Abstract
Mutations in the 81-bp rifampin resistance-determining region (RRDR) of the rpoB gene were analyzed by DNA sequencing of 50 Mycobacterium tuberculosis clinical isolates (44 resistant and 6 sensitive) from various parts of India. Fifty-three mutations of 18 different kinds, 17 point mutations and one deletion, were observed in 43 of 44 resistant isolates. Three novel mutations and three new alleles within the RRDR, along with two novel mutations outside the RRDR, are reported in this study.
Tuberculosis (TB), though curable, still remains a major killer disease worldwide. The magnitude of the problem is reflected in estimates of new cases, which are predicted to number around 10 million in the year 2000 and 12 million by 2005 (7). Global prevalence of infection due to Mycobacterium tuberculosis is 32%. Eighty percent of all new TB cases are found in around 22 countries, with more than half the cases occurring in 5 Southeast Asian countries (6). Thirty percent of the world's TB-infected population is in India. The enormity of the problem has increased with the emergence of multidrug-resistant (MDR) strains of M. tuberculosis. Dual infection with human immunodeficiency virus and MDR TB is a virtual death sentence in this era.
Rifampin (RIF) resistance serves as a surrogate marker for the detection of MDR TB, as 90% of Rifr isolates are also isoniazid resistant (5). RIF interferes with transcription and elongation of RNA by binding to the DNA-dependent RNA polymerase. It was observed that resistance to RIF follows a single-step, high-level resistance pattern in which mutations occur spontaneously at a frequency of 10−9. The genetic basis for RIF resistance in approximately 95% of the cases is due to mutations in an 81-bp RIF resistance-determining region (RRDR) of the rpoB gene, corresponding to codons 507 to 533 (Escherichia coli numbering system), which codes for the beta subunit of the RNA polymerase of M. tuberculosis.
Different groups of workers from diverse regions of the world (4, 8–12, 14, 16, 19, 20, 21, 23, 24) have thus far reported around 65 substitutions, 12 deletions, and 4 insertions in the RRDR of the rpoB gene. Only one of these earlier reports is concerned with Indian isolates, and in that study rpoB mutations were observed in three Rifr isolates (8). Determination of the mutation patterns among large numbers of isolates from different parts of India is essential, since this would help not only in the design of a suitable diagnostic method for rapid detection of MDR TB but also in the identification of any hot-spot regions in the country for proper implementation of TB control programs. It would also help in understanding whether mutated alleles arise independently or due to the spread of a particular genotype. Moreover, it is well known that clinical isolates from southern India are very different from isolates from other parts of the world. The former have lower virulence in guinea pigs (2), higher susceptibility to hydrogen peroxide (13, 18) and thiophene-2-carboxylic acid hydrazide (TCH), lower sensitivity to p-aminosalicylic acid (17) and thioacetazone, and an appreciably higher proportion of phage type I (1). It is of interest to determine whether these southern Indian isolates show any different kinds of mutations in the RRDR region of the rpoB gene.
DNA sequencing of RRDR.
The following are the results of DNA sequencing of the RRDR of 44 Rifr isolates (35 of which were isolated from patients from southern India) and 6 Rifs isolates, all obtained from seven states in India.
(i) M. tuberculosis isolates.
Forty-four Rifr and six Rifs strains were isolated from 50 patients from seven states in India. Of the 44 Rifr strains, the numbers isolated from each state were as follows: Andhra Pradesh (AP), 6; Delhi, 1; Goa, 3; Kerala, 4; Karnataka, 5; Sikkim, 5; and Tamil Nadu (TN), 20. Of the six Rifs strains, four were obtained from patients from TN, while two were from AP. The drug susceptibility patterns of these isolates are shown in Table 1.
TABLE 1.
Drug susceptibility patterns of MDR M. tuberculosis strains isolated in India
INH, isoniazid; EMB, ethambutol; STR, streptomycin.
R, resistant.
(ii) Determination of sensitivity to RIF.
Conventional indirect susceptibility testing was done using Lowenstein-Jensen medium, and RIF sensitivity was determined by the MIC method. Resistance was defined by a MIC equal to or greater than 128 mg/liter.
(iii) Sequencing of the RRDR of the rpoB gene.
The RRDR of the rpoB gene was sequenced after amplification by PCR to analyze the mutations associated with RIF resistance. Template DNA for PCR was obtained using cetyltrimethylammonium bromide (22). PCR was performed using primers rpo3 (5′ CAGACGTTGATCAACATCCG 3′) and rpo4 (5′ TACGGCGTTTCGATGAAC 3′) to generate a 305-bp product, which was purified and used for sequencing. A T7 Sequenase v 2.0 PCR product sequencing kit (Amersham Life Science) and primer TR9 (5′ TCGCCGCGATCAAGGAGT 3′) were used for manual sequencing. In brief, 1 μl of exonuclease I (10 U/μl) and 1 μl of shrimp alkaline phosphatase (2 U/μl) were added to 7 μl of PCR amplification mixture, and the mixture was incubated at 37°C for 15 min. The reaction mixture was inactivated by heating to 80°C for 15 min. To 9 μl of treated PCR product, 1 μl of primer (10 pmol/μl) was added, and the mixture was incubated at 100°C for 3 min, followed by snap cooling. To this ice-cold annealed DNA mixture, 2 μl of 5× T7 Sequenase reaction buffer, 1 μl of 0.1 M dithiothreitol, 2 μl of a 1:10-diluted labeling mixture, 5 μCi of [35S]dATP, and 2 μl of T7 Sequenase v 2.0 DNA polymerase were added, and the mixture was incubated at room temperature for 2 min. Aliquots of this mixture (3.5 μl each) were added to termination tubes containing 2.5 μl of each termination mixture (G, A, T, and C) and incubated at 37°C for 10 min. The reaction was stopped by adding 4 μl of stop solution. Samples were heated to 75°C for 2 min and then loaded onto a sequencing gel. An automated sequencer (ABI Prism model 377 version 3.0) was used with primer TR8 (5′ TGCACGTCGCGGACCTCCA 3′) to confirm the sequence in reverse order. The data obtained were compared with the sequence obtained from the database at the Sanger Centre using the BLAST program (www.sanger.ac.uk).
Molecular analysis.
The MIC of RIF for all 44 Rifr isolates was greater than 128 mg/liter, while the MIC of RIF for all six Rifs isolates was less than 32 mg/liter. DNA sequence analysis of the 44 Rifr isolates showed that 39 had a single mutation, two had triple mutations, and two had quadruple mutations in the 81-bp RRDR of the rpoB gene. One isolate did not contain any mutation. Fifty-three mutations of 18 different kinds, 17 point mutations and one deletion, were observed (Table 2). Two isolates from TN that contained four mutations had a novel mutation of GCG to GCA at codon 532. In addition, one of the two TN isolates had mutations at codon 518 (AAC to CAC), codon 531 (TCG to TTG), and codon 533 (CTG to CTT), while the other had mutations at codon 511 (CTG to ATG), codon 512 (AGC to AGG), and codon 526 (CAC to GAC). One isolate from AP that contained a triple mutation had two mutations in codon 516 (GAC to AAA) and another at codon 531 (TCG to TTG). This particular isolate also contained a mutation outside the RRDR (CCC to CAC at codon 535). The other isolate that contained a triple mutation was from TN and had mutations at codon 508 (ACC to AGC), codon 512 (AGC to AGG), and codon 526 (CAC to GAC). In Karnataka, four isolates had the same mutation, TCG to TTG, at codon 531, while the fifth exhibited a mutation at codon 526, CAC to GAC. None of the sensitive strains contained any mutation.
TABLE 2.
Distribution of mutations by state found in the RRDR of the rpoB gene in Rifr M. tuberculosis isolates from India
| Codon no. | Mutation | Distribution by statea |
|---|---|---|
| 508 | ACC → AGCb | TN, 1 |
| 511 | CTG → CCG | K, 1; TN, 1 |
| CTG → ATG | TN, 1 | |
| 512 | AGC → AGGc | TN, 2 |
| 513 | CAA → AAA | G, 1 |
| 516 | GAC → TACc | S, 1 |
| GAC → AAAd | AP, 1 | |
| 517 | CAG → delbe | AP, 1 |
| 518 | AAC → CAC | TN, 1 |
| 526 | CAC → CTC | S, 1; TN, 2 |
| CAC → TAC | AP, 1; K, 1 | |
| CAC → GAC | KA, 1; TN, 2 | |
| CAC → CGC | AP, 1 | |
| CAC → ACC | AP, 1 | |
| 531 | TCG → TGG | S, 1; TN, 1 |
| TCG → TTG | S, 2; G, 2; K, 2; KA, 4; AP, 2; D, 1; TN, 13 | |
| 532 | GCG → GCAb | TN, 2 |
| 533 | CTG → CTTc | TN, 1 |
KA, Karnataka; S, Sikkim; G, Goa; D, Delhi; K, Kerala.
Novel mutation.
New allele.
Double mutations in the same codon.
del, deletion.
Eighteen different types of mutations were identified in 44 Rifr M. tuberculosis clinical isolates. Ninety-five percent of these were point mutations involving 10 codons, while only one isolate had a deletion. The codons most frequently involved in mutation were codon 531 (frequency, 53%) and codon 526 (19%). Among the different mutations seen at codon 531, the mutation of TCG (Ser) to TTG (Leu) occurred at a very high frequency, 49%. Matsiota-Bernard et al. (10) and Pozzi et al. (14) found similarly high frequencies of this particular mutation, 56 and 59%, respectively. Though mutation of CAC to GAC at codon 526 occurred at frequencies of 30% in Italian isolates (14) and 19% in Greek isolates (10), we found a low frequency of this mutation, 6%. Previous workers have reported a wide range of frequencies for these particular mutations at codon 531 (20 to 71%) and codon 526 (0 to 30%) (14). Ramaswamy and Musser (15) showed frequencies of 41 and 36% for various mutations occurring at codons 531 and 526, respectively, in 478 isolates obtained from various parts of the world. Billington et al. (3) observed that mutants isolated more frequently in clinical practice have a higher mean relative fitness and the prevalence of each mutant type depends on its ability to survive. This might be the reason for the higher occurrence of the mutation of TCG to TTG at codon 531 in isolates in the present study from all the states except those from AP, where codon 526 was the most involved. Five different types of mutations were seen at codon 526 (Fig. 1). Hirano et al. (8) reported mutations in Indian isolates at codon 531 (TCG to TGG) and codon 526 (CAC to GAC). No mutation was observed in the RRDR of the six Rifs isolates analyzed in this study. One isolate, although resistant to RIF, did not show any mutation. Kapur et al. (9) also reported such observations. The RIF resistance in these isolates may be due either to the presence of a mutation elsewhere in this gene or to some other resistance mechanism.
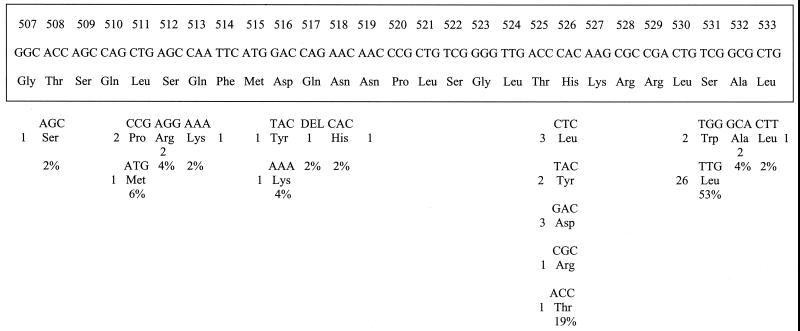
FIG. 1.
Mutations in the RRDR of the rpoB gene of M. tuberculosis Indian isolates. The bottom panel shows the mutated codons with corresponding amino acids. The original sequence is shown boxed. Numbers to the left of or below amino acid designations indicate numbers of isolates showing the mutation, while percentages denote the frequencies of occurrence of mutations at the particular codon.
Mutations at codon 532 from GCG (Ala) to GCA (Ala) in two isolates and at codon 508 from ACC (Thr) to AGC (Ser) and deletion at codon 517 alone of CAG (Gln) have not been reported previously. New alleles reported in this study include mutations from AGC (Ser) to AGG (Arg) at codon 512, GAC (Asp) to AAA (Lys) at codon 516, and CTG (Leu) to CTT (Leu) at codon 533. New mutations outside the RRDR were also seen in two isolates (GGG to GAG at codon 534 and CCC to CAC at codon 535). Previous workers have also reported mutations outside the RRDR (GAG to GCG at codon 504 [16], GAG to GAT at codon 541 [14], TCG to GCG at codon 553 [14], and ATC to TTC at codon 572 [24]).
In contrast to the results obtained by Taniguchi et al. (19), our results showed high MICs for isolates with mutations at codon 516 or codon 533. In fact, a high MIC (>128 mg/liter) was demonstrated even for an isolate with a silent mutation at codon 532.
Forty-three Rifr isolates in the present study either showed a substitution(s) or a deletion in the RRDR. New alleles as well as novel mutations within as well as outside the RRDR have been found in this study. The majority of the mutations map to codon 531, followed by codon 526. There was no specific geographical clustering of isolates in terms of mutations. Though the occurrence of mutations at codon 526 was high in AP compared to that in other states, analysis with a higher sample number needs to be done to arrive at a definitive conclusion.
Despite the large number of mutations already reported in other studies, the evidence of new mutations in this study indicates that mutations continue to arise, probably due to the ability of M. tuberculosis to adapt to drug exposure.
In this study, molecular analysis of the 81-bp RRDR of the rpoB gene of 50 M. tuberculosis clinical isolates from various parts of India was carried out and the mutations were identified. Three novel mutations and three new alleles within the RRDR and two novel mutations outside the RRDR were identified. Although the Indian isolates exhibited few distinct characteristics, the pattern of mutations in the 81-bp RRDR is similar to that reported for the majority of clinical isolates in different parts of the world.
Tests like Inno-LiPA Rif.TB (Innogenetics, Zwijndrecht, Belgium) rapidly identify clinical isolates as members of the M. tuberculosis complex and determine the presence of point mutations within the RRDR of the rpoB gene. Such tests basically exploit the principle of hybridization, and the probes for such tests are designed according to the expected mutations. If the expected mutations or their frequencies of occurrence are different in different countries, such molecular diagnostic tests will have to be slightly modified. Further, knowledge of the mutations present in these isolates would help in the use of rpoB genotyping as an epidemiological tool for Rifr M. tuberculosis isolates. rpoB genotyping can also be used for discrimination of Rifr M. tuberculosis isolates with identical IS6110 fingerprints. Therefore, the new mutations reported here need to be considered in the development of new molecular diagnostic methods to be implemented in India. This study, though confirming the universal pattern of mutations in rpoB, also reveals the presence of novel mutations and new alleles among Indian isolates.
Nucleotide sequence accession numbers.
The sequences with novel mutations found in this study are deposited in EMBL under accession numbers AJ297922, AJ297923, and AJ297924; those with mutations in the new alleles are deposited under accession numbers AJ297925, AJ297926, and AJ297927; and those with mutations outside RRDR are deposited under accession numbers AJ297928 and AJ297929.
Acknowledgments
We thank Nalini Sundarmohan, V. Kripa, and S. K. Vasan for technical assistance and William R. Jacobs, Jr., Howard Hughes Medical Institute at Albert Einstein College of Medicine, Bronx, N.Y., for providing the facilities for automated sequencing.
The Senior Research Fellowship extended by the Council of Scientific and Industrial Research, Government of India, to C. Mani is gratefully acknowledged.
REFERENCES
- 1.Bhatia A L. Characteristics of Indian tubercle bacilli. Ind J Chest Dis. 1961;3:147–153. [Google Scholar]
- 2.Bhatia A L, Csillag A, Mitchison D A, Selkon J B, Somasundaram P R, Subbaiah T V. The virulence in the guinea-pig of tubercle bacilli isolated before treatment from South Indian patients with pulmonary tuberculosis. 2. Comparison with virulence of tubercle bacilli from British patients. Bull W H O. 1961;25:313–322. [PMC free article] [PubMed] [Google Scholar]
- 3.Billington O J, McHugh T D, Gillespie S H. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnabella V, Martiniuk F, Kinney D, Bacerdo M, Bonk S, Hanna B, et al. Isolation of the gene for the β subunit of RNA polymerase from rifampin-resistant Mycobacterium tuberculosis and identification of new mutations. Am J Respir Cell Mol Biol. 1994;11:639–643. doi: 10.1165/ajrcmb.11.6.7946393. [DOI] [PubMed] [Google Scholar]
- 5.Drobniewski F A, Wilson S M. The rapid diagnosis of isoniazid and rifampin resistance in Mycobacterium tuberculosis—a molecular story. J Med Microbiol. 1998;47:189–196. doi: 10.1099/00222615-47-3-189. [DOI] [PubMed] [Google Scholar]
- 6.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 7.Heifets L B, Cangelosi G A. Drug susceptibility testing of Mycobacterium tuberculosis: a neglected problem at the turn of the century. Int J Tuberc Lung Dis. 1999;3:564–581. [PubMed] [Google Scholar]
- 8.Hirano K, Abe C, Takahashi M. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J Clin Microbiol. 1999;37:2663–2666. doi: 10.1128/jcm.37.8.2663-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsiota-Bernard P, Vrioni G, Marinis E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J Clin Microbiol. 1998;36:20–23. doi: 10.1128/jcm.36.1.20-23.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan Nair C, Mackay-Scollay E M, Ramachandran K, Selkon J B, Tripathy S P, Mitchison D A, et al. Virulence in the guinea-pig and susceptibility to hydrogen peroxide of isoniazid-sensitive tubercle bacilli from South Indian patients. Tubercle. 1964;45:345–353. doi: 10.1016/s0041-3879(64)80048-9. [DOI] [PubMed] [Google Scholar]
- 14.Pozzi G, Meloni M, Iona E, Orrù G, Thoresen O F, Ricci M L, Oggioni M R, Fattorini L, Orefici G. rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J Clin Microbiol. 1999;37:1197–1199. doi: 10.1128/jcm.37.4.1197-1199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 16.Schilke K, Weyer K, Bretzel G, Amthor B, Brandt J, Sticht-Groh V, Fourie P B, Haas W H. Universal pattern of rpoB gene mutations among multidrug-resistant isolates of Mycobacterium tuberculosis complex from Africa. Int J Tuberc Lung Dis. 1999;3:620–626. [PubMed] [Google Scholar]
- 17.Selkon J B, Subbaiah T V, Bhatia A L, Radhakrishna S, Mitchison D A. A comparison of the sensitivity to p-aminosalicylic acid of tubercle bacilli from South Indian and British patients. Bull W H O. 1960;23:599–611. [PMC free article] [PubMed] [Google Scholar]
- 18.Subbaiah T V, Mitchison D A, Selkon J B. The susceptibility of hydrogen peroxide of Indian and British isoniazid-sensitive and isoniazid-resistant tubercle bacilli. Tubercle. 1960;41:323–333. [Google Scholar]
- 19.Taniguchi H, Aramaki H, Nikaido Y, Mizuguchi Y, Nakamura M, Koga T, Yoshida S. Rifampin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol Lett. 1996;144:103–108. doi: 10.1111/j.1574-6968.1996.tb08515.x. [DOI] [PubMed] [Google Scholar]
- 20.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, et al. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 21.Valim A R M, Rossetti M L R, Ribeiro M O, Zaha A. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from Brazil. J Clin Microbiol. 2000;38:3119–3122. doi: 10.1128/jcm.38.8.3119-3122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA finger printing: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen L K W, Leslie D, Coloe P J. Bacteriological and molecular analysis of rifampin-resistant Mycobacterium tuberculosis strains isolated in Australia. J Clin Microbiol. 1999;37:3844–3850. doi: 10.1128/jcm.37.12.3844-3850.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]