Abstract
Diseases caused by trypanosomatids are serious public health concerns in low-income endemic countries. Leishmaniasis is presented in two main clinical forms, visceral leishmaniasis—caused by L. infantum and L. donovani—and cutaneous leishmaniasis—caused by many species, including L. major, L. tropica and L. braziliensis. As for certain other trypanosomatids, sexual reproduction has been confirmed in these parasites, and formation of hybrids can contribute to virulence, drug resistance or adaptation to the host immune system. In the present work, the capability of intraclonal and interspecies genetic exchange has been investigated using three parental strains: L. donovani, L. tropica and L. major, which have been engineered to express different fluorescent proteins and antibiotic resistance markers in order to facilitate the phenotypic selection of hybrid parasites after mating events. Stationary and exponential-phase promastigotes of each species were used, in in vitro experiments, some of them containing LULO cells (an embryonic cell line derived from Lutzomyia longipalpis). Several intraclonal hybrids were obtained with L. tropica as crossing progenitor, but not with L. donovani or L. major. In interspecies crossings, three L. donovani x L. major hybrids and two L. donovani x L. tropica hybrids were isolated, thereby demonstrating the feasibility to obtain in vitro hybrids of parental lines causing different tropism of leishmaniasis. Ploidy analysis revealed an increase in DNA content in all hybrids compared to the parental strains, and nuclear analysis showed that interspecies hybrids are complete hybrids, i.e. each of them showing at least one chromosomal set from each parental. Regarding kDNA inheritance, discrepancies were observed between maxi and minicircle heritage. Finally, phenotypic studies showed either intermediate phenotypes in terms of growth profiles, or a decreased in vitro infection capacity compared to the parental cells. To the best of our knowledge, this is the first time that in vitro interspecies outcrossing has been demonstrated between Leishmania species with different tropism, thus contributing to shed light on the mechanisms underlying sexual reproduction in these parasites.
Author summary
Neglected Tropical Diseases (NTDs) represent a serious threat to humans, especially for those living in poor or developing countries. Leishmanianiosis is considered a zoonotic NTD transmitted by the bite of female phlebotomine sandflies, and is manifested mainly as a visceral form (caused by L. infantum and L. donovani) and a cutaneous form (caused by many species including L. major, L. tropica and L. braziliensis). Although it is now known that sexual reproduction occurs in these parasites, more studies are necessary to characterize the ability of Leishmania to generate hybrids, which may represent an important mechanism for virulence, drug resistance or adaptation to the host immune system. Therefore, several experiments were conducted to generate either intraclonal or interspecies hybrids in vitro. Results demonstrated that hybrids can be formed even with outcrosses between parasites causing visceral and cutaneous forms of the disease. Characterization of hybrids in terms of ploidy, kDNA content, growth rate and infection capacity provide important information about sexual reproduction in these parasites.
Introduction
Major pathogenic trypanosomatids, Trypanosoma brucei (responsible for sleeping sickness in Africa), Trypanosoma cruzi (responsible for Chagas disease in South America) and Leishmania spp. are single-celled protists causing some of the most neglected and deadly transmissible diseases produced by eukaryotes in low-income countries. Leishmania is a digenetic parasite transmitted to humans by species of sandflies of the genus Phlebotomus and Lutzomyia, which maintain the zoonotic cycle [1,2]. In the human host, Leishmania causes different disorders: i) cutaneous leishmaniasis, produced among other species by L. major and L. tropica in the Old World and by L. braziliensis in the New World, is the less severe form and may lead to the formation of disfiguring sores and scars in exposed parts of the body [3]; ii) mucocutaneous leishmaniasis is a more severe presentation that can destroy the mucous membranes of mouth and nose [4]; iii) visceral leishmaniasis is responsible for hepato-splenomegaly and can be fatal if left untreated. It is produced by L. donovani and L. infantum in the Old World, and by L. infantum chagasi in the New World, and can derive into a rare skin form of the disease called post-kala azar dermal leishmaniasis when the antimony therapy fails [5,6].
The mode of reproduction of these species is no longer just a basic biology issue, but a source of information that can provide explanations to important clues regarding parasite virulence, drug resistance or adaptation to the host immune system. Nowadays, it is unquestionable that sexual reproduction and related processes such as gene recombination and chromosome segregation, takes place in trypanosomatids. As for other eukaryotes, meiosis-specific genes for the formation of gametes by reductive division during meiosis as well as their cytoplasmic fusion (syngamy) during mating, have been described in trypanosomatids [7–9]. The debate is now focused on determining how frequent is sex and what is the impact exerted on species and natural populations of these parasites from an evolutionary point of view [10–15].
Since the late 80s, there is confirmed evidence that, under controlled laboratory conditions, different strains of T. brucei are capable of generating viable hybrids by genetic exchange [16]. It was observed that filial hybrid generations inherited gene markers from each parent strain. In addition, this genetic exchange took place in the salivary glands of tsetse flies [16,17]. The filial generation shared characteristics from both parental strains (biparental inheritance), but some specific characteristics such as the occurrence of a proportion of filial hybrids with variable ploidy [18], or the existence of mating types capable of self- and non-self-recognition, were also reported [19]. Finally, thanks to the use of transgenic fluorescent strains, mating phenomena and gamete production could be observed [20–22].
The results obtained with Leishmania spp. are more recent and confirm those obtained with other trypanosomatids. The use of antibiotic resistance markers served to detect the first interclonal genetic exchange event between different strains of L. major in the presence of either the natural vector Phlebotomus duboscqi, or the permissive Lutzomyia longipalpis sandfly. Similar to T. brucei, a percentage of the hybrid generation had an unexpectedly higher ploidy than that of the parental strains [23,24]. Furthermore, viable hybrids that partly retained the visceralization capacity in mice were obtained between two identical clones of L. infantum (self-mating or selfing) [25]. Moreover, hybrids were also produced between strains of different species, such as L. infantum and L. major, whose filial hybrid generation retained part of the characteristics of each parent [26].
Until 2020, genetic exchange events in Leishmania had only been demonstrated in the midgut of the permissive Lu. Longipalpis or non-permissive Ph. papatasi and Ph. duboscqi sand flies fed with infected blood. However, a recent work demonstrated that these events could also take place between promastigotes in the absence of the insect vector in axenic conditions [27], and might be possible even between amastigotes within the macrophage phagolysosome [28]. These new findings could significantly facilitate this type of study.
Other important results obtained from genetic exchange processes were related to the inheritance of extrachromosomal kinetoplast DNA (kDNA). kDNA is an intricate mesh of both maxicircles (few in number and equivalent to mitochondrial DNA) and minicircles (very numerous and responsible for the editing of parasite mRNAs) [29]. Genetic exchange experiments with different strains of T. brucei showed that unlike minicircles [30], the DNA content of the maxicircles from filial hybrids seems to have uniparental origin, although recent results suggest the early biparental inheritance of maxicircles [31].
Encouraged by the importance of genetic exchange in Leishmania we have proceeded to perform interspecies and intraclonal hybridization studies between lines of L. donovani, L. tropica and L. major. These experiments have been conducted in the absence of the vector, but assessing the effect of an embryonic cell line from the permissive vector Lu. longipalpis (LULO cells) as feed layer for hybridization experiments. The inheritance of the hybrids obtained from each of the parental strains, their ploidy, kDNA composition, viability and infectivity in macrophages have been analysed to contribute to decipher the mechanism behind reproduction in Leishmania.
Materials and methods
Ethics statement
Experiments in this study involving animals have been carried out in accordance with Spanish and European Union legislation (RD 53/2013 and 2010/63/EU, respectively). The protocols used in this study have been approved by the Ethics Committee of the University of León (Spain), Project license number OEBA-ULE-007-2019.
Animals and infections
Six-to-eight week old female Balb/c mice, purchased from Janvier Laboratories (St Berthevin Cedex, France) were used to maintain the Leishmania strains. Mice were infected intraperitoneally with 5x108 metacyclic promastigotes of different Leishmania strains (see below) and after 6 weeks, animals were euthanized. Livers, spleens and lymph nodes were aseptically processed in order to recover the parasites as previously described [32,33].
Parasites and growth curves
Parental strains, L. donovani MHOM/ET/67/HU3, also known as LV9 or L-82 (kindly provided by Dr. Philippe Loiseau, Université Paris-Sud, Paris, France), L. major LV39c5 (RHO/SU/59/P) (kindly provided by Dr. Stephen M Beverley, Washington University School of Medicine, Saint Louis, USA), L. tropica MRAT/IQ/72/ADHANIS1 (kindly provided by Dr. José M. Requena, CBM, UAM, Madrid, Spain) and hybrid parasites were cultured at 26°C in M199 medium (Sigma), supplemented with 25 mM HEPES pH 6.9, 10 mM glutamine, 7.6 mM hemin, 0.1 mM adenosine, 0.01 mM folic acid, 1x RPMI 1640 vitamin mix (Sigma), 10% (v/v) heat-inactivated fetal bovine serum (FBS), and antibiotic mixture (200 U/mL penicillin and 200 μg/mL streptomycin). Stationary-phase parasites were harvested after 3 days in this phase, while exponential-phase promastigotes were collected 3 days after starting the culture. Both exponential and stationary phase promastigotes were confirmed microscopically before performing the experiments.
To prepare growth curves, parental and hybrid lines were seeded at a density of 106 promastigotes/mL in complete M199 medium and parasite numbers were determined daily on a Z1 Beckman Coulter.
LULO cell cultures
LULO cells were kindly provided by Dr. Felio Jesús Bello (Universidad de la Salle, Sede Norte, Bogotá, Colombia). LULO is an adherent embryonic cell line derived from the permissive sandfly Lutzomyia longipalpis [34]. LULO cells were cultured in monolayer at 26°C in L-15:Grace´s (1:1) medium (both from FisherScientific) supplemented with 10% FBS and antibiotic mixture until reaching confluence between passages. Interaction between parental promastigotes and LULO cells was tested by seeding 2x104 LULO cells in 8-well Ibidi chambers and adding promastigotes 24 hours later at a 1:5 ratio. Interaction was visualized by confocal microscopy. For this purpose, parasites were stained with Hoechst 33342 (Fisher Scientific) during 30 minutes at 37°C and images were acquired on a Zeiss LSM800 confocal microscope.
Generation of transgenic lines of Leishmania expressing fluorescent marker genes
To perform in vitro crossing experiments, one of the transgenic lines was generated previously in our lab, namely L. major mCh HYG (expressing the gene encoding mCherry (mCh) and the hygromicin resistance cassette (HYG)). In addition to this transgenic line, some new fluorescent cell lines were generated after electroporation of the corresponding plasmids (see below) and further selection of the parasites on semisolid medium containing specific antibiotic markers (see below) [35].
On the one hand, L. donovani mCh HYG and L. tropica mCh HYG were obtained after electroporation of these strains with pLEXSY-mCh-HYG construct [33] and subsequent selection of transfectants in medium containing hygromycin (200 μg/mL). On the other hand, electroporation of promastigotes of L. tropica with pLEXSY-iRFP-PAC [32] and further selection of transfectants in medium with puromycin (200 μg/mL), gave rise to L. tropica iRFP PAC, which constitutively expresses the iRFP gene. Finally, electroporation of promastigotes of L. donovani, L. major and L. tropica with the construct pLEXSY-CTN-PAC (see below) and selection of transfectants in medium with puromycin (200 μg/mL) gave rise to L. donovani CTN PAC, L. major CTN PAC and L. tropica CTN PAC, respectively, which produce the citrine (CTN) protein. For the construct of pLEXSY-CTN-PAC (S1 Fig), the gene encoding CTN was amplified by PCR from the plasmid pLEXSY-CTN-HYG [25] using the primers RBF921, which contains the sequences for XhoI and BglII at the 5´-end, and RBF614, the latter containing the sequences for NotI at the 5´-end (Table 1). The fragment obtained (CTN; 720 bp) included the kozak sequence for improved expression of the fluorescence protein. Then, it was digested with XhoI and NotI, and cloned in a pBluescript SK (Agilent), thus forming pSK-CTN fragment. In the next step, plasmids pSK-CTN and pLEXSY-hyg2 (Jena Bioscience) were digested with BglII and NotI, and the fragment CTN was cloned in the pLEXSY-hyg2 vector, thus obtaining pLEXSY-hyg2-CTN. Finally, pLEXSY-CTN-PAC was generated after digestion of pLEXSY-hyg2-CTN with SpeI and NotI in order to replace the fragment HYG-utr2 with the fragment PAC-HSP70. This fragment was obtained from pLEXSY-iRFP-70-PAC after digestion with SpeI and NotI, and contains the HSP70 downstream region, which has been reported to help increase the expression of the reporter gene included in pLEXSY vectors [32].
Table 1. Primers used in this work.
| Product name | Primers | Product size (bp) | Use |
|---|---|---|---|
| Citrine [25] | RBF921, FW: ccgCTCGAGgaAGATCTCCACCATGGTGAGCAAGGGCGAGG RBF614, RV: ataagaatGCGGCCGCTTACTTGTACAGCTCGTCCATG |
720 | ORF cloning |
| Hygromycine [33] | RBF646, FW: ATGAAAAAGCCTGAACTCACC RBF647, RV: CTATTCCTTTGCCCTCGGAC |
1025 | Characterization of transformants |
| Puromycin [32] | RBF774, FW: ATGACCGAGTACAAGCCCACG RBF775, RV: TCAGGCACCGGGCTTGCGG |
720 | Characterization of transformants |
| mCherry | TOX43, FW: TGGCCATCATCAAGGAGTTCA TOX44, RV: CCCTCGGCGCGTTCGT |
711 | Characterization of transformants |
| Citrine | TOX41, FW: CGAGGAGCTGTACACCGGG TOX42, RV: ACGAACTCCAGCAGGACCAT |
717 | Characterization of transformants |
| iRFP [32] | RBF843, FW: ATGGCGGAAGGATCCGTCGC RBF844, RV: TCACTCTTCCATCACGCCGATC |
950 | Characterization of transformants |
| Calmodulin-binding | TOX59, FW: CTGAACCGGATGCAGATTCGTGAG TOX60, RV: CTTGTTGCGTGTCGTCTCGATCTG |
786 | Genomic SNP-CAPs |
| ABC transporter | TOX61, FW: ATGGACTTGGTGCAGCTGCAGCGA TOX62, RV: GATGTCATCGCACAGGGACGTAATG |
2091 | Genomic SNP-CAPs |
| Sec20 | TOX63, FW: GGACCAGGCGCTACACGAGCTGAT TOX64, RV: GACACGCGCAGCAGCAGATCATCG |
618 | Genomic SNP-CAPs |
| Rad9 | TOX65, FW: GGCTGAGCGACGGCATCGAGATCAG TOX66, RV: CCTCGTTGGCGGAGGTGTAGGTGC |
1956–1959 | Genomic SNP-CAPs |
| Asparaginase | TOX69, FW: CGTCGCCGGATACCTGACGGAGC TOX70, RV: GGTGATTTCGCCTCGCAGATTCTG |
1015 | Genomic SNP-CAPs |
| Paraflagellar rod protein | TOX67, FW: ATGAGCATCGCTGCGGACATGGCGT TOX68, RV: CTACTCGGTGATCTGTCGCACCGTC |
1800 | Genomic SNP-CAPs |
| Ribosomal protein L22p/L17e | TOX55, FW: ATGCCGAAGCCAGCCCCTCGGTAC TOX56, RV: CAGCAGTGATGGCGGGCACGTCCA |
829 | Genomic SNP-CAPs |
| Fumarate hydratase (Fumerase) | TOX45, FW: ATGTCTCTGTGCGACCA TOX46, RV: TCACGCAAGCGTCTTCG |
1707 | Genomic SNP-CAPs |
| Aquaglyceroporin 1 | TOX51, FW: GGCTACGCGAGTATGTTGCC TOX52, RV: AAGACATACAAGAACATGCCGA |
688–736 | Genomic SNP-CAPs |
| 2-oxoisovalerate dehydrogenase beta subunit | TOX49, FW: AGCGTTCAACCGAGTGAGTT TOX50, RV: AAGTGCACCACGGACTTGAT |
1025 | Genomic SNP-CAPs |
| A2 [36] | L2: TTGGCAATGCGAGCGTCACAGTC R3: CAACGCGTACGATAATGCCACA |
230/480 | Genomic amplification |
| ACTIN | TOX83, FW: AATGGCTGACAACGAGCAGA TOX84, RV: CACTTGTTGTGCACGATGCT |
1125 | Genomic sequencing |
| ITS [37] | (LITSR); TOX75, FW: CTGGATCATTTTCCGATG (L5.8S); TOX76, RV: TGATACCACTTATCGCACTT |
420 | Strain confirmation |
| ND7 [38] | TOX97, FW: GTGCATTTATGCGTTTATTAATGTG TOX98, RV: ACAACATCAACATTACCAATAACTGC |
800 | Maxicircle SNP-CAPs |
| CYTB [26] | TOX99, FW: AGCGGAGAGRARAGAAAAGG TOX100, RV:: GYTCRCAATAAAATGCAAATC |
618 | Maxicircle SNP-CAPs |
| ND5 [26] | TOX103, FW: GAYGCDATGGAAGGACCDAT TOX104, RV: CCACAYAAAAAYCAYAANGAACA |
456 | Maxicircle SNP-CAPs |
| 12sRNA [26] | TOX101, FW: AACTARTGAWGGCACAGTTGTTCT TOX102, RV: ACCCAACTAACGAATTGCWTTT |
818 | Maxicircle SNP-CAPs |
| MINICIRCLES 1 (MIN 1) [39] | TOX73, FW: CCAGTTTCCCGCCCCG (KDNA-F) TOX74, RV: GGGGTTGGTGGTGTAAAATAG (KDNA-R) |
780 | Minicircle amplification |
| MINICIRCLES 2 (MIN 2) [40] | TOX75, FW: TAATATAGTGGGCCGCGCAC TOX76, RV: CCGACATGCCTCTGGGTAGG |
<250–1000 | Minicircle amplification |
The correct integration of both the reporter and antibiotic resistance cassettes in the Leishmania genome was tested by PCR. In addition, fluorescent signal was checked by flow cytometry (MACSQuant Analyzer 10) and confocal microscopy. For this purpose, 2x104 exponential phase promastigotes were collected, washed twice in 1xPBS and placed in 8-well Ibidi chambers previously treated with poly-L-lysine (Sigma). Parasites were stained with Hoechst 33342 (Fisher Scientific) during 30 minutes at 37°C and images were acquired on a Zeiss LSM800 confocal microscope.
Crossing experiments
In vitro hybrids were generated by mixing equal numbers (2.5x106) of parental promastigotes (exponential or stationary-phase promastigotes), either on empty 96 well plates or on 96 well plates previously seeded with confluent LULO cells. Several days after mixing parental cell lines (3, 6 or 9 days, depending on the experiment) parasites were transferred into 48-well plates including fresh complete M199 medium containing the selection antibiotics (puromycin and hygromycin, 100 μg/mL each), in order to select the double resistant hybrids generated due to genetic exchange.
Analysis of DNA content
Ploidy of hybrids and parental strains was determined through the analysis of their total DNA content. Briefly, 1x106 promastigotes were collected, washed twice in 1xPBS and fixed by adding 70% ethanol dropwise in vortex. Samples were kept at -20°C during at least 24 hours and up to 1 week until analysis. For this, fixed cells were washed again in 1xPBS twice, stained with propidium iodide (40 μg/mL) and treated with 200 μg/mL RNase (Sigma) at 37°C for 30 minutes. Analysis was carried out on MACSQuant Analyzer 10 flow cytometry equipment. Data were analysed with FlowJo v10 software. L. major Friedlin (largely diploid) and L. braziliensis Mb2904 (approximately 3n) were used as ploidy-content reference cell lines in order to compare them with hybrids and parental strains.
Identification of parental Leishmania strains
The characterization of parental Leishmania strains was performed by sequencing ITS1 (Internal Transcribed Spacer 1 region between the ssu and 5–8 rRNA genes)[37], using the primers described in Table 1.
Single Nucleotide Polymorphisms Cleaved Amplification Polymorphic site (SNP-CAPs)
Analysis of specific loci placed on different chromosomes and kinetoplastid DNA in the interspecific hybrids (L. donovani mCH HYG x L. major CTN PAC and L. donovani mCh HYG x L. tropica iRFP PAC) and in their parental strains was carried out by SNP-CAPs using the SNP2CAPS tool described by Thiel et al., (2004) [41]. Detailed description of the genes, primers, SNPs, restriction enzymes used for this purpose and the size of the predicted products in each Leishmania strain can be found in Table 2.
Table 2. Genes tested by SNP-CAPs.
The gene name, gene identification (ID), location, the name of forward and reverse primers used for PCR amplification (sequence of these primers can be found in Table 1), SNP position, as well as the expected band sizes after digestion with the suitable restriction enzymes, are shown.
| Gene name | ID | Location | Primers | SNP position | RE | Predicted products (bp) | ||
|---|---|---|---|---|---|---|---|---|
| L. donovani | L. major | L. tropica | ||||||
| Calmodulin-binding | LdBPK.01.2.000260 LMJLV39_010007500 LTRL590_010007500 |
Chr. 1 | TOX59 TOX60 |
608 | MscI (MlsI) | 608 + 178 | 786 | 786 |
| ABC Transporter | LdBPK.03.2.000150 LMJLV39_030006600 LTRL590_030006400 |
Chr. 3 | TOX61 TOX62 |
206 | NruI | 2091 | 1885 + 206 | n.a |
| PmlI | 2091 | n.a | 1812 + 279 | |||||
| Sec20 | LdBPK.11.2.000300 LMJLV39_110008400 LTRL590_110008700 |
Chr. 11 | TOX63 TOX64 |
158 + 204 | BcnI (NciI) | 414 + 158 + 46 | 618 | 618 |
| Rad9 | LdBPK.15.2.001040 LMJLV39_150016800 LTRL590_150016300 |
Chr. 15 | TOX65 TOX66 |
938 | MluCI (TasI) | 1018 + 938 | 1956 | n.a |
| 491 | AvaII (Eco47I) | 1956 | n.a | 1468 + 491 | ||||
| cytoplasmic l-asparaginase i-like protein | LdBPK.15.2.000440 LMJLV39_150009800 LTRL590_150009200 |
Chr. 15 | TOX69 TOX70 |
832 | EcoRV (Eco32I) | 832 + 183 | 1015 | 1015 |
| Paraflagellar rod protein | LdBPK.16.2.001510 LMJLV39_160021000 LTRL590_160021500 |
Chr. 16 | TOX67 TOX68 |
925 | XhoI | 1800 | 925 + 875 | n.a |
| 1441 | NruI | 1441 + 359 | n.a | 1800 | ||||
| Ribosomal protein L22p/L17e | LdBPK.22.2.000700 LMJLV39_220013900 LTRL590_220012400 |
Chr. 22 | TOX55 TOX56 |
593 | MluCI (TasI) | 593 + 236 | 829 | 829 |
| Fumarate hidratase | LdBPK.29.2.002080 LMJLV39_290027300 LTRL590_290026600 |
Chr. 29 | TOX45 TOX46 |
935 | MluCI (TasI) | 1707 | 955 + 772 | 955 + 772 |
| Aquaglyceroporin 1 | LdBPK.31.2.000030 LMJLV39_310005100 MG797692.1 |
Chr. 31 | TOX51 TOX52 |
279 | MluCI (TasI) | 457 + 279 | 736 | 688 |
| 2-oxoisovalerate dehydrogenase beta subunit | Ld .35.2.209420 LMJLV39_350005400 LTRL590_350005500 |
Chr. 35 | TOX49 TOX50 |
609 | HpaI (KspAI) | 609 + 416 | 1025 | 1025 |
| CYTB |
FJ416603.1 MK514113.1 MN904525.1 |
Maxicircle | TOX99 TOX100 |
618 | TatI | 420 + 140 + 60 | 252 + 228 + 141 | n.a. |
| NdeI | 621 | n.a. | 525 + 96 | |||||
| ND5 |
FJ416603.1 MK514113.1 MN904525.1 |
Maxicircle | TOX103 TOX104 |
456 | BglII | 464 | 258 + 206 | n.a. |
| EcoRI | 335 + 129 | n.a. | 464 | |||||
Determination of inheritance in polyploid hybrids
In order to determine the origin of the extra chromosome sets in the different hybrids, we based on the experiment carried out by Romano et al in 2014 [26]. Simulated 3n and 4n hybrids were created by mixing parental DNA in 1:1, 1:2 and 2:1 ratios (for the L. donovani x L. major hybrids) and 1:1, 1:3 and 3:1 ratios (for the L. donovani x L. tropica hybrids). Then, several loci were analysed by SNP-CAPs as indicated above. Subsequently, the ratio obtained between the largest band digested with the restriction enzyme belonging to one parental allele, and the uncut band corresponding to the allele of the other parental, was calculated using the Gene Tools 4.3.9 (Syngene) software. The ratio obtained in simulated 3n and 4n was compared to that obtained by the hybrids, thus allowing us to classify the hybrids according to one of the control ratios by statistical analysis using k-means analysis in SPSS COR statistical software.
In vitro infections
The ability of intraclonal and interspecies hybrids to infect in vitro was tested in RAW murine macrophages. For this purpose, 5x104 RAW cells were seeded in 8-well Ibidi chambers and infected in a proportion of 1:10 with promastigotes. After 2 hours, the co-culture was washed three times in PBS. Three days after the infection, samples were stained with Hoechst 33342 (Fisher Scientific) during 30 minutes at 37°C, and images were acquired on a Zeiss LSM800 confocal microscope.
The in vitro infective competence of hybrids vs. parents was determined using bone marrow-derived macrophages (BMMs) obtained from Balb/c mice. Briefly, after euthanasia, the animals were dissected and the femur and tibia were cleaned and separated in sterile conditions using scissors and forceps. The heads of the bones were cut off, and 5 mL of 1xHBSS (Hanks Balanced Salt Solution) were injected through the bone channel with a 25G syringe. The undifferentiated cells were collected [42] and frozen. When needed, cells were thawed and the protocol proposed by Legarda was followed [43]. Briefly, cells were placed in 5 mL of differentiation medium, which contains RPMI supplemented with 10% FBS and 30% L929 supernatant (L929 fibroblast cells produce rM-CSF produce rM-CSF [recombinant macrophage colony stimulating factor]) in a small petri dish to allow them to attach, and 24 hours later, they were collected and seeded in 10 mL of differentiation medium in a 10-cm non-treated petri dish. On day 4, 5 mL of differentiation medium were added and BMMs were ready for parasite infection on day 7. Amastigotes of each parental and hybrid strains were added to differentiated-macrophages at 1:10 ratio for 6 hours and maintained at 37°C. Later, non-internalised parasites were washed twice with PBS. After 3 days, samples were fixed in 100% methanol and stained with Giemsa. Then, pictures were acquired with a Nikon microscope and 100 macrophages per parasite-cell-line were analysed for the presence of amastigotes, which were counted in three independent experiments.
Results and discussion
Generation of engineered parental cell lines
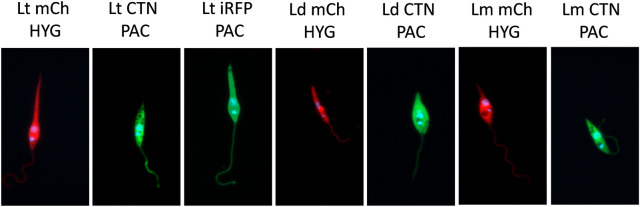
In order to ease the phenotypic analysis of the parental and hybrid strains, in addition to L. major mCh HYG, which had been already generated in our laboratory and was able to stably express the gene encoding mCh and the hygromycin resistance cassette [33], we prepared six other novel transgenic strains (see Materials and methods). After the analysis of these transgenic strains by PCR to confirm the integration of both the reporter and antibiotic resistance cassettes, and analysis by flow cytometry (MACSQuant Analyzer 10), the fluorescent parasites were visualized with different excitation wavelengths by confocal microscopy (Fig 1).
Fig 1. Engineered Leishmania parental strains.
Representative confocal microscopy pictures of promastigotes of the parental fluorescent strains of Leishmania used for the intraclonal and interspecies outcrossings. Lt mCh HYG: L. tropica mCh HYG (561 nm); Lt CTN PAC: L. tropica CTN PAC (488 nm); Lt iRFP PAC: L. tropica iRFP PAC (640 nm); Ld mCh HYG: L. donovani mCh HYG (561 nm); Ld CTN PAC: L. donovani CTN PAC (488 nm); Lm mCh HYG: L. major mCh HYG (561 nm); Lm CTN PAC: L. major CTN PAC (488 nm). The wavelength refers to the excitation laser.
Formation and characterization of intraclonal hybrid strains in vitro
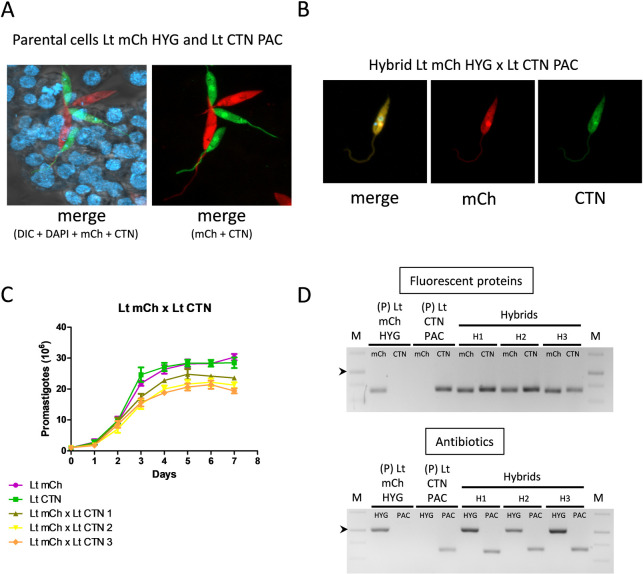
Stationary-phase promastigotes of L. tropica mCh HYG and L. tropica CTN PAC were mixed in the presence of a feed layer of cells isolated from Lu. longipalpis embryos (LULO cells) (Fig 2A), and the double selection of antibiotics (puromycin and hygromycin) was started at three different time points: days 3, 6 and 9 post-mixing. Two different experiments were carried out, and as shown in Table 3, the highest ratio of hybrid formation was achieved when antibiotic selection was performed at day 3 post-mixing, which could be related to direct competition between the newly-formed hybrid and the parental promastigotes. If hybrids are formed relatively soon after parental contact, and antibiotic selection is added at late times (9 days), development of the newly-formed hybrid could be impaired regarding parental cells due to biological competition mechanisms, thus avoiding the recovery of hybrids at late time. However, if selection is performed soon after hybrid formation, competition with parental strains would be lower, since the latter would die, thereby promoting the growth of hybrids bearing antibiotic resistance cassettes in the genome.
Fig 2. Characterization of intraclonal hybrids.
(A) Confocal microscopy showing the interaction between LULO cells and parental cell lines of L. tropica before mating. (B) Representative image of confocal microscopy of one hybrid clon of L. tropica mCh HYG x L. tropica CTN PAC and fluorescence emission in the mCh (561 nm) and CTN (488 nm) channels. (C) Growth curves (number of promastigotes in culture during 7 days) of the parental L. tropica mCh HYG (Lt mCh) and L. tropica CTN PAC (Lt CTN), and three representative intraclonal hybrids (Lt mCh x Lt CTN 1, 2 and 3). (D) Agarose gels of the electrophoresis of the PCR products obtained after the amplification of the genes encoding fluorescent proteins (mCh and CTN) and antibiotic resistance (HYG or PAC) in the parental (P) and three representative intraclonal hybrids (H). The arrowheads indicate the 1000 bp band in the DNA molecular weight marker (M).
Table 3. Intraclonal hybrid formation in L. tropica.
| Addition of antibiotics (days post-mixing) | Percentage of positive wells | |
|---|---|---|
|
L. tropica mCh HYG x L. tropica CTN PAC |
3 days | Exp. 1: 2.34% (9/384) |
| Exp. 2: 10.67% (41/384) | ||
| 6 days | Exp. 1: 1.3% (5/384) | |
| Exp. 2: 2.6% (10/384) | ||
| 9 days | Exp. 1: 0.78% (3/384) | |
| Exp. 2: 0% (0/384) |
These hybrids have yellow color (using both red and green channels for microscopic visualization) (Fig 2B), are resistant to puromycin and hygromycin, and show lower growth rate (lower number of promastigotes during the stationary phase) than the parental lines (Fig 2C). To confirm the inheritance of DNA from parental strains in the hybrid filial strains, we amplified the genes conferring antibiotic resistance and the genes encoding fluorescent proteins using the primers listed in Table 1. Fig 2D shows agarose gels of the amplification products of the L. tropica strain before mating and in three hybrids. These results clearly show that unlike the parental strains, the hybrids contain genes from both parents either for antibiotic resistance or for fluorescence emission, thus confirming that genetic exchange events occurred during the outcrossing.
Typically, the outcrossing experiments carried out in trypanosomatids have been performed in the midgut of the vector’s digestive tract between promastigotes [23–26,44,45]. These experiments consisted of feeding the insect vectors with blood infected with both strains of parental promastigotes to be crossed, with mating taking place in the midgut of the insect (in the case of Leishmania). A recent paper, however, described the mating of two different strains of L. tropica promastigotes in axenic conditions [27]. The yield of hybrids obtained, although significant, was much lower than that obtained in the presence of the insect [45]. Considering these results, we performed intraclonal mating experiments in vitro using a feed layer of LULO cells, which were used to mimic the conditions within the insect midgut. L. tropica was initially chosen as model microorganism due to the fact that this species has a greater mating success in flies [45]. Stationary-phase promastigotes were selected because they are less mobile and allowed us to maintain the culture for up to 9 days without disturbing the LULO cell monolayer.
We also attempted to generate intraclonal hybrids for other Leishmania species, namely L. major (L. major mCh HYG x L. major CTN PAC) and L. donovani (L. donovani mCh HYG x L. donovani CTN PAC). Based on our previous results with L. tropica, we used stationary-phase promastigotes and LULO cells, and the addition of selection antibiotics was carried out on day 3 after mixing. Unfortunately, no hybrids were obtained under these conditions. Therefore, we decided to modify the promastigote phase of growth and use exponential-phase promastigotes in addition to stationary-phase promastigotes in the absence of LULO cells in an attempt to increase the probability of success, since as previously reported, mating-competent forms of Leishmania can also occur in axenic conditions [27]. Despite these modifications, we could not obtain intraclonal hybrids between these two Leishmania species. These results confirm the better capacity of L. tropica to generate mating-competent forms compared to L. major [45] and, as shown in this work, to L. donovani as well.
Formation and characterization of interspecies hybrid strains in vitro
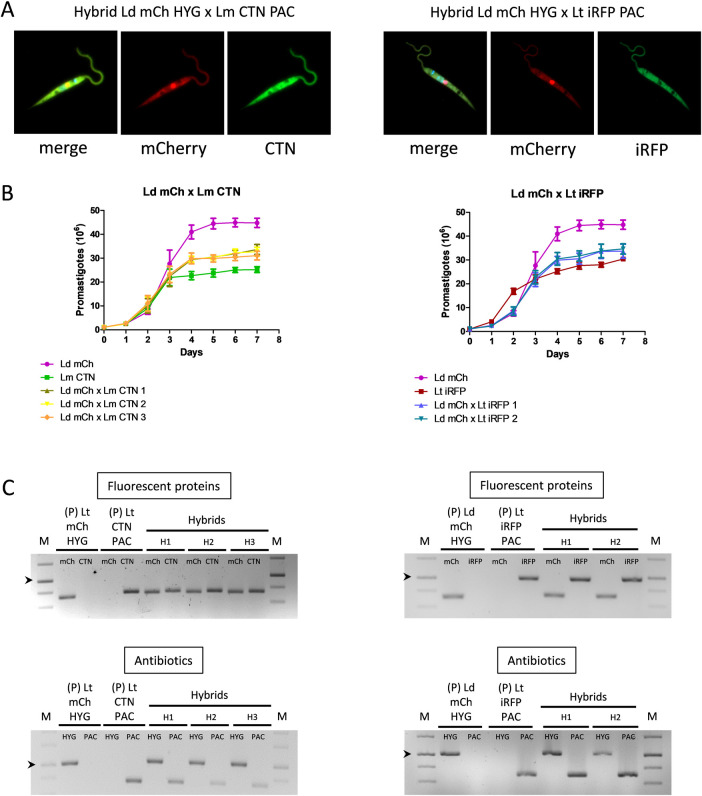
Interspecies hybrid formation was also attempted between L. donovani x L. major, L. donovani x L. tropica and L. major x L. tropica, using either exponential or stationary-phase promastigotes and in the presence and absence of LULO cells. Two different experiments were performed and addition of antibiotics was carried out on day 3 after mixing. As shown in Table 4, three hybrids (0.78% success) were obtained between stationary-phase promastigotes of L. donovani mCh HYG x L. major CTN PAC, which have yellow color (using red and green channels for visualization) (Fig 3A left panel) and are resistant to both hygromycin and puromycin. On the other hand, two hybrids (0.52% success) were formed in the absence of LULO cells between exponential-phase promastigotes of L. donovani mCh HYG x L. tropica iRFP PAC, which have yellow color (using red and far red channels for visualization) (Fig 3A right panel) and are resistant to both antibiotics. No hybrid progeny was obtained after crossing L. tropica x L. major, which can be related to the low success rate observed for the in vitro mating. Indeed, interspecies crosses occurred with a low success rate, and regardless the promastigote type stage (exponential or stationary phase) or the presence of an insect cellular feed layer, which confirms that LULO cells are not strictly required for in vitro formation of hybrids.
Table 4. Interspecies crossings performed with L. donovani, L. major and L. tropica, and the success rate (percentage of hybrids obtained) using different mating parental lines, different promastigote stages and in the presence and absence of LULO cells.
| Crossing experiments | L. tropica iRFP PAC x L. major mCh HYG | L. donovani mCh HYG x L. tropica iRFP PAC | L. donovani mCh HYG x L. major CTN PAC |
|---|---|---|---|
| Exponential-phase promastigotes | Exp1: 0% (0/384) | Exp 1: 0.52% (2/384) | Exp 1: 0% (0/384) |
| Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | |
| Exponential-phase promastigotes+ LULO cells | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) |
| Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | |
| Stationary-phase promastigotes | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) |
| Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | |
| Stationary-phase promastigotes + LULO cells | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) | Exp 1: 0% (0/384) |
| Exp 2: 0% (0/384) | Exp 2: 0% (0/384) | Exp 2: 0.78% (3/384) |
Fig 3. Characterization of interspecific hybrids.
(A, left panel) Representative merged images of confocal microscopy and fluorescence emission in the mCh (561 nm) and CTN (488 nm) channels of one hybrid clone obtained after the outcrossing of L. donovani mCh HYG x L. major CTN PAC (Ld mCH HYG x Lm CTN PAC. (A, right panel) Representative merged images of confocal microscopy and fluorescence emission in the mCh (561 nm) and iRFP (640 nm) channels of one hybrid clone obtained after the outcrossing of L. donovani mCh HYG x L. tropica iRFP PAC (Ld mCH HYG x Lt iRFP PAC. (B, left panel) Growth curves (number of promastigotes in culture during 7 days) of the parental strains L. donovani mCh HYG (Ld mCh) and L. major CTN PAC (Lm CTN) and the interspecies hybrids Ld mCh x Lm CTN 1, 2 and 3. (B, right panel) Growth curves (number of promastigotes in culture during 7 days) of the parental strains L. donovani mCh HYG (Ld mCh) and L. tropica iRFP PAC (Lt iRFP), and the interspecies hybrids Ld mCh x Lt iRFP 1 and 2. (C, left panel) Agarose gels of the electrophoresis of the PCR products obtained after the amplification of the genes encoding fluorescent proteins (mCh and CTN) and antibiotic resistance (HYG or PAC) in the parental strains (P) and in the three interspecies hybrids (H) obtained after the outcrossing of L. donovani mCh HYG x L. major CTN PAC. (C, right panel) Agarose gels of the electrophoresis of the PCR products obtained after the amplification of the genes encoding fluorescent proteins (mCh and iRFP) and antibiotic resistance (HYG or PAC) in the parental strains (P) and in the two interspecies hybrids (H) resulting from the outcrossing of L. donovani mCh HYG x L. tropica iRFP PAC. The arrowheads indicate the 1000 bp band in the DNA molecular weight marker (M).
L. donovani mCh HYG x L. major CTN PAC and L. donovani mCh HYG x L. tropica iRFP PAC hybrids showed a growth profile in between their parental strains; the highest parasite number being provided by the parental L. donovani mCh HYG and the lowest parasite number being provided by the other parental strains (i.e. L. major CTN PAC and L. tropica iRFP PAC) (Fig 3B). Inheritance of the parental antibiotic resistance and reporter genes in the hybrid filial strains was confirmed by PCR amplification using the primers listed in Table 1. Fig 3C shows agarose gels of the amplification products in the parental and hybrid strains, thus confirming inheritance of the above-mentioned genes.
To the best of our knowledge, this is the first report describing in vitro interspecies outcrossing in Leishmania, and in this specific case, between species with different tropism; i.e. visceral (L. donovani) and cutaneous (L. tropica and L. major).
Ploidy profile of hybrids
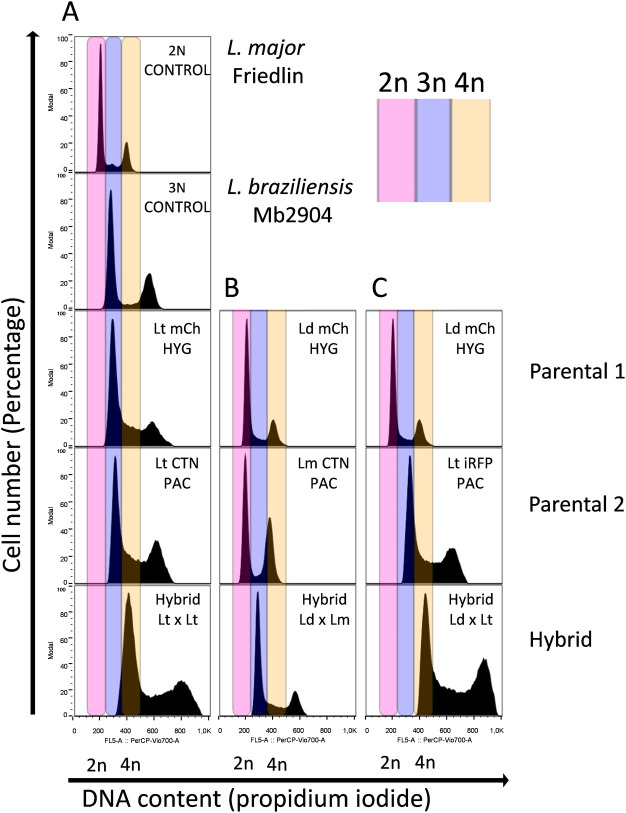
Once the hybrids were identified phenotypically, namely by their dual antibiotic resistance and mixed color detected by confocal microscopy, the ploidy of both the parental and filial generations was determined by flow cytometry in order to assess whether reductive or additive chromosome processes might have occurred after mating.
Fig 4A shows a representative chart of flow cytometry of the result of L. tropica mCh HYG x L. tropica CTN PAC crossing (only one hybrid is shown, since all the hybrids exhibited identical results). As indicated by flow cytometry, the ploidy of the parental L. tropica strain was 3n, whereas ploidy of the filial strains after in vitro mating was approximately 4n. Regarding interspecies hybrids, flow cytometry results indicated that the three hybrids obtained from L. donovani x L. major (both 2n) were 3n (Fig 4B), while the ploidy of the two hybrids obtained from the L. donovani x L. tropica offspring was 4n (Fig 4C).
Fig 4. Analysis of the ploidy profile.
(A) Representative chart of flow cytometry showing the ploidy of L. tropica mCh HYG (Lt mCh HYG), L. tropica CTN PAC (Lt CTN PAC) and one hybrid of the intraclonal crossing (Hybrid 1 Lt x Lt). (B) Representative chart of flow cytometry showing the ploidy of L. donovani mCh HYG (Ld mCh HYG), L. major CTN PAC (Lm CTN PAC) and one hybrid of the interspecies crossing (Hybrid 1 Ld x Lm). (C) Representative chart of flow cytometry showing the ploidy of L. donovani mCh HYG (Ld mCh HYG), L. tropica iRFP PAC (Lt iRFP PAC) and one hybrid of the interspecies crossing (Hybrid 1 Ld x Lt). L. major Friedlin (ploidy = 2n) and L. braziliensis Mb2904 (ploidy = 3n) were used as ploidy controls (2N CONTROL and 3N CONTROL, respectively) in order to assess the ploidy of the rest of the parasites.
These results indicate that either intraclonal or interspecies crossings give rise to additive chromosome events in the hybrid progeny. This phenomenon has been previously observed by other authors, who have obtained in their crosses a mixture of hybrids with variable ploidy, with the presence of 2n, 3n and even 4n hybrids from 2n parents, regardless of whether the hybrids were obtained in the sandfly or in vitro [23–27]. In our work, all hybrids having a DNA content of 4n resulted from at least one 3n parental strain (L. tropica). As far as we know, intraclonal or interespecies hybrids from 3n parentals have never been described before, either in sandflies or in vitro.
Genotypic characterization of interspecies hybrids
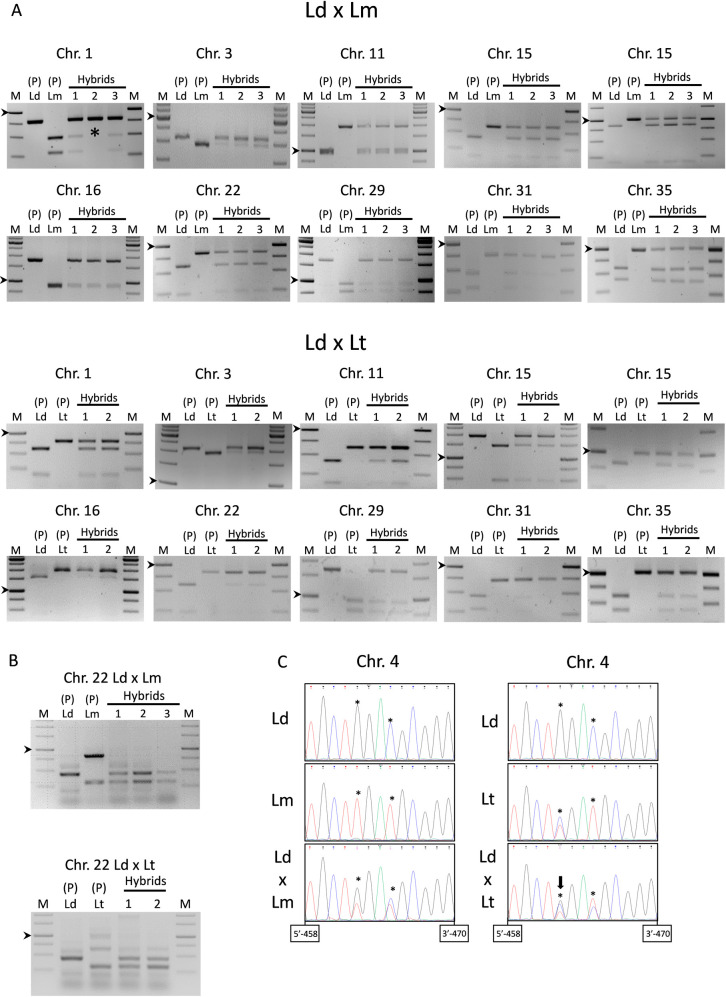
Genotypic characterization of the interspecies hybrids was initially performed by SNP-CAPs in ten genes encoding the following proteins: calmodulin-binding protein, ABC transporter, Sec20, Rad9, cytoplasmic L-asparaginase i-like protein, paraflagellar rod protein, ribosomal protein L22p/L17e, fumarate hydratase, aquaglyceroporin 1, and 2-oxoisovalerate dehydrogenase beta subunit, which are present in different Leishmania chromosomes as indicated in Table 2. For this purpose, these genes were amplified with specific primers and cut with suitable restriction enzymes (see Table 2 for details). Amplification and restriction analysis allowed us to characterize the SNPs present in the progenitor strains and in the interspecies hybrids resulting from crosses of L. donovani mCh HYG x L. major CTN PAC and L. donovani mCh HYG x L. tropica iRFP PAC (Fig 5A).
Fig 5. Characterization of the nuclear inheritance of the interspecies hybrids.
(A) Agarose gels of the SNP-CAPs analysis showing the DNA products after PCR amplification and enzymatic digestion of the ten genes present in nine different chromosomes. The electrophoretic profile of the parental lines [P (Ld, Lm)] and three hybrids [Hybrids (1, 2, 3)] resulting from the outcrossing of L. donovani mCh HYG x L. major CTN PAC is shown in the top panel, whereas the electrophoretic profile of the parental lines [P (Ld, Lt)] and two hybrids [Hybrids (1, 2)] resulting from the outcrossing of L. donovani mCh HYG x L. tropica iRFP PAC is shown in the bottom panel. Note the absence of the L. major allele of the gene located on chromosome 1 in hybrid number two (indicated with an asterisk). (B) Agarose gels of the PCR amplification of the A2 gene in the parental lines [P (Ld, Lm)] and three hybrids [Hybrids (1, 2, 3)] resulting from the outcrossing of L. donovani mCh HYG x L. major CTN PAC (top panel), and in the parental lines [P (Ld, Lt)] and two hybrids [Hybrids (1, 2)] resulting from the outcrossing of L. donovani mCh HYG x L. tropica iRFP PAC (bottom panel). The arrowheads indicate the 1000 bp band in the DNA molecular weight marker (M). (C) Fluorograms showing the sequence of part of the actin gene and some SNPs (marked with an asterisk) in the parental strains L. donovani mCh HYG (Ld), L. major CTN PAC (Lm) and L. tropica iRFP PAC (Lt), and one representative hybrid of the outcrossing between L. donovani mCh HYG x L. major CTN PAC (Ld x Lm) and L. donovani mCh HYG x L. tropica iRFP PAC (Ld x Lt). Note the triple nucleotide peaks in the fluorograms of L. donovani x L. tropica hybrid (arrow in the SNP located at position 462 in the right panel).
After amplification and restriction analyses it can be observed that the bands corresponding to both progenitors are visible in the three hybrids of L. donovani x L. major outcrossing, which would correspond to a biparental inheritance produced by a genetic exchange after mating. As an exception we can highlight the absence of the L. major allele of the gene located on chromosome 1 in one of the hybrids (Fig 5A top panel) that can be due to a loss of heterozygosity and/or aneuploidy in Leishmania, as it has been previously reported [26]. Similar conclusions are raised from the crossing of L. donovani x L. tropica (Fig 5A bottom panel), where the biparental inheritance of the hybrids appears again represented by the bands corresponding to the enzymatic restriction of the amplified gene of both progenitors.
We also analysed the composition of A2 gene in the hybrids after mating. The interest of the product of A2 gene is because it has been involved in the organic tropism of visceralizing Leishmania strains including L. donovani [46]. In this species, the A2 gene is represented by a multicopy cluster (tandem-repeats) in chromosome 22 and encodes a family of A2 proteins with 10-amino acid repeated sequences. However, a single and truncated pseudogene is present in the species with cutaneous tropism, namely L. major and L. tropica [46,47]. For the amplification of this gene, we used the primers L2/R3 (Table 1), which were previously described by Garin et al. (2005) [46] and later used by Romano et al. (2014) [26]. As we can see in Fig 5B, the parental L. donovani strain shows a main band near 500 bp, whereas L. major and L. tropica show a main band over 250 bp, which correspond to the pattern amplified by these primers in these species [46,48]. The presence of the main amplification bands that are visible in both parental strains and in the hybrid clones from both crosses, further demonstrates biparental inheritance (Fig 5B).
Finally, nuclear inheritance was also demonstrated by sequencing the actin gene located on chromosome 4, which was amplified using primers TOX83 and TOX84 (Table 1). Sequence analysis (Fig 5C) of several regions confirmed the inheritance of the SNPs located at positions 462 and 465 of the actin gene (from the initial ATG codon) in the hybrids obtained from both interspecies outcrossings. The parental L. tropica strain turned out to be heterozygous for some SNPs. This led to the visualization of triple nucleotide peaks in the sequencing fluorograms of L. donovani x L. tropica hybrids (Fig 5C right panel), which inherited at least two copies of the parental L. tropica and at least one copy of the parental L. donovani actin gene.
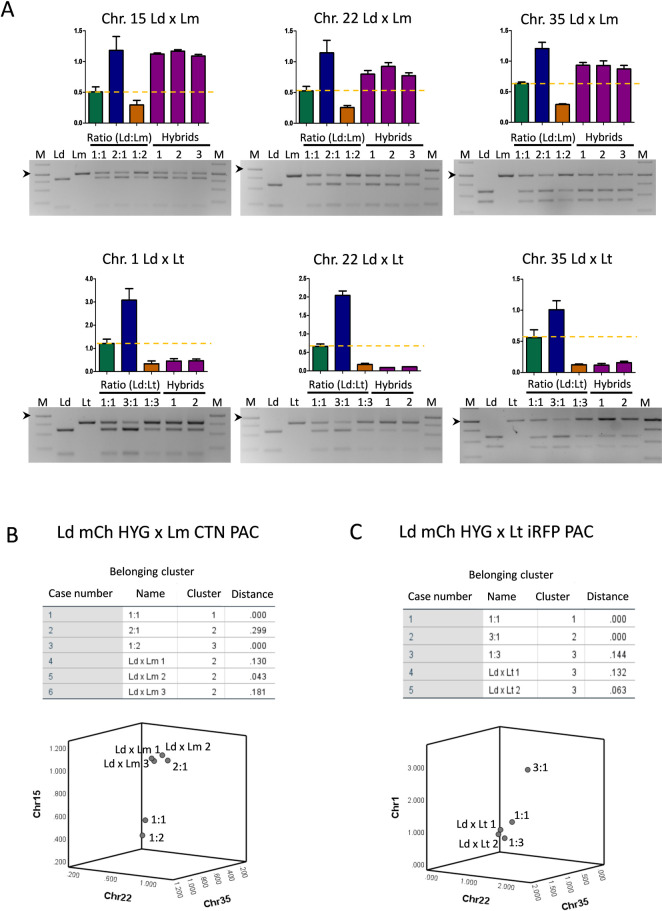
The origin of the extra chromosome sets in the different hybrids was determined as indicated in the Materials and methods section. Fig 6A shows the SNP-CAPs digestion products for different genes encoding the following proteins: calmodulin-binding protein (chromosome 1), cytoplasmic L-asparaginase i-like protein (chromosome 15), ribosomal protein L22p/L17e (chromosome 22) and 2-oxoisovalerate dehydrogenase beta subunit (chromosome 35) (see Table 2 for details), and the ratios between the intensity of the upper band from the parental L. donovani and the uncut band from the other parental (L. major or L. tropica) in the corresponding hybrid clones. Results indicated that the hybrids obtained from L. donovani x L. major crosses, inherited the extra trisomic chromosomes 15, 22 and 35 from L. donovani (Fig 6A top panel). On the contrary, the hybrids obtained from L. donovani x L. tropica crossing seem to have inherited the extra chromosomes 1, 22 and 35 from L. tropica (Fig 6A botton panel). This is consistent with the results represented in Fig 5C, which showed that at least two copies of the actin gene would have been inherited from L. tropica. Statistical analysis of the ratios showed that L. donovani x L. major hybrids are grouped within the category 2:1 (L. donovani:L. major) (Fig 6B), whereas L. donovani x L. tropica hybrids are grouped within the category 1:3 (L. donovani:L. tropica) (Fig 6C), thus confirming the inheritance of extrachromosome sets from L. donovani, in the case of L. donovani x L. major hybrids (3n), or L. tropica in the case of L. donovani x L. tropica hybrids (4n).
Fig 6. Characterization of the inheritance of extra chromosome sets in the interspecies hybrid clones.
(A) Agarose gels of the SNP-CAPs analysis showing the DNA products after PCR amplification and enzymatic digestion of three loci present in chromosomes 2, 22 and 35. The top panel shows the electrophoretic profile of L. donovani mCh HYG (Ld), L. major CTN PAC (Lm) and three hybrids (Ld x Lm), and the ratios between the intensity of the upper band from the parental L. donovani and the uncut upper band from the other parental L. major in the corresponding hybrid clones. Simulated 3n were created by mixing parental DNA in 1:1, 1:2 and 2:1 ratio (L. donovani:L. major). The bottom panel shows the electrophoretic profile of L. donovani mCh HYG (Ld), L. tropica iRFP PAC (Lt) and two hybrids (Ld x Lt), and the ratios between the intensity of the upper band from the parental L. donovani and the uncut upper band from the other parental L. tropica in the corresponding hybrid clones. Simulated 4n were created by mixing parental DNA in 1:1, 1:3 and 3:1 ratio (L. donovani:L. tropica). The arrowhead indicates 1000 bp in the DNA molecular weight marker (M). (B, C) Statistical representation by k-means analysis in SPSS of the extrachromosome inheritance of L. donovani mCh HYG x L. major CTN PAC (Ld mCh HYG x Lm CTN PAC) hybrids, which are clustered together with ratio 2:1 (LdxLm) (B), and L. donovani mCh HYG x L. tropica iRFP PAC (Ld mCh HYG x Lt iRFP PAC) hybrids, the latter being clustered together with ratio 1:3 (LdxLt) (C).
The differences in ratios for each locus may reflect mosaic aneuploidy in the progeny. This process has been demonstrated in several Leishmania species [49], and has been suggested to represent a unique source of adaptability for the parasites to new environment [50].
Extranuclear DNA inheritance of interspecies hybrid clones
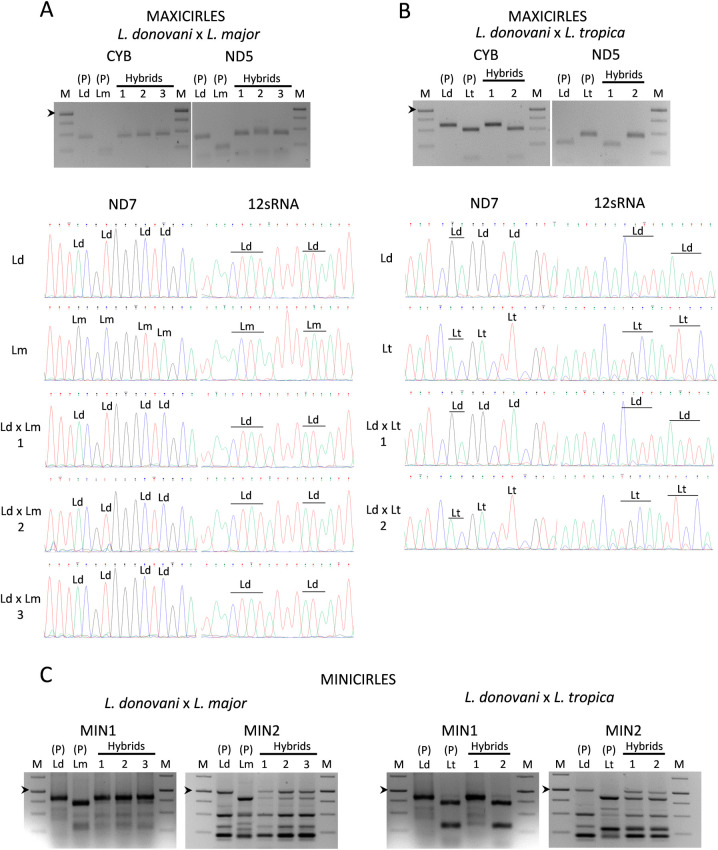
The inheritance of maxi and minicircles of extrachromosomal kDNA in the interspecies hybrids was also analysed. For the study of the inheritance of maxicircles, four genes (Cyb, ND5, ND7 and 12sRNA) were analysed. Inheritance of Cyb and ND5 was assessed by SNP-CAPs (see Table 2 for details). As seen in the top panel of Fig 7A, interspecies hybrid clones of L. donovani mCh HYG x L. major CTN PAC only exhibit the restriction pattern of the parental L. donovani strain for these two genes. In the case of the L. donovani mCh HYG x L. tropica iRFP PAC interspecies outcrossing, one of the hybrid clones showed a restriction pattern for Cyb and ND5 similar to that observed for L. donovani, whereas the other hybrid clone showed a behavior similar to the other parental (i.e. L. tropica) (Fig 7B top panel). In order to confirm this phenomenon, the analysis of the inheritance of ND7 and 12sRNA was performed by sequencing these genes after amplification with the primers TOX97, TOX98 and TOX101, TOX102 (Table 1). Fluorograms indicated that in the three hybrid clones L. donovani mCh HYG x L. major CTN PAC, the SNPs were inherited from the parental L. donovani strain (Fig 7A bottom panel), whereas one of the hybrid clones of the L. donovani mCh HYG x L. tropica iRFP PAC outcrossing inherited the SNPs from L. donovani and the other hybrid clone inherited the SNPs from L. tropica (Fig 7B bottom panel). These results indicate that inheritance of maxicircles is uniparental or alternatively, that genetic information contained in maxicircles from one of the parental strains can be lost by genetic drift, as it has been previously suggested in T. brucei [31].
Fig 7. Characterization of the inheritance of kDNA in the interspecies hybrid clones.
(A, B, top panels) Agarose gels of the SNP-CAPs analysis showing the DNA products after PCR amplification and enzymatic digestion of Cyb and ND5 genes. The electrophoretic profile of the parental lines [P (Ld, Lm)] and three hybrids [Hybrids (1, 2, 3)] resulting from the outcrossing of L. donovani mCh HYG (Ld) x L. major CTN PAC (Lm) (A), or the parental lines [P (Ld, Lt)] and two hybrids [Hybrids (1, 2)] resulting from the outcrossing L. donovani mCh HYG (Ld) x L. tropica iRFP PAC (Lt) (B), are shown. (A, B bottom panels) Fluorograms showing the sequence of part of the ND7 and 12 sRNA genes with some SNPs, which have been indicated as Ld (L. donovani) and Lm (L. major) in the parental strains L. donovani mCh HYG (Ld), L. major CTN PAC (Lm) and in their clonal hybrids (LdxLm) 1, 2 and 3 (A), or as Ld (L. donovani) and Lt (L. tropica) in the parental strains L. donovani mCh HYG (Ld), L. tropica iRFP PAC (Lt) and in their clonal hybrids (LdxLt) 1 and 2 (B). (C) Agarose gels showing the band profile after amplification of a conserved region (MIN1) or a variable region (MIN2) of minicircles in the parental lines [P (Ld, Lm)] and three hybrids [Hybrids (1, 2, 3)] resulting from the outcrossing of L. donovani mCh HYG (Ld) x L. major CTN PAC (Lm) (left panel), or in the parental lines [P (Ld, Lt)] and two hybrids [Hybrids (1, 2)] resulting from the outcrossing L. donovani mCh HYG (Ld) x L. tropica iRFP PAC (Lt) (right panel). The arrowhead indicates 1000 bp in the DNA molecular weight marker (M).
For the study of the inheritance of minicircles, PCR experiments were conducted with primers TOX73 and TOX 74, which amplify near 780 bp from a conserved region (MIN1) in minicircle kDNA in Leishmania spp. [39] (Table 1), and with primers TOX75 and TOX76 (Table 1), the latter amplifying a variable region of minicircles (MIN2) and generating multi-sized products [40]. As shown in Fig 7C, hybrids from either L. donovani mCh HYG x L. major CTN PAC or L. donovani mCh HYG x L. tropica iRFP PAC exhibited an amplification pattern of MIN1 that included bands from both parental strains, although with different intensity. In the case of L. donovani mCh HYG x L. major CTN PAC, the most intense band in the three hybrid clones corresponds to that inherited from L. donovani, which correlates well with the results obtained with the inheritance of maxicircles.
The pattern for MIN2 in hybrids from either L. donovani mCh HYG x L. major CTN PAC or L. donovani mCh HYG x L. tropica iRFP PAC, provided a more precise picture of the presence of bands from both parental strains, thereby suggesting the biparental inheritance of minicircles.
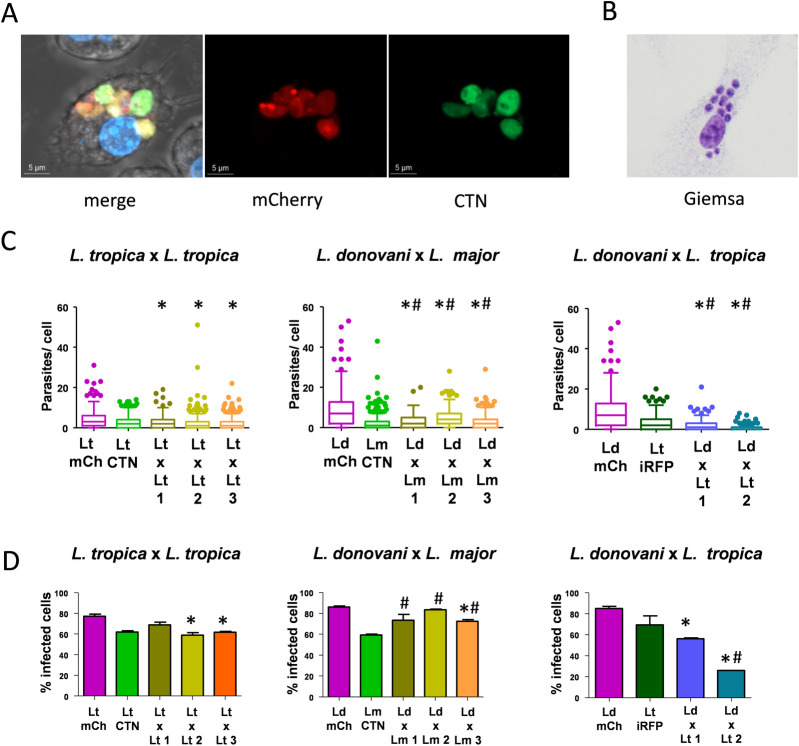
Infectivity of intraclonal and interspecies hybrids
In vitro infection of hybrid clones obtained from different mating experiments was evaluated in BMMs. The objective of this experiment was the phenotypic comparison using amastigotes from the hybrids and the parental species under the same infection conditions. The ability of hybrids to infect macrophages was first checked in RAW cells by confocal microscopy (Fig 8A) because BMMs show high autofluorescence that interferes with visualization of amastigotes. Once the ability of these parasites to infect macrophages was confirmed in vitro, the infectivity of amastigotes was tested in BMMs derived from Balb/c mice by microscopy analysis after Giemsa staining (Fig 8B). The number of parasites infecting each BMMs was determined after the analysis of 100 BMMs in triplicate and represented for each hybrid (Fig 8C). Results indicate that amastigotes of all the intraclonal hybrids showed a reduction in their infection ability regarding L. tropica mCh HYG. Interspecies hybrids L. donovani mCh HYG x L. major CTN PAC showed infectivity values between both parental strains. However, amastigotes of interspecies hybrids L. donovani mCh HYG x L. tropica iRFP PAC exhibited reduced infectivity values in comparison with both parental strains (Fig 8C). Regarding the number of macrophages that hybrid and parental parasites were able to infect (Fig 8D), intraclonal hybrids L. tropica mCh HYG x L. tropica CTN PAC 2 and 3 showed a significant decrease regarding the parental L. tropica mCh HYG. Infection with amastigotesof the interspecies hybrids L. donovani mCh HYG x L. major CTN PAC did not show significant differences regarding L. donovani mCh HYG, although a significant increase in the percentage of infected BMMs was observed regarding the other parental strain L. major CTN PAC. The exception was hybrid 3, which exhibited a significant decrease in the percentage of infected BMMs regarding L. donovani mCh HYG. Infection with either amastigotes of the interspecies hybrids L. donovani mCh HYG x L. tropica iRFP PAC, in general, provided a reduction in the percentage of infected BMMs regarding both parental strains, this decrease being more patent in the case of interspecies hybrid 2 (Fig 8D).
Fig 8. Infectivity of intraclonal and interspecies hybrids.
(A) Representative image of confocal microscopy of a RAW cell infected with one hybrid of L. donovani mCh HYG x L. major CTN PAC. Merge and fluorescent pictures in the mCh (561 nm) and CTN (488 nm) channels are shown. (B) Representative image of BMMs infected with amastigotes of one hybrid of L. donovani mCh HYG x L. major CTN PAC and stained with Giemsa. (C) Box plots representing the number of amastigotes present within each BMM after in vitro infection withamastigotes of L. tropica mCh HYG (Lt mCh), L. tropica CTN PAC (Lt CTN), three intraclonal hybrids L. tropica mCh HYG x L. tropica CTN PAC (Lt x Lt 1, 2 and 3), L. donovani mCh HYG (Ld mCh), L. major CTN PAC (Lm CTN) three interspecies hybrids L. donovani mCh HYG x L. major CTN PAC (Ld x Lm 1, 2 and 3), L. tropica iRFP PAC (Lt iRFP), and two interspecies hybrids L. donovani mCh HYG x L. tropica iRFP PAC (Ld x Lt 1 and 2). (D) Bar graphs representing the percentage of infected BMMs after in vitro infection withamastigotes of L. tropica mCh HYG (Lt mCh), L. tropica CTN PAC (Lt CTN), three intraclonal hybrids L. tropica mCh HYG x L. tropica CTN PAC (Lt x Lt 1, 2 and 3), L. donovani mCh HYG (Ld mCh), L. major CTN PAC (Lm CTN) three interspecies hybrids L. donovani mCh HYG x L. major CTN PAC (Ld x Lm 1, 2 and 3), L. tropica iRFP PAC (Lt iRFP), and two interspecies hybrids L. donovani mCh HYG x L. tropica iRFP PAC (Ld x Lt 1 and 2). Differences were considered as significant when p<0,05 and in a Kruskal-Wallist test (C) or in a one-way ANOVA analysis (D) (represented with an asterisk regarding the first parental line and with a hash regarding the second parental line).
Hybrid promastigotes were also able to infect mice, and amastigotes were recovered from the spleens of infected mice. The recovered parasites maintained double antibiotic resistance, thus confirming stability of the hybrids upon in vivo infection.
In conclusion, we have demonstrated in this work that outcrossing in Leishmania can also occur under in vitro conditions, even between species of different tropism and even if a parental strain contains a ploidy of 3n. In addition, the resultant 4n hybrids are stable after in vivo infection.
With the data obtained, the mechanism underlying sexual reproduction in the crosses could be related with meiosis in which the hybrids contain one allele of each parent for the markers analysed. On the other hand, polyploidy hybrids are frequently formed after outcrossings of Trypanosoma and Leishmania species. However, it has been reported that most of those crosses correspond to a meiotic cycle [7,9,51]. This could be explained by the existence of a tetraploid meiotic cycle in which one of the parental nucleus does not undergo meiosis after cell fusion [45], or alternatively, by the fusion of meiotic intermediates with gametes [8].
These results contribute to the understanding of sexual reproduction in this trypanosomatid and suggest that mechanisms of virulence and drug resistance can be acquired by Leishmania through this process.
Supporting information
(TIF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
C.G.C (LE255-16) and B.D.A (LE208-17) are recipients of Junta de Castilla y León (JCyL) and European Social Found (ESF)’s Fellowships Scheme for Doctoral Training Programs. This research was funded by MINECO; SAF2017-83575-R to RMR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: A review. F1000Res. 2017;6:750. doi: 10.12688/f1000research.11120.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telleria EL, Martins-Da-Silva A, Tempone AJ, Traub-Cseko YM. Leishmania, microbiota and sand fly immunity. Parasitology. 2018;145(10): 1336–1353. doi: 10.1017/S0031182018001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caridha D, Vesely B, van Bocxlaer K, Arana B, Mowbray CE, Rafati S, et al. Route map for the discovery and pre-clinical development of new drugs and treatments for cutaneous leishmaniasis. Int J Parasitol Drugs Drug Resist. 2019;11: 106–117. doi: 10.1016/j.ijpddr.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abadías-Granado I, Diago A, Cerro PA, Palma-Ruiz AM, Gilaberte Y. Cutaneous and mucocutaneous leishmaniasis. Actas Dermosifiliogr. 2021;S0001<-7310(21)00108. doi: 10.1016/j.ad.2021.02.008 [DOI] [PubMed] [Google Scholar]
- 5.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366(9496): 1561–1577. doi: 10.1016/S0140-6736(05)67629-5 [DOI] [PubMed] [Google Scholar]
- 6.Zijlstra EE, Musa AM, Khalil EAG, El Hassan IM, El-Hassan AM. Post-kala-azar dermal leishmaniasis. Lancet Infect Dis. 2003;3(2): 87–98. doi: 10.1016/s1473-3099(03)00517-6 [DOI] [PubMed] [Google Scholar]
- 7.Peacock L, Ferris V, Sharma R, Sunter J, Bailey M, Carrington M, et al. Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc Natl Acad Sci U S A. 2011;108: 3671–3676. doi: 10.1073/pnas.1019423108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peacock L, Kay C, Farren C, Bailey M, Carrington M, Gibson W. Sequential production of gametes during meiosis in trypanosomes. Commun Biol. 2021;4(1): 555. doi: 10.1038/s42003-021-02058-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peacock L, Bailey M, Carrington M, Gibson W. Meiosis and haploid gametes in the pathogen Trypanosoma brucei. Curr Biol. 2014;24(2): 181–186. doi: 10.1016/j.cub.2013.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson W, Stevens J. Genetic exchange in the Trypanosomatidae. Adv Parasitol. 1999;43: 1–46. doi: 10.1016/s0065-308x(08)60240-7 [DOI] [PubMed] [Google Scholar]
- 11.de Meeûs T, Balloux F. Clonal reproduction and linkage disequilibrium in diploids: A simulation study. Infect Genet Evol. 2004;4(4): 345–351. doi: 10.1016/j.meegid.2004.05.002 [DOI] [PubMed] [Google Scholar]
- 12.Prugnolle F, De Meeus T. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity. 2010;104(2): 135–140. doi: 10.1038/hdy.2009.128 [DOI] [PubMed] [Google Scholar]
- 13.Ramírez JD, Llewellyn MS. Reproductive clonality in protozoan pathogens—Truth or artefact? Mol Ecol. 2014;23(17): 4195–4202. doi: 10.1111/mec.12872 [DOI] [PubMed] [Google Scholar]
- 14.Rougeron V, de Meeûs T, Ouraga SK, Hide M, Bañuls AL. “Everything you always wanted to know about sex (but were afraid to ask)” in Leishmania after two decades of laboratory and field analyses. PLoS Pathog; 2010;6(8):e1001004. doi: 10.1371/journal.ppat.1001004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rougeron V, De MeeÛs T, Bañuls AL. A primer for Leishmania population genetic studies. Trends Parasitol. 2015;31(2): 52–59. doi: 10.1016/j.pt.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 16.Jenni L, Marti S, Schweizer J, Betschart B, Le Page RWF, Wells JM, et al. Hybrid formation between African trypanosomes during cyclical transmission. Nature. 1986;322(6075): 173–175. doi: 10.1038/322173a0 [DOI] [PubMed] [Google Scholar]
- 17.Gibson W, Whittington H. Genetic exchange in Trypanosoma brucei: Selection of hybrid trypanosomes by introduction of genes conferring drug resistance. Mol Biochem Parasitol. 1993;60(1): 19–26. doi: 10.1016/0166-6851(93)90024-r [DOI] [PubMed] [Google Scholar]
- 18.Gibson W, Garside L, Bailey M. Trisomy and chromosome size changes in hybrid trypanosomes from a genetic cross between Trypanosoma brucei rhodesiense and T. b. brucei. Mol Biochem Parasitol. 1992;51(2): 189–199. doi: 10.1016/0166-6851(92)90069-v [DOI] [PubMed] [Google Scholar]
- 19.Peacock L, Ferris V, Bailey M, Gibson W. Mating compatibility in the parasitic protist Trypanosoma brucei. Parasit Vectors. 2014;7: 78. doi: 10.1186/1756-3305-7-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bingle LEH, Eastlake JL, Bailey M, Gibson WC. A novel GFP approach for the analysis of genetic exchange in trypanosomes allowing the in situ detection of mating events. Microbiology. 2001;147(Pt 12): 3231–3240. doi: 10.1099/00221287-147-12-3231 [DOI] [PubMed] [Google Scholar]
- 21.Gibson W, Peacock L, Ferris V, Williams K, Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasit Vectors. 2008;1(1): 4. doi: 10.1186/1756-3305-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock L, Ferris V, Bailey M, Gibson W. Intraclonal mating occurs during tsetse transmission of Trypanosoma brucei. Parasit Vectors. 2009;2(1): 43. doi: 10.1186/1756-3305-2-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N, Lawyer P, et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324(5924): 265–268. doi: 10.1126/science.1169464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inbar E, Akopyants NS, Charmoy M, Romano A, Lawyer P, Elnaiem DEA, et al. The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 2013;9(7):e1003672. doi: 10.1371/journal.pgen.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvo-Álvarez E, Álvarez-Velilla R, Jiménez M, Molina R, Pérez-Pertejo Y, Balaña-Fouce R, et al. First evidence of intraclonal genetic exchange in trypanosomatids using two Leishmania infantum fluorescent transgenic clones. PLoS Negl Trop Dis. 2014;8(9):e3075. doi: 10.1371/journal.pntd.0003075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romano A, Inbar E, Debrabant A, Charmoy M, Lawyer P, Ribeiro-Gomes F, et al. Cross-species genetic exchange between visceral and cutaneous strains of Leishmania in the sand fly vector. Proc Natl Acad Sci U S A. 2014;111(47): 16808–16813. doi: 10.1073/pnas.1415109111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louradour I, Ferreira TR, Ghosh K, Shaik J, Sacks D. In vitro generation of Leishmania hybrids. Cell Rep. 2020;31(2):107507. doi: 10.1016/j.celrep.2020.03.071 [DOI] [PubMed] [Google Scholar]
- 28.Telittchenko R, Descoteaux A. Study on the occurrence of genetic exchange among parasites of the Leishmania mexicana complex. Front Cell Infect Microbiol. 2020;10: 607253. doi: 10.3389/fcimb.2020.607253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin RH, Lai DH, Zheng LL, Wu J, Lukeš J, Hide G, et al. Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen. Parasit Vectors. 2015;8:665. doi: 10.1186/s13071-015-1281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson W, Garside L. Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol Biochem Parasitol. 1990;42(1): 45–53. doi: 10.1016/0166-6851(90)90111-x [DOI] [PubMed] [Google Scholar]
- 31.Turner CMR, Hide G, Buchanan N, Tait A. Trypanosoma brucei: Inheritance of kinetoplast DNA maxicircles in a genetic cross and their segregation during vegetative growth. Exp Parasitol. 1995;80(2): 234–241. doi: 10.1006/expr.1995.1029 [DOI] [PubMed] [Google Scholar]
- 32.Calvo-Álvarez E, Stamatakis K, Punzón C, Álvarez-Velilla R, Tejería A, Escudero-Martínez JM, et al. Infrared fluorescent imaging as a potent tool for in vitro, ex vivo and in vivo models of visceral leishmaniasis. PLoS Negl Trop Dis. 2015;9(3): e0003666. doi: 10.1371/journal.pntd.0003666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo-Álvarez E, Guerrero NA, Álvarez-Velilla R, Prada CF, Requena JM, Punzón C, et al. Appraisal of a Leishmania major strain stably expressing mCherry fluorescent protein for both in vitro and in vivo studies of potential drugs and vaccine against cutaneous leishmaniasis. PLoS Negl Trop Dis. 2012;6(11): e1927. doi: 10.1371/journal.pntd.0001927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rey GJ, Ferro C, Bello FJ. Establishment and characterization of a new continuous cell line from Lutzomyia longipalpis (Diptera: Psychodidae) and its susceptibility to infections with arboviruses and Leishmania chagasi. Mem Inst Oswaldo Cruz. 2000;95(1): 103–110. doi: 10.1590/s0074-02762000000100017 [DOI] [PubMed] [Google Scholar]
- 35.Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128(2): 217–228. doi: 10.1016/s0166-6851(03)00079-3 [DOI] [PubMed] [Google Scholar]
- 36.McCall LI, Matlashewski G. Involvement of the Leishmania donovani virulence factor A2 in protection against heat and oxidative stress. Exp Parasitol. 2012;132(2): 109–115. doi: 10.1016/j.exppara.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 37.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HDFH, Presber W, et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47(1): 349–358. doi: 10.1016/s0732-8893(03)00093-2 [DOI] [PubMed] [Google Scholar]
- 38.Kaufer A, Ellis J, Stark D. Identification of clinical infections of Leishmania imported into Australia: Revising speciation with polymerase chain reaction-RFLP of the kinetoplast maxicircle. Am J Trop Med Hyg. 2019;101: 590–601. doi: 10.4269/ajtmh.19-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha RF, Menezes E V., Xavier AREO, Royo VA, Oliveira DA, Júnior AFM, et al. Standardization of a molecular method for epidemiologic identification of Leishmania strains. Genet Mol Res. 2016;15(4): doi: 10.4238/gmr.15048854 [DOI] [PubMed] [Google Scholar]
- 40.Brenière SF, Telleria J, Bosseno MF, Buitrago R, Bastrenta B, Cuny G, et al. Polymerase chain reaction-based identification of New World Leishmania species complexes by specific kDNA probes. Acta Trop. 1999;73(3): 283–293. doi: 10.1016/s0001-706x(99)00025-x [DOI] [PubMed] [Google Scholar]
- 41.Thiel T, Kota R, Grosse I, Stein N, Graner A. SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 2004;32(1): e5. doi: 10.1093/nar/gnh006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Quan N. Immune cell isolation from mouse femur bone marrow. Bio Protoc. 2015;5(20): e1631. doi: 10.21769/bioprotoc.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legarda D, Ting AT. Analysis of necroptosis in bone marrow-derived macrophages. Methods Mol Biol. 2018;1857: 63–70. doi: 10.1007/978-1-4939-8754-2_6 [DOI] [PubMed] [Google Scholar]
- 44.Sadlova J, Yeo M, Seblova V, Lewis MD, Mauricio I, Volf P, et al. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One. 2011;6(5): e19851. doi: 10.1371/journal.pone.0019851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inbar E, Shaik J, Iantorno SA, Romano A, Nzelu CO, Owens K, et al. Whole genome sequencing of experimental hybrids supports meiosis-like sexual recombination in Leishmania. PLoS Genet. 2019;15(5): e1008042. doi: 10.1371/journal.pgen.1008042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garin YJF, Meneceur P, Pratlong F, Dedet JP, Derouin F, Lorenzo F. A2 gene of Old World cutaneous Leishmania is a single highly conserved functional gene. BMC Infect Dis. 2005;5:18. doi: 10.1186/1471-2334-5-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCall LI, Matlashewski G. Localization and induction of the A2 virulence factor in Leishmania: Evidence that A2 is a stress response protein. Mol Microbiol. 2010;77: 518–530. doi: 10.1111/j.1365-2958.2010.07229.x [DOI] [PubMed] [Google Scholar]
- 48.Oliveira LF, Schubach AO, Martins MM, Passos SL, Oliveira R V., Marzochi MC, et al. Systematic review of the adverse effects of cutaneous leishmaniasis treatment in the New World. Acta Trop. 2011;118(2): 87–96. doi: 10.1016/j.actatropica.2011.02.007 [DOI] [PubMed] [Google Scholar]
- 49.Sterkers Y, Lachaud L, Crobu L, Bastien P, Pagès M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol. 2011;13(2): 274–283. doi: 10.1111/j.1462-5822.2010.01534.x [DOI] [PubMed] [Google Scholar]
- 50.Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P, Pagès M. Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol Microbiol; 2012;86(1): 15–23. doi: 10.1111/j.1365-2958.2012.08185.x [DOI] [PubMed] [Google Scholar]
- 51.Shaik J, Dobson D, Sacks D, Beverley SM. Leishmania sexual reproductive strategies as resolved through computational methods designed for aneuploid genomes. Genes; 2021; 12(2): 167. doi: 10.3390/genes12020167 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.