Abstract
Vaults are ubiquitous ribonucleoprotein particles involved in a diversity of cellular processes, with promising applications as nanodevices for delivery of multiple cargos. The vault shell is assembled by the symmetrical association of multiple copies of the major vault protein that, initially, generates half vaults. The pairwise, anti-parallel association of two half vaults produces whole vaults. Here, using a combination of vault recombinant reconstitution and structural techniques, we characterized the molecular determinants for the vault opening process. This process commences with a relaxation of the vault waist, causing the expansion of the inner cavity. Then, local disengagement of amino-terminal domains at the vault midsection seeds a conformational change that leads to the aperture, facilitating access to the inner cavity where cargo is hosted. These results inform a hitherto uncharacterized step of the vault cycle and will aid current engineering efforts leveraging vault for tailored cargo delivery.
Cryo-EM reveals the mechanism of vault opening by disruption of D39 symmetry.
INTRODUCTION
Cytoplasmic segmentation allows cells to derive essential tasks into specialized compartments. Especially in eukaryotes, this evolutionary strategy bestowed cells with a network of highly interconnected compartments that maximized fitness and allowed cellular specialization (1). The concomitant gain on efficiency brought by compartmentalization came at the cost of the need of transport systems, dedicated to shuffling cargo between compartments. The diversity and reach of cellular transport systems are large, with multiple and redundant routes for distribution of the various and specialized cargo (2). The use of protein “cages” assembled by the self-association of proteins is a widespread and cost-effective solution for the encapsulation and delivery of payload in cells (3). Vault particles are cargo delivery nanodevices self-assembled from polypeptides with wide cellular distribution (Fig. 1A) (4). Vaults present an ample phylogenetic presence across eukaryotes, hinting at an essential biological role (Fig. 1A), which, however, remains poorly understood (5, 6).
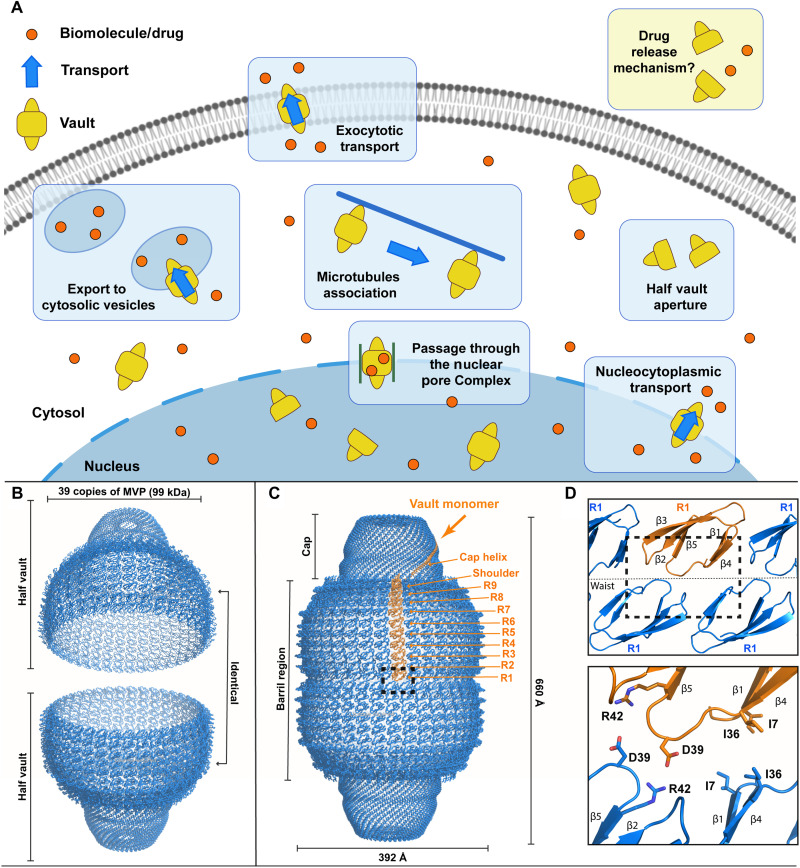
Fig. 1. Suggested cellular vault functions, general architecture, and MVP domain organization.
(A) Schematics representing proposed vault functions related to transport of variable cargo in the cell. (B) Schematics showing two half vault particles with dimensions indicated. (C) Structure of whole vaults with one MVP monomer colored orange (PDB ID: 4HL8) with its constituent domains explicitly labeled. (D) Top: Detailed view of R1 domains located at the interface of the two half vaults. Bottom: Putative residues involved in the stabilization of the whole vault state.
Mature vaults consist of the association of two half vaults into a particle with a molecular weight of around 10 MDa and with overall dimensions of 40 nm × 40 nm × 70 nm (Fig. 1, B and C) (7). The major vault protein (MVP) is the main component of vaults, with a mass of around 100 kDa (8). Self-oligomerization of multiple MVP monomers generates half vaults (Fig. 1, B and C), which will eventually associate into whole vaults (9). In mammals, additional components can be found in mature vaults, like the 193-kDa vault poly(adenosine 5′-diphosphate) ribose polymerase (VPARP) protein (10) or the 290-kDa telomerase-associated protein 1 (TEP1) (11). In addition, small untranslated RNAs have been found to be associated with vault particles, with numbers varying among species (12, 13).
Structures of native vaults and truncated versions of MVP monomers were obtained by x-ray crystallography techniques, revealing the architectural basis of the particle and the domains integrated in the MVP monomer (Fig. 1, C and D) (14, 15). The MVP monomer is thus organized into 12 domains, with 9 structural repeat domains at the N terminus (R domains, R1 to R9; Fig. 1C, orange) followed by an α/β shoulder domain, a cap helix, and a C-terminal cap-ring domain. Thirty-nine copies of MVP self-associate in a symmetrical arrangement, generating a half vault (Fig. 1B).
The vault particle architecture immediately suggests a built-in mechanism for cargo encapsulation and delivery, namely, a cycle of vault opening/closing by coupling/uncoupling of half vault pairs. The ability of half vaults to associate into whole vaults and disengage following a functional opening/closing cycle was demonstrated early, and a putative pH-dependent mechanism mediating disassembly was proposed (16). However, in vivo experiments demonstrated that vault particles are capable of half vault exchange at neutral pH (17), which would imply a pH-independent mechanism for vault opening/closing. Efforts on clarifying the role of pH in the opening/closing cycle led to studies by atomic force microscopy (AFM) and complementary biophysical techniques, which revealed that acidic pH weakens the MVP-MVP lateral association rather than the vault midsection (18). Overall, these results suggest a scenario where vaults explore multiple conformational states, with built-in dynamics integrated by numerous axis of variation: half vault association/dissociation, whole vault enlargement/contraction (19), and the fracture/recuperation ability observed in the vault shell (20) are all described vault dynamics.
With the aim to delineate the molecular principles governing the opening/closing cycle, we leveraged a combination of biophysical/structural techniques with the production of recombinant mutant vaults. This approach revealed precise insights into how vault particles operate.
RESULTS
Vaults populate multiple states in solution
Single-particle cryo–electron microscopy (cryo-EM) analysis with image processing and in silico classification techniques revealed multiple vault populations in solution. Three different vault states were identified, corresponding to a fully closed (engaged) vault, the hitherto uncharacterized half vault state, and an additional whole vault state poised at the beginning of the opening cycle.
On a first instance and to obtain a high-resolution reconstruction using recombinant vault particles, we determined the cryo-EM structure of the whole vault particle imposing D39 symmetry (fig. S1). This high-resolution map [with an overall resolution of 3.7 Å using the Fourier shell correlation (FSC) 0.143 criteria] exhibited features compatible with the reported resolution, with side chains clearly visible in the best areas of the map (Fig. 2A). This map also allowed confident model building and model refinement for all MVP monomers and their domains, including domains R1 to R9, the α/β shoulder, and the 42-turn cap helix (fig. S2). This high-resolution vault reconstruction was used as a reference to analyze the differences when compared to the crystal structure of native vault particles (14, 15) and to the cryo-EM structure of recombinant vaults solved previously (19).
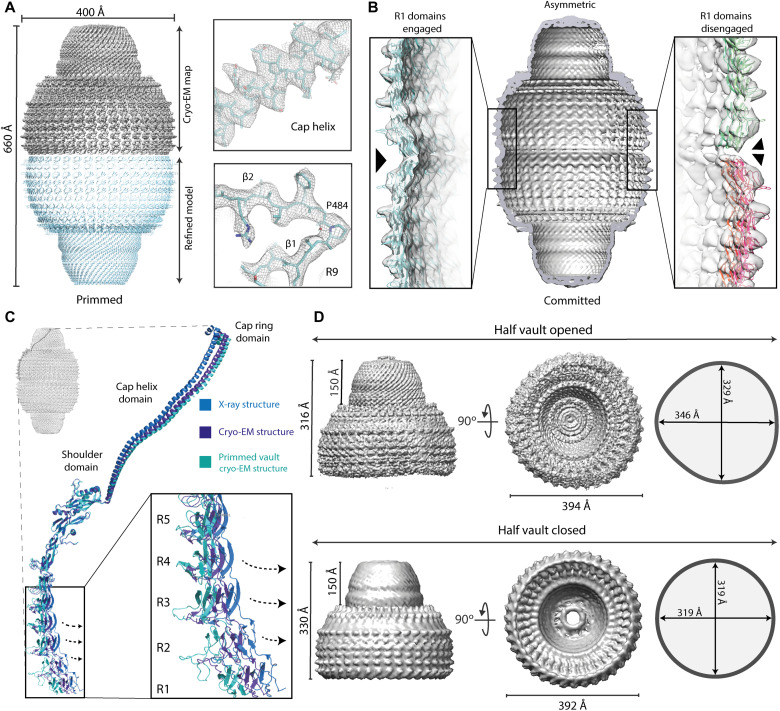
Fig. 2. Cryo-EM structures of multiple vault conformations in solution.
(A) Top left: Cryo-EM density for the high-resolution vault reconstruction with D39 symmetry imposed. Bottom: The corresponding refined atomic model derived from this map. Clear side chains could be identified in the map, especially around the cap helix and the R9 domain (right). (B) A second class of whole vault particles showed an asymmetrical configuration, with R1 domains from top and bottom half vaults engaged on one side (left) and disengaged at the other side (right). (C) While the overall conformation of MVP monomers is maintained in both symmetrical and asymmetrical reconstructions, the orientation of domains R1 to R5 relative to the N-terminal helix is changed, indicating a built-in structural variability in the whole vault particle affecting the volume of the internal cavities. (D) Top: A third reconstruction of free half vaults was identified in the datasets, exhibiting class distinctive architectural features, with a noncircular, oval contour. Dimensions for the cap and the waist cavity for the half vault reconstruction are indicated in two orthogonal views. For comparison, similar views from a map derived from the x-ray structure of the whole vault are shown at the bottom (PDB ID: 4HL8).
A global analysis of the volumes of the internal cavities defined by the assembled whole vault particle revealed further differences when compared with the crystalized structure of native particles. These differences encompass an expansion in the diameter of the barrel region in our cryo-EM reconstruction (~1.5% wider), with an average diameter of 400 Å (332-Å waist diameter), while in the native, crystallized particles, this value is 392 Å (318-Å waist diameter). However, the main architecture of the vault is generally maintained, especially at the cap region. This suggests that fully assembled vaults exhibit an intrinsic plasticity affecting the shape and volume of the enclosed internal cavity defined by MVP monomers. Further analysis, comparing the relative disposition of individual domains within MVP monomers, revealed the origin of this plasticity, which can be explained by variations in two regions of the vault: a change on the degree of curvature of the vault barrel and a distinctive orientation of the cap helices (Fig. 2C). These changes globally affect the architecture of the particle and are explained by local repositioning of MVP domains. Thus, the curvature of the barrel domains is caused by changes around Gln378, within the loop connecting domains R7 and R8. The change in the trajectory of these barrel domains, when compared to the crystalized state, induces a “relaxed” conformation of the whole barrel region. For the cap helix, a pivoting movement around residue Pro645, at the base of the helix, can be noted when compared to the crystal structure. A small change at the base of the helix is amplified at the distal end, as a small change at the base is leveraged due to the long length of the helix. These differences recapitulate previously reported discrepancies between vault cryo-EM [Protein Data Bank (PDB) ID: 6BP8, “conformation 1”] and x-ray (PDB ID: 4HL8) structures. Both our cryo-EM model and that reported by Ding et al. (19) are similar and different from the crystal structure, but differences between cryo-EM reconstructions themselves can also be noted, especially in the magnitude on the displacements of the regions previously described. Our reconstruction exhibits a wider, expanded cavity, with a relaxed conformation of the barrel domain.
Maximum likelihood classification methods implemented in RELION (21) allowed the identification of a group of particles deviating from the symmetrical arrangement of the “closed” whole vault state. This deviation was evident in the region of the waist, and we could characterize it aligning this class of particles without imposing any symmetrical condition (fig. S3). The obtained cryo-EM map showed an overall monomer organization similar to the one described for the symmetrical reconstruction. However, differences could be identified around domains R1 and R2 in only one region of the waist (Fig. 2B). This local, asymmetric distortion is a consequence of disruptions on the reciprocal R1 domain interactions from opposite half vaults (Fig. 2B, right). However, the remainder of the MVP monomers maintain a similar configuration as in the symmetric, closed vault particle state described above (Fig. 2B, left).
Cryo-EM structures of half vaults in solution
The large size of the dataset collected was instrumental to localized additional vault states. Visual inspection of the aligned micrographs and the reference-free two-dimensional (2D) class averages revealed the presence of particles that resemble half vaults (fig. S1). Visualization of top views revealed a distortion of the circular shape of the vault waist, suggesting an oval contour configuration rather than a circular one as in the closed whole vault state. A variety of views, in multiple orientations, allowed us to obtain the 3D reconstruction of these particles (fig. S1). The resulting map revealed structural changes responsible for the oval waist contour, which are not restricted to the waist, as they also affect the central region of the vault barrel. The main structural variation found is a reorganization of the MVP monomers in three clusters of 13 monomers each, while the half vault overall configuration is maintained.
The overall fold of the MVP monomers in the context of the half vault is similar to that of the entire particle (Fig. 2D, top). The individual N-terminal domains from R3 to R9 of the barrel, the shoulder, and the cap helix and cap ring at the top of the structure match those of the whole vault state. However, a flexibility locus within the half vault state was identified around domains R1 and R2, which could not be modeled due to the lack of ordered density in this region.
The existence of differences in the waist region between the isolated half vault and the whole “closed” vault is also evident (Fig. 2D). These differences affect not only the shape of the waist but also its dimensions. The radius of the circular waist of the closed conformation is 319 Å, whereas, in the half vault, the waist, adopting the mentioned oval conformation, exhibits a maximum radius of 346 Å and a minimum of 329 Å. When compared with the crystal structure, the half vault radius is also 14 Å shorter than the equivalent region on the crystal structure.
The structure of isolated half vaults revealed previously uncharacterized dynamics in the half vault state, especially for MVP domains R1 and R2. This conformational freedom will be restricted in the assembled vault, where R1 domains from top and bottom half vaults engage in reciprocal interactions.
A cluster of residues in MVP R1 domain is critical for vault assembly and stabilization
To further characterize the vault opening/closing cycle, we took advantage of a recombinant system to produce site-specific mutants. Our structural analysis by cryo-EM revealed unexpected dynamics at the edge of the half vault (MVP R1 domains), suggesting a critical role for this region in the regulation of vault opening. We designed mutants targeting specific residues of the MVP R1 domain and characterize the impact these mutations have on the stability of vaults (fig. S4). The high-resolution crystallographic structure of truncated MVPs with seven N-terminal domains R1 to R7 (22) provided clues regarding the identity of R1 residues involved in the stability of vaults (Fig. 1D). A salt bridge established between the universally conserved residues Asp39 and Arg42 appeared to be a relevant contact. In addition, a cluster of hydrophobic residues (Ala6, Ile7, and Ile36) seemed to form a large contact surface. We thus designed three different types of mutants: a D39A mutant to disrupt the single salt bridge, a I7K mutant to interfere with the hydrophobic cluster, and, lastly, a quadruple mutant, D39K/I7N/E4K/E5K, adding additional mutations flanking residue I7 to fully abolish this interaction surface. How these mutations affect vault stability was monitored by three different and complementary experimental approaches: negative staining EM (Fig. 3A), AFM (Fig. 3B), and differential scanning fluorimetry (DSF; Fig. 3C).
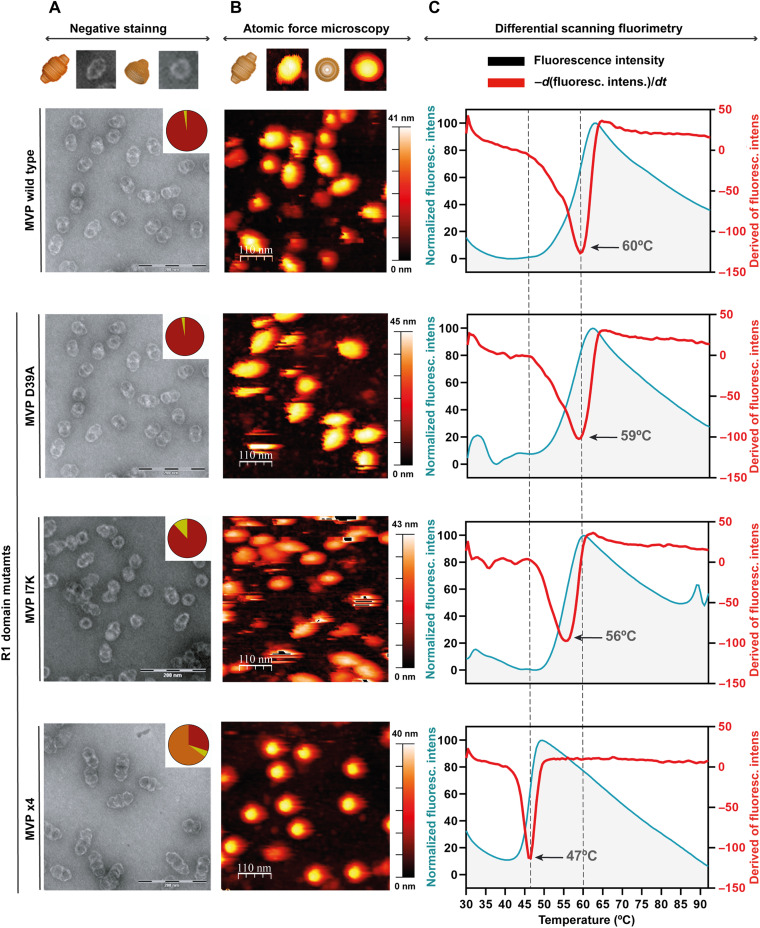
Fig. 3. Mutant vaults revealed the importance of R1 domain residues facing the half vault interface.
(A) Representative EM images of wild-type vaults and the three mutant vaults in the R1 domain described in the main text. The top diagram assists in the identification of the views seen on the micrographs. The top right corner of each micrograph includes a pie chart, showing the distribution of whole (red) versus half vaults (yellow). The quadruple mutant causes a catastrophic effect, interfering with vault assembly and yielding nonfunctional, pseudo-filament arrangements of whole vaults and half vault multimers (orange in the pie chart). (B) Corresponding AFM views of wild-type and mutant vaults. On top, a diagram with common whole vault and half vault views as seen in AFM. All R1 MVP mutants exhibit an altered behavior when compared to wild-type vaults under AFM conditions. (C) Fluorescent scanning anisotropy traces obtained at increasing temperatures from left to right for wild-type vaults and the described mutants. The I7K mutant and, especially, the quadruple mutant, with Tm of 56° and 47°, respectively, show the negative impact on whole vault stability when mutating MVP R1 residues facing the half vault interface.
Recombinant D39A mutant vaults exhibit a behavior by negative staining very similar to the wild type with ~98% of particles assembled as whole vaults. Quantification of whole versus half vaults per micrography for this mutant revealed 725 whole particles and 11 halves (22 halves counted in total), yielding 1.49% of particles as half vaults. Although the D39A substitution implies the loss of an electrostatic interaction, the hydrophobic contact between the two A39 side chains remains (fig. S5).
The MVP I7K mutant, designed to disrupt the hydrophobic cluster formed by residues Ala6, Ile7, and Ile36, clearly affect the whole vault/half vault ratio, increasing the fraction of half vaults to 12%.
The quadruple mutant showed an unexpected behavior when imaged by negative staining: A supramolecular structure resembling a filament of half vaults was visible. These “filaments” are constituted by one entire vault particle capped with one, two, or even three vault halves clustered on top of each other. Quantification across all micrographs revealed 30% of particles as whole particles, 5% as half vaults, and 65% of particles forming half vault filaments with one (313 particles), two (81 particles), or three halves (two particles). The experimental conditions used (see Materials and Methods) seemed to favor an alternative half vault/half vault arrangement in this mutant, involving contacts between the vault waist of one particle and the cap helix and shoulder domains of its partner (fig. S6).
Complementary to negative staining analysis, we performed an in-depth AFM characterization with the aim to confirm the imaging results via an orthogonal approach (Fig. 3B and table S1). Mutant vaults were attached to freshly cleaved ordered pyrolytic graphite, with measurements made in jumping mode, revealing vaults in three different orientations: whole particles lying on the barrel (reclined particles), whole particles lying on the cap, and half vaults adsorbed through the waist (20). To investigate the local effects caused by the introduced mutations, we performed individual nanoindentations on the reclined particles in each mutant to find differences in the force needed to break the particle in the two halves for the wild-type MVP and for the mutants. The single-indentation assay consists of perpendicularly indenting an individual vault with the tip. The tip is attached to a cantilever, whose bending is proportional to the force applied to the vault when it deforms (23). When the force applied to the vault reaches a certain value, the particle breaks, but the fracture never occurs at the midsection between the two halves; it, however, happens along the MVP subunits in the vault barrel, in the direction of the aligned MVPs (20). Single-indentation assays performed with different MVP mutants showed that, in all cases, the fracture occurred in the same direction, hampering the study of the effect of these mutations for the formation of half vaults. Alternatively, AFM assays are also capable of providing information about the stiffness of the particle through the calculation of the elastic constant (Ks) and breaking force (24). However, no measurable differences were observed between the Ks values for the wild type and mutants.
AFM imaging also allows an alternative quantification of vault population distribution. This approach revealed 75% of particles as whole particles and 25% as halves for wild-type vaults. This ratio slightly changed to 69.2% of whole vaults for the D39A substitution with 30.8% of half vaults. The I7K mutation affected this ratio, decreasing the percentage of whole vaults to 57% with 43% of halves. Last, the quadrupole MVP mutant exhibited a marked decrease in the number of whole vaults with only 6% of vaults found in this conformation.
We lastly assayed the stability of mutants and wild-type vaults by DSF (Fig. 3C). Fluorescence emission of wild-type MVP and mutants was recorded from 30° to 95°C, and the first derivative was calculated for each experiment (Fig. 3C, right). DSF data revealed changes in the Tm values, depending on the substitution introduced: Wild-type MVP exhibits a Tm of 60°C, very close to that of the D39A mutant (Tm = 59°C). However, vaults containing the I7K mutation affecting the hydrophobic cluster exhibit a 4° decrease in the Tm value (Tm = 56°C). Last, consistent with negative staining and AFM results, the quadruple MVP mutant exhibits the largest variation on Tm when compared to the wild type, Tm = 47°C.
DISCUSSION
Understanding the molecular determinants that govern vault dynamics is of utmost importance not only for deciphering the role these particles exert in multiple cellular processes but also for optimizing their use as engineered molecular transporters. Here, we have used a combination of recombinant vault reconstitution with cryo-EM and complementary biophysical methods to characterize the different states vault particles navigate along their functional cycle. The integrated results suggest a mechanistic model for the vault opening-closing cycle with functional implications for cargo encapsulation, transport, and delivery.
Fully engaged, closed whole vaults can be described by two levels of self-assembly. A first level consists of lateral associations of multiple MVP monomers to form half vaults. The reciprocal stabilization, in an anti-parallel configuration of two half vaults interacting through R1 domains, generates a whole vault particle. The self-association of half vaults represents the second level of self-assembly. In the whole vault state, an internal large cavity is defined, which is isolated from the exterior and hosts cargo. The molecular principle exploited by vaults as nanodevice transporters relies on the regulated transit from half to wholly assembled states that controls access to the internal cavity. The closed symmetrical vaults determined in the present work compared to those previously solved (7, 15, 19) revealed certain conformational plasticity in this state. This variability can be explained by concerted changes in the degree of curvature of the repeat domains forming the vault barrel, leading to variations in the width of the internal cavity, with the largest movements concentrated in the R1 domains (Fig. 2C).
The identification of a new class of fully assembled but locally distorted vaults (Fig. 2B and fig. S3) indicates the presence of an additional nonsymmetrical intermediate in the vault assembly/disassembly process, in which R1 domains locally engage and disengage on reciprocal contacts from opposite half vaults while maintaining the overall assembled vault architecture (Fig. 2B). The weakening of the MVP-MVP lateral contacts as a consequence of the vault barrel expansion would probably facilitate the structural alterations found in these particles (Fig. 2B). The intrinsic plasticity of the vault shell, seen in our structural comparisons, was also observed in vaults directly imaged in intact cells (25). Using whole-cell electron cryo-tomography, Woodward and colleagues described vaults located in the extreme periphery of the cytoplasm where they mainly associate with granule-like structures and actin (25) . In some cases, vaults were visualized opening at their waist to interact with the associated granules, showing a conformation closely related to that we have described here for the fully assembled, locally distorted particles (Fig. 2B and fig. S3). Hence, the multiple conformations that recombinant vaults exhibit in solution—some of them characterized in this work—reflect the behavior of intracellular vaults when imaged in their natural environment.
Furthermore, the vault stabilization studies performed here, by combining site-directed mutagenesis and different biophysical approaches, confirmed the importance of the hydrophobic and electrostatic interactions mediated by the MVP N-terminal residues E4, E5, and I7 in the vault assembly process. These results correlate well with previous published data (26) where the substitution of amino acids at the MVP N terminus by histidines generates unstable vault particles prone to disassembly. Collectively, these results highlight the crucial role that the MVP N terminus plays for the correct assembly of the vault particle as well as in opening regulation.
Mechanistic model for vault aperture
The above summarized results prompt to hypothesize a mechanistic model for how wholly assembled vaults transition from a closed, engaged state to an open, two-halves state with the concomitant exposure of the enclosed cavity (Fig. 4).
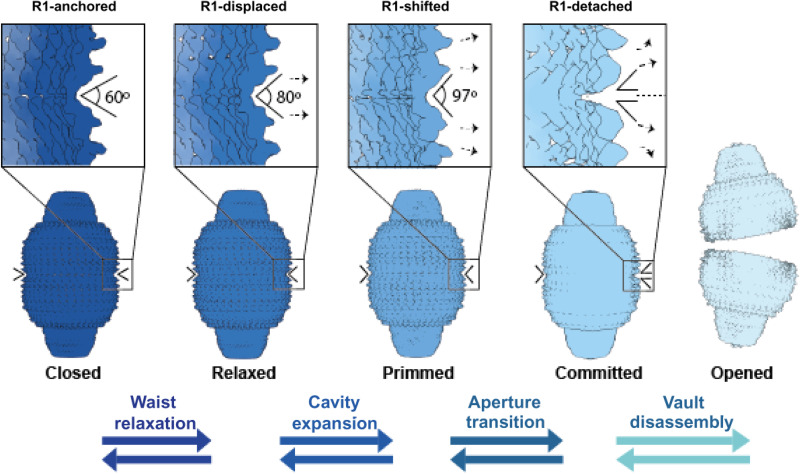
Fig. 4. Hypothetical model for the stepwise process of vault aperture.
Transitioning from a closed to an opened state is a critical process in the functional cycle of vault particles. Fully assembled, functionally committed to cargo delivery vaults exhibit a symmetrical configuration with all MVP R1 domains, from top and bottom half vaults, fully engaged in reciprocal interactions (left, closed). The tight set of reciprocal interactions via R1 domains restricts the flexibility of MVPs, rigidifying the waist cavity and allowing a stable inner compartment needed for cargo delivery. The opening process commences with a waist relaxation, caused by local displacement of R1 domains of specific MVPs, yielding a localized distortion that increases the angle between the R1 domains involved to around 80° (“Relaxed”). Local distortions of R1 domain interactions are progressively increased, primming the vault towards opening (“Primmed”). When the angle between these R1 domains exceeds 97° (“Committed”), the localized distortion can be easily transmitted beyond the area where it started, generating a “gap” between R1 domains that will eventually trigger full separation of the two half vaults (“Opened”). Reversible arrows indicate the likely interconversion of these conformational states as demonstrated by their coexistence in solution.
Fully engaged vaults exhibit global dynamics while maintaining a closed internal cavity with all R1 domains anchored in reciprocal contacts between half vaults (Fig. 4, left, closed). Stable interactions between R1 domains are implemented via a highly conserved cluster of residues strategically located at the junction of several R1 domains from top and bottom half vaults. Mutation of these residues affects the stability of the vault, affecting the second level of self-assembly, namely, the association of two half vaults into whole vaults. However, disrupting this cluster of residues seems to not affect the assembly of MVP monomers into half vaults (first level of self-assembly).
In whole vaults, the cargo hosting cavity is contracted to its minimal volume. Through relaxation of the vault waist, the cavity expands, with a concomitant repositioning of R1 domains (Fig. 4, relaxed). Vault reconfiguration via waist expansion continues until the inner cavity expansion reaches a point where vaults are committed to opening (Fig. 4, primmed). Then, local disruptions of the reciprocal R1 interactions on a subset of MVP monomers seed a conformational change that will eventually be transmitted to the whole vault (Fig. 4, committed). The opening process finishes when the whole vault is disassembled into two half vaults, exposing the inner cavity where cargo is hosted (Fig. 4, right, opened).
Ongoing engineering efforts to leverage vault scaffolding for targeted drug delivery demands a precise understanding of the molecular logic governing the functional cycle of vaults. Here, we delineated the principles conducting vault disassembly, uncovering the pivotal role played by a set of residues at the half vault interface and the localized origin of the distortion leading toward opening. With remarkable reduced working principles for assembly, cargo encapsulation, and disassembly, vaults represent a promising avenue in the development of tailored systems for drug delivery. We hope that the analysis presented here will both assist in the engineering efforts to leverage vault scaffolding for biotechnological applications and inspire future efforts aimed at understanding the role played by vaults in regular physiology, an aspect not completely understood at present.
MATERIALS AND METHODS
Production of recombinant vaults and MVP mutants
MVP mutants were generated by introducing single point mutations by polymerase chain reaction (PCR) using standard protocols. Oligonucleotides used to mutagenize the MVP gene cloned in the plasmid pFB_MVP (MVP wild-type gene inserted in the pFastBac HT A vector; Invitrogen) for each mutant are described in table S2. Site-directed mutagenesis was performed using the Herculase II Fusion DNA polymerase (Agilent Technologies) and treated with Dpn I for 2 hours at 37°C after a PCR cycle. Plasmids were purified using a plasmid purification kit (QIAGEN Plasmid Miniprep Kit). The resulting plasmids were subjected to nucleotide sequencing to assess correct incorporation of the mutations in the MVP sequence. For the quadruple mutant (Asp39 → Lys/Ile7 → Asn/Glu4 → Lys/Glu5 → Lys), mutations were introduced in three consecutive PCR steps. After each PCR cycle, the plasmid was treated with Dpn I, as described above, and subjected to nucleotide sequencing.
Recombinant baculovirus
Generation of a recombinant baculovirus (rBV) containing full-length MVP was performed with a DNA fragment containing the MVP sequence, flanked by Nco I and Kpn I restriction sites generated by PCR. The DNA fragment was digested with Nco I and Kpn I and inserted into the multiple cloning site of the baculovirus transfer vector pFastBac HTA (Invitrogen, Carlsbad, CA), previously digested with the same restriction enzymes. The resulting plasmid, pFB_MVP, was subjected to nucleotide sequencing to assess the correctness of the inserted MVP sequence and was then used to produce the corresponding rBV using the Bac-to-Bac system according to the manufacturer’s instructions (Invitrogen). In the case of the MVP mutants created for this study, we introduced the desired mutation over the pFB_MVP, checked the corresponding mutant MVP sequence, and proceeded in the exact way to obtain the rBV (20).
Production and purification of recombinant vaults
HighFive cells (Invitrogen) were infected with rBVs at a multiplicity of infection of 5 plaque-forming units per cell. Cells were harvested at 72 hours after infection, washed using phosphate-buffered saline, and pelleted with a 5-min centrifugation at 3000 rpm (using a A-4-38 rotor). This pellet was resuspended in 6 ml of buffer A [75 mM NaCl, 50 mM tris (pH 7.4), 1.5 mM MgCl2, and 1 mM dithiothreitol] plus 1% NP-40 and protease inhibitors (protease inhibitor cocktail tablets; Roche, Basel, Switzerland) and maintained on ice for 30 min. The resuspended pellet was sonicated, and cellular debris was removed by centrifugation at 10,000 rpm (using an F-34-6-38 rotor) for 30 min. The supernatant was applied to 4 ml of buffer A with 25% sucrose and centrifuged at 37,000 rpm (using a SW41Ti rotor) for 2.5 hours. The resulting pellet was resuspended in 600 μl of buffer A and centrifuged for 1 min at 13,000 rpm (using an F-45-6-30 rotor). The supernatant was applied to a 25 to 50% sucrose gradient in buffer A and centrifuged for 45 min at 40,000 rpm (using a SW41Ti rotor). The gradient was fractionated and then analyzed by SDS–polyacrylamide gel electrophoresis and negative-stain EM. Last, fractions enriched in recombinant vaults were concentrated to 5 mg/ml using a centrifugal filter device (Centricon YM-100, Merck Millipore, Billerica, MA). With the mutant MVP vaults, we follow the same protocol to purify the particles with the punctual mutations (20).
Atomic force microscopy
AFM was used to study the mechanical differences between the different MVP mutant vaults. Measurements were performed with an AFM microscope (Nanotec Electrónica, Madrid, Spain) operating in Jumping mode plus (27). In this mode, the tip displaces laterally when it is far from the sample, and images are taken by performing force–versus–Z-piezo displacement curves at all points. Most images were taken with a maximal force of ~75 pN, which leads to average heights of 38 ± 2 nm and 35.5 ± 2 nm for reclining particles lying on the barrels and half vaults, respectively; these values agree with the dimensions obtained by x-ray 35 and 40 nm for reclining vaults (14). Rectangular silicon nitride cantilevers (RC800PSA, Olympus, Center Valley, PA) with a nominal spring constant of 0.05 N/m were used and calibrated by Sader’s method. The experiments were carried out under physiological conditions: One 20-μl drop of stock solution, consisting of vault particles under TMS (50 mM Tris (pH7.4), 0.75 mM MgCl2, 75 mM NaCl) buffer conditions, was incubated on a fresh highly ordered pyrolytic graphite surface (ZYA quality; NT-MDT, Tempe, AZ) and, after 30 min, washed with buffer until a volume of 60 μl was reached. The tip was also prewetted with a 20-μl drop of buffer (20).
Differential scanning fluorimetry
Thermal shift assays of the different MVP mutant vaults were performed to study the difference in the stability of the particle. After the initial tests, we realized that the best results were obtained with relatively high quantities of protein (around 50 μg of protein at a concentration of 1 mg/ml). The experiment was carried out with 50 μg of MVP wild type or mutant and 7.5 μl of SYPRO™ Orange (Life Technologies) in a final volume of 50 μl in a 96-well thin-wall PCR plate (Bio-Rad). The SYPRO solution had been diluted 1:17 in buffer A following the manufacturer’s instructions. The mix was read in a Bio-Rad IQ5 multicolor real-time PCR detection system. The program used was a 1° per minute step gradient from 30° to 95°. Fluorescence changes in the wells of the plates were monitored simultaneously with a charge-coupled device (CCD) camera. The wavelengths for excitation and emission were 290 and 575 nm, respectively.
Negative staining EM
Transmission electron microscopy (TEM) of uranyl acetate–stained vaults was used to check alterations in the overall particle morphology due to the mutations inserted in the MVP sequence. Purified vaults were absorbed onto ionized carbon-coated copper EM grids for 1 min at room temperature. Following sample adsorption, the nonadsorbed sample was removed, using a filter paper. Grids were then positioned on top of a 70-μl 2% uranyl acetate drop for 10 s at room temperature, then on another drop of acetate uranyl acetate for another 10 s, and dried on filter paper after. The samples were viewed with a Tecnai Spirit microscope (TEM) (FEI, Eindhoven, The Netherlands) equipped with a LaB6 cathode. TEM images were acquired at 120 kV with a 1376 × 1024 pixel CCD camera (FEI, Eindhoven, The Netherlands).
Cryo-EM and movie processing
For cryo-EM, an aliquot of 3 μl of recombinant vault sample was applied during 15 s to each EM grid with holey gold film. After that, the grid was blotted with Vitrobot (FEI, Eindhoven, The Netherlands) in 100% humidity for 3 s and then plunged into liquid ethane to vitrify the sample. Movies were obtained in Titan Krios 300-kV EM (FEI, Eindhoven, The Netherlands) equipped with a Gatan K2 direct electron detection camera at ×81,000. The pixel size was measured to 1.78 Å on the specimen scale. We used an electron dose rate of 8 electrons per pixel per second, and each movie contains 30 frames recorded in 12 s. Image stacks in each movie were aligned with MotionCor2 (28). The 30 frames in each stack were averaged to obtain a dose weight of 30 e−/ A2. The whole dataset has 3407 movies.
A second dataset was collected at pixel size 1.07 Å per pixel, corresponding to a magnification of ×130,000. We used an electron dose rate of 7.4 electrons per pixel per second, and each movie contains 40 frames recorded in 8 s.
Data processing and 3D reconstruction
Aligned micrographs were used for the contrast transfer function (CTF) estimation using Gctf (29), with defocus values ranging from −1.5 to −3.5 μm. A total of 34,822 particles were automatically picked using RELION (21) with a box of 400 × 400 pixels of size. All picked particles were classified using RELION in a 2D classification process (Class2D). The classes with no interpretable features were discarded. A total of 9793 particles were selected for 3D classifications (Class3D) to calculate the closed vault map and 7539 particles were selected to calculate the half vault map. The initial model for Class3D was generated from a previous atomic model (PDB: 4HL8) of rat vault low-filtered to 60 Å resolution to eliminate the risk of model bias. In the closed vault reconstruction, particles were subjected to consecutive Class3D and Refine3D jobs and additionally to CTF refinement and particle polishing. This class was refined separately with RELION by applying D39 symmetry. To enhance the signal, a mask was generated around the whole vault, obtaining a map after RELION postprocessing of 3.7 Å. The resolution was determined based on a “gold standard” FSC coefficient of 0.143.
A model of the vault particle (PDB ID: 4HL8) was docked into the map using UCSF Chimera (30), and Coot (31) was used to manually adjust these initial models. An initial round of refinement was performed in Phenix (32) using real-space refinement.
In the case of the half vault, Class3D analysis allowed to calculate a map that presented a reasonable resolution (around 14 Å). This half vault class was refined separately with RELION by applying C1 symmetry. To enhance the signal, a mask was generated around the whole half vault, obtaining a map after RELION postprocessing of 9.3 Å. The resolution was determined based on a gold standard FSC coefficient of 0.143.
The vault in committed conformation was obtained by the data analysis of the second dataset. A total of 207,815 particles were automatically picked using RELION (21) with a 700 × 700 pixels box. All picked particles were classified using RELION in a 2D classification process (Class2D). A total of 11,666 particles were selected for further 3D classifications (Class3D) that yielded the vault reconstruction in committed conformation. This class was refined separately without imposing any symmetrical condition, obtaining a map of 7.9 Å. The resolution was determined based on a gold standard FSC coefficient of 0.143.
Acknowledgments
We are indebted to I. S. Fernández for technical advice in cryo-EM data collection and processing, fruitful discussions, and critically reading the manuscript. We also thank R. Fernández-Leiro for technical support and helpful discussions as well as E. Nogales for critically reading the manuscript. Cryo-EM data were collected at Simons Electron Microscopy Center (New York, USA) and at the European Synchrotron Radiation Facility (Grenoble, France) beamline CM01, with assistance of G. Effantin.
Funding: This work was supported by the Spanish Ministry for Science and Innovation (grants BIO2017-83906-P and PID2020-117976GB-I00), the Generalitat de Catalunya (grant 2017 SGR 01192), and the CSIC (grant 20202CEX003).
Author contributions: P.G. performed the EM and cryo-EM experiments, analyzed data, discussed results, prepared figures, and revised the manuscript. M.G.-A. performed the production and purification of wild type and vaults and helped in data analysis. A.L. and P.J.d.P. designed and performed AFM experiments and helped in manuscript preparation. A.C. and J.Q.-A. designed the MVP mutants, performed the DSC experiments, and helped to prepare the manuscript. N.V. conceived experiments, analyzed the data, discussed results, and drafted and revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The cryo-EM maps and atomic models have been deposited in the EMDB/PDB. Accession codes are indicated in Table 1.
Table 1. Cryo-EM data collection, refinement, and validation statistics.
RMS, root mean square; EMDB, Electron Microscopy Data Bank.
|
Data collection
and processing |
Primmed vault |
Committed
vault |
Half vault |
| Magnification | ×81,000 | ×130,000 | ×81,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) |
30 | 35 | 30 |
| Defocus range (μm) |
−1.5/−3.5 | −1.5/−3.5 | −1.5/−3.5 |
| Pixel size (Å) | 1.78 | 1.07 | 1.78 |
| Symmetry imposed |
D39 | C1 | C1 |
| Initial particle images (n) |
34,822 | 207,815 | 34,822 |
| Final particle images (n) |
9793 | 11,666 | 7539 |
| Map resolution (Å) |
3.7 | 7.9 | 9.3 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) |
3–8 | 7–13 | 8–14 |
| Refinement | |||
| Initial model used (PDB code) |
4HL8 | – | – |
| B factors (Å2) | −156.45 | – | – |
| RMS deviations | |||
| Bond lengths (Å) |
0.07 | – | – |
| Bond angles (°) | 0.81 | – | – |
| Validation | |||
| MolProbity score |
20.27 | – | – |
| Ramachandran plot |
|||
| Favored (%) | 88.24 | – | – |
| Allowed (%) | 11.25 | – | – |
| Outliers (%) | 0.52 | – | – |
| Databases | |||
| EMDB | EMD-13478 | EMD-13483 | EMD-13482 |
| PDB | 7PKR | – | – |
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
REFERENCES AND NOTES
- 1.Rothman J. E., Felix Hoppe-Seyler Lecture 1996. Mechanisms of intracellular protein transport. Biol. Chem. Hoppe Seyler 377, 407–410 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Aridor M., Hannan L. A., Traffic jam: A compendium of human diseases that affect intracellular transport processes. Traffic 1, 836–851 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Cocucci E., Kim J. Y., Bai Y., Pabla N., Role of passive diffusion, transporters, and membrane trafficking-mediated processes in cellular drug transport. Clin. Pharmacol. Ther. 101, 121–129 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hamill D. R., Suprenant K. A., Characterization of the sea urchin major vault protein: A possible role for vault ribonucleoprotein particles in nucleocytoplasmic transport. Dev. Biol. 190, 117–128 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Berger W., Steiner E., Grusch M., Elbling L., Micksche M., Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci. 66, 43–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frascotti G., Galbiati E., Mazzucchelli M., Pozzi M., Salvioni L., Vertemara J., Tortora P., The vault nanoparticle: A gigantic ribonucleoprotein assembly involved in diverse physiological and pathological phenomena and an ideal nanovector for drug delivery and therapy. Cancers (Basel). 13, 707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka H., Tsukihara T., Structural studies of large nucleoprotein particles, vaults. Proc. Japan Acad. Ser. B Phys. Biol. Sci. 88, 416–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kedersha N. L., Rome L. H., Isolation and characterization of a novel ribonucleoprotein particle: Large structures contain a single species of small RNA. J. Cell Biol. 103, 699–709 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephen A. G., Raval-Fernandes S., Huynh T., Torres M., Kickhoefer V. A., Rome L. H., Assembly of vault-like particles in insect cells expressing only the major vault protein. J. Biol. Chem. 276, 23217–23220 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Kickhoefer V. A., Siva A. C., Kedersha N. L., Inman E. M., Ruland C., Streuli M., Rome L. H., The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146, 917–928 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kickhoefer V. A., Stephen A. G., Harrington L., Robinson M. O., Rome L. H., Vaults and telomerase share a common subunit, TEP1. J. Biol. Chem. 274, 32712–32717 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Kickhoefer V. A., Searles R. P., Kedersha N. L., Garber M. E., Johnson D. L., Rome L. H., Vault ribonucleoprotein particles from rat and bullfrog contain a related small RNA that is transcribed by RNA polymerase III. J. Biol. Chem. 268, 7868–7873 (1993). [PubMed] [Google Scholar]
- 13.Stadler P. F., Chen J. J. L., Hackermüller J., Hoffmann S., Horn F., Khaitovich P., Kretzschmar A. K., Mosig A., Prohaska S. J., Qi X., Schutt K., Ullmann K., Evolution of vault RNAs. Mol. Biol. Evol. 26, 1975–1991 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H., Kato K., Yamashita E., Sumizawa T., Zhou Y., Yao M., Iwasaki K., Yoshimura M., Tsukihara T., The structure of rat liver vault at 3.5 angstrom resolution. Science 323, 384–388 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Casañas A., Querol-Audí J., Guerra P., Pous J., Tanaka H., Tsukihara T., Verdaguer N., Fita I., New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach. Acta Crystallogr. Sect. D Biol. Crystallogr. 69, 1054–1061 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Goldsmith L. E., Yu M., Rome L. H., Monbouquette H. G., Vault nanocapsule dissociation into halves triggered at low pH. Biochemistry 46, 2865–2875 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Yang J., Kickhoefer V. A., Ng B. C., Gopal A., Bentolila L. A., John S., Tolbert S. H., Rome L. H., Vaults are dynamically unconstrained cytoplasmic nanoparticles capable of half vault exchange. ACS Nano 4, 7229–7240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llauró A., Guerra P., Kant R., Bothner B., Verdaguer N., De Pablo P. J., Decrease in pH destabilizes individual vault nanocages by weakening the inter-protein lateral interaction. Sci. Rep. 6, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding K., Zhang X., Mrazek J., Kickhoefer V. A., Lai M., Ng H. L., Yang O. O., Rome L. H., Zhou Z. H., Solution structures of engineered vault particles. Structure 26, 619–626.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llauró A., Guerra P., Irigoyen N., Rodríguez J. F., Verdaguer N., De Pablo P. J., Mechanical stability and reversible fracture of vault particles. Biophys. J. 106, 687–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querol-Audí J., Casañas A., Usón I., Luque D., Castón J. R., Fita I., Verdaguer N., The mechanism of vault opening from the high resolution structure of the N-terminal repeats of MVP. EMBO J. 28, 3450–3457 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanovska I. L., de Pablo P. J., Ibarra B., Sgalari G., MacKintosh F. C., Carrascosa J. L., Schmidt C. F., Wuite G. J. L., Bacteriophage capsids: Tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. 101, 7600–7605 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baclayon M., Roos W. H., Wuite G. J. L., Sampling protein form and function with the atomic force microscope. Mol. Cell. Proteomics 9, 1678–1688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodward C. L., Mendonça L. M., Jensen G. J., Direct visualization of vaults within intact cells by electron cryo-tomography. Cell. Mol. Life Sci. 72, 3401–3409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mrazek J., Toso D., Ryazantsev S., Zhang X., Zhou Z. H., Fernandez B. C., Kickhoefer V. A., Rome L. H., Polyribosomes are molecular 3D nanoprinters that orchestrate the assembly of vault particles. ACS Nano 8, 11552–11559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortega-Esteban A., Horcas I., Hernando-Pérez M., Ares P., Pérez-Berná A. J., San Martín C., Carrascosa J. L., De Pablo P. J., Gómez-Herrero J., Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid. Ultramicroscopy 114, 56–61 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2