ABSTRACT
Type I interferons (IFN-Is) play a key role in host defense against virus infection, but porcine reproductive and respiratory syndrome virus (PRRSV) infection does not effectively activate IFN-I response, and the underlying molecular mechanisms are poorly characterized. In this study, a novel transcription factor of the heme oxygenase-1 (HO-1) gene, homeobox A3 (HOXA3), was screened and identified. Here, we found that HOXA3 was significantly increased during PRRSV infection. We demonstrated that HOXA3 promotes PRRSV replication by negatively regulating the HO-1 gene transcription, which is achieved by regulating IFN-I production. A detailed analysis showed that PRRSV exploits HOXA3 to suppress beta interferon (IFN-β) and IFN-stimulated gene (ISG) expression in host cells. We also provide direct evidence that the activation of IFN-I by HO-1 depends on its interaction with IRF3. Then we further proved that a deficiency of HOXA3 promoted the HO-1-IRF3 interaction and subsequently enhanced IRF3 phosphorylation and nuclear translocation in PRRSV-infected cells. These data suggest that PRRSV uses HOXA3 to negatively regulate the transcription of the HO-1 gene to suppress the IFN-I response for immune evasion.
IMPORTANCE Porcine reproductive and respiratory syndrome (PRRS), caused by PRRSV, causes significant worldwide economic losses in the pork industry. HOXA3 is generally considered to be an important molecule in the process of body development and cell differentiation. Here, we found that a novel transcription factor of the HO-1 gene, HOXA3, can negatively regulate the transcription of the HO-1 gene and play an important role in the suppression of IFN-I response by PRRSV. PRRSV induces the upregulation of HOXA3, which can negatively regulate HO-1 gene transcription, thereby weakening the interaction between HO-1 and IRF3 for inhibiting the type I IFN response. This study extends the function of HOXA3 and provides new insights into the PRRSV immune evasion mechanism.
KEYWORDS: PRRSV, HOXA3, HO-1, IRF3, IFN-I, regulatory transcription, immune evasion
INTRODUCTION
Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most economically important viruses affecting the swine industry worldwide, resulting in significant economic losses each year (1–3). Porcine reproductive and respiratory syndrome (PRRS) is caused by PRRSV, which is an enveloped, single-stranded RNA virus belonging to the genus Arterivirus, family Arteriviridae, and order Nidovirales (4). Current vaccination strategies cannot effectively control PRRSV infection because of its high antigenic heterogeneity (5, 6), its replication in and destruction of lung alveolar macrophages (7, 8), and its observed antibody-dependent enhancement (9, 10). Therefore, a better understanding of host-PRRSV interactions will provide more effective strategies and references for preventing and controlling PRRSV (11).
Our previous studies have already shown that HO-1 can effectively inhibit the infection of PRRSV by carbon monoxide (CO) and biliverdin (BV) (12, 13). HO-1 is a key cytoprotective, antioxidant, and anti-inflammatory molecule (14). Most of the physiological functions of HO-1 are related to the enzyme activity in heme catabolism, which plays a cellular protective role through the downstream metabolites, including CO, BV, and iron (Fe2+) (15, 16). Enhancing HO-1 expression inhibits the replication of many viruses. For instance, HO-1 can inhibit infection of human immunodeficiency virus (HIV) (17), hepatitis B virus (HBV) (18), hepatitis C virus (HCV) (19), Ebola virus (EBOV) (20), human respiratory syncytial virus (HRSV) (21), and dengue virus (DENV) (22). Multiple pathways can activate HO-1 expression, but it is regulated mainly at the transcriptional level. Nuclear factor-erythroid 2 (Nrf2), heat shock factor (HSF), nuclear factor-κB (NF-κB), and activator protein-1 (AP-1) have been reported to regulate the transcription of the HO-1 gene, and there are still many potential HO-1 gene transcription factors that have not been discovered (23), especially those that play an important role in the process of viral infection.
HOXA3 is a member of the HOX gene family, which encode master regulator transcription factors that specify segmental identity along the anterior-posterior axis (23, 24). The combination of tissue-specific HOX proteins and other transcription factors lead to the specific activation of downstream genes, including other transcription factors and signaling pathway components (25). According to reports, HOXA3 has been implicated in patterning, cell migration, proliferation, apoptosis, and differentiation (26). Except for the typical functions in embryonic development (27, 28), HOXA3 is a necessary transcription factor for thymic development. Hoxa3+/−Pax1−/− compound mutants have dysplastic thymus and fewer CD4+/8+T thymocytes (29). HOXA3 treatment significantly reduced the number of inflammatory cells recruited into the wound, promoted stem cell mobilization and recruitment, and inhibited the gene expression of the proinflammatory NF-κB signaling pathway to weaken the excessive inflammatory response in diabetic mice (30). Further studies showed that HOXA3 could promote the transformation of macrophages from M1-like to M2-like phenotypes (31). Meanwhile, HOXA3 is also involved in the development and progression of cancer cells; chicken gga-miR-130a targets HOXA3 and MDFIC and inhibits the proliferation and migration of Marek’s lymphoma cells (32). HOXA3 promotes tumor growth in human colon cancer by activating the EGFR/Ras/Raf/MEK/ERK signaling pathway (33). But the role of HOXA3 in virus replication has not been reported.
In this study, we demonstrated that HOXA3 could negatively regulate transcription of the HO-1 gene, and PRRSV induced HOXA3 to attenuate transcription of the HO-1 gene. The reduction of HO-1 expression attenuates phosphorylation and nuclear translocation of IRF3 and inhibits type I interferon (IFN-I) activation. Collectively, these findings define a previously unknown function of HOXA3 and establish a mechanism by which PRRSV uses HOXA3 to negatively regulate the transcription of the HO-1 gene to suppress the IFN-I response for immune evasion.
RESULTS
HOXA3 is a novel transcription factor of the HO-1 gene.
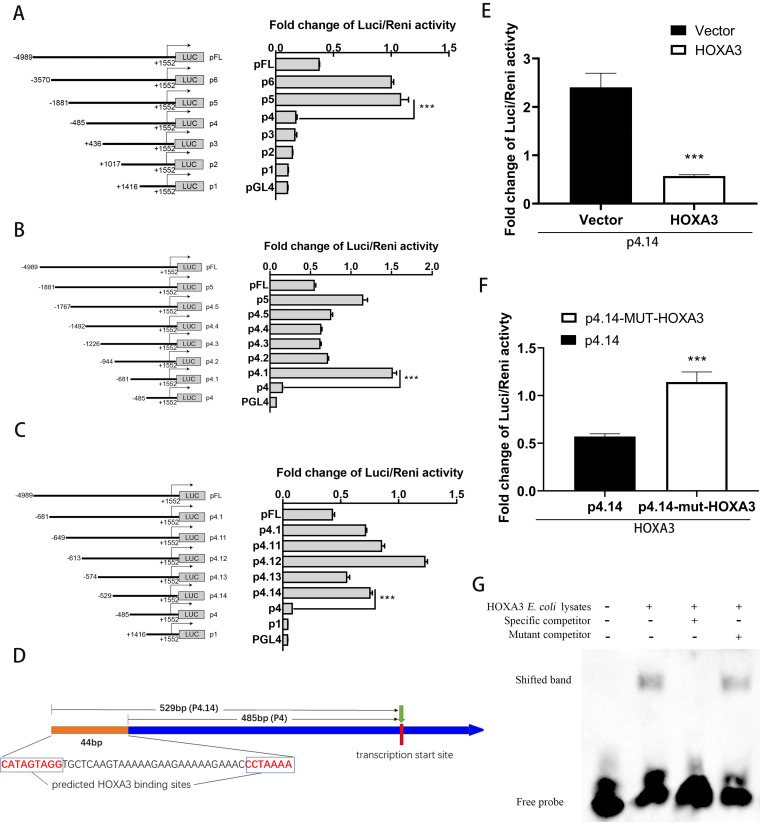
The expression of many genes in eukaryotic cells is regulated generally by transcription factors (34). Although some transcription factors of the HO-1 gene have been verified, there are potential HO-1 transcription factors that have not been reported. A series of truncation plasmids containing a variety of fragments of the HO-1 gene promoter region was constructed to find new transcription factors of the HO-1 gene, and potential transcriptional regulatory regions were determined by the dual-luciferase reporter gene assay (Fig. 1A to C). The region causing a significant change in enzyme activity was first identified between 486 and 1,881 bp before the transcription start site (TSS) of the HO-1 gene (Fig. 1A). We then narrowed the range down to 486 to 681 bp before the TSS of the HO-1 gene (Fig. 1B) and finally positioned the 486- to 529-bp region in the same way (Fig. 1C). In this 44-bp section, we used website Gene-regulation Match to predict transcription factors that might bind at this location, and we found that HOXA3 might be a potential transcription factor of the HO-1 gene (Fig. 1D).
FIG 1.
HOXA3 is a new transcription factor of the HO-1 gene. (A to C) A series of truncation plasmids of pGL4 luciferase reporter assays containing a variety of fragments of the HO-1 gene promoter region was constructed. HEK293T cells were transfected with plasmids for 24 h, and cells were lysed to detect different truncated promoter regions of HO-1 gene luciferase activity. (D) Schematic illustration of HOXA3 binding site in the promoter region of the HO-1 gene. (E) HEK293T cells were cotransfected with pGL4-p4.14 and pCAGEN-HOXA3-myc for 36 h; the empty vector of pCAGEN-myc was used as the control. The luciferase activity was measured with a dual-luciferase assay. (F) HEK293T cells were cotransfected with pGL4-p4.14-mut-HOXA3 and pCAGEN-myc-HOXA3 for 36 h; the plasmid of pGL4-p4.14 was used as the control. The luciferase activity was measured with a dual-luciferase assay. (G) EMSA. Lane 1, biotin‐labeled WT probe. Lane 2, biotin‐labeled WT probe +HOXA3 E. coli lysates. Lane 3, biotin‐labeled WT probe + HOXA3 E. coli lysates + 100‐fold molar excess of biotin‐unlabeled specific competitor probe. Lane 4, biotin‐labeled WT probe + HOXA3 E. coli lysates + 100‐fold molar excess of biotin‐unlabeled mutant competitor probe. The details were described in the Materials and Methods. Results are expressed as means ± SD of three independent replicates. Statistically significant values were denoted as follows: ***, P < 0.001.
For further validation, overexpression of HOXA3 in HEK293T cells significantly repressed the luciferase reporter activity of pGL4-p4.14 firefly luciferase reporter plasmids (Fig. 1E), which suggests that HOXA3 may negatively regulate the transcription of the HO-1 gene. When the potential binding site of HOXA3 was mutated, this inhibitory effect of HOXA3 on the luciferase reporter activity of the pGL4-p4.14 plasmid was reversed (Fig. 1F). As expected, an electrophoretic mobility shift assay (EMSA) confirmed that HOXA3 could bind the HO-1 gene promoter region in vitro (Fig. 1G). These data suggest that HOXA3 is a new transcription factor of HO-1 and negatively regulates HO-1 gene transcription.
HOXA3 negatively regulates HO-1 transcription and promotes PRRSV replication.
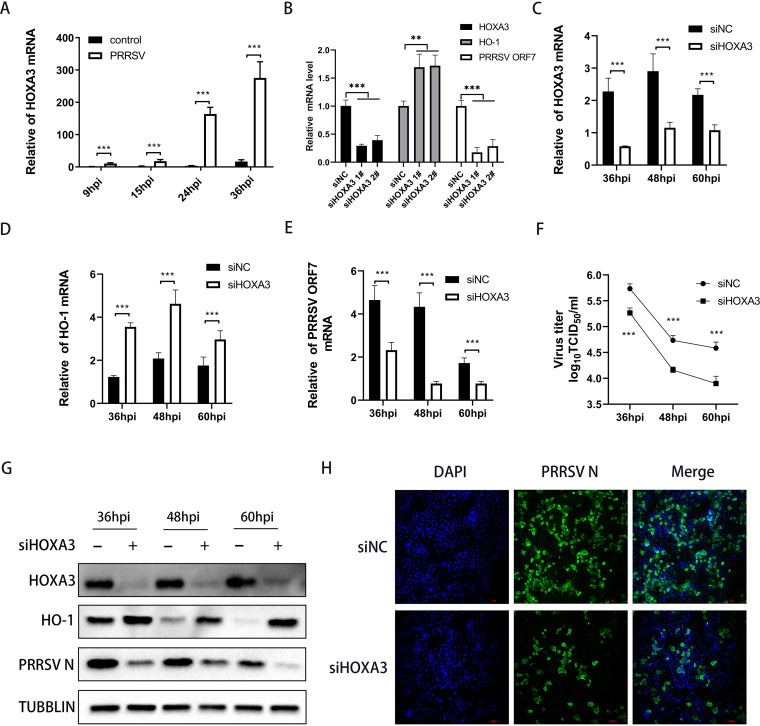
The function of HOXA3 had not been investigated in the field of the virus. To delve into the potential function of HOXA3 in PRRSV infection, we infected the main target cells in pigs, namely, porcine alveolar macrophages (PAMs), and then analyzed HOXA3 expression. As shown in Fig. 2A, HOXA3 mRNA was significantly elevated in PRRSV-infected PAMs, especially at middle and late stages of PRRSV infection. To further investigate whether HOXA3 influenced the replication of PRRSV, we designed two small interfering RNAs (siRNAs) targeting HOXA3. Both siRNAs potently downregulated the protein levels of HOXA3 and suppressed the replication of PRRSV (Fig. 2B). The no. 1 siRNA was used for the experiments described below. Knocking down HOXA3 significantly enhanced HO-1 expression (Fig. 2C, D, and G) and resulted in a marked decrease in PRRSV ORF7 mRNA and N protein (Fig. 2E and G) at more PRRSV infection time points, and the viral titers of PRRSV were also decreased (Fig. 2F). Meanwhile, immunofluorescence staining showed similar results (Fig. 2H).
FIG 2.
Knocking down HOXA3 upregulated HO-1 expression and inhibited PRRSV replication. (A) PAMs were infected with PRRSV at an MOI of 0.1. Samples were collected at 9, 15, 24, and 36 hpi; RT-qPCR determined HOXA3 mRNA levels. (B) RT-qPCR analysis of HOXA3, HO-1, and PRRSV ORF7 mRNA in MARC-145 cells transfected with negative control siRNA (siNC) or HOXA3 siRNA (siHOXA3) no. 1 and 2 for 24 h followed by infection with PRRSV for 36 h. (C to E) MARC-145 cells were transfected with siHOXA3 no 1 or siNC of 60 nM for 24 h before infection with PRRSV at an MOI of 0.1. Samples were collected at 36, 48, and 60 hpi. HOXA3 (C), HO-1 (D), and ORF7 (E) mRNA levels were determined by RT-qPCR. (F) Cell culture supernatants were collected at the indicated times. The TCID50 assay was performed to determine the levels of supernatant virus production. (G) Protein levels of HOXA3, HO-1, PRRSV N, and tubulin at the indicated time points were measured by Western blotting. (H) The expression of the N protein was determined by IFA at 36 hpi, with MARC-145 cells transfected with siNC included as a control. Results are expressed as means ± SD of three independent replicates. Statistically significant values were denoted as follows: ***, P < 0.001.
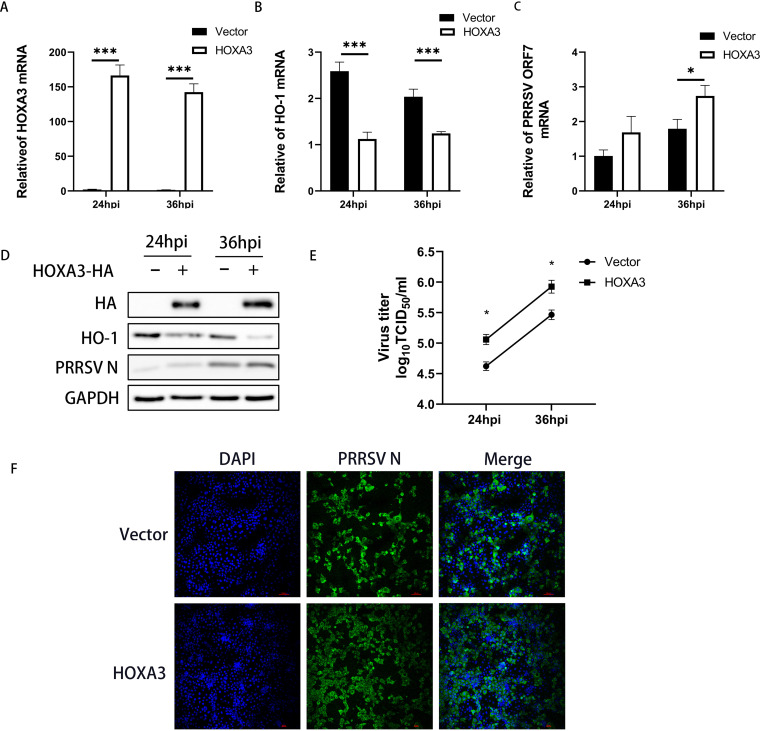
The above results indicate that HOXA3 may be able to promote PRRSV replication. To test this hypothesis, we constructed a MARC-145 cell line stably expressing HOXA3 by using lentiviral vectors (Fig. 3A and D). Then, these cell lines were infected with PRRSV at a multiplicity of infection (MOI) of 0.1. The results showed that the overexpression of HOXA3 reduced the expression of HO-1 (Fig. 3C and D) and promoted PRRSV replication at 24 and 36 hours postinfection (hpi) (Fig. 3C to E). Immunofluorescence staining demonstrated more intuitively the promotion of PRRSV replication by overexpression of HOXA3 (Fig. 3F). Overall, these data suggest that PRRSV induced HOXA3 to decrease HO-1 expression for promoting its replication.
FIG 3.
Overexpressing HOXA3 downregulated HO-1 expression and promoted PRRSV replication. The lentivirus-mediated overexpression method was used to enhance the expression of HOXA3 in MARC-145 cells. (A to C) MARC-145-HOXA3 cells were infected with PRRSV at an MOI of 0.1. Samples were collected at 24 and 36 hpi. HOXA3 (A), HO-1 (B), and ORF7 (C) mRNA levels were determined by RT-qPCR. (D) The expression of HOXA3-HA, HO-1, PRRSV N, and GAPDH were detected by Western blotting. (E) Cell culture supernatants were collected at the indicated times. TCID50 assay was performed to determine the levels of supernatant virus production. (F) The expression of the N protein was determined by IFA at 36 hpi, with WT MARC-145 cells infected with PRRSV included as a control. Results are expressed as means ± SD of three independent replicates. Statistically significant values were denoted as follows: *, P < 0.05; ***, P < 0.001.
HOXA3 negatively regulates IFN-I expression upon PRRSV infection.
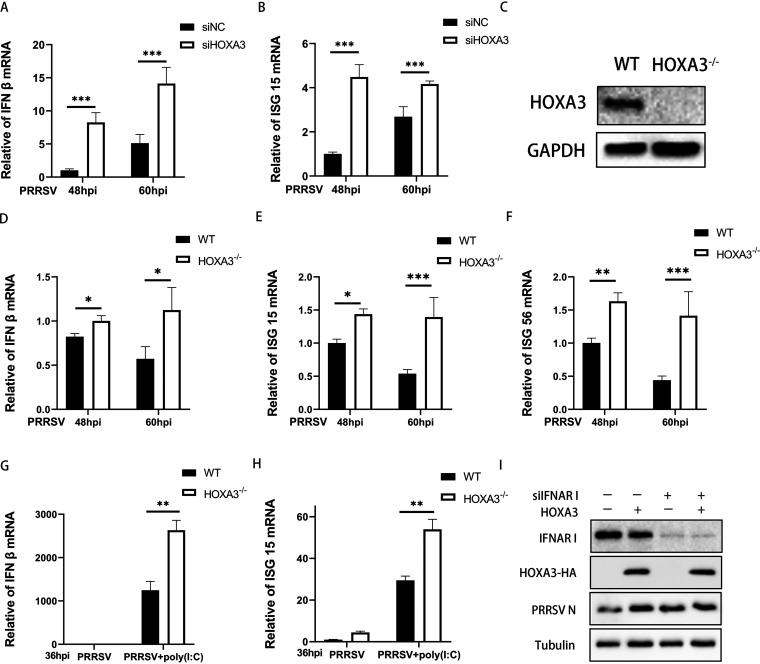
IFN-I can significantly reduce PRRSV replication, but PRRSV inhibits IFN-I induction (35). Because PRRSV uses HOXA3 to promote replication, we wanted to know whether this process is achieved by inhibiting IFN-I responses. The results showed that IFN-β mRNA levels were remarkably increased in HOXA3-knockdown MARC-145 cells with PRRSV infection (Fig. 4A). The deficiency of HOXA3 also significantly increased the PRRSV-induced expression of IFN-stimulated gene 15 (ISG15) mRNA (Fig. 4B).
FIG 4.
Deficiency of HOXA3 increased IFN-I expression in PRRSV infection. (A and B) MARC-145 cells were transfected with siHOXA3 no. 1 or siNC of 60 nM for 24 h before infection with PRRSV at an MOI of 0.1. Samples were collected at 48 and 60 hpi. IFN-β (A) and ISG15 (B) mRNA levels were determined by RT-qPCR. (C) CRISPR-Cas9 technology was used to construct HOXA3−/−-MARC-145 cells, and the expressions of HOXA3 and GAPDH were detected by Western blotting in the WT and HOXA3−/−-MARC-145 cell lines. (D to F) HOXA3−/−-MARC-145 cells were infected with PRRSV at an MOI of 0.1. Cells were collected at 48 and 60 hpi. IFN-β (D), ISG15 (E), and ISG56 (F) mRNA levels were determined by RT-qPCR. (G and H) RT-qPCR analysis of IFN-β and ISG15 mRNA in HOXA3−/−-MARC-145 cells transfected with poly(I·C) (500 ng/ml) for 12 h followed by infection with PRRSV for 36 h. (I) MARC-145-HOXA3 and WT MARC-145 cells were transfected with siIFNAR1 or siNC of 60 nM followed by infection with PRRSV for 36 h at an MOI of 0.1. The expression of HOXA3-HA, PRRSV N, and tubulin was detected by Western blotting. Results are expressed as means ± SD of three independent replicates. Statistically significant values were denoted as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To evaluate the role of HOXA3 more accurately in negative regulation of IFN-I induction in PRRSV infection, the HOXA3 knockout (KO) cell line was constructed by CRISPR-Cas9. Western blot results showed that HOXA3 was knocked out completely in MARC-145 cells (Fig. 4C). Relative to wild-type (WT) cells, the IFN-β mRNA level was increased in HOXA3 KO cells infected by PRRSV (Fig. 4D). The expression levels of some ISGs were also measured with reverse transcriptase quantitative PCR (RT-qPCR); the knockout of HOXA3 increases the expression of ISG15 and ISG56 mRNA (Fig. 4E and F). We also evaluated the effect of KO-HOXA3 on the poly(I·C)-triggered transcriptional induction of the IFN-β and ISG gene and showed consistent results (Fig. 4G and H). These results suggest that HOXA3 may promote PRRSV infection through inhibition of IFN-I signaling. To prove this hypothesis, we knocked down IFNAR1, a key molecule in the IFN-I signaling in MARC-145 cell lines stably expressing HOXA3; the difference caused by HOXA3 in PRRSV infectivity was eliminated (Fig. 4I). In summary, PRRSV induces HOXA3 to negatively regulate the IFN-I response during infection.
The inhibition of IFN-I by HOXA3 depends on the interaction of HO-1 and IRF3 in PRRSV infection.
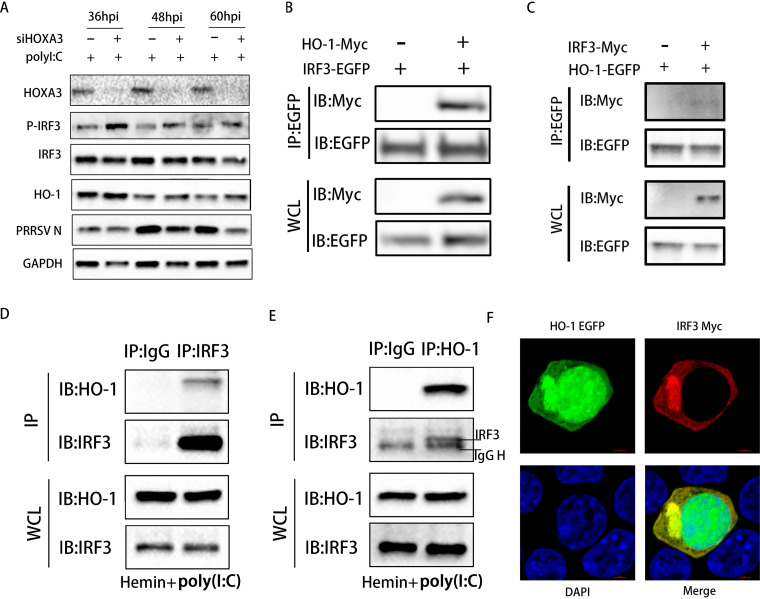
IRF3 plays a key role in inducing IFN-I and the antiviral response (36). HO-1 is reported to be necessary to activate IRF3 to modulate IFN-I in virus-infected macrophages (37). Activation of IRF3 depends on virus-induced C-terminal phosphorylation events on Ser396 (36, 38, 39). To explore the influence of HOXA3 negative regulation of HO-1 gene transcription on the activation of IRF3 in PRRSV infection, we examined the phosphorylation of IRF3 (Ser396) induced by PRRSV infection in MARC-145 cells after knockdown of HOXA3. Phosphorylation of IRF3 was significantly reduced or even disappeared at the middle and late stages of PRRSV infection even with poly(I·C) stimulation (Fig. 5A). However, when HOXA3 was knocked down, the phosphorylation of IRF3 was significantly enhanced at the same time as HO-1 was increased, but there was no change in the total IRF3 protein (Fig. 5A).
FIG 5.
Negative regulation of IFN-I responses by HOXA3 is achieved through reducing the interaction between HO-1 and IRF3. (A) MARC-145 cells were transfected with siNC and siHOXA3 no. 1 for 24 h and infected with PRRSV at an MOI of 0.1. Cells were collected at 36, 48, and 60 hpi after transfection with poly(I·C) (500 ng/ml) for 12 h. The expression of HOXA3, p-IRF3, IRF3, HO-1, PRRSV N, and GAPDH were detected by Western blotting. (B) HEK293T cells transfected with pCAGEN-HO-1-Myc and pEGFP-C1-IRF3 for 36 h were immunoprecipitated (IP) with an anti-EGFP antibody. (C) HEK293T cells transfected with pCAGEN-IRF3-Myc and pEGFP-C1-HO-1 for 36 h were immunoprecipitated with or anti-EGFP antibody. (D and E) MARC-145 cells treated with hemin (100 μM) for 24 h and transfected with poly(I·C) (500 ng/ml) for 12 h were immunoprecipitated with anti-IRF3 antibody (D) or anti-HO-1 antibody (E). (F) Confocal microscopy of HEK293T cells cotransfected with plasmids encoding EGFP-tagged HO-1 and Myc-tagged IRF3 stained with Alexa Fluor 555-conjugated anti-Myc antibody (red). The 4′,6-diamidino-2-phenylindole (DAPI) serves as a marker for nuclei (blue).
Since the increase of HO-1 expression enhances the phosphorylation of IRF3, we hypothesize that the interaction of HO-1 and IRF3 causes the activation of IRF3. We overexpressed the enhanced green fluorescent protein (EGFP)-tagged IRF3 along with Myc-tagged HO-1 in HEK293T cells. The coimmunoprecipitation (co-IP) assay was performed with an anti-EGFP antibody. As shown in Fig. 5B, there was a direct interaction between HO-1 and IRF3. The reverse co-IP assay also proved the same result (Fig. 5C). MARC-145 cells were treated with hemin to induce endogenous HO-1 expression and poly(I·C) to activate the IRF3. An interaction between endogenous HO-1 and endogenous IRF3 was demonstrated by co-IP and reverse co-IP assay (Fig. 5D and E). To test whether HO-1 and IRF3 share similar subcellular locations, we conducted an indirect immunofluorescence assay (IFA). As shown in Fig. 5F, both EGFP-tagged HO-1 and Myc-tagged IRF3 were expressed in HEK293T cells, and the two proteins colocalized in the cytoplasm. MARC-145 cells were infected with PRRSV for 48 h to verify the endogenous interaction between IRF3 and HO-1. Cell lysates were obtained, and some HO-1-interacting proteins were identified using immunoprecipitation-liquid chromatography-tandem mass spectrometry (IP/LC-MS/MS) analysis (Table 1). Among these proteins, IRF3 is a known HO-1-interacting protein. These results confirmed that HOXA3 inhibits HO-1 expression and indirectly reduces the interaction between HO-1 and IRF3 to inhibit the activation of IRF3.
TABLE 1.
Summary of the HO-1-interacting proteins identified by mass spectrometry
| Protein identified | Description | Species | Organism indentifier | Gene name | Protein existence | Sequence version | No. of peptides | Score |
|---|---|---|---|---|---|---|---|---|
| ACTB | Actin, cytoplasmic 1 | Chlorocebus aethiops | 9534 | ACTB | 2 | 1 | 12 | 323.31 |
| IRF3 | IRF tryptophan pentad repeat domain-containing protein | Chlorocebus sabaeus | 60711 | IRF3 | 4 | 1 | 12 | 86.422 |
| XRCC5 | X-ray repair cross-complementing protein 5 | Chlorocebus sabaeus | 60711 | XRCC5 | 3 | 1 | 7 | 62.511 |
| MYO9B | Myosin IXB | Chlorocebus sabaeus | 60711 | MYO9B | 3 | 1 | 4 | 56.2 |
| TPI1 | Triosephosphate isomerase | Chlorocebus sabaeus | 60711 | TPI1 | 3 | 1 | 3 | 100.03 |
The deficiency of HOXA3 promotes IRF3 nuclear translocation.
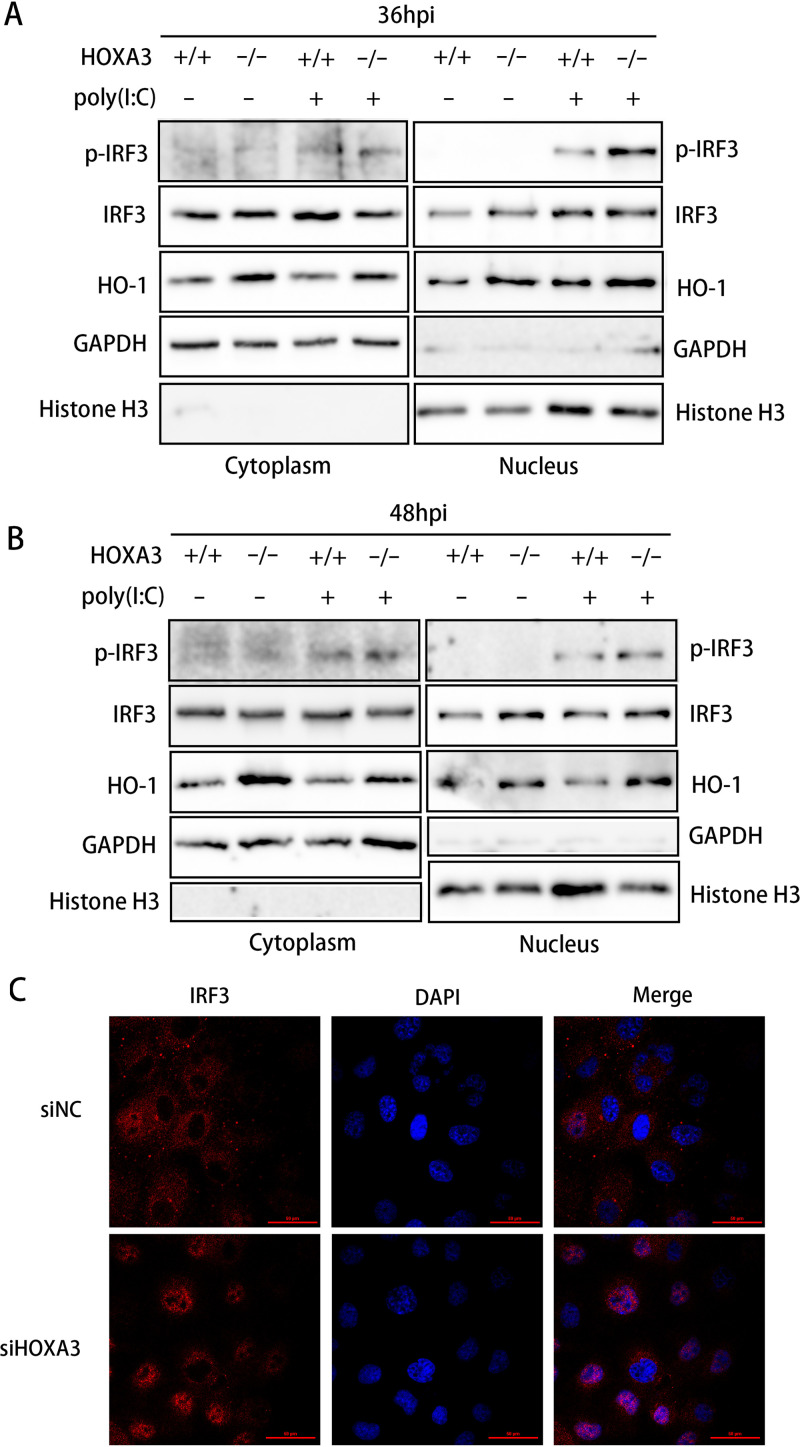
It is generally accepted that virus infection causes IRF3 activation in three main steps, namely, phosphorylation, dimerization, and translocation, from the cytoplasm into the nucleus (40). IRF3, a transcription factor involved in type I IFN production and ISG expression, needs to be transported into the nucleus to play a role in transcriptional regulation. Nuclear and cytoplasmic proteins were extracted from WT and HOXA3 KO cells with poly(I·C) treatment and PRRSV infection, and knocking out HOXA3 significantly increased the HO-1 level in the cytoplasm and the nucleus (Fig. 6A and B). The increase in HO-1 expression also promoted the nuclear translocation of IRF3 (Fig. 6A and B). We used IFA to analyze the nuclear translocation of IRF3 more intuitively and accurately. According to the results of IFA, PRRSV infection does not cause extensive IRF3 translocation from the cytoplasm into the nucleus (Fig. 6C). However, knocking down HOXA3 significantly increased the proportion of IRF3 moving into the nucleus (Fig. 6C). These results suggested that the deficiency of HOXA3 promotes IRF3 nuclear translocation.
FIG 6.
Deficiency of HOXA3 promoted nuclear translocation of IRF3. (A and B) HOXA3−/−-MARC-145 cells were infected with PRRSV at an MOI of 0.1. Cells were collected at 36 and 48 hpi after transfection of poly(I·C) for 12 h. Cytoplasmic and nuclear extracts were prepared and subjected to Western blotting. The expressions of p-IRF3, IRF3, HO-1, GAPDH, and histone H3 were detected. (C) MARC-145 cells transfected with siNC and siHOXA3 no. 1 for 24 h were infected with PRRSV at an MOI of 0.1 for 48 h. The subcellular localization of IRF3 was determined by IRF3 intracellular staining, and confocal fluorescence images were captured.
DISCUSSION
HO-1 induction is an effective way used by cells to neutralize a variety of stress conditions (41), such as hypoxia; oxidative stress; and the presence of cytokines, lipopolysaccharides (LPS), and heavy metals, in biological systems (42). Many signaling molecules and transcription factors can regulate HO-1 expression in cells, such as AP-1, NF-κB, and Nrf2. Their upstream kinases, including mitogen-activated protein kinases (MAPKs), play an important regulatory role in HO-1 gene induction (43). A large amount of data now support the role of MAPK cascades in signal-mediated HO-1 gene activation. Other signaling molecules, such as phosphatidylinositol 3-kinase (PI3K); tyrosine kinases; and protein kinases A, B, C, and G, play a role in HO-1 induction (44). However, HO-1 expression is regulated mainly at the transcriptional level and governed by response elements (REs) localized in the promoter 5′ flanking region of the HO-1 gene (44). Our study found a new transcription factor of the HO-1 gene that negatively regulates HO-1 transcription (Fig. 1 and 3). Previous studies have shown that Nrf2 is crucial for HO-1 transcriptional regulation, and HO-1 is not inducible in Nrf2 null mice (45). Upon cellular stress, such as proinflammatory stimuli or the presence of heme analogs, the Nrf2 protein dissociates from Keap1 and then phosphorylates and enters the nucleus; the binding of the complex Nrf2/MAF to an antioxidant responsive element (ARE) in the Hmox1 gene promoter site activates the transcription of HO-1 (46). Conversely, we identified that the binding motif of HOXA3 in the HO-1 gene promoter region was not an ARE. In other words, the transcriptional regulation of HOXA3 on HO-1 may not be affected by oxidative stress. HOXA3 was significantly upregulated in PRRSV infection (Fig. 2A), but it did not cause a change in NRF2 (the result is not shown). These results suggest that cells infected with PRRSV cannot actively activate HO-1 transcription to resist viral infection. Instead, the virus induces HOXA3 to inhibit HO-1 transcription and promote its replication.
HO-1 is a stress-induced and cytoprotective enzyme expressed in most cell types in the organism. The end products of the HO-1 enzymatic reaction are iron (Fe2+), BV, biliverdin (BR), and CO. Each of them plays an antiviral role in replicating different endowments. Fe2+ inhibits viral replication by binding to Mg2+ binding sites on the HCV RNA polymerase (47). BV is a potent HCV NS3/4A protease inhibitor, which inhibits HCV replication (48). Our previous study also found the inhibitory effect of BV on PRRSV (13). BR can inhibit the viral replication of HIV, herpes simplex virus type-1, and enterovirus (49, 50). CO is an important immune regulatory factor. We found in our previous research that CO decreases the replication ability of PRRSV by suppressing the activation of the cyclic GMP/protein kinase G and NF-κB signaling pathway (12). More and more research have been reported on the regulation of HO-1 on innate immunity. Mouse macrophages that knock out HO-1 weaken the innate immune response (37). CoPP, as a specific activator of HO-1, decreased HRSV and influenza A virus (IAV) replication through increasing the production of IFN-α/β (21, 51). Also, a variety of HO-1 activators show antiviral activity through the activation/restoration of IFNs (52–54), but the specific mechanism was not elaborated. In the manuscript, we enhanced HO-1 transcription by knockdown or knockout of HOXA3 and significantly enhanced the expression of IFN-β and ISGs with PRRSV infection (Fig. 4). We further explored the regulation mechanism of HO-1 on IFN-I and found that the interaction between HO-1 and IRF3 is critical (Fig. 5). Based on our research, we have clarified the regulatory effect of HO-1 on PRRSV, whether from the downstream products of HO-1 or the regulation of HO-1 on IFN-I (Fig. 7).
FIG 7.
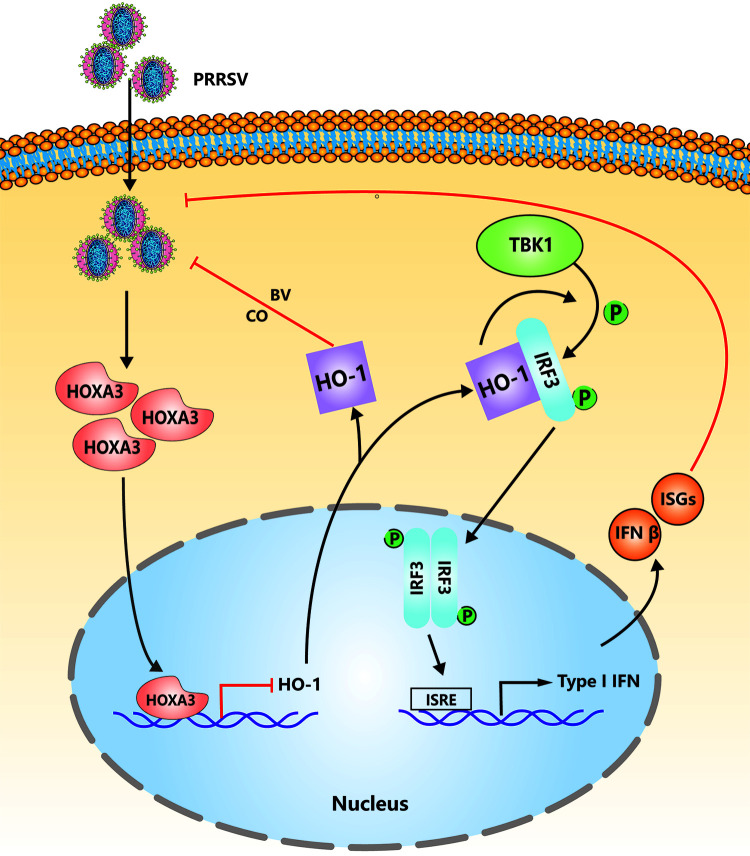
The model of the roles of HOXA3 in inhibition of type I IFN signaling upon PRRSV infection. To replicate better, PRRSV induces the upregulation of HOXA3 to reduce HO-1 expression, thereby inhibiting type I IFN responses by impairing the interaction between HO-1 and IRF3.
Viral infection activates various signaling cascades that enable IFN-I expression against the virus. PRRSV has developed multiple mechanisms to escape the host immune system by modulating innate immunity during coevolution with hosts. PRRSV nsp4 cleaves NF-κB essential modulator (NEMO) at multiple sites which impairs IFN-β production (55). PRRSV nsp11 antagonizes IFN-I by targeting IRF9 via a NendoU activity-independent mechanism (56), and PRRSV nsp11 can also induce STAT2 degradation to inhibit IFN-activated signaling (57). We found that IFN-β and ISGs were significantly upregulated after a deficiency of HOXA3 (Fig. 4). There is no doubt that IRF3 plays a central role in inducing IFN-I and the antiviral response (36). Research has shown that NF-κB and AP-1 were activated by PRRSV infection, whereas the activity of transcription factor IRF3 was significantly inhibited (58); PRRSV can also inhibit IRF3 phosphorylation (59, 60). These studies indicated that inhibition of IFN-β by PRRSV occurs through a mechanism involving IRF3 activity, which was consistent with the results in our experiment (Fig. 5A). The enhancement of IRF3 phosphorylation by HOXA3 knockdown is caused by enhancing the interaction between HO-1 and IRF3 (Fig. 5). To find the specific area of interaction between HO-1 and IRF3, we truncated the HO-1 protein for two parts, and one of them includes its enzyme activity region, but each of them lost its interaction with IRF3 (data are not shown). It has also been reported that the use of the HO-1 enzyme activity inhibitor in the process of influenza virus infection does not affect the activation of IRF3 by HO-1 (51), suggesting that HO-1 enzyme activity may not affect IRF3 phosphorylation and that the HO-1 protein in complete conformation is crucial in this process.
Altogether, we found a novel molecular mechanism by which PRRSV induces a new transcription factor, HOXA3, to negatively regulate HO-1 transcription to inhibit IFN-I production by affecting the interaction between HO-1 and IRF3 (a proposed model illustrated in Fig. 7). Combined with our previous research, the underlying mechanism by which PRRSV and host cells interact with each other through HO-1 was revealed (Fig. 7). These findings provide new insights for understanding molecular mechanisms by which PRRSV regulates host innate immune responses, which is helpful for developing new antiviral strategies against PRRSV infection.
MATERIALS AND METHODS
Reagents and antibodies.
Hemin and poly(I·C) were obtained from Sigma-Aldrich. The stock solutions of hemin were prepared in 0.2 M NaOH, neutralized to pH 7.4, and stored at −80°C until used. Anti-HO-1(5853) and anti-Myc-Tag (4947) antibodies were purchased from Cell Signaling Technology. Anti-phospho-IRF3 (Ser396) was purchased from Beyotime. Anti-HO-1(11312-1-AP) was purchased from Proteintech. Anti-HOXA3(117919) was purchased from LifeSpan BioSciences. Anti-GAPDH (ab 0037) and anti-GFP-Tag (ab 0005) were purchased from Abways Technology. Horseradish peroxidase (HRP)-conjugated goat anti-mouse (PA1-86717) and rabbit IgG (SA1-9510) were purchased from Thermo Fisher Scientific. HRP-conjugated mouse anti-rabbit IgG light-chain-specfic (LCS) antibody (A25022) was purchased from Abbkine Scientific, and anti-PRRSV N was from our laboratory (Northwest A&F University, Shaanxi, China).
Cells and viruses.
PAMs were obtained by postmortem lung lavage of 6-week-old pigs using a lung lavage technique as described previously (12) and were maintained in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. MARC-145 cells (an African green monkey kidney cell line) and HEK293T cells (a human embryonic kidney cell line) were purchased from China Center for Type Culture Collection (CCTCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% FBS and penicillin-streptomycin (Life Technologies). All cells were cultured and maintained at 37°C with 5% CO2. All animal work was done in strict accordance with the guidelines of the NWAFU Research Ethics Committee.
A highly pathogenic PRRSV (HP-PRRSV) strain, GD-HD (GenBank identifier [ID] KP793736.1), was used in this study.
Genes and plasmids.
A 6,553-bp sequence region (+1552 to −4989 from transcription start site) from a swine HO-1 gene was truncated and amplified to clone into a pGL4.10 vector (Fig. 1A to C). The binding site of HOXA3 was predicted using Gene-regulation Match (http://gene-regulation.com/pub/programs.html) in 44-bp region (−485 to −529 from transcription start site) (Fig. 1D). The mutant luciferase construct changed the sequence of the HOXA3 binding site. The sequence after mutation was as follows: 5′‐ACTGCTAGGGTGCTCAAGTAAAAAGAAGAAAAAGAAACCCTGGCC‐3′.
Swine HOXA3 was cloned into the Lenti-hemagglutinin (HA) vector and pCAGEN-myc vector. Swine HO-1 was cloned into the pEGFP-C1 vector. Swine IRF3 was cloned into the pCAGEN-myc vector. The empty vector is our laboratory stock. The prokaryotic expression vector pCOLD-SUMO (HaiGene) was used to express the soluble HOXA3 protein.
Lentiviral production and infection.
For constructs expressing HOXA3-HA and HOXA3 single guide RNA (sgRNA), to produce the lentivirus, the recombinant packaging plasmids were cotransfected with Lenti-HA-HOXA3 or LentiCRISPR v2-sgHOXA3 plasmids into HEK-293T cells, and culture supernatants containing the virus were collected 48 and 72 h after transfection. For infection with lentivirus, MARC-145 cells were cultured with lentivirus solution for 24 h in the presence of 15 μg/ml puromycin (Sigma).
Real-time quantitative PCR.
The experiments were performed as described previously (12, 13), with siRNA specific to HOXA3 and control siRNA synthesized by RiboBio. Total RNA containing PRRSV RNA was extracted from cells using the RNAiso Plus reagent (TaKaRa); cDNA was generated from total RNA using HiScript II Q RT SuperMix (Vazyme) following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using the ChamQ SYBR qPCR master mix (Vazyme). The primer sequences used for RT-qPCR were as follows: MARC-145 HOXA3 forward, 5′-CAGAATGCCAGCAGCAACC-3′; MARC-145 HOXA3 reverse, 5′-CGCAGCTCTCGCCTGA-3′; porcine HOXA3 forward, 5′-AGTACAAGAAGGATCAGAAGG-3′; porcine HOXA3 reverse, 5′-CGCTGTTCACCAGAGAAT-3′; PRRSV ORF7 forward, 5′-AGATCATCGCCCAACAAAAC-3′; PRRSV ORF7 reverse, 5′-GACACAATTGCCGCTCACTA-3′; porcine HPRT1 forward, 5′-TGGAAAGAATGTCTTGATTGTTGAAG-3′; porcine HPRT1 reverse, 5′-ATCTTTGGATTATGCTGCTTGACC-3′; MARC-145 β-actin forward, 5′-GAGAAGCTGTGCTACGTCGC-3′; MARC-145 β-actin reverse, 5′-CCAGACAGCACTGTGTTGGC-3′; MARC-145 IFN-β forward, 5′-TGCTCTCCTGTTGTGCTTCTC-3′; MARC-145 IFN-β reverse, 5′-CTGCGGCTGCTTAATTTCCTC-3′; MARC-145 ISG15 forward, 5′-CACCGTGTTCATGAATCTGC-3′; and MARC-145 ISG15 reverse, 5′-CTTTATTTCCGGCCCTTGAT-3′.
Virus titration.
Viral replication was determined by titration as described previously (13). MARC-145 cells were seeded in a 96-well plate 16 h before virus infection. Viral supernatants were prepared by serial dilutions, and a 100-μl solution was added to each well in replicates of eight once cell confluence reached 80%. Six days after infection, the 50% tissue culture infective dose (TCID50) was calculated using the Reed-Muench method.
Electrophoretic mobility shift assays.
Lysates of transformed Escherichia coli that measured the protein concentration by bicinchoninic acid (BCA) protein assay kit were used for the DNA-protein conjugation reaction. We synthesized a biotin-labeled probe for the electrophoretic mobility shift assay (EMSA) (Sangon Biotec). The WT probe sequence was 5′‐CATAGTAGGTGCTCAAGTAAAAAGAAGAAAAAGAAACCCTAAAA‐3′. The mutant competitor probe sequence was 5′‐CAGCTGCTGTGCTCAAGTAAAAAGAAGAAAAAGAAACCCTGGCC‐3′.
Then, we executed the DNA-protein binding reactions using the EMSA gel-shift kit (Beyotime; GS009) following the manufacturer’s instructions at 25°C. Finally, the reaction mixture was separated through 4% nondenaturing PAGE gels and developed.
LC-MS/MS analysis.
To identify potential HO-1-binding proteins, MARC-145 cells were infected with HP-PRRSV for 36 h. Cells were collected, and HO-1 was immunoprecipitated using an anti-HO-1 antibody and protein A agarose at 4°C. The IP products were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using the Q Exactive HF mass spectrometer (Thermo Fisher Scientific) and Dionex Ultimate 3000 (Thermo Fisher Scientific, USA) instruments. MS data were analyzed using MaxQuant software (https://www.maxquant.org/). MS assays were conducted by Nanning MHelixProTech Co., Ltd. (China).
Coimmunoprecipitation and immunoblot analysis.
The cells grown in the 10-cm dishes were transfected with the various plasmids for 36 h or treated with reagents. Cells were harvested and lysed in NP-40 lysis buffer (Beyotime) containing protease inhibitor. For immunoprecipitation assays, the lysates were immunoprecipitated with IgG or the appropriate antibodies, protein A/G agarose beads (TransGen Biotech) were added to the samples for 2 h at 4°C, and the precipitants were washed three times with lysis buffer. The cell lysates (10-μg protein per sample) were then separated by SDS-PAGE and electrotransferred onto a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked with Tris-buffered saline containing 0.05% Tween 20 and 5% nonfat dried milk and then followed by incubation at 4°C overnight with primary antibodies. The membranes were washed in Tris-buffered saline with Tween 20 (TBST). HRP-conjugated goat anti-mouse, goat anti-rabbit IgG, or anti-rabbit IgG LCS was used as the secondary antibody. The reactions were visualized using an ECL reagent.
IFA and confocal microscopy.
An immunofluorescence assay (IFA) was performed as described previously (13) with the following modifications. Cells were fixed for 10 min with 4% paraformaldehyde and then were permeabilized for 15 min with 0.25% Triton X-100 in phosphate-buffered saline (PBS). After blockade of nonspecific binding by incubation of cells for 1 h with 1% BSA in PBS, coverslips were incubated with the appropriate primary antibodies (identified above) and then sequentially with corresponding secondary antibodies, and then were visualized with a Nikon A1 confocal microscope.
Luciferase reporter assay.
Luciferase reporter assays were processed as previously described (61) with some modifications. In brief, HEK293T cells were seeded in a 24-well plate 24 h before transfection, and the monolayer cells were transfected with 0.2 μg of a pCAGEN-HOXA3-Myc vector, 0.2 μg of a HO-1 promoter-driven luciferase reporter plasmid, and 0.01 μg of an internal control pRL-TK reporter plasmid. The empty vector plasmids were used in the whole transfection process to ensure the cells received the same amounts of total plasmids. At 36 h posttransfection, the firefly and Renilla luciferase activities of transfected cells were determined with a dual-luciferase reporter assay (Promega) following the manufacturer’s instructions.
Statistical analysis.
All experiments were performed with at least three independent replicates. The experimental data obtained in this study were analyzed and plotted using the software GraphPad Prism, and significant differences were analyzed using either the Student’s t test or one-way analysis of variance (ANOVA). The following values were considered statistically significant: *, P < 0.05; **, P < 0.01; and ***, P < 0.001; error bars indicate means ± standard deviations (SD).
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (31772764 and 32172846), the China Agriculture Research System of MOF and MARA, the Science Foundation for Distinguished Young Scholars of Shaanxi Province (2021JC-18), the Open Project of the State Key Laboratory of Veterinary Etiological Biology (SKLVEB2020KFKT017), the Chinese Academy of Agricultural Science and Technology Innovation Project (CAAS-ASTIP-JBGS-20210602), the Youth Innovation Team of Shaanxi Universities, and the Fundamental Research Funds for the Central Universities (2452021154).
Contributor Information
Haixue Zheng, Email: haixuezheng@163.com.
Shuqi Xiao, Email: xiaoshuqi@nwsuaf.edu.cn.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Lunney JK, Benfield DA, Rowland RR. 2010. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res 154:1–6. 10.1016/j.virusres.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderon Diaz JA, Fitzgerald RM, Shalloo L, Rodrigues da Costa M, Niemi J, Leonard FC, Kyriazakis I, Garcia Manzanilla E. 2020. Financial analysis of herd status and vaccination practices for porcine reproductive and respiratory syndrome virus, swine influenza virus, and Mycoplasma hyopneumoniae in farrow-to-finish pig farms using a bio-economic simulation model. Front Vet Sci 7:556674. 10.3389/fvets.2020.556674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieuwenhuis N, Duinhof TF, van Nes A. 2012. Economic analysis of outbreaks of porcine reproductive and respiratory syndrome virus in nine sow herds. Vet Rec 170:225. 10.1136/vr.100101. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 5.Brar MS, Shi M, Hui RK, Leung FC. 2014. Genomic evolution of porcine reproductive and respiratory syndrome virus (PRRSV) isolates revealed by deep sequencing. PLoS One 9:e88807. 10.1371/journal.pone.0088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng CL, Tian ZJ, Zhang WC, Zhang HL, Zhai HY, An TQ, Peng JM, Ye C, Sun L, Wang Q, Sun Y, Li L, Zhao HY, Chang D, Cai XH, Zhang GH, Tong GZ. 2014. Characterization of two newly emerged isolates of porcine reproductive and respiratory syndrome virus from Northeast China in 2013. Vet Microbiol 171:41–52. 10.1016/j.vetmic.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Lu Q, Bai J, Zhang L, Liu J, Jiang Z, Michal JJ, He Q, Jiang P. 2012. Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J Proteome Res 11:2890–2903. 10.1021/pr201266z. [DOI] [PubMed] [Google Scholar]
- 8.Genini S, Delputte PL, Malinverni R, Cecere M, Stella A, Nauwynck HJ, Giuffra E. 2008. Genome-wide transcriptional response of primary alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. J Gen Virol 89:2550–2564. 10.1099/vir.0.2008/003244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon KJ, Wu LL, Zimmerman JJ, Hill HT, Platt KB. 1996. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol 9:51–63. 10.1089/vim.1996.9.51. [DOI] [PubMed] [Google Scholar]
- 10.Bao D, Wang R, Qiao S, Wan B, Wang Y, Liu M, Shi X, Guo J, Zhang G. 2013. Antibody-dependent enhancement of PRRSV infection down-modulates TNF-alpha and IFN-beta transcription in macrophages. Vet Immunol Immunopathol 156:128–134. 10.1016/j.vetimm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Crisci E, Moroldo M, Vu Manh TP, Mohammad A, Jourdren L, Urien C, Bouguyon E, Bordet E, Bevilacqua C, Bourge M, Pezant J, Pleau A, Boulesteix O, Schwartz I, Bertho N, Giuffra E. 2020. Distinctive cellular and metabolic reprogramming in porcine lung mononuclear phagocytes infected with type 1 PRRSV strains. Front Immunol 11:588411. 10.3389/fimmu.2020.588411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang A, Zhao L, Li N, Duan H, Liu H, Pu F, Zhang G, Zhou EM, Xiao S. 2017. Carbon monoxide inhibits porcine reproductive and respiratory syndrome virus replication by the cyclic GMP/protein kinase G and NF-kappaB signaling pathway. J Virol 91:e01866-16. 10.1128/JVI.01866-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang A, Duan H, Li N, Zhao L, Pu F, Huang B, Wu C, Nan Y, Du T, Mu Y, Zhao Q, Sun Y, Zhang G, Hiscox JA, Zhou EM, Xiao S. 2017. Heme oxygenase-1 metabolite biliverdin, not iron, inhibits porcine reproductive and respiratory syndrome virus replication. Free Radic Biol Med 102:149–161. 10.1016/j.freeradbiomed.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Atef Y, El-Fayoumi HM, Abdel-Mottaleb Y, Mahmoud MF. 2017. Quercetin and tin protoporphyrin attenuate hepatic ischemia reperfusion injury: role of HO-1. Naunyn Schmiedebergs Arch Pharmacol 390:871–881. 10.1007/s00210-017-1389-9. [DOI] [PubMed] [Google Scholar]
- 15.Ryter SW, Choi AM. 2009. Heme oxygenase-1/carbon monoxide: from metabolism to molecular therapy. Am J Respir Cell Mol Biol 41:251–260. 10.1165/rcmb.2009-0170TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryter SW, Choi AM. 2016. Targeting heme oxygenase-1 and carbon monoxide for therapeutic modulation of inflammation. Transl Res 167:7–34. 10.1016/j.trsl.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill AJ, Kovacsics CE, Vance PJ, Collman RG, Kolson DL. 2015. Induction of heme oxygenase-1 deficiency and associated glutamate-mediated neurotoxicity is a highly conserved HIV phenotype of chronic macrophage infection that is resistant to antiretroviral therapy. J Virol 89:10656–10667. 10.1128/JVI.01495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hosel M, Schirmacher P, Tiegs G. 2007. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology 133:1156–1165. 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Kah J, Volz T, Lutgehetmann M, Groth A, Lohse AW, Tiegs G, Sass G, Dandri M. 2017. Haem oxygenase-1 polymorphisms can affect HCV replication and treatment responses with different efficacy in humanized mice. Liver Int 37:1128–1137. 10.1111/liv.13347. [DOI] [PubMed] [Google Scholar]
- 20.Hill-Batorski L, Halfmann P, Neumann G, Kawaoka Y. 2013. The cytoprotective enzyme heme oxygenase-1 suppresses Ebola virus replication. J Virol 87:13795–13802. 10.1128/JVI.02422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinoza JA, Leon MA, Cespedes PF, Gomez RS, Canedo-Marroquin G, Riquelme SA, Salazar-Echegarai FJ, Blancou P, Simon T, Anegon I, Lay MK, Gonzalez PA, Riedel CA, Bueno SM, Kalergis AM. 2017. Heme oxygenase-1 modulates human respiratory syncytial virus replication and lung pathogenesis during infection. J Immunol 199:212–223. 10.4049/jimmunol.1601414. [DOI] [PubMed] [Google Scholar]
- 22.Tseng CK, Lin CK, Wu YH, Chen YH, Chen WC, Young KC, Lee JC. 2016. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci Rep 6:32176. 10.1038/srep32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burke AC, Nelson CE, Morgan BA, Tabin C. 1995. Hox genes and the evolution of vertebrate axial morphology. Development 121:333–346. 10.1242/dev.121.2.333. [DOI] [PubMed] [Google Scholar]
- 24.Krumlauf R. 1994. Hox genes in vertebrate development. Cell 78:191–201. 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 25.Rezsohazy R, Saurin AJ, Maurel-Zaffran C, Graba Y. 2015. Cellular and molecular insights into Hox protein action. Development 142:1212–1227. 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- 26.Chojnowski JL, Masuda K, Trau HA, Thomas K, Capecchi M, Manley NR. 2014. Multiple roles for HOXA3 in regulating thymus and parathyroid differentiation and morphogenesis in mouse. Development 141:3697–3708. 10.1242/dev.110833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manley NR, Capecchi MR. 1995. The role of Hoxa-3 in mouse thymus and thyroid development. Development 121:1989–2003. 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- 28.Chisaka O, Capecchi MR. 1991. Regionally restricted developmental defects resulting from targeted disruption of the mouse homeobox gene hox-1.5. Nature 350:473–479. 10.1038/350473a0. [DOI] [PubMed] [Google Scholar]
- 29.Su DM, Manley NR. 2000. Hoxa3 and pax1 transcription factors regulate the ability of fetal thymic epithelial cells to promote thymocyte development. J Immunol 164:5753–5760. 10.4049/jimmunol.164.11.5753. [DOI] [PubMed] [Google Scholar]
- 30.Mace KA, Restivo TE, Rinn JL, Paquet AC, Chang HY, Young DM, Boudreau NJ. 2009. HOXA3 modulates injury-induced mobilization and recruitment of bone marrow-derived cells. Stem Cells 27:1654–1665. 10.1002/stem.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Sadoun H, Burgess M, Hentges KE, Mace KA. 2016. Enforced expression of Hoxa3 inhibits classical and promotes alternative activation of macrophages in vitro and in vivo. J Immunol 197:872–884. 10.4049/jimmunol.1501944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han B, Lian L, Li X, Zhao CF, Qu LJ, Liu CJ, Song JZ, Yang N. 2016. Chicken gga-miR-130a targets HOXA3 and MDFIC and inhibits Marek’s disease lymphoma cell proliferation and migration. Mol Biol Rep 43:667–676. 10.1007/s11033-016-4002-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XX, Liu GW, Ding L, Jiang T, Shao SH, Gao Y, Lu Y. 2018. HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell Biochem 119:2864–2874. 10.1002/jcb.26461. [DOI] [PubMed] [Google Scholar]
- 34.Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu CL. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110. 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 35.Miller LC, Laegreid WW, Bono JL, Chitko-McKown CG, Fox JM. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch Virol 149:2453–2463. 10.1007/s00705-004-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu SQ, Cai X, Wu JX, Cong Q, Chen X, Li T, Du FH, Ren JY, Wu YT, Grishin NV, Chen ZJJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:1217. 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 37.Tzima S, Victoratos P, Kranidioti K, Alexiou M, Kollias G. 2009. Myeloid heme oxygenase-1 regulates innate immunity and autoimmunity by modulating IFN-beta production. J Exp Med 206:1167–1179. 10.1084/jem.20081582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porritt RA, Hertzog PJ. 2015. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol 36:150–160. 10.1016/j.it.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Schwanke H, Stempel M, Brinkmann MM. 2020. Of keeping and tipping the balance: host regulation and viral modulation of IRF3-dependent IFNB1 expression. Viruses-Basel 12:733. 10.3390/v12070733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Z, Zhang MX, Tang Z, Zhang Q, Ye J, Xiong TC, Zhang ZD, Zhong B. 2020. USP22 promotes IRF3 nuclear translocation and antiviral responses by deubiquitinating the importin protein KPNA2. J Exp Med 217:e20191174. 10.1084/jem.20191174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryter SW, Alam J, Choi AM. 2006. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev 86:583–650. 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 42.Waza AA, Hamid Z, Ali S, Bhat SA, Bhat MA. 2018. A review on heme oxygenase-1 induction: is it a necessary evil. Inflamm Res 67:579–588. 10.1007/s00011-018-1151-x. [DOI] [PubMed] [Google Scholar]
- 43.Farombi EO, Surh YJ. 2006. Heme oxygenase-1 as a potential therapeutic target for hepatoprotection. J Biochem Mol Biol 39:479–491. 10.5483/bmbrep.2006.39.5.479. [DOI] [PubMed] [Google Scholar]
- 44.Immenschuh S, Ramadori G. 2000. Gene regulation of heme oxygenase-1 as a therapeutic target. Biochem Pharmacol 60:1121–1128. 10.1016/s0006-2952(00)00443-3. [DOI] [PubMed] [Google Scholar]
- 45.Chan KM, Kan YW. 1999. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc Natl Acad Sci USA 96:12731–12736. 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espinoza JA, Gonzalez PA, Kalergis AM. 2017. Modulation of antiviral immunity by heme oxygenase-1. Am J Pathol 187:487–493. 10.1016/j.ajpath.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Fillebeen C, Rivas-Estilla AM, Bisaillon M, Ponka P, Muckenthaler M, Hentze MW, Koromilas AE, Pantopoulos K. 2005. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C Virus. J Biol Chem 280:9049–9057. 10.1074/jbc.M412687200. [DOI] [PubMed] [Google Scholar]
- 48.Zhu ZW, Wilson AT, Luxon BA, Brown KE, Mathahs MM, Bandyopadhyay S, McCaffrey AP, Schmidt WN. 2010. Biliverdin inhibits hepatitis C virus nonstructural 3/4A protease activity: mechanism for the antiviral effects of heme oxygenase? Hepatology 52:1897–1905. 10.1002/hep.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santangelo R, Mancuso C, Marchetti S, Di Stasio E, Pani G, Fadda G. 2012. Bilirubin: an endogenous molecule with antiviral activity in vitro. Front Pharmacol 3:36. 10.3389/fphar.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu XM, Durante ZE, Peyton KJ, Durante W. 2016. Heme oxygenase-1-derived bilirubin counteracts HIV protease inhibitor-mediated endothelial cell dysfunction. Free Radic Biol Med 94:218–229. 10.1016/j.freeradbiomed.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma LL, Zhang P, Wang HQ, Li YF, Hu J, Jiang JD, Li YH. 2019. Heme oxygenase-1 agonist CoPP suppresses influenza virus replication through IRF3-mediated generation of IFN-alpha/beta. Virology 528:80–88. 10.1016/j.virol.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 52.Zhu ZW, Mathahs MM, Schmidt WN. 2013. Restoration of type I interferon expression by heme and related tetrapyrroles through inhibition of NS3/4A protease. J Infect Dis 208:1653–1663. 10.1093/infdis/jit338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu JS, Chen WC, Tseng CK, Lin CK, Hsu YC, Chen YH, Lee JC. 2016. Sulforaphane suppresses hepatitis C virus replication by up-regulating heme oxygenase-1 expression through PI3K/Nrf2 pathway. PLoS One 11:e0152236. 10.1371/journal.pone.0152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong M, Wang H, Ma L, Yan H, Wu S, Gu Z, Li Y. 2019. DMO-CAP inhibits influenza virus replication by activating heme oxygenase-1-mediated IFN response. Virol J 16:21. 10.1186/s12985-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen JY, Wang D, Sun Z, Gao L, Zhu XY, Guo JH, Xu SG, Fang LR, Li K, Xiao SB. 2019. Arterivirus nsp4 antagonizes interferon beta production by proteolytically cleaving NEMO at multiple sites. J Virol 93:e00385-19. 10.1128/JVI.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Chen JY, Yu CL, Zhu XY, Xu SG, Fang LR, Xiao SB. 2019. Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J Virol 93:e00623-19. 10.1128/JVI.00623-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang LP, He J, Wang R, Zhang XH, Lin SL, Ma ZX, Zhang YJ. 2019. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus induces STAT2 degradation to inhibit interferon signaling. J Virol 93:e01352-19. 10.1128/JVI.01352-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo R, Xiao SB, Jiang YB, Jin H, Wang D, Liu ML, Chen HC, Fang LR. 2008. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol Immunol 45:2839–2846. 10.1016/j.molimm.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol 84:1574–1584. 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren Y, Khan FA, Pandupuspitasari NS, Li S, Hao X, Chen X, Xiong J, Yang L, Fan M, Zhang S. 2016. Highly pathogenic porcine reproductive and respiratory syndrome virus modulates interferon-beta expression mainly through attenuating interferon-regulatory factor 3 phosphorylation. DNA Cell Biol 35:489–497. 10.1089/dna.2016.3283. [DOI] [PubMed] [Google Scholar]
- 61.Xiao S, Wang X, Ni H, Li N, Zhang A, Liu H, Pu F, Xu L, Gao J, Zhao Q, Mu Y, Wang C, Sun Y, Du T, Xu X, Zhang G, Hiscox JA, Goodfellow IG, Zhou EM. 2015. MicroRNA miR-24-3p promotes porcine reproductive and respiratory syndrome virus replication through suppression of heme oxygenase-1 expression. J Virol 89:4494–4503. 10.1128/JVI.02810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]