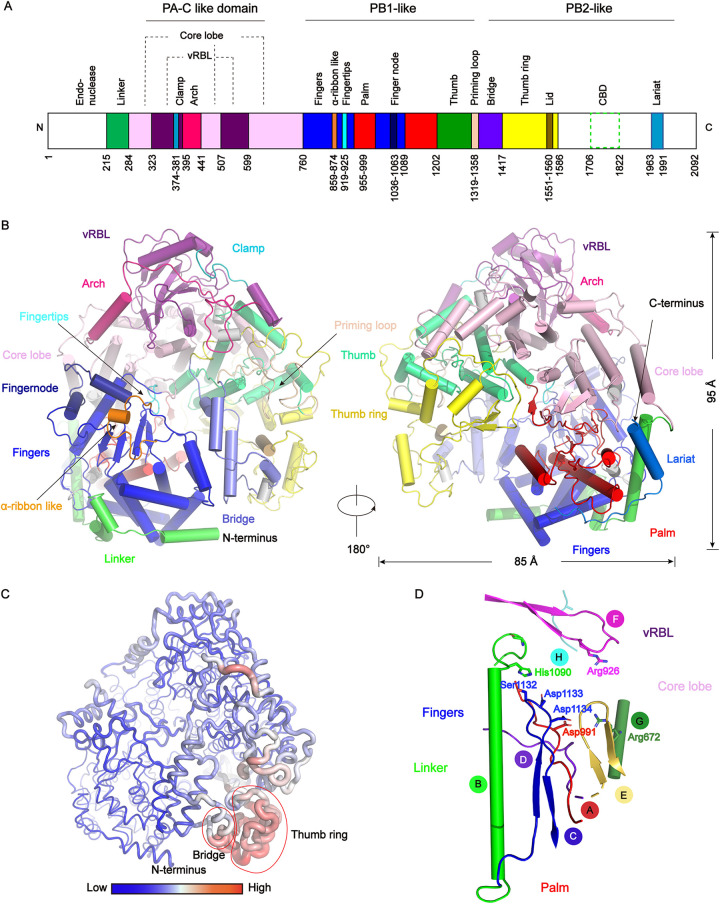
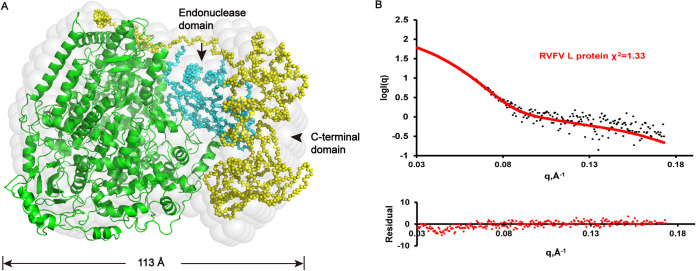
We performed single particle cryo-EM analysis of the RVFV L protein and obtained one 3D reconstruction at 3.6 Å resolution (Fig. 2, Table 1). Our solved L protein structure had ∼1400 residues, corresponding to the PA-C, PB1, and N-terminal part of PB2 (PB2-N) of influenza virus (17) (Fig. 3A). The overall shape of the L protein structure resembled a figure-"6” (Fig. 2F). The dimensions of the RVFV L protein structure are about 85 Å x 75 Å x 95 Å (Fig. 3B). Although we successfully obtained the full-length protein, we did not obtain a complete cryo-EM density containing the C-terminal cap-binding domain (CBD) (19) and the endonuclease domain (residues 1–214). To know the locations of these two domains in the RVFV L protein structure, we used the pure, monodisperse, and monomeric full-length RVFV L protein to perform SAXS experiments and obtained a low-resolution structure in solution (Fig. 4). Using the rigid body fitting of the recently published endonuclease domain and C-terminal domain of the PB2-like region of SFTSV (22) and the above RVFV L protein structure, we obtained an integrated RVFV L protein model (Fig. 4A), which fit well with the SAXS-derived envelope (Fig. 4B).
FIG 2.
Cryo-EM reconstruction of RVFV L protein. (A) Raw image of RVFV L protein particles in vitreous ice recorded by K3 camera at defocus range of −1.5 to 2.5 μm. Scale bar 100 nm. (B) Power spectrum of image (A) and the white circle indicated the spatial frequency corresponding to 3.19 Å estimated by the Gctf program. (C) Representative 2D classes. The box size of each class was 172.8 Å. (D) Fourier shell correlation (FSC) curves of the postprocessed 3D map, following the gold standard refinement. (E) Euler angle distribution. (F) Local resolution was performed with Relion3.0.8. (G) The flowthrough of the data processing used to get the final map.
TABLE 1.
Cryo-EM data collection, refinement, and validation statistics
| RVFV L | |
|---|---|
| Data collection and processing | |
| Magnification | 64,000 |
| Voltage (kV) | 300 |
| Electron exposure (e-/Å2) | 50 |
| Defocus range (μm) | -1.0 to −3.0 |
| Pixel size (Å) | 1.08 |
| Symmetry imposed | C1 |
| Final particle images (no.) | 554K |
| Map resolution (Å) | 3.6 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 3.5−4.7 |
| Refinement | |
| Initial model used (PDB code) | 6L42 |
| Map sharpening B factor (Å2) | −180 |
| Model composition | |
| Nonhydrogen atoms | 9,819 |
| Protein residues | 1400 |
| B factors (Å2) | |
| Protein | 103.86 |
| R.m.s deviation | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.078 |
| Validation | |
| MolProbity score | 1.28 |
| Clashscore | 5.25 |
| Poor rotamers (%) | 0.00 |
| Ramachandran plot | |
| Favored (%) | 100 |
| Allowed (%) | 0.00 |
| Disallowed (%) | 0.00 |
FIG 3.
Overall structure of the RVFV L protein and conserved motifs. (A) Schematic representation of the monomeric RVFV L protein domain structure. The white areas are missing in the determined structure but present in the purified protein. (B) Cartoon representation of the RVFV L protein. The structure is colored by domains using the same color code as in (A). 310 helices are colored in gray. (C) The RVFV L protein B-factor map. A larger radius and red color represent high B-factor values and a smaller radius and blue color represent low B-factor values. (D) The arrangement of the conserved RdRp motifs in the RVFV active site colored in red, green, blue, purpleblue, yelloworange, magenta, forest, and teal for motifs A–H, respectively.
FIG 4.
Superposition of low-resolution ab initio model and rigid body model. (A) The ab initio model is shown as light-gray spheres. PA-like, PB1-like, and PB2-like N-terminal domains of RVFV L protein are colored in green. The simulated endonuclease domain and the C-terminal domain of the PB2-like region are colored in cyan and yellow, respectively. (B) Upper: Comparison of SAXS experimental data and calculated scattering profiles. Experimental data are represented in black dots. The data calculated from the RVFV L protein (1.25 mg/ml) is in red. Lower: Error-weighted residual difference plot.
We superimposed the structure of RVFV L protein with other L proteins of order Bunyavirales by the DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/). The RMSD values are 2.5 Å (PDB 6L42, SFTSV), 3.9 Å (PDB 6Z8K, LACV), 4.5 Å (PDB 6KLD, MACV), 4.3 Å (PDB 6KLC, LASV), respectively. The first domain at the RVFV L protein N terminus is the endonuclease domain (residues 1–214) which plays an essential role in cleaving 5′ m7G caps from host mRNAs during the cap-snatching process (14, 28). The catalytic center of RdRp is constituted of fingers, thumb, and palm domains. The fingers domain (residues 760–858, 874–918, 925–954, 1000–1035, 1063–1088) displays a “sandwich” shape and is comprised of upper and lower ɑ-helix layers with a β-sheet in the middle (Fig. 3B). The key residues of the thumb domain can promote template translocation following the first polymerization reaction through conformational rearrangement (29). In brief, the fingers, palm, and thumb domains probably collaborate to promote vRNA template and NTP binding. The C-terminal lariat circles the large globular core and folds back to stabilize the fingers’ structure. In summary, the active site of the L protein polymerase is relatively stable and conservative, while the structures of the thumb ring and bridge regions are flexible (Fig. 3C).
Conserved motifs of RdRp.
Common and unique features of viral RNA-dependent polymerase motifs (motifs A-F) that contain key residues for the enzymatic function have been identified in many RNA-dependent polymerases (30). Like L proteins from many RNA viruses, RVFV L protein has six conserved structural motifs (motifs A-F) (31–33) that cooperate with viral RNA template, NTPs, and metal ions to promote the polymerization of nucleotide triphosphates (Fig. 3D). RVFV L protein also has two single-stranded negative-sense RNA virus specific motifs, G and H like LACV L and SFTSV L (22, 34, 35) (Fig. 3D). Motifs A to E are within the palm domain while motif F (913-HICLFKKQQHGGLREIYV-930), composed of two β-strands connected by a loop, constitutes part of the fingertips (Fig. 3D). The position distribution of motifs A-F is similar to that of positive-strand and double-stranded RNA viruses, and the overall structure of the polymerase is assembled into a cupped right hand (29). Trp995 of motif A sits in a strategic position where it can maintain L protein structural stability locally. Motif C is the active center of RdRp. These motifs and functional regions participate in the most critical steps for the correct recognition and incorporation of ribonucleotides.
The fingertips of RVFV L protein.
The fingertips subdomain (Fig. 5), mostly composed of the conserved motif F, extends from the fingers domain to the thumb domain, and is located directly above RdRps catalytic center. According to previous reports, during the process of NTPs binding to RdRp, the positively charged Lys and Arg side chains in the fingertips subdomain can interact with the triphosphate moiety, which also interacts with both divalent metal ions in the active center (Fig. 5) (36). Moreover, the fingertips subdomain can bind 5′ viral RNA by basic residues (17, 34). Based on the RVFV L protein structural analysis, we identified that the key residues (Lys918, Arg926) (Fig. 5E) for its vitality are positioned in the middle loop region, right at the junction of the template entry channel and NTP entry channel. Therefore, it is reasonable to consider that the fingertips subdomain is crucial for binding both template and NTPs. The previously reported structure of the apo FluCPol harbors highly unstable fingertips (Fig. 5C), whereas the same region in the 5′ vRNA-bound FluAPol and FluBPol structure is stable (17, 37). For the LACV (order Bunyavirales) L protein (34), 5′ vRNA induces conformational changes of the fingertips and the adjacent finger extension, further stabilizing them. Moreover, the C-terminal lariat solved in the L protein structure further stabilized the fingers’ structure. The finger extension is oriented in a conformation that provides enough space to hold the fingertip. Residues Tyr760, Lys762, Asn763, and Glu766 (finger extension domain) are highly conserved and have especially strong interactions with the residues (Lys918, Lys919, Gln920, Gln921) in the fingertips subdomain (Fig. 5F).
FIG 5.
The fingertips loops of several RdRp polymerases. The B-factor maps for (A) Machupo virus (MACV, PDB 6KLD) (61), (B) Lassa Virus (LASV, PDB 6KLC), (C) Influenza C (FluC, PDB 5D98) (62), (D) La Crosse encephalitis virus (LACV) (PDB 6Z8K) (21), (E) RVFV L. (A, B, E) Fingertips are conservative, while FluC’s and LACV’s fingertips are unstable, and their RNA synthesis initiation requires 5′ vRNA. (F) Interaction of residues between fingertips and finger extension. The residue on the fingertips and the finger extension are colored green and yellow, respectively.
Conservation analysis for RVFV L protein.
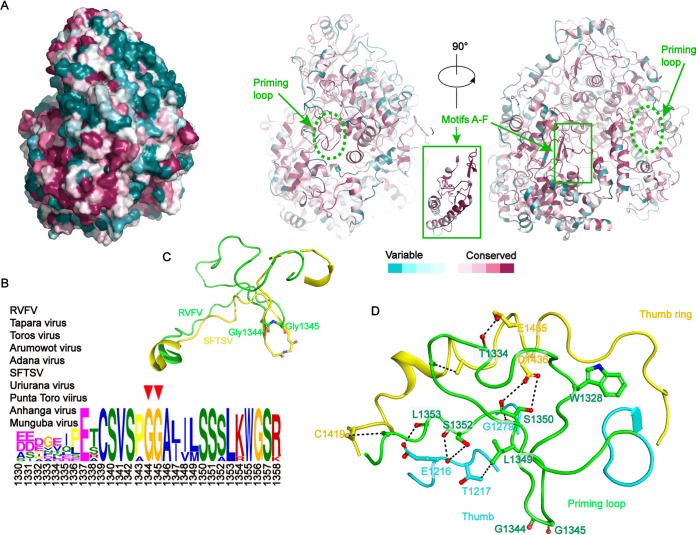
We performed sequence alignment of L proteins from a set of 10 representative viruses from the family Phenuiviridae and analyzed the conservation differences among the various domains (38). The most conserved regions were the motifs A–F of RNA viruses (Fig. 6A). Based on this sequence alignment, and conservation analysis of the overall sequence of the polymerase on the Consurf website, the residues on the polymerase surface were identified as variable and the residues around the polymerase active center and the four access channels (Fig. 7) were highly conserved and corresponded to the motifs A–F (Fig. 6A). The conserved residues around the channels were mainly positively charged residues, reflecting their roles in nucleic acids’ interaction during viral genome replication and transcription.
FIG 6.
Conservation analysis for RVFV L protein. (A) Sequence conservation analysis. The left panel displayed in surface mode is positioned at the same view as the middle panel. Two different views of the RVFV L protein were displayed in ribbon mode and colored based on the residue conservation among family Phenuiviridae. The bottom color code bar is from low conservation (cyan) to high conservation (purple). Figures are colored using the Consurf server (http://consurf.tau.ac.il/2016/) based on a multiple sequence alignment, including the sequences of Tapara virus (YP_009346035), Toros virus (YP_009246447), Arumowot virus (YP_009010958.1), Adana virus (YP_009227127.1), severe fever with thrombocytopenia syndrome virus (SFTSV) (YP_006504091.1), Uriurana virus (YP_009346036.1), Punta Toro virus (YP_009512941.1), Anhanga virus (API68876.1), and Munguba virus (YP_009346010.1). (B) Sequence conservation in the priming loop. Plotting was performed with The MEME Suite software (40). (C) The priming loop and connecting regions are shown as ribbons colored in green (RVFV) and yellow (SFTSV and PDB 6L42) (22). Two glycine residues (Gly1344 and Gly1345), which are important for an early stage of RNA synthesis, are shown as sticks. (D) Structural demonstration shows the interaction sites of the priming loop (green) and thumb (cyan) and thumb ring domains (yellow).
FIG 7.
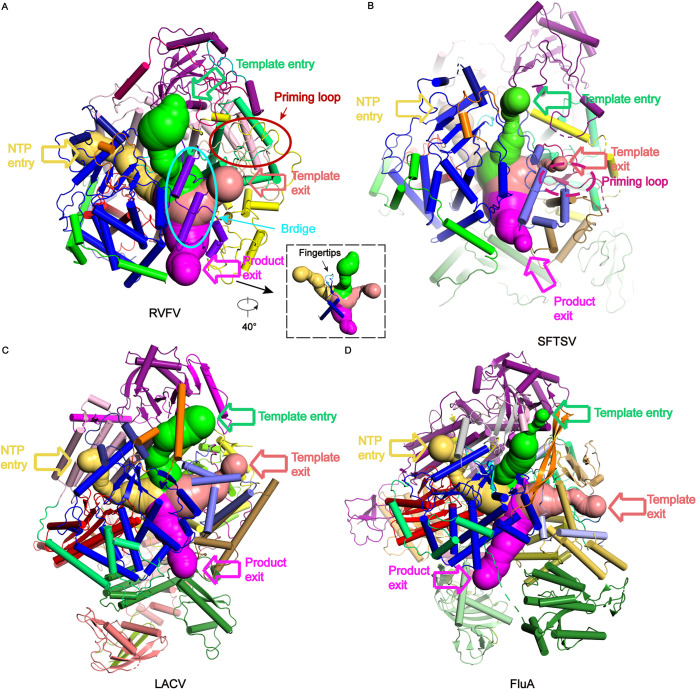
The entrance and exit channels of RVFV, SFTSV, LACV, and FluA polymerases. The template entry, NTP entry, template exit, and product exit channels of (A) RVFV L protein, (B) SFTSV L protein (PDB 6L42), (C) LACV L protein (PDB 6Z8K), (D) Influenza A polymerase (FluA) (PDB 6T0V) are colored in limegreen, yelloworange, salmon, and magenta, respectively. The colors of the RVFV L domains are same as those used in Fig. 2A A 40° rotated view of the RVFV L’s channels is shown inset.
The priming loop (residues 1319–1358) is a fundamental element that generally stabilizes the first “priming” nucleotide of the product during replication initiation but is not essential for internal initiation and transcription (39). MEME-ChIP analysis revealed that the two glycine residues (Gly1344 and Gly1345) at the tip of the priming loop are conserved (40, 41), and structural comparison with SFTSV (22, 23) showed that these residues might be essential for identifying specific transcription template (Fig. 6B and C). However, a structural comparison between RVFV and SFTSV L proteins demonstrated a distinct difference in their priming loops (Fig. 6C). The RVFV L protein priming loop is located next to the template exit tunnel and extrudes from the polymerase active center cavity to the surface of the protein, forming a compact interaction with the thumb domain and thumb ring domain (Fig. 6D and 7A). The priming loop of SFTSV L protein extends to the polymerase active center, so the template exit tunnel is smaller in the pre-initial state of replication (Fig. 7B). The L protein thumb ring domain residues (Cys1419, Glu1435, Asp1436) are tightly bound with several priming loop residues (Thr1334, Ile1335, Glu1337), and the thumb domain residues (Glu1216, Thr1217, Gly1279, Phe1315) act on the other part of the priming loop residues (Trp1328, Leu1349, Ser1350, Ser1352, Leu1353) (Fig. 6D). These two domains strictly regulate the conformation of the priming loop to adapt to different stages of viral replication or transcription (Fig. 6D). The tight cooperation of these two domains provides a larger space for template exiting (Fig. 7A). According to the structural analysis of the complex of influenza virus and LACV (21, 42, 43), we speculate that the observed priming loop conformation may be in a preparative state for de novo initiation. The disordered conformation is analogous to that observed in the SFTSV L apo structure (22), but different from that of the LACV L elongation-mimicking state structure (21).
Comparison of four channels.
The catalytic residues of the polymerases from negative-strand RNA viruses are at the center of an enclosed cavity connected to the exterior by four channels: an incorporated template entry channel, template exit channel, NTP entry channel, and product exit channel (Fig. 7). The configurations of the four tunnels of RVFV L protein are similar to those described for SFTSV and LACV L proteins, and for Flu A polymerase (Fig. 7) (17, 22, 34). The template entry channel is mainly composed of vRBL, core lobe, and thumb domains. The conserved Trp513 aromatic residue, located on an α-helix of the vRBL domain adjacent to the core lobe domain, is critical for stabilizing the vRBL domain and core lobe domain by interacting with Phe581, Pro328, Val517, Leu654, and Leu658. Residues Ser1411, Leu1412, and Ser1413 (bridge domain), located in the middle of the template entry channel, are involved in identifying the sequence-specific template entry process. Similar to that in SFTSV polymerase, the ɑ-ribbon in RVFV polymerase is distant from the template entry channel. The NTP entry channel is mainly composed of the core lobe domain and fingers domain, and the fingers domain separates the NTP entry and template entry channels (Fig. 7A). The product exit channel is surrounded by linker, fingers, thumb, and bridge domains.
DISCUSSION
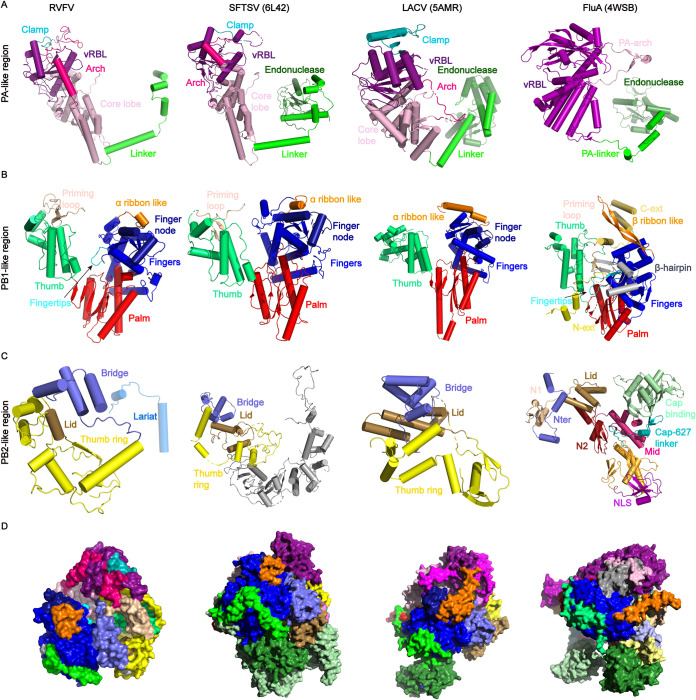
According to the World Health Organization, RVF is one of the top 20 global pandemic and epidemic diseases (https://www.who.int/emergencies/diseases/en/). Here, after optimizing the expression and purification of RVFV L protein, we solved the first L protein structures of the genus Phlebovirus. Comparison of the RdRp core structure with those previously reported in SFTSV, LACV, and Orthomyxoviridae (influenza) (Fig. 8), revealed that the RdRp core is typically conserved. The C-terminal domain is tightly integrated into the thumb domain of the polymerase core. All domains contribute to the formation of the channels that facilitate template entry and exit, NTP entry, and product exit (Fig. 7). Our results reveal that the RVFV polymerase has a compact core structure and is a flexible multidomain protein.
FIG 8.
Comparison of subdomains in the RdRp region. For overall structure comparison, parts of structures of RVFV L protein, SFTSV L protein (PDB 6L42), LACV L protein (PDB 5AMR), and Influenza A polymerases (PDB 4WSB) were shown side by side. Regions corresponding to influenza virus PA-like region (A), PB1-like region (B), PB2-like region (C), and overall architectures (D). Colors correspond to Fig. 3A
RVFV replication and transcription occur in the cytoplasm. This is unlike the influenza virus, in which ribonucleoprotein (RNPs) complex are transported into the nucleus for replication and transcription (44). Replication is a primer-independent process. First, a replication intermediate called cRNA (cRNA) is synthesized, and then cRNA is used as a template for replication. The priming loop conformation of RVFV L is distinctly different from those of other L proteins of bunyaviruses (Fig. 6C). Then, the priming loop recruits the first nucleotide to bind to the vRNA and provides sufficient space in the active center for the replication product (21). Consistent with the replication initiation mechanism in SFTSV (23), called prime and realign, initiation of replication in RVFV might occur at several positions in the 3′ vRNA template.
Our in vitro experiments showed that the RNA synthesis product was not detected when using the 5′ and 3′ ends of any of or all of the three viral genome segments as transcription templates. The primary product in these assays was a dinucleotide and further synthesis was largely hindered likely by the usage of the fluorophore-labeled UTP. When only one template RNA was added, the L protein was capable of RNA synthesis (Fig. 1D). Therefore, the RVFV L protein can initiate de novo RNA synthesis in the absence of 5′ vRNA, and this activity is notably enhanced by the addition of the 5′ vRNA. This is different from the RNA synthesis mode of most bunyaviruses that require both 5′ vRNA and 3′ vRNA for successful initiation (17, 34). Interestingly, 5′ vRNA can significantly increase the RNA polymerase initiation activity. This is because the basic residues (Lys918 and Arg926) in the fingertips subdomain (Fig. 5D and E) might bind the 5′ viral RNA and stabilize the RdRp core structure.
Transcription of RVFV genomes is a primer-dependent process that involves capturing host capped RNAs using a cap-snatching mechanism. During cap-snatching, short capped fragments are snatched by the CBD (19) and host mRNAs are cleaved at a position close to the 5′ cap by the endonuclease domain. Although we successfully obtained the full-length protein and added RNA in the sample preparation, we did not obtain a complete cryo-EM density. The CBD might need capped vRNA to stabilize the conformation. SAXS method is a typical method to uncover the structure in solution. A low-resolution structure of the full-length RVFV L protein was obtained (Fig. 4). The integrated RVFV L protein model shows that the endonuclease domain and the CBD are close to each other, which is supported by their functions: the CBD captures RNA and passes it to the endonuclease domain for cutting to be used as transcription primer.
At present, viral RdRps are promising drug targets. Thus, our structure provides an important basis for developing effective antivirals for protecting humans and animals exposed to, and infected with, RVFV.
MATERIALS AND METHODS
Small scale expression.
The codon-optimized sequence for RVFV (strain: ZH-501, GenBank: DQ375406.1) L protein was synthesized and cloned (using the seamless assembly cloning method) into a modified pPICZ expression vector with a C-terminal 6 × His tag without/with a GFP tag (EcoR I/Nco I) under the AOX1 promoter. Each single colony was inoculated into 5 ml of YPD medium (1% yeast extract, 2% peptone, 2% glycerol) in a 50-ml centrifuge tube and incubated at 30°C in a shaking incubator (170 rpm) until the culture reached an OD600 = 6 (approximately 24 h). The cells were harvested by centrifugation at 3,000 × g for 2 min at 4°C. The supernatant was decanted, and the cell pellets were resuspended in approximately 5 ml of BMM medium (100 mM potassium phosphate, pH 6.0, 1.34% YNB, 1% methanol) to induce expression. Methanol was added to a final concentration of 1% every 24 h to maintain induction. Cell pellets were analyzed for protein expression by Western blot or GFP fluorescence detection. The extremely low level of wild-type L protein expression makes it impossible to obtain the protein by purification. To improve protein expression, we tried expressing several different mutants which were located away from the endonuclease active center. We found that expression of the D103A mutant protein was significantly increased, so this site was mutated in all of our proteins. In the manuscript, this protein is referred to as wild type because the mutant has endonuclease activity (Fig. 1).
Protein expression and purification.
His tagged L protein was expressed in the X-33 Pichia strain in BMM medium at 28°C for 48 h after induction with 1% methanol. After pelleting, cells were resuspended in 50 mM Tris-HCl pH 8.5, 1 M NaCl, 5% glycerol, 20 mM imidazole and 5 mM benzamidine hydrochloride. The cells were lysed in the presence of 1 mM protease inhibitor (phenylmethyl sulfonyl fluoride [Invitrogen]) using a high pressure cell disruption system and the crude lysate was centrifuged at 47,000 × g for 50 min at 4°C to isolate the supernatant. The protein was purified from the soluble fraction by Ni affinity chromatography (GenScript), and was washed using a buffer containing 20 mM imidazole, 50 mM Tris-HCl pH 8.5, 1 M NaCl, 5% glycerol, and 5 mM benzamidine hydrochloride to eliminate nonspecific proteins. The elution buffer contained 50 mM imidazole. The eluted protein was concentrated using an Amicon Ultra centrifugal filter (Millipore). The protein was further purified by size exclusion chromatography using a Superdex 200 10/300 GL (GE Healthcare) column equilibrated in 20 mM BTP (1,3-Bis [tris [hydroxymethyl] methylamino] propane) pH 8.5, 300 mM NaCl, and 2 mM β-ME. Collected protein fractions were concentrated to 2 mg/ml and stored at 4°C for further use.
The proteins containing a 6×His tag alone were used as the basis of Pichia protein expression screening. However, this method is cumbersome, the detection is not sensitive, and it requires considerable time to screen the strains expressing the protein. We improved the expression screening using a GFP-6×His tag. The GFP fluorescence screening method was more convenient, simple to operate, and highly sensitive. GFP-positive cells were resuspended in 50 mM Tris-HCl pH 8.5, 500 mM NaCl, 5% glycerol, 20 mM imidazole, and 5 mM benzamidine hydrochloride. The lysate was centrifuged at 47,000 × g for 50 min to remove cell debris and was loaded onto Ni affinity chromatography (GenScript). The elution buffer was the same as the washing buffer (20 mM imidazole, 50 mM Tris-HCl, pH 8.5, 500 mM NaCl, 5% glycerol, and 5 mM benzamidine hydrochloride) but contained an additional 50 mM imidazole. After changing the buffer to remove the imidazole, the GFP-6×His Tags were removed by the tobacco etch virus protease at a 1:1 (wt/wt, protease/protein) ratio at 4°C overnight. Then, the mixture was purified using Ni affinity chromatography (GenScript). Fractions containing L protein were collected and applied to a Superdex 200 10/300 GL (GE Healthcare) column equilibrated in 50 mM Tris-HCl pH 8.5, 500 mM NaCl, and 5% glycerol. The fractions were flash-frozen, and stored at –80°C. All protein purification procedures were performed at 4°C.
Endonuclease assay.
The FAM-5′-Poly-A RNA 30 mer was chemically synthesized (Sangon Biotech). Reaction mixtures containing RVFV protein and 0.45 μM FAM-5′-Poly-A RNA were performed in a volume of 20 μl with 0.5 U/μl RNasin (Promega), 100 mM HEPES pH 7.0, 100 mM NaCl, 5 mM dithiothreitol (DTT), and divalent cations as indicated in the figure legends, and incubated at 30°C for a specified period of time. The reaction was stopped by adding an equivalent volume of RNA loading buffer (95% formamide, 0.5 mM EDTA, 0.025% SDS, and 0.025% xylene cyanol) and heating the samples at 100°C for 3 min. Products were separated by electrophoresis on denaturing 7 M urea, 20% polyacrylamide (acrylamide:bisacrylamide is 19:1) Tris-borate-EDTA gels. Fluorescence was visualized using a ChemiDoc MP (BIO-RAD).
Polymerase initiation assay.
For the nucleotide-incorporation assay, 0.37 μM RVFV polymerase was mixed with 1 μΜ 20 nt 5′ end of the RVFV M genome segment (5′ M: 5′-ACACAAAGACGGUGCAUUAA-3′) or/and 1 μM 20 nt template RNA (5′-UGUGUUUCUGGCCACGUUGA-3′), NTPs (25 μM ATP, 25 μM GTP, 25 μM CTP and 16.25 μM UTP) and 8.75 μM fluorescein-12-UTP (Sigma) in assay buffer (50 mM Tris-HCl pH 7.0, 2.5 mM DTT, 5 mM MnCl2/MgCl2). Reaction mixtures were incubated at 30°C for 3 h and samples were analyzed on a 20% acrylamide, 7 M urea denaturing gels. The fluorescence was visualized in a ChemiDoc MP (BIO-RAD).
Cryo-EM sample preparation and data collection.
The two vRNA oligonucleotides (5′ vRNA: 5′-GGUGCA-3′ and 3′: vRNA 5′-UGCACCGGUCUUUGUGU-3′) were annealed by heating at 95°C for 5 min followed by cooling down on bench at room temperature to form partially double-stranded RNA. These two oligonucleotides are conserved 3′ and 5′ vRNA ends of viral genome and the selection is similar to that described for La Crosse encephalitis virus (LACV) (21, 34). This partially dsRNA was then mixed with L protein in a 1.1:1 molar ratio at 4°C for 3 h and the complex was ultimately resolved on Superdex 200 10/300 GL in a gel filtration buffer (50 mM BTP pH 8.5, 300 mM NaCl). Aliquots of 4 μl purified RVFV L protein at a concentration of ∼0.4 mg/ml were applied to glow-discharged holey carbon Cu grids (Quantifoil R1.2/1.3). Then these grids were flash-cooled in liquid ethane using an FEI Vitrobot Mark IV. After initial screening, the well-prepared cryo-specimens were transferred onto an FEI Titan Krios transmission electron microscope operated at 300 kV for data collection. The microscope was equipped with a GIF-Quantum energy filter, which was used with a slit width of 20 eV. Images were recorded using a K3 Summit direct electron counting camera (Gatan Inc.) at a calibrated magnification of 64,000 (pixel size of 1.08 Å). The defocus range of the data sets was roughly −1.0 to −3.0 μm.
Image processing.
Beam induced motion and anisotropic magnification were corrected using the MotionCor2 program (45). The initial contrast transfer function (CTF) parameters were estimated by the Gctf program (46, 47) at the micrograph level and the micrographs with the estimated resolution limit worse than 5 Å were discarded. The selected Gctf star file was imported into RELION 3.0.8 program (48) and all subsequent particle picking, classification, and reconstruction progress were performed in RELION 3.0.8.
For the RVFV L protein data set, about 5,090,000 particles were automatically selected from about 3,700 micrographs. After two rounds of extensive 2D classification, a subset of 3,380,000 particles were selected and further subjected to 3D classification with an initial model that was produced by the RELION program and low-pass-filtered to a resolution of 40 Å. Because of the large number of particles, we chose to split the particles into 3 groups and did the 3D classification separately. We used the same procedure of 3D classification on all the 3 groups separately. During the 3D classification, we used an angular sampling interval of 7.5 degree and did not perform local angular search. After all the three groups were executed with primary 3D classification, we joined the particles into a total data set of 1,184,890 particles to do further 3D classification. After two rounds of 3D classification, a reconstruction from 554,102 particles displayed clear features of secondary structural elements. We then performed 3D refinement from the ∼550,000 particles, yielding a reconstruction at a resolution of 3.84 Å. After postprocessing, we got a final map at 3.6 Å resolution as determined by the Fourier shell correlation (FSC) 0.143 cutoff value.
Model building.
The SFTSV L structure was used to guide manual building of RVFV L by Chainsaw (49). Manual model building was carried out using Coot (50) and refinement of the coordinates was performed using Phenix (51).
Small-angle X-ray scattering.
The overall model of the full-length RVFV L protein was obtained with the solved RVFV L (without the Lariat), and the simulated endonuclease domain and the PB2 C-terminal domain built from the structure of SFTSV (PDB 6L42) using the SASREF (52) with the distance restriction between the linker domain and the Lariat.
Small-angle X-ray scattering measurements were performed on beamline BL19U2 at the SSRF following previously published methods (53, 54). The protein concentration is from 1.25 mg/ml to 5 mg/ml. Briefly, all proteins were subjected to size exclusion chromatography in a buffer containing 20 mM BTP pH 8.5, 300 mM NaCl. The BioXTAS RAW (55) software was used for data processing. GNOM (56) provided the pair distribution function P(r) of the particle, the maximum size Dmax and the Porod volume. The 20 individual ab initio reconstructions were generated with DAMMIN (57), averaged using DAMAVER (58), and aligned using SUPCOMB (59). The structures were visualized using PyMOL (60).
Data availability.
Cryo-EM reconstructions and atomic models generated during this study are available at wwPDB and EMBD (https://www.rcsb.org; http://emsearch.rutgers.edu) under accession codes PDB: 7EEI and EMD-31077.
ACKNOWLEDGMENTS
We thank J. L. Lei, Y. J. Xu, and T. Yang for their support in cryo-EM and high-performance computation in the National Protein Science Facility (Beijing) at Tsinghua University. We thank J. Wen and X. M. Li for their friendly help in cryo-EM sample screening in the National Protein Science Facility (Beijing) at Tsinghua University. We thank Y. Li and C. Liu for cryo-EM data set collection at Shuimu Inc., Beijing. We thank the staff of beamlines BL19U2 at the Shanghai Synchrotron Radiation Facility for the excellent technical assistance.
This work was supported financially by the National Key Research and Development Program of China (2018YFE0113100 and 2016YFA0501100 to H.-W.W.), the National Natural Science Foundation of China (31825009 to H.-W.W. and 31872713, 32071210, and 31800629), the Open Fund of the State Key Laboratory of Pathogenic Microbial Biosafety (SKLPBS1834), and the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (2021SKLAB6-12). We declare no conflict of interest.
Contributor Information
Hong-Wei Wang, Email: hongweiwang@tsinghua.edu.cn.
Zhongzhou Chen, Email: chenzhongzhou@cau.edu.cn.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Freiberg AN, Sherman MB, Morais MC, Holbrook MR, Watowich SJ. 2008. Three-dimensional organization of Rift Valley fever virus revealed by cryoelectron tomography. J Virol 82:10341–10348. 10.1128/JVI.01191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komoda K, Narita M, Yamashita K, Tanaka I, Yao M. 2017. Asymmetric Trimeric Ring Structure of the Nucleocapsid Protein of Tospovirus. J Virol 91:e01002-17. 10.1128/JVI.01002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reguera J, Gerlach P, Rosenthal M, Gaudon S, Coscia F, Gunther S, Cusack S. 2016. Comparative Structural and Functional Analysis of Bunyavirus and Arenavirus Cap-Snatching Endonucleases. PLoS Pathog 12:e1005636. 10.1371/journal.ppat.1005636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eifan S, Schnettler E, Dietrich I, Kohl A, Blomstrom AL. 2013. Non-structural proteins of arthropod-borne bunyaviruses: roles and functions. Viruses 5:2447–2468. 10.3390/v5102447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salekwa LP, Wambura PN, Matiko MK, Watts DM. 2019. Circulation of Rift Valley fever virus antibody in cattle during inter-epizootic/epidemic periods in selected regions of Tanzania. Am J Trop Med Hyg 101:459–466. 10.4269/ajtmh.18-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick R, Bouloy M. 2005. Rift Valley fever virus. Curr Mol Med 5:827–834. 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- 7.Guardado-Calvo P, Atkovska K, Jeffers SA, Grau N, Backovic M, Pérez-Vargas J, de Boer SM, Tortorici MA, Pehau-Arnaudet G, Lepault J, England P, Rottier PJ, Bosch BJ, Hub JS, Rey FA. 2017. A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion. Science 358:663–667. 10.1126/science.aal2712. [DOI] [PubMed] [Google Scholar]
- 8.Golnar AJ, Turell MJ, LaBeaud AD, Kading RC, Hamer GL. 2014. Predicting the mosquito species and vertebrate species involved in the theoretical transmission of Rift Valley fever virus in the United States. PLoS Negl Trop Dis 8:e3163. 10.1371/journal.pntd.0003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vloet RPM, Vogels CBF, Koenraadt CJM, Pijlman GP, Eiden M, Gonzales JL, van Keulen LJM, Wichgers Schreur PJ, Kortekaas J. 2017. Transmission of Rift Valley fever virus from European-breed lambs to Culex pipiens mosquitoes. PLoS Negl Trop Dis 11:e0006145. 10.1371/journal.pntd.0006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen SS, Alvarez J, Bicout DJ, Calistri P, Depner K, Drewe JA, Garin-Bastuji B, Rojas JLG, Schmidt CG, Michel V, Chueca MÁM, Roberts HC, Sihvonen LH, Stahl K, Calvo AV, Viltrop A, Winckler C, Bett B, Cetre-Sossah C, Chevalier V, Devos C, Gubbins S, Monaco F, Sotiria-Eleni A, Broglia A, Abrahantes JC, Dhollander S, Stede YV, Zancanaro G. 2020. Rift Valley fever - epidemiological update and risk of introduction into. EFSA J 18:e06041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Ma T, Wu Y, Chen Z, Zeng H, Tong Z, Gao F, Qi J, Zhao Z, Chai Y, Yang H, Wong G, Bi Y, Wu L, Shi R, Yang M, Song J, Jiang H, An Z, Wang J, Yilma TD, Shi Y, Liu WJ, Liang M, Qin C, Gao GF, Yan J. 2019. Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus. Nat Microbiol 4:1231–1241. 10.1038/s41564-019-0411-z. [DOI] [PubMed] [Google Scholar]
- 12.Balenghien T, Cardinale E, Chevalier V, Elissa N, Failloux AB, Jean Jose Nipomichene TN, Nicolas G, Rakotoharinome VM, Roger M, Zumbo B. 2013. Towards a better understanding of Rift Valley fever epidemiology in the south-west of the Indian Ocean. Vet Res 44:78. 10.1186/1297-9716-44-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lokugamage N, Freiberg AN, Morrill JC, Ikegami T. 2012. Genetic subpopulations of Rift Valley fever virus strains ZH548 and MP-12 and recombinant MP-12 strains. J Virol 86:13566–13575. 10.1128/JVI.02081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gogovi GK, Almsned F, Bracci N, Kehn-Hall K, Shehu A, Blaisten-Barojas E. 2019. Modeling the Tertiary Structure of the Rift Valley Fever Virus L Protein. Molecules 24:1768. 10.3390/molecules24091768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balkhy HH, Memish ZA. 2003. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents 21:153–157. 10.1016/s0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- 16.Terasaki K, Murakami S, Lokugamage KG, Makino S. 2011. Mechanism of tripartite RNA genome packaging in Rift Valley fever virus. Proc Natl Acad Sci USA 108:804–809. 10.1073/pnas.1013155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 18.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 19.Gogrefe N, Reindl S, Gunther S, Rosenthal M. 2019. Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PLoS Pathog 15:e1007829. 10.1371/journal.ppat.1007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferron F, Li Z, Danek EI, Luo D, Wong Y, Coutard B, Lantez V, Charrel R, Canard B, Walz T, Lescar J. 2011. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog 7:e1002030. 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arragain B, Effantin G, Gerlach P, Reguera J, Schoehn G, Cusack S, Malet H. 2020. Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes. Nat Commun 11:3590. 10.1038/s41467-020-17349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Liu L, Liu A, Yan L, He Y, Shen S, Hu M, Guo Y, Liu H, Liu C, Lu Y, Wang P, Deng F, Rao Z, Lou Z. 2020. Structure of severe fever with thrombocytopenia syndrome virus L protein elucidates the mechanisms of viral transcription initiation. Nat Microbiol 5:864–871. 10.1038/s41564-020-0712-2. [DOI] [PubMed] [Google Scholar]
- 23.Vogel D, Thorkelsson SR, Quemin ERJ, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Günther S, Cusack S, Grünewald K, Rosenthal M. 2020. Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res 48:5749–5765. 10.1093/nar/gkaa253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerome H, Rudolf M, Lelke M, Pahlmann M, Busch C, Bockholt S, Wurr S, Gunther S, Rosenthal M, Kerber R. 2019. Rift Valley fever virus minigenome system for investigating the role of L protein residues in viral transcription and replication. J Gen Virol 100:1093–1098. 10.1099/jgv.0.001281. [DOI] [PubMed] [Google Scholar]
- 25.Holm T, Kopicki JD, Busch C, Olschewski S, Rosenthal M, Uetrecht C, Gunther S, Reindl S. 2018. Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses. J Biol Chem 293:19686–19698. 10.1074/jbc.RA118.004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar G, Cuypers M, Webby RR, Webb TR, White SW. 2021. Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism. Nucleic Acids Res 49:1609–1618. 10.1093/nar/gkaa1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Lu G, Zhang B, Gong P. 2015. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J Virol 89:249–261. 10.1128/JVI.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouloy M, Weber F. 2010. Molecular biology of rift valley Fever virus. Open Virol J 4:8–14. 10.2174/1874357901004020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te Velthuis AJ. 2014. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci 71:4403–4420. 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacome R, Becerra A, Ponce de Leon S, Lazcano A. 2015. Structural Analysis of Monomeric RNA-Dependent Polymerases: evolutionary and Therapeutic Implications. PLoS One 10:e0139001. 10.1371/journal.pone.0139001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong P, Peersen OB. 2010. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA 107:22505–22510. 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruenn JA. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res 31:1821–1829. 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerlach P, Malet H, Cusack S, Reguera J. 2015. Structural Insights into Bunyavirus Replication and Its Regulation by the vRNA Promoter. Cell 161:1267–1279. 10.1016/j.cell.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK. 2002. The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324:47–62. 10.1016/s0022-2836(02)01033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkataraman S, Prasad B, Selvarajan R. 2018. RNA Dependent RNA Polymerases: insights from Structure, Function and Evolution. Viruses 10:76. 10.3390/v10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, Hart DJ, Beck M, Cusack S. 2016. Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol Cell 61:125–137. 10.1016/j.molcel.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Te Velthuis AJ, Robb NC, Kapanidis AN, Fodor E. 2016. The role of the priming loop in Influenza A virus RNA synthesis. Nat Microbiol 1:16029. 10.1038/nmicrobiol.2016.29. [DOI] [PubMed] [Google Scholar]
- 40.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 41.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell 162:314–327. 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandzik JM, Kouba T, Karuppasamy M, Pflug A, Drncova P, Provaznik J, Azevedo N, Cusack S. 2020. A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase. Cell 181:877–893. 10.1016/j.cell.2020.03.061. [DOI] [PubMed] [Google Scholar]
- 43.Kouba T, Drncova P, Cusack S. 2019. Structural snapshots of actively transcribing influenza polymerase. Nat Struct Mol Biol 26:460–470. 10.1038/s41594-019-0232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Te Velthuis AJW, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han Y, Yan C, Fishbain S, Ivanov I, He Y. 2018. Structural visualization of RNA polymerase III transcription machineries. Cell Discov 4:40. 10.1038/s41421-018-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Yang F, Ling S, Lv P, Zhou Y, Fang W, Sun W, Zhang L, Shi P, Tian C. 2020. Single-particle cryo-EM structural studies of the β(2)AR-Gs complex bound with a full agonist formoterol. Cell Discov 6:45. 10.1038/s41421-020-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng J, Li N, Wang X, Hu J, Zhai Y, Gao N. 2020. Structural insight into the assembly and conformational activation of human origin recognition complex. Cell Discov 6:88. 10.1038/s41421-020-00232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein N. 2008. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr 41:641–643. 10.1107/S0021889808006985. [DOI] [Google Scholar]
- 50.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petoukhov MV, Svergun DI. 2005. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89:1237–1250. 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y, Zhang Q, Lang Y, Liu Y, Dong X, Chen Z, Tian W, Tang J, Wu W, Tong Y, Chen Z. 2017. Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation. J Mol Cell Biol 9:220–230. 10.1093/jmcb/mjx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tian Z, Li X, Li M, Wu W, Zhang M, Tang C, Li Z, Liu Y, Chen Z, Yang M, Ma L, Caba C, Tong Y, Lam HM, Dai S, Chen Z. 2020. Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms. Cell Discov 6:17. 10.1038/s41421-020-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen SS, Møller M, Gillilan RE. 2012. High-throughput biological small-angle X-ray scattering with a robotically loaded capillary cell. J Appl Crystallogr 45:213–223. 10.1107/S0021889812000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svergun D. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25:495–503. 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 57.Svergun DI. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76:2879–2886. 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Volkov VV, Svergun DI. 2003. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36:860–864. 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozin MB, Svergun DI. 2001. Automated matching of high- and low-resolution structural models. J Appl Crystallogr 34:33–41. 10.1107/S0021889800014126. [DOI] [Google Scholar]
- 60.Lam WW, Siu SW. 2017. PyMOL mControl: manipulating molecular visualization with mobile devices. Biochem Mol Biol Educ 45:76–83. 10.1002/bmb.20987. [DOI] [PubMed] [Google Scholar]
- 61.Peng R, Xu X, Jing J, Wang M, Peng Q, Liu S, Wu Y, Bao X, Wang P, Qi J, Gao GF, Shi Y. 2020. Structural insight into arenavirus replication machinery. Nature 579:615–619. 10.1038/s41586-020-2114-2. [DOI] [PubMed] [Google Scholar]
- 62.Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart DI, Grimes JM, Fodor E. 2015. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527:114–117. 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Cryo-EM reconstructions and atomic models generated during this study are available at wwPDB and EMBD (https://www.rcsb.org; http://emsearch.rutgers.edu) under accession codes PDB: 7EEI and EMD-31077.