Abstract
An fljB-negative, multidrug-resistant Salmonella enterica serovar 4,5,12:i:− phage type DT U302 strain (resistant to ampicillin, chloramphenicol, sulfonamide, gentamicin, streptomycin, tetracycline, and sulfamethoxazole-trimethoprim) emerged and spread in Spain in 1997. Sequences specific for Salmonella serovar Typhimurium and phage type DT 104 and U302 were present in this atypical Salmonella strain, suggesting that it is a monophasic Salmonella serovar Typhimurium variant.
Salmonella enterica is responsible for the majority of food-borne cases of disease worldwide. The epidemiological marker generally used for the identification of S. enterica is serotyping. Knowledge of serovar distributions allows the detection of new serovars, an increase in the frequency of already known serovars, and the geographical and temporal distributions of serovars.
An atypical S. enterica subsp. enterica strain of serovar 4,5,12:i:− emerged and spread in Spain in 1997 (5). The main strain characteristics are lysis by phage 10, recently incorporated into the Salmonella serovar Typhimurium phage typing scheme (phage type DT U302) (1); resistance to ampicillin, chloramphenicol, sulfonamide, gentamicin, streptomycin, tetracycline, and sulfamethoxazole-trimethoprim; and the absence of second-phase flagellar antigen by detection by either slide agglutination or specific PCR amplification (5).
According to the Kauffman-White scheme, the strain could be either a Salmonella serovar Typhimurium strain (serovar 4,5,12:i:1,2), a serovar Lagos strain (serovar 4,5,12:i:1,5), a monophasic variant, or a new serovar (9). Monophasic Salmonella strains could represent ancestral forms which did not acquire a second flagellar antigen or the necessary switching mechanism during evolution. Alternatively, they could originate as mutants of biphasic strains which have lost the switching mechanism, either the fliC or the fljB flagellar gene, or the ability to express one of these genes (4).
Until 1981, Salmonella serovar Typhimurium was the most frequently isolated serovar in Spain. Today, it is the second most frequently isolated serovar (12). Salmonella serovar Lagos has never been detected in Spain. If Salmonella serovar 4,5,12:i:− is a monophasic variant, it is most likely a Salmonella serovar Typhimurium variant.
The selective advantage of multidrug resistance can probably be one of the factors that has influenced the extension of this strain in Spain, which became the fourth most common serovar during the period from 1998 to 2000 (12, 13).
These Salmonella serovar 4,5,12:i:− isolates possess two or three small cryptic plasmids together with a large 140-kb spvC (Salmonella plasmid virulence gene)-positive or 120-kb spvC-negative plasmid (2). The 140- or 120-kb plasmid could be derived from the Salmonella serovar Typhimurium-associated 90-kb virulence plasmid by the acquisition of class I integrons (7).
To determine the reason why second-phase flagellar antigens were not expressed in these isolates, PCR amplification of a selection of these monophasic strains was carried out to selectively amplify the total or partial second-phase (fljB) flagellar gene. PCR was performed as described previously with primers sense-56, sense-60, antisense-58, and antisense-83 (14) and sense-F1, antisense-R5, antisense-R6, antisense-R7, and antisense-R1 (6). No amplification of the 18 different possible fragments of the fljB gene was obtained. This result indicates the absence of the fljB gene.
To determine if Salmonella serovar 4,5,12:i:− is a Salmonella serovar Typhimurium monophasic variant, two type-specific sequences of Salmonella serovar Typhimurium and phage type DT 104 and U302, respectively, were investigated.
Thirteen multidrug-resistant (resistant to ampicillin, chloramphenicol, sulfonamide, gentamicin, streptomycin, tetracycline, and sulfamethoxazole-trimethoprim) Salmonella serovar 4,5,12:i:− (fljB- negative) strains were selected. One Salmonella serovar Typhimurium phage type LT2, two Salmonella serovar Typhimurium phage type DT 104, and two Salmonella serovar Typhimurium phage type DT U302 strains were selected as control strains. Nineteen different Salmonella serovars were selected as negative controls (Table 1).
TABLE 1.
Serogroup, serovar, and PCR results for 37 isolates of S. enterica used in the present study
| Serogroup | Serovar | No. of strains | Size (by) of fliB-fliA intergenic region | Presence of 162-bp amplicona |
|---|---|---|---|---|
| B | 4, 5, 12:i:− | 13 | 1,000 | + |
| Typhimurium LT2 | 1 | 1,000 | − | |
| Typhimurium DT 104 | 2 | 1,000 | + | |
| Typhimurium DT U302 | 2 | 1,000 | + | |
| Lagos | 1 | 250 | − | |
| Agona | 1 | 250 | − | |
| Brandenburg | 1 | 250 | − | |
| Derby | 1 | 250 | − | |
| 4, 12:b:− subsp. II | 1 | 250 | − | |
| C1 | Infantis | 1 | 250 | − |
| Mbandaka | 1 | 250 | − | |
| Mikawasima | 1 | 250 | − | |
| Montevideo | 1 | 250 | − | |
| Ohio | 1 | 250 | − | |
| Rissen | 1 | 250 | − | |
| Virchow | 1 | 250 | − | |
| C2 | Goldcoast | 1 | 250 | − |
| Hadar | 1 | 250 | − | |
| Muenchen | 1 | 250 | − | |
| D | Enteritidis | 1 | 250 | − |
| E1 | Anatum | 1 | 250 | − |
| E4 | Senftenberg | 1 | 250 | − |
| O:61 | 61:lv:z subsp. IIIb | 1 | 250 | − |
+, present; −, absent.
Salmonella serovar Typhimurium strains harbor a specific IS200 fragment within the flagellin gene cluster. The IS200 fragment is located downstream of the fliB gene and upstream of the noncoding fliA gene region. This location is Salmonella serovar Typhimurium specific (3).
The fliB-fliA intergenic regions of all Salmonella strains tested were amplified with two primers, primers FFLIB (5′-CTGGCGACGATCTGTCGATG-3′) and RFLIA (5′-GCGGTATACAGTGAATTCAC-3′), that anneal with the last fliB gene nucleotides and the first fliA gene nucleotides, respectively. The primers were designed by using already published sequences (3, 8). PCR amplification was performed with a Ready-to-Go system (Amersham Pharmacia Biotech Inc.). Five microliters of a strain suspension boiled for 10 min was used as a template. PCR amplification was as follows: denaturation at 95°C for 5 min; 30 cycles of 95°C for 1 min, 58°C for 1 min, and 72°C for 2 min; and 1 final extension cycle at 72°C for 7 min. Fragments were separated in a 3% agarose gel by unidirectional electrophoresis. The fragments were visualized by staining with ethidium bromide.
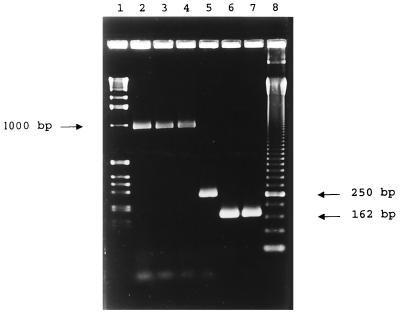
PCR amplification of the fliB-fliA intergenic region of Salmonella serovar 4,5,12:i:− generated a 1,000-bp fragment, similar to the fragments from Salmonella serovar Typhimurium phage types LT2, DT 104, and DT U302. All 19 different Salmonella serovars tested, including Salmonella serovar Lagos, generated a 250-bp fragment, similar as those described previously (3) (Fig. 1).
FIG. 1.
PCR amplification products of Salmonella isolates analyzed. Lanes 2 through 5, fliB-fliA intergenic region of Salmonella serovar 4,5,12:i:−, serovar Typhimurium phage type LT2, serovar Typhimurium phage type DT U302, and serovar Brandenburg (3), respectively; lanes 6 and 7, phage type DT 104 and U302 162-bp specific amplicon of Salmonella serotype 4,5,12:i:− and serotype Typhimurium phage type DT U302, respectively (10); lane 1, molecular weight marker X (Roche Molecular Biochemicals); lane 8, 50-bp ladder (Amersham Pharmacia Biotech Inc.).
The amplification products from four Salmonella serovar 4,5,12:i:− strains and Salmonella serovar Typhimurium phage type LT2 were purified and sequenced by using primers FFLIB and RFLIA with an Applied Biosystems ABI PRISM377 DNA Sequenator and the Taq Dye Deoxy Terminator cycle sequencing kit (Applied Biosystems/Perkin-Elmer). Sequence analysis was performed with PC-Gene software (Intelligenics Inc.). All five sequences were identical and harbored the Salmonella serovar Typhimurium IS200-specific fragment already described (3). The IS200 element obtained (i) started 38 nucleotides downstream of the stop codon of the fliB gene, (ii) lacked a terminal inverted repeat, (iii) had two additional base pairs at the 5′ end, and (iv) had an additional C at nucleotide 1542.
PCR amplification of a 162-bp Salmonella serovar Typhimurium phage type DT 104- and U302-specific region has been used as a rapid technique for the identification of both phage types. This fragment was also obtained from 15 Salmonella strains belonging to 6 different serovars among the 36 Salmonella serovars studied. This fragment can be considered specific for Salmonella serovar Typhimurium phage types DT 104 and U302 (10).
Because bacteriophages are serovar specific and because Salmonella serovar Typhimurium phage 10 was able to lyse Salmonella serovar 4,5,12:i:−, the strain would probably be a monophasic variant of Salmonella serovar Typhimurium phage type DT U302.
All Salmonella 4,5,12:i:− and Salmonella serovar Typhimurium phage type DT 104 and U302 strains tested amplified a 162-bp fragment (Fig. 1). Salmonella serovar Typhimurium phage type LT2, Salmonella serovar Lagos, and 18 other Salmonella serovars tested did not express any fragment, as expected. Fragments of 162 bp from four Salmonella 4,5,12:i:− strains were sequenced, with sequences identical to those of Salmonella serovar Typhimurium phage types DT 104 and U302 published previously being obtained (10).
The present study demonstrates the presence of the IS200-IV fragment (11) in the intergenic fliB-fliA flagellin cluster region in all Salmonella serovar 4,5,12:i:− strains tested. This fragment is located at the same position at which it is located in Salmonella serovar Typhimurium and has the same sequence as Salmonella serovar Typhimurium (3). It was also possible to amplify a 162-bp fragment that was similar in size and sequence to those from Salmonella serovar Typhimurium phage types DT 104 and U302.
We conclude that the emerging fljB-negative Salmonella serovar 4,5,12:i:− phage type DT U302 strain resistant to ampicillin, chloramphenicol, sulfonamide, gentamicin, streptomycin, tetracycline, and sulfamethoxazole-trimethoprim should be considered a Salmonella serovar Typhimurium 5+ monophasic variant.
REFERENCES
- 1.Anderson E S, Ward L R, Saxe M J, de Sa J D. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd E F, Hartl D L. Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics. 1998;149:1183–1190. doi: 10.1093/genetics/149.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnens A P, Stanley J, Sack R, Hunziker P, Brodard I, Nicolet J. The flagellin N-methylase gene fliB and an adjacent serovar-specific IS200 element in Salmonella typhimurium. Microbiology. 1997;143(Pt 5):1539–1547. doi: 10.1099/00221287-143-5-1539. [DOI] [PubMed] [Google Scholar]
- 4.Burnens A P, Stanley J, Sechter I, Nicolet J. Evolutionary origin of a monophasic Salmonella serovar, 9,12:l,v:−, revealed by IS200 profiles and restriction fragment polymorphisms of the fljB gene. J Clin Microbiol. 1996;34:1641–1645. doi: 10.1128/jcm.34.7.1641-1645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echeita M A, Aladuena A, Cruchaga S, Usera M A. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:− strain in Spain. J Clin Microbiol. 1999;37:3425. doi: 10.1128/jcm.37.10.3425-3425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echeita M A, Usera M A. Rapid identification of Salmonella spp. phase 2 antigens of the H1 antigenic complex using “multiplex PCR.”. Res Microbiol. 1998;149:757–761. doi: 10.1016/s0923-2508(99)80022-9. [DOI] [PubMed] [Google Scholar]
- 7.Guerra B, Soto S M, Argüelles J M, Mendoza M C. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:−] Antimicrob Agents Chemother. 2001;45:1305–1308. doi: 10.1128/AAC.45.4.1305-1308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikebe T, Iyoda S, Kutsukake K. Structure and expression of the fliA operon of Salmonella typhimurium. Microbiology. 1999;145(Pt 6):1389–1396. doi: 10.1099/13500872-145-6-1389. [DOI] [PubMed] [Google Scholar]
- 9.Popoff M Y, Le Minor L. Antigenic formulas of the Salmonella serovars, 7th revision. WHO Collaborating Centre for Reference Research on Salmonella. Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 10.Pritchett L C, Konkel M E, Gay J M, Besser T E. Identification of DT104 and U302 phage types among Salmonella enterica serotype Typhimurium isolates by PCR J. Clin Microbiol. 2000;38:3484–3488. doi: 10.1128/jcm.38.9.3484-3488.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanderson K E, Sciore P, Liu S L, Hessel A. Location of IS200 on the genomic cleavage map of Salmonella typhimurium LT2 J. Bacteriol. 1993;175:7624–7628. doi: 10.1128/jb.175.23.7624-7628.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usera M A, Aladueña A, Diez R, de la Fuente M, Gutierrez F, Cerdán R, Echeita A. Análisis de las cepas de Salmonella spp aisladas de muestras clínicas de origen humano en España en el año 1999 (I) Bol Epidemiol Semanal. 2000;8:45–48. [Google Scholar]
- 13.Usera M A, Aladueña A, Diez R, de la Fuente M, Gutierrez F, Cerdán R, Echeita A. Análisis de las cepas de Salmonella spp aisladas de muestras de origen no humano en España en el año 1999. Bol Epidemiol Semanal. 2000;8:133–144. [Google Scholar]
- 14.Vanegas R A, Joys T M. Molecular analyses of the phase-2 antigen complex 1,2,.. of Salmonella spp. J Bacteriol. 1995;177:3863–3864. doi: 10.1128/jb.177.13.3863-3864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]