Abstract
Women live longer than men but experience greater disability and a longer period of illness as they age. Despite clear sex differences in aging, the impact of pregnancy and its complications, such as preeclampsia, on aging is an underexplored area of geroscience. This review summarizes our current knowledge about the complex links between pregnancy and age-related diseases, including evidence from epidemiology, clinical research, and genetics. We discuss the relationship between normal and pathological pregnancy and maternal aging, using preeclampsia as a primary example. We review the results of human genetics studies of preeclampsia, including genome wide association studies (GWAS), and attempted to catalogue genes involved in preeclampsia as a gateway to mechanisms underlying an increased risk of later life cardio- and neuro- vascular events. Lastly, we discuss challenges in interpreting the GWAS of preeclampsia and provide a functional genomics framework for future research needed to fully realize the promise of GWAS in identifying targets for geroprotective prevention and therapeutics against preeclampsia.
1. Introduction
The world’s population is rapidly aging and women currently hold a longevity advantage compared to men. The current life expectancy at birth for women is 74.7 years compared to 69.9 years for men. In 2019 the global population of adults aged 65 or older was 703 million and in 2050 this number is projected to more than double to 1.5 billion. From these expected 1.5 billion persons aged 65 or older, 54 percent are projected to be women. It is also predicted that persons aged 80 or over will rise from 143 million to 426 million from 2019–2050 and that women will comprise 59 per cent of this aged population in 2050 (UnitedNations, 2019). Unfortunately, this longevity advantage for women does not come without consequence. Though women generally outlive men, they have a longer period of disability and illness at the end of life. Women’s tendency to outlive men but with more disease and worse health late-in-life has been labeled the “male-female, health-survival paradox” (Archer et al., 2018; Gordon et al., 2017; Oksuzyan et al., 2008).
Age is the single greatest risk factor for a majority of chronic diseases (Kennedy et al., 2014), including dementia, cancer, cardiovascular disease, and stroke, resulting in an increasing burden of death and disability worldwide. Most people over 65 in the US have 1–3 chronic diseases (Marengoni et al., 2011). Both age and sex are considered well established determinants of multimorbidity (Marengoni et al., 2011; Violan et al., 2014). Women are disproportionately affected by age-related diseases (Austad and Fischer, 2016). For example, dementia is the 5th leading cause of death in women worldwide compared to the 10th in men (WorldHealthOrganization, 2020). Women suffer from higher morbidity than men due to acute and chronic physical and psychiatric diseases (Almagro et al., 2020).
Reproductive factors, including age at menarche, reproductive lifespan, and age at menopause have been implicated in sex differences in aging (Lockhart et al., 2017; Shadyab et al., 2017). There is overwhelming evidence that female reproductive aging influences lifespan and diverse health outcomes in cardiovascular, skeletal, and metabolic systems as well as various cancer types (Beral et al., 2012; Green et al., 2012; Muka et al., 2016; Pelucchi et al., 2007; Rahman et al., 2015; Shen et al., 2017; Sullivan et al., 2015). Reproductive lifespan, defined as the time interval between menarche and menopause, has also been associated with decreased morbidity and mortality. However, the impact of pregnancy and its complications, particularly preeclampsia, on aging in women remains a surprisingly unexplored and often overlooked area of aging science.
The goal of this review is to summarize what is known about the links between pregnancy, its adverse outcomes, e.g preeclampsia, and age-related diseases, from epidemiology, clinical research and functional genomics; to summarize the current understanding of the relationship between normal and pathological pregnancy and maternal aging; and to provide a framework for future research needed to fill gaps in knowledge.
2. Pregnancy and age-related diseases.
2.1. Costs of reproduction
Natural selection favors successful reproduction, even at a cost to health at older ages (Williams, 1957). The evolutionary theory of aging, termed antagonistic pleiotropy, predicts the genetic trade-off between early life fitness and late life mortality and is compatible with the disposable soma theory of aging (Jasienska, 2020; Kirkwood and Austad, 2000). The disposable soma theory posits tradeoffs between somatic maintenance and reproduction (Kirkwood, 1990), resulting to an accumulation of damage, faster senescence, and lower post-reproductive survival (Kirkwood, 2002; Kirkwood and Rose, 1991). These trade-offs have been ubiquitously observed in animals in nature and under laboratory conditions (Austad and Hoffman, 2018). However, it has been also demonstrated that reproduction and lifespan can be uncoupled. For example, certain mutations and manipulations that increase lifespan do not reduce fecundity or fertility in C. elegans and Drosophila melanogaster (Dillin et al., 2002; Gems et al., 1998; Johnson and Hutchinson, 1993; Kenyon et al., 1993; Mason et al., 2018) and there is no direct relationship between lifespan and reproduction in birds and mammals in captivity (Ricklefs and Cadena, 2007; Tarin et al., 2014).
In humans, the evidence for the evolutionary trade-off is also mixed. In a study of Utah residents, higher parity, defined as the number of live births, and shorter birth intervals are associated with higher fitness costs, as evidenced by decreasing parental survival (Penn and Smith, 2007). These relationships were stronger in mothers than fathers, even after accounting for childbirth mortality. As parity has fallen over the 19th and early 20th centuries, female life span increased, while male life span remained stabled in the same Utah database. These results support costs of reproduction as sex-specific influence on females (Bolund et al., 2016). However, a study of the Tsimane forager-horticulturalists in the Bolivian Amazon shows that greater parity and shorter birth intervals are associated with higher weight, body mass index, and body fat percentage, but not with biomarkers of nutrition or immune activation (Gurven et al., 2016). Studies on the costs of reproduction should consider other factors such as socioeconomic status, social support, and breastfeeding as well as pregnancy (Jasienska, 2020).
Many changes and adaptations of the maternal immune system are required to allow successful implantation and subsequent development of the fetus. Early trophoblast invasion and spiral artery remodeling relies heavily on the maternal immune system for its success. Specifically, the maternal uterine decidual plays an essential role in protecting the embryo from being attacked by maternal immune cells and ultimately forms the maternal side of the placenta. The decidua contains a high number of immune cells that allow maternal immune tolerance leading to implantation and appropriate trophoblast migration through the production of cytokines and angiogenic factors. Importantly, they assure that invasion is neither too shallow nor too deep resulting in a normal pregnancy. Alternatively, immune dysregulation can result in shallow trophoblast invasion resulting in placental ischemia which is hypothesized to lead to preeclampsia later in gestation (Geldenhuys et al., 2018).
The imbalance in immune function consists of increased proinflammatory immune cells and cytokines and decreased regulatory immune cells and cytokines resulting in an uncontrolled and chronic state of inflammation contributing to the pathophysiology of preeclampsia (Eddy et al., 2018). This state of inflammation will augment oxidative stress and stimulates the release of hypoxia-induced anti-angiogenic factors including soluble fms-like tyrosine kinase-1 (sFlt-1) and soluble endoglin (sEng) which are associated with the induction of hypertension.
Although it is clear that there is an inflammatory component to preeclampsia the underlying causes of the initial altered immune regulation are unknown as is the long-term effect of this on the mother. Further details of the immunology associated with preeclampsia are beyond the scope of this chapter and are reviewed in a commentary by Cornelius (Cornelius, 2018).
2.2. Parity and age-related diseases.
Parity is a reproductive factor which may contribute to sex differences in risk of age-related diseases due to social factors, hormonal influences, and physiological stress of pregnancy or its complications such as preeclampsia. The effects of parity on long term maternal age-related diseases remain unclear. A case-control study found that having at least one child was associated with higher risk of Alzheimer disease, an age-related neurodegenerative disorder, in women but not in men (Ptok et al., 2002); however, the risk did not appear to be dose-dependent (i.e. higher in women with higher parity). Another retrospective study showed an association between higher parity and both higher odds and earlier onset of Alzheimer disease (Colucci et al., 2006). Similarly, a neuropathological study found that having more children correlated with more Alzheimer disease pathology in women, but not in men (Beeri et al., 2009). In contrast, in the Georgia Centenarian Study, an aging cohort, women with higher parity had later menopause and greater longevity (Lockhart et al., 2017). However, reverse causality may have played a role in these results, in that women with longer reproductive lifespans might be more likely to have more children (given the lack of readily available contraception during their childbearing years), and longer reproductive lifespan might be a marker of overall slower aging. Supporting these results, a recent retrospective cohort study using medical records data from Kaiser found that markers of a shorter reproductive lifespan, including later menarche, early menopause, or hysterectomy, were associated with higher dementia risk (Gilsanz et al., 2019).
Mid-life cognition, which is associated with dementia risk, may also benefit from the social connections that come with parenting (Frustenberg, 2005; Read and Grundy, 2017) for both mothers and fathers. Previous studies in the United Kingdom and Mexico have found that low (0–1 births) and high (≥ 3 births) parity are related to poor midlife (age 40 – 64 years) and later life (≥ 65 years) cognition, compared to a middle range of parity (2–3 births). The role of education in this relationship may depend on social context (Read and Grundy, 2017; Saenz et al., 2021). Becoming a mother at age > 35 years has been associated with better mid- to late-life cognition in the United Kingdom (Read and Grundy, 2017), but in a sample from rural Louisiana, teenage motherhood was associated with better midlife cognition (Harville et al., 2020). Other studies have found a lack of association between parity or age at first birth and cognition in mothers (Ilango et al., 2019), or even that parous men and women had better cognition then their nulliparous peers (Harville et al., 2020; Ning et al., 2020).
Parity may be protective against some types of cancer, another age-related disease: many studies have shown an inverse relationship between parity and cancer risk in women, including lung cancer (Yin et al., 2020), ovarian cancer (Sung et al., 2016), some types of breast cancer (Lambertini et al., 2016), and endometrial cancer (Troisi et al., 2018). However, whether these associations are due to parity-influenced changes in aging is unknown. The associations between parity and cardiovascular diseases and stroke are similarly conflicting. In the UK Biobank, parous men and women were more likely to experience cardiovascular disease or mortality compared to nulliparous men and women (Magnus et al., 2017). As results were similar for men and women, social, behavioral, lifestyle and biological processes may contribute to cardiovascular profile in relation to parenthood (Lacey et al., 2017; Magnus et al., 2017). Complicating the relationship further, some epidemiological studies have demonstrated a U-or J-shaped relationship between parity and age-related diseases. In a Swedish population-based cohort of 1.3 million women, parity was associated with incident maternal cardiovascular disease in a J-shaped pattern, with 2 births representing the lowest risk category (Parikh et al., 2010). The United States population-based National Health and Nutrition Examination Survey (NHANES) found a U-shaped relationship between parity and validated biological markers of aging in post-menopausal, but not pre-menopausal women (Shirazi et al., 2020).
When accidental and infectious etiologies are removed, mortality is perhaps the ultimate age-related disease. There are many studies examining the associations between pregnancy and age of death. In the Women’s Health Initiative (WHI), women with age at first birth ≥ 25 years had a higher odds of survival than women with age at first birth < 25 years. Complicating the picture, in the United States, the relationship between pregnancy history and mortality varies by race. In Black women, parity in WHI was not related to survival to 90 years, though in White women, a history of 2–4 term pregnancies was associated with higher odds of survival to age 90 years compared to women with 1 term pregnancy (Shadyab et al., 2017). In the US National Longitudinal Survey of Mature Women, nulliparous Black women had longer post-reproductive lives than parous Black women, but parous White women lived longer than nulliparous White women (Reyes et al., 2018). Having ≥ 6 children was related to lower mortality in Black women, though the protective effects diminished with age. A similar relationship was evident for White women only after adjusting for age at first birth, social, economic, and health characteristics (Spence and Eberstein, 2009), supporting the weathering hypothesis of differing maternal health and mortality consequences of childbearing by age as a physical manifestation of social inequality (Geronimus, 1996).
The studies discussed in this section are summarized in the Supplemental Table 1.
2.3. Pregnancy: a human model of aging, with a twist.
Pregnancy itself has been proposed as a natural human model of aging (Giller et al., 2020). Senescence at the cellular level plays a critical role in syncytiotrophoblast formation, placental implantation, embryonic development and parturition (Cox and Redman, 2017). The metabolic stress of pregnancy mimics many of the mechanisms thought to contribute to aging, including oxidative stress, inflammation, changes in DNA methylation and shortening of telomeres (Giller et al., 2020; Iqbal et al., 2018; Pollack et al., 2018). However, in contrast to aging, after healthy pregnancy women undergo a period of “rejuvenation” and repair in the weeks and months postpartum (Giller et al., 2020). In a small study using high-resolution brain imaging and a validated brain age index, women’s brains at 6 weeks postpartum were estimated to be five years younger than the same women’s brains in the first days postpartum (Luders et al., 2018). These findings support a restoration/rejuvenation effect after giving birth. Evidence from translational research supports the hypothesis that pregnancy may lead to permanent alteration of injury repair pathways in women. In a mouse model, multiparity had a protective effect after stroke: despite having features of higher vascular risk, multiparous mice had smaller brain infarcts after experimental stroke, and better stroke recovery courses. The effect was dose dependent, with better functional recovery in the mice with more litters (Ritzel et al., 2017). The authors hypothesized that these protective effects might be mediated by fetal microchimerism, or stem-cell-like fetal cells that cross the placenta and persist in maternal bone marrow and brain for decades. In this mouse model, microchimeric cells migrated to the brains of parous females after stroke and took on endothelial properties with pro-angiogenic and anti-inflammatory effects (Ritzel et al., 2017).
3. Preeclampsia and age-related diseases.
3.1. Epidemiology of preeclampsia and age-related diseases.
Preeclampsia, a multisystem, placentally-mediated hypertensive disorder affecting 2–8% of pregnancies (Abalos et al., 2013), is characterized by systemic inflammation, global endothelial dysfunction, increased oxidative stress, and vascular damage. Preeclampsia occurs only in pregnancy, but has a robust association with future age-related cardiovascular disease, including hypertension, ischemic heart disease, chronic kidney disease, and stroke (Wu et al., 2017). Women with a history of early-onset preeclampsia, before 34 weeks of gestation, are at even higher risk; one study showed a more than 9-fold increased risk of cardiovascular death in women with a history of early-onset preeclampsia (Mongraw-Chaffin et al., 2010). Shared risk factors such as hypertension and obesity may account for some of this epidemiologic association. However, emerging evidence demonstrates that the effects of preeclampsia may include lasting endothelial and cardiac dysfunction, which may contribute to cerebrovascular disease (Aouache et al., 2018; Berends et al., 2008; Granger et al., 2018; Hod et al., 2015). Cancer may also be associated with preeclampsia history: the Jerusalem Perinatal Study, a large registry-based study, found that women with a history of preeclampsia had 23% higher risk of any cancer, 37% higher risk of breast cancer, and more than double the risk of ovarian cancer (Calderon-Margalit et al., 2009). Of note, a meta-analysis of cohort studies did not show an association between preeclampsia and higher risk of breast cancer (Sun et al., 2018).
The relationship between preeclampsia and dementia is less well characterized, but multiple studies have suggested a long-term deleterious effect on cognition in women with a history of preeclampsia (Adank et al., 2021; Basit et al., 2018; Elharram et al., 2018; Postma et al., 2016; Postma et al., 2014). Small studies have demonstrated increased signs of cerebral small vessel disease on brain magnetic resonance imaging in middle aged women with a history of preeclampsia (Siepmann et al., 2017; Soma-Pillay et al., 2017). A prospective population-based Danish cohort study reported a 3-fold greater risk of vascular dementia in women with a history of preeclampsia (Basit et al., 2018). Analysis of the Swedish Medical Birth Register found that history of preeclampsia was associated with a more modest increased risk for vascular dementia of 63% (Andolf et al., 2020). Preeclampsia’s detrimental effects on cognition may be mediated by the increased risk of chronic hypertension after preeclampsia, since there is a strong positive association between elevated blood pressure and cognitive change (Gottesman et al., 2014; Iadecola et al., 2016; Muela et al., 2017). Thus, the relationship between preeclampsia and age-related cognitive decline could reflect a common underlying pathophysiology of maternal microvascular disease. In addition, preeclampsia may share some direct pathophysiological pathways with Alzheimer disease pathology: urine from women with preeclampsia exhibits congophilia, protein misfolding, and defective amyloid processing (Buhimschi et al., 2014).
3.2. Preeclampsia: placental aging gone wrong?
Preeclampsia is thought to result in part from abnormal implantation of the placenta, resulting in placental hypoxia, increased oxidative stress and premature placental aging(Sultana et al., 2017). This mechanism predominates in early-onset preeclampsia (Farladansky-Gershnabel et al., 2019), the form most highly associated with future maternal age-related cardiovascular disease (Mongraw-Chaffin et al., 2010). In contrast, late onset preeclampsia (after 34 weeks gestation) may reflect poor maternal adaptation to the high cardiometabolic demand of pregnancy (Thilaganathan, 2017), leading to placental oxidative stress and a similar clinical syndrome. Supporting this is the observation that maternal age is itself a risk factor for the development of gestational hypertension and preeclampsia (Gaillard et al., 2011; Sheen et al., 2018). A study in vervet monkeys reared in matrilineal colonies showed elevated blood pressure and inflammation in the third trimester in older mothers, suggesting that the aging process may impair maternal adaptation to pregnancy even under controlled environmental conditions (Plant et al., 2020). Evidence supports the hypothesis that accelerated placental aging leads to poor pregnancy outcomes, including preeclampsia, intrauterine growth restriction, and preterm delivery (Kohlrausch and Keefe, 2020). At the cellular level, placentas from preeclamptic pregnancies show signs of accelerated senescence including telomere shortening, altered mitochondrial autophagy, and activation of inflammatory and stress-associated signaling pathways (Manna et al., 2019).
4. Does preeclampsia “accelerate” vascular aging?
Preeclampsia is clearly associated with age-related diseases, particularly cardiovascular disease. Women who successfully “tolerate” and adapt to pregnancy without developing the preeclampsia syndrome may represent a more resilient phenotype. However, preeclampsia results in widespread maternal vascular endothelial damage that may persist long after the pregnancy is over (Barr et al., 2021; Kvehaugen et al., 2011). This may help explain the link between preeclampsia and maternal aging (Seals et al., 2011). Vascular endothelial dysfunction is a prominent feature of the aging process and is a hallmark of a wide array of cardiovascular diseases including hypertension, heart failure, chronic kidney disease, atherosclerosis and cerebrovascular disease (Herrera et al., 2010). Proposed mechanisms of age-related vascular endothelial dysfunction include oxidative stress, decreased endothelial nitric oxide bioavailability, increased production of endothelium-derived vasoconstrictive factors, compromised repair of endothelial cells, and increased expression of pro-inflammatory cytokines, all of which contribute to cellular senescence (Herrera et al., 2010; Wang et al., 2020). Similar mechanisms of vascular endothelial dysfunction are well described in preeclampsia pathophysiology (Myatt and Webster, 2009). Thus, it is biologically plausible that the widespread endothelial dysfunction observed during and after preeclampsia may contribute to premature vascular endothelial aging and early onset of age-related cardiovascular disease.
DNA damage has been implicated in the aging process as early as the 1960s (Vijg, 2021) and studies continue to suggest that it may play a central role in aging (Dai and Guo, 2021; Schumacher et al., 2021; Vijg and Dong, 2020). Intriguingly, there may also be a link between DNA damage and preeclampsia. One study found that mononuclear leukocyte DNA damage was significantly increased in mildly preeclamptic women and their offspring compared to controls (Hilali et al., 2013). A different study found that DNA damage was significantly more common in preeclamptic placentas and, notably, that this DNA damage was specifically localized to the maternal side of the placenta as opposed to the fetal (Tadesse et al., 2014). Hence, it is plausible that the DNA damage present in preeclamptic patients may further contribute to the acceleration of aging and age-related diseases.
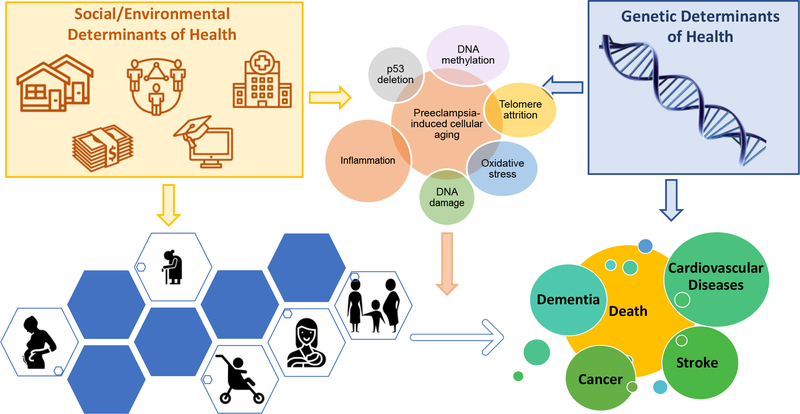
The complex web of overlapping factors that may contribute to the impact of pregnancy and preeclampsia on aging are illustrated in Figure 1.
Figure 1.
Interconnected environmental, social, and innate factors affecting women’s states of health and disease in pregnancy and beyond that may alter risk for aging and age-related diseases.
5. Genetics of preeclampsia.
The heritability of preeclampsia is estimated to be approximately ~55% (Cnattingius et al., 2004; Esplin et al., 2001; Gray et al., 2018b; Thomsen et al., 2015). The maternal genetic effect attributing to heritability is believed to be roughly ~35% and the fetal ~20% (Cnattingius et al., 2004; Gray et al., 2018b). The recent meta-analysis of genome-wide association studies (GWAS) (Steinthorsdottir et al., 2020) reported the maternal and fetal heritability for Europe as 38.1% and 21.3%, respectively, while those for Central Asia were found to be a bit higher—54.4% and 42.5% for maternal and fetal, respectively. Thus, preeclampsia has been the focus of numerous medical-genetic studies, including both candidate gene and GWAS, to identify genetic variants that influence the risk of developing preeclampsia.
5.1. The candidate gene approach
The candidate gene approach is based on prior biological knowledge of preeclampsia and consists of genotyping of single nucleotide polymorphisms (SNPs) in, or sequencing of, a known locus/gene or multiple loci/genes of interest to identify genetic variants associated with preeclampsia. Candidate studies for preeclampsia are summarized in Table 1. Of note, these studies are limited by small sample sizes and reported p values do not reach the canonically accepted genome-wide significance threshold of p<5e-8.
Table 1:
Candidate Approach of Preeclampsia (PE)
| Candidate(s) | Study participants | Associated SNP(s) | P-values | OR | Nature of variant(s) | Mapped Gene(s) | Reference |
|---|---|---|---|---|---|---|---|
|
| |||||||
| rs2234693 and rs9340799 in ESR1 | 119 severe PE cases and 103 controls; Caucasian women from Hungary | homozygous T-A haplotype carriers of rs2234693 and rs9340799 | 0.003 | 4.36 | intronic | ESR1 | (Molvarec et al., 2007) |
| rs1800629 in TNF-α, rs1800896 in IL-10, rs1800795 in IL-6, and rs2430561 in IFN-γ | 116 severe PE cases and 107 normotensive pregnant and 58 non-pregnant controls; women from Brazil | rs2430561 | <0.001 | not reported | intronic | IFN-γ | (Pinheiro et al., 2015) |
| C3 selected as candidate gene after primary complement screen | Primary complement screen: 259 severe PE cases and 426 controls; women from Finland C3 candidate screen: 960 severe PE cases and 760 controls; women from Finland |
rs2287845 rs366510 rs2287848 |
0.038 0.039 0.041 |
1.158 1.158 1.155 |
intronic intronic intronic |
C3 C3 C3 |
(Lokki et al., 2017) |
| STOX1 | 373 early-onset PE cases and 500 controls; Turkish women | rs1341667 | 0.03 | 1.45 | missense (Tyr153His) | STOX1 | (Pinarbasi et al., 2020) |
| rs1800896, rs1800871, rs180072 in IL-10 | 177 early-onset PE cases and 182 controls; women from Mongolia | rs1800872 | 0.04 | 1.67 | promoter region | IL-10 | (Song and Zhong, 2015) |
| rs1800629 in TNF-α | 14 PE cases and 12 controls; women from UK | rs1800629 | <0.05 | not reported | promoter region | TNF-α | (Chen et al., 1996) |
| rs1799724 in TNF-α | 133 PE cases and 115 controls; women from Finland | rs1799724* | 0.03 | 0.45 | promoter region | TNF-α | (Heiskanen et al., 2002) |
| rs2430561 in IFN-γ and rs1800896, rs1800871, and rs1800872 in IL-10 | 134 cases and 164 controls; women from Iran | rs1800896 | 0.045 | 1.45 | promoter region | IL-10 | (Kamali-Sarvestani et al., 2006) |
| rs1800629 in TNF-α, rs1800795 in IL-6, rs1800896 in IL-10 | 101 cases and 95 controls; women from Turkey | rs1800896 | 0.04 | 1.52 | promoter region | IL-10 | (Vural et al., 2010) |
| rs2243250 in IL-4, rs4986790 in TLR2, rs3918242 in MMP-9 | 117 PE cases and 146 controls; women from UK | rs2243250 | 0.055 | 1.534 | promoter region | IL-4 | (Fraser et al., 2008) |
| 71 SNPs in candidate genes (CXCR4, HNMT, KYNU, RPRM, ACVR1, ACVR1C, ACVR2A) at 2q22-23 locus on chromosome 2 | 1,139 PE cases and 2,269 controls; Norwegian women | rs1014064 rs17742134 rs1424941 rs2161983 rs3768687 rs3764955 |
0.0184 0.0214 0.0171 0.0196 0.0214 0.0327 |
0.86 1.17 1.18 0.86 0.86 0.87 |
intronic intronic intronic intronic intronic intronic |
ACVR2A ACVR2A ACVR2A ACVR2A ACVR2A ACVR2A |
(Roten et al., 2009) |
5.1.1. Genes involved in inflammation
Disruption of well-controlled immune functions is known to lead to preeclampsia. Preeclampsia is associated with an increase of inflammatory cytokines and a decrease in anti-inflammatory cytokines, both of which are thought to contribute to the promotion of the inflammatory state during preeclampsia (Harmon et al., 2016). Specifically, significantly higher levels of IL-2, TNF-α, and IFN-γ, and lower levels of IL-4 and IL-10, have been identified in preeclamptic patients (Darmochwal-Kolarz and Oleszczuk, 2014). Hence, several candidate gene studies focus on these inflammatory and anti-inflammatory cytokines. Several studies have reported associations for SNPs within the promoter region of TNF-α, a proinflammatory cytokine gene. The first study was conducted by Chen et al. (Chen et al., 1996) and the same polymorphism (rs1800629) was also found to be associated with preeclampsia in a later study by Mohajertehran et al (Mohajertehran et al., 2012). Interestingly, a protective variant was also found in the TNF-α promoter (Heiskanen et al., 2002), suggesting that regulation of TNF-α gene expression can modulate genetic risk of preeclampsia.
Aside from TNF-α, SNPs in the other cytokines namely IL-10, IL-4, IL-27, IL-1β, and IL-1α, were also reported in candidate gene studies (Table 1). A study conducted in India of 194 preeclamptic women and 194 controls showed that the levels of pro-inflammatory cytokines (TNF-α, IL-6) were significantly increased in placental tissues and serum samples while anti-inflammatory cytokines (IL-4, IL-10) were significantly decreased (Aggarwal et al., 2019). Pinheiro et al. found that the IFN-γ rs2430561 SNP was associated with severe preeclampsia in a Brazilian cohort of women (Pinheiro et al., 2015). Additionally, IL-10 rs1800872 SNP was associated with early-onset preeclampsia (Song and Zhong, 2015) and the rs1800896 SNP in the same gene was found to be associated with preeclampsia (Kamali-Sarvestani et al., 2006). For IL-4 two different SNPs were identified from two separate studies—the rs2243250 SNP and the VNTR polymorphism (Fraser et al., 2008; Salimi et al., 2014). Another cytokine implicated in association with preeclampsia is IL-1 which consists of two pro-inflammatory forms, IL-1alpha (IL-1α) and IL-1beta (IL-1β), that activate the inflammatory process (Di Paolo and Shayakhmetov, 2016). SNPs in both IL-1α and IL-1β were found to be associated with preeclampsia in cohorts of women from Sri Lanka, North Africa, and China (Andraweera et al., 2016; Hamid et al., 2020; Li et al., 2014). Aside from SNPs in IL-1α, Andraweera et al. also identified a SNP in MBL1, rs1800450, with a protective effect.
SNPs in IL-27 have also been associated with preeclampsia. IL-27 belongs to the IL-12 family of heterodimeric cytokines. This family of cytokines helps the differentiation and maintenance of Th1 subset cells (Yoshida et al., 2009). Additionally, levels of IL-27 and its receptor were found to be elevated in trophoblastic cells from the placenta of patients with preeclampsia (Yin et al., 2014). Liu et al. found that the CC genotype of rs153109 was associated with increased risk of preeclampsia while the CT genotype appeared to have a protective effect (Liu et al., 2016). Chen et al. also found a protective effect for rs153109 but reported significantly reduced pre-eclampsia susceptibility with the AG/GG genotypes of rs153109. It is plausible Chen et al. made an error in reporting genotypes since dbSNP reports the alleles for rs153109 as T>C (Chen et al., 2016). Chen et al. also reported that the IL-27 missense SNP rs17855750 is associated with mild preeclampsia risk.
SNPs in the maternal C3 complement gene were found to be associated with severe preeclampsia in a candidate gene study (Table 1). A more recent candidate gene study found that two SNPs in the fetal CD46 complement gene were also significantly associated with preeclamptic pregnancies relative to controls. This study by Banadakoppa et al.(Banadakoppa et al., 2020) also identified several complotypes consisting of fetal CD46 variants and maternal C3 or CHF variants that were significantly associated with preeclampsia. Increased C3 expression in the aging brain has been previously shown (Cribbs et al., 2012) and a study by Shi et al.(Shi et al., 2015) demonstrated that C3 deficient mice were protected from synapse loss compared to their WT counterparts as they aged. Furthermore, Wu et al. (Wu et al., 2019) found that C3 protein levels were increased in the brain and cerebrospinal fluid of Alzheimer’s disease patients. They also found that deleting C3 in mouse models of amyloidosis rescued plaque-associated synapse loss and deleting C3 in mouse models of tauopathy reduced neuron loss and brain atrophy.
Inflammasomes are involved in the process of pathogen clearance and sterile inflammation. They are large multi-protein complexes that play key roles in the production of the pivotal inflammatory cytokines. Recent evidence indicates that the inflammasome is involved in preeclampsia (Shirasuna et al., 2020; Socha et al., 2020). SNPs in the inflammasome genes were found to be associated with preeclampsia in candidate gene studies (Table 1). A SNP in CARD8 was identified by Wang et al. in a population of Han Chinese women and a SNP in NLRP1 was identified by Pontillo et al. in a population of Brazilian women (Pontillo et al., 2015; Wang et al., 2015).
5.1.2. Genes identified in family studies
STOX1 was identified as a preeclampsia susceptibility gene by genome-wide linkage analysis followed by sequencing of the confirmed region in a Dutch familial cohort (van Dijk et al., 2005). The same SNP identified in this gene, rs1341667 (Tyr153His), was also found to be associated with early-onset preeclampsia in a Turkish population (Pinarbasi et al., 2020) (Table 1). The rs1341667 SNP is in the DNA binding domain of the STOX1A (longest isoform of STOX1) transcription factor and is shown to negatively regulate trophoblast invasion by upregulation of the cell–cell adhesion protein α-T-catenin (CTNNA3) (van Dijk et al., 2010). A later study by this same group suggests that this variant may not only play a role in preeclampsia but also preterm birth by illustrating that the STOX1 Tyr153His mutation in extravillous trophoblast may have a significant negative effect on utero-vascular remodeling—a shared compromised process in preeclampsia and preterm birth (Dunk et al., 2021). For functional validation the group utilized primary extravillous trophoblast (EVT) explant and placental decidual coculture models as well as EVT-like cell lines which were used for transfection of the variant. EVT and transfected EVT-like cells with the STOX1 Tyr153His mutation secreted lower levels of IL-6, and IL-8, and higher XCL16 (chemokine [C-X-C motif] ligand 16) and TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) compared to control wild-type EVT cell and EVT-like cells. A study utilizing the STOX1A-overexpressing mice found that genes upregulated in endothelial cells of the transgenic mice were involved in inflammation and cellular stress (Miralles et al., 2019), implicating the functional role of STOX1A in preeclampsia etiology.
The ACVR2A gene was identified as a preeclampsia risk gene in Australian/New Zealand family studies (Fitzpatrick et al., 2009; Moses et al., 2006). Follow up studies found an association between SNPs in ACVR2A and preeclampsia in Norwegian and Brazilian populations (Ferreira et al., 2015; Roten et al., 2009) (Table 1). ACVR2A encodes activin A type II receptor (ActRIIA) which, along with Activin A receptor, type IB (Alk-4), plays an important role in the establishment of pregnancy, especially during decidualization, trophoblast invasion and placentation processes (Florio et al., 2010).
5.1.3. Genes associated with blood pressure
SNPs within PLEKHG1 have been associated with blood pressure traits (i.e., both systolic and diastolic blood pressure) in multi-ethnic GWAS (Franceschini et al., 2013). A study by Gray et al. identified a maternal SNP in the PLEKHG1 gene associated with increased risk of preeclampsia and reported a p-value of 5.9e-7 which is close to approaching canonical GWAS significance (Gray et al., 2018a). The genetic deletion of catechol O-methyltransferase (COMT) in mice produces a preeclampsia-like phenotype such as hypertension, proteinuria, and histological changes, consistent with human pathological features (Palmer et al., 2011). The functional COMT missense SNP, which results in the substitution of a valine for a methionine (Val158Met) was found to be associated with preeclampsia in two separate candidate gene studies where the study subjects for one were Korean women and the other Chinese women. The Met(A)-allele of this SNP is associated with a 3- to 4-fold decrease in COMT enzyme activity (Lotta et al., 1995) and pregnant mice deficient in catechol-O-methyltransferase (COMT) show a preeclampsia-like phenotype resulting from an absence of 2-methoxyoestradiol (2-ME) (Kanasaki et al., 2008).
5.1.4. Genes involved in angiogenic balance
A hallmark characteristic of preeclampsia, especially early-onset preeclampsia, is angiogenic imbalance. This imbalance results from reduced bioavailability of vascular endothelial growth factor (VEGF) and placental growth factor (PlGF) due to an inappropriately upregulated sFLT-1 acting as a decoy receptor (Eddy et al., 2018). A study by Turanov and colleagues explored RNAi-based modulation of sFLT-1 as a potential treatment of preeclampsia in animal models. They found that targeting sFLT-1 via siRNAs resulted in a decrease of circulating sFLT-1 in pregnant mice. In a baboon model of preeclampsia, a single dose of siRNAs successfully suppressed sFLT-1 overexpression as well as ameliorated clinical manifestations of preeclampsia, specifically hypertension and proteinuria (Turanov et al., 2018). SNPs associated with preeclampsia in FLT1, the receptor for VEGFA, VEGFB, and placental growth factor (PIGF) were found across several studies as were SNPs in VEGFA, VEGFB, and VEGFC (Amosco et al., 2016; Hamid et al., 2020; Kikas et al., 2020; Srinivas et al., 2010) (Table 1).
5.1.5. MTHFR
Homocysteine concentration is increased in preeclampsia and it negatively correlates with plasma folate concentration (Powers et al., 1998). Hyperhomocysteinemia leads to vascular and metabolic changes, an established risk factor for endothelial disorders. The human MTHFR gene encodes methylenetetrahydrofolate reductase (MTHFR), a key enzyme in folate and homocysteine metabolism. Thomsen et al. identified a preeclampsia associated MTHFR SNP—rs17367504— with a protective effect by a study done in Norwegian women and reported one of the most significant p values from the other candidate gene studies reported in Table 1 (p=3.53e-5)(Thomsen et al., 2017) (Table 1). Moreover, the G allele of this SNP has also been associated with lower blood pressure (Newton-Cheh et al., 2009).
5.1.6. ESR1
Estrogen is one of the main pregnancy hormones produced by the placenta and stimulates angiogenesis in the uterus during the reproductive cycle. Estrogen receptor α (ESR1) plays an important role in the adaptation of increased uterine blood flow during gestation (Chen et al., 2012). The ESR1 gene encodes a ligand-activated transcription factor that can be activated by estrogen and in its absence by growth factors. Molverac et al. found that haplotype carriers of the ESR1 rs2234693 and rs9340799 SNPs showed an increased risk of severe preeclampsia (Molvarec et al., 2007) (Table 1). A preceding study had linked the same estrogen receptor 1 (ESR1) haplotype to risk of myocardial infraction in postmenopausal women but not in men (Schuit et al., 2004).
5.2. The GWAS approach
Complex traits such as preeclampsia are those for which both genetics and environment contribute to the variance in the population. For most complex traits, a large number of distinct genetic loci influence the phenotypic variability. Over the past decade, genome-wide association studies (GWAS) have become the standard approach to assess the genetic contribution of complex traits. These hypothesis-free genome-wide scans have delivered many novel discoveries with sufficient power to detect small effects at common SNPs. However, as summarized in Table 2 (Johnson et al., 2012; McGinnis et al., 2017; Steinthorsdottir et al., 2020; Zhao et al., 2013; Zhao et al., 2012), the majority of SNPs identified in the preeclampsia GWAS do not reach the genome-wide significance threshold of p<5e-8, suggesting that GWAS of preeclampsia is still in its infancy. An exception is the study by McGinnis et al. (McGinnis et al., 2017). This was the first GWAS of preeclamptic offspring and reported that a SNP within the enhancer element near the FLT1 gene encoding vascular endothelial growth factor receptor 1 (VEGFR1) was associated with preeclampsia risk. A recent study by the same group (Steinthorsdottir et al., 2020) identified more SNPs (rs1421085, rs259983, rs1918975, rs1458038, rs10774624, rs4769612) with the GWAS significance. The SNP reported from the cohort of offspring born from preeclamptic pregnancies (rs4769612) is in linkage disequilibrium (LD) with the most significant SNP (rs4769613) previously reported by the same group. SNPs in LD with one another are often inherited together due to co-segregation during meiotic recombination (Cano-Gamez and Trynka, 2020). This new study also had a much larger sample size that was comprised of both European and Asian women as opposed to just European women in their previous study. In fact, the study by Steinthorsdottir et al. (Steinthorsdottir et al., 2020) was the largest regarding sample size of all the GWAS reported. The SNPs reported from the maternal genome (rs1421085, rs259983) have also been reported to associate with blood pressure (BP) in previous studies. Interestingly, rs4769612 was found to be associated with the offspring genome, providing strong support for genetic variants near FTL1 associated with preeclampsia in offspring of such pregnancies.
Table 2:
GWAS Approach of Preeclampsia (PE)
| Study participants | SNPs | P-values | OR | Nature of variant(s) | Mapped gene(s) | Reference |
|---|---|---|---|---|---|---|
|
| ||||||
| 538 PE cases and 540 controls; Caucasian women from Australia | rs7579169 rs12711941 |
3.58e-7 4.26e-7 |
1.57 1.56 |
intergenic intergenic |
INHBB INHBB |
(Johnson et al., 2012) |
| 177 PE cases and 116 controls; Caucasian women from Iowa | rs1426409 rs17686866 rs9831647 rs10743565 |
2.58e-2 (allelic); 3.14e-6(genotypic) 1.97e-5(allelic); 3.80e-6(genotypic) 1.65e-2(allelic); 9.36e-6(genotypic) 5.00e-6(allelic); 1.64e-5(genotypic) |
not reported | intergenic intronic intronic intronic |
KIAA1239 ESRRG LMCD1 IFLTD1 |
(Zhao et al., 2012) |
| 21 PE cases and 1,049 controls; Afro-Caribbean women from Barbados 50 PE cases and 1,207 controls; women of European ancestry 62 PE cases and 661 controls; Hispanic women |
rs11617740 rs7322722 rs17412740 |
7.3e-7 (Afro-Carribean) 1.23e-6 (European) 2.14e-6 (Hispanic) |
16.69 2.93 6.08 |
intronic intronic intergenic |
FGF14 MYCBP2 LZTS1 |
(Zhao et al., 2013) |
| GWAS + replication n = 4,380 cases and 310,238 controls; offspring from European women | (infant) rs4769613 rs12050029 rs149427560 |
5.40e-11 3.00e-6 4.10e-5 |
1.22 1.20 1.30 |
enhancer enhancer intergenic |
Flt-1 Flt-1 Flt-1 |
(McGinnis et al., 2017) |
| meta -analysis of eight previously unreported GWAS consisting of 9,515 PE cases and 157,719 controls; women from Europe and Central Asia. Follow-up with additional samples of European and Kazakh origin (2,635 PE cases; 6,379 controls) (maternal) meta-analysis consisting of 6,775 offspring born of PE complicated pregnancies and 375,372 controls; women from Europe and Central Asia. (infant) | (maternal) rs1421085 rs259983 rs1918975 rs1458038 rs10774624 (infant) rs4769612 |
1.2e-9 2.9e-10 1.2e-8 1.2e-8 1.7e-8 4.3e-14 |
1.11 1.17 1.10 1.11 1.11 1.19 |
intronic intronic intronic intergenic intronic intergenic |
FTO ZNF831 MECOM PRDM8, FGF5 LINC02356 Flt-1 |
(Steinthorsdottir et al., 2020 |
While we indicate mapped genes of GWAS SNPs in Table 2, it is challenging to identify causal genes that underlie genetic association as we described in section 6.
6. Functional genomics of preeclampsia
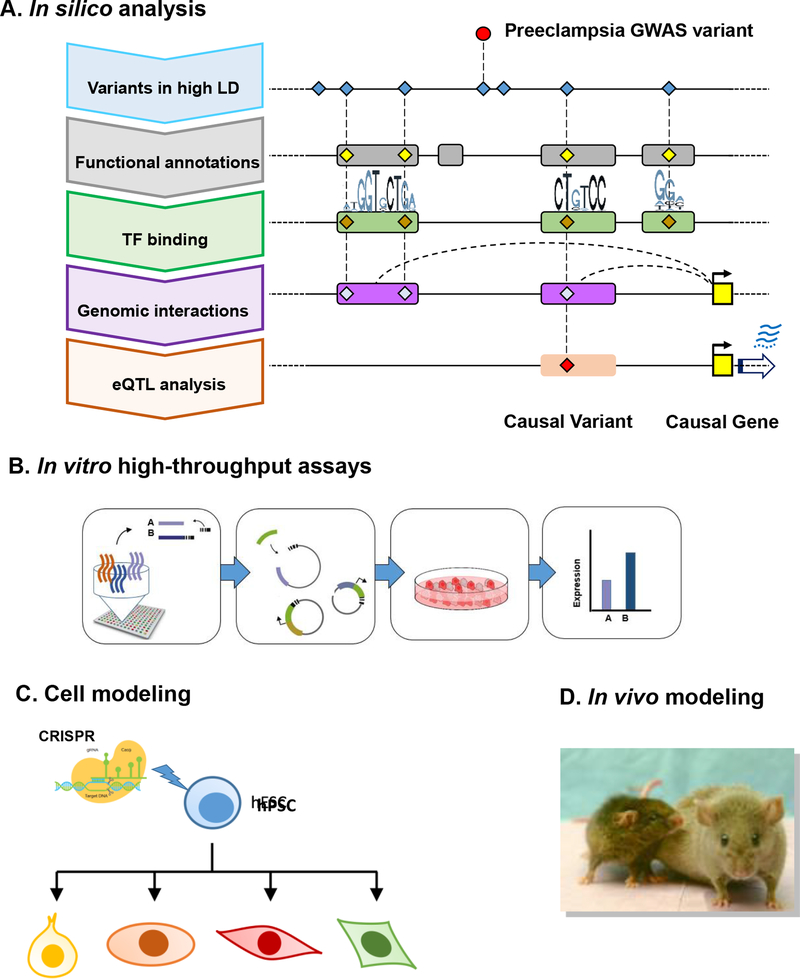
Figure 2 illustrates a multi-faceted challenge in identification of causal variants and causal genes of preeclampsia starting with GWAS signal and provides approaches to realize the full promise of GWAS in revealing new biological insights and novel targets for intervention. We will discuss current progress towards in silico and experimental identification, and validation of causal variants that interfere with enhancer function and causal genes that are dysregulated by these variants, thereby conferring preeclampsia risk.
Figure 2.
Functional genomics of preeclampsia illustrating challenges in interpreting GWAS. (A) In silico analysis to identify candidate causal variants and genes of preeclampsia informed by GWAS. (B) High-throughput screens such as a massive parallel reporter assay (MPRA) to investigate a large number of regulatory variants within a single experiment for identification and prioritization of functional variants. (C) Cell modeling using CRISPR-engineered pluripotent stem cell followed by differentiation into multiple cell types to study cell type-specific functional impact of variants in vitro. (D) Mouse modeling to study the functional impact of variants in vivo.
6.1. Challenges in identifying causal genes and causal variants of preeclampsia risk
All of the GWAS SNPs found to be associated with preeclampsia occur in the non-coding regions of the genome (Table 2). This is in line with the fact that a vast majority of GWAS SNPs associated with human diseases occur outside of protein coding regions (Cano-Gamez and Trynka, 2020). These results suggest that disease associated GWAS variants confer disease risk by altering functional DNA elements in the genome that regulate gene expression. Variation in gene expression has been shown to be highly heritable and a significant determinant of human disease susceptibility (Cookson et al., 2009). Indeed, disease-associated SNPs are found to be significantly enriched in transcriptional regulatory elements (Altshuler et al.; Corradin et al., 2014; Harismendy et al.; Hnisz et al., 2013; Lee et al., 2013; Maurano et al., 2012; Musunuru et al., 2010; Stitzel et al., 2010; Trynka et al., 2013), indicating that GWAS SNPs contribute to disease risk by altering gene expression. However, the most prevalent regulatory element in the human genome, i.e. enhancer, is capable of activating transcription of their target genes at great distances independent of the location, and a single enhancer can regulate multiple target genes (Jin et al., 2013). Therefore, identifying true causal genes of a non-coding variant is challenging. A common empirical practice is to assign the non-coding GWAS variants to the nearest gene, which may not necessarily reflect the real situation and can be misleading (Li et al., 2012; Sanyal et al., 2012). The best example is the obesity-related variants identified within the intronic regions of the FTO gene, which turned out to regulate two distal genes, IRX3 and IRX5, located a megabase away from the variants, but not the FTO gene itself, in both humans and mice (Smemo et al., 2014). Until this discovery, significant efforts over a number of years were wasted to study the role of FTO in obesity/T2D in mouse model studies (Church et al., 2010; Dina et al., 2007; Fischer et al., 2009; Frayling et al., 2007; Grunnet et al., 2009; Scuteri et al., 2007; Wahlen et al., 2008). Of note, a SNP in the FTO locus was also found to be associated with preeclampsia by GWAS (Table 2).
Furthermore, due to extended LD in the genome, multiple variants in an associated region can be identified as significant and hence identifying truly causal variants is highly challenging. For example, a causal SNP in the FTO locus was found to be a linked SNP within an intron of FTO, which disrupts the binding of a transcriptional repressor, ARID5B (Claussnitzer et al., 2015). This alteration in binding of the repressor leads to inappropriate enhancer activation during adipocyte differentiation, and a doubling of IRX3 and IRX5 expression, resulting in a shift in adipocyte cell fate (Claussnitzer et al., 2015). The FTO example demonstrates the significant challenge in the post-genomic era in identifying truly causal GWAS variants and elucidating how they cause dysregulation of target gene expression.
6.2. Identification of mechanisms underlying genetic association of preeclampsia
Since GWAS detect only statistical associations, not functional signals, it is difficult to predict mechanisms underlying genetic association. All of preeclampsia GWAS risk variants represent genetic variations in the non-coding regions of the genome (Table 2), suggesting a significant role of gene regulatory changes on preeclampsia risk. Functional roles of causal variants on preeclampsia risk are difficult to predict due to incomplete knowledge of non-coding regulatory elements, their mechanisms of action, and the cellular states and processes in which they function, let alone the identification of truly causal variants and their target genes. Lack of such knowledge is a major impediment to understand the molecular mechanisms of preeclampsia and to develop strategies against the debilitating condition.
Non-coding variants may play regulatory roles for gene expression through multiple mechanisms: variants in promoters can directly impact transcription initiation (Nurnberg et al., 2012); variants in intronic and in untranslated region (UTR) can potentially affect the property of mRNAs, leading to altered stability or splicing patterns; and variants may alter function or expression of multiple classes of non-coding RNAs (ncRNAs), including long non-coding RNAs (lncRNAs) and small RNAs such as micro RNAs (miRNAs) and small nucleolar RNAs (snoRNAs) (Hrdlickova et al., 2014).
6.2.1. Role of enhancers
Many recent studies show that non-coding GWAS variants are significantly enriched in cell type-specific transcriptional enhancer regions (Altshuler et al.; Corradin et al., 2014; Harismendy et al.; Hnisz et al., 2013; Lee et al., 2013; Maurano et al., 2012; Musunuru et al., 2010; Stitzel et al., 2010; Trynka et al., 2013), especially within enhancers specific to disease-relevant cell types (Javierre et al., 2016; Kundaje et al., 2015). Enhancers are the principle regulatory components of the genome that enable cell-type and cell-state specificities of gene expression. Enhancers were initially defined as DNA elements that act over a distance to positively regulate expression of protein encoding target genes, independent of orientation and direction with respect to the target gene promoters (Banerji et al., 1981). Enhancers have emerged as major points of integration of intra- and extra-cellular signals associated with development, homeostasis and disease, resulting in context-specific transcriptional outputs (Levine, 2010).
The human genome is estimated to encode ~1 million enhancer elements and distinct sets of approximately 30,000 – 40,000 enhancers are active in a particular cell type (Bernstein et al., 2012; Roadmap Epigenomics et al., 2015), vastly outnumbering protein-coding genes and promoters. Enhancer activation entails the presence of specific recognition sequences required for the cooperative recruitment of transcription factors (TFs) that initially activate and subsequently permit signal-dependent regulation of gene expression in a spatial and temporal fashion (Heinz et al., 2010). By contrast, genetic variations in enhancer sequences that alter TF binding would predispose to ‘improper’ gene expression and ultimately susceptibility to diseases (Harismendy et al., 2011; Musunuru et al., 2010). The enhancer-bound TFs facilitate chromatin accessibility by recruitment of nucleosome remodelling complexes with the core 80–120 basepairs representing the sites for binding of the activating/regulatory TFs. Cell-specific enhancer activation is driven by combinatorial actions of lineage-determining (LDTFs) and signal-dependent transcription factors (SDTFs)(Heinz et al., 2015), and genetic variation affecting enhancer function is a major determinant of differences in cell-specific gene expression between individuals (Tak and Farnham, 2015). By defining the role of a specific enhancer implicated by a preeclampsia risk association, it is possible to infer the identities of regulatory factors and environmental signals the cell is receiving that might impact preeclampsia.
6.2.2. Enhancer variants and causal genes
A significant challenge in understanding and predicting gene expression is to define the target genes of transcriptional enhancers. Since enhancers are capable of activating transcription of their target genes at great distances, the definite way to identify targets of enhancers is through the detection of enhancer-promoter interaction loops. Indeed, the recent ability to develop contact maps on a genome-wide scale indicates that many enhancer-like regions skip over the nearest gene and make contacts with more distant targets. Therefore, an accurate interpretation of the effects of non-coding genetic variation requires methods that allow correct assignment of regulatory elements with their target genes. Chromatin conformation capture (3C) and its derivatives such as HiC, Chromatin Interaction Analysis with Paired-End-Tag sequencing (ChIA-PET), Hi-ChIP, and Proximity ligation assisted chromatin immunoprecipitation (PLAC-seq) (Fang et al., 2016; Rao et al., 2014; Wei et al., 2006) have enabled open-ended mapping of interactions between enhancers and cognate genes in a chromosome-wide fashion to permit a greater understanding of enhancer regulatory networks (Calo and Wysocka, 2013; Catarino and Stark, 2018). Through comparative analyses, a locus where alterations of regulatory loop interactions are associated with GWAS variants and disease conditions can be identified. Cell type- and allele-specific effects of GWAS SNPs on enhancer-promoter interactions can be also elucidated, providing insight into how causal non-coding risk SNPs induce alterations in interaction with the causal target genes.
6.3. Functional validation of causal genes and variants of preeclampsia
Active and poised enhancers are found within open chromatin and contain transcription factor (TF) binding sites, and are enriched for various histone marks (Heintzman et al., 2009). Computational methods, like ChromHMM (Ernst and Kellis, 2012), use these features to identify enhancers. It has been unclear to what extent the marker signals indicate true enhancer activity as opposed to regulatory potential (Sur and Taipale, 2016), necessitating extensive experimental validation studies of predicted enhancers and target genes. Therefore, functional studies are needed in order to elucidate cell-specific regulatory activity and the affected causal gene(s) (Figure 2B–D). This is one of the most critical, but often lagged behind, steps to test the functional relevance of the non-coding variants detected in GWAS and to establish the causality of GWAS variants in conferring disease risks.
6.3.1. In silico analysis to identify causal variants and causal genes of preeclampsia
Figure 2A illustrates a conceptual framework for in silico identification of causal variants and causal genes of preeclampsia starting with GWAS signal. Genomic annotation of regulatory elements such as enhancers has been greatly facilitated by the development of high-throughput methods, providing surrogate markers for regulatory activity at an unprecedented resolution (Andersson et al., 2014; Consortium, 2012; Kim et al., 2010; Wang et al., 2011). Enhancers are typically characterized by the presence of histone modifications (detected by ChIP-seq) such as H3K27Ac and H3K4me1/2 (Heintzman et al., 2007). Notably, the H3K27Ac positive enhancers showed high enhancer activity and co-occupancy with lineage--determining transcription factors (LDTFs) (Creyghton et al., 2010). Thus, it has been proposed that H3K27Ac distinguishes active enhancers from the primed or poised enhancers. Binding of transcription factors to enhancers results in depletion of nucleosomes, making the region detectable by DNase-seq and ATAC-seq (Buenrostro et al., 2015; Song and Crawford, 2010). The enhancers are transcription units themselves, and enhancer DNA is transcribed bidirectionally, resulting in the generation of enhancer RNAs (eRNAs) that are usually unstable. Specifically, it is active enhancers that produce eRNAs (Kim et al., 2010; Wang et al., 2011), which are critical for the recruitment of transcription and splicing complexes, driving their gene-regulatory function (Engreitz et al., 2016; Joung et al., 2017). Expression of enhancer RNAs (eRNAs) can be detected by deep RNA-seq, Global Run-On Sequencing (Gro-Seq) or Cap Analysis Gene Expression (CAGE) (Andersson et al., 2014; Core et al., 2014; Wu et al., 2014). Recent studies suggested that eRNAs could play a role in chromatin looping for interaction with the target gene promoter (Yang et al., 2013). Finally, enhancers are hypomethylated at CpG dinucleotides, and hence can be detected by bisulfite sequencing (Wiench et al., 2011).
By integrating the genome-wide epigenome signals with GWAS signals (Figure 2A) based on rigorous statistical modeling, adopting genomic annotations and estimating probabilities of causality for regulatory elements and variants, one can identify or substantially narrow the set of functional preeclampsia SNPs and their target genes that are candidates for affecting preeclampsia risk. Functional variants will be those that reside in the active regulatory elements such as enhancers that are active in cell- and tissue- types relevant for preeclampsia pathophysiology. By connecting enhancer activity and target gene transcription by analysis of expression quantitative trait loci (eQTL) and chromatin interactions, one can investigate how functional variants induce their alterations and to what extent preeclampsia SNPs exert functional effects on target gene expression in each cell- and tissue- type. Ultimately, integrative analysis of preeclampsia cases versus controls will be needed to provide insights into the dysregulated enhancers/targets in each cell type of patients compared to controls as well as the transcription factors underlying the enhancer dysregulation.
6.3.2. High-throughput assays
The central strategy is to employ assays that will determine if causal enhancers and SNPs prioritized from the in silico analysis actually impair regulatory function and dysregulate cognate target genes, and to understand the underlying mechanisms. With the advent of high-throughput sequencing and genome-editing, it is now more feasible to accomplish functional studies on a genomic scale. Here we highlight technologies that can be used in functional analysis of preeclampsia SNPs. The massive parallel reporter assay (MPRA) allows examination of a large number of enhancers and enhancer variants within a single experiment (Arnold et al., 2013; Inoue and Ahituv, 2015; Melnikov et al., 2012). Typically, thousands of candidate enhancer regions are synthesized and cloned into a mammalian expression vector, where the co-synthesized barcodes or the enhancers themselves are transcribed as identifiers for each construct. The mixed reporter library is then transfected to a cell line, and the vector DNA and the reporter RNA transcripts are individually collected and sequenced. The enhancer activity of the constructs can then be represented by the read count ratio between RNA and DNA. Several groups have reported success in using MPRA to identify causal GWAS variants (Tewhey et al., 2016; Ulirsch et al., 2016) (Figure 2B). To overcome the limitations of enhancer reporter assays, one can transiently silence causal enhancers harboring preeclampsia SNPs using CRISPRi technology(Gilbert et al., 2013; Laufer and Singh, 2015; Thakore et al., 2015). By interfering with enhancer activity in the context of the genome, one can investigate the endogenous role of the causal enhancer preeclampsia SNPs on cognate target gene expression. This approach will expedite the selection of high-probability causal enhancers harboring preeclampsia SNPs before performing the more challenging CRISPR-mediated genome engineering.
6.3.3. Cell modeling
Since enhancer function depends on the local chromatin context, genome-editing tools are indispensable for studies of enhancer mechanisms in the endogenous genome. CRISPR/Cas9 is a recently developed technology that allows efficient and scalable targeted genome-editing. CRISPR/Cas9 recognizes target sequence by binding to a roughly 20 basepair-long complementary guide RNA (gRNA), allowing highly cost-efficient and simplified assay designs. While the CRISPR/Cas9-mediated gene editing has revolutionized the functional validation of enhancers, the throughput of the method is typically low, requiring prioritization of candidate variants by other approaches. One exception, though, is that if the phenotypic outcome is either related to cellular survival or detectable by cell sorting, it will be possible to design CRISPR-based screening assays by creating complex viral libraries and infecting cells with low density. The principles of such screening assays have been well demonstrated by several studies performing Genome-Scale CRISPR Knock-Out (GeCKO) (Hart et al., 2015; Shalem et al., 2014). In addition to the throughput, another concern, when performing genome-editing, is the choice of a suitable model. Generation of knock-in cells by CRISPR in human pluripotent stem cells, such as embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) followed by differentiation into cell types relevant to preeclampsia will be an ideal approach to establish the causality of candidate functional variants in dysregulating target genes in the context of the endogenous chromatin landscape (Figure 2C).
6.3.4. In vivo modeling
Since preeclampsia is a complex trait, in vivo studies should be intuitively preferred. However, performing studies using animal models is usually limited due to poor conservation of non-coding sequences between species. Studies of enhancers across species suggested their conservation at a functional level rather than nucleotide sequence (Fisher et al., 2006; Stergachis et al., 2014). Therefore, although modeling the effect of a particular variant could be difficult, the underlying functional enhancer would be more likely conserved and available for in vivo studies. Epigenetic annotations of regulatory elements in model organisms, such as mouse ENCODE, are available to search for enhancer candidates (Celniker et al., 2009; Mouse et al., 2012). In silico analysis will lead to identification of orthologous enhancers in the syntenic regions of mouse genome and one can harness the power of mouse genetics to investigate the impact of the enhancers harboring preeclampsia risk alleles in vivo. To test the functional impact of candidate causal enhancers in vivo using mouse models, one can genetically engineer mice to mimic the variant effects by mutagenizing preeclampsia enhancers highly conserved between human and mice at the levels of sequence homology and enhancer activity in the right cell- and tissue-types and investigate their impact on transcriptional outputs in multiple cell types and phenotypes relevant to preeclampsia. The in vivo modeling will ultimately reveal the casual role of GWAS variants in vivo for clinical relevance.
6.3.5. Functional validation studies of preeclampsia risk variants
To the best of our knowledge, there has yet to be a study describing the functional effects of a preeclampsia GWAS SNP. Closest studies in identifying causal variants have been made in regards to FOXD1 mutations and Intrauterine growth restriction (IUGR), a disorder frequently associated with early-onset preeclampsia (Huppertz, 2008). A study conducted by Quintero-Ronderos et al. (Quintero-Ronderos et al., 2019) utilized luciferase assays as the basis for functional experiments and identified a novel and rare variant in an IUGR patient. This variant, rs750578392, led to increased C3 transcriptional activity and increased PIGF transcriptional activity. Quintero-Ronderos et al. also found a previously studied mutation from the work of Laissue et al. (Laissue et al., 2016) in a patient affected by both preeclampsia and IUGR. The Laissue et al. study found that the rs917127030 variant was unable to activate the PIGF promoter in luciferase assays. PIGF levels are often found to be decreased in preeclamptic women (Chau et al., 2017).
7. Summary and perspectives
There are clear differences between men and women in overall lifespan and disability and disease as they age. Despite clear sex differences in aging, the impact of pregnancy and its complications, such as preeclampsia, on aging is an underexplored area of aging research. One of the most significantly distinct female life events is pregnancy and the need for women to adapt their physiology to maintain a successful gestation. Despite our significant understanding of the acute changes that are required during pregnancy, we know very little about the longer-term impact of this adaptation. In this review, we have tried to disentangle the complex interactions between pregnancy and its adverse outcomes, preeclampsia, and maternal aging and aging-related diseases from the perspective of epidemiology to current functional genomics.
From an epidemiologic standpoint, it remains uncertain whether pregnancy itself slows, hastens or prolongs the aging process. The data are complex, and it is difficult to separate the physiologic impact of pregnancy from secondary effects such as child rearing or socioeconomic weathering. However, what appears to be certain is that having an adverse response to pregnancy will identify a group of women at risk for age-related diseases, potentially presenting important mid-life clues about the subsequent aging process. In particular, specific pregnancy related events such as preeclampsia and gestational diabetes lead to an increased risk of later life cardio- and neuro- vascular events that occur at an earlier age than women with an uncomplicated pregnancy and are frequently preceded by a prolonged asymptomatic period. Understanding the physiologic underpinnings of this has the potential to reveal clues to the overall aging process in women.
Since preeclampsia is a major risk factor of major age-related diseases in women, understanding the fundamental mechanisms of preeclampsia has a transformative potential to improve women’s health in the aging population. This is in line with the conceptual framework of “geroscience”, highlighting the need for targeting the main risk factor for all chronic diseases, the aging process itself, to combat all the diseases of old age simultaneously. From the few hints that are available, endothelial dysfunction appears to have a prominent role in both preeclampsia and aging (Aouache et al., 2018; Berends et al., 2008; Granger et al., 2018; Hod et al., 2015). However, the precise nature of the underlying factors precipitating the endothelial cell dysfunction and ultimately triggering the biological cascade(s) leading to both the acute and longer term pathologic vascular changes is unknown. Whether a primary, perhaps genetic, defect underlies both the abnormal response to pregnancy and the subsequent accelerated aging or alternatively, a primary pregnancy induced endothelial injury from an episode of preeclampsia triggers the cardiovascular disease, is unknown. There are clues that support either hypothesis.
A number of GWAS and candidate studies point to genes involved in endothelial function supporting an underlying genetic etiology. It is equally reasonable to surmise that the excessive maternal cardiovascular responses associated with placental insufficiency and leading to preeclampsia, such as oxidative stress; inflammation; changes in DNA methylation; and shortening of telomeres (Figure 1), may trigger irreversible endothelial damage which may contribute to premature vascular endothelial aging and early onset of age-related cardiovascular disease (Giller et al., 2020; Iqbal et al., 2018; Pollack et al., 2018). At present, convincing data confirming either etiology is limited. The similarities between preeclampsia and cardiovascular aging suggest that studying this relationship could be revealing in understanding more chronic aging processes. This will require emphasis on cell-specific regulatory activity; be it response to injury or inherent dysfunction. Similarly, treatments presently suggested to prevent or ameliorate many of the cardiovascular sequelae of preeclampsia, such as statins, low dose aspirin, and metformin, may have value in delaying aging (Brownfoot et al., 2015; Brownfoot et al., 2015 July; Smith et al., 2020; Ma’ayeh et al., 2020).
While variants identified by GWAS confer a rather small increase in risk individually, recent meta-analysis has shown that as a group, targets based on evidence from GWAS-associated loci are twice as likely to be therapeutically valid as those that are not (Nelson et al., 2015). Thus, it is important to delineate the mechanisms underlying preeclampsia-associated variants at a molecular level. Preeclampsia GWAS have shown that all associated variants reside in the non-coding regions of the genome, suggesting that gene regulatory changes contribute to preeclampsia risk. To identify truly causal non-coding variants and their affected target genes remains challenging but is a critical step to translate the genetic associations to molecular mechanisms, and ultimately clinical applications. Biological insights can then be utilized to improve clinical outcomes, including developing effective strategies for disease prevention and/or therapeutics.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abalos E, Cuesta C, Grosso AL, Chou D, Say L, 2013. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol 170, 1–7. [DOI] [PubMed] [Google Scholar]
- Adank MC, Hussainali RF, Oosterveer LC, Ikram MA, Steegers EAP, Miller EC, Schalekamp-Timmermans S, 2021. Hypertensive Disorders of Pregnancy and Cognitive Impairment: A Prospective Cohort Study. Neurology 96, e709–e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal R, Jain AK, Mittal P, Kohli M, Jawanjal P, Rath G, 2019. Association of pro- and anti-inflammatory cytokines in preeclampsia. J Clin Lab Anal 33, e22834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro P, Ponce A, Komal S, de la Asuncion Villaverde M, Castrillo C, Grau G, Simon L, de la Sierra A, 2020. Multimorbidity gender patterns in hospitalized elderly patients. Plos One 15, e0227252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES, 2008. Genetic mapping in human disease. Science 322, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amosco MD, Villar VA, Naniong JM, David-Bustamante LM, Jose PA, Palmes-Saloma CP, 2016. VEGF-A and VEGFR1 SNPs associate with preeclampsia in a Philippine population. Clin Exp Hypertens 38, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Consortium F, Forrest AR, Carninci P, Rehli M, Sandelin A, 2014. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolf E, Bladh M, Moller L, Sydsjo G, 2020. Prior placental bed disorders and later dementia: a retrospective Swedish register-based cohort study. Bjog-Int J Obstet Gy 127, 1090–1099. [DOI] [PubMed] [Google Scholar]
- Andraweera PH, Dekker GA, Jayasekara RW, Dissanayake VH, Roberts CT, 2016. Polymorphisms in the inflammatory pathway genes and the risk of preeclampsia in Sinhalese women. J Matern Fetal Neonatal Med 29, 1072–1076. [DOI] [PubMed] [Google Scholar]
- Aouache R, Biquard L, Vaiman D, Miralles F, 2018. Oxidative Stress in Preeclampsia and Placental Diseases. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer CR, Recker M, Duffy E, Hosken DJ, 2018. Intralocus sexual conflict can resolve the male-female health-survival paradox. Nat Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A, 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074–1077. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE, 2016. Sex Differences in Lifespan. Cell Metab 23, 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad SN, Hoffman JM, 2018. Is antagonistic pleiotropy ubiquitous in aging biology? Evol Med Public Health 2018, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banadakoppa M, Balakrishnan M, Yallampalli C, 2020. Common variants of fetal and maternal complement genes in preeclampsia: pregnancy specific complotype. Sci Rep 10, 4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W, 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299–308. [DOI] [PubMed] [Google Scholar]
- Barr LC, Pudwell J, Smith GN, 2021. Postpartum microvascular functional alterations following severe preeclampsia. Am J Physiol Heart Circ Physiol. [DOI] [PubMed] [Google Scholar]
- Basit S, Wohlfahrt J, Boyd HA, 2018. Pre-eclampsia and risk of dementia later in life: nationwide cohort study. BMJ 363, k4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeri MS, Rapp M, Schmeidler J, Reichenberg A, Purohit DP, Perl DP, Grossman HT, Prohovnik I, Haroutunian V, Silverman JM, 2009. Number of children is associated with neuropathology of Alzheimer’s disease in women. Neurobiol Aging 30, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beral V, Bull D, Pirie K, Reeves G, Peto R, Skegg D, LaVecchia C, Magnusson C, Pike MC, Thomas D, Hamajima N, Hirose K, Tajima K, Rohan T, Friedenreich CM, Calle EE, Gapstur SM, Patel AV, Coates RJ, Liff JM, Talamini R, Chantarakul N, Koetsawang S, Rachawat D, Marcou Y, Kakouri E, Duffy SW, Morabia A, Schuman L, Stewart W, Szklo M, Coogan PF, Palmer JR, Rosenberg L, Band P, Coldman AJ, Gallagher RP, Hislop TG, Yang P, Cummings SR, Canfell K, Sitas F, Chao P, Lissowska J, Horn-Ross PL, John EM, Kolonel LM, Nomura AMY, Ghiasvand R, Hu J, Johnson KC, Mao Y, Callaghan K, Crossley B, Goodill A, Green J, Hermon C, Key T, Lindgard I, Liu B, Pirie K, Reeves G, Collins R, Doll R, Peto R, Bishop T, Fentiman IS, De Sanjose S, Gonzaler CA, Lee N, Marchbanks P, Ory HW, Peterson HB, Wingo P, Ebeling K, Kunde D, Nishan P, Hopper JL, Eliassen H, Gajalakshmi V, Martin N, Pardthaisong T, Silpisornkosol S, Theetranont C, Boosiri B, Chutivongse S, Jimakorn P, Virutamasen P, Wongsrichanalai C, Neugut A, Santella R, Baines CJ, Kreiger N, Miller AB, Wall C, Tjonneland A, Jorgensen T, Stahlberg C, Pedersen AT, Flesch-Janys D, Hakansson N, Cauley J, Heuch I, Adami HO, Persson I, Weiderpass E, Magnusson C, Chang-Claude J, Kaaks R, McCredie M, Paul C, Skegg DCG, Spears GFS, Iwasaki M, Tsugane S, Anderson G, Daling JR, Hampton J, Hutchinson WB, Li CI, Malone K, Mandelson M, Newcomb P, Noonan EA, Ray RM, Stanford JL, Tang MTC, Thomas DB, Weiss NS, White E, Izquierdo A, Viladiu P, Fourkala EO, Jacobs I, Menon U, Ryan A, Cuevas HR, Ontiveros P, Palet A, Salazar SB, Aristizabal N, Cuadros A, Tryggvadottir L, Tulinius H, Riboli E, Andrieu N, Bachelot A, Le MG, Bremond A, Gairard B, Lansac J, Piana L, Renaud R, Clavel-Chapelon F, Fournier A, Touillaud M, Mesrine S, Chabbert-Buffet N, Boutron-Ruault MC, Wolk A, Torres-Mejia G, Franceschi S, Romieu I, Boyle P, Lubin F, Modan B, Ron E, Wax Y, Friedman GD, Hiatt RA, Levi F, Kosmelj K, Primic-Zakelj M, Ravnihar B, Stare J, Ekbom A, Erlandsson G, Persson I, Beeson WL, Fraser G, Peto J, Hanson RL, Leske MC, Mahoney MC, Nasca PC, Varma AO, Weinstein AL, Hartman ML, Olsson H, Goldbohm RA, van den Brandt PA, Palli D, Teitelbaum S, Apelo RA, Baens J, de la Cruz JR, Javier B, Lacaya LB, Ngelangel CA, La Vecchia C, Negri E, Marubini E, Ferraroni M, Pike MC, Gerber M, Richardson S, Segala C, Gatei D, Kenya P, Kungu A, Mati JG, Brinton LA, Freedman M, Hoover R, Schairer C, Ziegler R, Banks E, Spirtas R, Lee HP, Rookus MA, van Leeuwen FE, Schoenberg JA, Graff-Iversen S, Selmer R, Jones L, McPherson K, Neil A, Vessey M, Yeates D, Mabuchi K, Preston D, Hannaford P, Kay C, McCann SE, Rosero-Bixby L, Gao YT, Jin F, Yuan JM, Wei HY, Yun T, Zhiheng C, Berry G, Booth JC, Jelihovsky T, MacLennan R, Shearman R, Hadjisavvas A, Kyriacou K, Loisidou M, Zhou X, Wang QS, Kawai M, Minami Y, Tsuji I, Lund E, Kumle M, Stalsberg H, Shu XO, Zheng W, Monninkhof EM, Onland-Moret NC, Peeters PHM, Katsouyanni K, Trichopoulou A, Trichopoulos D, Tzonou A, Baltzell KA, Dabancens A, Martinez L, Molina R, Salas O, Alexander FE, Anderson K, Folsom AR, Gammon MD, Hulka BS, Millikan R, Chilvers CED, Lumachi F, Bain C, Schofield F, Siskind V, Rebbeck TR, Bernstein LR, Enger S, Haile RW, Paganini-Hill A, Ross RK, Ursin G, Wu AH, Yu MC, Ewertz DM, Clarke EA, Bergkvist L, Anderson GL, Gass M, O’Sullivan MJ, Kalache A, Farley TMM, Holck S, Meirik O, Fukao A, Factors CGH, Factors CGH, Grp SHNHSIIR, 2012. Menarche, menopause, and breast cancer risk: individual participant meta-analysis, including 118 964 women with breast cancer from 117 epidemiological studies. Lancet Oncol 13, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends AL, de Groot CJ, Sijbrands EJ, Sie MP, Benneheij SH, Pal R, Heydanus R, Oostra BA, van Duijn CM, Steegers EA, 2008. Shared constitutional risks for maternal vascular-related pregnancy complications and future cardiovascular disease. Hypertension 51, 1034–1041. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolund E, Lummaa V, Smith KR, Hanson HA, Maklakov AA, 2016. Reduced costs of reproduction in females mediate a shift from a male-biased to a female-biased lifespan in humans. Sci Rep 6, 24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, 2015. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhimschi IA, Nayeri UA, Zhao G, Shook LL, Pensalfini A, Funai EF, Bernstein IM, Glabe CG, Buhimschi CS, 2014. Protein misfolding, congophilia, oligomerization, and defective amyloid processing in preeclampsia. Sci Transl Med 6, 245ra292. [DOI] [PubMed] [Google Scholar]
- Calderon-Margalit R, Friedlander Y, Yanetz R, Deutsch L, Perrin MC, Kleinhaus K, Tiram E, Harlap S, Paltiel O, 2009. Preeclampsia and subsequent risk of cancer: update from the Jerusalem Perinatal Study. Am J Obstet Gynecol 200, 63 e61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J, 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez E, Trynka G, 2020. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front Genet 11, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino RR, Stark A, 2018. Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev 32, 202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH, Consortium, m., 2009. Unlocking the secrets of the genome. Nature 459, 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau K, Hennessy A, Makris A, 2017. Placental growth factor and pre-eclampsia. J Hum Hypertens 31, 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wilson R, Wang SH, Zheng HZ, Walker JJ, McKillop JH, 1996. Tumour necrosis factor-alpha (TNF-alpha) gene polymorphism and expression in pre-eclampsia. Clin Exp Immunol 104, 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Sheehan PM, Brennecke SP, Keogh RJ, 2012. Vessel remodelling, pregnancy hormones and extravillous trophoblast function. Mol Cell Endocrinol 349, 138–144. [DOI] [PubMed] [Google Scholar]
- Chen P, Gong Y, Pu Y, Wang Y, Zhou B, Song Y, Wang T, Zhang L, 2016. Association between polymorphisms in IL-27 gene and pre-eclampsia. Placenta 37, 61–64. [DOI] [PubMed] [Google Scholar]
- Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Bruning JC, Nolan PM, Ashcroft FM, Cox RD, 2010. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet 42, 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson PA, Hsu YH, Drucker DJ, Mellgren G, Hui CC, Hauner H, Kellis M, 2015. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. The New England journal of medicine 373, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnattingius S, Reilly M, Pawitan Y, Lichtenstein P, 2004. Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study. Am J Med Genet A 130A, 365–371. [DOI] [PubMed] [Google Scholar]
- Colucci M, Cammarata S, Assini A, Croce R, Clerici F, Novello C, Mazzella L, Dagnino N, Mariani C, Tanganelli P, 2006. The number of pregnancies is a risk factor for Alzheimer’s disease. Eur J Neurol 13, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Consortium EP, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M, 2009. Mapping complex disease traits with global gene expression. Nat Rev Genet 10, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT, 2014. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46, 1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelius DC, 2018. Preeclampsia: From Inflammation to Immunoregulation. Clin Med Insights Blood Disord 11, 1179545X17752325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sal Lari R, Lupien M, Markowitz S, Scacheri PC, 2014. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome research 24, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Redman C, 2017. The role of cellular senescence in ageing of the placenta. Placenta 52, 139–145. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R, 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, Cotman CW, 2012. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J Neuroinflammation 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Guo X, 2021. Decoding and rejuvenating human ageing genomes: Lessons from mosaic chromosomal alterations. Ageing Res Rev 68, 101342. [DOI] [PubMed] [Google Scholar]
- Darmochwal-Kolarz D, Oleszczuk J, 2014. The critical role of Th17 cells, Treg cells and co-stimulatory molecules in the development of pre-eclampsia. Dev Period Med 18, 141–147. [PubMed] [Google Scholar]
- Di Paolo NC, Shayakhmetov DM, 2016. Interleukin 1alpha and the inflammatory process. Nat Immunol 17, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C, 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834. [DOI] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P, 2007. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39, 724–726. [DOI] [PubMed] [Google Scholar]
- Dunk CE, van Dijk M, Choudhury R, Wright TJ, Cox B, Leavey K, Harris LK, Jones RL, Lye SJ, 2021. Functional Evaluation of STOX1 (STORKHEAD-BOX PROTEIN 1) in Placentation, Preeclampsia, and Preterm Birth. Hypertension 77, 475–490. [DOI] [PubMed] [Google Scholar]
- Eddy AC, Bidwell GL 3rd, George EM, 2018. Pro-angiogenic therapeutics for preeclampsia. Biol Sex Differ 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elharram M, Dayan N, Kaur A, Landry T, Pilote L, 2018. Long-Term Cognitive Impairment After Preeclampsia: A Systematic Review and Meta-analysis. Obstet Gynecol 132, 355–364. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES, 2016. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M, 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplin MS, Fausett MB, Fraser A, Kerber R, Mineau G, Carrillo J, Varner MW, 2001. Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med 344, 867–872. [DOI] [PubMed] [Google Scholar]
- Fang R, Yu M, Li G, Chee S, Liu T, Schmitt AD, Ren B, 2016. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res 26, 1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farladansky-Gershnabel S, Gal H, Kidron D, Krizhanovsky V, Amiel A, Sukenik-Halevy R, Biron-Shental T, 2019. Telomere Homeostasis and Senescence Markers Are Differently Expressed in Placentas From Pregnancies With Early- Versus Late-Onset Preeclampsia. Reprod Sci 26, 1203–1209. [DOI] [PubMed] [Google Scholar]
- Ferreira LC, Gomes CE, Araujo AC, Bezerra PF, Duggal P, Jeronimo SM, 2015. Association between ACVR2A and early-onset preeclampsia: replication study in a Northeastern Brazilian population. Placenta 36, 186–190. [DOI] [PubMed] [Google Scholar]
- Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC, Ruther U, 2009. Inactivation of the Fto gene protects from obesity. Nature 458, 894–898. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS, 2006. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312, 276–279. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick E, Johnson MP, Dyer TD, Forrest S, Elliott K, Blangero J, Brennecke SP, Moses EK, 2009. Genetic association of the activin A receptor gene (ACVR2A) and preeclampsia. Mol Hum Reprod 15, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio P, Gabbanini M, Borges LE, Bonaccorsi L, Pinzauti S, Reis FM, Boy Torres P, Rago G, Litta P, Petraglia F, 2010. Activins and related proteins in the establishment of pregnancy. Reprod Sci 17, 320–330. [DOI] [PubMed] [Google Scholar]
- Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, Johnson AD, Young JH, Rice K, Duan Q, Chen F, Li Y, Tang H, Fornage M, Keene KL, Andrews JS, Smith JA, Faul JD, Guangfa Z, Guo W, Liu Y, Murray SS, Musani SK, Srinivasan S, Velez Edwards DR, Wang H, Becker LC, Bovet P, Bochud M, Broeckel U, Burnier M, Carty C, Chasman DI, Ehret G, Chen WM, Chen G, Chen W, Ding J, Dreisbach AW, Evans MK, Guo X, Garcia ME, Jensen R, Keller MF, Lettre G, Lotay V, Martin LW, Moore JH, Morrison AC, Mosley TH, Ogunniyi A, Palmas W, Papanicolaou G, Penman A, Polak JF, Ridker PM, Salako B, Singleton AB, Shriner D, Taylor KD, Vasan R, Wiggins K, Williams SM, Yanek LR, Zhao W, Zonderman AB, Becker DM, Berenson G, Boerwinkle E, Bottinger E, Cushman M, Eaton C, Nyberg F, Heiss G, Hirschhron JN, Howard VJ, Karczewsk KJ, Lanktree MB, Liu K, Liu Y, Loos R, Margolis K, Snyder M, Asian Genetic Epidemiology Network, C., Psaty BM, Schork NJ, Weir DR, Rotimi CN, Sale MM, Harris T, Kardia SL, Hunt SC, Arnett D, Redline S, Cooper RS, Risch NJ, Rao DC, Rotter JI, Chakravarti A, Reiner AP, Levy D, Keating BJ, Zhu X, 2013. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet 93, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser R, Walker JJ, Ekbote UV, Martin KL, McShane P, Orsi NM, 2008. Interleukin-4 −590 (C>T), toll-like receptor-2 +2258 (G>A) and matrix metalloproteinase-9 −1562 (C>T) polymorphisms in pre-eclampsia. BJOG 115, 1052–1056; discussion 1056. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI, 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frustenberg FF, 2005. Banking on families: How families generate and distribute social capital. Journal of Marriage and Family 67, 809–821. [Google Scholar]
- Gaillard R, Bakker R, Steegers EA, Hofman A, Jaddoe VW, 2011. Maternal age during pregnancy is associated with third trimester blood pressure level: the generation R study. Am J Hypertens 24, 1046–1053. [DOI] [PubMed] [Google Scholar]
- Geldenhuys J, Rossouw TM, Lombaard HA, Ehlers MM, Kock MM, 2018. Disruption in the Regulation of Immune Responses in the Placental Subtype of Preeclampsia. Front Immunol 9, 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL, 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150, 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronimus AT, 1996. Black/white differences in the relationship of maternal age to birthweight: a population-based test of the weathering hypothesis. Soc Sci Med 42, 589–597. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giller A, Andrawus M, Gutman D, Atzmon G, 2020. Pregnancy as a model for aging. Ageing Res Rev 62, 101093. [DOI] [PubMed] [Google Scholar]
- Gilsanz P, Lee C, Corrada MM, Kawas CH, Quesenberry CP Jr., Whitmer RA, 2019. Reproductive period and risk of dementia in a diverse cohort of health care members. Neurology 92, e2005–e2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EH, Peel NM, Samanta M, Theou O, Howlett SE, Hubbard RE, 2017. Sex differences in frailty: A systematic review and meta-analysis. Exp Gerontol 89, 30–40. [DOI] [PubMed] [Google Scholar]
- Gottesman RF, Schneider AL, Albert M, Alonso A, Bandeen-Roche K, Coker L, Coresh J, Knopman D, Power MC, Rawlings A, Sharrett AR, Wruck LM, Mosley TH, 2014. Midlife hypertension and 20-year cognitive change: the atherosclerosis risk in communities neurocognitive study. JAMA Neurol 71, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger JP, Spradley FT, Bakrania BA, 2018. The Endothelin System: A Critical Player in the Pathophysiology of Preeclampsia. Curr Hypertens Rep 20, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KJ, Kovacheva VP, Mirzakhani H, Bjonnes AC, Almoguera B, DeWan AT, Triche EW, Saftlas AF, Hoh J, Bodian DL, Klein E, Huddleston KC, Ingles SA, Lockwood CJ, Hakonarson H, McElrath TF, Murray JC, Wilson ML, Norwitz ER, Karumanchi SA, Bateman BT, Keating BJ, Saxena R, 2018a. Gene-Centric Analysis of Preeclampsia Identifies Maternal Association at PLEKHG1. Hypertension 72, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KJ, Saxena R, Karumanchi SA, 2018b. Genetic predisposition to preeclampsia is conferred by fetal DNA variants near FLT1, a gene involved in the regulation of angiogenesis. Am J Obstet Gynecol 218, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Roddam A, Pirie K, Kirichek O, Reeves G, Beral V, Collaborators MWS, 2012. Reproductive factors and risk of oesophageal and gastric cancer in the Million Women Study cohort. Brit J Cancer 106, 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet LG, Nilsson E, Ling C, Hansen T, Pedersen O, Groop L, Vaag A, Poulsen P, 2009. Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes 58, 2402–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurven M, Costa M, Ben T, Stieglitz J, Beheim B, Eid Rodriguez D, Hooper PL, Kaplan H, 2016. Health costs of reproduction are minimal despite high fertility, mortality and subsistence lifestyle. Sci Rep 6, 30056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid HM, Abdalla SE, Sidig M, Adam I, Hamdan HZ, 2020. Association of VEGFA and IL1beta gene polymorphisms with preeclampsia in Sudanese women. Mol Genet Genomic Med 8, e1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu X-D, Topol EJ, Rosenfeld MG, Frazer KA, 2011. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature 470, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Cornelius DC, Amaral LM, Faulkner JL, Cunningham MW Jr., Wallace K, LaMarca B, 2016. The role of inflammation in the pathology of preeclampsia. Clin Sci (Lond) 130, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J, 2015. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Harville EW, Guralnik J, Romero M, Bazzano LA, 2020. Reproductive History and Cognitive Aging: The Bogalusa Heart Study. Am J Geriatr Psychiatry 28, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B, 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B, 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–318. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK, 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK, 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen J, Romppanen EL, Hiltunen M, Iivonen S, Mannermaa A, Punnonen K, Heinonen S, 2002. Polymorphism in the tumor necrosis factor-alpha gene in women with preeclampsia. J Assist Reprod Genet 19, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera MD, Mingorance C, Rodriguez-Rodriguez R, Alvarez de Sotomayor M, 2010. Endothelial dysfunction and aging: an update. Ageing Res Rev 9, 142–152. [DOI] [PubMed] [Google Scholar]
- Hilali N, Kocyigit A, Demir M, Camuzcuoglu A, Incebiyik A, Camuzcuoglu H, Vural M, Taskin A, 2013. DNA damage and oxidative stress in patients with mild preeclampsia and offspring. Eur J Obstet Gynecol Reprod Biol 170, 377–380. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA, 2013. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hod T, Cerdeira AS, Karumanchi SA, 2015. Molecular Mechanisms of Preeclampsia. Cold Spring Harb Perspect Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlickova B, de Almeida RC, Borek Z, Withoff S, 2014. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta 1842, 1910–1922. [DOI] [PubMed] [Google Scholar]
- Huppertz B, 2008. Placental origins of preeclampsia: challenging the current hypothesis. Hypertension 51, 970–975. [DOI] [PubMed] [Google Scholar]
- Iadecola C, Yaffe K, Biller J, Bratzke LC, Faraci FM, Gorelick PB, Gulati M, Kamel H, Knopman DS, Launer LJ, Saczynski JS, Seshadri S, Zeki Al Hazzouri A, American Heart Association Council on, H., Council on Clinical, C., Council on Cardiovascular Disease in the, Y., Council on, C., Stroke, N., Council on Quality of, C., Outcomes, R., Stroke, C., 2016. Impact of Hypertension on Cognitive Function: A Scientific Statement From the American Heart Association. Hypertension 68, e67–e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilango SD, McEvoy LK, Laughlin GA, Bergstrom J, Barrett-Connor E, Kritz-Silverstein D, 2019. Pregnancy history and cognitive aging among older women: the Rancho Bernardo Study. Menopause 26, 750–757. [DOI] [PubMed] [Google Scholar]
- Inoue F, Ahituv N, 2015. Decoding enhancers using massively parallel reporter assays. Genomics 106, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S, Lockett GA, Holloway JW, Arshad SH, Zhang H, Kaushal A, Tetali SR, Mukherjee N, Karmaus WJJ, 2018. Changes in DNA Methylation from Age 18 to Pregnancy in Type 1, 2, and 17 T Helper and Regulatory T-Cells Pathway Genes. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasienska G, 2020. Costs of reproduction and ageing in the human female. Philos Trans R Soc Lond B Biol Sci 375, 20190615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnstrom K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, Fraser P, 2016. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384.e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B, 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Brennecke SP, East CE, Goring HH, Kent JW Jr., Dyer TD, Said JM, Roten LT, Iversen AC, Abraham LJ, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Laivuori H, Group FS, Austgulen R, Blangero J, Moses EK, 2012. Genome-wide association scan identifies a risk locus for preeclampsia on 2q14, near the inhibin, beta B gene. Plos One 7, e33666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, Hutchinson EW, 1993. Absence of strong heterosis for life span and other life history traits in Caenorhabditis elegans. Genetics 134, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng Y-Y, Yoon CH, Boehm JS, Lander ES, Zhang F, 2017. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamali-Sarvestani E, Kiany S, Gharesi-Fard B, Robati M, 2006. Association study of IL-10 and IFN-gamma gene polymorphisms in Iranian women with preeclampsia. J Reprod Immunol 72, 118–126. [DOI] [PubMed] [Google Scholar]
- Kanasaki K, Palmsten K, Sugimoto H, Ahmad S, Hamano Y, Xie L, Parry S, Augustin HG, Gattone VH, Folkman J, Strauss JF, Kalluri R, 2008. Deficiency in catechol-O-methyltransferase and 2-methoxyoestradiol is associated with pre-eclampsia. Nature 453, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F, 2014. Geroscience: Linking Aging to Chronic Disease. Cell 159, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- Kikas T, Inno R, Ratnik K, Rull K, Laan M, 2020. C-allele of rs4769613 Near FLT1 Represents a High-Confidence Placental Risk Factor for Preeclampsia. Hypertension 76, 884–891. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME, 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood, 1990. The disposable soma theory of aging, in Genetic Effects on Aging II, edited by Harrison DE, Caldwell NJ, Telford Press. 9–19. [Google Scholar]
- Kirkwood TB, 2002. Evolution of ageing. Mech Ageing Dev 123, 737–745. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Austad SN, 2000. Why do we age? Nature 408, 233–238. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Rose MR, 1991. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond B Biol Sci 332, 15–24. [DOI] [PubMed] [Google Scholar]
- Kohlrausch FB, Keefe DL, 2020. Telomere erosion as a placental clock: From placental pathologies to adverse pregnancy outcomes. Placenta 97, 101–107. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, 2015. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvehaugen AS, Dechend R, Ramstad HB, Troisi R, Fugelseth D, Staff AC, 2011. Endothelial function and circulating biomarkers are disturbed in women and children after preeclampsia. Hypertension 58, 63–69. [DOI] [PubMed] [Google Scholar]
- Lacey RE, Kumari M, Sacker A, McMunn A, 2017. Age at first birth and cardiovascular risk factors in the 1958 British birth cohort. J Epidemiol Community Health 71, 691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue P, Lakhal B, Vatin M, Batista F, Burgio G, Mercier E, Santos ED, Buffat C, Sierra-Diaz DC, Renault G, Montagutelli X, Salmon J, Monget P, Veitia RA, Mehats C, Fellous M, Gris JC, Cocquet J, Vaiman D, 2016. Association of FOXD1 variants with adverse pregnancy outcomes in mice and humans. Open Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini M, Santoro L, Del Mastro L, Nguyen B, Livraghi L, Ugolini D, Peccatori FA, Azim HA Jr., 2016. Reproductive behaviors and risk of developing breast cancer according to tumor subtype: A systematic review and meta-analysis of epidemiological studies. Cancer Treat Rev 49, 65–76. [DOI] [PubMed] [Google Scholar]
- Laufer BI, Singh SM, 2015. Strategies for precision modulation of gene expression by epigenome editing: an overview. Epigenetics & chromatin 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Espéli M, Anderson CA, Linterman MA, Pocock JM, Williams NJ, Roberts R, Viatte S, Fu B, Peshu N, Hien TT, Phu NH, Wesley E, Edwards C, Ahmad T, Mansfield JC, Gearry R, Dunstan S, Williams TN, Barton A, Vinuesa CG, Consortium UIG, Parkes M, Lyons PA, Smith KGC, 2013. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 155, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, 2010. Transcriptional enhancers in animal development and evolution. Current biology : CB 20, R754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y, 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu M, Zong J, Tan P, Wang J, Wang X, Ye Y, Liu S, Liu X, 2014. Genetic variations in IL1A and IL1RN are associated with the risk of preeclampsia in Chinese Han population. Sci Rep 4, 5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li Y, Yao Y, Li H, Liang H, Xin M, Wang L, Zhao L, Lin J, Liu S, 2016. Polymorphisms of the IL27 gene in a Chinese Han population complicated with pre-eclampsia. Sci Rep 6, 23029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart PA, Martin P, Johnson MA, Shirtcliff E, Poon LW, 2017. The Relationship of Fertility, Lifestyle, and Longevity Among Women. J Gerontol A Biol Sci Med Sci 72, 754–759. [DOI] [PubMed] [Google Scholar]
- Lotta T, Vidgren J, Tilgmann C, Ulmanen I, Melen K, Julkunen I, Taskinen J, 1995. Kinetics of human soluble and membrane-bound catechol O-methyltransferase: a revised mechanism and description of the thermolabile variant of the enzyme. Biochemistry 34, 4202–4210. [DOI] [PubMed] [Google Scholar]
- Luders E, Gingnell M, Poromaa IS, Engman J, Kurth F, Gaser C, 2018. Potential Brain Age Reversal after Pregnancy: Younger Brains at 4–6Weeks Postpartum. Neuroscience 386, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus MC, Iliodromiti S, Lawlor DA, Catov JM, Nelson SM, Fraser A, 2017. Number of Offspring and Cardiovascular Disease Risk in Men and Women: The Role of Shared Lifestyle Characteristics. Epidemiology 28, 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna S, McCarthy C, McCarthy FP, 2019. Placental Ageing in Adverse Pregnancy Outcomes: Telomere Shortening, Cell Senescence, and Mitochondrial Dysfunction. Oxid Med Cell Longev 2019, 3095383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marengoni A, Angleman S, Melis R, Mangialasche F, Karp A, Garmen A, Meinow B, Fratiglioni L, 2011. Aging with multimorbidity: A systematic review of the literature. Ageing Res Rev 10, 430–439. [DOI] [PubMed] [Google Scholar]
- Mason JS, Wileman T, Chapman T, 2018. Lifespan extension without fertility reduction following dietary addition of the autophagy activator Torin1 in Drosophila melanogaster. Plos One 13, e0190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA, 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis R, Steinthorsdottir V, Williams NO, Thorleifsson G, Shooter S, Hjartardottir S, Bumpstead S, Stefansdottir L, Hildyard L, Sigurdsson JK, Kemp JP, Silva GB, Thomsen LCV, Jaaskelainen T, Kajantie E, Chappell S, Kalsheker N, Moffett A, Hiby S, Lee WK, Padmanabhan S, Simpson NAB, Dolby VA, Staines-Urias E, Engel SM, Haugan A, Trogstad L, Svyatova G, Zakhidova N, Najmutdinova D, Consortium F, Consortium G, Dominiczak AF, Gjessing HK, Casas JP, Dudbridge F, Walker JJ, Pipkin FB, Thorsteinsdottir U, Geirsson RT, Lawlor DA, Iversen AC, Magnus P, Laivuori H, Stefansson K, Morgan L, 2017. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat Genet 49, 1255–1260. [DOI] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG Jr., Kinney JB, Kellis M, Lander ES, Mikkelsen TS, 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Collinot H, Boumerdassi Y, Ducat A, Duche A, Renault G, Marchiol C, Lagoutte I, Bertholle C, Andrieu M, Jacques S, Mehats C, Vaiman D, 2019. Long-term cardiovascular disorders in the STOX1 mouse model of preeclampsia. Sci Rep-Uk 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohajertehran F, Tavakkol Afshari J, Rezaieyazdi Z, Ghomian N, 2012. Association of single nucleotide polymorphisms in the human tumor necrosis factor-alpha and interleukin 1-beta genes in patients with pre-eclampsia. Iran J Allergy Asthma Immunol 11, 224–229. [PubMed] [Google Scholar]
- Molvarec A, Ver A, Fekete A, Rosta K, Derzbach L, Derzsy Z, Karadi I, Rigo J Jr., 2007. Association between estrogen receptor alpha (ESR1) gene polymorphisms and severe preeclampsia. Hypertens Res 30, 205–211. [DOI] [PubMed] [Google Scholar]
- Mongraw-Chaffin ML, Cirillo PM, Cohn BA, 2010. Preeclampsia and cardiovascular disease death: prospective evidence from the child health and development studies cohort. Hypertension 56, 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses EK, Fitzpatrick E, Freed KA, Dyer TD, Forrest S, Elliott K, Johnson MP, Blangero J, Brennecke SP, 2006. Objective prioritization of positional candidate genes at a quantitative trait locus for pre-eclampsia on 2q22. Mol Hum Reprod 12, 505–512. [DOI] [PubMed] [Google Scholar]
- Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, Giste E, Johnson A, Zhang M, Balasundaram G, Byron R, Roach V, Sabo PJ, Sandstrom R, Stehling AS, Thurman RE, Weissman SM, Cayting P, Hariharan M, Lian J, Cheng Y, Landt SG, Ma Z, Wold BJ, Dekker J, Crawford GE, Keller CA, Wu W, Morrissey C, Kumar SA, Mishra T, Jain D, Byrska-Bishop M, Blankenberg D, Lajoie BR, Jain G, Sanyal A, Chen KB, Denas O, Taylor J, Blobel GA, Weiss MJ, Pimkin M, Deng W, Marinov GK, Williams BA, Fisher-Aylor KI, Desalvo G, Kiralusha A, Trout D, Amrhein H, Mortazavi A, Edsall L, McCleary D, Kuan S, Shen Y, Yue F, Ye Z, Davis CA, Zaleski C, Jha S, Xue C, Dobin A, Lin W, Fastuca M, Wang H, Guigo R, Djebali S, Lagarde J, Ryba T, Sasaki T, Malladi VS, Cline MS, Kirkup VM, Learned K, Rosenbloom KR, Kent WJ, Feingold EA, Good PJ, Pazin M, Lowdon RF, Adams LB, 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muela HC, Costa-Hong VA, Yassuda MS, Moraes NC, Memoria CM, Machado MF, Macedo TA, Shu EB, Massaro AR, Nitrini R, Mansur AJ, Bortolotto LA, 2017. Hypertension Severity Is Associated With Impaired Cognitive Performance. J Am Heart Assoc 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muka T, Oliver-Williams C, Kunutsor S, Laven JSE, Fauser BCJM, Chowdhury R, Kavousi M, Franco OH, 2016. Association of Age at Onset of Menopause and Time Since Onset of Menopause With Cardiovascular Outcomes, Intermediate Vascular Traits, and All-Cause Mortality A Systematic Review and Meta-analysis. Jama Cardiol 1, 767–776. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ, 2010. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt L, Webster RP, 2009. Vascular biology of preeclampsia. J Thromb Haemost 7, 375–384. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, Sanseau P, 2015. The support of human genetic evidence for approved drug indications. Nat Genet 47, 856–860. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Wellcome Trust Case Control, C., Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB, 2009. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41, 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning K, Zhao L, Franklin M, Matloff W, Batta I, Arzouni N, Sun F, Toga AW, 2020. Parity is associated with cognitive function and brain age in both females and males. Sci Rep 10, 6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg ST, Rendon A, Smethurst PA, Paul DS, Voss K, Thon JN, Lloyd-Jones H, Sambrook JG, Tijssen MR, HaemGen C, Italiano JE Jr., Deloukas P, Gottgens B, Soranzo N, Ouwehand WH, 2012. A GWAS sequence variant for platelet volume marks an alternative DNM3 promoter in megakaryocytes near a MEIS1 binding site. Blood 120, 4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuzyan A, Juel K, Vaupel JW, Christensen K, 2008. Men: good health and high mortality. Sex differences in health and aging. Aging Clin Exp Res 20, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K, Saglam B, Whitehead C, Stock O, Lappas M, Tong S, 2011. Severe early-onset preeclampsia is not associated with a change in placental catechol O-methyltransferase (COMT) expression. Am J Pathol 178, 2484–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh NI, Cnattingius S, Dickman PW, Mittleman MA, Ludvigsson JF, Ingelsson E, 2010. Parity and risk of later-life maternal cardiovascular disease. Am Heart J 159, 215–221 e216. [DOI] [PubMed] [Google Scholar]
- Pelucchi C, Galeone C, Talamini R, Bosetti C, Montella M, Negri E, Franceschi S, La Vecchia C, 2007. Lifetime ovulatory cycles and ovarian cancer risk in 2 Italian case-control studies. Am J Obstet Gynecol 196, 83–85. [DOI] [PubMed] [Google Scholar]
- Penn DJ, Smith KR, 2007. Differential fitness costs of reproduction between the sexes. Proc Natl Acad Sci U S A 104, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinarbasi E, Cekin N, Bildirici AE, Akin S, Yanik A, 2020. STOX1 gene Y153H polymorphism is associated with early-onset preeclampsia in Turkish population. Gene 754. [DOI] [PubMed] [Google Scholar]
- Pinheiro MB, Gomes KB, Ronda CR, Guimaraes GG, Freitas LG, Teixeira-Carvalho A, Martins-Filho OA, Dusse LM, 2015. Severe preeclampsia: association of genes polymorphisms and maternal cytokines production in Brazilian population. Cytokine 71, 232–237. [DOI] [PubMed] [Google Scholar]
- Plant M, Armstrong C, Ruggiero A, Sherrill C, Uberseder B, Jeffries R, Nevarez J, Jorgensen MJ, Kavanagh K, Quinn MA, 2020. Advanced maternal age impacts physiologic adaptations to pregnancy in vervet monkeys. Geroscience 42, 1649–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack AZ, Rivers K, Ahrens KA, 2018. Parity associated with telomere length among US reproductive age women. Hum Reprod 33, 736–744. [DOI] [PubMed] [Google Scholar]
- Pontillo A, Reis EC, Bricher PN, Vianna P, Diniz S, Fernandes KS, Chies JA, Sandrim V, 2015. NLRP1 L155H Polymorphism is a Risk Factor for Preeclampsia Development. Am J Reprod Immunol 73, 577–581. [DOI] [PubMed] [Google Scholar]
- Postma IR, Bouma A, de Groot JC, Aukes AM, Aarnoudse JG, Zeeman GG, 2016. Cerebral white matter lesions, subjective cognitive failures, and objective neurocognitive functioning: A follow-up study in women after hypertensive disorders of pregnancy. J Clin Exp Neuropsychol 38, 585–598. [DOI] [PubMed] [Google Scholar]
- Postma IR, de Groot JC, Aukes AM, Aarnoudse JG, Zeeman GG, 2014. Cerebral white matter lesions and perceived cognitive dysfunction: the role of pregnancy. Am J Obstet Gynecol 211, 257 e251–255. [DOI] [PubMed] [Google Scholar]
- Powers RW, Evans RW, Majors AK, Ojimba JI, Ness RB, Crombleholme WR, Roberts JM, 1998. Plasma homocysteine concentration is increased in preeclampsia and is associated with evidence of endothelial activation. Am J Obstet Gynecol 179, 1605–1611. [DOI] [PubMed] [Google Scholar]
- Ptok U, Barkow K, Heun R, 2002. Fertility and number of children in patients with Alzheimer’s disease. Arch Womens Ment Health 5, 83–86. [DOI] [PubMed] [Google Scholar]
- Quintero-Ronderos P, Jimenez KM, Esteban-Perez C, Ojeda DA, Bello S, Fonseca DJ, Coronel MA, Moreno-Ortiz H, Sierra-Diaz DC, Lucena E, Barbaux S, Vaiman D, Laissue P, 2019. FOXD1 mutations are related to repeated implantation failure, intrauterine growth restriction and preeclampsia. Mol Med 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman I, Akesson A, Wolk A, 2015. Relationship between age at natural menopause and risk of heart failure. Menopause 22, 12–16. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read SL, Grundy EMD, 2017. Fertility History and Cognition in Later Life. J Gerontol B Psychol Sci Soc Sci 72, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes AM, Hardy M, Pavalko E, 2018. Race Differences in Linking Family Formation Transitions to Women’s Mortality. J Health Soc Behav 59, 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs RE, Cadena CD, 2007. Lifespan is unrelated to investment in reproduction in populations of mammals and birds in captivity. Ecol Lett 10, 867–872. [DOI] [PubMed] [Google Scholar]
- Ritzel RM, Patel AR, Spychala M, Verma R, Crapser J, Koellhoffer EC, Schrecengost A, Jellison ER, Zhu L, Venna VR, McCullough LD, 2017. Multiparity improves outcomes after cerebral ischemia in female mice despite features of increased metabovascular risk. Proc Natl Acad Sci U S A 114, E5673–E5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, 2015. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roten LT, Johnson MP, Forsmo S, Fitzpatrick E, Dyer TD, Brennecke SP, Blangero J, Moses EK, Austgulen R, 2009. Association between the candidate susceptibility gene ACVR2A on chromosome 2q22 and pre-eclampsia in a large Norwegian population-based study (the HUNT study). Eur J Hum Genet 17, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz JL, Diaz-Venegas C, Crimmins EM, 2021. Fertility History and Cognitive Function in Late Life: The Case of Mexico. J Gerontol B Psychol Sci Soc Sci 76, e140–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi S, Mohammadoo-Khorasani M, Yaghmaei M, Mokhtari M, Moossavi M, 2014. Possible association of IL-4 VNTR polymorphism with susceptibility to preeclampsia. Biomed Res Int 2014, 497031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J, 2012. The long-range interaction landscape of gene promoters. Nature 489, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuit SC, Oei HH, Witteman JC, Geurts van Kessel CH, van Meurs JB, Nijhuis RL, van Leeuwen JP, de Jong FH, Zillikens MC, Hofman A, Pols HA, Uitterlinden AG, 2004. Estrogen receptor alpha gene polymorphisms and risk of myocardial infarction. JAMA 291, 2969–2977. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Pothof J, Vijg J, Hoeijmakers JHJ, 2021. The central role of DNA damage in the ageing process. Nature 592, 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR, 2007. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS genetics 3, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DR, Jablonski KL, Donato AJ, 2011. Aging and vascular endothelial function in humans. Clin Sci (Lond) 120, 357–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadyab AH, Gass ML, Stefanick ML, Waring ME, Macera CA, Gallo LC, Shaffer RA, Jain S, LaCroix AZ, 2017. Maternal Age at Childbirth and Parity as Predictors of Longevity Among Women in the United States: The Women’s Health Initiative. Am J Public Health 107, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen JJ, Wright JD, Goffman D, Kern-Goldberger AR, Booker W, Siddiq Z, D’Alton ME, Friedman AM, 2018. Maternal age and risk for adverse outcomes. Am J Obstet Gynecol 219, 390 e391–390 e315. [DOI] [PubMed] [Google Scholar]
- Shen L, Song L, Li H, Liu B, Zheng X, Zhang L, Yuan J, Liang Y, Wang Y, 2017. Association between earlier age at natural menopause and risk of diabetes in middle-aged and older Chinese women: The Dongfeng-Tongji cohort study. Diabetes Metab 43, 345–350. [DOI] [PubMed] [Google Scholar]
- Shi Q, Colodner KJ, Matousek SB, Merry K, Hong S, Kenison JE, Frost JL, Le KX, Li S, Dodart JC, Caldarone BJ, Stevens B, Lemere CA, 2015. Complement C3-Deficient Mice Fail to Display Age-Related Hippocampal Decline. J Neurosci 35, 13029–13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasuna K, Karasawa T, Takahashi M, 2020. Role of the NLRP3 Inflammasome in Preeclampsia. Front Endocrinol (Lausanne) 11, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi TN, Hastings WJ, Rosinger AY, Ryan CP, 2020. Parity predicts biological age acceleration in post-menopausal, but not pre-menopausal, women. Sci Rep 10, 20522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepmann T, Boardman H, Bilderbeck A, Griffanti L, Kenworthy Y, Zwager C, McKean D, Francis J, Neubauer S, Yu GZ, Lewandowski AJ, Sverrisdottir YB, Leeson P, 2017. Long-term cerebral white and gray matter changes after preeclampsia. Neurology 88, 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez-Skarmeta JL, Nobrega MA, 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socha MW, Malinowski B, Puk O, Dubiel M, Wicinski M, 2020. The NLRP3 Inflammasome Role in the Pathogenesis of Pregnancy Induced Hypertension and Preeclampsia. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma-Pillay P, Suleman FE, Makin JD, Pattinson RC, 2017. Cerebral white matter lesions after pre-eclampsia. Pregnancy Hypertens 8, 15–20. [DOI] [PubMed] [Google Scholar]
- Song L, Crawford GE, 2010. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc 2010, pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhong M, 2015. Association between Interleukin-10 gene polymorphisms and risk of early-onset preeclampsia. Int J Clin Exp Pathol 8, 11659–11664. [PMC free article] [PubMed] [Google Scholar]
- Spence NJ, Eberstein IW, 2009. Age at first birth, parity, and post-reproductive mortality among white and black women in the US, 1982–2002. Soc Sci Med 68, 1625–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas SK, Morrison AC, Andrela CM, Elovitz MA, 2010. Allelic variations in angiogenic pathway genes are associated with preeclampsia. Am J Obstet Gynecol 202, 445 e441–411. [DOI] [PubMed] [Google Scholar]
- Steinthorsdottir V, McGinnis R, Williams NO, Stefansdottir L, Thorleifsson G, Shooter S, Fadista J, Sigurdsson JK, Auro KM, Berezina G, Borges MC, Bumpstead S, Bybjerg-Grauholm J, Colgiu I, Dolby VA, Dudbridge F, Engel SM, Franklin CS, Frigge ML, Frisbaek Y, Geirsson RT, Geller F, Gretarsdottir S, Gudbjartsson DF, Harmon Q, Hougaard DM, Hegay T, Helgadottir A, Hjartardottir S, Jaaskelainen T, Johannsdottir H, Jonsdottir I, Juliusdottir T, Kalsheker N, Kasimov A, Kemp JP, Kivinen K, Klungsoyr K, Lee WK, Melbye M, Miedzybrodska Z, Moffett A, Najmutdinova D, Nishanova F, Olafsdottir T, Perola M, Pipkin FB, Poston L, Prescott G, Saevarsdottir S, Salimbayeva D, Scaife PJ, Skotte L, Staines-Urias E, Stefansson OA, Sorensen KM, Thomsen LCV, Tragante V, Trogstad L, Simpson NAB, Laivuori H, Morgan L, Aripova T, Casas JP, Dominiczak AF, Walker JJ, Thorsteinsdottir U, Iversen AC, Feenstra B, Lawlor DA, Boyd HA, Magnus P, Laivuori H, Zakhidova N, Svyatova G, Stefansson K, Morgan L, Laivuori H, Heinonen S, Kajantie E, Kere J, Kivinen K, Pouta A, Morgan L, Pipkin FB, Kalsheker N, Walker JJ, Macphail S, Kilby M, Habiba M, Williamson C, O’Shaughnessy K, O’Brien S, Cameron A, Redman CWG, Farrall M, Caulfield M, Dominiczak AF, 2020. Genetic predisposition to hypertension is associated with preeclampsia in European and Central Asian women. Nat Commun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K, Thurman RE, Vong S, Bates D, Neri F, Diegel M, Giste E, Dunn D, Vierstra J, Hansen RS, Johnson AK, Sabo PJ, Wilken MS, Reh TA, Treuting PM, Kaul R, Groudine M, Bender MA, Borenstein E, Stamatoyannopoulos JA, 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott LJ, Margulies EH, Boehnke M, Furey TS, Crawford GE, Collins FS, 2010. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell metabolism 12, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SD, Lehman A, Thomas F, Johnson KC, Jackson R, Wactawski-Wende J, Ko M, Chen Z, Curb JD, Howard BV, 2015. Effects of self-reported age at nonsurgical menopause on time to first fracture and bone mineral density in the Women’s Health Initiative Observational Study. Menopause 22, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana Z, Maiti K, Aitken J, Morris J, Dedman L, Smith R, 2017. Oxidative stress, placental ageing-related pathologies and adverse pregnancy outcomes. Am J Reprod Immunol 77. [DOI] [PubMed] [Google Scholar]
- Sun M, Fan Y, Hou Y, Fan Y, 2018. Preeclampsia and maternal risk of breast cancer: a meta-analysis of cohort studies. J Matern Fetal Neonatal Med 31, 2484–2491. [DOI] [PubMed] [Google Scholar]
- Sung HK, Ma SH, Choi JY, Hwang Y, Ahn C, Kim BG, Kim YM, Kim JW, Kang S, Kim J, Kim TJ, Yoo KY, Kang D, Park S, 2016. The Effect of Breastfeeding Duration and Parity on the Risk of Epithelial Ovarian Cancer: A Systematic Review and Meta-analysis. J Prev Med Public Health 49, 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Taipale J, 2016. The role of enhancers in cancer. Nat Rev Cancer 16, 483–493. [DOI] [PubMed] [Google Scholar]
- Tadesse S, Kidane D, Guller S, Luo T, Norwitz NG, Arcuri F, Toti P, Norwitz ER, 2014. In vivo and in vitro evidence for placental DNA damage in preeclampsia. Plos One 9, e86791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, Farnham PJ, 2015. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics & chromatin 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin JJ, Gomez-Piquer V, Garcia-Palomares S, Garcia-Perez MA, Cano A, 2014. Absence of long-term effects of reproduction on longevity in the mouse model. Reprod Biol Endocrinol 12, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK, Andersen KG, Mikkelsen TS, Lander ES, Schaffner SF, Sabeti PC, 2016. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell 165, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA, 2015. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12, 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilaganathan B, 2017. Placental syndromes: getting to the heart of the matter. Ultrasound Obstet Gynecol 49, 7–9. [DOI] [PubMed] [Google Scholar]
- Thomsen LC, McCarthy NS, Melton PE, Cadby G, Austgulen R, Nygard OK, Johnson MP, Brennecke S, Moses EK, Bjorge L, Iversen AC, 2017. The antihypertensive MTHFR gene polymorphism rs17367504-G is a possible novel protective locus for preeclampsia. J Hypertens 35, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen LC, Melton PE, Tollaksen K, Lyslo I, Roten LT, Odland ML, Strand KM, Nygard O, Sun C, Iversen AC, Austgulen R, Moses EK, Bjorge L, 2015. Refined phenotyping identifies links between preeclampsia and related diseases in a Norwegian preeclampsia family cohort. J Hypertens 33, 2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisi R, Bjorge T, Gissler M, Grotmol T, Kitahara CM, Myrtveit Saether SM, Ording AG, Skold C, Sorensen HT, Trabert B, Glimelius I, 2018. The role of pregnancy, perinatal factors and hormones in maternal cancer risk: a review of the evidence. J Intern Med 283, 430–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S, 2013. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet 45, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turanov AA, Lo A, Hassler MR, Makris A, Ashar-Patel A, Alterman JF, Coles AH, Haraszti RA, Roux L, Godinho B, Echeverria D, Pears S, Iliopoulos J, Shanmugalingam R, Ogle R, Zsengeller ZK, Hennessy A, Karumanchi SA, Moore MJ, Khvorova A, 2018. RNAi modulation of placental sFLT1 for the treatment of preeclampsia. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Nandakumar SK, Wang L, Giani FC, Zhang X, Rogov P, Melnikov A, McDonel P, Do R, Mikkelsen TS, Sankaran VG, 2016. Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell 165, 1530–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UnitedNations, 2019. World Population Ageing 2019: Highlights. United Nations. [Google Scholar]
- van Dijk M, Mulders J, Poutsma A, Konst AA, Lachmeijer AM, Dekker GA, Blankenstein MA, Oudejans CB, 2005. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat Genet 37, 514–519. [DOI] [PubMed] [Google Scholar]
- van Dijk M, van Bezu J, van Abel D, Dunk C, Blankenstein MA, Oudejans CB, Lye SJ, 2010. The STOX1 genotype associated with pre-eclampsia leads to a reduction of trophoblast invasion by alpha-T-catenin upregulation. Hum Mol Genet 19, 2658–2667. [DOI] [PubMed] [Google Scholar]
- Vijg J, 2021. From DNA damage to mutations: All roads lead to aging. Ageing Res Rev 68, 101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Dong X, 2020. Pathogenic Mechanisms of Somatic Mutation and Genome Mosaicism in Aging. Cell 182, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violan C, Foguet-Boreu Q, Flores-Mateo G, Salisbury C, Blom J, Freitag M, Glynn L, Muth C, Valderas JM, 2014. Prevalence, Determinants and Patterns of Multimorbidity in Primary Care: A Systematic Review of Observational Studies. Plos One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlen K, Sjolin E, Hoffstedt J, 2008. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. Journal of lipid research 49, 607–611. [DOI] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD, 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu M, Liu Z, Niu Z, Liu S, 2015. The Association of CARD8 rs2043211 Polymorphism with Preeclampsia in the Chinese Han Population. Gynecol Obstet Invest 80, 193–198. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lim R, Nie G, 2020. Elevated circulating HtrA4 in preeclampsia may alter endothelial expression of senescence genes. Placenta 90, 71–81. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y, 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219. [DOI] [PubMed] [Google Scholar]
- Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL, 2011. DNA methylation status predicts cell type-specific enhancer activity. EMBO J 30, 3028–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC, 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411. [Google Scholar]
- WorldHealthOrganization, 2020. The top 10 causes of death. World Health Organization. [Google Scholar]
- Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA, Visel A, 2014. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS genetics 10, e1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Haththotuwa R, Kwok CS, Babu A, Kotronias RA, Rushton C, Zaman A, Fryer AA, Kadam U, Chew-Graham CA, Mamas MA, 2017. Preeclampsia and Future Cardiovascular Health: A Systematic Review and Meta-Analysis. Circ Cardiovasc Qual Outcomes 10. [DOI] [PubMed] [Google Scholar]
- Wu T, Dejanovic B, Gandham VD, Gogineni A, Edmonds R, Schauer S, Srinivasan K, Huntley MA, Wang YY, Wang TM, Hedehus M, Barck KH, Stark M, Ngu H, Foreman O, Meilandt WJ, Elstrott J, Chang MC, Hansen DV, Carano RAD, Sheng M, Hanson JE, 2019. Complement C3 Is Activated in Human AD Brain and Is Required for Neurodegeneration in Mouse Models of Amyloidosis and Tauopathy. Cell Rep 28, 2111–+. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin N, Zhang H, Luo X, Ding Y, Xiao X, Liu X, Shan N, Zhang X, Deng Q, Zhuang B, Qi H, 2014. IL-27 activates human trophoblasts to express IP-10 and IL-6: implications in the immunopathophysiology of preeclampsia. Mediators Inflamm 2014, 926875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Zhu Z, Hosgood HD, Lan Q, Seow WJ, 2020. Reproductive factors and lung cancer risk: a comprehensive systematic review and meta-analysis. BMC Public Health 20, 1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nakaya M, Miyazaki Y, 2009. Interleukin 27: a double-edged sword for offense and defense. J Leukoc Biol 86, 1295–1303. [DOI] [PubMed] [Google Scholar]
- Zhao L, Bracken MB, DeWan AT, 2013. Genome-wide association study of pre-eclampsia detects novel maternal single nucleotide polymorphisms and copy-number variants in subsets of the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study cohort. Ann Hum Genet 77, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Triche EW, Walsh KM, Bracken MB, Saftlas AF, Hoh J, Dewan AT, 2012. Genome-wide association study identifies a maternal copy-number deletion in PSG11 enriched among preeclampsia patients. BMC Pregnancy Childbirth 12, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.