Abstract
Recent in crystallo reaction intermediates have detailed how nucleic acid hydrolysis occurs in the RNA ribonuclease H1 (RNase H1), a fundamental metalloenzyme involved in maintaining the human genome. At odds with the previous characterization, these in crystallo structures unexpectedly captured multiple metal ions (K+ and Mg2+) transiently bound in the vicinity of the two-metal-ion active site. Using multi-microsecond all-atom molecular dynamics and free-energy simulations, we investigated the functional implications of the dynamic exchange of multiple K+ and Mg2+ ions at the RNase H1 reaction center. We found that such ions are timely positioned at non-overlapping locations near the active site, at different stages of catalysis, being crucial for both reactants’ alignment and leaving group departure. We also found that this cation trafficking is tightly regulated by variations of the solution’s ionic strength and is aided by two conserved second-shell residues, E188 and K196, suggesting a mechanism for the cations’ recruitment during catalysis. These results indicate that controlled trafficking of multi-cation dynamics, opportunely prompted by second-shell residues, is functionally essential to the complex enzymatic machinery of the RNase H1. These findings revise the current knowledge on the RNase H1 catalysis and open new catalytic possibilities for other similar metalloenzymes including, but not limited to, CRISPR-Cas9, group II intron ribozyme and the human spliceosome.
Keywords: Cations, second-shell residues, positive-charges trafficking, nucleic acid processing, catalysis, molecular dynamics
Graphical Abstract
Introduction
Recently, structural and biophysical studies have revealed that additional metal ions (viz., K+ and Mg2+) are transiently engaged in the vicinity of the well-recognized two-metal-ion catalytic site of enzymes like DNA/RNA polymerases,1,2 endo- and exo-nucleases,3-6 and even type II topoisomerase.7-9 All these metalloenzymes are fundamental for the expression and maintenance of RNA and DNA within the cell,10-12 and are often targeted to treat human diseases, from cancer to viral and bacterial infections.13-16
Remarkably, transient K+ and Mg2+ ions have been often captured unexpectedly close to conserved second-shell protein residues, which have been thereby proposed to favor metal recognition and recruitment from the bulk.17 Such positive ions at the metal-aided catalytic site during the processing of nucleic acids were suggested to contribute actively to the overall catalytic process,1,18-24 although their exact functional and dynamic role remains only partially understood.
In this context, the prototypical nucleic acid-processing enzyme RNA ribonuclease H1 (RNase H1, Figure 1A), which cleaves phosphodiester bond in RNA:DNA hybrids being responsible for removing the Okazaki fragments during DNA replication,25 has been extensively characterized via biophysical26-32 and computational33-35 studies.
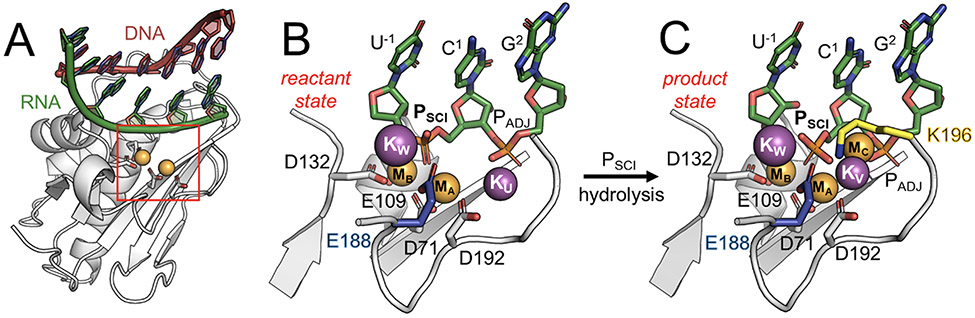
Figure 1.
RNase H1 catalytic intermediates captured by time-resolved X-ray crystallography.35 (A) Overview of the RNase H1 (white) in complex with an RNA:DNA hybrid (green:red). (B) Catalytic site before RNA hydrolysis (i.e., reactant state, PDBid: 6DO9). The catalytic residues forming the DEDD motif (D71, E109, D132 and D192; white) and the second shell residue E188 (blue) are represented as sticks. RNA nucleotides (green) including the scissile phosphate (PSCI) and its adjacent phosphate (PADJ) are also shown. The two catalytic magnesium ions MA-MB (orange) and the additional potassium ions KW and KU (purple) are shown as spheres. (C) Catalytic site upon RNA hydrolysis (i.e., product state, PDBid: 6DOX). Here, KU is replaced by the third divalent metal ion MC (orange), or by the monovalent KV (purple), while the second-shell residue K196 (yellow) can directly interact with the scissile phosphate.
Nevertheless, only recent data, obtained using time-resolved X-ray crystallography,36 have shown that the catalysis of RNase H1 is accompanied by the trafficking of both mono (K+) and divalent (Mg2+) metal ions, transiently bound in the vicinity of the two-metal-ion catalytic site, at different stages of catalysis (Figure 1B-C). Indeed, the new X-ray structures of the Bacillus Halodurans RNase H1 in complex with an RNA:DNA hybrid substrate have captured the in crystallo intermediates of the catalysis (Figure 1B-C). The active site displays both catalytic Mg2+ ions (MA-MB) properly located and coordinated by first-shell conserved carboxylates, referred to as the DEDD motif (D71, E109, D132, D192).
These novel findings support the possible functional role of such transitory metal ions.1,18-23 Intriguingly, prior RNA hydrolysis, two K+ ions are captured at the active site, at different times (Figure 1B). The X-ray structure solved 40 seconds (40s) after the in crystallo incubation shows that a K+ ion locates at the “U” site (and is hereafter named KU, PDBid: 6DMV, Figure S1A).36 In this position, KU binds to the catalytic D192 and the phosphate adjacent (PADJ) to the scissile group (PSCI). Notably, at this stage, no reaction is observed as the Michaelis-Menten complex is not adequately formed. The distance between Pro-Sp oxygen (OSp) of the scissile phosphate and the catalytic MB is 2.6Å (dMB–OSp in Figure S1A, close-up view). Subsequently, after 120s, a second K+ ion locates at the “W” site (viz., KW, Figure 1B; PDBid: 6DO9),36 where it is directly coordinated to the second-shell residue E188 and to PSCI, in agreement with previous proposal from classical molecular simulations of this enzyme.34 Notably, at this stage, the scissile phosphate results properly positioned for the catalysis, profiting of the increased vicinity to MB (with the dMB–OSp = 2.2Å, Figure S1B, close-up view). Then, upon RNA hydrolysis, while KW is still present at the active site (Figure 1C), the occupancy of KU decreases with product formation. Indeed, after 360s, a third K+ ion locates very close to the “U” site, at the “V” site (viz., KV, Figure 1C; PDBid: 6DOX),36 eventually substituting KU. Moreover, at high concentrations of divalent metals [M2+], a third Mg2+ ion is captured to directly coordinate the scissile phosphate at the “C” site (viz., MC, Figure 1C; PDBid: 6DPD).36 The in crystallo catalysis has also shown that the RNase H1 activity depends on the ionic strength of the reaction buffer.36 Indeed, the formation of reaction products increases with the concentration of both K+ and Mg2+ cations (i.e., [K+] and [Mg2+]), although an excess of ions leads to the so-called “attenuation effect”, which reduces RNA hydrolysis.
Interestingly, catalysis is impaired for the single mutants E188A or K196A, in presence of Mg2+.36 The X-ray structures of such mutants have revealed an altered active site architecture as compared to the wild type (wt) RNase H1 upon RNA hydrolysis, with only partial product formation. In the product structure of the E188A mutant, no K+ ion locates at the “W” site (Figure S2A), in contrast to what observed in the wt form, indicating that E188 directly impacts the presence of K+ at the active site. On the other hand, in the structure of the K196A mutant (Figure S2B), one K+ ion is captured at the “W” site, as in the wt RNase H1. Nonetheless, in absence of K196, the products are misfolded due to a slight rotation of the scissile phosphate, in contrast to what was observed in the reactive center of the wt form. Most importantly, no other MC locates at the active site in this K196A mutant.
Such a wealth of structural and biochemical evidence suggests a functional role of multiple metal ions (K+ and Mg2+) transiently bound in the vicinity of the two-metal-ion active site in RNase H1, where metal trafficking and exact localization at the reaction center seems critical for catalysis.36 Indeed, this mechanistic hypothesis would justify the stage-dependent location of additional mono and divalent metals during catalysis, as observed in the in crystallo intermediates of RNase H1.
To test this hypothesis and examine the dynamics of this complex step-wise and metal-aided enzymatic process, we applied extensive all-atom molecular dynamics (MD) and free-energy simulations (>27 μs, in total) on multiple systems of RNase H1, at different stages of the catalysis and ionic strengths of the solution. Our results identify an ordered dynamics of multiple cations and their controlled trafficking at the RNase H1 active site as a functional element of the enzymatic strategy to prompt RNA hydrolysis.
Results
Two transient K+ ions are alternatively located at the reactants for catalysis.
According to structural data,36 the RNase H1 catalytic site can contain three K+ ions, namely KU, KW, and KV, which are located at the respective binding site “U”, “W”, and “V” (Figure 1). The in crystallo data also show that this extended K+ ion cluster is transiently formed and disrupted during catalysis. Only KU is initially bound (at 40s, when no reaction is observed, PDBid: 6DMV).36 Then, in the reactant state, KU occupancy decreases, while KW locates at the active site (at 120s, PBBid: 6DO9; at 200s, PDBid: 6DOB). Finally, KW and KV are found in the products (at 360s PDBid: 6DOX).36
To investigate K+ ion's dynamical recruitment and binding at the catalytic site, we first performed two MD simulations of ~1 us each, where both KU and KW were removed from the reactant state (PDBid: 6DO9). Such simulations were run at the reference and optimal concentration for catalysis (in line with the experimental conditions of 6mM [Mg2+] and 200mM [K+]). First, we noted that even in the absence of K+ ions, the overall catalytic architecture is maintained well (see RMSD in Figure S3). Monitoring the spatial distribution and number of K+ ions in a sphere of 6 Å centered on the scissile phosphate (PSCI, Figure 2A), we found that a transient K+ ion can be intermittently located at the W-site (i.e., KW). In this position, KW sits at ~2.77 ± 0.24 Å from PSCI, (Figure S3), in excellent agreement with X-ray data (PDBid: 6DO9, KW at = 2.5 Å). On the other hand, K+ ions do not occupy the “U” site, which remains empty (Figure 2A and S4A).
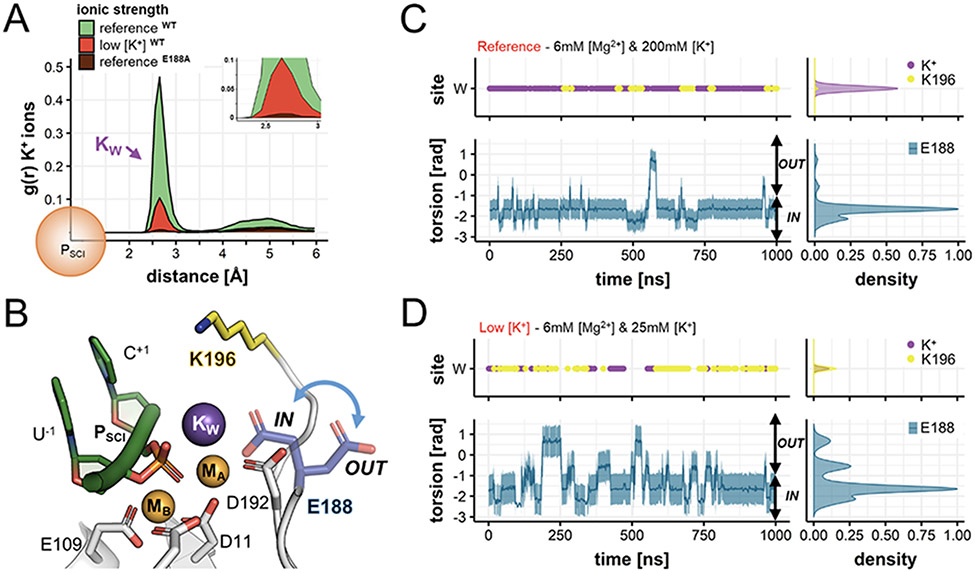
Figure 2.
Occupancy of positive charges (i.e., K+ ions and K196) and conformational dynamics of E188 in the reactant state. (A) Spatial distribution and number of K+ ions around the scissile phosphate (PSCI) of wild type RNase H1, computed during MD simulations as the radius of gyration, g(r), for K+ ions at reference (green) and low (red) K+ concentrations. In both cases, K+ ions are mainly found at ~2.77 ± 0.24 Å from the scissile phosphate, thus located at the “W” site. The g(r) for K+ is also computed during MD simulations of the E188A mutant performed at reference concentration (brown). Here, the “W” site is never occupied by K+ ions. (B) Conformational change of the second-shell residue E188 (blue arrow), pointing inside the active site (E188IN) and outside (E188OUT). (C) Occupancy of the “W” site (upper panel) by K+ ions (purple) and by K196 (yellow); and variation of the torsional angle E188-θ (lower panel, computed between the N-Cα-Cβ-Cδ atoms) along MD simulations at reference [K+]. Normalized densities are on the right. At optimal ionic strength, the binding of K+ is favored and E188 is stabilized in the E188IN conformation. (D) Same descriptors as in (C) for MD simulations at low [K+]. Here, the binding of KW is disfavored, substituted by K196, and the inner ↔ outer swings of E188 are more frequent.
We also found that the spontaneous binding of KW is associated with a conformational change of the second-shell residue E188 (Figure 2B), which can assume two main orientations, pointing towards the active site as in the X-ray structure (viz., E188IN), and outside (viz., E188OUT) toward the bulk. By monitoring the variation of the torsional angle E188-θ (formed by the E188 atoms N-Cα-Cβ-Cδ, Figure 2C, lower panel), we found that E188 mainly samples the E188IN state when KW locates at the “W” site (shown in the upper panel). On the other hand, E188 samples the E188OUT state when the “W” site is not occupied by KW. This correlation, which is also observed in the simulation replicas (Figure S6), suggests that the inner ↔ outer conformational swing of E188 can favor the recruitment of K+ ions from the bulk to the “W” site. Importantly, these findings are also in agreement with biochemical data showing that catalysis is impaired in the E188A mutant.36 While the active site architecture of this mutant is highly similar to the wt RNase H1 (Figure S2A), the only structural difference is the lack of KW (PDBid: 6DPO).36 Accordingly, we further evaluated the role of E188 for the dynamic recruitment of KW, and performed two additional MD simulations (1us, each). First, we simulated the E188A mutant at optimal ionic strength. As a result, no K+ ions bind at the active site (Figure 2A), at odds with what was observed for the wt RNase H1. This result confirms that the presence of the second-shell E188 promotes the binding of KW. Then, we performed MD simulations of the wt RNase H1, restraining the E188 residue in the “out” conformation. Here, the occupancy of the “W” site by K+ ions was dramatically reduced (Figure S6C), suggesting that the inner ↔ outer conformational swing of E188 is fundamental to guarantee the recruitment and proper positioning of KW, which thus appears crucial for catalysis.
As noted above, K+ ions do not occupy the “U” site during such MD simulations. To further corroborate this observation, a third simulation replica has been performed by locating one K+ ion at the “U” site, based on the coordinates of the in crystallo RNase H1 intermediate obtained as soon as 40s after incubation (Figure S1A, PDBid: 6DMV).36 In this simulation, such K+ ion spontaneously departs from the “U” site (Figure S4B), just after the equilibration phase, when the PSCI approaches the MA-MB reactive center. That is, the distance between MB and the Pro-Sp oxygen (OSp) of the scissile phosphate reaches dMB–OSp = 1.9 ± 0.13 Å. Notably, these findings align with the occupancy trend observed for KU in the series of X-ray structures obtained at different times from incubation. Indeed, the occupancy of KU (KUOcc) progressively reduces when the scissile phosphate gets properly positioned for catalysis and approaches MB. In detail, at dMB–OSp = 2.6 Å, KUOcc = 0.35 (at 40s, PDBid:6DMV); at dMB–OSp = 2.2 Å, KUOcc = 0.25 (at 120s, PDBid:6DO9); and at dMB–OSp = 2.1 Å, KUOcc = 0.10 (200s, PDBid:6DOB). Thus, MD simulations agree with structural evidence, supporting that the proper positioning of the substrate favors KU departure.
Interestingly, the in crystallo data also revealed that low [K+] lead to reduced product formation in RNase H1.36 We simulated such conditions with three more ~1μs-long MD runs at low K+ concentration (i.e., 6mM [Mg2+] and 25mM of [K+], according to experimental conditions), with both “U” and “W” sites empty. This allowed us to investigate whether a diminished K+ ionic strength could affect the recruitment of K+ ions at the W site, as well as the dynamical role of E188 in this process. As a result, also at low [K+], we observed null occupancy of the “U” site (Figures 2A and S4A). Moreover, the recruitment of KW occurs less frequently compared to the simulations at the optimal [K+] (paragraph above), as shown by the decreased occupancy of the “W” site by K+ ions (Figure 2A). Most notably, E188 more frequently samples the E188OUT conformation, pointing toward the solvent, searching for a transient K+ ion from the bulk (Figure 2D, lower panel). In this regard, however, it is worth noting that the lack of KW is often compensated by the positively charged side chain of the second-shell residue K196, which at times accesses the “W” site, with almost the same frequency of a K+ ion (Figure 2D, upper panel). Overall, MD runs at low [K+] indicate that when K+ does not access the “W” site, E188 preferably assumes the E188OUT state, at odds with MD runs at high [K+], where the E188IN conformation is mostly observed and K+ occupies the “W” site. These data corroborate that the binding of KW is associated with the conformational change of E188, which could act as a KW functional recruiter.
To further understand the influence of K+ concentration on the E188-mediated recruitment of KW and to estimate the energetics of such process, we performed free-energy simulations using well-tempered metadynamics at both optimal (200mM) and low (25mM) [K+] (Figure S7). The torsional angle E188-θ was used as a collective variable to sample multiple inner ↔ outer conformational swings of this residue in both directions. At optimal [K+], the free-energy profile for such a conformational switch, when going from E188IN to E188OUT conformations, showed a barrier of ~6.2 kcal mol−1, while at low [K+] was ~3.7 kcal mol−1 (Figure S7). Additionally, the E188OUT to E188IN conformational swing displayed a barrier of ~3.2 kcal mol−1 at optimal [K+], while at low [K+] it was ~2.4 kcal mol−1 (Figure S7). Such low barriers are consistent with the conformational changes observed in our equilibrium simulations. Additionally, the slightly smaller barriers at low [K+], compared to the barrier at high [K+], is in line with the result from unbiased MD, overall indicating that a more frequent conformational change of E188 may thus favor the recruitment of one K+ ion, from the bulk to the “W” site.
Finally, four MD simulations of the reactant were performed at high [Mg2+] (80mM [Mg2+] and 75mM [K+], three MD runs of ~1 us each), which was experimentally shown to reduce RNA hydrolysis through the so-called attenuation effect.36 In these simulations, despite E188 is stabilized in the E188IN conformation, no K+ ions are captured at “W” site (Figure S8). This is because an additional Mg2+ metal ion locates at ~4.53 ± 0.11 Å from the PSCI (Figure S5), close to the “W” site (viz., MW) and directly binds E188, closing the accessibility of the “W” site to K+ ions (Figure S8E). In this way, this third Mg2+ ion hampers the dynamic recruitment of KW, preventing the formation of the K+ metal cluster for catalysis at high [Mg2+]. This finding may thus represent an explanation of the attenuation effect.28,37 Notably, these results were confirmed through multiple MD runs using three different non-bonded models for Mg2+ (Figure S8). Indeed, despite Mg2+ parametrization can affect the MC-E188 binding mode,38 and the dynamics of E188 (Figure S8A-D, lower panel), still high [Mg2+] reduces the E188 inner ↔ outer conformational switch, regardless the ions’ model. In turn, the conformational preference of E188 prevents the E188-mediated recruitment of KW for catalysis.
Taken together, MD simulations of the reactant state reveal an ordered dynamics of cations, which agrees well and adds further clarification to recent hypotheses based on experimental data.36 Accordingly, the catalytic site of RNase H1 initially contains only KU, which is released just upon the reactants' alignment, so prone to catalysis – in general agreement with the mechanistic proposal by Samara and Yang.36 In addition, the simulations reveal an inner ↔ outer swing of the second-shell residue E188, which is suggested here to critically favor the recruitment of one additional K+ ion at the “W” site to form the reactant RNase H1 active site. In this way, this sequence of dynamic events, which we show to be optimally controlled by the ionic strength in solution, leads to a competent catalytic state (as in PDBid: 6DOB).36
Multi K+ and Mg2+ trafficking enhances product formation.
We performed MD simulations of RNase H1 in the product state to evaluate if and how the ionic distribution in the vicinity of the active site is altered upon RNA hydrolysis. The starting configuration included both catalytic metals MA-MB and the additional KW, as observed after 600s of incubation when the RNA hydrolysis reached a plateau (PDBid: 6DOG).36 MD simulations have been performed in analogy with those of the reactant state (previous section), considering an initial reference, optimal ionic strength of 6mM [Mg2+] and 200mM [K+].
In these simulations, KW stably coordinates the scissile phosphate PSCI by locating at the “W” site (Figure 3A, upper panel and S9A). Moreover, one additional K+ ion transiently locates at the “V” site (i.e., in the vicinity of the catalytic site, but in a distinct position compared to the “W” and “U” sites). In this position, KV interacts with PSCI (Figure 4A), together with KW. Importantly, this transient KV ion superimposes well with the Rb+ ion observed in the X-ray data (PDBid: 6DOX).36 Interestingly, the simulations show that the K196 side chain can also reversibly access the “W” site, when empty.
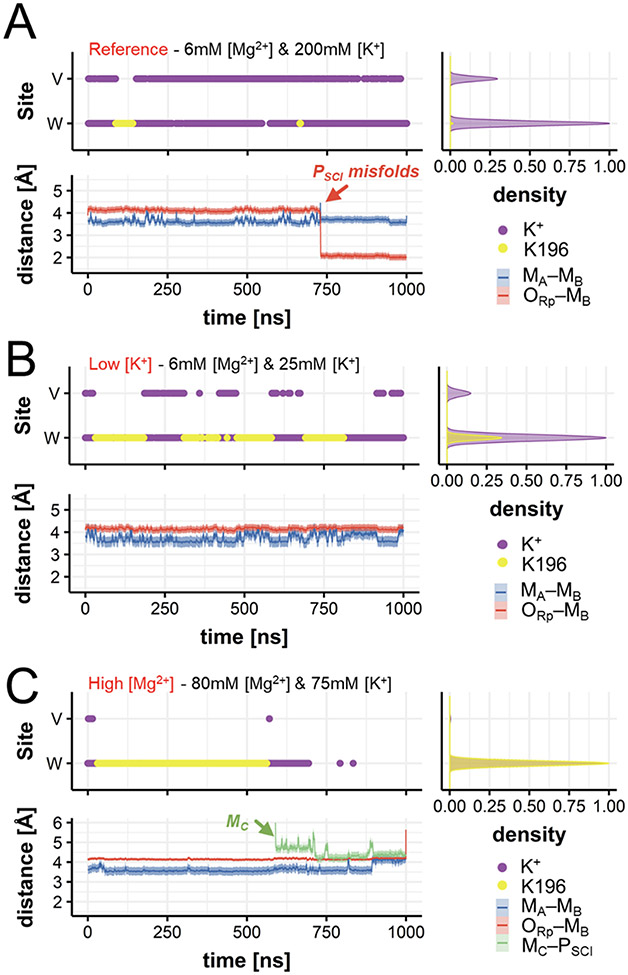
Figure 3.
Occupancy of positive charges (viz., K+ ions and K196) and conformational dynamics of the products. (A) Occupancy of the W and the V sites (upper panel) by K+ ions (purple) and by the K196 side chain (yellow); and distances (lower panel) between the catalytic ions MA–MB (blue trace), and between the pro-Rp oxygen of the scissile phosphate and MB (ORp–MB, red trace) during MD at optimal concentrations. Normalized densities are on the right. (B-C) Same descriptors as in (A) for MD simulations at low [K+] (B) and at high [Mg2+] (C). At low [K+], the interaction between K196 and the scissile phosphate is more frequent (Figure 4B). At high [Mg2+], MC approaches the scissile phosphate (green trace of MC–PSCI), while the MA–MB distance increases, preluding the release of the products (Figure 4C).
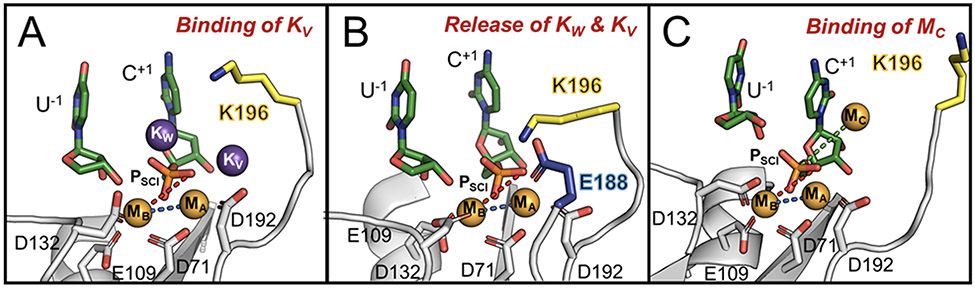
Figure 4.
Snapshots of the RNase H1 active site upon catalysis. (A) In the product state, KW is stably coordinated at the active site, while KV is only transiently observed. (B) KW and KV can exchange their position with K196, which directly coordinates the scissile phosphate preventing its misfolding. (C) The third additional MC binds the scissile phosphate and promotes the increase of the distance between the two catalytic MA–MB ions for product release.
Three additional ~1μs-long MD runs considered a lower [K+] (i.e., 6mM [Mg2+] and 25mM of [K+]). Here, a more frequent interchange between K+ and K196 was observed (Figure 3B, upper panel). At lower [K+], the “W” and “V” sites are often not occupied by K+, compared to their occupancy at higher [K+]. Consequently, K196 is more often found to interact directly with the phosphate leaving group (Figure 4B). This results in a stable configuration of the PSCI (Figure 3B, lower panel). On the other hand, when K196 more rarely accesses the “W” site, a partial misfold of the PSCI is observed (Figure 3A, lower panel). These findings, which are well reproduced in the simulation replicas (Figure S9), suggest that K196 could be quite relevant for the precise accommodation and preliminary departure of the solvent-exposed leaving nucleotides. Indeed, K196 can, at times, interchange with KW and KV. As a result, the correct positioning of the phosphate of the leaving group for departure is maintained (as observed in the PDBid: 6DOG, captured right before leaving group departure).36 Notably, the positively-charged side chain of K196 locates at the W-site (at ~2.7 Å from PSCI) to H-bond the leaving group only if both KW and KV ions have departed from the catalytic site (see Figure S9, where the only presence of KW still preserves the hydrolyzed leaving group in place, at the two-metal-ion center).
Molecular simulations of the products have also been performed at increased [Mg2+] (i.e., 80mM of Mg2+ and 75mM of K+, five MD runs of ~1μs each) in line with the experimental conditions. These simulations show that K196 can be replaced by a transient third Mg2+ ion, which locates at the “C” site (viz., MC, Figures 4C). The approach of MC consistently occurred in MD replicas, conducted using three different non-bonded models for Mg2+ (Figures 3C, lower panel and S9C-D). This finding matches the structural data obtained at high divalent metals concentration (≥ 16mM), which identified such MC in a very similar position, close to the leaving group (PDBid: 6DPD).36 Moreover, upon binding of MC, the internuclear MA–MB distance is increased (from 3.72 ± 0.11 Å to ~4.11 ± 0.07 Å, Figures 3C, lower panel), reflecting the destabilization of the active site’s architecture to prelude the products’ exit. Thus, such a transient third ion seems recruited in the product state to favor the leaving group departure, as seen in other nucleic acid processing enzymes.17,39,40
It is important to recall that MD of the reactants at high [Mg2+] have also shown that a third Mg2+ ion can locate in the vicinity of the catalytic site (Figures S5 and S8E). In that case, however, the third Mg2+ ion locates close to the “W” site and binds E188, hampering the dynamic recruitment of KW and preventing the formation of the K+ metal cluster for catalysis. Hence, taken together, the simulations at high [Mg2+] support the idea that a third MC ion could be recruited in the product state to favor the leaving group departure.17,39,40
These results are also in line with the structure of the K196A mutant (Figure S2B; PDBid: 6DPM).36 Here, MC is missing, suggesting that the K196A mutation would somehow impair product stabilization, disfavoring MC recruitment for leaving group departure. To test this hypothesis, we carried out two additional MD runs of the K196A mutant, at both low K+ (i.e., 6mM [Mg2+] and 25mM [K+]) and high Mg2+ (i.e., 80mM [Mg2+] and 75mM [K+]) concentrations (~300 ns and ~1 μs, respectively). These simulations confirm that the K196A mutant does not lead to MC recruitment, further sustaining the crucial role of K196 for product stabilization and MC binding (Figure S11).
In summary, molecular simulations of the product state indicate that a controlled metal trafficking at the catalytic site plays a critical role in RNA hydrolysis and product release. Upon RNA hydrolysis, the potassium metal cluster breaks, while K196 contributes to product stabilization before the third divalent MC ion can further favor product release (vide infra).
Third Mg2+ trafficking for product release.
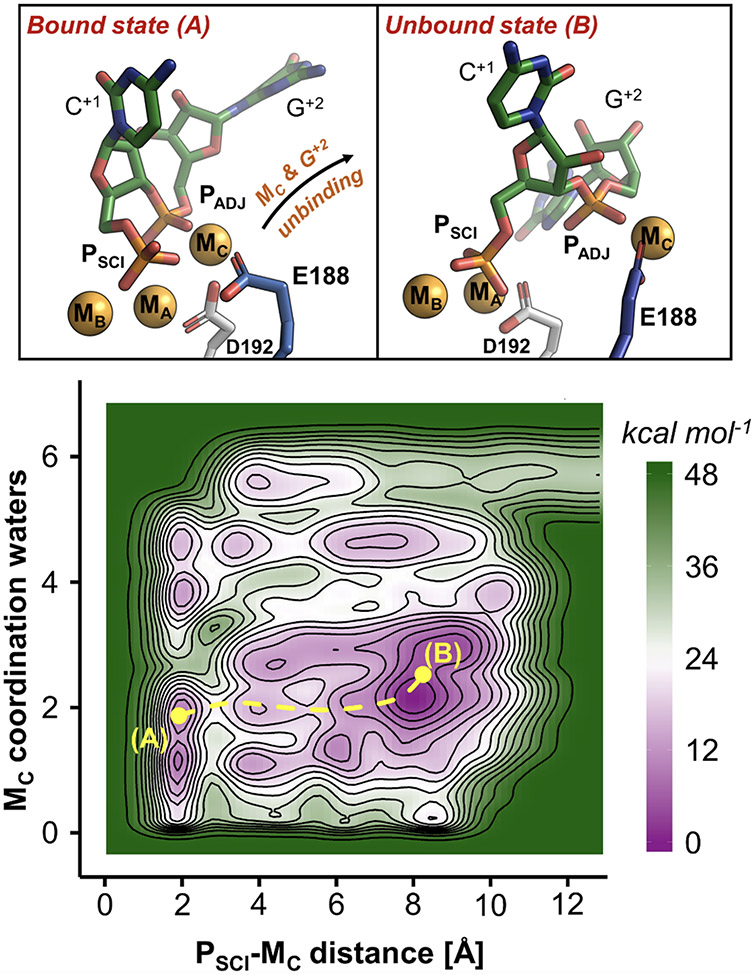
In the products, upon binding of MC, the internuclear MA–MB distance increased (Figure 3C), suggesting that the active site might be prone to release the reaction products, as observed in other two-metal-ion enzymes.17,39,40 To further investigate how the unbinding of the third MC may facilitate product exit, we performed well-tempered metadynamics simulations of the product state, which comprehends an extended metal cluster formed by MA-MB, MC, and KW. The gaussian-shaped potential was deposited using two collective variables (CVS): (i) the coordination number of the MC ion, which traced the number of water molecules in its first-shell; and (ii) the distance between MC and the scissile phosphate (PSCI–MC).
Metadynamics identifies two main metastable states (viz., A and B, shown in Figure 5, upper panel), which correspond to the two main minima of the computed free-energy surface (FES, Figure 5, lower panel). In state A, MC and bridges the scissile and adjacent phosphates (PSCI and PADJ), while the catalytic D192 and the second-shell E188 establish a monodentate interaction with MC. In state B, MC loses the coordination with D192 and gets bidentate coordination with the E188 carboxylic moiety. At this point, the rotation of E188 shuttles the MC metal out of the active site, together with PADJ, now located ~9 Å away from MA-MB. Notably, the base G+2 and the active site are completely exposed to the bulk, and the distance between the catalytic MA-MB, reaches ~4.7 Å, indicating the opening of the active site for product release. The free-energy barrier estimated on the minimum free-energy path for this process is ~15 kcal mol−1, with the unbound state B favored of ~6 kcal mol−1 with respect to the bound state A.
Figure 5.
Role of MC and E188 in the product release. Upper panel: intermediates states (A and B) of the RNase H1 after phosphodiester bond cleavage, identified via free-energy simulations. In state A, MC is coordinated by E188. Upon rotation of this residue (state B), MC is released with the departure of the adjacent phosphate (PADJ). Lower panel: free-energy surface describing the product release mechanism, indicating the two intermediate A and B. The energy scale is in kcal mol−1, contours are traced every 3 kcal mol−1.
We also investigated the role of E188 in the product state via additional metadynamics runs of the E188A mutant (Figure S2A). Remarkably, in these simulations, MC preserves the coordination with the adjacent phosphate PADJ and D192 (state A’; Figure S12). However, the absence of the E188 carboxylic moiety, which in the wt RNase H1 functions as an anchoring point for the exit of MC (Figure 5), strongly disfavors the release mechanism (state B’, Figure S12). Indeed, in the E188A mutant, MC needs first to be partially hydrated before being released together with the products, as shown by minimum free energy path connecting the state A’ with B’ (Figure S12). Importantly, in this case, the estimated energetic barrier for MC unbinding and leaving group departure is ~35 kcal mol−1 (Figure S12), more than twice compared to the one obtained for the wt RNase H1. Moreover, in the absence of E188, the bound state A’ results favored the unbound state B’ of ~18 kcal mol−1, as opposed to what was observed in the wt RNase H1.
These results suggest that MC acts in concert with the second-shell residue E188 to promote the release of the reaction products.
Discussion
Controlled cations trafficking and enzymatic strategy in RNase H1.
The RNase H1 enzyme is a fundamental endonuclease involved in DNA replication.25 Recently, time-resolved X-ray crystallography revealed the transient presence of multiple K+ and Mg2+ ions at the active site of RNase H1, at different stages of catalysis.36 Here, extensive molecular dynamics (MD) and free-energy simulations (over a sampling of >27 μs) characterized the functional dynamics of this newly determined heteronuclear metal cluster before and upon enzymatic catalysis. Our simulations capture an ordered coordination of motion of transient metal ions and second-shell residues for catalysis. We show that four biding sites in the vicinity of the RNase H1 catalytic core, namely the “U”, “W”, “V”, and the “C” sites (Figure 1), are neatly but only intermittently occupied by K+ and Mg2+ metals during catalysis, in agreement with experiments.36
We observed that, before RNA hydrolysis (i.e., in the reactant state), a K+ ions cluster is dynamically formed to favor substrate alignment and, possibly, activation for catalysis. At first, one K+ ion transiently locates at the “U” site (viz., KU) and assists in forming the catalytically competent fold of the RNase H1 active site. This KU is spontaneously released into the bulk only after the proper coordination between the scissile phosphate and the two catalytic MA-MB ions (Figure S4). At this point, the inner ↔ outer conformational swing of the second-shell residue E188 favors the recruitment and the precise positioning of a second K+ ion at the “W” site (viz., KW, Figure 2), as also observed for enzymes such as human Exo1.17 In RNase H1, this KW directly contacts the scissile phosphate to favor the correct positioning of the reactants, prompting catalysis. Such KW can depolarize the phosphodiester bond, thus promoting the catalytic activation of the reactants.1,20
Importantly, we found that this chain of dynamic (un)binding events of K+ ions is significantly affected by the reaction buffer's ionic strength. In fact, in agreement with in crystallo data showing that catalysis is reduced at low concentrations of K+ (i.e., [K+]),36 MD simulations performed at low [K+] show that the E188-mediated recruitment of KW occurs less frequently (Figure 2D). Accordingly, the inner ↔ outer conformational swing of E188 is energetically favored at low [K+] with a barrier of ~3.7 kcal mol−1 vs. ~6.2 kcal mol−1 higher [K+] (Figure S7). The crucial role observed here for the E188-mediated recruitment of KW is also sustained by both biochemical and structural evidence. Indeed, alanine mutation of E188 reduces the catalytic efficiency of RNase H1.36 As well, X-ray structures have shown that such mutation displays no KW at the active site (Figure S2A), at odds with the wt RNase H1.36 In line with this evidence, our MD simulations show that the recruitment of KW is dramatically hampered when E188 is restrained in its “out” conformation (Figure S6C), as well as when E188 is mutated into an alanine (Figure 2A). These findings support the E188-mediated mechanism for KW recruitment, which ensures the reactants' proper formation. Notably, these results further corroborate the recruitment role of second-shell residues for two-metal-ion catalysis, such as recently reported for human Exo1, ExoG, Exo-λ and other 5’-exonucleases.17
Upon RNA hydrolysis, we observe an additional K+ ion intermittently located at the so-called “V” site, in the vicinity of the catalytic core. This KV ion coordinates the scissile phosphate PSCI, together with KW (Figures 3A and 4A), as also shown by in crystallo data (PDBid: 6DOX).36 We note that these KW/KV ions can interchange their distinct position with the second-shell residue K196, near PSCI (Figure 3B and 4B). Indeed, such residues favor the stabilization of the PSCI-centered reaction products. The functional role of K196 in products’ stabilization is also sustained by the X-ray data of the catalytically impaired K196A mutant of RNase H1 (PDBid: 6DPM),36 in which the products are misfolded and no third Mg2+ ion (MC) is resolved. Accordingly, MD simulations of this mutant show that the absence of this lysine induces the product’s misfolding and hampers the binding of MC (Figure S11). On the other hand, in the wt RNase H1, leaving group departure occurs when the K+ ions and K196 are ultimately displaced by a third Mg2+, which spontaneously sits at the “C” site (viz., MC) and binds PSCI (Figure 4C). At this point, the internuclear MA–MB distance is increased (from 3.72 ± 0.11 Å to ~4.11 ± 0.07 Å), as the overall active-site structure evolves towards the products (Figures 3C and 4C). This is in line with the X-ray structure showing that the distance between the MA–MB increases with product release (PDBid: 2G8V), from 3.7 Å to 4.8 Å.41 From this point, the leaving group departure is expected to occur.
To further investigate the products’ release, metadynamics simulations show that MC is involved in a step-wise product release mechanism. In detail, MC initially bridges together PSCI and its adjacent phosphate, PADJ (Figure 5, state A). In this conformation, MC functions as a positively-charged anchor for E188. At this point, the swing of E188 occurs with the simultaneous departure of the movable MC, together with the reaction products (Figure 5, state B), as also observed for other metalloenzymes.19,22,42-44 This process occurs with an estimated energy barrier of ~15 kcal mol−1 (Figure 5). Metadynamics simulations of the E188A mutant also confirm such a functional role of E188 for product release. In this case, the estimated energetic barrier for the departure of the leaving group was ~35 kcal mol−1 (Figure S12), which is more than 2-fold higher than that in the wt RNaseH1. In this regard, we note that in the human RNase H1, E188 corresponds to H264. Based on both structural and biochemical analysis,28,29,41 this histidine residue has been proposed to favor, together with the mobile C-terminal loop, the release of the products by altering the coordination of MA ion (Figure S14).
Such a dynamically controlled trafficking of positive charges at RNase H1 active site is finely dependent on the specific ion concentration, as displayed in the X-ray experiments. In fact, MD of the reactant performed at higher [Mg2+] showed a third Mg2+ ion that spontaneously locates at the “W” site (viz., MW differently located than MC, Figure S5), closing the accessibility of the “W” site to K+ ions. In this location, MW interacts with E188 (Figure S8E), hampering the E188-mediated recruitment of KW and preventing the formation of the K+ metal cluster for catalysis. This mechanistic finding is of particular interest in light of the so-called “attenuation effect”.28,37 Indeed, the precise location of this third Mg2+ at the “W” site precludes the formation of a catalytic metal cluster, thereby representing an explanation for the reduced RNA hydrolysis at high [Mg2+].
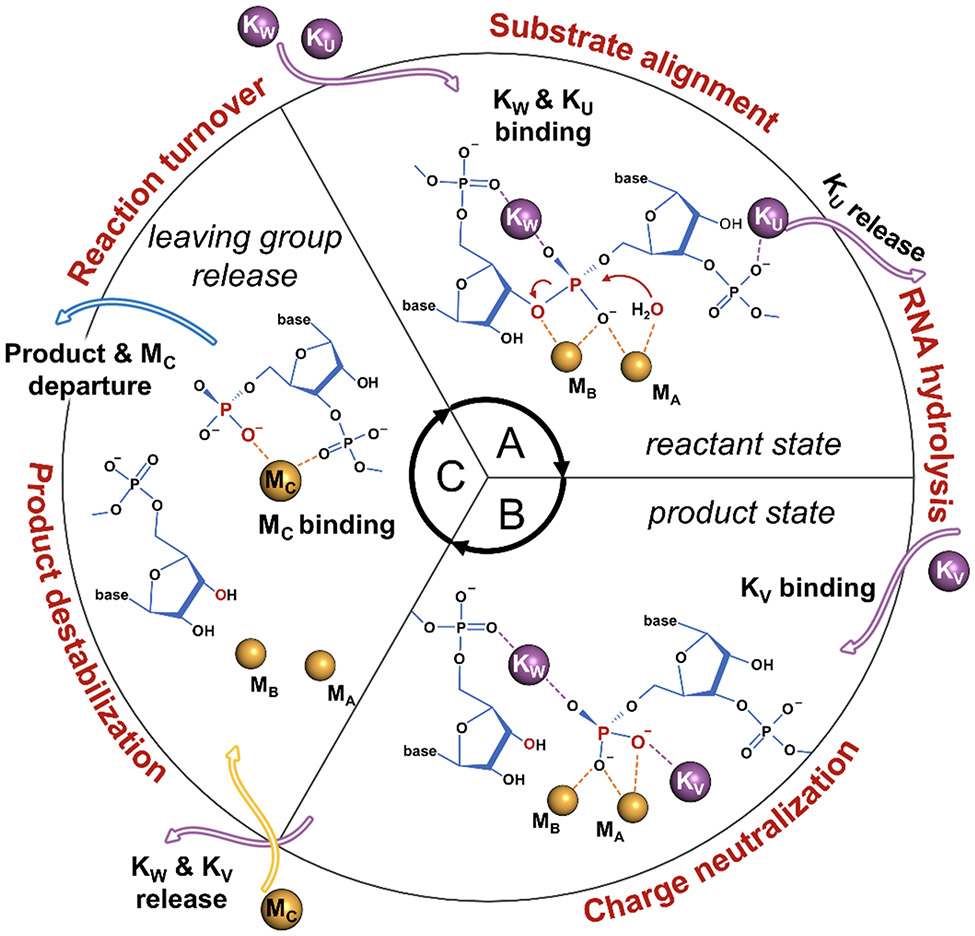
In summary, within the limits of the employed computational approach, our simulations provide a dynamic characterization of metal ion trafficking and its interplay with surrounding residues prior and upon catalysis. These results qualitatively describe an enzymatic strategy in which an extended and heteronuclear cations cluster (K+ and Mg2+ ions) is dynamically formed and disrupted, with the aid of second-shell residues. This mechanism prompts the processing of RNA:DNA hybrids in RNase H1 (Figure 6). That is, the RNase H1 cation trafficking starts with the binding of KW and KU, which prompt the alignment of the substrate (Figure 6A). At this point, KU is released, and the RNA hydrolysis can occur. Upon hydrolysis (i.e., product state, Figure 6B), KW is assisted by another KV in neutralizing the negatively charged product. Finally, the exchange of these two K+ ions with a third Mg2+ ion (viz., MC) contributes to the product destabilization, leading to the leaving group's release together with MC (Figure 6C) that precedes the reaction turnover and the continuation of catalysis. Ab-initio simulations will now be needed to evaluate the mechanistic and energetic implications of trafficking and strategic localization of such cations for the chemical step of RNA hydrolysis at the reaction center. Nonetheless, our classical simulations indicate that the controlled trafficking of cations, observed over a multi-microsecond sampling, is crucial to promote the formation of catalytic states that just precede and follow the enzymatic reaction for substrate hydrolysis.
Figure 6.
Controlled cations trafficking favors RNase H1 catalysis. (A) In the reactant state, KW and KU prompt the alignment of the substrate. At this point, KU is released and the RNA hydrolysis can occur. (B) Upon hydrolysis (i.e., product state), the negatively charged product is neutralized by KW and another KV ion. (C) The ultimate exchange of these two K+ ions with a third Mg2+ ion (viz. MC) promotes the destabilization of the products, which are then released together with MC to induce the reaction turnover and the continuation of catalysis.
Similarity of binding and trafficking of metals in other nucleic acid-processing metalloenzymes.
It has been recently shown that conserved and positively-charged residues are strategically located at the active core of, e.g., DNA or RNA polymerases and nucleases, at conserved sites, expanding the two-metal-ion functional architecture of these enzymes.18,20,40 On this basis, we used structural analyses to investigate whether the mechanistic insights herein provided may be shared among other nucleic acid processing enzymes. We found that the binding sites “U”, “W”, and “V”, transiently occupied by K+ ions in RNase H1, overlap well with second-shell positions occupied by positively-charged residues in other enzymes. In fact, the RNase H1 KW and KU/KV, respectively, well superimpose with, e.g., R92 and K85/G2(NH) in human exonuclease 1 (PDBid: 5V06, Figure S14A),5 or with H539 and R557 in HIV-1 reverse transcriptase (PDBid: 6BSH, Figure S14B),45 as well as with R100 and K93/G2(NH) in the endonuclease FEN1 (PDBid: 5UM9, Figure S14C).46
Structural analysis of FEN-1 bringing an alanine mutation of R100 (PDBid: 5KSE) revealed that, in the absence of R100, an additional metal ion is captured in the same putative “W” site before the catalysis.46 Notable similarities also arise from our recent studies investigating the structure and the catalysis of the CRISPR-Cas9 genome editing system. We found that the K970 and R972 residues within the catalytic RuvC domain also transiently engage the so-called “W” and “U/V” binding sites at specific catalytic steps (Figure S14D).47,48 Together, these findings support that the trafficking of positive charges at conserved regions of several metalloenzymes is strategically designed and controlled to favor catalysis. In this context, we note that only one positively-charged residue is present in the vicinity of the RNase H1 active site (i.e., K196), contrary to what is found in several other two-metal-ion enzymes.18,49 This observation may be coupled to the highly solvent-exposed localization of the RNase H1 active site when compared to those mentioned above and second-shell basic residues-rich enzymes (see Figure S15). This may be why the RNase H1 active site shows such prominent trafficking of multi-cations, in the lack of multiple strategically located positively-charged residues in the surrounding of the enzymatic reaction center. That is, multi-cations binding at the catalytic site may counterbalance the lack of multiple second-shell positively-charged residues. As a matter of fact, this would not represent the first example of its kind. Indeed, a similarly extended, heteronuclear cluster of movable cations has been captured at the catalytic site of the group II introns ribozymes (two K+, K1-K2; and two Mg2+ ions, M1-M2), RNA-based enzymes that perform RNA hydrolysis in the absence of (positively-charged) aminoacids.50,51 In group II introns, the position of these ions has been shown to be conserved for catalysis, in analogy with positively-charged residues in proteins.18,52 Importantly, we have recently demonstrated that such a structured metal cluster needs to be dynamically formed and broken to guarantee the proper formation of the introns’ catalytic core and favor the release of the reaction products, which matches well what we uncover herein for RNase H1.53 Moreover, a recent high-resolution cryo-EM map of the human spliceosome,54 which is structurally and chemically related to the group II introns, has revealed the presence of the same conserved heteronuclear metal cluster at its active site, in strike agreement with a model system proposed years before.18,50 Together, this evidence suggests that the controlled trafficking of cations may indeed be a common enzymatic strategy to trigger and assist phosphoryl transfer reactions in living cells, which has been evolutionary conserved and optimized in proteins. In light of these findings and observations, this study shows that finely regulated and controlled trafficking of multiple cations is an essential feature for the enzymatic-mediated hydrolysis of nucleic acids.
Conclusions
In conclusion, we have used extended atomistic molecular dynamics and free-energy simulations to investigate the cations trafficking recently observed in crystallo reaction intermediates during catalysis of the RNA ribonuclease H1 (RNase H1) enzyme.36 Our results illustrate a finely regulated trafficking of multiple and diverse cations with functional implications at the reaction center of RNase H1. In agreement with the recent mechanistic hypotheses and experimental data,36 our findings corroborate and sensibly expand the recent notion of an extended two-metal-ion architecture for nucleic acid-processing enzymes.18 Such controlled metal ion trafficking in the vicinity of the catalytic reaction center seems critically designed to aid catalysis in RNase H1, and possibly other similar nucleic-acid processing enzymes. Taken together, these findings may encourage further investigations related to enzyme engineering and drug discovery.
Material and methods
Here, the catalytic intermediates of the RNase H1 captured by time-resolved X-ray have been object of extensive molecular dynamics (MD) and enhanced sampling free-energy simulations, with the goal of characterizing the dynamics of cation trafficking prior to and upon RNA hydrolysis. Cation trafficking has also been investigated considering the second shell E188A and K196A mutations and three different ionic strengths of the solution. Overall, we collected a total of >27 μs of MD multi-replica simulations.
Structural models.
To perform MD simulations, six systems were modeled based on the recently time-resolved X-ray structures of Bacillus Halodurans RNase H1 in complex with an RNA/DNA hybrid construct.36 First, the reactant state of the wild type (wt) RNase H1, as obtained from the PDBid: 6DOG (occupancy C), was considered (Figure 1B). Here, only the crystallized metals MA-MB were included in the starting configuration to verify spontaneous binding events of K+ ions at the “U”, and “W” site. Second, the reactant state of the wt RNase H1 included the ions MA-MB and KU, as in PDBid: 6DMV, to verify the stability of the KU bound state. Third, the reactant state of the E188A mutant included MA-MB, as in PDBid: 6DPO, to verify the effect of the mutation for the recruitment of KW. Fourth, we considered the product state of the wt RNAse H1, modeled upon the PDBid: 6DOG (occupancy B), including the crystalized ions MA, MB, and KW (Figure 1C). Fifth, the product state of the K196A mutant, built on the PDBid: 6DPM, included the crystallized metals MA, MB, and KW (Figure S2B). Sixth, the product state of the E188A mutant, as obtained from the PDBid: 6DPO, included the crystallized metals MA, MB, and MC (Figure S2A). All the systems were solvated with a layer of water molecules of 15Å. According to crystallization experiments, each system was simulated at multiple ionic strengths to characterize the effect of the concentrations of the ions on their dynamics. Specifically, Mg2+, K+ and Cl− ions were added into the simulation boxes to match three different ionic strengths: (i) reference, optimal concentrations, 6mM [Mg2+] and 200mM [K+]; (ii) low K+ concentrations, 6mM [Mg2+] and 25mM [K+]; (iii) high Mg2+ concentration, 80mM [Mg2+] and 75mM [K+].
Molecular simulations.
The RNase H1 protein was parametrized with the AMBER force-field ff14SB,55 while the RNA and the DNA of the hybrid were parametrized with AMBER force-field RNA.OL356-59 and DNA.OL15.60 For the K+ metal ions, the parameters of Joung and Cheatham were used.61,62 The Mg2+ ions were systematically modelled according to both 12-6 (Aqvist63 and Allnér64) and 12-6-4 (Panteva65) non-bonded fixed point charge models, to verify that the observed results were independent from the chosen set of parameters.38,66 Last, the two catalytic Mg2+ ions MA-MB were modelled following the atom-in-molecule charges’ partitioning scheme,67 to consider also charge transfer between the ions and the first-shell coordination residues. The TIP3P model was used for water molecule.68 AMBER69,70 was used to perform Langevin71 MD simulations, using an integration time step of 2 fs. The temperature was set at 300K using a collision frequency γ = 1 per picosecond, while the constant 1 atm pressure was controlled through Berendsen barostat72 with a relaxation time of 2 ps.
We used the same simulation protocol for all systems. First, we performed an energy minimization to relax the water molecules. At this point, positional restraints of 300 kcal mol·Å2 were imposed to all the heavy atoms of the systems, including the metals. Then, we used a series of NVT and NPT simulations to smoothly thermalize the systems and remove the positional restraints. First, the systems were heated up with one NVT simulations of ~600 ps, using the same positional restraints as used during the energy minimization. Subsequently, the positional restraints were progressively halved with a series of two NVT simulations of ~200 ps each. Then, two additional simulations in the isothermal-isobaric ensemble (NPT) of ~200 ps were performed by further halving the positional restraints, while a third NPT run of ~2 ns was performed without any positional restraints to relax the density of the systems to ~1.01 g cm−3. Last, we performed multiple-replicas production runs to collect overall ~20 μs of simulation time. Specifically, we collected: ~9 μs for the pre-reactive state of the wild type, 9 replicas; ~9 μs for the post-reactive state of the wild type, 9 replicas; ~1.3 μs for the post-reactive state of the K196A mutants, 2 replicas.
Metadynamics.
Two independent metadynamics simulations were performed to characterize the conformational change of E188 in the pre-reactive state. To ensure the convergence of the calculations, we used the well-tempered variant73, setting the temperature at 300K and the bias factor at 8. The gaussian-shaped potential (height = 1.2 kJ mol−1, width = 0.35 rad.) was added with a frequency of 0.5 ps on the torsional angle E188-θ (between the E188 atoms N-Cα-Cβ-Cδ). Overall, we collected ~400 ns at both high and low K+ concentrations.
Additional well-tempered metadynamics simulations were performed to elucidate the release of the MC metal and the reaction products. The post-reactive state of both the wild type and the E188A mutant was used as starting configurations for the two independent simulations. Here, the temperature was set to 300K and the bias factor was set to 15. The coordination number of the MC, which defines the number of water molecules in the first coordination shell, and the distance between the scissile phosphate and the MC were used as collective variables (CV1 and CV2, respectively). The gaussian-shaped potential was added with a frequency of 0.2 ps on both the CV1 (height = 0.3 kJ mol−1, width = 0.1) and CV2 (height = 0.3 kJ mol−1, width = 0.5 Å).
Supplementary Material
Acknowledgments
The manuscript was written through contributions of all authors. We thank all members of the Palermo and De Vivo Labs for helpful discussions. Research reported in this publication was supported by the National Science Foundation under Grant Number CHE-1905374 [to G.P.] and by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number R01GM141329 [to G.P.]. Computer time has been awarded by XSEDE [grant No. TG- MCB160059] and by NERSC [grant No. M3807 to GP]. MDV thanks the Italian Association for Cancer Research (AIRC) for financial support [IG 23679].
Footnotes
Supporting Information
Supplementary Figures reporting: (i) analysis of structural data (Figures S1, S2, S15 and S16), (ii) in-depth analysis of the results obtained from multiple simulations replicas (Figures S3-S6, S8-S11) and (ii) results from metadynamics simulations (Figures S7, S12-S14). The Supporting Information is available free of charge at https://pubs.acs.org.
Competing interests
The authors declare no competing financial interest.
References
- (1).Gao Y; Yang W Capture of a Third Mg2+ Is Essential for Catalyzing DNA Synthesis. Science 2016, 352 (6291), 1334–1337. 10.1126/science.aad9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Nakamura T; Zhao Y; Yamagata Y; Hua YJ; Yang W Watching DNA Polymerase η Make a Phosphodiester Bond. Nature 2012, 487 (7406), 196–201. 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Uson ML; Carl A; Goldgur Y; Shuman S Crystal Structure and Mutational Analysis of Mycobacterium Smegmatis FenA Highlight Active Site Amino Acids and Three Metal Ions Essential for Flap Endonuclease and 5 Exonuclease Activities. Nucleic Acids Res. 2018, 46 (8), 4164–4175. 10.1093/nar/gky238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Prieto J; Redondo P; Merino N; Villate M; Montoya G; Blanco FJ; Molina R Structure of the I-SceI Nuclease Complexed with Its DsDNA Target and Three Catalytic Metal Ions. Acta Crystallogr. Sect. Struct. Biol. Commun 2016, 72, 473–479. 10.1107/S2053230X16007512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Shi Y; Hellinga HW; Beese LS Interplay of Catalysis, Fidelity, Threading, and Processivity in the Exo-and Endonucleolytic Reactions of Human Exonuclease I. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (23), 6010–6015. 10.1073/pnas.1704845114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang W Nucleases: Diversity of Structure, Function and Mechanism; 2011; Vol. 44. 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Palermo G; Cavalli A; Klein ML; Alfonso-Prieto M; Dal Peraro M; De Vivo M Catalytic Metal Ions and Enzymatic Processing of DNA and RNA. Acc. Chem. Res 2015, 48 (2), 220–228. 10.1021/ar500314j. [DOI] [PubMed] [Google Scholar]
- (8).Palermo G; Stenta M; Cavalli A; Dal Peraro M; De Vivo M Molecular Simulations Highlight the Role of Metals in Catalysis and Inhibition of Type II Topoisomerase. J. Chem. Theory Comput 2013, 9 (2), 857–862. 10.1021/ct300691u. [DOI] [PubMed] [Google Scholar]
- (9).Schmidt BH; Burgin AB; Deweese JE; Osheroff N; Berger JM A Novel and Unified Two-Metal Mechanism for DNA Cleavage by Type II and IA Topoisomerases. Nature 2010, 465 (7298), 641–644. 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Nishino T; Morikawa K Structure and Function of Nucleases in DNA Repair: Shape, Grip and Blade of the DNA Scissors. Oncogene 2002, 21 (58 REV. ISS. 8), 9022–9032. 10.1038/sj.onc.1206135. [DOI] [PubMed] [Google Scholar]
- (11).Potapov V; Fu X; Dai N; Corrêa IR; Tanner NA; Ong JL Base Modifications Affecting RNA Polymerase and Reverse Transcriptase Fidelity. Nucleic Acids Res. 2018, 46 (11), 5753–5763. 10.1093/nar/gky341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Pavlov YI; Shcherbakova PV; Rogozin IB Roles of DNA Polymerases in Replication, Repair, and Recombination in Eukaryotes. Int. Rev. Cytol 2006, 255 (06), 41–132. 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- (13).Pommier Y Drugging Topoisomerases: Lessons and Challenges. ACS Chem. Biol 2013, 8 (1), 82–95. 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Arencibia JM; Brindani N; Franco-Ulloa S; Nigro M; Kuriappan JA; Ottonello G; Bertozzi SM; Summa M; Girotto S; Bertorelli R; Armirotti A; De Vivo M Design, Synthesis, Dynamic Docking, Biochemical Characterization, and in Vivo Pharmacokinetics Studies of Novel Topoisomerase II Poisons with Promising Antiproliferative Activity. J. Med. Chem 2020, 63 (7), 3508–3521. 10.1021/acs.jmedchem.9b01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zheng L; Jia J; Finger LD; Guo Z; Zer C; Shen B Functional Regulation of FEN1 Nuclease and Its Link to Cancer. Nucleic Acids Res. 2011, 39 (3), 781–794. 10.1093/nar/gkq884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Riccardi L; Genna V; De Vivo M Metal–Ligand Interactions in Drug Design. Nat. Rev. Chem 2018, 2 (7), 100–112. 10.1038/s41570-018-0018-6. [DOI] [Google Scholar]
- (17).Donati E; Genna V; De Vivo M Recruiting Mechanism and Functional Role of a Third Metal Ion in the Enzymatic Activity of 5′ Structure-Specific Nucleases. J. Am. Chem. Soc 2020, 142 (6), 2823–2834. 10.1021/jacs.9b10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Genna V; Colombo M; De Vivo M; Marcia M Second-Shell Basic Residues Expand the Two-Metal-Ion Architecture of DNA and RNA Processing Enzymes. Structure 2018, 26 (1), 40–50.e2. 10.1016/j.str.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Perera L; Freudenthal BD; Beard WA; Shock DD; Pedersen LG; Wilson SH; Broyde S Requirement for Transient Metal Ions Revealed through Computational Analysis for DNA Polymerase Going in Reverse. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (38), E5228–E5236. 10.1073/pnas.1511207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Genna V; Donati E; De Vivo M The Catalytic Mechanism of DNA and RNA Polymerases. ACS Catal. 2018, 8 (12), 11103–11118. 10.1021/acscatal.8b03363. [DOI] [Google Scholar]
- (21).Genna V; Carloni P; De Vivo M A Strategically Located Arg/Lys Residue Promotes Correct Base Paring during Nucleic Acid Biosynthesis in Polymerases. J. Am. Chem. Soc 2018, 140 (9), 3312–3321. 10.1021/jacs.7b12446. [DOI] [PubMed] [Google Scholar]
- (22).Stevens DR; Hammes-Schiffer S Exploring the Role of the Third Active Site Metal Ion in DNA Polymerase η with QM/MM Free Energy Simulations. J. Am. Chem. Soc 2018, 140 (28), 8965–8969. 10.1021/jacs.8b05177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yang W; Weng PJ; Gao Y A New Paradigm of DNA Synthesis: Three-Metal-Ion Catalysis. Cell Biosci. 2016, 6 (1), 1–7. 10.1186/s13578-016-0118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Borišek J; Magistrato A An Expanded Two-Zn 2+ -Ion Motif Orchestrates Pre-MRNA Maturation in the 3′-End Processing Endonuclease Machinery . ACS Catal. 2021, 11 (7), 4319–4326. 10.1021/acscatal.0c05594. [DOI] [Google Scholar]
- (25).Broccoli S; Rallu F; Sanscartler P; Cerritelli SM; Crouch RJ; Drolet M Effects of RNA Polymerase Modifications on Transcription-Induced Negative Supercoiling and Associated R-Loop Formation. Mol. Microbiol 2004, 52 (6), 1769–1779. 10.1111/j.1365-2958.2004.04092.x. [DOI] [PubMed] [Google Scholar]
- (26).Cerritelli SM; Crouch RJ Ribonuclease H: The Enzymes in Eukaryotes. FEBS J. 2009, 276 (6), 1494–1505. 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tadokoro T; Kanaya S Ribonuclease H: Molecular Diversities, Substrate Binding Domains, and Catalytic Mechanism of the Prokaryotic Enzymes. FEBS J. 2009, 276 (6), 1482–1493. 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- (28).Nowotny M; Gaidamakov SA; Crouch RJ; Yang W Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. Cell 2005, 121 (7), 1005–1016. 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- (29).Nowotny M; Gaidamakov SA; Ghirlando R; Cerritelli SM; Crouch RJ; Yang W Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Mol. Cell 2007, 28 (2), 264–276. 10.1016/j.molcel.2007.08.015. [DOI] [PubMed] [Google Scholar]
- (30).Yang W; Hendrickson WA; Crouch RJ; Satow Y Structure of Ribonuclease H Phased at 2 Å Resolution by MAD Analysis of the Selenomethionyl Protein. Science 1990, 249 (4975), 1398–1405. 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- (31).Nowotny M; Cerritelli SM; Ghirlando R; Gaidamakov SA; Crouch RJ; Yang W Specific Recognition of RNA/DNA Hybrid and Enhancement of Human RNase H1 Activity by HBD. EMBO J. 2008, 27 (7), 1172–1181. 10.1038/emboj.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Champoux JJ; Schultz SJ Ribonuclease H: Properties, Substrate Specificity and Roles in Retroviral Reverse Transcription. FEBS J. 2009, 276 (6), 1506–1516. 10.1111/j.1742-4658.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).De Vivo M; Dal Peraro M; Klein ML Phosphodiester Cleavage in Ribonuclease H Occurs via an Associative Two-Metal-Aided Catalytic Mechanism. J. Am. Chem. Soc 2008, 130 (33), 10955–10962. 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ho MH; De Vivo M; Dal Peraro M; Klein ML Understanding the Effect of Magnesium Ion Concentration on the Catalytic Activity of Ribonuclease H through Computation: Does a Third Metal Binding Site Modulate Endonuclease Catalysis? J. Am. Chem. Soc 2010, 132 (39), 13702–13712. 10.1021/ja102933y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Rosta E; Nowotny M; Yang W; Hummer G Catalytic Mechanism of RNA Backbone Cleavage by Ribonuclease H from Quantum Mechanics/Molecular Mechanics Simulations. J. Am. Chem. Soc 2011, 133 (23), 8934–8941. 10.1021/ja200173a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Samara NL; Yang W Cation Trafficking Propels RNA Hydrolysis. Nat. Struct. Mol. Biol 2018, 25 (8), 715–721. 10.1038/s41594-018-0099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Keck JL; Goedken ER; Marqusee S Activation/Attenuation Model for RNase H. J. Biol. Chem 1998, 273 (51), 34128–34133. 10.1074/jbc.273.51.34128. [DOI] [PubMed] [Google Scholar]
- (38).Panteva MT; Giambaşu GM; York DM Comparison of Structural, Thermodynamic, Kinetic and Mass Transport Properties of Mg2+ Ion Models Commonly Used in Biomolecular Simulations. J. Comput. Chem 2015, 36 (13), 970–982. 10.1002/jcc.23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Genna V; Vidossich P; Ippoliti E; Carloni P; De Vivo M A Self-Activated Mechanism for Nucleic Acid Polymerization Catalyzed by DNA/RNA Polymerases. J. Am. Chem. Soc 2016, 138 (44), 14592–14598. 10.1021/jacs.6b05475. [DOI] [PubMed] [Google Scholar]
- (40).Genna V; Gaspari R; Dal Peraro M; De Vivo M Cooperative Motion of a Key Positively Charged Residue and Metal Ions for DNA Replication Catalyzed by Human DNA Polymerase-η. Nucleic Acids Res. 2016, 44 (6), 2827–2836. 10.1093/nar/gkw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Nowotny M; Yang W Stepwise Analyses of Metal Ions in RNase H Catalysis from Substrate Destabilization to Product Release. EMBO J. 2006, 25 (9), 1924–1933. 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Golosov AA; Warren JJ; Beese LS; Karplus M The Mechanism of the Translocation Step in DNA Replication by DNA Polymerase I: A Computer Simulation Analysis. Structure 2010, 18 (1), 83–93. 10.1016/j.str.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Da LT; Pardo Avila F; Wang D; Huang X A Two-State Model for the Dynamics of the Pyrophosphate Ion Release in Bacterial RNA Polymerase. PLoS Comput. Biol 2013, 9 (4). 10.1371/journal.pcbi.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yoon H; Warshel A Simulating the Fidelity and the Three Mg Mechanism of Pol η and Clarifying the Validity of Transition State Theory in Enzyme Catalysis. Proteins Struct. Funct. Bioinforma 2017, 85 (8), 1446–1453. 10.1002/prot.25305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Tian L; Kim MS; Li H; Wang J; Yang W Structure of HIV-1 Reverse Transcriptase Cleaving RNA in an RNA/DNA Hybrid. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (3), 507–512. 10.1073/pnas.1719746115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Tsutakawa SE; Thompson MJ; Arvai AS; Neil AJ; Shaw SJ; Algasaier SI; Kim JC; Finger LD; Jardine E; Gotham VJB; Sarker AH; Her MZ; Rashid F; Hamdan SM; Mirkin SM; Grasby JA; Tainer JA Phosphate Steering by Flap Endonuclease 1 Promotes 5′-Flap Specificity and Incision to Prevent Genome Instability. Nat. Commun 2017, 8 (May), 1–14. 10.1038/ncomms15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Palermo G Structure and Dynamics of the CRISPR-Cas9 Catalytic Complex. J. Chem. Inf. Model 2019, 59 (5), 2394–2406. 10.1021/acs.jcim.8b00988. [DOI] [PubMed] [Google Scholar]
- (48).Casalino L; Nierzwicki Ł; Jinek M; Palermo G Catalytic Mechanism of Non-Target DNA Cleavage in CRISPR-Cas9 Revealed by Ab Initio Molecular Dynamics. ACS Catal. 2020, 10, 13596–13605. 10.1021/acscatal.0c03566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Vidossich P; Castañeda Moreno LE; Mota C; de Sanctis D; Miscione G. Pietro; De Vivo M Functional Implications of Second-Shell Basic Residues for DUTPase DR2231 Enzymatic Specificity. ACS Catal. 2020, 13825–13833. 10.1021/acscatal.0c04148. [DOI] [Google Scholar]
- (50).Marcia M; Pyle AM Visualizing Group II Intron Catalysis through the Stages of Splicing. Cell 2012, 151 (3), 497–507. 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Casalino L; Palermo G; Rothlisberger U; Magistrato A Who Activates the Nucleophile in Ribozyme Catalysis? An Answer from the Splicing Mechanism of Group II Introns. J. Am. Chem. Soc 2016, 138 (33), 10374–10377. 10.1021/jacs.6b01363. [DOI] [PubMed] [Google Scholar]
- (52).Genna V; Marcia M; De Vivo M A Transient and Flexible Cation-ΠInteraction Promotes Hydrolysis of Nucleic Acids in DNA and RNA Nucleases. J. Am. Chem. Soc 2019, 141 (27), 10770–10776. 10.1021/jacs.9b03663. [DOI] [PubMed] [Google Scholar]
- (53).Manigrasso J; Chillón I; Genna V; Vidossich P; Somarowthu S; Pyle AM; De Vivo M; Marcia M Visualizing Group II Intron Dynamics between the First and Second Steps of Splicing. Nat. Commun 2020, 11 (1). 10.1038/s41467-020-16741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Wilkinson ME; Fica SM; Galej WP; Nagai K Structural Basis for Conformational Equilibrium of the Catalytic Spliceosome. Mol. Cell 2021, 1–14. 10.1016/j.molcel.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput 2015, 11 (8), 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Darian E; Gannett PM Application of Molecular Dynamics Simulations to Spin-Labeled Oligonucleotides. J. Biomol. Struct. Dyn 2005, 22 (5), 579–593. 10.1080/07391102.2005.10507028. [DOI] [PubMed] [Google Scholar]
- (57).Cieplak P; Kollman PA; Wang J; Cieplak P; Kollman PA How Well Does a Restrained Electrostatic Potential (RESP) Model Perform in Calculating Conformational Energies of Organic and Biological Molecules? J. Comput. Chem 2000, 21, 1049–1074. [Google Scholar]
- (58).Pérez A; Marchán I; Svozil D; Sponer J; Cheatham TE; Laughton CA; Orozco M Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys. J 2007, 92 (11), 3817–3829. 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zgarbová M; Otyepka M; Šponer J; Mládek A; Banáš P; Cheatham TE; Jurečka P Refinement of the Cornell et Al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput 2011, 7 (9), 2886–2902. 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Galindo-Murillo R; Robertson JC; Zgarbová M; Šponer J; Otyepka M; Jurečka P; Cheatham TE Assessing the Current State of Amber Force Field Modifications for DNA. J. Chem. Theory Comput 2016, 12 (8), 4114–4127. 10.1021/acs.jctc.6b00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Joung IS; Cheatham TE Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112 (30), 9020–9041. 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Joung S; Cheatham TE Molecular Dynamics Simulations of the Dynamic and Energetic Properties of Alkali and Halide Ions Using Water-Model-Specific Ion Parameters. J. Phys. Chem. B 2009, 113 (40), 13279–13290. 10.1021/jp902584c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Åqvist J Ion-Water Interaction Potentials Derived from Free Energy Perturbation Simulations. J. Phys. Chem 1990, 94 (21), 8021–8024. 10.1021/j100384a009. [DOI] [Google Scholar]
- (64).Allnér O; Nilsson L; Villa A Magnesium Ion-Water Coordination and Exchange in Biomolecular Simulations. J. Chem. Theory Comput 2012, 8 (4), 1493–1502. 10.1021/ct3000734. [DOI] [PubMed] [Google Scholar]
- (65).Panteva MT; Giambaşu GM; York DM Force Field for Mg2+, Mn2+, Zn2+, and Cd2+ Ions That Have Balanced Interactions with Nucleic Acids. J. Phys. Chem. B 2015, 119 (50), 15460–15470. 10.1021/acs.jpcb.5b10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Casalino L; Palermo G; Abdurakhmonova N; Rothlisberger U; Magistrato A Development of Site-Specific Mg2+-RNA Force Field Parameters: A Dream or Reality? Guidelines from Combined Molecular Dynamics and Quantum Mechanics Simulations. J. Chem. Theory Comput 2017, 13 (1), 340–352. 10.1021/acs.jctc.6b00905. [DOI] [PubMed] [Google Scholar]
- (67).Dal Peraro M; Spiegel K; Lamoureux G; De Vivo M; DeGrado WF; Klein ML Modeling the Charge Distribution at Metal Sites in Proteins for Molecular Dynamics Simulations. J. Struct. Biol 2007, 157 (3), 444–453. 10.1016/j.jsb.2006.10.019. [DOI] [PubMed] [Google Scholar]
- (68).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys 1983, 79 (2), 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- (69).Case DA; Belfon K, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham III TE, Cruzeiro VWD, Darden TA, Duke RE, Giambasu G, Gilson MK, Gohlke H, Goetz AW, Harris R, Izadi S, Izmailov SA, Kasavajhala K, Kovalenko A, Krasny R, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Man V, Merz KM, Miao Y, Mikhailovskii O, Monard G, Nguyen H, Onufriev A, Pan F, Pantano S, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Skrynnikov NR, Smith J, Swails J, Walker RC, Wang J, Wilson L, Wolf RM, Wu X, Xiong Y, Xue Y, York DM and Kollman PA AMBER 2018. Univ. California, San Fr. [Google Scholar]
- (70).Lee TS; Cerutti DS; Mermelstein D; Lin C; Legrand S; Giese TJ; Roitberg A; Case DA; Walker RC; York DM GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model 2018, 58 (10), 2043–2050. 10.1021/acs.jcim.8b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Turq P; Lantelme F; Friedman HL Brownian Dynamics: Its Application to Ionic Solutions. J. Chem. Phys 1976, 66 (7), 3039–3044. 10.1063/1.434317. [DOI] [Google Scholar]
- (72).Berendsen HJC; Postma JPM; Van Gunsteren WF; Dinola A; Haak JR Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys 1984, 81 (8), 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- (73).Barducci A; Bussi G; Parrinello M Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett 2008, 100 (2), 1–4. 10.1103/PhysRevLett.100.020603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.