Abstract
Unlike all other biological molecules that are degraded and replaced if damaged, DNA must be repaired as chromosomes cannot be replaced. Indeed, DNA endures a wide variety of structural damage that need to be repaired accurately to maintain genomic stability and proper functioning of cells and to prevent mutation leading to disease. Given that the genome is packaged into chromatin within eukaryotic cells, it has become increasingly evident that the chromatin context of DNA both facilitates and regulates DNA repair processes. In this review, we discuss mechanisms involved in removal of histones (chromatin disassembly) from around DNA lesions, by histone chaperones and chromatin remodelers, that promotes accessibility of the DNA repair machinery. We also elaborate on how the deposition of core histones and specific histone variants onto DNA (chromatin assembly) during DNA repair promotes repair processes, the role of histone post translational modifications in these processes and how chromatin structure is reestablished after DNA repair is complete.
1. Introduction
The packaging of eukaryotic nuclear genomes together with histone proteins into the structure known as chromatin was long considered an inconvenient hurdle for nuclear processes to navigate. Chromatin comprises an array of nucleosomes which have approximately 1.75 turns of DNA around the outside of an octamer of two molecules of each core histone, H2A, H2B, H3 and H4, and this compaction of the DNA into chromatin limits accessibility to the DNA. Although the structure of the nucleosome has been known for over 20 years [1], we have only recently begun to understand how the packaging of the genome into nucleosomes facilitates and tightly regulates DNA repair. A variety of changes in the chromatin accompany and regulate multiple DNA repair processes. Here we review what we consider to be the ultimate way to provide access to the DNA template to promote DNA repair, which is disassembly of nucleosomes involving removal of histones from the DNA by histone chaperones. Histone chaperones are key players in histone metabolism involved in escorting histones and mobilizing them in and out of chromatin [2,3]. The list of known histone chaperones has been growing significantly, but only a few have been associated with the DNA damage response [1,2]. During chromatin disassembly and assembly by histone chaperones, they are assisted by ATP-dependent nucleosome remodelers which use energy of ATP hydrolysis to evict / exchange histones [3]. Other chromatin alterations also play an important role in facilitating DNA repair that will only be covered here where they impinge on chromatin disassembly / reassembly and have been reviewed extensively elsewhere. These include histone post-translational modifications (PTMs) to recruit repair proteins and to modulate chromatin compaction [4], nucleosome remodeling to slide histone octamers out of the way of the DNA lesion [5], and incorporation of histone variants [6]. Following such chromatin changes that facilitate repair processes, it is important that the chromatin is restored to its original pre-damage configuration after repair is completed so that the epigenetic information can be reinstated, and normal cellular processes can resume. This is achieved in part by chromatin assembly and will also be discussed here.
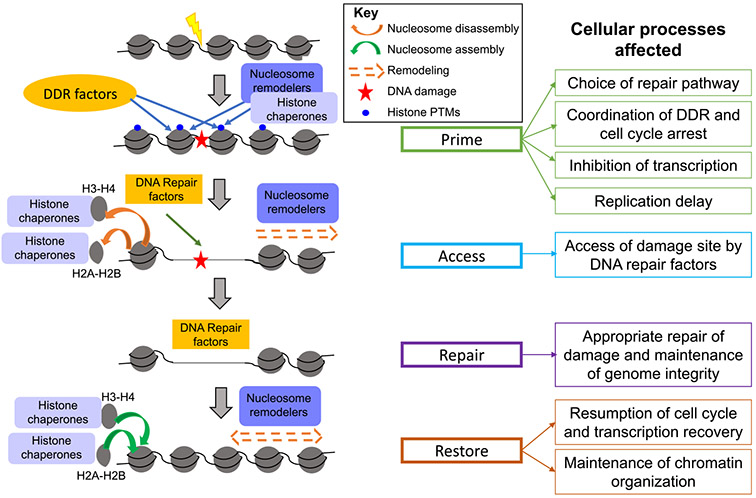
2. The “prime-access-repair-restore” model
One of the major constraints that DNA repair pathways encounter is the organization of the genome into chromatin which causes DNA lesions to be buried within a relatively inaccessible nucleoprotein structure. Repair factors require access to sites of damage to successfully perform their roles, and one way in which cells address this issue is by temporarily evicting histones from the proximity of the damage, by chromatin disassembly, to allow DNA repair proteins to localize to the lesion and perform their repair functions. After repair, and equally important, the nucleosomes are reassembled to restore the original chromatin state. Initially, the “access-repair-restore” model was put forth to explain this cellular strategy of temporarily removing the chromatin obstruction near damage sites to facilitate repair processes and then ultimately restoring the original chromatin conformation [7-9]. This preliminary model then went on to be modified into the “prime-access-repair-restore” model as roles and interactions of the chromatin with repair factors became better understood [1,10] (Fig. 1). Importantly, this modified model incorporated the idea that histones and nucleosomes do not act merely as obstacles to DNA repair but play important roles by “priming” the repair pathways in multiple ways, such as recruiting specific proteins and imposing regulatory constraints, and consequently influencing the choice and efficiency of downstream repair processes. The “access” step is then mediated by histone chaperones and ATP-dependent nucleosome remodelers that create a local “clearance” by disassembling nucleosomes or sliding the histone octamer away from the DNA lesion, which then allows repair factors to localize to these sites and perform their functions. Finally, the “restore” step occurs in which chromatin organization is returned to its original state and is mediated by histone chaperones that deposit histones post-repair. Below we discuss different chromatin disassembly and reassembly strategies that cells use and their mechanisms involved in various DNA repair pathways.
Fig. 1.
The Prime-Access-Repair model. The Prime-Access-Repair-Restore model explains how DNA is repaired in a chromatin context, as explained in the text. Only a subset of the histone PTMs are represented (those involved in recruitment on DDR factors, histone chaperones and chromatin remodelers).
3. Nucleosome disassembly and reassembly during DNA repair
3.1. Homologous recombination
One of the major pathways that cells use to repair double strand breaks (DSBs) in the DNA is homologous recombination (HR), where breaks are repaired using a homologous DNA template. DSBs are one of the most deleterious kinds of damage that DNA can undergo, which can then lead to mutations or genome rearrangements if the breaks are repaired improperly and cell death if they are left unrepaired. Early steps of HR involve resecting one of the strands of DNA on either side of the DSB in the 5′ to 3′ direction. Resection is initiated by the Mre11-Rad50-Xrs2 (MRX) complex and Sae2 in yeast creating short 3′ overhangs, and long-range resection is then carried out by the Exo1 and Dna2 nucleases, the latter’s activity being promoted by the Sgs1-Top3-Rmi1 (STR) helicase-topoisomerase complex [11,12]. The 3′ single-strand DNA (ssDNA) generated during resection is protected from degradation by binding of Replication Protein A (RPA), which is then displaced by Rad51, with the aid of yeast Rad52, from the 3′ ends of the ssDNA. This leads to a strand invasion process in which the Rad51 nucleoprotein filament invades a stretch of homologous double strand DNA (dsDNA) by displacing the complementary strand to form a displacement loop (D-loop). DNA synthesis mediated by polymerases and the replication processivity clamp PCNA then takes place at the 3′ end of the invading strand using the homologous donor strand as a template, followed by resolution of the interlinked DNA joint molecules. There exist multiple sub-pathways of HR such as the canonical double strand break repair (DSBR) pathway, the synthesis-dependent strand annealing (SDSA) pathway and the single strand annealing (SSA) pathway. For detailed reviews on mechanisms of HR and its sub-pathways, see [13,14].
3.2. Nucleosome disassembly during HR
Chromatin dynamics at the site of the DSB appears to play critical roles in facilitating the repair via HR. Immediately after the formation of a DSB, a variety of chromatin modifications followed by reorganization and disassembly of nucleosomes occurs in the vicinity of the break to facilitate and provide access to repair factors. Early studies in S. cerevisiae showed that histone levels decrease in the vicinity of a DSB as inferred from chromatin immunoprecipitation (ChIP) experiments followed by quantitative PCR (qPCR) of DNA regions surrounding the DNA break [15-17]. These studies also showed increased susceptibility of DNA near a DSB to digestion by micrococcal nuclease, and taken together, indicated compromised nucleosome integrity and likely histone eviction near a DNA break. Similar eviction of histones surrounding a DSB has been observed in mammalian systems and was found to be dependent on the Mre11-Rad50-Nbs1 (MRN) complex (MRX in yeast) and the DNA damage response ataxia-telangiectasia (ATM) kinase [18,19]. Consistent with earlier work, a more recent study in budding yeast performing MNase digestion followed by qPCR showed that nucleosome occupancy around a break is reduced by more than 75% due to 5′ to 3′ resection at the break [20]. Moreover, a high resolution MNase-seq genome mapping in yeast demonstrated that nucleosomes in the immediate vicinity (~200 bp) of an inducible site-specific DSB were removed rapidly, followed by a more gradual reorganization and eviction of nucleosomes up to ~8 kb on either side of the break in an MRX-dependent manner [21]. In agreement with the above studies, recent work in budding yeast showed using strand-specific ChIP-seq that ssDNA binding proteins such as RPA, but not histones, are detected near an induced site-specific DSB in a strand-specific manner, pointing to a model where histones are evicted near a DSB and are not found as a persistent species on resected ssDNA [22]. Analogous to nucleosome dynamics in DSB repair in somatic cells, nucleosomes surrounding DSBs generated during meiosis in yeast have also been shown to be disassembled after programmed breaks are formed across the genome [23]. Finally, although relatively underexplored, linker histones appear to be evicted from the vicinity of DSBs as well. A study in human cell lines showed that the linker histone isoform H1.2 suppresses ATM activity, protecting chromatin from aberrant ATM loading and activation, and upon DNA damage, undergoes rapid PARP1-dependent chromatin dissociation through poly-ADP-ribosylation (PARylation) of its C terminus and further proteasomal degradation to permit full activation of ATM [24]. This is consistent with earlier work in budding yeast which showed that the linker histone H1 inhibits HR and its depletion increased resistance to DNA damaging drugs [25]. A similar study in mouse embryonic cells also showed that depletion of the linker histone H1 caused hyper-resistance to DNA damage by increased activation of damage related signaling pathways and checkpoints [26]. Taken together, these studies demonstrate using various techniques that histones are evicted during HR from the vicinity of the DNA break.
The precise mechanisms and players involved in nucleosome disassembly in the vicinity of a break and their coordination and crosstalk with repair processes are not fully understood. The use of site specific nucleases such as the HO endonuclease in budding yeast and the I-PpoI and AsiSI nucleases in mammalian cells has enabled histone occupancy to be measured around DSBs by ChIP analysis, greatly enhancing the ability to detect and study chromatin disassembly and assembly around DSBs [18,27,28]. Histone removal appears to be closely intertwined with DNA end resection. Multiple studies in yeast and mammalian cells have shown that components of the yeast MRX (or MRN in mammals) complex that initiate short range resection, are also required for the eviction of histones surrounding a break [15,18,19,29] (Fig. 2). Various studies have also shown roles for ATP-dependent nucleosome remodelers that mediate DNA end resection also promote chromatin disassembly. For example, nucleosome remodelers such as Fun30 and INO80 in yeast that are required to facilitate DNA end resection, also facilitate removal of histone H3 from around DSBs undergoing HR [19,30-32] (Fig. 2). Due to its central position in the nucleosome if H3 is removed by default the entire nucleosome is disassembled. Additionally, knocking down either the p400 ATPase, which belongs to the INO80 family of ATP-dependent nucleosome remodelers, or Rad51, with which it interacts to form a complex, leads to a decrease in histone loss around DSBs in human cells [33]. Micrococcal nuclease mapping of nucleosomes around the yeast HO lesion revealed that the yeast ATP-dependent nucleosome remodeler RSC also mobilizes nucleosomes to promote HR [16]. In the absence of any of these factors, HR is defective. As such, the evidence for nucleosome mobilization and disassembly during HR, and its requirement for HR, is convincing.
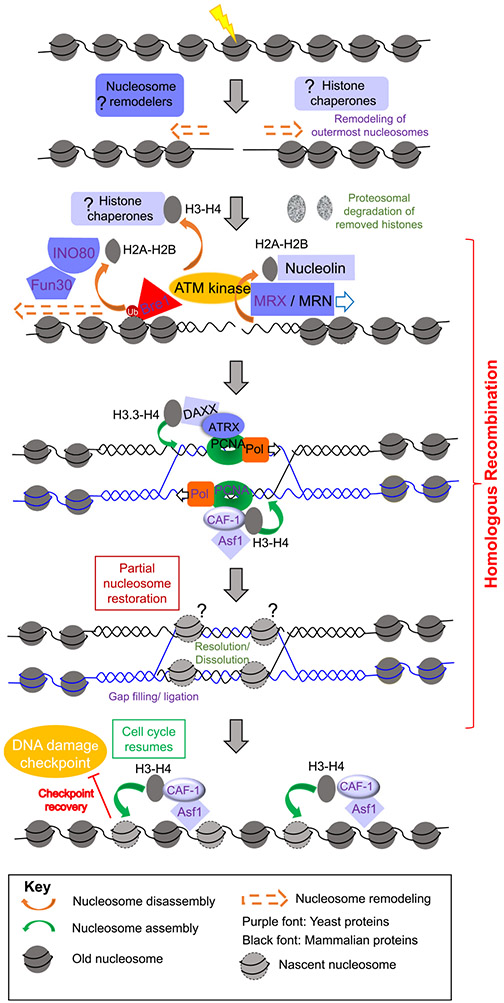
Fig. 2.
Chromatin disassembly and reassembly during homologous recombination. After DSB induction, a single nucleosome at the DSB is disassembled and the two adjacent nucleosomes reposition themselves away from the DSB, prior to DNA end resection. Subsequently, extensive chromatin disassembly around the DSB occurs concomitant with DNA end resection during HR, and the removed histones are degraded. Chromatin also appears to be reassembled during HR to promote various steps during HR, and once HR is complete chromatin is reestablished fully and the cell cycle checkpoint is inactivated via checkpoint recovery. The question marks indicate likely events, but that have not yet been proven to occur. The font color corresponds to either yeast or mammalian systems, as indicated in the key.
One aspect that was not completely clear, however, is if nucleosome removal near a break precedes 5′ to 3′ resection or is a consequence of the latter. In this regard, the studies by Shim et al. indicate a 30 min delay in DNA end resection compared to nucleosome mobilization, [16], while other studies find chromatin disassembly, as detected by ChIP analysis, and end resection to be kinetically inseparable [17]. In vitro studies have shown that resection by S. cerevisiae Sgs1-Dna2 and Exo1 is blocked by nucleosomes, yet Exo1 resects quickly and with high processivity in vivo near meiotic breaks, indicating that nucleosomes are destabilized, or histones are removed prior to end resection [23,34]. Furthermore, while ATP-dependent nucleosome remodelers such as INO80 and Fun30 have been shown to be important for the removal of histones such as H3 and histone variants such as H2AZ from around a DSB in yeast, Fun30 has also been shown to promote resection by both Sgs1-Dna2 and Exo1 pathways and INO80 promotes DNA end resection, raising the possibility that nucleosome remodeling and eviction by Fun30 and INO80 facilitate subsequent end resection [30,35-37]. Consistently a recent study in budding yeast showed using strand-specific ChIP-seq that reduced histone eviction in the absence of RSC and SWI/SNF complexes led to a decrease in resection [22]. The answer to this question was finally provided by a recent high resolution spatiotemporal micrococcal nuclease mapping analysis of nucleosome occupancy and positioning upon induction of an unrepairable HO lesion [38]. This study found that immediately after induction of the DSB, one nucleosome flanking the break disappears while the adjacent nucleosome on either side of the break repositioned themselves further from the DSB in a manner independent of DNA end resection. This is followed by MRX mediated end resection and concomitant disassembly of nucleosomes up to 8 kb away from the DSB. As such, the initial local changes to the chromatin structure at a DSB occur in a manner independent of end resection, but the bulk disassembly of histones around a DSB is dependent on end resection [38] (Fig. 2).
The histone chaperones that mediate chromatin disassembly during HR are less clear. At least for DSBs induced within the rDNA loci, knocking down both isoforms A and B of human chaperone anti-silencing function 1 (ASF1) was shown to attenuate eviction of histones H3-H4 from sites of HR, while knocking down the nucleolar protein nucleolin attenuated eviction of H2A-H2B from sites of HR in the nucleolus [19] (Fig. 2). Other factors that regulate histone eviction during HR include PTMs of histones, such as ubiquitination of H2B by the ubiquitin ligase Bre1, that was shown to be important for its eviction from the vicinity of DSBs in yeast [39]. After eviction of histones from around DSBs, their fate appears to be degradation as treatment of yeast with a radiomimetic drug Zeocin that induces global DSBs, causes a 20–40% loss of total core histone proteins (H2A, H2B, H3, H4) from yeast cells [40]. Levels of the histone variant H2A.Z, however, remained relatively stable. This global histone loss was shown to be from chromatin and was proteasome mediated in addition to being dependent on the DNA damage checkpoint and the INO80 nucleosome remodeler, and functionally enhances chromatin mobility and the homology search [40].
3.3. Chromatin reassembly during HR
While disassembly of nucleosomes is important for allowing access of break sites to repair factors, reassembly of nucleosomes is important for restoring the chromatin conformation of the damage sites to their original state and resumption of the cell cycle. Nucleosome assembly during HR has been shown to be coupled with DNA synthesis and is mediated by histone chaperones such as Chromatin Assembly Factor 1 (CAF-1) in yeast, in a PCNA-interaction dependent manner [41-44] (Fig. 2). Aiming to understand the precise role of such nucleosome reassembly, multiple studies have shown that chromatin assembly mediated by histone chaperones Asf1 and CAF-1 is not required for repairing the DSB per se in budding yeast but is needed for turning off the DNA damage cell cycle checkpoint, termed checkpoint recovery (Fig. 2) [17,45,46]. Acetylation of H3 K56 by Asf1 and the histone acetyl transferase Rtt109, is required to reassemble nucleosomes on repaired DNA and turn off the DNA damage checkpoint in yeast after HR [17]. Furthermore, the role of Asf1 in chromatin assembly in yeast can be bypassed by a mimetic of acetylation of H3 K56, which is presumably because this acetylation increases the ability of Asf1 to transfer histones H3-H4 to CAF-1 which in turn transfers the H3-H4 to newly-synthesized DNA [17,47]. CAF-1 and Asf1 mediated chromatin reassembly after HR in yeast promotes removal of Rad51, and the checkpoint sensors Ddc1 and Ddc2, to enable checkpoint recovery in budding yeast [46].
While all the evidence indicates that ongoing chromatin assembly is not required for HR in budding yeast, chromatin assembly appears to be intrinsically required for HR per se in fission yeast and metazoans. DNA synthesis dependent assembly of nucleosomes in fission yeast have been shown to stabilize joint molecules formed during HR occurring at replication forks and prevent disassembly of D-loops by the RecQ helicase Rqh1 [44,48]. In mammalian cells, chromatin assembly by both ASF1 and CAF-1 are required for Rad51 loading onto ssDNA, and in their absence DNA end resection and RPA coating extends extensively [49]. Another recent study providing evidence of repair synthesis coupled chromatin assembly, showed in mammalian cells that the ATP-dependent nucleosome remodeler ATRX along with the histone chaperone DAXX and PCNA function to deposit the histone variant H3.3 during G2, and in the process facilitated long range DNA synthesis and sister chromatid exchange [50] (Fig. 2). An important question that emerges is how final steps of repair such as joint molecule resolution or dissolution, gap filling, and ligation occur in the presence of nucleosomes that are deposited earlier in a manner coupled to the DNA synthesis step. DNA synthesis coupled nucleosome deposition during HR in yeast was shown to only partially restore the nucleosome occupancy level in a cell cycle independent manner after HR, and further restoration of nucleosome occupancy to levels seen before DSB initiation was only seen much later after resumption of the cell cycle [20]. It is tempting to speculate that such a mechanism of partial restoration of nucleosomes during HR stabilizes joint molecules while still allowing sufficient access for HR factors to complete final repair steps, following which the cell cycle resumes and nucleosome deposition restores its occupancy to pre-break levels (Fig. 2).
3.4. Non-homologous end joining
A second major pathway (and the predominant pathway of choice in mammalian cells) that is utilized to repair DSBs is non-homologous end joining (NHEJ), where the two ends of a DSB are ligated together in a flexible manner, allowing a variety of DNA end configurations to be repaired. During NHEJ, the Ku70–80 complex (KU) binds to free DSB DNA ends soon after a break is formed, providing a platform for the recruitment of a variety of factors involved in NHEJ and simultaneously suppressing Exo1 and Dna2-mediated end resection that promotes HR [12,51,52]. KU and the MRX/MRN complex act antagonistically to each other to regulate the level of resection and thereby influence the repair pathway choice between NHEJ and HR. Additionally, KU recruits downstream NHEJ factors such as the ligase complex Dnl4-Lif1 (LIG4-XRCC4 in mammals) that enables the ligation of the DSB ends and Nej1 (XLF in mammals) which apart from stabilizing KU at DSB ends, inhibits Dna2 and stimulates the ligase activity of Dnl4-Lif1 [53,54]. Interestingly, the MRX/MRN complex is known to be important for both NHEJ and HR: its structural features are important for tethering of the DSB ends, and the catalytic activity is required for resection [55].
3.5. Nucleosome disassembly during NHEJ
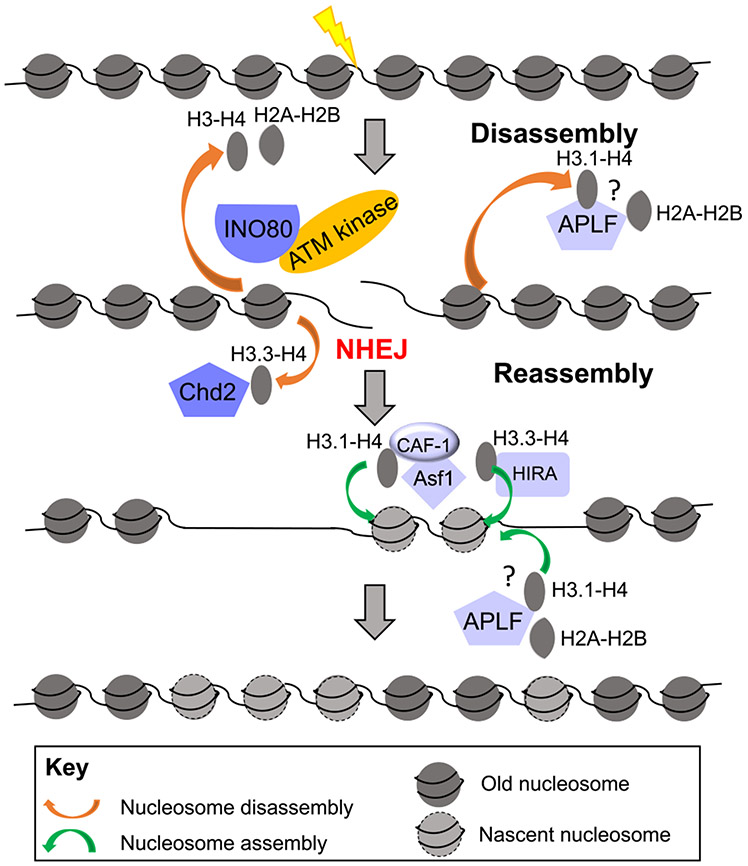
Like HR, repair of a DSB by NHEJ necessitates disassembly of nucleosomes at least locally to provide access to repair factors. In this regard, Goldstein et al. observed that only H2A-H2B was lost from regions surrounding a break in the rDNA locus in G1-arrested mammalian cells which was likely going through repair by NHEJ as HR is downregulated in G1 phase cells and the NHEJ factor XRCC4 was recruited to the break [19]. Meanwhile DSBs in cycling cells showed loss of all four core histones near the break and was accompanied by recruitment of the ssDNA binding protein RPA, indicating that the repair in the cycling cells was by homologous recombination. In contrast, Li and Tyler showed in mammalian cells that DSBs at euchromatic loci that were repaired by NHEJ experienced a complete loss of all four core histones within a 750 bp range on either side from the DNA ends in a manner independent of end resection, but dependent on INO80 and the ATM kinase [56] (Fig. 3). In the absence of INO80 and ATM, NHEJ was compromised, supporting a model where localized finite nucleosome disassembly allows the access of NHEJ factors, while at the same time avoiding the atleast transient loss of epigenetic information that accompanies extensive nucleosome loss. The reasons for the discrepancy in these studies findings, as to whether H2A-H2B or all four core histones gets removed during NHEJ, are unclear, although the former study examined chromatin disassembly during NHEJ at heterochromatin regions while the latter examined chromatin disassembly during NHEJ at euchromatin regions. Recently in mammalian cells, a DNA repair factor APLF, that provides a scaffold for NHEJ factors, has been shown to bind both H2A-H2B and (H3-H4)2 and have histone chaperone activity [57-59]. This raises the possibility that APLF may function in transiently disassembling and storing histones while NHEJ factors repair the DSB, and subsequently may reassemble the nucleosomes back onto the ligated DNA after the completion of repair [59].
Fig. 3.
Chromatin disassembly and reassembly during non-homologous end joining. Chromatin is disassembled during NHEJ to promote DNA repair, and chromatin is reassembled after NHEJ, as described in the text.
3.6. Nucleosome reassembly after NHEJ
Our current understanding of nucleosome reassembly after NHEJ is incomplete but appears to involve the so called “replication-dependent” chromatin assembly pathway despite no known occurrence of significant DNA synthesis during NHEJ. Early genetic epistasis analysis indicated that the “replication-dependent” CAF-1 histone chaperone complex is involved in both HR and NHEJ mediated repair of DSBs in budding yeast [42]. Additionally, CAF-1 was shown to be induced upon generation of DSBs in quiescent human cells and was shown to colocalize with the NHEJ factor XRCC4 [60]. Moreover, PCNA, a known interactor of CAF-1, has also been shown to interact with the KU heterodimer in human cells [61,62]. However, more recent high resolution MNase-seq approaches probing NHEJ repair in yeast have indicated that the local chromatin architecture is restored rapidly via nucleosome reassembly in a manner independent of DNA replication in G1 arrested yeast cells, suggesting that repair-coupled but replication-independent chromatin reassembly is involved in restoring nucleosome patterns surrounding DSBs following NHEJ repair [38]. Strikingly, the nucleosomes return to their original pre-lesion locations on the DNA after repair by NHEJ [38]. Why full chromatin assembly after HR of the HO lesion requires passage through the cell cycle [20] while chromatin assembly after NHEJ of the HO lesion does not require DNA replication [38], despite both processes involving Asf1 and CAF-1 mediated chromatin assembly, is unclear. It is possible that both replication-dependent (i.e., CAF-1 dependent assembly of H3.1-H4 onto DNA) and replication-independent (i.e., HIRA/DAXX dependent assembly of H3.3-H4 onto DNA) mechanisms of nucleosome assembly play roles in restoring nucleosome integrity at the repair site after the completion of NHEJ. Consistent with this idea, Li and Tyler demonstrated using human cells that nucleosome reassembly around DSBs that were repaired by NHEJ was facilitated by the replication-independent HIRA histone chaperone as well as the replication-dependent CAF-1 histone chaperone in an inter-dependent manner, in conjunction with ASF1 [56] (Fig. 3). The exact mechanisms by which such replication-dependent histone reassembly pathways may facilitate NHEJ where there is hardly any DNA synthesis is yet to be explored, as are the mechanisms of coordination between the various reassembly modes. Finally, apart from core histones, certain histone variants also seem to play a dynamic role near DSBs. For instance, the histone H3.3 variant was shown to be deposited near DSBs by the chromatin remodeler CHD2 and plays a role in recruiting NHEJ core factors such as KU and XRCC4 in mammalian cells [63]. Similarly, the incorporation of H2A.Z facilitates the loading of Ku70, and its subsequent removal by the histone chaperone ANP32e is important for completion of NHEJ [64,65]. Clearly chromatin disassembly and assembly both facilitate the access / recruitment of the repair machinery during NHEJ.
3.7. Mismatch repair
Polymerase misincorporation errors during DNA replication are repaired by the mismatch repair (MMR) pathway, and defects in this pathway lead to accumulation of mutations in the cell. In budding yeast, various kinds of mismatches are initially recognized by the MutS homolog heterodimers Msh2-Msh6 (MutSα) or Msh2-Msh3 (Mutsβ), which then enter a sliding-clamp conformation that allows their diffusion on DNA [66,67]. The MutS homologs then recruit MutL homolog heterodimers, primarily Mlh1-Pms1, whose endonuclease activity is then activated by PCNA resulting in nicking of the newly synthesized strand of DNA in an ATP-dependent manner. Subsequently, these nicks may act as loading sites of Exo1 whose exonuclease activity results in excision of the newly synthesized DNA strand across the mismatch, or an alternate Polδ / Polε-dependent pathway involving strand-displacement synthesis may remove the nascent strand containing the mismatch. Finally, DNA polymerases re-synthesize the excised DNA, thus correcting the mismatch.
3.8. A delicate tug of war between chromatin assembly and mismatch repair
Advances have been made in our understanding of how the chromatin context closely interacts and communicates with the MMR machinery to regulate DNA repair. DNA replication is tightly coupled with nucleosome disassembly ahead of the replication fork and reassembly behind the fork that enables genome stability maintenance and epigenetic inheritance. Several histone chaperones such as CAF-1, Asf1, Rtt106 and FACT have been implicated in nucleosome assembly after DNA replication [47,68-72]. Chromatin disassembly ahead of the replication fork is a poorly understood process. Regardless, after replication-dependent chromatin disassembly, when a mismatch is detected following a misincorporation event during replication, the cell must ensure that it is repaired before nucleosomes are assembled on the newly replicated DNA. Recent advances have been made in understanding interactions between histone assembly and mismatch repair, and how these two processes regulate each other.
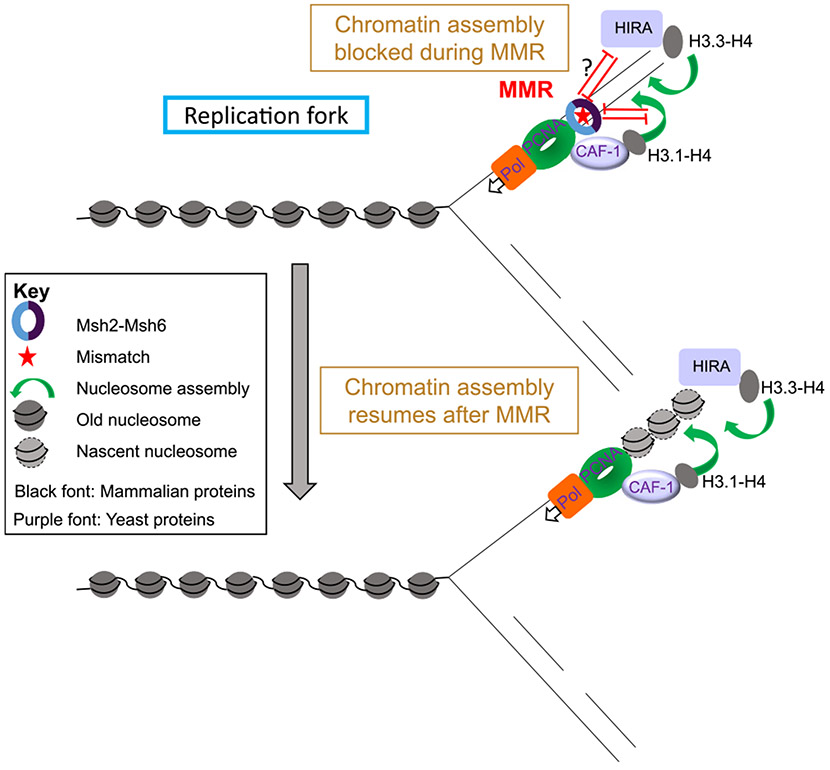
It appears that the MMR machinery actively impairs chromatin assembly over the mismatch. On one hand, nucleosomes have been shown in vitro to have an inhibitory effect on mismatch recognition, ATPase, or ADP-binding, and sliding of human MutSα when interacting with DNA heteroduplexes [73,74]. Nucleosome deposition has also been shown to protect discontinuous mismatch-containing strands from excessive degradation by the MMR apparatus [75,76]. On the other hand, several studies have demonstrated using reconstituted biochemical systems that human MutSα inhibits CAF-1 and ASF1A-H3-H4 dependent formation of H3-H4 tetramers on DNA in the presence of mismatches [75-77] (Fig. 4). Consistent with the mutually inhibitory roles of MMR and nucleosome deposition, CAF-1 was shown to physically interact with mismatch repair factors MutSα and PCNA, which also interact with each other [41,47,77-79]. Finally, it was also shown using Xenopus egg extracts as a model system that nucleosomes are excluded from a > 1-kb region surrounding a mismatch in a MutSα-dependent but MutL homolog-independent manner, countering CAF-1 and HIRA mediated nucleosome assembly [80]. Together these studies point to a model in which replication coupled nucleosome assembly and MMR are in a delicate tug of war to ensure that genomic stability and epigenetic inheritance are maintained.
Fig. 4.
The interplay between chromatin disassembly / reassembly during mismatch repair. Replication-dependent chromatin assembly after DNA replication is halted when a mismatch is encountered while other factors promote the accuracy of MMR, as described in the text. Nucleosomes are deposited on DNA after mismatch is corrected.
On the other side of the coin, chromatin assembly factors in turn regulate mismatch repair (Fig. 4). Consistent with its interaction with members of the MMR pathway, CAF-1 was shown to suppress a parallel activity of the MMR machinery, namely the cytotoxic response to Sn1-type methylating agents [81]. Other histone chaperones have also been implemented in regulating MMR. For example, Terui et al. showed data suggesting that the histone chaperone FACT along with the chromatin remodeling ATPase Smarcad1 assists nucleosome exclusion from the surrounding DNA in the presence of a mismatch [80]. Additionally, CAF-1 and Rtt106 were shown using genetic assays to suppress heteroduplex rejection, a pathway in which MutS homolog proteins promote the fidelity of HR by identifying mismatches during strand invasion and unwinding the heteroduplex by recruiting the STR complex [82,83]. Further in vivo studies are required to get a better understanding of the interplay between nucleosome deposition and MMR.
3.8.1. Excision repair
Base excision repair (BER) is the primary mechanism that removes base lesions that result from oxidation, alkylation, deamination, and spontaneous hydrolysis [84]. So far, about 100 nucleobase lesions have been identified in vitro, and many of them have been identified in cellular DNA [85]. Given the fact that ~104 nucleobase lesions occur per cell per day, cells need to efficiently repair them to maintain genomic integrity and prevent mutagenesis [85,86]. Several enzymes are required for BER, including a DNA glycosylase, apurinic/apyrimidinic (AP) endonuclease, gap-filling DNA polymerase, and DNA ligase [87]. These enzymes are coordinated in a sequential manner to ensure efficient repair and prevent accumulation of deleterious BER intermediates such as AP sites or strand breaks [88].
Additionally, environmental factors such as UV radiation and chemotherapeutic drugs (e.g., cisplatin) can induce bulky lesions, which cause significant distortion to the structure of the DNA double helix. Bulky lesions are generally more harmful to the cell than base damage, because they can effectively block DNA replication and transcription [89]. The major repair pathway for bulky lesions is nucleotide excision repair (NER), which is a multi-step process comprising ~30 repair proteins [89]. Mutations of many NER genes are associated with severe human diseases such as Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) [90]. XP patients are at extremely high risk of developing skin cancers, while CS patients are characterized by neurological degeneration and premature aging. Collectively, excision repair (ER), including BER and NER, acts as an essential ‘first-line’ defense mechanism in the cell to maintain genome stability by removing a wide spectrum of common DNA lesions. On the other hand, ER is a double-edged sword for chemotherapy. DNA adducts induced by chemotherapeutic agents may be removed by ER factors in cancer cells. The removal of these lethal DNA adducts in tumors can compromise chemotherapy and cause drug resistance [91].
ER proteins have to recognize and remove damage in all chromatin, at different levels of chromatin compaction [92]. Therefore, a central challenge of DNA repair studies in eukaryotes is to understand how repair factors cope with chromatin to efficiently repair DNA damage and avoid mutations. Biochemical data indicates that even the first level of chromatin compaction – the nucleosome – can significantly inhibit BER and NER [93,94]. These studies suggest that DNA damage in nucleosome-occupied DNA is more likely to persist and potentially cause mutations. In addition to affecting repair, the nucleosome structure has also been shown to modulate the formation of certain types of DNA lesions (e.g., UV photoproducts) [92].
4. Nucleotide excision repair
Genome-wide surveys of NER activity revealed that UV damage repair is strongly modulated by the chromatin state [95,96]. Specifically, fast NER preferentially occurs in open chromatin regions characterized with DNase I hypersensitivity and active histone PTMs. In contrast, slow repair is observed in closed and transcriptionally inactive chromatin [95,97]. The impact of chromatin states on NER is correlated with the mutation density in the melanoma genome. Closed chromatin regions, which usually are repaired less efficiently by NER, are associated with high somatic mutations in melanomas while lower mutation density is found in accessible chromatin regions, indicating that accessibility to the NER machinery is generally reduced by the closed chromatin state, independent of the damage type [95,98], [96,99].
4.1. Nucleosome disassembly during NER
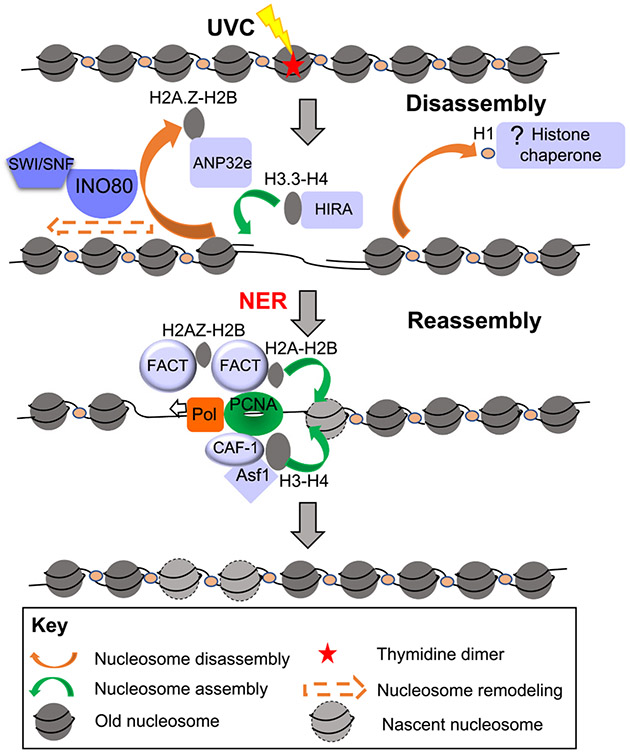
It has not yet been demonstrated that core histones are disassembled during NER although ANP32e evicts histone variant H2A.Z from the chromatin and histone H1 is also evicted from UV-damage chromatin during NER repair (Fig. 5) [100,101]. It is possible that cores histones are disassembled during NER, because factors that reassemble chromatin after NER have been identified and space will be required to allow their reassembly (see below). Furthermore, the histone variant H2A.X is deposited at NER sites by FACT histone chaperone [100], which is likely to depend on the concomitant removal of H2A at sites of UV damage. Several ATP-dependent nucleosome remodelers promote NER, but whether this is via promoting nucleosome disassembly per se has not been shown. In mammalian cells, both SWI/SNF and INO80 remodeling factors stimulate NER as their down-regulation confers hypersensitivity to UVC, associated with inefficient removal of UV damage and impaired recruitment of early/intermediate NER factors [102-106]. The coupling between remodelers and NER factors is further supported by co-immunoprecipitation experiments revealing interactions between the SWI/SNF complex subunits BRG1 and SNF5/INI1 with XPC [104, 106] and between INO80 and DDB1 [102]. Human ISWI isoform SMARCA5/SNF2H and the binding partners ACF1 and WSTF are rapidly recruited to UV-C induced DNA damage to specifically facilitate CSB proteins binding and to promote lesion-stalled transcription recovery [107].
Fig. 5.
Chromatin disassembly and reassembly during nucleotide excision repair. Chromatin remodelers facilitate NER, while histones / histone variants are assembled during or after NER as described in the text. Mammalian proteins are shown here.
Histone PTMs appear to promote access of the NER machinery, potentially via promoting chromatin disassembly. Acetylation was the first histone PTM shown to promote UV-damaged chromatin accessibility and to stimulate NER, as reported in yeast and mammalian cells [108]. Histones H2A, H3 and H4 are ubiquitylated in the course of NER in mammals [109-114]. By examining H3 and H2A extractability from damaged chromatin in vitro and in vivo, histone ubiquitylation was shown to destabilize nucleosomal organization, suggesting that histone ubiquitylation during NER could facilitate access to damaged chromatin in vivo by promoting histone displacement from damaged nucleosomes [110,115]. The mechanisms for how this modification are established in response to UVC damage and coupled with NER are still under investigation. Several E3 ubiquitin ligases acting at different steps of the NER pathway have been identified as histone modifiers. The E3 ubiquitin ligase complex RBX1 (Ring-BoX 1)-Cul4 (Cullin 4)-DDB1-DDB2 (DNA Damage Binding protein), a key player in UVC damage detection, was shown to ubiquitylate H2A in vitro and in vivo [109,111]. This complex is also involved in H3 and H4 ubiquitylation stimulated by UVC irradiation [110]. In addition, H2A was found to be ubiquitylated by the ubiquitin ligase RNF2 in a manner dependent on the NER factor XPA [112]. The ubiquitin ligase RNF8 also modifies H2A upon formation of ssDNA, an NER intermediate resulting from lesion processing [114]. While the multiplicity of E3 ubiquitin ligases involved in modifying H2A complicates the analysis of its function in the NER pathway, it clearly underlines a critical role of this modified histone in this process.
Somewhat surprisingly, there also appears to be chromatin assembly of H3.3-H4 even during repair of UVC damage by NER. This was revealed by HIRA depositing H3.3 onto chromatin during NER, recruited via a ubiquitin-dependent segregase VCP [116,117]. The function of this H3.3 incorporation during NER is unclear because it is not required for NER nor for transcription recovery after DNA repair [116,117]. However, HIRA functions in a histone-chaperone independent manner to promote transcriptional restart after UV repair [117]. Perhaps the H3.3 incorporation during NER reflects the dynamic nature of the chromatin around the DNA lesions. Newly synthesized H2A is incorporated at sites of NER by the FACT histone chaperone and this is required for transcriptional restart after NER [118].
4.2. Nucleosome reassembly after NER
Once NER is complete, chromatin assembly after NER is mediated by CAF-1 dependent assembly of newly synthesized H3-H4, as shown both biochemically and in cells using fluorescently tagged histones. [119-121,122]. Consistent with a role in chromatin assembly after DNA repair, CAF-1 is not required per se for efficient repair of UV lesions or for the recruitment of NER factors to damage in human cells [123]. As is the case during DNA replication, the direct interaction of CAF-1 with the polymerase sliding clamp PCNA helps tether CAF-1 to sites of UV repair [41,119]. ASF1 functions synergistically with CAF-1 to assemble nucleosomes during NER in vitro and helps turning off the DNA damage checkpoint after UV irradiation both in yeast and mammalian cells [45,124,125]. ASF1 acts as a donor of new histones for CAF-1 in chromatin restoration coupled to NER. Another attractive possibility is that ASF1 could also be involved in old histone recycling at damage sites as described at the replication fork [126]. H2A ubiquitylation has also been proposed to occur after repair synthesis and to be dependent on the H3.1 histone chaperone CAF-1 [113]. Histone ubiquitylation, reported to destabilize nucleosomes, might help ATP-dependent nucleosome remodelers to reposition newly formed nucleosomes and could thus be an important player in chromatin restoration upon UVC irradiation. Future studies may also give more insights into the composition of nucleosomes formed upon repair-coupled chromatin restoration by determining whether histone variants other than H3.1 get deposited at NER sites.
5. Base excision repair
General inhibition of BER by nucleosomes has been observed in vitro using purified BER enzymes and reconstituted nucleosomes [127]. By changing the damage location to different rotational positions on the nucleosome (e.g., either facing “in” towards the globular core of the nucleosome or “out” from the globular core), it has been shown that BER of uracil residues is significantly modulated by the nucleosomal rotational settings, presumably because uracil lesions at “out” rotational settings are more easily recognized by the uracil-DNA glycosylase [128,129]. This periodic BER pattern in nucleosomes may in some cases promote a similar mutation periodicity in certain types of human cancer, such as esophageal cancer and gastric cancer [130]. Low BER activity at “in” rotational positions could promote the persistence of unrepaired base damage at inward facing positions, thereby promoting mutagenesis at these positions in cancers. In contrast, the high BER activity at “out” positions may reduce mutation accumulation at outward rotational settings in nucleosomes.
Genome-wide mapping of base damage has provided new insights into BER kinetics in chromatin. Mapping alkylation lesions remaining after repair indicated that BER is strongly modulated by nucleosome organization in gene coding regions. When ~5000 yeast genes were aligned at their transcription start sites (TSS), there was a striking repair periodicity that correlated with the stereotypic nucleosome positioning (e.g., +1, +2, and so on nucleosomes [131]). BER is elevated in nucleosome-depleted regions (NDR) in gene promoters and also in nucleosomal linker DNA, but repressed within nucleosomes, particularly near the nucleosome dyad axis, indicating that the translational setting of the DNA lesion affects its repair efficiency in vivo [131]. Consistent with BER being less efficient at the DNA at the center of the nucleosome, the mutation density is elevated near the nucleosome center after treatment with alkylating agents, suggesting that the reduced repair of alkylation damage near the central dyad promotes mutagenesis [131].
5.1. Nucleosome dynamics during BER
Our current knowledge of the factors that helps the BER machinery repair DNA lesions within the nucleosome context in vivo is rudimentary. However, we know that the histone chaperone FACT participates in BER through its “co-remodeling” activity and acts in concert with the RSC ATP-dependent nucleosome remodeler to facilitate the removal of uracil from DNA, indicating that FACT could promote the repair of DNA damage in the initial step of BER [132]. The findings of chromatin remodelers cooperating with histone chaperones highlight the sophisticated mechanisms utilized by cells to regulate BER.
There is increasing evidence implicating the participation of other ATP-dependent nucleosome remodelers in BER. In biochemical studies, the activity of BER factors such as OGG1, APE1, and pol β are stimulated by the SWI/SNF complex [133]. A similar observation has been made that the barrier imposed by histones is greatly alleviated by ISW1 and ISW2 complexes for pol β mediated DNA synthesis [134]. Furthermore, an overall increase in BER, especially at inward-facing sites, is achieved by RSC [132,135]. The participation of chromatin remodelers in modulating BER has also been indicated by in vivo studies, although detailed mechanisms need to be further elucidated. One member of the SWI/SNF family of ATP-dependent chromatin remodelers, CSB (Cockayne syndrome group B), has been shown to function in the repair of 8-oxoG lesions [136,137]. Furthermore, CSB co-localizes with OGG1 after γ-radiation, and its knockdown and mutations result in a reduced level of OGG1 expression and 8-oxoG repair [137]. Depletion of subunits of RSC and INO80 complexes increases the sensitivity of yeast cells to alkylating agents [138]. Specifically, for RSC it has been shown that yeast lacking STH1, a critical ATPase subunit of the RSC complex, exhibit genome-wide BER deficiency in removing alkylated bases and their chromatin shows reduced accessibility to micrococcal nuclease digestion in the absence of STH1 [138].
Histone PTMs also regulate BER in a manner that likely functions to influence the accessibility of the DNA within the nucleosome, either directly or indirectly. Histone PTMs enriched toward the 5’ end of active genes (e.g., H3K14ac, H3K4me3) appear to promote BER at the DNA regions near where they exit and enter the nucleosome (the nucleosome edges), but paradoxically repress repair at the dyad, while PTMs that are enriched toward the 3’ end of genes (e.g., H3K36me3 and H3K79me3) display the opposite trend [131,139]. Previous studies have suggested that H3K14ac can promote repair in part by enhancing binding of the ATP-dependent nucleosome remodeler RSC to lesion-containing nucleosomes [140]. RSC-remodeled nucleosomes may have increased DNA accessibility near the nucleosome edges, which could explain the faster BER toward the DNA ends of H3K14ac nucleosomes [131]. On the other hand, H3K14ac has been shown to strongly inhibit the gap-filling activity of DNA polymerase β in BER near the nucleosomal dyad, which may contribute to the reduced BER at the dyad in H3K14ac-enriched nucleosomes in vivo [131,141]. Although these studies provide a potential mechanism for how histone PTMs affect BER, clearly further studies are needed.
6. Perspectives and future directions
Most of our current understanding of how chromatin impacts DNA repair has come from distinct technical advances that have allowed the field to (1) biochemically reconstitute MMR, NER and BER on chromatin templates, (2) measure repair efficiency within cells upon depletion of chromatin modulating factors, (3) induce site specific DSBs with endonucleases within cells, (4) examine incorporation of histones via SNAP-tag-based pulse-chase imaging, a powerful technique that allows tracking new or old histones in live cells and quantifying their turnover [142] and (5) detect localization of factors to repair foci, or sites of localized UVC irradiation within cells, or by ChIP to induced DSBs. Further improvements in our understanding of the interplay between chromatin dynamics and genome stability will necessitate development of new technologies. An important question that is yet to be well understood is how local and global nucleosome disassembly processes are coordinated after DNA damage [40,143] and what mechanisms relay damage-induced cellular responses from a local to a global level. Additionally, such loss of histones has been implicated in increased chromatin mobility that facilitates the repair process [143,144]. Several studies have subsequently been aimed at understanding mechanisms by which decreased histone levels promote increased chromatin mobility. For example, it was shown in yeast that global nucleosome loss resulted in decompaction of chromatin and ultimately resulted in increased global chromatin mobility that facilitated HR [144]. Another study showed using live cell microscopy and computational simulations that DNA damage led to increased chromatin subdiffusion and intrachromosomal distances, and provided evidence that genome-wide chromatin stiffening (mediated in part by H2A phosphorylation and leading to decreased chromosome folding) that occurred as a consequence of DNA damage was, at least in part, the basis of increased chromatin mobility [145]. A related question that warrants further investigation is how nucleosome loss during HR may be mediated at the template DNA in order to better facilitate strand invasion and stabilize the joint molecules. Such loss of nucleosomes from the template DNA may be simply achieved as a part of the global nucleosome disassembly process as discussed above or at a more local scale by specific signaling cascades ensuing from recognition of template sequences by the broken DNA during strand invasion. Another important area that remains to be fully explored is how mechanisms of nucleosome disassembly and reassembly may be differently regulated in euchromatic and heterochromatic genomic regions. Such context-specific regulation would seem necessary given that heterochromatic regions contain highly repetitive DNA sequences that may result in DNA breaks being repaired inappropriately if nucleosome disassembly isn’t regulated appropriately. Such regulation in heterochromatin may be facilitated by specific heterochromatic histone PTMs upon DNA damage and/or heterochromatic proteins interacting with and regulating histone chaperones following damage. In this regard, a recent study found that UV induced damage in pericentromeric heterochromatin in mammalian cells leads to heterochromatin compaction changes and linker histone H1 (and possibly to a lesser extent H3.3 and H2B) removal from chromatin facilitated by the UV damage sensor DDB2 [101]. Interestingly, heterochromatin-specific histone PTMs were found to be maintained despite massive unfolding of heterochromatin following damage and was found to play a key role in deposition of new histones in the damaged DNA. The authors found that the deposition of new H3 histones in damaged heterochromatin was executed by histone chaperones such as H3.1-H4 deposition mediated by CAF-1, that has also been implicated in euchromatic DNA repair, and H3.3-H4 deposition mediated by HIRA, that was shown to accumulate on damaged heterochromatin in a manner comparable to damaged euchromatin, and possibly to a lesser extent DAXX, that was shown to be specifically recruited to damaged heterochromatin but not euchromatin.
Another important area that remains is “how is the epigenetic information restored after DNA repair?” We know that newly synthesized histones are incorporated after DSB repair and UV repair. This incorporation of newly synthesized histones challenges the maintenance of epigenetic information because newly synthesized histones are known to bear PTMs that differ from parental nucleosomal histones [146]. Thus, deposition of new histones could lead to substantial changes in the chromatin in repaired regions that if not reversed, would alter the transcriptional output of the repaired regions. Revealing how this happens would require approaches to determine histone PTM patterns specifically over the repaired DNA at multiple time points before and after repair at known genomic locations. We know from DNA replication that the parental histone PTMs are diluted 2-fold during replication, and it takes up to a cycle to modify the new histones to mirror the parental histones, while other modifications are added to both the parental and new histones continuously [147]. Notably, the dynamics of pre-existing histones and other variants also needs to be considered, and histone deposition of H3.3 at earlier steps in the NER process cannot be excluded. Further investigation of histone variant dynamics coupled to repair in vivo should now be possible by exploiting SNAP-tagging. Advances in our knowledge promise to further highlight the intertwined dance of histone chaperones and repair factors to ensure the maintenance of genome integrity.
Acknowledgements
NIH R35 GM139816 and JKT, RO1 CA95641.
Footnotes
Conflicts of interest
None of the authors have any Conflicts of interest to declare.
CRediT authorship contribution statement
UJ and Z-J S wrote the review (half each) and JKT edited the review.
This article is part of the special issue Maintenance of Active Genome Integrity.
This Special Issue is edited by Sukesh R. Bhaumik.
References
- [1].Soria G, Polo SE, Almouzni G, Prime, repair, restore: the active role of chromatin in the DNA damage response, Mol. Cell 46 (6) (2012) 722–734. [DOI] [PubMed] [Google Scholar]
- [2].Ransom M, Dennehey BK, K Tyler J, Chaperoning histones during DNA replication and repair, Cell 140 (2) (2010) 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Clapier CR, Iwasa J, Cairns BR, Peterson CL, Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes, Nat. Rev. Mol. Cell. Biol 18 (7) (2017) 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ferrand J, Plessier A, Polo SE, Control of the chromatin response to DNA damage: histone proteins pull the strings, Semin. Cell Dev. Biol 113 (2021) 75–87. [DOI] [PubMed] [Google Scholar]
- [5].Rother MB, van Attikum H, DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance, Philos Trans. R. Soc. Lond. B Biol. Sci 372 (2017) 1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Clouaire T, Legube G, A snapshot on the Cis chromatin response to DNA double-strand breaks, Trends Genet. 35 (5) (2019) 330–345. [DOI] [PubMed] [Google Scholar]
- [7].Smerdon MJ, DNA repair and the role of chromatin structure, Curr. Opin. Cell Biol 3 (3) (1991) 422–428. [DOI] [PubMed] [Google Scholar]
- [8].Green CM, Almouzni G, When repair meets chromatin. First in series on chromatin dynamics, EMBO Rep. 3 (1) (2002) 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Groth A, Rocha W, Verreault A, Almouzni G, Chromatin challenges during DNA replication and repair, Cell 128 (4) (2007) 721–733. [DOI] [PubMed] [Google Scholar]
- [10].Polo SE, Almouzni G, Chromatin dynamics after DNA damage: the legacy of the access-repair-restore model, DNA Repair 36 (2015) 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cejka P, DNA end resection: nucleases team up with the right partners to initiate homologous recombination, J. Biol. Chem 290 (38) (2015) 22931–22938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Symington LS, Mechanism and regulation of DNA end resection in eukaryotes, Crit. Rev. Biochem. Mol. Biol 51 (3) (2016) 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mehta A, Haber JE, Sources of DNA double-strand breaks and models of recombinational DNA repair, Cold Spring Harb. Perspect. Biol 6 (9) (2014), a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Symington LS, Rothstein R, Lisby M, Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae, Genetics 198 (3) (2014) 795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tsukuda T, Fleming AB, Nickoloff JA, Osley MA, Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae, Nature 438 (7066) (2005) 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE, RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin, Mol. Cell Biol 27 (5) (2007) 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK, Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair, Cell 134 (2) (2008) 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Berkovich E, Monnat RJ Jr., Kastan MB, Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair, Nat. Cell Biol 9 (6) (2007) 683–690. [DOI] [PubMed] [Google Scholar]
- [19].Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB, Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair, Proc. Natl. Acad. Sci. U.S.A 110 (42) (2013) 16874–16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tsabar M, Hicks WM, Tsaponina O, Haber JE, Re-establishment of nucleosome occupancy during double-strand break repair in budding yeast, DNA Repair 47 (2016) 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tripuraneni V, Memisoglu G, MacAlpine HK, Tran TQ, Zhu W, Hartemink AJ, Haber JE, MacAlpine DM, Local nucleosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair in the absence of DNA replication, Genome Res. 31 (5) (2021) 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Peritore M, Reusswig KU, Bantele S, Straub T, Pfander B, Strand-specific ChIP-seq at DNA breaks distinguishes ssDNA versus dsDNA binding and refutes single-stranded nucleosomes, Mol. Cell 81 (8) (2021) 1841–1853 e4. [DOI] [PubMed] [Google Scholar]
- [23].Mimitou EP, Yam ad a S, Keeney S, A global view of meiotic double-strand break end resection, Science 355 (6320) (2017) 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Liu C, Wang L, Xu X, Zhao Y, Wang H, Yang Y, Zhu WG, Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair, Cell Res. 28 (7) (2018) 756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Downs JA, Kosmidou E, Morgan A, Jackson SP, Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone, Mol. Cell 11 (6) (2003) 1685–1692. [DOI] [PubMed] [Google Scholar]
- [26].Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O, Global chromatin compaction limits the strength of the DNA damage response, J. Cell Biol 178 (7) (2007) 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G, High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome, EMBO J. 29 (8) (2010) 1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Haber JE, In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases, Bioessays 17 (7) (1995) 609–620. [DOI] [PubMed] [Google Scholar]
- [29].Tripuraneni V, et al. , Local nucleosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair in the absence of DNA replication. bioRxiv, 2019: p. 866673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].van Attikum H, Fritsch O, Gasser SM, Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks, EMBO J. 26 (18) (2007) 4113–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bantele SCS, Pfander B, Nucleosome remodeling by Fun30(SMARCAD1) in the DNA damage response, Front. Mol. Biosci 6 (2019) 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kobayashi J, Fujimoto H, Sato J, Hayashi I, Burma S, Matsuura S, Chen DJ, Komatsu K, Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway, PLoS One 7 (11) (2012), e49245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y, The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks, J. Cell Biol 199 (7) (2012) 1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Adkins NL, Niu H, Sung P, Peterson CL, Nucleosome dynamics regulates DNA processing, Nat. Struct. Mol. Biol 20 (7) (2013) 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE, The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation, Mol. Cell Biol 32 (22) (2012) 4727–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, van Attikum H, Llorente B, The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection, Nature 489 (7417) (2012) 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G, The Fun30 nudeosome remodeller promotes resection of DNA double-strand break ends, Nature 489 (7417) (2012) 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tripuraneni V, Memisoglu G, MacAlpine HK, Tran TQ, Zhu W, Hartemink AJ, Haber JE, MacAlpine DM, Local nudeosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair in the absence of DNA replication, Gen. Res 31 (5) (2021) 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zheng S, Li D, Lu Z, Liu G, Wang M, Xing P, Wang M, Dong Y, Wang X, Li J, Zhang S, Peng H, Ira G, Li G, Chen X, Bre1-dependent H2B ubiquitination promotes homologous recombination by stimulating histone eviction at DNA breaks, Nucleic Acids Res. 46 (21) (2018) 11326–11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hauer MH, Seeber A, Singh V, Thierry R, Sack R, Amitai A, Kryzhanovska M, Eglinger J, Holcman D, Owen-Hughes T, Gasser SM, Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates, Nat. Struct. Mol. Biol 24 (2) (2017) 99–107. [DOI] [PubMed] [Google Scholar]
- [41].Shibahara K, Stillman B, Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin, Cell 96 (4) (1999) 575–585. [DOI] [PubMed] [Google Scholar]
- [42].Linger J, Tyler JK, The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents, Genetics 171 (4) (2005) 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].K Lewis L, Karthikeyan G, Cassiano J, Resnick MA, Reduction of nudeosome assembly during new DNA synthesis impairs both major pathways of double-strand break repair, Nucleic Acids Res. 33 (15) (2005) 4928–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pietrobon V, Fréon K, Hardy J, Costes A, Iraqui I, Ochsenbein F, Lambert SA, The chromatin assembly factor 1 promotes Rad51-dependent template switches at replication forks by counteracting D-loop disassembly by the RecQ-type helicase Rqh1, PLoS Biol. 12 (10) (2014), e1001968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim JA, Haber JE, Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete, Proc. Natl. Acad. Sci. U.S.A 106 (4) (2009) 1151–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Diao LT, Chen CC, Dennehey B, Pal S, Wang P, Shen ZJ, Deem A, Tyler JK, Delineation of the role of chromatin assembly and the Rtt101Mms1 E3 ubiquitin ligase in DNA damage checkpoint recovery in budding yeast, PLoS One 12 (7) (2017), e0180556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z, Acetylation of histone H3 lysine 56 regulates replication-coupled nudeosome assembly, Cell 134 (2) (2008) 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hardy J, Dai D, Ait Saada A, Teixeira-Silva A, Dupoiron L, Mojallali F, Fréon K, Ochsenbein F, Hartmann B, Lambert S, Histone deposition promotes recombination-dependent replication at arrested forks, PLoS Genet. 15 (10) (2019), e1008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huang TH, Fowler F, Chen CG, Shen ZJ, Sleckman B, Tyler JK, The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 loading onto ssDNA during homologous recombination in human cells, Mol Cell 69 (5) (2018) 879–892 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Juhász S, Elbakry A, Mathes A, Löbrich M, ATRX promotes DNA repair synthesis and sister chromatid exchange during homologous recombination, Mol. Cell 71 (1) (2018) 11–24 e7. [DOI] [PubMed] [Google Scholar]
- [51].Frit P, Ropars V, Modesti M, Charbonnier JB, Calsou P, Plugged into the Ku-DNA hub: The NHEJ network, Prog. Biophys. Mol. Biol 147 (2019) 62–76. [DOI] [PubMed] [Google Scholar]
- [52].Marini F, Rawal CC, Liberi G, Pellicioli A, Regulation of DNA double strand breaks processing: focus on barriers, Front. Mol. Biosci 6 (2019) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Emerson CH, Bertuch AA, Consider the workhorse: nonhomologous end-joining in budding yeast, Biochem. Cell Biol 94 (5) (2016) 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mojumdar A, Sorenson K, Hohl M, Toulouze M, Lees-Miller SP, Dubrana K, Petrini J, Cobb JA, Nej1 interacts with Mre11 to regulate tethering and Dna2 binding at DNA double-strand breaks, Cell Rep. 28 (6) (2019) 1564–1573.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tisi R, Vertemara J, Zampella G, Longhese MP, Functional and structural insights into the MRX/MRN complex, a key player in recognition and repair of DNA double-strand breaks, Comput. Struct. Biotechnol. J 18 (2020) 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li X, Tyler JK, Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly, Elife (2016) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen-Hughes T, Ahel I, DNA repair factor APLF is a histone chaperone, Mol. Cell 41 (1) (2011) 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hammel M, Yu Y, Radhakrishnan SK, Chokshi C, Tsai MS, Matsumoto Y, Kuzdovich M, Remesh SG, Fang S, Tomkinson AE, Lees-Miller SP, Tainer JA, An intrinsically disordered APLF links Ku, DNA-PKcs, and XRCC4-DNA ligase IV in an extended flexible non-homologous end joining complex, J. Biol. Chem 291 (53) (2016) 26987–27006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Corbeski I, Dolinar K, Wienk H, Boelens R, van Ingen H, DNA repair factor APLF acts as a H2A-H2B histone chaperone through binding its DNA interaction surface, Nucleic Acids Res. 46 (14) (2018) 7138–7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nabatiyan A, Szuts D, Krude T, Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells, Mol. Cell Biol 26 (5) (2006) 1839–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Balajee AS, Geard CR, Chromatin-bound PCNA complex formation triggered by DNA damage occurs independent of the ATM gene product in human cells, Nucleic Acids Res. 29 (6) (2001) 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pospiech H, Rytkonen AK, Syvaoja JE, The role of DNA polymerase activity in human non-homologous end joining, Nucleic Acids Res. 29 (15) (2001) 3277–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot A, Pines A, Vertegaal A, Jacobs J, Shah GM, van Attikum H, PARP1 Links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining, Mol Cell 61 (4) (2016) 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD, Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair, Mol. Cell 48 (5) (2012) 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gursoy-Yuzugullu O, Ayrapetov MK, Price BD, Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair, Proc. Natl. Acad. Sci. U.S.A 112 (24) (2015) 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kunkel TA, Erie DA, Eukaryotic mismatch repair in relation to DNA replication, Annu. Rev. Genet 49 (2015) 291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Goellner EM, Chromatin remodeling and mismatch repair: access and excision, DNA Repair 85 (2020), 102733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Smith S, Stillman B, Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro, Cell 58 (1) (1989) 15–25. [DOI] [PubMed] [Google Scholar]
- [69].Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT, The RCAF complex mediates chromatin assembly during DNA replication and repair, Nature 402 (6761) (1999) 555–560. [DOI] [PubMed] [Google Scholar]
- [70].Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z, Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing, Proc. Natl. Acad. Sci. U.S.A 102 (38) (2005) 13410–13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yang J, Zhang X, Feng J, Leng H, Li S, Xiao J, Liu S, Xu Z, Xu J, Li D, Wang Z, Wang J, Li Q, The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly, Cell Rep. 16 (12) (2016) 3414. [DOI] [PubMed] [Google Scholar]
- [72].Zhang W, Feng J, Li Q, The replisome guides nucleosome assembly during DNA replication, Cell Biosci. 10 (2020) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Li F, Tian L, Gu L, Li GM, Evidence that nudeosomes inhibit mismatch repair in eukaryotic cells, J. Biol. Chem 284 (48) (2009) 33056–33061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC, Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice, Nat. Struct. Mol. Biol 17 (8) (2010) 932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kadyrova LY, Blanko ER, Kadyrov FA, CAF-I-dependent control of degradation of the discontinuous strands during mismatch repair, Proc. Natl. Acad. Sci. U.S.A 108 (7) (2011) 2753–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rodriges Blanko E, Kadyrova LY, Kadyrov FA, DNA mismatch repair interacts with CAF-1- and ASF1A-H3-H4-dependent histone (H3-H4)2 tetramer deposition, J. Biol. Chem 291 (17) (2016) 9203–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Schöpf B, Bregenhorn S, Quivy JP, Kadyrov FA, Almouzni G, Jiricny J, Interplay between mismatch repair and chromatin assembly, Proc. Natl. Acad. Sci. U.S.A 109 (6) (2012) 1895–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA, Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes, J. Biol. Chem 275 (47) (2000) 36498–36501. [DOI] [PubMed] [Google Scholar]
- [79].Flores-Rozas H, Clark D, Kolodner RD, Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex, Nat. Genet 26 (3) (2000) 375–378. [DOI] [PubMed] [Google Scholar]
- [80].Terui R, Nagao K, Kawasoe Y, Taki K, Higashi TL, Tanaka S, Nakagawa T, Obuse C, Masukata H, Takahashi TS, Nucleosomes around a mismatched base pair are excluded via an Msh2-dependent reaction with the aid of SNF2 family ATPase Smarcad1, Genes Dev. 32 (11–12) (2018) 806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kadyrova LY, Dahal BK, Kadyrov FA, The major replicative histone chaperone CAF-1 suppresses the activity of the DNA mismatch repair system in the cytotoxic response to a DNA-methylating agent, J. Biol. Chem 291 (53) (2016) 27298–27312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chakraborty U, Alani E, Understanding how mismatch repair proteins participate in the repair/anti-recombination decision, FEMS Yeast Res. 16 (2016) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Chakraborty U, Mackenroth B, Shalloway D, Alani E, Chromatin modifiers alter recombination between divergent DNA sequences, Genetics 212 (4) (2019) 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Baute J, Depicker A, Base excision repair and its role in maintaining genome stability, Crit. Rev. Biochem. Mol. Biol 43 (4) (2008) 239–276. [DOI] [PubMed] [Google Scholar]
- [85].Gates KS, An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals, Chem. Res. Toxicol 22 (11) (2009) 1747–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Friedman JI, Stivers JT, Detection of damaged DNA bases by DNA glycosylase enzymes, Biochemistry 49 (24) (2010) 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Krokan HE, Bjørås M, Base excision repair, Cold Spring Harb. Perspect. Biol 5 (4) (2013), a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH, A review of recent experiments on step-to-step “hand-off’ of the DNA intermediates in mammalian base excision repair pathways, Mol. Biol 45 (4) (2011) 586–600. [PMC free article] [PubMed] [Google Scholar]
- [89].Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD, DNA repair: from molecular mechanism to human disease, DNA Repair 5 (8) (2006) 986–996. [DOI] [PubMed] [Google Scholar]
- [90].Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH, Understanding nucleotide excision repair and its roles in cancer and ageing, Nat. Rev. Mol. Cell. Biol 15 (7) (2014) 465–481. [DOI] [PubMed] [Google Scholar]
- [91].Martin LP, Hamilton TC, Schilder RJ, Platinum resistance: the role of DNA repair pathways, Clin. Cancer Res 14 (5) (2008) 1291–1295. [DOI] [PubMed] [Google Scholar]
- [92].Smerdon MJ, Conconi A, Modulation of DNA damage and DNA repair in chromatin, Prog. Nucleic Acid Res. Mol. Biol 62 (1999) 227–255. [DOI] [PubMed] [Google Scholar]
- [93].Rodriguez Y, Smerdon MJ, The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes, J. Biol. Chem 288 (19) (2013) 13863–13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hara R, Mo J, Sancar A, DNA damage in the nucleosome core is refractory to repair by human excision nuclease, Mol. Cell Biol 20 (24) (2000) 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Adar S, Hu J, Lieb JD, Sancar A, Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis, Proc Natl Acad Sci U S A 113 (15) (2016) E2124–E2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Hu J, Adebali O, Adar S, Sancar A, Dynamic maps of UV damage formation and repair for the human genome, Proc. Natl. Acad. Sci. U.S.A. 114 (26) (2017) 6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hanawalt PC, Spivak G, Transcription-coupled DNA repair: two decades of progress and surprises, Nat. Rev. Mol. Cell Biol 9 (12) (2008) 958–970. [DOI] [PubMed] [Google Scholar]
- [98].Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence M, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA, Sunyaev SR, Cell-of-origin chromatin organization shapes the mutational landscape of cancer, Nature 518 (7539) (2015) 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Li W, Hu J, Adebali O, Adar S, Yang Y, Chiou YY, Sancar A, Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo [a]pyrene, Proc. Natl. Acad. Sci. U.S.A 114 (26) (2017) 6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Piquet S, Le Parc F, Bai SK, Chevallier O, Adam S, Polo SE, The histone chaperone FACT coordinates H2A.X-dependent signaling and repair of DNA damage, Mol. Cell 72 (5) (2018) 888–901 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fortuny A, Chansard A, Caron P, Chevallier O, Leroy O, Renaud O, Polo SE, Imaging the response to DNA damage in heterochromatin domains reveals core principles of heterochromatin maintenance, Nat. Commun 12 (1) (2021) 2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L, INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway, Proc. Natl. Acad. Sci. U.S.A 107 (40) (2010) 17274–17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gong F, Fahy D, Liu H, Wang W, Smerdon MJ, Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage, Cell Cycle 7 (8) (2008) 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA, Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex, J. Biol. Chem 284 (44) (2009) 30424–30432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang L, Zhang Q, Jones K, Patel M, Gong F, The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage, Cell Cycle 8 (23) (2009) 3953–3959. [DOI] [PubMed] [Google Scholar]
- [106].Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA, Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation, Mol. Cell Biol 29 (23) (2009) 6206–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ay din ÖZ, Marteijn JA, Ribeiro-Silva C, López A. Rodríguez, Wijgers N, Smeenk G, van Attikum H, Poot RA, Vermeulen W, Lans H, Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription, Nucleic Acids Res. 42 (13) (2014) 8473–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Reed SH, Nucleotide excision repair in chromatin: damage removal at the drop of a HAT, DNA Repair 10 (7) (2011) 734–742. [DOI] [PubMed] [Google Scholar]
- [109].Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS, The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites, Proc. Natl. Acad. Sci. U.S.A 103 (8) (2006) 2588–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y, Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage, Mol. Cell 22 (3) (2006) 383–394. [DOI] [PubMed] [Google Scholar]
- [111].Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapić-Otrin V, The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A, Cancer Res. 68 (13) (2008) 5014–5022. [DOI] [PubMed] [Google Scholar]
- [112].Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP, DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A, Genes Dev. 20 (10) (2006) 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA, Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites, DNA Repair. 8 (2) (2009) 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W, Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response, J. Cell Biol 186 (6) (2009) 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Lan L, Nakajima S, Kapetanaki MG, Hsieh CL, Fagerburg M, Thickman K, Rodriguez-Collazo P, Leuba SH, Levine AS, Rapić-Otrin V, Monoubiquitinated histone H2A destabilizes photolesion-containing nucleosomes with concomitant release of UV-damaged DNA-binding protein E3 ligase, J. Biol. Chem 287 (15) (2012) 12036–12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Adam S, Polo SE, Almouzni G, Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA, Cell 155 (1) (2013) 94–106. [DOI] [PubMed] [Google Scholar]
- [117].Bouvier D, Ferrand J, Chevallier O, Paulsen MT, Ljungman M, Polo SE, Dissecting regulatory pathways for transcription recovery following DNA damage reveals a non-canonical function of the histone chaperone HIRA, Nat. Commun 12 (1) (2021) 3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dinant G, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, Fousteri M, Houtsmuller AB, Vermeulen W, Marteijn JA, Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage, Mol. Cell 51 (4) (2013) 469–479. [DOI] [PubMed] [Google Scholar]
- [119].Moggs JG, Grandi P, Quivy JP, Jónsson ZO, Hübscher U, Becker PB, Almouzni G, A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage, Mol. Cell Biol 20 (4) (2000) 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Martini E, Roche DM, Marheineke K, Verreault A, Almouzni G, Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells, J. Cell. Biol 143 (3) (1998) 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Green GM, Almouzni G, Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo, EMBO J. 22 (19) (2003) 5163–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gaillard PHL, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G, Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I, Cell 86 (6) (1996) 887–896. [DOI] [PubMed] [Google Scholar]
- [123].Polo SE, Roche D, Almouzni G, New histone incorporation marks sites of UV repair in human cells, Cell 127 (3) (2006) 481–493. [DOI] [PubMed] [Google Scholar]
- [124].Mello JA, Silljé HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G, Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway, EMBO Rep. 3 (4) (2002) 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Battu A, Ray A, Wani AA, ASF1A and ATM regulate H3K56-mediated cell-cycle checkpoint recovery in response to UV irradiation, Nucleic Acids Res. 39 (18) (2011)7931–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G, Regulation of replication fork progression through histone supply and demand, Science 318 (5858) (2007) 1928–1931. [DOI] [PubMed] [Google Scholar]
- [127].Rodriguez Y, Hinz JM, Smerdon MJ, Accessing DNA damage in chromatin: preparing the chromatin landscape for base excision repair, DNA Repair 32 (2015) 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Olmon ED, Delaney S, Differential ability of five DNA glycosylases to recognize and repair damage on nucleosomal DNA, ACS Chem. Biol 12 (3) (2017) 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hinz JM, Rodriguez Y, Smerdon MJ, Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme, Proc. Natl. Acad. Sci. U.S.A 107 (10) (2010) 4646–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pich O, Muiños F, Sabarinathan R, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N, Somatic and germline mutation periodicity follow the orientation of the DNA minor groove around nucleosomes, Cell 175 (4) (2018) 1074–1087. e18. [DOI] [PubMed] [Google Scholar]
- [131].Mao P, Brown AJ, Male EP, Mieczkowski PA, Smerdon MJ, Roberts SA, Wyrick JJ, Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity, Genome Res. 27 (10) (2017) 1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Charles Richard JL, Shukla MS, Menoni H, Ouararhni K, Lone IN, Roulland Y, Papin C, Simon E. Ben, Kundu T, Hamiche A, Angelov D, Dimitrov S, FACT assists base excision repair by boosting the remodeling activity of RSC, PLoS Genet. 12 (7) (2016), e1006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D, ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes, Mol. Cell Biol 27 (17) (2007) 5949–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Nakanishi S, Prasad R, Wilson SH, Smerdon M, Different structural states in oligonucleosomes are required for early versus late steps of base excision repair, Nucleic Acids Res. 35 (13) (2007) 4313–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D, Base excision repair of 8-oxoG in dinucleosomes, Nucleic Acids Res. 40 (2) (2012) 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Tuo J, Müftüoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM Jr, Rodriguez H, Dizdaroglu M, Bohr VA, The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA, J Biol. Chem 276 (49) (2001) 45772–45779. [DOI] [PubMed] [Google Scholar]
- [137].Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA, Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein, DNA Repair 1 (11) (2002) 913–927. [DOI] [PubMed] [Google Scholar]
- [138].Czaja W, Mao P, Smerdon MJ, Chromatin remodelling complex RSC promotes base excision repair in chromatin of Saccharomyces cerevisiae, DNA Repair 16 (2014) 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N, High-resolution chromatin dynamics during a yeast stress response, Mol. Cell 58 (2) (2015) 371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Duan MR, Smerdon MJ, Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin), J. Biol. Chem 289 (12) (2014) 8353–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, Smerdon MJ, Site-specific acetylation of histone H3 decreases polymerase β activity on nucleosome core particles in vitro, J. Biol. Chem 291 (21) (2016) 11434–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LE, Almouzni G, Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity, Mol. Cell 44 (6) (2011) 928–941. [DOI] [PubMed] [Google Scholar]
- [143].Hauer MH, Gasser SM, Chromatin and nucleosome dynamics in DNA damage and repair, Genes Dev. 31 (22) (2017) 2204–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Cheblal A, Challa K, Seeber A, Shimada K, Yoshida H, Ferreira HC, Amitai A, Gasser SM, DNA damage-induced nucleosome depletion enhances homology search independently of local break movement, Mol. Cell 80 (2) (2020) 311–326 e4. [DOI] [PubMed] [Google Scholar]
- [145].Herbert S, Brion A, Arbona JM, Lelek M, Veillet A, Lelandais B, Parmar J, Fernández FG, Almayrac E, Khalil Y, Birgy E, Fabre E, Zimmer C, Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast, EMBO J 36 (17) (2017) 2595–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G, PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state, Mol. Cell 24 (2) (2006) 309–316. [DOI] [PubMed] [Google Scholar]
- [147].Alabert C, K Barth T, Reverón-Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A, Two distinct modes for propagation of histone PTMs across the cell cycle, Genes Dev. 29 (6) (2015) 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]