ABSTRACT
The chikungunya virus has spread globally with a remarkably high attack rate. Infection causes arthralgic sequelae that can last for years. Nevertheless, there are no specific drugs or vaccines to contain the virus. Understanding the biology of the virus, such as its replication cycle, is a powerful tool to identify new drugs and comprehend virus-host interactions. Even though the chikungunya virus has been known for a long time (it was first described in 1952), many aspects of the replication cycle remain unclear. Furthermore, part of the cycle is based on observations of other alphaviruses. In this study, we used electron and scanning microscopy, as well as biological assays, to analyze and investigate the stages of the chikungunya virus replication cycle. Based on our data, we found infection cellular activities other than those usually described for the chikungunya virus replication cycle, i.e., we show particles enveloping intracellularly without budding in a membrane-delimited morphogenesis area, and we also observed virion release by membrane protrusions. Our work provides novel details regarding the biology of chikungunya virus and fills gaps in our knowledge of its replication cycle. These findings may contribute to a better understanding of virus-host interactions and support the development of antivirals.
IMPORTANCE The understanding of virus biology is essential to containing virus dissemination, and exploring the virus replication cycle is a powerful tool to do this. There are many points in the biology of the chikungunya virus that need to be clarified, especially regarding its replication cycle. Our incomplete understanding of chikungunya virus infection stages is based on studies with other alphaviruses. We systematized the chikungunya virus replication cycle using microscopic imaging in the order of infection stages, as follows: entry, replication, protein synthesis, assembly/morphogenesis, and release. The imaging evidence shows novel points in the replication cycle of enveloping without budding, as well as particle release by cell membrane protrusion.
KEYWORDS: chikungunya virus, electron microscopy, replication cycle, life cycle, membrane protrusions
INTRODUCTION
Chikungunya virus (CHIKV) emergence was reported in 1952 in the Tanganyika territory district, in the current territory of Tanzania, Africa, as the cause of a disease associated with fever and severe joint pain in humans (1, 2). The Chikungunya virus species belongs to the realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Martellivirales, family Togaviridae and genus Alphavirus, according to the International Committee on Taxonomy of Viruses (ICTV) (3). The virion is ∼70 nm, with an envelope originating from the host cell membrane that coats an ∼40 nm capsid containing an ∼11.8-kb positive single-stranded RNA molecule (4–6).
It has been previously noted that the CHIKV replication cycle occurs faster than other RNA arboviruses, and the first virions are formed about 5 h postinfection (hpi) in vertebrate cells (7, 8). Virus entry into the host cell is clathrin mediated (9, 10). At early time points after infection (about 2 h), CHIKV induces the formation of vesicular structures, termed spherules, formed by the CHIKV replication complex (nonstructural proteins [nsP1 to nsP4]) in association with the cell membrane; later in infection, spherules internalize, creating compartments known as cytopathic vacuoles type I (CPV-I) (11). After structural protein synthesis and assembly in the cytoplasm, immature particles pass through the endoplasmic reticulum-Golgi pathway. Capsid proteins, together with host vesicles, form cytopathic structures, termed cytopathic vacuoles type II (CPV-II) (12). Virions obtain envelopes from the host cell membrane after release by budding (13).
Most studies describe the CHIKV replication cycle based on alphaviruses in general, mostly on the Sindbis virus (14). However, several characteristics differentiate species between alphaviruses, namely, nucleotide/amino acid sequences, disease association, and vector and host association (15). To date, the entire CHIKV replication cycle remains unclear. Specific stages such as entry, assembly, maturation, budding process, and particularly the final processes of the cycle are incompletely resolved (16, 17). Also, protein receptors, proteins involved in virus production, and the role of CPV-II are still unknown (14, 17). Several works have provided electron microscopy evidence of CHIKV (17–19), but to our knowledge, there are no studies that have provided an in-depth morphological analysis of CHIKV replication cycle stages. These studies would help us understand the biology and replication cycle of this virus (20).
Here, we confirmed previously described data, added new images of replication cycle stages, and provided novel details about CHIKV replication cycle stages. Through electron and scanning microscopy, as well as immunofluorescent and biological assays, we analyzed CHIKV cell infection events. Using electron microscopy, we observed membrane protrusions engulfing viral particles in the first steps of infection, suggestive of macropinocytosis, as suggested elsewhere (21). However, the use of endocytic pathway inhibitors reinforced the importance of clathrin-mediated vesicles as a major entry pathway for CHIKV. We report that CHIKV particles can obtain an envelope intracellularly in a morphogenesis-specific area. Furthermore, we observed a release process similar to actin tail, with viral particles exiting from cell membrane protrusions. This study provides structured and well-characterized images in an attempt to shed new light on CHIKV biology.
RESULTS
Entry—Chikungunya virus enters Vero cells by a process involving coated vesicles and protrusions.
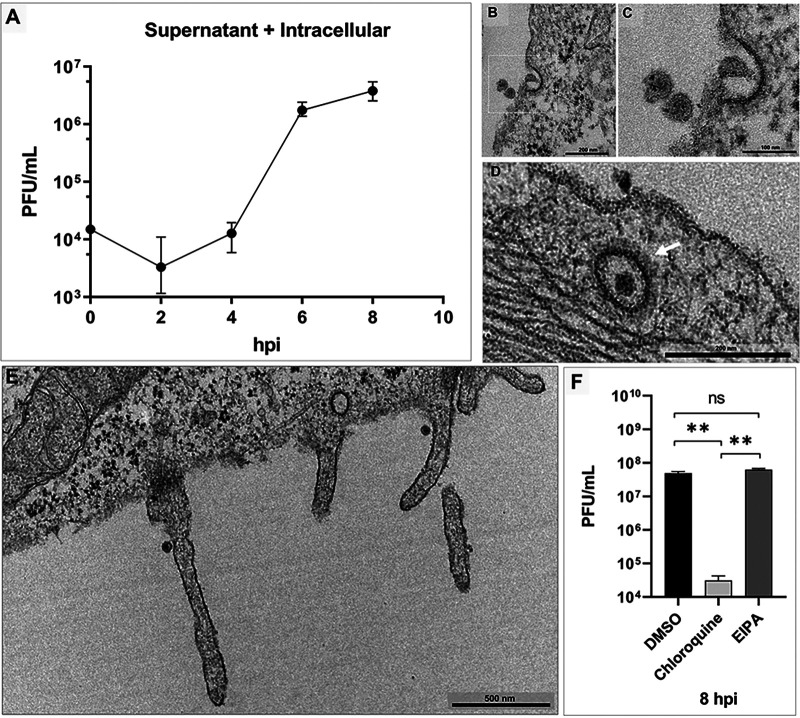
To analyze the CHIKV pattern of propagation in vitro, we performed a one-step growth curve in Vero cells at multiplicity of infection (MOI) of 5.0 with five time points. Titer was determined for particles in the supernatant and intracellular environment together by plaque assay (Fig. 1A). We confirmed a previous study on alphavirus growth that reported virion production starting from about 5 h postinfection (hpi) (22) and presenting rapid growth in these cells. We observed that viral progeny started to be released at 6 hpi (Fig. 1A).
FIG 1.
Chikungunya virus one-step growth curve and cell entry by coated vesicles and protrusions. Vero cells infected with chikungunya virus (CHIKV) at a multiplicity of infection (MOI) of 5.0 were assessed at 30 min postinfection (mpi) (B, C, D and E). (A) One-step growth curve. Three CHIKV particles (B) and one entering (C) starting an endosome. (D) A formed endosome coated with clathrin (white arrow) at a high MOI. (E) Membrane protrusions. (E) At early time points, two protrusions are present, each one with a single CHIKV particle. (F) Dimethyl sulfoxide (DMSO), 5-(N-ethyl-N-isopropyl) amiloride (EIPA), and chloroquine pretreatment for 1 h and infected at an MOI of 5.0 at 8 hpi. The images show electron micrographs with scale bars. Error bars indicate standard deviations. *, P < 0.05 (two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Bars, 200 nm (B); 100 nm (C); 200 nm (D); 500 nm (E).
It has been demonstrated that alphaviruses penetrate host cells mainly by clathrin-mediated endocytosis, although there is evidence that CHIKV can enter muscle cells by macropinocytosis (21). To evaluate entry events, based on previous studies (23), we obtained electron micrographs of Vero cells infected at an MOI of 5.0 at 30 min postinfection (mpi) (Fig. 1B to E). We observed that CHIKV particles entered Vero cells by two distinct approaches. In one of them, i.e., classical entry by clathrin-mediated endocytosis, virion penetration occurred through membrane invagination to form an endocytic coated vesicle in a Vero cell (Fig. 1B to D), and clathrin molecules surrounded the vesicle in formation (magnified) (Fig. 1C). Erasmus and colleagues showed the same patterns of clathrin-coated vesicles containing virus particle in electron micrographs (22). In the other approach, CHIKV entry occurred by protrusions that could be related to macropinocytosis events. At a high MOI (5.0) and at early time points after infection (30 min), two protrusions (lamellipodia) ∼700 nm in size were observed, with a single CHIKV particle in each one (Fig. 1E). We also observed this structure at a low MOI, with single virion particles and multiple particles inducing the formation of membrane protrusions (data not shown). In every 25 infected cells, we observed four macropinocytic projections (16%).
We analyzed the influence of entry pathway inhibitors to CHIKV in Vero cells. Vero cells were pretreated for 1 h with 60 μM 5-(N-ethyl-N-isopropyl) amiloride (EIPA), a specific macropinocytosis inhibitor; 100 μM chloroquine, an acidic-related endocytic pathway inhibitor; and dimethyl sulfoxide (DMSO) as the control. Afterward, cells were infected with CHIKV and titer was determined at 8 hpi. We observed a significant reduction (P = 0.0066) after treatment with chloroquine compared to DMSO treatment and a nonsignificant difference between EIPA and DMSO treatments (P = 0.1263) (Fig. 1F). Therefore, although we observed particle entry associated with protrusions, we could not confirm that macropinocytosis has a significant role in CHIKV entry into Vero cells.
Structures of replication—spherules appear in different regions of the cell and distinct formations of CPV-I.
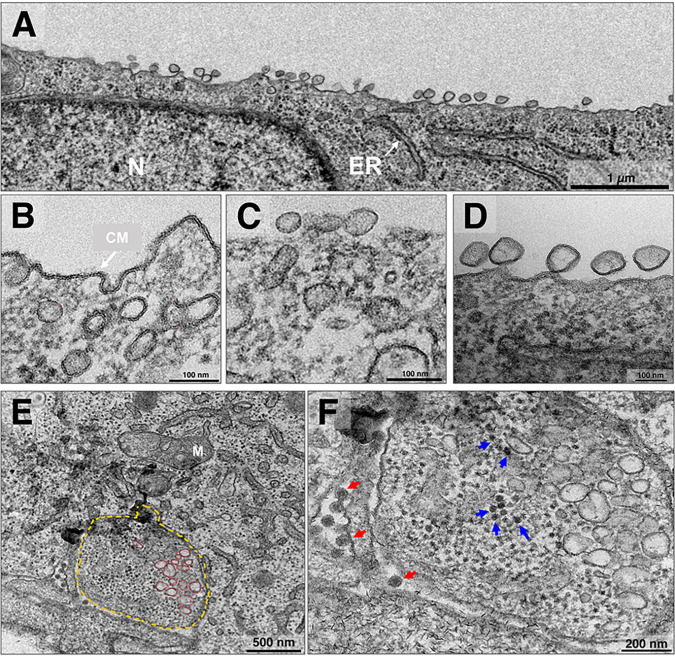
The nonstructural proteins (nsPs) constitute the replication complex of alphaviruses and form a membrane structure known as a spherule, which anchors to the cell membrane externally through a narrow neck (24). This structure produces an intermediary double-stranded RNA inside (25). Previous studies have shown micrographs of spherules for other alphaviruses (26, 27) and for CHIKV (28). However, we observed CHIKV spherules with various conformations in infected cells. These spherule conformations appeared at both time points, i.e., at 4 hpi with an MOI of 5.0 (Fig. 2A to D) and at 8 hpi with an MOI of 0.1 (Fig. 2E and F) in infected Vero cells. In most of the cells, these structures, with mixed sizes (∼60 to 100 nm), were found to be concentrated in one of the sides of the cell covering an extended part of the cell membrane (Fig. 2A). However, we found some spherules concentrated internally next to the cell membrane (Fig. 2B) and both internally and externally on the cell membrane (Fig. 2C). These distributions are atypical compared to those in other studies (13, 26) that have shown the standard distribution of spherules, anchored by the neck on the surface of the cell membrane (Fig. 2D). Generally, in alphaviruses, these spherules internalize in the late phase of infection, forming CPV-I, and continue RNA production throughout the infection. Interestingly, in CHIKV infection, it has been observed that most of the spherules remain on the cell surface for a longer time (29). Here, we found CPV-I (∼1.3 μm) housing disordered spherules (Fig. 2E) and capsid proteins (Fig. 2F). Curiously, this differs from the CPV-I pattern described previously, in which the spherules coated the vesicle internally and there were no capsids present (18).
FIG 2.
Chikungunya virus spherule distribution and CPV-I structure. Vero cells were infected with chikungunya virus (CHIKV) at a multiplicity of infection (MOI) of 5.0 were assessed at 4 h postinfection (hpi) (A to D) and at an MOI of 0.1 at 8 hpi by transmission electron microscopy (TEM) (E and F). (A) Spherule arrangement in the cell membrane. (B to D) Varied spherule arrangement. (B) Spherules of ∼60 to 100 nm in size next to the cell membrane, starting an externalization or internalizing. (C) Three external spherules and two internal spherules. (C and D) Spherules distributed over the cell membrane. (E and F) Cytopathic vacuole type 1 (CPV-I). (E) Internalized spherules (red), and the membrane of CPV-I (in yellow) formed after spherule internalization (in red). (F) Also present at higher magnification in CPV-I capsid proteins (blue arrow) and externally formed CHIKV particles (red arrow). N, nucleus; ER, endoplasmic reticulum; CM, cell membrane; M, mitochondria. Images show electron micrographs with scale bars. Bars, 1 μm (A); 100 nm (B); 100 nm (C); 100 nm (D); 500 nm (E); and 200 nm (F).
Chikungunya virus forms different types of cytopathic vacuoles in Vero cells.
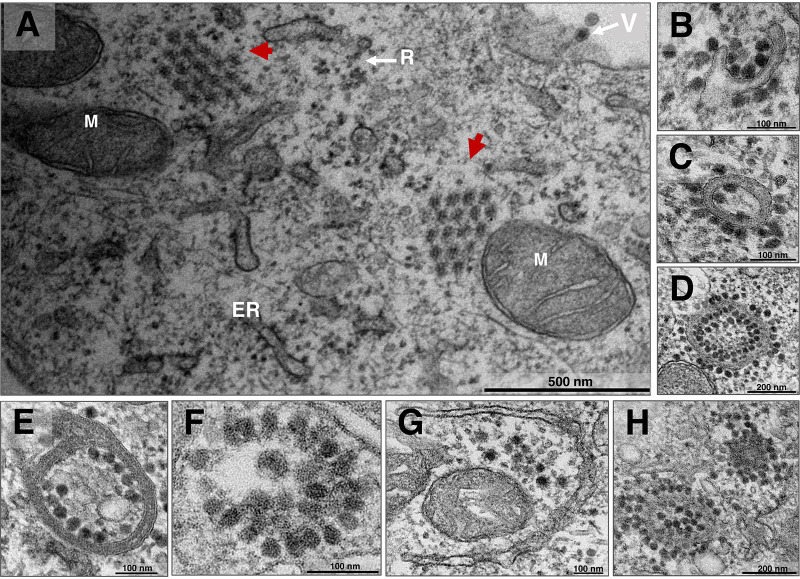
CHIKV produces many particles, intensely exploiting the host cell protein machinery. The structural protein nsP2 coordinates host translation shutoff, prioritizing viral protein translation (30). Structural proteins have been described as an organized distribution for CHIKV (18). This association, described as a “honeycomb,” is present as a morphogenesis area (31) or a release strategy (18). Higashi and colleagues previously showed structural proteins establishing different cytopathic structures and producing membraned vacuoles (19). Here, we confirm the presence of these vacuoles and introduce other types; also, we show the honeycomb structure of capsid precursors. In Vero cells infected with CHIKV at 8 hpi and an MOI of 5.0, coordinated capsid proteins established a honeycomb structure (Fig. 3A, red arrows). Additionally, the virus produced structures with a hand-like shape, containing capsids (Fig. 3B), and structures with a closed membrane shape (Fig. 3C and D). These closed, membraned structures could contain a few (Fig. 3C) or multiple capsids (Fig. 3D). CHIKV also produced other patterns differing in compartment number (Fig. 3E), capsid distribution, and membrane absence (Fig. 3F) or presence (Fig. 3G). More than one structure can be present in the same cell (Fig. 3H).
FIG 3.
Chikungunya virus capsid protein arrangement and cytopathic structures. Vero cells were infected with chikungunya virus (CHIKV) at late infection at a multiplicity of infection (MOI) of 5.0 and assessed at 8 h postinfection (hpi) by transmission electron microscopy (TEM). (A) Honeycomb arrangement capsid protein structures (red arrows) were observed. (B to D) Cytopathic hand-like structure formation in the process of closure (B), closed with some capsids (C), and closed with a large amount of CHIKV capsids (D). (E to H) CHIKV infection with varied cytopathic structures. M, mitochondria; ER, endoplasmic reticulum; R, ribosomes; V, mature CHIKV particle. Electron micrographs are shown with scale bars. Bars, 500 nm (A); 100 nm (B); 100 nm (C); 200 nm (D); 100 nm (E); 100 nm (F); 100 nm (G); 200 nm (H).
Morphogenesis—chikungunya virus can acquire an envelope without budding.
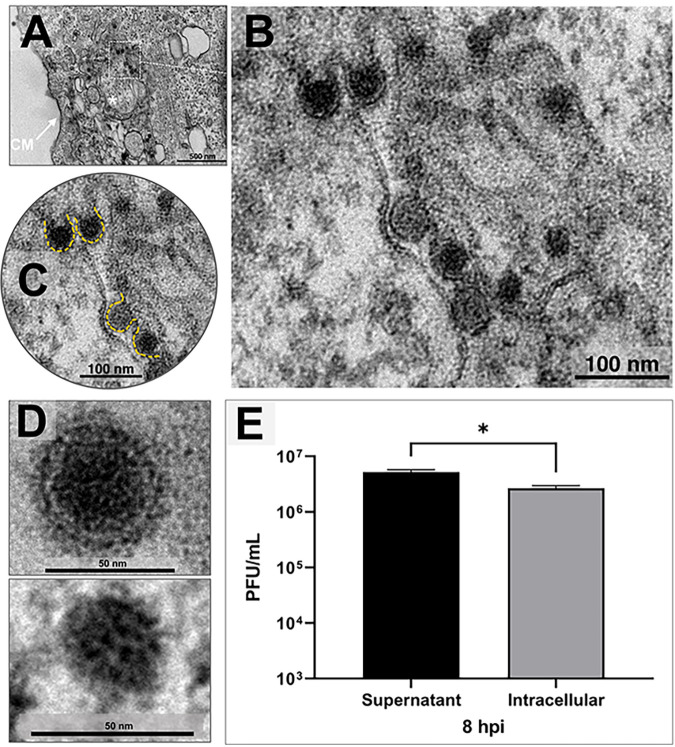
Alphaviruses obtain the cell membrane’s lipid envelope by budding, where the immature virus can become a virion (14). We assessed images of a synchronous CHIKV infection in Vero cells and, strikingly, we observed some viral particles acquiring an envelope without budding release. These particles obtained envelopes in a membrane-delimited morphogenesis area at 8 hpi and an MOI of 5.0 (Fig. 4A and B). For comparison, we show the highlighted envelopes (Fig. 4C) and two CHIKV particles, a mature CHIKV particle of ∼60 nm and a nucleocapsid without an envelope of ∼35 nm (Fig. 4D).
FIG 4.
Chikungunya virus assembly. (A to D) Vero cells were infected with chikungunya virus (CHIKV) at a multiplicity of infection (MOI) of 5.0 and assessed at 8 h postinfection (hpi) by transmission electron microscopy (TEM). (A to C) A morphogenesis area with CHIKV particles acquiring an envelope intracellularly. Magnification of image shown in panel A shows capsids with and without envelopes in progression (B), with new envelopes highlighted in yellow (C). (D) Two CHIKV particles, a complete mature CHIKV particle and a core capsid without envelope. (E) Plaque assay showing the difference between virions in the supernatant and intracellular virions at 8 hpi at an MOI of 5.0. CM, cell membrane. An asterisk indicates the morphogenesis area. Electron micrographs are shown with scale bars. Error bars indicate standard deviations. *, P < 0.05 (two-tailed Student’s t test). Data are representative of three independent experiments with similar results. Bars, 500 nm (A); 100 nm (B); 100 nm (C); and 50 nm (D).
In the next step, to evaluate if the intracellular enveloped particles could be infectious, we performed a one-step growth curve at an MOI of 5.0 by titrating both (i) the supernatant and (ii) the intracellular viruses at 8 hpi, from the same pool of infected cells. Interestingly, a large number of infectious particles was detected in the intracellular space (Fig. 4E).
Release—Vero cells are induced to produce protrusions during the release step of chikungunya virus in addition to virus budding.
Alphaviruses release by budding from host cells (32), but the complete final steps of CHIKV infection remain unclear (16, 17). To evaluate the release process, we infected Vero cells with CHIKV at a high MOI (5.0) and analyzed cells at late time points (8 hpi). First, we observed by electron microscopy the already described virus release by budding from the cell membrane. In Fig. 5A to C, we observed particles being released in different steps of the budding process.
FIG 5.
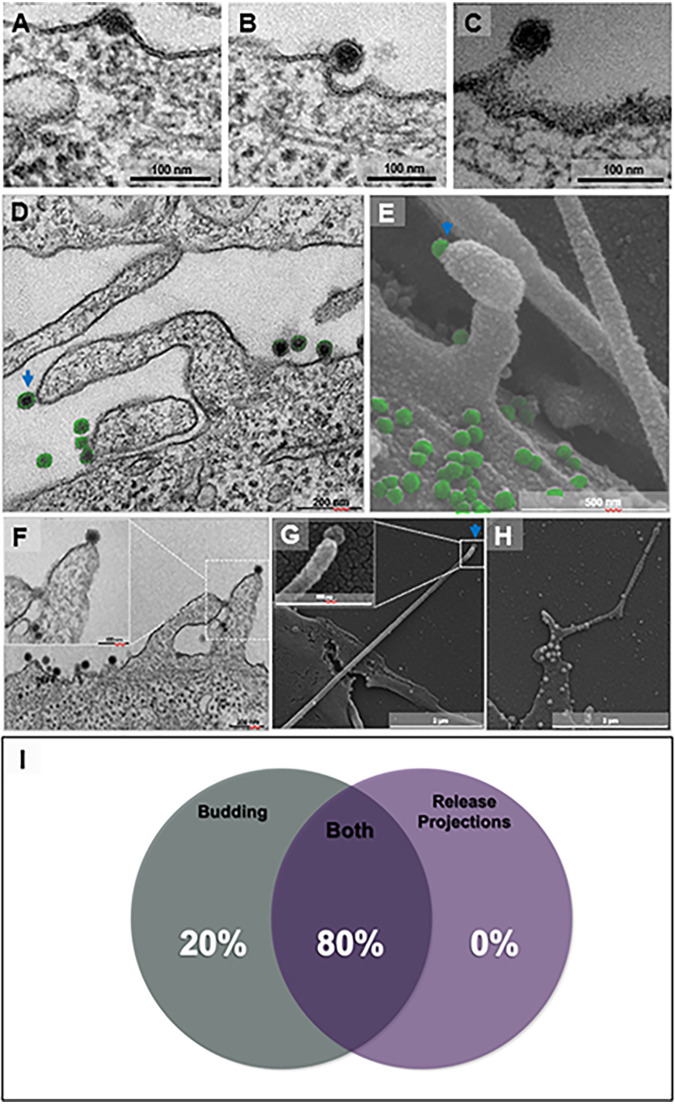
Chikungunya virus release by cellular protrusions in addition to budding. Vero cells were infected with chikungunya virus (CHIKV) at a multiplicity of infection (MOI) of 5.0 at 8 hpi (A to D and F) by transmission electron microscopy (TEM). Furthermore, for scanning electron microscopy (SEM), cells were infected with CHIKV at an MOI of 5.0 and assessed at 8 hpi (E, G, and H). (A to C) Release of CHIKV by budding, acquiring an early envelope from the cell membrane (A), finishing envelope formation (B), and almost completing release (C). (D to H) Release by cell membrane protrusions. A protrusion with a single CHIKV particle (blue arrow) on the tip and other particles near the cell surface (in green) in a transmission micrograph (D) and a scanning micrograph (E). The transmission micrograph shows two projections with a single CHIKV particle at the tip (F). The scanning micrograph shows a long actin tail with a single particle at the tip (G, blue arrow). The scanning micrograph shows multiple particles over the cell surface and some particles along the tail with a particle at the tip (H). Percentage of presence of release protrusions and budding present in 25 infected cells (I). Electron micrographs are shown with scale bars. Bars, 100 nm (A); 100 nm (B); 100 nm (C); 200 nm (D); 500 nm (E); 200 nm (F); 2 μm (G); 2 μm (H).
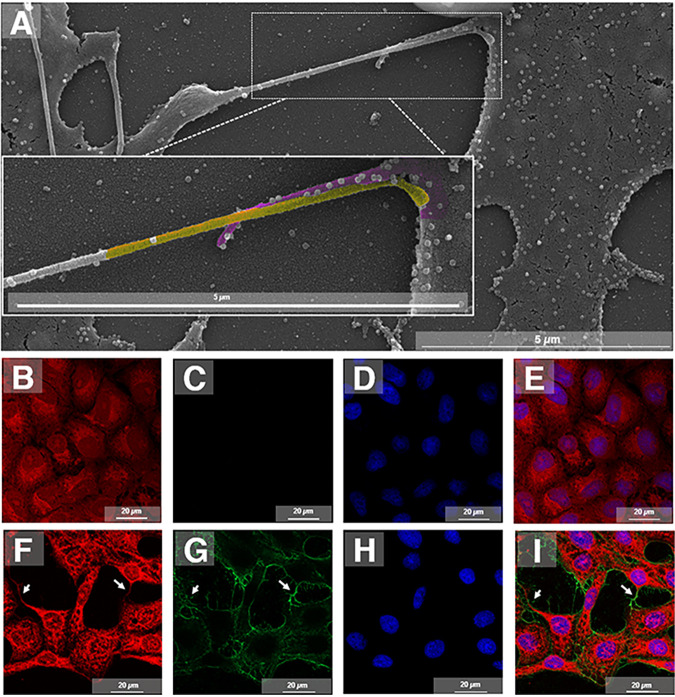
Surprisingly, we also observed by transmission and scanning electron microscopy (TEM and SEM) particles being released from host cells by cell membrane protrusions (Fig. 5D and E). We found such protrusions carrying single particles at the apical tip (Fig. 5D, blue arrow). Notably, in this mode of release, viruses seem to leave the cell host by a process like budding, but somehow induce the formation of a protrusion (Fig. 5F). In addition, using scanning electron microscopy (SEM), we found atypically long protrusions that contained a single particle at the tip (Fig. 5G) and many particles distributed along the protrusion (Fig. 5H). About 80% of the observed cells presented particles being released by both budding in regular flat cell membranes and associated with protrusions (Fig. 5I). Some of those protrusions containing particles were observed, by SEM, to attach to other infected cells (Fig. 6A). By immunofluorescence (IF), infected cells, at an MOI of 5 and at 8 hpi, appeared to be connected by projections labeled by anti-CHIKV antibodies (Fig. 6B to I).
FIG 6.
Virus cell distribution and connection between cells by cell membrane protrusions. Vero cells were infected with chikungunya virus (CHIKV) at a multiplicity of infection (MOI) of 5.0 and assessed at 8 hpi by scanning electron microscopy (SEM). Two actin tails from different cells (cell one, yellow; cell two, purple) connected with multiple viruses along with the protrusions (A). Vero cells were infected with CHIKV at an MOI of 5.0 and assessed at 8 hpi by immunofluorescence microscopy. (B to E) Control cells mock-infected with the cytoskeleton stained with Evans blue (in red) (B), labeled with Alexa Fluor 488 (C), nuclei stained with DAPI (in blue) (D), and merged images (E). (F to I) Immunofluorescence of CHIKV infection shows the cytoskeleton stained with Evans blue (in red) (F), the virus distribution over the cell surface and connections labeled with Alexa Fluor 488 (in green) (G), nuclei stained with DAPI (in blue) (H), and colocalization of the virus and cell connections (I). White arrows indicate the connections between cells. Immunofluorescent images are shown with scale bars. Bars, 5 μm (A); 20 μm (B to I).
DISCUSSION
There are intriguing, obscure questions and answers to explore regarding the biology of many viruses in the virosphere. Several of these questions have been raised about the CHIKV replication cycle, mainly because of the vast number of hosts and cell types CHIKV can infect. CHIKV has been known since 1952, but its biology remains incompletely understood (14). Here, we provide evidence of structures and CHIKV replication cycle stages that confirm classical findings, but also open up questions about unclear points. These data provide organized images regarding the CHIKV replication cycle obtained by electron and scanning micrography and confirmed by biological assays. In our data, we observed that early points of infection correspond to 3 to 4 hpi and late points begin at 8 hpi, both at a high MOI.
The CHIKV entry mechanism was classically described based on alphavirus studies that show clathrin-mediated endocytosis requiring low pH for successful infection (33). Although clathrin-mediated entry occurs for CHIKV, macropinocytic entry was recently observed in muscle cells (21). To date, this novel entry process is still unconfirmed in other cell lines. A considerable number of DNA or RNA viruses with or without an envelope utilize macropinocytosis to enter the host cell (34). Some of these viruses include vaccinia virus (35), adenovirus (36), echovirus 1 (37), human immunodeficiency virus (38), and rubella virus (39). These viruses form a macropinosome with a variable size and shape between 0.5 to 10 μm (34). Here, even though we have shown electron micrographs of CHIKV particles entering into Vero cells associated with protrusions (Fig. 1E), the treatment of cells with the macropinocytosis inhibitor EIPA did not affect virus titers at 8 hpi. Taken together, our results indicate that the main mechanism of entry is clathrin mediated in Vero cells (Fig. 1F). However, future experiments with different concentrations of EIPA could be useful to better investigate this entry pathway. Rapid replication of CHIKV in cells could be investigated further, and it could perhaps utilize other entry mechanisms because of the vast array of cell types and hosts (vertebrate/invertebrate) infected by CHIKV (40, 41). Despite this, more in-depth studies are necessary to explore the precise times and specific triggers of these protrusions in CHIKV infection.
Early in infection, i.e., about 2 h postinfection, translation by host ribosomal nsP1 to nsP4 forms the replication complex and starts RNA replication. This complex and the cell membrane compose an evagination with a flask shape, called a spherule (13). Spherules are found in the outer part of the cell membrane, and the replication complex is located in the narrow neck. This external localization helps an RNA double-stranded intermediary, produced by nsPs, to evade pattern recognition receptors such as the retinoic acid-inducible gene I (RIG-I) (14). During the infection course, these spherules are internalized and form a vacuolar structure termed CPV-I that contains lysosomal and endosomal markers (11). This CPV-I forms a large vesicular structure surrounded on the surface internally by spherules that continue RNA production (18). Intriguingly, here, we detected some spherules distributed in different positions, namely, next to the cell membrane and within the cytoplasm (Fig. 2B) and combined within and outside the cytoplasm (Fig. 2C), besides the usual arrangement (Fig. 2A and D). In Fig. 2B and C, it is possible that those spherules are undergoing internalization in a CPV-I formation. We also found a different presentation of CPV-I presenting polarization of spherules and disarranged capsid proteins within the vacuole (Fig. 2E and F). The CPV-I encountered in this work may represent an initial form of the CHIKV morphogenesis area, as shown in Fig. 4A (asterisk), although more studies with specific membrane markers need to be performed to confirm this.
During the protein synthesis stage, CHIKV produces cytopathic structures comprising mainly capsid (C) protein (19). Late in infection, structural proteins form CPV-II, a vacuolar structure housing envelope (E) proteins and coated with capsids that are believed to release by budding (14). These structures are produced in both vertebrate and invertebrate cell infection (18). Here, we present many additional images of cytopathic vacuole structures presenting membranes or not and with capsid proteins arranged in different conformations (Fig. 3). Besides these structures, capsids can form a honeycomb structure, as seen in giant viruses (31, 42) and also seen with CHIKV, but only in the late phase, as mature virions are released by exocytosis (18). We believe that CPV-II structural variation may represent an alternative form that can enhance virus assembly and even promote alternative release, such as the exocytosis of multiple particles. However, to confirm this hypothesis, studies evaluating precise time points with synchronous infection in sequence are necessary to visualize this final step.
It has been described that alphaviruses only acquire an envelope by budding from the cell membrane. There is no other way reported, as far as we know (33). Surprisingly, we found microscopic evidence that raises questions about the CHIKV enveloping process (Fig. 4B and C). For the first time, to our knowledge, CHIKV particles were found acquiring an envelope intracellularly in Vero cells (Fig. 4B). Our biological assay confirmed the infectivity of these intracellular mature viruses, since a high number of intracellular particles could form plaques compared to supernatant virions at 8 hpi (considering the whole flask content) (Fig. 4E). We propose that CHIKV also can obtain an envelope and become mature before leaving the host cell. This process may be an alternative mechanism of particle maturation, in which the most immature viruses (only capsids) are released by budding. This alternative maturation pathway could produce different virus particles, as seen in vaccinia virus, which produces intracellular mature virions (IMV), intracellular enveloped virions (IEV), cell-associated enveloped virions (CEV), and extracellular enveloped virions (EEV) (43). More experiments employing membrane markers and budding inhibitors should be carried out to understand this mechanism.
In addition, in the final stage of infection, interestingly, we observed another different feature during CHIKV release, namely the formation of cell membrane protrusions containing particles (Fig. 5G). These protrusions are similar to actin tails (Fig. 5D and E), which are common in other viruses such as vaccinia virus; however, further analyses are necessary to reveal the nature of those protrusions. These structures carry a single particle in the tip of the tail (44). These extensions were visualized at a late phase of infection at a high MOI (5.0) at 8 hpi as by TEM (Fig. 5D and F) and by SEM (Fig. 5E and G and H). Curiously, we did not find an infected cell forming projections toward an uninfected cell; we only found different connected cells, which had already been infected, connected by these protrusions carrying multiple particles (Fig. 6A). About 80% of the observed infected cells presented particles budding both in flat membrane regions and associated with such protrusions (Fig. 5I). We believe this process can potentiate virus dissemination between cells; we also believe that large CHIKV plaques may be due to the participation of these protrusions, as occurs with vaccinia virus in which a mutant virus that is unable to polymerize actin tails forms only small plaques (45).
There are still some unanswered questions about CHIKV biology, and a vast field needs to be explored. This study provides new images of confirmed classic patterns and found novel structures regarding CHIKV replication cycle stages, namely, intracellularly enveloped infectious particles and virion release by cell membrane protrusions. Future studies and strategies are necessary and will further increase our comprehension of CHIKV infection. Using a high MOI and different infection time points, we provide microscopic evidence of novel mechanisms other than classical ones involved in the CHIKV replication cycle. We hope to clarify points in the CHIKV replication cycle and contribute to basic science in the extensive field of virology to better understand virus behavior and host pathogenesis.
MATERIALS AND METHODS
Cell culture, virus growth curve, one-step growth curve, and titration.
Vero cells (ATCC CCL-81) were cultured in Eagle’s minimum essential medium (MEM; Gibco, USA) supplemented with 5% (vol/vol) heat-inactivated fetal bovine serum (FBS) (Cultilab, Campinas, SP, Brazil) and antibiotics (40 μg/mL streptomycin, 200 U/mL penicillin, and 1.5 μg/mL amphotericin B [Fungizone]) and incubated in 5% CO2 at 37°C. The virus used in this work was CHIKV belonging to the ECSA genotype and strain BHI1762H804917 and was a generous gift from Maurício Lacerda Nogueira from the Faculty of Medicine of São José do Rio Preto–FAMERP, São Paulo, Brazil. This study is registered on the SISGEN platform (number A3FA3B4).
To perform the one-step growth curve (synchronous infection), Vero cells (approximately 1 × 106 cells per 25-cm2 flask) were infected with CHIKV at an MOI of 5.0 and incubated at 37°C with 5% CO2. The adsorption was performed for 1 h; the volumes were then completed to 4 mL with MEM. The supernatants, phosphate-buffered saline (PBS), and or monolayers were collected at preselected times (2, 4, 6, and 8 hpi). For the supernatant curve, 200 μL of the supernatant was collected and titrated (described below). Furthermore, for the intracellular samples, after washing with 500 μL PBS four times and without any remaining PBS present, 300 μL of MEM was aliquoted and frozen in a −70°C ultrafreezer; these monolayers were frozen and thawed three times, scraped with tips, and collected in 1.5-mL microtubes. These tubes were centrifuged at 13,000 × g for 5 min, and the supernatant was collected in 0.6-mL microtubes. After that, the contents of these tubes were titrated. The titration was performed by the plaque assay with the viral suspension, using the serial dilution method (10−1 to 10−6), on a monolayer of Vero cells grown in 24-well plates (1.5 × 105 cells per well) and with uninfected wells maintained as the control. After 1 h of adsorption at 37°C, 1 mL of 199 medium was added per well, supplemented with 2% FBS and containing 1.5 carboxymethylcellulose (CMC; Synth). Plates were incubated at 37°C in an atmosphere of 5% CO2 for 2 days, then 1 mL of a 10% formalin solution was added for 24 h at room temperature (RT) for fixation. The next day, the solution was discarded. Afterward, the monolayer was stained with a solution containing 1% (wt/vol) of crystal violet in PBS to count the lysis plaques, and the viral titer was expressed as the number of PFU per milliliter. Graphs were constructed using Prism version 7 for Windows (GraphPad Software).
Inhibitor assays.
Vero cells were seeded in T25 culture flasks (25 cm2) with minimum essential medium (MEM) supplemented with 5% (vol/vol) heat-inactivated FBS (Cultilab, Campinas, SP, Brazil), antibiotics (40 μg/mL streptomycin and 200 U/mL penicillin) and 1.5 μg/mL amphotericin B. The cells were incubated in 5% CO2 at 37°C until they reached 90% confluence (1.5 million cells). The cells were then treated for 1 h with dimethyl sulfoxide (DMSO) as a control; 60 μM 5-(N-ethyl-N-isopropyl) amiloride (EIPA); or 100 μM chloroquine (two biological replicates for each inhibitor and control). Afterward, the cells were washed three times with PBS 1× and were infected with CHIKV at an MOI of 5.0 with adsorption for 1 h. Cells were then washed with PBS 1× and MEM with 5% FBS was added. At 8 hpi, the flask contents were collected and added to a 24-well plate with three replicates, and the results were analyzed.
Transmission electron microscopy and scanning electron microscopy.
For transmission electron microscopy (TEM), Vero cells were infected as described in the previous section at an MOI of 5.0 and fixed at various time points postinfection (30 mpi, 4 hpi, and 8 hpi for an MOI of 5.0) with 2.5% glutaraldehyde in a 0.1 M sodium phosphate buffer for 2 h at RT. Cells were postfixed with 2% osmium tetroxide and embedded in Epon resin. Ultrathin sections were then analyzed under TEM (Spirit BioTwin FEI, 120 kV) at the Center of Microscopy of UFMG, Brazil.
For scanning electron microscopy (SEM), Vero cells infected at an MOI of 5.0 were adhered to round glass coverslips covered with poly-l-lysine and at 8 hpi fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 h at RT. The samples were then washed three times with 0.1 M cacodylate buffer and postfixed with 1.0% osmium tetroxide for 1 h at RT. After the second fixation, the samples were washed three times with 0.1 M cacodylate buffer and immersed in 0.1% tannic acid for 20 min. The samples were then washed in cacodylate buffer and dehydrated by serial passages in ethanol solutions at concentrations ranging from 35% to 100%. Samples were then subjected to critical point drying using CO2, placed on stubs, and metallized with a 5-nm gold layer. The analyses were completed using scanning electronic microscopy (FEG Quanta 200 FEI) at the Center of Microscopy of UFMG, Brazil.
Immunofluorescence assay.
For immunofluorescence (IF) assay, 1.5 × 105 Vero cells were cultured on round glass coverslips, and after incubation for 24 h at 37°C 5% CO2, the cells were infected at an MOI of 5.0. After 8 hpi, cells were fixed with acetone for 10 min at −20°C. After fixation, cells were then incubated with 3% bovine serum albumin (BSA)-PBS for 30 min, followed by three rinses with PBS. After that, cells were stained with polyclonal anti-CHIKV whole-particle antibodies produced in our animal facility (institutional animal care and use committee [IACUC] number 7/2019) in mice (diluted 1:1,000 in PBS) at 4°C overnight, followed by three rinses with PBS. After a 1-h incubation with Alexa Fluor 488-conjugated anti-mouse IgG (diluted 1:400 in PBS) at 37°C, followed by three rinses with PBS, the cells were also incubated with 4,6-diamino-2-phenylindole (DAPI, diluted 1:1,000 in PBS; Sigma, São Paulo, Brazil) for 1 h at RT. After three rinses with PBS, the cells were incubated with Evans Blue (Thermo Scientific) for 1 h at RT. Uninfected cells (control) were also fixed and prepared as described above. Fluorescently labeled cells were observed using an Axio Imager Z2-Apotome 2 microscope (Zeiss) at the Center of Microscopy of UFMG, Brazil. Zen Lite software from Zeiss microscopy was used for image processing.
ACKNOWLEDGMENTS
We thank our colleagues from Grupo de Transdução de Sinal (GTS), the Laboratório de Vírus, and the Center of Microscopy of UFMG for their admirable scientific support.
We also thank FAPEMIG, PRPq-UFMG, CNPq, and CAPES for financial support.
J.S.A. and C.A.B. are CNPq researchers.
Contributor Information
Jônatas S. Abrahão, Email: jonatas.abrahao@gmail.com.
Cláudio A. Bonjardim, Email: cbonjardim29@gmail.com.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Lumsden WHR. 1955. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952–1953 II. General description and epidemiology. Trans R Soc Trop Med Hyg 49:33–57. 10.1016/0035-9203(55)90081-X. [DOI] [PubMed] [Google Scholar]
- 2.Robinson MC. 1955. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R Soc Trop Med Hyg 49:28–32. 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Dolja VV, Krupovic M, Varsani A, Yutin N, Zerbini MK. 2019. Create a megataxonomic framework, filling all principal taxonomic ranks, for realm Riboviria. ICTV report no. 2019.006G. 10.13140/RG.2.2.20234.21444. [DOI]
- 4.Chen L, Wang M, Zhu D, Sun Z, Ma J, Wang J, Kong L, Wang S, Liu Z, Wei L, He Y, Wang J, Zhang X. 2018. Implication for alphavirus host-cell entry and assembly indicated by a 3.5Å resolution cryo-EM structure. Nat Commun 9:5326. 10.1038/s41467-018-07704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y-N, Deng C-L, Li J-Q, Li N, Zhang Q-Y, Ye H-Q, Yuan Z-M, Zhang B. 2019. Infectious chikungunya virus (CHIKV) with a complete capsid deletion: a new approach for a CHIKV vaccine. J Virol 93:e00504-19. 10.1128/JVI.00504-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metz SW, Gardner J, Geertsema C, Le TT, Goh L, Vlak JM, Suhrbier A, Pijlman GP. 2013. Effective chikungunya virus-like particle vaccine produced in insect cells. PLoS Negl Trop Dis 7:e2124. 10.1371/journal.pntd.0002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin DE. 2013. Alphaviruses, p 655. In Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 8.Chain MM, Doane FW, McLean DM. 1966. Morphological development of chikungunya virus. Can J Microbiol 12:895–900. 10.1139/m66-122. [DOI] [PubMed] [Google Scholar]
- 9.Ooi YS, Stiles KM, Liu CY, Taylor GM, Kielian M. 2013. Genome-wide RNAi screen identifies novel host proteins required for alphavirus entry. PLoS Pathog 9:e1003835-10. 10.1371/journal.ppat.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wichit S, Hamel R, Bernard E, Talignani L, Diop F, Ferraris P, Liegeois F, Ekchariyawat P, Luplertlop N, Surasombatpattana P, Thomas F, Merits A, Choumet V, Roques P, Yssel H, Briant L, Missé D. 2017. Imipramine inhibits chikungunya virus replication in human skin fibroblasts through interference with intracellular cholesterol trafficking. Sci Rep 7:3145. 10.1038/s41598-017-03316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spuul P, Balistreri G, Kääriäinen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J Virol 84:7543–7557. 10.1128/JVI.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanova L, Schlesinger MJ. 1993. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol 67:2546–2551. 10.1128/JVI.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin J, Galaz-Montoya JG, Sherman MB, Sun SY, Goldsmith CS, O’Toole ET, Ackerman L, Carlson L-A, Weaver SC, Chiu W, Simmons G. 2018. Neutralizing antibodies inhibit chikungunya virus budding at the plasma membrane. Cell Host Microbe 24:417–428.e5. 10.1016/j.chom.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva LA, Dermody TS. 2017. Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest 127:737–749. 10.1172/JCI84417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey LL, Powers A, Weaver SC. 2018. ICTV virus taxonomy profile: Togaviridae. J Gen Virol 99:761–762. 10.1099/jgv.0.001072. [DOI] [PubMed] [Google Scholar]
- 16.Schnierle BS. 2019. Cellular attachment and entry factors for chikungunya virus. Viruses 11:1078. 10.3390/v11111078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solignat M, Gay B, Higgs S, Briant L, Devaux C. 2009. Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393:183–197. 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen KC, Kam Y-W, Lin RPT, Ng MM-L, Ng LFP, Chu JHJ. 2013. Comparative analysis of the genome sequences and replication profiles of chikungunya virus isolates within the East, Central and South African (ECSA) lineage. Virol J 10:169. 10.1186/1743-422X-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higashi N, Matsumoto A, Tabata K, Nagatomo Y. 1967. Electron microscope study of development of chikungunya virus in green monkey kidney stable (VERO) cells. Virology 33:55–69. 10.1016/0042-6822(67)90093-1. [DOI] [PubMed] [Google Scholar]
- 20.Buzón P, Maity S, Roos WH. 2020. Physical virology: from virus self-assembly to particle mechanics. Wiley Interdiscip Rev Nanomed Nanobiotechnol 12:e1613. 10.1002/wnan.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua C, Lee R, Hussain KM, Chu JJH. 2019. Macropinocytosis dependent entry of chikungunya virus into human muscle cells. PLoS Negl Trop Dis 13:e0007610-19. 10.1371/journal.pntd.0007610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erasmus JH, Auguste AJ, Kaelber JT, Luo H, Rossi SL, Fenton K, Leal G, Kim DY, Chiu W, Wang T, Frolov I, Nasar F, Weaver SC. 2017. A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat Med 23:192–199. 10.1038/nm.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crill WD, Roehrig JT. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol 75:7769–7773. 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartholomeeusen K, Utt A, Coppens S, Rausalu K, Vereecken K, Ariën KK, Merits A. 2018. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol 92:e00152-18. 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. 2016. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 84:11679–11695. 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75:3873–3884. 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellström K, Kallio K, Utt A, Quirin T, Jokitalo E, Merits A, Ahola T. 2017. Partially uncleaved alphavirus replicase forms spherule structures in the presence and absence of RNA template. J Virol 91:e00787-17. 10.1128/JVI.00787-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thaa B, Biasiotto R, Eng K, Neuvonen M, Götte B, Rheinemann L, Mutso M, Utt A, Varghese F, Balistreri G, Merits A, Ahola T, McInerney GM. 2015. Differential phosphatidylinositol-3-kinase-Akt-mTOR activation by Semliki Forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J Virol 89:11420–11437. 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frolov I, Schlesinger S. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol 68:1721–1727. 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borges I, Rodrigues RAL, Dornas FP, Almeida G, Aquino I, Bonjardim CA, Kroon EG, La Scola B, Abrahão JS. 2019. Trapping the enemy: Vermamoeba vermiformiscircumvents Faustovirus mariensis dissemination by enclosing viral progeny inside cysts. J Virol 93:e00312-19. 10.1128/JVI.00312-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown RS, Wan JJ, Kielian M. 2018. The alphavirus exit pathway: what we know and what we wish we knew. Viruses 10:89. 10.3390/v10020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost M-C, Fsihi H, Frenkiel M-P, Blanchet F, Afonso PV, Ceccaldi P-E, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saïb A, Rey FA, Arenzana-Seisdedos F, Desprès P, Michault A, Albert ML, Schwartz O. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog 3:e89. 10.1371/journal.ppat.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat Cell Biol 11:510–520. 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 35.Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535. 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 36.Meier O, Boucke K, Hammer SV, Keller S, Stidwill RP, Hemmi S, Greber UF. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol 158:1119–1131. 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krieger SE, Kim C, Zhang L, Marjomaki V, Bergelson JM. 2013. Echovirus 1 entry into polarized Caco-2 cells depends on dynamin, cholesterol, and cellular factors associated with macropinocytosis. J Virol 87:8884–8895. 10.1128/JVI.03415-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maréchal V, Prevost M-C, Petit C, Perret E, Heard J-M, Schwartz O. 2001. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol 75:11166–11177. 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J-Y, Bowden DS. 2000. Rubella virus replication and links to teratogenicity. Clin Microbiol Rev 13:571–587. 10.1128/CMR.13.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard E, Solignat M, Gay B, Chazal N, Higgs S, Devaux C, Briant L. 2010. Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS One 5:e11479. 10.1371/journal.pone.0011479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoornweg TE, van Duijl-Richter MKS, Nuñez NVA, Albulescu IC, van Hemert MJ, Smit JM. 2016. Dynamics of chikungunya virus cell entry unraveled by single-virus tracking in living cells. J Virol 90:4745–4756. 10.1128/JVI.03184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung C-S, Chen C-H, Ho M-Y, Huang C-Y, Liao C-L, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J Virol 80:2127–2140. 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cudmore S, Cossart P, Griffiths G, Way M. 1995. Actin-based motility of vaccinia virus. Nature 378:636–638. 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W-H, Wilcock D, Smith GL. 2000. Vaccinia virus F12L protein is required for actin tail formation, normal plaque size, and virulence. J Virol 74:11654–11662. 10.1128/jvi.74.24.11654-11662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]