ABSTRACT
The persistence of cells latently infected with HIV-1, named the latent reservoir, is the major barrier to HIV-1 eradication, and the formation and maintenance of the latent reservoir might be exacerbated by activation of the immunoinhibitory pathway and dysfunction of CD8+ T cells during HIV-1 infection. Our previous findings demonstrated that prophylactic vaccination combined with PD-1 blockade generated distinct immune response profiles and conferred effective control of highly pathogenic SIVmac239 infection in rhesus macaques. However, to our surprise, herein we found that a therapeutic vaccination in combination with PD-1 blockade resulted in activation of the viral reservoir, faster viral rebound after treatment interruption, accelerated AIDS progression, and, ultimately, death in chronically SIV-infected macaques after antiretroviral therapy (ART) interruption. Our study further demonstrated that the SIV provirus was preferentially enriched in PD-1+CD4+ T cells due to their susceptibility to viral entry, potent proliferative ability, and inability to perform viral transcription. In addition, the viral latency was effectively reactivated upon PD-1 blockade. Together, these results suggest that PD-1 blockade may be a double-edged sword for HIV-1 immunotherapy and provide important insight toward the rational design of immunotherapy strategies for an HIV-1 cure.
IMPORTANCE As it is one of the most challenging public health problems, there are no clinically effective cure strategies against HIV-1 infection. We demonstrated that prophylactic vaccination combined with PD-1 blockade generated distinct immune response profiles and conferred better control of highly pathogenic SIVmac239 infection in rhesus macaques. In the present study, to our surprise, PD-1 blockade during therapeutic vaccination accelerated the reactivation of latent reservoir and AIDS progression in chronically SIV-infected macaques after ART interruption. Our study further demonstrated that the latent SIV provirus was preferentially enriched in PD-1+CD4+ T cells because of its susceptibility to viral entry, inhibition of SIV transcription, and potent ability of proliferation, and the viral latency was effectively reactivated by PD-1 blockade. Therefore, PD-1 blockade might be a double-edged sword for AIDS therapy. These findings provoke interest in further exploring novel treatments against HIV-1 infection and other emerging infectious diseases.
KEYWORDS: rhesus macaques, HIV, SIV, latent viral reservoir, programmed cell death protein 1 (PD-1) blockade, therapeutic vaccination
INTRODUCTION
As one of the most challenging public health problems, there are no clinically effective cure strategies against human immunodeficiency virus type 1 (HIV-1) infection/AIDS, even nearly four decades after its discovery. Antiretroviral therapy (ART) can effectively control but cannot cure HIV-1 infection. The latent replication-competent HIV-1 provirus has been recognized as the major obstacle to HIV-1 cure, and another critical question is to identify the specific biomarker of latent HIV-infected cell subsets (1, 2). Recent studies have shown that cell subsets expressing immunoinhibitory molecules, such as cytotoxic T lymphocyte antigen 4 (CTLA-4), programmed cell death 1 (PD-1), and lymphocyte-activation gene 3 (LAG3), are enriched to harbor HIV-1 provirus and, therefore, are intrinsically linked to the main source of the latent reservoir in HIV-1 patients (3, 4). As a result, it is of great interest to develop strategies to modulate the latent reservoir status by targeting immunoinhibitory signaling pathways.
Immunoinhibitory pathways are extensively involved in the pathogenesis of infectious diseases and the response to vaccination. PD-1, in conjunction with a variety of immunoinhibitory molecules, including CTLA-4, T cell immunoglobulin and mucin-domain-containing-3 (Tim3), T cell immunoreceptor with Ig and ITIM domains (TIGIT), and LAG3, is usually upregulated on HIV-specific CD4+ T cells, which have been previously thought to cause the exhaustion and dysfunction of immune cells in HIV-1 patients (5–7). Among them, PD-1/PD-L1 has been studied as a promising target in the field of immunotherapy against cancer and infectious diseases. Numerous studies have demonstrated that blockade of the PD-1/PD-L1 pathway can restore the immune functions of exhausted T cells in infected individuals (8, 9). Both our recent work and studies by others have shown that in vivo PD-1 blockade in combination with prophylactic vaccination effectively augment and sustain the magnitude, polyfunctionality, and proliferation of simian immunodeficiency virus (SIV)-specific CD8+ T cell responses and substantially improve protective efficacy against pathogenic SIVmac239 infection in rhesus macaques (10, 11). Recently, PD-1 blockade in combination vaccination in ART-treated macaques provided therapeutic benefit against SIV infection (12, 13). However, the exact interactions between PD-1 signal modulation and latent reservoir status have not yet been fully elucidated, especially in chronically SIV-infected macaques aimed to evaluate the time to viral rebound after ART interruption.
In this study, based on our established SIV-infected macaque model, we investigated how PD-1 blockade affected the immunogenicity and protective efficacy of an adenovirus-vectored therapeutic SIV vaccine and whether PD-1 signal modulation was correlated with a viral rebound in chronically SIV-infected macaques after ART interruption. We also further investigated the mechanisms underlying the preferential formation and maintenance of latent reservoirs in PD-1+CD4+ T cells. Overall, these findings provide insights to develop potential therapeutic strategies against HIV-1 infection by targeting PD-1-expressing reservoir cells.
RESULTS
Exacerbated AIDS progression by PD-1 blockade during therapeutic vaccination in chronically SIV-infected macaques after ART interruption.
Our previous study indicated that PD-1 blockade during prophylactic vaccination improved immune protection against SIV challenge in macaques (10). However, it remains unknown how PD-1 blockade affects the status of the latent viral reservoir during therapeutic vaccination in chronically SIV-infected macaques after ART interruption. Therefore, 12 chronically SIV-infected macaques were evenly assigned into three groups: αPD-1 blockade + therapeutic vaccine group, therapeutic vaccine-only group, and control group (Table S1). All macaques were pretreated with ART for 3 weeks to reduce the interference of free SIV particles. The ART regime was given as a daily subcutaneous injection of 9-(2-[R]-[Phosphonomethoxy]propyl) adenine (PMPA, 30 mg/kg) and emtricitabine (FTC, 20 mg/kg). To remove the possible impact of ART on the following studies, we discontinued ART in all macaques. At weeks 0 and 4, 8 of the macaques were immunized with the SIV vaccine as previously described (14). Among them, 4 macaques were treated with PD-1 antibody (GB226) every 2 weeks from −1 to 7 weeks post-ART discontinuation (WPAD) (Fig. 1A). Our previous study reported that the pharmacokinetic parameters of the PD-1 antibody (through intravenous injection) in macaques and the half-life of the PD-1 monoclonal antibody (GB226) used in the study in macaques was about 2 weeks (10). We, therefore, chose week 7 (the last time of PD-1 antibody administration) as the key time point to evaluate the potential effect.
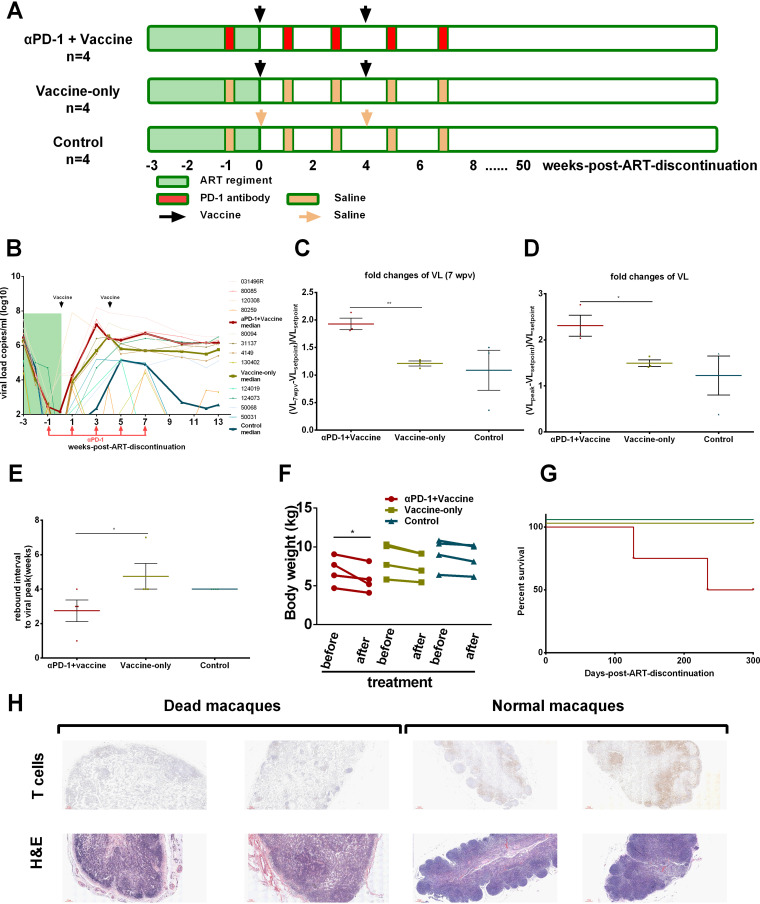
FIG 1.
Exacerbated AIDS progression by PD-1 blockade during therapeutic vaccination in chronically SIV-infected macaques after ART interruption. (A) Twelve macaques were evenly assigned into three groups: the PD-1 blockade + therapeutic vaccine group, therapeutic vaccine-only group, and control group (n = 4 per group). All macaques were pretreated with antiretroviral therapy (ART) from −3 WPAD to 0 WPAD. Eight macaques were immunized with the SIV vaccine at 0 and 4 WPAD. Among them, four macaques were intravenously injected with an anti-PD-1 antibody every 2 weeks from −1 to 7 WPAD. Macaques from other groups were injected with saline at the same time points. (B) The dynamics of plasma viral load of all rhesus macaques during this study. Data are presented as the median with an interquartile range. (C) Fold changes in plasma viral load (VL) in 7 WPAD relative to VLsetpoint (VL-1WPAD). P values were obtained from the unpaired t test. (D) Fold changes in VLpeak relative to VLsetpoint among three groups. P values were obtained from the unpaired t test. (E) The rebound interval to the viral peak among three groups. P values were obtained from Dunn’s multiple-comparison test. (F) Changes in body weight of macaques before and after treatment. P values were obtained from Tukey’s multiple-comparison test. (G) Comparisons of survival of macaques following different interventions. (H) To further confirm the clinical and pathological changes of the experimental macaques, the lymphoid tissues, such as inguinal lymph nodes, were collected from either healthy macaques or dead macaques because of AIDS. Hematoxylin-eosin (H&E) staining was then performed to examine the pathological changes, and the in situ distribution of T cells in lymphoid tissue was also detected by immunohistochemistry technique. WPAD represents weeks post ART discontinuation. All average data are presented as mean ± standard error of the mean (s.e.m.) (*, P < 0.05; **, P < 0.01).
To monitor how SIV replication was affected by PD-1 blockade during therapeutic vaccination, the dynamics of plasma viral load in chronically SIV-infected macaques were tested by real-time quantitative PCR. In our SIV-infected macaque model, the plasma viral load was effectively suppressed to undetectable levels during ART. As we previously described (10, 14), the viral load in SIV-infected macaques fluctuated to a large extent after ART discontinuation, rebounded to a peak, gradually decreased, and then fluctuated several times to finally reach a stable undulating state (setpoint). To our surprise, the plasma viral load in the αPD-1 blockade + vaccine group rebounded to a higher level than that in the other two groups (Fig. 1B). The median fold change of the 7 WPAD plasma viral load (pVL) relative to the setpoint in the αPD-1 blockade + vaccine group was 1.82-fold, accounting for half of the fold change in the vaccine-only group (Fig. 1C). The median fold change of pVL from setpoint to peak in the αPD-1 blockade + vaccine group (2.08-fold) was also higher than that in the vaccine-only group (1.42-fold) and control group (1.60-fold) (Fig. 1D). Furthermore, the median rebound interval of pVL from setpoint to peak was 3 weeks in the αPD-1 blockade + vaccine group, which was faster than that in the other two groups at 4 and 7.5 weeks (Fig. 1E). These data suggested that PD-1 blockade during therapeutic vaccination exacerbated viral rebound in chronically SIV-infected macaques after ART interruption.
More importantly, all four monkeys in the αPD-1 blockade + vaccine group developed obvious simian AIDS progression, as defined by clinical symptoms of diarrhea, wasting and body weight loss, and high viral load (Fig. 1F). Of note, two macaques in the αPD-1 blockade + vaccine group died because of AIDS symptoms during the experimental period (50% survival rate), while all eight macaques in the other two groups survived throughout the study (100% survival rate) (Fig. 1G). In addition, we also conducted a study to investigate the effect of PD-1 blockade alone, and observed an accelerated AIDS progression in chronically SIV-infected macaques after ART interruption (75% survival rate), although a long-term observation should be further monitored. To further confirm the clinical and pathological changes in these experimental macaques, immunohistochemistry and hematoxylin-eosin (H&E) staining were performed to examine the pathological changes and the in situ distribution of T cells in lymphoid tissues, such as inguinal lymph nodes (Fig. 1H). Overall, these findings indicated that PD-1 blockade accelerated the reactivation of the latent reservoir and AIDS progression in chronically SIV-infected macaques after ART interruption.
Exacerbated AIDS progression was not caused by additional cytotoxicity of PD-1 antibody in SIV-infected macaques.
We next investigated the mechanism responsible for this exacerbated AIDS progression by PD-1 blockade. Our previous study showed that the GB226 antibody exerted cross-reactivity with rhesus macaque PD-1 proteins in vitro (10), and we further confirmed its effect on blocking PD-1 signaling in vivo in chronically SIV-infected macaques. The GB226 antibody used in this study has been extensively verified to block the PD-1/PD-L1 signaling pathway, including some clinical trials in China (2016L10520) and Australia (NCT03053466). Consistent with these data, the frequency of PD-1 expression on the surface of CD3+ T lymphocytes dropped sharply below the detectable sensitivity in the αPD-1 blockade + vaccine group throughout GB226 treatment, while PD-1 expression remained high in the vaccine-only and control groups (Fig. 2A). A similar phenomenon was observed in CD4+ T lymphocytes (Fig. 2B) and CD8+ T lymphocytes (Fig. 2C). These data demonstrated that the GB226 antibody effectively blocked the PD-1 signal on T cells in chronically SIV-infected macaques.
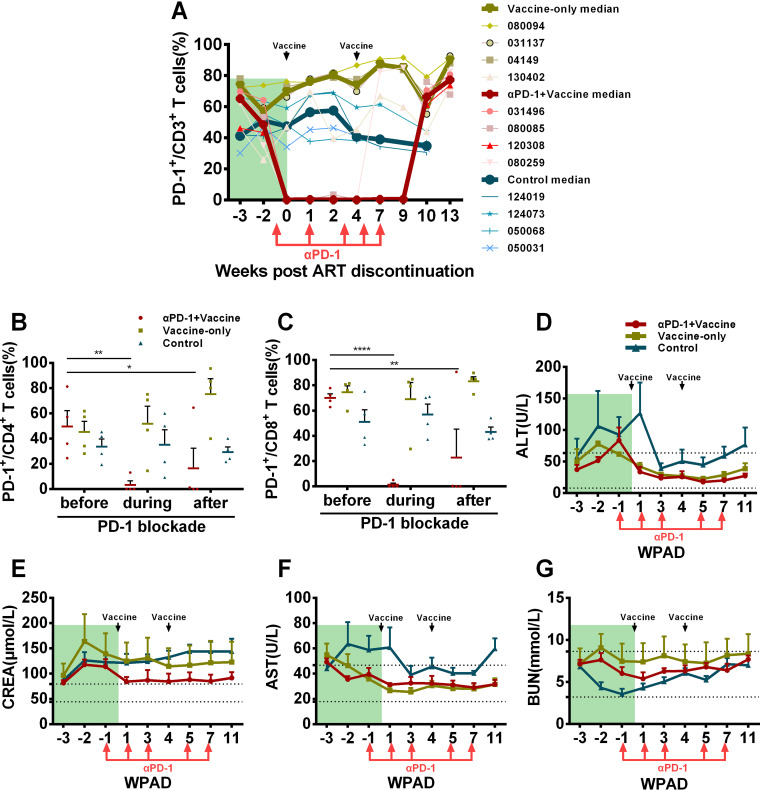
FIG 2.
The GB226 antibody effectively blocks PD-1 signaling on the T lymphocyte surface in SIV-infected macaques without additional toxicity. To assess the effect of the GB226 antibody on PD-1 signaling blockade in rhesus macaques, the expression of PD-1 on CD3+, CD4+, and CD8+ T cells was monitored by flow cytometry. (A) The percentages of PD-1+/CD3+ T cells throughout the study. (B) The frequencies of PD-1+CD4+/CD4+ T cells before (−3 WPAD), during (2 WPAD) and after PD-1 blockade (9 WPAD). P values were obtained from Tukey’s multiple-comparison test. (C) The ratios of PD-1+CD8+/CD8+ T cells before (−3 WPAD), during (2 WPAD) and after PD-1 blockade (9 WPAD). Bars reflect the median. P values were obtained from Tukey’s multiple-comparison test. (D) The concentrations of alanine aminotransferase (ALT), (E) aspartate aminotransferase (AST), (F) creatinine (CREA), and (G) urea nitrogen (BUN) in rhesus macaque plasma were monitored throughout the study. Data are presented as the median with an interquartile range. WPAD represents weeks post ART discontinuation. All average data are presented as mean ± s.e.m. (* P < 0.05, ** P < 0.01, **** P < 0.0001).
One concern for PD-1 blockade therapy is its potential side effects in vivo. We, therefore, wondered whether the GB226 antibody exerted additional cytotoxicity that could damage the physiological status of experimental macaques and subsequently exacerbate AIDS progression. To evaluate tolerance to the GB226 antibody in vivo, all experimental macaques were subjected to blood biochemistry, complete blood count, and physical examinations throughout the study period. As shown in Fig. 2D to G, the concentrations of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) during PD-1 blockade were similar to those in the other two groups, suggesting that neither GB226 antibody nor AVIP immunization caused additional liver function toxicity. The creatinine (CREA) content in these animals was slightly higher than the normal value, but the CREA content of the plasma in the αPD-1 blockade + vaccine group was lower than that in the other groups. In addition, the concentration of blood urea nitrogen (BUN) in all animals was within the normal range during this study, indicating that the GB226 antibody did not cause additional damage to kidney function (Fig. 2D to G). The ratio of AST/ALT and the concentration of glucose (GLU) were also within the normal range, indicating that these macaques had a good nutritional status (Fig. S1A to B). The concentration of lactate dehydrogenase (LDH) in macaques given ART was slightly higher than the normal value, but it showed a decreasing trend after the GB226 antibody treatment (Fig. S1C). The complete blood count also showed that there was no significant difference in the cell subpopulation composition after GB226 treatment, although the proportions of lymphocytes (“LYMPH#”) and monocytes (“MONO#”) in the blood of macaques in the αPD-1 blockade + vaccine group were higher than those of the vaccine-only group, which indicated that more robust immune responses were elicited in these macaques (Fig. S1D to S). Together, these data indicated that the GB226 antibody effectively blocked PD-1 signaling in chronically SIV-infected macaques with no additional toxicity to liver and kidney function.
Immune modulation by PD-1 blockade in chronically SIV-infected macaques.
Challenging issues related to chronic HIV-1 infection are persistent inflammation and excessive immune activation, which are deleterious for immune reconstruction in these patients. We next assessed how PD-1 antibody administration affected immune activation and inflammation in chronically SIV-infected macaques. We found that GB226 antibody treatment significantly increased CD3+ T cell numbers in both the early and late treatment periods. Interestingly, compared to the other two groups, the CD4+ T cell number significantly increased earlier at 1 WPAD, and the CD8+ T cell number significantly increased at 7 WPAD in the αPD-1 blockade + vaccine group (Fig. 3A).
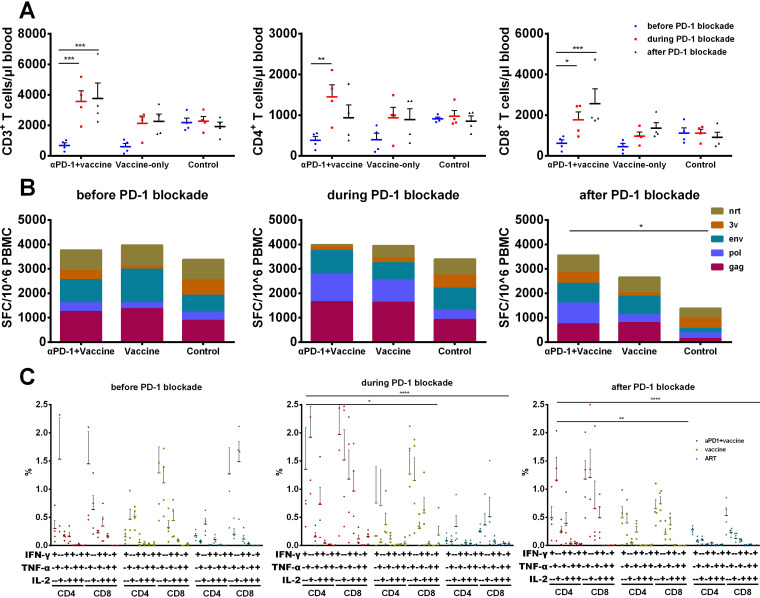
FIG 3.
Immune modulation by PD-1 blockade in SIV chronically infected macaques. (A) The numbers of CD3+, CD4+, and CD8+ T cells were detected by flow cytometry at −2, 1, and 7 WPAD, respectively. P values were obtained from Tukey’s multiple-comparison test. (B) SIV antigen-specific IFN-γ secreting cells were tested by ELISPOT assay at −1, 4, and 9 WPAD. P values were obtained from Tukey’s multiple-comparison test. (C) The frequency of IFN-γ, IL-2, and TNF-α-secreting CD4+ and CD8+ T cells was detected by intracellular cytokine staining (ICS) assay at −2, 7, and 13 WPAD. P values were obtained from Tukey’s multiple-comparison test. WPAD represented for weeks post ART discontinuation. All average data are presented as mean ± s.e.m. (* P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001).
To assess the in vivo effect on the SIV-specific immune response by PD-1 blockade, interferon-gamma (IFN-γ)-mediated enzyme-linked immunosorbent spot assay (ELISPOT) and intracellular cytokine staining (ICS) assays were performed throughout the study period. The initial (−1 WPAD) SIV antigen-specific IFN-γ spot-forming cells (SFCs) in the three groups were at similar levels, while the SIV structural protein (Gag, Pol, and Env)-specific SFCs maintained a high level in the αPD-1 blockade + vaccine group during and after PD-1 blockade administration (Fig. 3B). Consistent with the above results, SIV-specific cytokine production of CD4+ and CD8+ T lymphocytes before PD-1 blockade was also similar among the three groups based on the ICS assay, but higher levels of polyfunctional cytokine production were found in the αPD-1 blockade + vaccine group during and after PD-1 blockade than in the other two groups (Fig. 3C). Taken together, PD-1 blockade during therapeutic vaccination partially restored the immune function of T lymphocytes in chronically SIV-infected macaques.
Transcriptomics of PD-1+CD4+ T cells facilitated the determination of latent HIV/SIV infection.
Because CD4+ T cells were recognized as the main component of the HIV/SIV reservoir, we next wondered whether the GB226 antibody would play a role in reversing the latent SIV reservoir and subsequently lead to the acceleration of AIDS progression in our study. Despite some reports on PD-1 expression and HIV latency ex vivo, we did not find any RNA-seq data concerning this topic in rhesus macaques. Based on the above findings in SIV-infected macaques, we, conducted RNA-seq analysis of PD-1+CD4+ and PD-1−CD4+ T cells to further clarify these observations for the first time in rhesus macaques.
CD4+ T cells from chronically SIV-infected rhesus macaques were isolated and sorted into two subsets depending on PD-1 expression by flow cytometry, and then a global landscape of transcriptional profiling between PD-1+CD4+ and PD-1−CD4+ T subsets was obtained by RNA-seq. The gene expression signature was drastically different between these subsets, with a total of 5447 transcripts showing differential gene expression (false discovery rate [FDR]-adjusted P < 0.05 and log2 change [FC] > 0) (Fig. 4A and B). The differentially expressed genes (DEGs) were analyzed, including the top 30 significant terms based on gene ontology (GO) enrichment analysis and the top 20 enriched pathways by KEGG analyses. As expected, immune and transcription-related signaling pathways were enriched in our analysis (Fig. 4C).
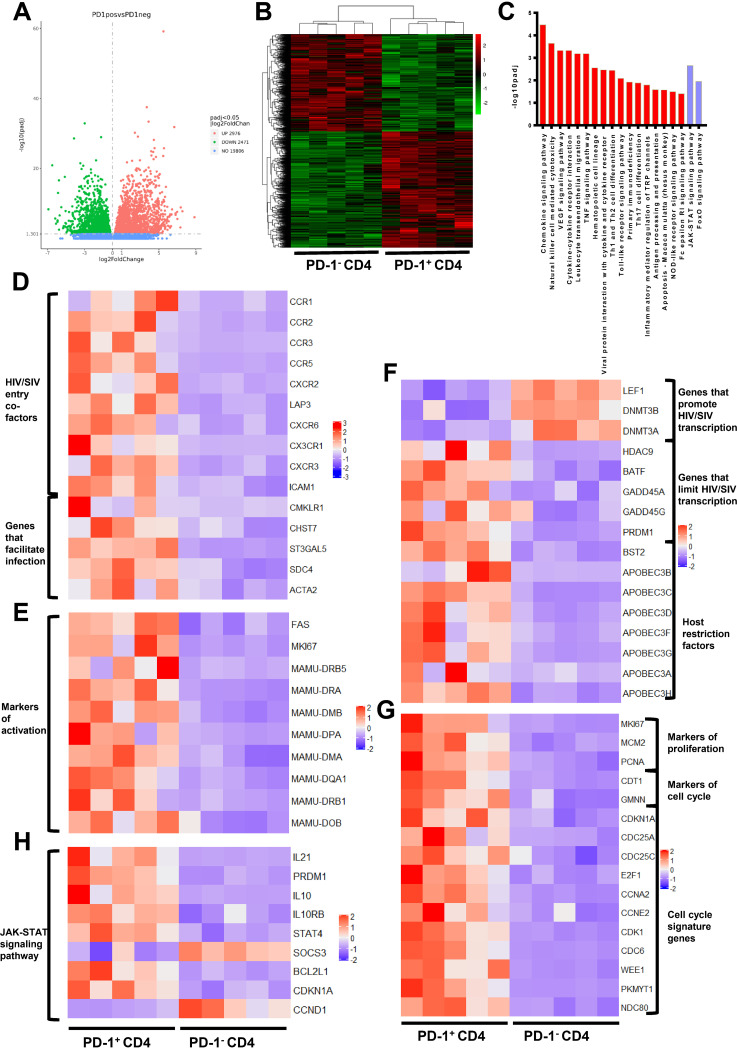
FIG 4.
Transcriptional profiling of PD-1+CD4+ T cells is beneficial for determining the latent status of HIV/SIV infection. (A) Volcano plot displaying genes detected by RNA-seq. Green dots represent the downregulated genes, and red dots indicate the upregulated genes in PD-1+CD4+ T cells. Green and red dots represent genes with a false discovery rate (FDR)-adjusted P < 0.05 and log2 change (FC) > 0; blue dots indicate genes with a FDR adjusted P > 0.05. (B) Hierarchical clustering and heatmap of the differentially expressed genes (DEGs) between PD-1+CD4+ and PD-1−CD4+ T cells. The displayed DEGs had |log2 FC| ≥0-fold changes in gene expression intensity and a FDR adjusted P value < 0.05 relative to the control cohort. (C) Top enriched immune and transcription-related pathways based on KEGG pathway analysis. Red bars indicate the enriched pathways in PD-1+CD4+ cells, and the purple bars represent the enriched pathways in PD-1−CD4+ cells. (D to G) Heatmap of the functional annotations of DEGs between PD-1+CD4+ and PD-1−CD4+ T cells. (H) Heatmap of DEGs in the JAK-STAT signaling pathway between PD-1+CD4+ and PD-1−CD4+ T cells.
Interestingly, compared with PD-1−CD4+ T cells, the expression levels of HIV/SIV entry-related factors, such as CCR5, LAP3 (also known as CXCR4), CXCR6, CX3CR1, CXCR3, and ICAM1 (15–17), were upregulated in PD-1+CD4+ T cells. We also observed an upregulation of CMKLR1, CHST7, ST3GAL5, SDC4, and ACTA2, which have been reported to facilitate HIV entry into target cells (18–20), suggesting that PD-1+CD4+ T cells were more susceptible to HIV/SIV acquisition (Fig. 4D). In addition, activation-related genes (MKI67, FAS, and major histocompatibility complex class II family) were significantly increased in PD-1+CD4+ T cells (Fig. 4E), which is consistent with the greater sensitivity of activated cells to HIV/SIV infection (21).
More importantly, compared with PD-1−CD4+ T cells, we found that genes associated with transcription of the HIV/SIV genome were significantly changed in PD-1+CD4+ T cells. For example, genes like LEF-1, DNMT3B, and DNMT3A, which might play a role in promoting HIV/SIV transcription (22, 23), were downregulated. In contrast, genes like HDAC9, BATF (a negative regulator of AP-1), GADD45A, GADD45G, and PRDM1 (gene name for Blimp-1), which might play a role in inhibiting HIV/SIV transcription (24–26), were upregulated (Fig. 4F). In addition, the transcriptional levels of host restriction factors, including BST2 (also referred to as tetherin, CD317, or HM1.24) and the APOBEC family (27, 28), were elevated in PD-1+CD4+ T cells (Fig. 4F). These data suggested that the microenvironment of PD-1+CD4+ T cells might lead to suppression of transcription of the integrated HIV/SIV genome.
We also observed that cell proliferation markers, including PCNA, MCM2, and MKI67 (29), and cell cycle markers, including CTD1, GMNN, CDKN1A, CDC25A, CDC25C, E2F1, and PRDM1 (30), were highly expressed in PD-1+CD4+ T cells (Fig. 4G). KEGG analyses of the DEGs indicated that the higher proliferative activity of PD-1+CD4+ T cells was likely regulated by PRDM1 and interleukin-10 (IL-10) through the JAK-STAT signaling pathway (Fig. 4H). These data indicated that these PD-1+CD4+ T cells might contribute to the maintenance and turnover of the viral reservoir by undergoing cell proliferation. As a result, the microenvironment of PD-1+CD4+ T cells was favorable to HIV/SIV acquisition but unfavorable to its transcription after HIV/SIV entry and, therefore, facilitated latent infection.
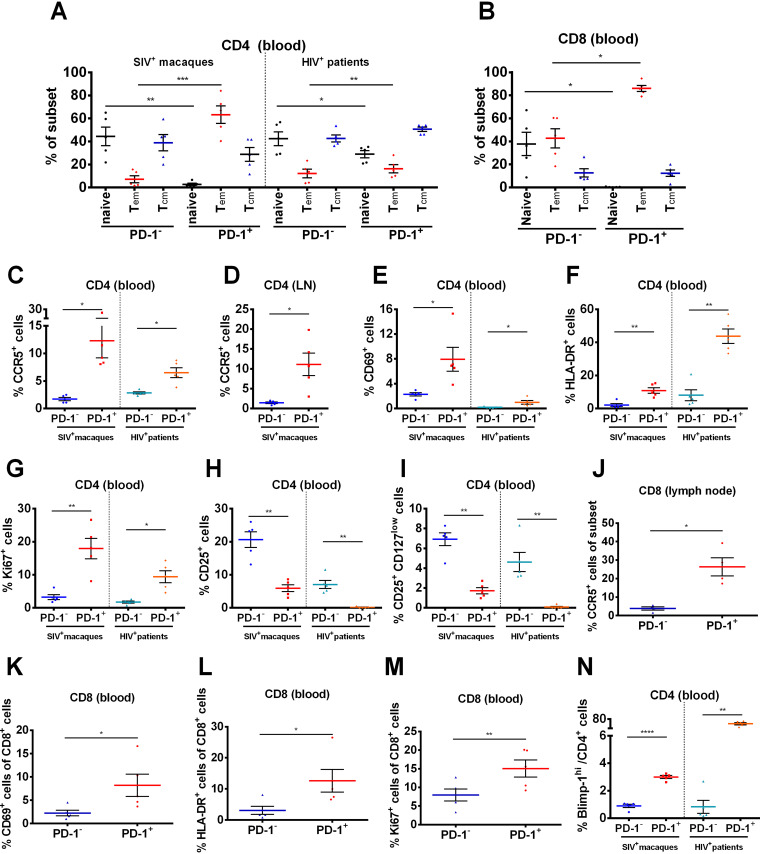
The phenotypic characteristics of PD-1+CD4+ T cells in peripheral blood and lymph nodes from chronically SIV-infected macaques and HIV-infected patients confirmed their potential to form the latent SIV reservoir.
To determine whether the RNA-seq results could reflect the real situation in vivo, we collected peripheral blood and lymph nodes from chronically SIV-infected macaques and HIV-infected patients and detected the expression levels of relevant markers, including CD28, CD95, CCR5, CD25, CD127, Ki-67, and Blimp-1. The phenotype of PD-1+CD4+ T cells represented a greater fraction of effector memory subsets (P < 0.01) with a lower fraction of naive cell subsets (P < 0.01) than that of PD-1−CD4+ T cells (Fig. 5A) and PD-1+CD8+ T cells (Fig. 5B). Consistent with the above RNA-seq analysis, PD-1+CD4+ T cells exhibited a higher level of CCR5 than PD-1−CD4+ T cells in both peripheral blood and lymphoid tissue (P < 0.01), supporting their susceptibility to HIV/SIV entry (Fig. 5C and D). Activation markers, such as CD69 (P < 0.01) and HLA-DR (P < 0.01), were significantly upregulated in PD-1+CD4+ T subsets in both SIV+ macaques and HIV+ patients, suggesting their active status and thus ability to promote viral infection (Fig. 5E and F). Moreover, the percentage of Ki-67+ cells was significantly higher in the PD-1+CD4+ T subset than in the PD-1−CD4+ T subset (P < 0.01), indicating the capacity for proliferation and self-renewal (Fig. 5G). In addition, the frequency of CD25+ and CD25+CD127low cells were significantly lower in PD-1+CD4+ T subsets than in PD-1−CD4+ T cells (P < 0.01) (Fig. 5H and I). Similar observations were obtained in PD-1+CD8+ T cells (Fig. 5J to M). In line with the RNA-seq analyses, the expression of Blimp-1 was significantly higher in PD-1+CD4+ T cells than in PD-1−CD4+ T cells (P < 0.01) (Fig. 5N), which is involved in the inhibition of HIV-1 transcriptional elongation and maintenance of the latent reservoir (25).
FIG 5.
Phenotypic characteristics of PD-1+CD4+, PD-1−CD4+ T cells and PD-1+CD8+, PD-1−CD8+ T cells in peripheral blood and lymph nodes from chronically SIV-infected macaques and HIV-infected patients. Blood and lymph nodes were collected from chronically SIV-infected macaques (n = 5) and HIV+ patients on ART (n = 5) to analyze the phenotypic characteristics of PD-1+CD4+ T cells and PD-1−CD4+ T cells. (A) Percentages of naive T cells (CD28+CD95−), effector memory T cells (Tem) (CD28−CD95+) and central memory T cells (Tcm) (CD28+CD95+) in PD-1+CD4+ and PD-1−CD4+ T cells in the peripheral blood of SIV-infected macaques and HIV-infected patients. (B) The percentage of naive, Tem and Tcm within PD-1+CD8+ and PD-1−CD8+ T cells. (C and D) Expression levels of CCR5 in PD-1+CD4+ and PD-1−CD4+ subsets in peripheral blood and lymph nodes (LNs). Expression levels of CD69 (E), HLA-DR (F), and Ki-67 (G) in PD-1+CD4+ and PD-1−CD4+ T cells in peripheral blood. Percentages of CD25+ (H) and CD25+CD127low (I) subsets in PD-1+CD4+ and PD-1−CD4+ T cells in peripheral blood. (J) The expression of CCR5 between PD-1+CD8+ and PD-1−CD8+ subtype in lymph nodes. The expression of CD69 (K), HLA-DR (L) and Ki-67 (M) between PD-1+CD8+ and PD-1−CD8+ T cells in peripheral blood. (N) Percentage of Blimp-1hi subsets in PD-1+CD4+ and PD-1−CD4+ T cells in peripheral blood. P values were obtained from paired t test. All average data are presented as mean ± s.e.m. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
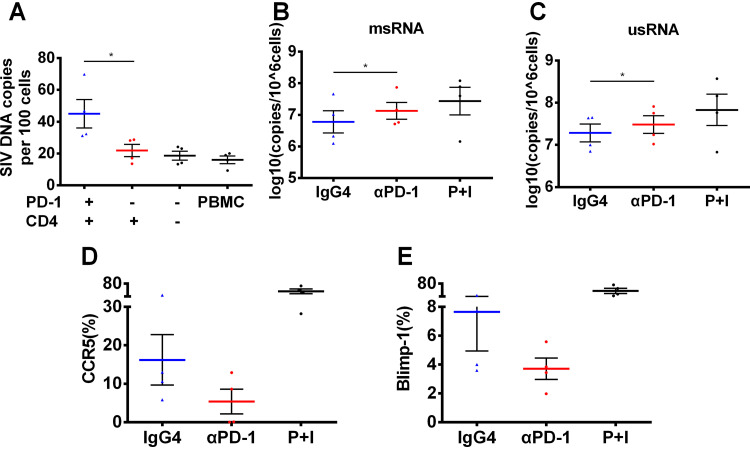
SIV proviruses are preferentially enriched in PD-1+CD4+ T cells and effectively reactivated by PD-1 blockade.
To further identify the preferential enrichment of SIV proviruses in PD-1+CD4+ T cells, the different cell subsets were sorted from four chronically infected rhesus macaques at 1 week after the interruption of 10-week long ART and then incubated with a CD3/CD28 antibody for 72 h. Cell-associated DNA was extracted and tested for SIV provirus. The level of SIV provirus in PD-1+CD4+ T cells was significantly higher than that in PD-1−CD4+ T cells (P < 0.05) (Fig. 6A). We next wondered whether PD-1 blockade might play a role in reactivating latent viral reservoirs. CD4+ T cells were sorted using a magnetic bead sorting kit from chronically SIV-infected macaques and incubated for 72 h with or without GB226 antibody. Cell-associated RNA, such as multiply spliced RNA (msRNA) and unspliced RNA (usRNA), is thought to be a sensitive parameter to represent the status of viral transcription and reactivation (31). We found that the median level of msRNA reached 6.96, which was higher than the 6.68 that was reached in the IgG4 (isotype control) group (Fig. 6B). The median level of usRNA reached 7.50, which was higher than the 7.32 that was reached in the isotype control group (Fig. 6C). These data suggested that the PD-1 antibody could act as a latency-reversing agent (LRA) in peripheral CD4+ T cells from chronically SIV-infected macaques. Furthermore, the expression level of CCR5 dramatically declined in CD4+ T cells after PD-1 blockade, with a median of 4.34 compared with 11.80 in the isotype control group (Fig. 6D). The median expression level of Blimp-1 was 3.64 in CD4+ T cells after PD-1 blockade, which was 37.9% less than that in the isotype control group (Fig. 6E). This result indicated that Blimp-1 might be a potential regulator of viral latency. Overall, the SIV provirus was preferentially enriched in PD-1+CD4+ T cells and effectively reactivated by PD-1 blockade.
FIG 6.
SIV proviruses are preferentially enriched in PD-1+CD4+ T cells and effectively reactivated by PD-1 blockade. (A) PBMCs from ART-treated SIV-infected macaques without detectable viremia (n = 4) were isolated, and the different cell subsets were isolated and incubated with CD3/CD28 antibodies for 72 h. Then, copies of the cell-associated SIV DNA were detected by quantitative PCR. P values were obtained from the ratio paired t test. The purified CD4+ T cells were sorted from chronically SIV-infected macaques (n = 4) and incubated with GB226 antibody or isotype IgG for 72 h, and then the cell-associated RNA was extracted to analyze the copies of two kinds of transcriptional SIV proviruses, multiply spliced RNA (msRNA) (B) and unspliced RNA (usRNA) (C), P values were obtained from paired t test. CD4+ T cells from chronically SIV-infected macaques were stimulated with GB226 or isotype IgG, and then the expression levels of CCR5 (D) and Blimp-1 (E) were detected by flow cytometry. P+I represent phorbol-12-myristate-13-acetate (40 ng/mL) and ionomycin (1000 ng/mL). All average data are presented as mean ± s.e.m. (* P < 0.05).
DISCUSSION
Viral latency is the major obstacle to eradicating HIV-1 infections, and how this latency is established and persists in HIV-1 patients are still areas of extensive investigation. Currently, many efforts are underway to target latent viral reservoirs to cure AIDS, including shock-and-kill, block-and-lock, CRISPR/Cas9-based gene editing, chimeric antigen receptor T-cell-based immunotherapy, and therapeutic vaccination (31–34). Recent evidence has shown that the status of viral latency might be affected by modulating immunoinhibitory pathways (35).
Several case reports have suggested that anti-PD-1 therapy for cancer patients with HIV-1 infection changes the level of cell-associated HIV-1 RNA and DNA copies (9, 35), implying that HIV-1 latency may be perturbed by targeting the PD-1 signaling pathway. In addition, targeting other immunosuppressive signals, including CTLA4, LAG3, and TIM3, has also been reported to alter HIV-1 latency (36, 37). However, a similar observation was not found in other case reports (38). Of note, the dosage of PD-1 antibody in these pilot trials was optimized for immunotherapy of malignant tumors, which may be one reason for the inconsistent results. To date, there is a lack of evidence to effectively modulate the viral latency status by targeting the immunoinhibitory signaling pathway. The present study, using a model of chronically SIV-infected macaques, provides the first report showing that in vivo PD-1 blockade during therapeutic vaccination exacerbates viral rebound and AIDS progression by reactivating viral latency after ART interruption.
Recent studies have shown that memory CD4+ T cells expressing PD-1 harbor greater proportions of genetically replication-competent proviruses than PD-1−CD4+ T cells (39), and PD-1+ follicular helper T cells in lymph nodes contribute to the persistent transcription of competent HIV-1 in long-term ART-treated aviremic AIDS patients (40, 41), indicating that PD-1-expressing cells contribute significantly to the formation of the latent reservoir. Nevertheless, the maintenance of latent reservoir might be exacerbated due to the immunoinhibitory microenvironment and dysfunction of cytotoxic CD8+ T cells in HIV-1 patients (38). Therefore, memory CD4+ T cells expressing PD-1 molecules are closely related to HIV-1 latency, and these immunoregulatory molecules may be ideal targets to eliminate latent HIV-1 reservoirs. These findings showing that PD-1+CD4+ T cells harbor more replication-competent proviruses than PD-1−CD4+ T cells are consistent with recent reports demonstrating a correlation between PD-1 expression and HIV latency (35, 41–43). However, the mechanism involved in the enrichment of proviruses in this cell subset has not been clarified.
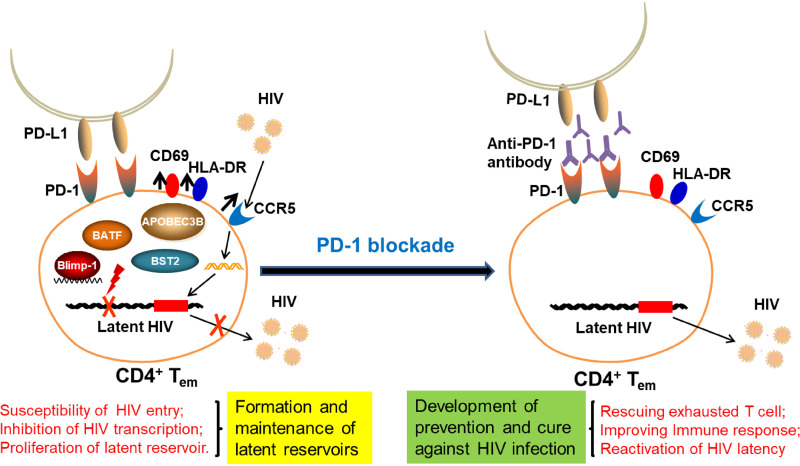
Based on the in vitro and in vivo findings in this study, we propose a pattern to illustrate the control of HIV/SIV infection by modulating the immune inhibitory pathway (Fig. 7). We found that PD-1+CD4+ T cells exhibited a higher level of CCR5 and markers of activation (CD69, HLA-DR), indicating that this subset might be more susceptible to HIV/SIV entry (21, 44). We also observed that Blimp-1 was highly expressed in PD-1+CD4+ T cells, which might inhibit transcription of the latent HIV/SIV promoter. A recent study has reported that Blimp-1 is an HIV-1 provirus transcriptional repressor in memory CD4+ T cells (25). In addition, a high level of Blimp-1 has been correlated with increased PD-1, CTLA-4, and CD160 expression in chronic HIV-1 infection (45). Moreover, these subsets exhibited potent proliferation with a higher level of Ki-67, which might facilitate the turnover and maintenance of the latent provirus. As a result, the PD-1+CD4+ T subsets might be preferentially enriched and form latent HIV/SIV reservoirs. Furthermore, when blocking PD-1 signaling in CD4+ T cells in SIV-infected macaques, these cells exhibited downregulation of Blimp-1 expression, causing transcription and reactivation of the latent provirus. Thus, Blimp-1 might be a key molecule regulating the latency of PD-1+CD4+ T cells and might serve as a target for reactivation of latent HIV/SIV. Further studies are needed to determine the key factor modulating viral latency by PD-1 blockade.
FIG 7.
Pattern to modulate the HIV/SIV latency by regulating immune inhibitory pathway.
One limitation of this pilot study is the relatively small numbers of macaques, and further studies involving more animals are needed to verify our findings. Nevertheless, these macaques were assigned carefully based on age, sex, and weight to minimize potential individual differences. Of note, the macaques included in this study had been infected with SIVmac239 for more than 6 years, and the plasma viral load ranged from 4.4 to 7.5 log when this study was initiated (Table S1). These animals had also experienced long-term ART and, thus, provided a good model to mimic the status of chronically infected HIV-1 patients.
Taken together, our findings demonstrated that PD-1 blockade might be a double-edged sword for HIV-1 immunotherapy, including immune modulation and viral latency reactivation. Of note, PD-1 blockade exacerbated viral rebound and AIDS progression in chronically SIV-infected macaques after ART interruption, and future studies are needed to test whether an anti-PD-1 antibody might be developed as a component of the shock-and-kill strategy. For instance, repeated courses of PD-1 blockade in combination with long-term ART and other therapeutic strategies might progressively decrease the size of the viral reservoir and eventually achieve a functional cure in chronically infected HIV-1 patients. This study provides important insights for the rational design of therapeutic strategies against HIV infection and other related diseases.
MATERIALS AND METHODS
Antibodies, peptides, viruses, and therapeutic vaccines.
Genor Biopharma Co., Ltd. (Pudong, Shanghai, China) kindly provided the PD-1 monoclonal antibody (GB226), which has been approved for clinical trials by both the China Food and Drug Administration (2016L10520) and Australia Therapeutic Goods Administration (NCT03053466).
The NIH HIV Reagent Program generously sent us SIVmac239-specific peptides as a gift. Peptides covering the entire amino acid sequences of SIV Gag, Pol, Env, Nef, Vif, Vpx, Vpr, Rev, and Tat were dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 0.4 mg per peptide per mL and used according to our previously published methods (46, 47).
We constructed replication-defective adenoviral vectors carrying the SIVmac239 Gag, Pol, and Env genes (Ad-SIVgpe) as described in our previously published methods (14). The cytomegalovirus (CMV) promoter was used to drive the transgenes in this recombinant adenoviral vector. These adenovirus vectored SIV vaccines were used as therapeutic vaccines in this study as previously described (14).
Peripheral blood mononuclear cells (PBMCs) of rhesus macaques were used to amplify and titrate the challenge virus of the SIVmac239 stock.
Assays for evaluating cellular immune responses.
Cellular immune assays, including IFN-γ ELISPOT, which represents total cellular immune responses, multicolor intracellular cytokine staining (ICS) showing the polyfunctionality of T lymphocytes, and carboxyfluorescein diacetate succinimidyl ester (CFSE) staining for ex vivo T cell proliferation, were conducted as previously described (46, 47). In brief, for the IFN-γ ELISPOT, freshly isolated PBMCs were added at 4 × 105 cells/well to 96-well plates containing immobilon-P membrane (Millipore) precoated with anti-monkey IFN-γ monoclonal antibody (BD Pharmingen). SIV peptide pools were added to stimulate cells for 20 to 24 h (h), and then a polyclonal anti-monkey IFN-γ biotinylated detector antibody (BD Pharmingen) was added. The next day, the plates were washed, and color was developed by incubation in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Pierce) for 10 min. Spots were counted using an ELISPOT reader (Bioreader 4000, BIOSYS, Germany), and data were reported as the number of spot-forming cells (SFCs) per million PBMCs. Concanavalin A stimulation was used as a positive-control in this assay.
For multicolor ICS (intracellular cytokine staining) assays, SIV peptides were added to stimulate one million PBMCs for 2 h, and then brefeldin A (BD Biosciences) was added for an additional 16 h. Subsequently, the cells were washed and stained for 30 min with anti-CD3-Pacific Blue (BD Biosciences), anti-CD4-phycoerythrin (PE)-CF594 (BD Biosciences), anti-CD8-allophycocyanin (APC)-Cy7 (BD Biosciences), anti-CD28-fluorescein isothiocyanate (FITC) (BD Biosciences), and anti-CD95-PE-Cy5 (BD Biosciences) antibodies. Next, the cells were suspended in 250 μL of Cytofix/Cytoperm solution (BD Pharmingen) for 20 min, washed with Perm/Wash solution (BD Pharmingen), and intracellularly stained with anti-IFN-γ-PE (BD Biosciences), anti-TNF-α-PE-Cy7 (BD Biosciences) and anti-IL-2-APC (BD Biosciences) for 30 min. Samples were analyzed with a BD LSRFortessaTM (BD Biosciences) instrument and FlowJo software (Tree Star, Inc.). Phorbol myristate acetate/ionomycin (PMA/I) stimulation was used as a positive-control in this assay. The antibodies used for analytical flow cytometry are listed in Table S2.
For the CFSE staining assay, PBMCs were incubated with 0.25 μM CFSE (Molecular Probes) at 37°C for 10 min and then stimulated with SIV peptides. Cells were harvested and stained with anti-CD3-Pacific Blue (BD Biosciences), anti-CD4-APC (BD Biosciences), and anti-CD8-APC-Cy7 (BD Biosciences) on day 6. Samples were analyzed with a BD LSRFortessaTM (BD Biosciences) instrument and FlowJo software (Tree Star, Inc.). The fraction of antigen-specific proliferating cells in response to antigen was calculated by subtracting the proportion of proliferating cells in unstimulated samples. Phorbol 12-myristate 13-acetate and ionomycin (PMA/I) stimulation was used as a positive-control in this assay.
For determination of T cell counts, the numbers of circulating CD4+ and CD8+ T lymphocytes were ascertained by Becton, Dickinson Trucount tubes. Samples were analyzed with Accuri C6 Plus (BD Biosciences) and FlowJo software (Tree Star, Inc.).
Assays for viral reactivation and phenotypic characteristic determination.
For viral reactivation by PD-1 blockade, CD4+ T cells were sorted using a nonhuman primate CD4+ T cell isolation kit (Miltenyi Biotec) following the standard magnetic cell sorting protocol. The purified CD4+ T cells were counted and suspended in 200 μL of medium (Roswell Park Memorial Institute (RPMI) 1640 with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin) in a 96-well U-bottom plate. Each well had 0.5 to 2 million CD4+ T cells. After stimulation with different compounds for 72 h, total cellular RNA was isolated using an EastepTM Super Total RNA Extraction kit (Promega) and eluted into 100 μL of RNase-free water. Aliquots of 10 μL RNA were directly reverse-transcribed following the standard procedure (BIO-RAD, transcription supermix for reverse transcription-quantitative real-time PCR [RT–qPCR]). Cell-associated complementary DNA (cDNA) or serial dilutions of cDNA standards were then subjected to two rounds of PCR amplification. The SIV viral multiply spliced transcripts (msRNA) were determined using three primers spanning Tat and Rev for seminested real-time PCR. The amplicon sizes were 130 bp for the first-round PCR and 103 bp for the second-round real-time PCR. The SIV viral unspliced transcripts (usRNA) were determined using two pairs of primers specific to a region within the SIV gag gene. The amplicon sizes were 227 bp for the first-round PCR and 176 bp for the second-round real-time PCR. The sequences of all primers used in this study are listed in Table S3.
For the phenotypic characteristic determination assay, CD4+ T cells were sorted and purified as described above, counted, and suspended in 200 μL of medium (RPMI 1640 with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin) in a 96-well U-bottom plate. Each well contained 0.5 to 2 million CD4+ T cells. After 72 h of stimulation with different compounds, the cells were stained with anti-CD3-Pacific Blue (BD Biosciences), anti-CD4-FITC (BD Biosciences), anti-CCR5-PE (BD Biosciences), and anti-Blimp-1-PE (BD Biosciences), and they were analyzed with a BD LSRFortessaTM (BD Biosciences) instrument using FlowJo software (Tree Star, Inc.). PMA/I (phorbol myristate acetate/ionomycin) stimulation was used as a positive-control in this assay.
Animals and ethics statement.
Chinese rhesus macaques (Macaca mulatta) were housed in the experimental animal center of Guangzhou Institutes of Biomedicine and Health (GIBH, Guangzhou, China). This study was carried out in accordance with the “Regulations for the Administration of Affairs Concerning Experimental Animals” by the State Council of the People's Republic of China, and the protocol was approved by the Institutional Animal Care and Use Committee of GIBH (IACUC permit number: 2019004). All procedures were performed by trained personnel under the supervision of veterinarians. Twelve monkeys chronically infected with SIVmac239 were used for in vivo experiments in three settings: PD-1 blockade combined with a vaccine, vaccine, and ART alone. The overall vaccination and challenge schedule is shown in Fig. 1. Briefly, 12 monkeys (5 to 15 years of age) weighing 4 to 10 kg were evenly assigned into three groups: (i) eight monkeys received PBS as the vaccine group and ART group, and four of them received rAd5-SIVgpe (1011 viral particle (vp) in 1 mL PBS buffer) through intramuscular injection at week 0 and week 4; (ii) four monkeys received genolimzumab injection (20 mg/kg) through intravenous injection every 2 weeks and rAd5-SIVgpe (1011 vp in 1 mL PBS buffer) through intramuscular injection at week 0 and week 4. At week −3 before the initial vaccination, animals were treated subcutaneously with ART as we previously described (31, 48). Sequential peripheral blood samples were collected to evaluate virological and immunological responses following the experimental schedule in Fig. 1A.
SIV viral RNA and DNA copy assays.
For SIV RNA quantification, we collected plasma samples according to standard protocols. As described previously, we used the QIAamp Viral RNA Minikit (Qiagen) to extract viral RNA from plasma and the QuantiTect SYBR green RT–PCR kit (Qiagen) to quantify viral RNA (10). The detection limits of this assay were 100 copies/mL plasma.
For SIV DNA quantification, different cell types were sorted from PBMCs using a FACS Aria IIU instrument. Then, the total cellular DNA was extracted from approximately 0.5 to 5 million cells using a QIAamp DNA Blood Minikit (Qiagen). PCR assays were performed with 200 ng samples of DNA, and the SIV viral DNA was determined using a pair of primers specific to a conserved region of the SIV gag gene as previously described (49). Quantitation was analyzed by comparison to the standard curve of SIV gag copies, and the β-actin gene was simultaneously amplified as a reference for input cell counts.
RNA sequencing (RNA-seq)-based transcriptional profiling experiments.
RNA-seq experiments were performed as previously described (50). In brief, a total amount of 1 μg RNA per sample was used to prepare cDNA libraries with the NEBNext UltraTM RNA Library Prep kit for Illumina (New England BioLabs). The quality and integrity of the tagged libraries were initially assessed on the Agilent Bioanalyzer 2100 system. The libraries were normalized, pooled, and then clustered using TruSeq PE Cluster kit v3-cBot-HS (Illumina). They were then imaged and sequenced on an Illumina NovaSeq platform, and 150-bp paired-end reads were generated. Raw data (raw reads) in fastq format were first processed using an in-house script. Clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from raw data. All downstream analyses were based on clean data of high quality. The filtered reads were aligned to the Macaca mulatta genome Ensembl GCA008058575.1_rheMacS_1.0 using Hisat2 v2.0.5. The mapped and aligned reads were quantified to obtain the gene-level counts using StringTie (v1.3.3b). Raw counts were processed using featureCounts v1.5.0-p3, and then the FPKM of each gene was calculated based on the length of the gene and the read count mapped to this gene. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1). The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P value <0.05 found by DESeq2 were assigned as differentially expressed.
Data analysis.
The flow cytometry data were analyzed using the Flowjo software, and graphical presentations were computed with the GraphPrism 7 software (GraphPad Software Inc., La Jolla, CA). The survival curves were analyzed by log-rank (Mantel-Cox) tests. Two-tailed P values were calculated, and differences were considered statistically significant when P < 0.05.
ACKNOWLEDGMENTS
We thank Yichu Liu, Xiangjie Feng, and Xuehua Zheng for their technical assistance with the monkey experiment. We appreciate Genor Biopharma for providing the PD-1 antibody, and the NIH HIV Reagent Program for providing the SIV peptide pools.
This work was supported by the National Natural Science Foundation of China (81971927 and 32000124), the National Science and Technology Major Project of China (2018ZX10731101-002), the Science and Technology Planning Project of Shenzhen City (20190804095916056, JCYJ20200109142601702, JSGG20200225152008136), the Natural Science Foundation of Guangdong Province (2019A1515110458, 2021A1515010456), the Municipal Health and Medical Cooperation Innovation Major Project of Guangzhou City (201704020219, 201803040002), and the China Postdoctoral Science Foundation (2019M663140, 2020T130150ZX). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Project design and supervision were done by C.S., L.C., P.L. Literature collection, original draft preparation, and rewriting were done by C.W., C.S., P.L. Y.H., J.Z. Experiments were performed by C.W., P.L., Y.H., J.Z., Y.Z., F.F., M.L. Data analysis was done by C.W., P.L., Y.H., J.Z., K.L., Y.C., Z.W., C.W., Z.H. Materials and reagents were contributed by L.C., G.L. All authors have read and agreed to the final version of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Pingchao Li, Email: li_pingchao@gibh.ac.cn.
Ling Chen, Email: chen_ling@gibh.ac.cn.
Caijun Sun, Email: suncaijun@mail.sysu.edu.cn.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 2.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, Chung YS, Penugonda S, Chipman J, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. 2016. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530:51–56. 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olesen R, Leth S, Nymann R, Østergaard L, Søgaard OS, Denton PW, Tolstrup M. 2016. Immune checkpoints and the HIV-1 reservoir: proceed with caution. J Virus Erad 2:183–186. 10.1016/S2055-6640(20)30463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porichis F, Kaufmann DE. 2012. Role of PD-1 in HIV pathogenesis and as target for therapy. Curr HIV/AIDS Rep 9:81–90. 10.1007/s11904-011-0106-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350–354. 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 6.Kaufmann DE, Kavanagh DG, Pereyra F, Zaunders JJ, Mackey EW, Miura T, Palmer S, Brockman M, Rathod A, Piechocka-Trocha A, Baker B, Zhu B, Le Gall S, Waring MT, Ahern R, Moss K, Kelleher AD, Coffin JM, Freeman GJ, Rosenberg ES, Walker BD. 2007. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol 8:1246–1254. 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- 7.Tian X, Zhang A, Qiu C, Wang W, Yang Y, Qiu C, Liu A, Zhu L, Yuan S, Hu H, Wang W, Wei Q, Zhang X, Xu J. 2015. The upregulation of LAG-3 on T cells defines a subpopulation with functional exhaustion and correlates with disease progression in HIV-infected subjects. J Immunol 194:3873–3882. 10.4049/jimmunol.1402176. [DOI] [PubMed] [Google Scholar]
- 8.Davar D, Wilson M, Pruckner C, Kirkwood JM. 2015. PD-1 blockade in advanced melanoma in patients with hepatitis C and/or HIV. Case Rep Oncol Med 2015:737389. 10.1155/2015/737389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guihot A, Marcelin AG, Massiani MA, Samri A, Soulié C, Autran B, Spano JP. 2018. Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann Oncol 29:517–518. 10.1093/annonc/mdx696. [DOI] [PubMed] [Google Scholar]
- 10.Pan E, Feng F, Li P, Yang Q, Ma X, Wu C, Zhao J, Yan H, Chen R, Chen L, Sun C. 2018. Immune protection of SIV challenge by PD-1 blockade during vaccination in Rhesus monkeys. Front Immunol 9:2415. 10.3389/fimmu.2018.02415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, Vanderford TH, Chennareddi L, Silvestri G, Freeman GJ, Ahmed R, Amara RR. 2009. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature 458:206–210. 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mylvaganam GH, Chea LS, Tharp GK, Hicks S, Velu V, Iyer SS, Deleage C, Estes JD, Bosinger SE, Freeman GJ, Ahmed R, Amara RR. 2018. Combination anti-PD-1 and antiretroviral therapy provides therapeutic benefit against SIV. JCI Insight 3. 10.1172/jci.insight.122940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman SA, Yagnik B, Bally AP, Morrow KN, Wang S, Vanderford TH, Freeman GJ, Ahmed R, Amara RR. 2021. PD-1 blockade and vaccination provide therapeutic benefit against SIV by inducing broad and functional CD8(+) T cells in lymphoid tissue. Sci Immunol 6:eabh3034. 10.1126/sciimmunol.abh3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C, Feng L, Zhang Y, Xiao L, Pan W, Li C, Zhang L, Chen L. 2012. Circumventing antivector immunity by using adenovirus-infected blood cells for repeated application of adenovirus-vectored vaccines: proof of concept in rhesus macaques. J Virol 86:11031–11042. 10.1128/JVI.00783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam S, Shimizu N, Hoque SA, Jinno-Oue A, Tanaka A, Hoshino H. 2013. CCR6 functions as a new coreceptor for limited primary human and simian immunodeficiency viruses. PLoS One 8:e73116. 10.1371/journal.pone.0073116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tardif MR, Tremblay MJ. 2003. Presence of host ICAM-1 in human immunodeficiency virus type 1 virions increases productive infection of CD4+ T lymphocytes by favoring cytosolic delivery of viral material. J Virol 77:12299–12309. 10.1128/jvi.77.22.12299-12309.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poggi A, Carosio R, Fenoglio D, Brenci S, Murdaca G, Setti M, Indiveri F, Scabini S, Ferrero E, Zocchi MR. 2004. Migration of V delta 1 and V delta 2 T cells in response to CXCR3 and CXCR4 ligands in healthy donors and HIV-1-infected patients: competition by HIV-1 Tat. Blood 103:2205–2213. 10.1182/blood-2003-08-2928. [DOI] [PubMed] [Google Scholar]
- 18.He FQ, Sauermann U, Beer C, Winkelmann S, Yu Z, Sopper S, Zeng AP, Wirth M. 2014. Identification of molecular sub-networks associated with cell survival in a chronically SIVmac-infected human CD4+ T cell line. Virol J 11:152. 10.1186/1743-422X-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samson M, Edinger AL, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms RW, Parmentier M. 1998. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol 28:1689–1700. . [DOI] [PubMed] [Google Scholar]
- 20.Iyengar S, Hildreth JE, Schwartz DH. 1998. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol 72:5251–5255. 10.1128/JVI.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zack JA, Kim SG, Vatakis DN. 2013. HIV restriction in quiescent CD4+ T cells. Retrovirology 10:37. 10.1186/1742-4690-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, Jones KA. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev 9:2090–2104. 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 23.Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J 20:2536–2544. 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colin L, Van Lint C. 2009. Molecular control of HIV-1 postintegration latency: implications for the development of new therapeutic strategies. Retrovirology 6:111. 10.1186/1742-4690-6-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaczmarek Michaels K, Natarajan M, Euler Z, Alter G, Viglianti G, Henderson AJ. 2015. Blimp-1, an intrinsic factor that represses HIV-1 proviral transcription in memory CD4+ T cells. J Immunol 194:3267–3274. 10.4049/jimmunol.1402581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Z, Liu R, Zhang H, Zhang S, Hu X, Tan J, Liang C, Qiao W. 2016. GADD45 proteins inhibit HIV-1 replication through specific suppression of HIV-1 transcription. Virology 493:1–11. 10.1016/j.virol.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 28.Yu Q, Chen D, König R, Mariani R, Unutmaz D, Landau NR. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem 279:53379–53386. 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 29.Juríková M, Danihel Ľ, Polák Š, Varga I. 2016. Ki67, PCNA, and MCM proteins: markers of proliferation in the diagnosis of breast cancer. Acta Histochem 118:544–552. 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Xouri G, Lygerou Z, Nishitani H, Pachnis V, Nurse P, Taraviras S. 2004. Cdt1 and geminin are down-regulated upon cell cycle exit and are over-expressed in cancer-derived cell lines. Eur J Biochem 271:3368–3378. 10.1111/j.1432-1033.2004.04271.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang Q, Feng F, Li P, Pan E, Wu C, He Y, Zhang F, Zhao J, Li R, Feng L, Hu F, Li L, Zou H, Cai W, Lehner T, Sun C, Chen L. 2019. Arsenic Trioxide Impacts Viral Latency and Delays Viral Rebound after Termination of ART in Chronically SIV-Infected Macaques. Adv Sci (Weinh) 6:1900319. 10.1002/advs.201900319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. 2011. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117:2791–2799. 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Zhao N, Berkhout B, Das AT. 2016. A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep 17:2819–2826. 10.1016/j.celrep.2016.11.057. [DOI] [PubMed] [Google Scholar]
- 34.Kessing CF, Nixon CC, Li C, Tsai P, Takata H, Mousseau G, Ho PT, Honeycutt JB, Fallahi M, Trautmann L, Garcia JV, Valente ST. 2017. In vivo suppression of HIV rebound by didehydro-cortistatin A, a “block-and-lock” strategy for HIV-1 treatment. Cell Rep 21:600–611. 10.1016/j.celrep.2017.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, Palmer S, Fromentin R, Chomont N, Sékaly RP, Cameron PU, Lewin SR. 2018. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 32:1491–1497. 10.1097/QAD.0000000000001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wightman F, Solomon A, Kumar SS, Urriola N, Gallagher K, Hiener B, Palmer S, McNeil C, Garsia R, Lewin SR. 2015. Effect of ipilimumab on the HIV reservoir in an HIV-infected individual with metastatic melanoma. AIDS 29:504–506. 10.1097/QAD.0000000000000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hryniewicz A, Boasso A, Edghill-Smith Y, Vaccari M, Fuchs D, Venzon D, Nacsa J, Betts MR, Tsai WP, Heraud JM, Beer B, Blanset D, Chougnet C, Lowy I, Shearer GM, Franchini G. 2006. CTLA-4 blockade decreases TGF-beta, IDO, and viral RNA expression in tissues of SIVmac251-infected macaques. Blood 108:3834–3842. 10.1182/blood-2006-04-010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, Truong R, Macedo AB, Bosque A, Kovacs C, Benko E, Piechocka-Trocha A, Wong H, Jeng E, Nixon DF, Ho YC, Siliciano RF, Walker BD, Jones RB. 2018. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 128:876–889. 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noto A, Procopio FA, Banga R, Suffiotti M, Corpataux JM, Cavassini M, Riva A, Fenwick C, Gottardo R, Perreau M, Pantaleo G. 2018. CD32(+) and PD-1(+) lymph node CD4 T Cells support persistent HIV-1 transcription in treated aviremic individuals. J Virol 92:e00901-18. 10.1128/JVI.00901-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. 2016. PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 22:754–761. 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- 42.Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, Hoh R, Deeks SG, Hazuda DJ, Lewin SR, Routy JP, Sékaly RP, Chomont N. 2019. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat Commun 10:814. 10.1038/s41467-019-08798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sékaly RP, Chomont N. 2016. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 12:e1005761. 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorry PR, Ancuta P. 2011. Coreceptors and HIV-1 pathogenesis. Curr HIV/AIDS Rep 8:45–53. 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- 45.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol 10:29–37. 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C, Zhang L, Zhang M, Liu Y, Zhong M, Ma X, Chen L. 2010. Induction of balance and breadth in the immune response is beneficial for the control of SIVmac239 replication in rhesus monkeys. J Infect 60:371–381. 10.1016/j.jinf.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Sun C, Chen Z, Tang X, Zhang Y, Feng L, Du Y, Xiao L, Liu L, Zhu W, Chen L, Zhang L. 2013. Mucosal priming with a replicating-vaccinia virus-based vaccine elicits protective immunity to simian immunodeficiency virus challenge in rhesus monkeys. J Virol 87:5669–5677. 10.1128/JVI.03247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Zhao J, Li R, Feng F, He Y, Li Y, Huang R, Li G, Yang H, Cheng G, Chen L, Ma F, Li P, Sun C. 2021. Modulation of antiviral immunity and therapeutic efficacy by 25-hydroxycholesterol in chronically SIV-Infected, ART-treated rhesus macaques. Virol Sin 1–13. 10.1007/s12250-021-00407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, Shields J, Parenteau L, Whitney JB, Abbink P, Ng'ang'a DM, Seaman MS, Lavine CL, Perry JR, Li W, Colantonio AD, Lewis MG, Chen B, Wenschuh H, Reimer U, Piatak M, Lifson JD, Handley SA, Virgin HW, Koutsoukos M, Lorin C, Voss G, Weijtens M, Pau MG, Schuitemaker H. 2015. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science 349:320–324. 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng F, Hao H, Zhao J, Li Y, Zhang Y, Li R, Wen Z, Wu C, Li M, Li P, Chen L, Tang R, Wang X, Sun C. 2021. Shell-mediated phagocytosis to reshape viral-vectored vaccine-induced immunity. Biomaterials 276:121062. 10.1016/j.biomaterials.2021.121062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3; Fig. S1<br>. Download jvi.01785-21-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)