Abstract
Rapid environmental change can lead to population extinction or evolutionary rescue. The global staple crop sorghum (Sorghum bicolor) has recently been threatened by a global outbreak of an aggressive new biotype of sugarcane aphid (SCA; Melanaphis sacchari). We characterized genomic signatures of adaptation in a Haitian breeding population that had rapidly adapted to SCA infestation, conducting evolutionary population genomics analyses on 296 Haitian lines versus 767 global accessions. Genome scans and geographic analyses suggest that SCA adaptation has been conferred by a globally rare East African allele of RMES1, which spread to breeding programs in Africa, Asia, and the Americas. De novo genome sequencing revealed potential causative variants at RMES1. Markers developed from the RMES1 sweep predicted resistance in eight independent commercial and public breeding programs. These findings demonstrate the value of evolutionary genomics to develop adaptive trait technology and highlight the benefits of global germplasm exchange to facilitate evolutionary rescue.
A staple crop being decimated by aphids was rescued by global sharing of a rare resistance allele from traditional varieties.
INTRODUCTION
Ongoing processes of global change, encompassing climate change, nutrient cycles, and pest outbreaks, are shaping the evolution of natural and agricultural ecosystems (1, 2). Intense selection pressure following environment changes may lead to the rapid decline or extinction of populations (3, 4). If a population is to persist under such strong selection, adaptive standing genetic variation must exist or adaptive de novo variation must arise on a sufficiently fast time scale (5). This population genetic phenomenon, evolutionary rescue, has become a focus of considerable empirical and theoretical study in ecology and conservation biology, since the current rate of global change could exceed the capacities of many populations to adapt (6, 7). Still, there is a lack of examples of evolutionary rescue occurring in the field and at large geographic scales (4). In agricultural systems, the spread of pests or emergence of new aggressive biotypes may lead to a reduction of crop diversity or a total loss of crop cultivation (8). Therefore, understanding and facilitating evolutionary rescue in agricultural systems is critical for global food security.
Populations of crops or wild species subjected to strong selection pressure may experience a major population bottleneck, resulting in a loss of genetic diversity (9). The level of diversity preserved in a population recovering from strong selection depends on the number of backgrounds on which the adaptive alleles emerge (10), which can determine the potential for future adaptation or genetic gain. Conversely, adaptation conferred by a beneficial variant derived from a single progenitor causes the removal of genetic diversity from the surviving population (10, 11). Evolutionary population genomics approaches using genome-wide polymorphism data from diverse germplasm can identify candidate loci for adaptive traits (12). While genome scans for selection have been widely used to identify putative adaptive alleles in crops (9, 13–16), they have not yet been used to identify trait-predictive markers for molecular breeding of stress-resilient varieties (17).
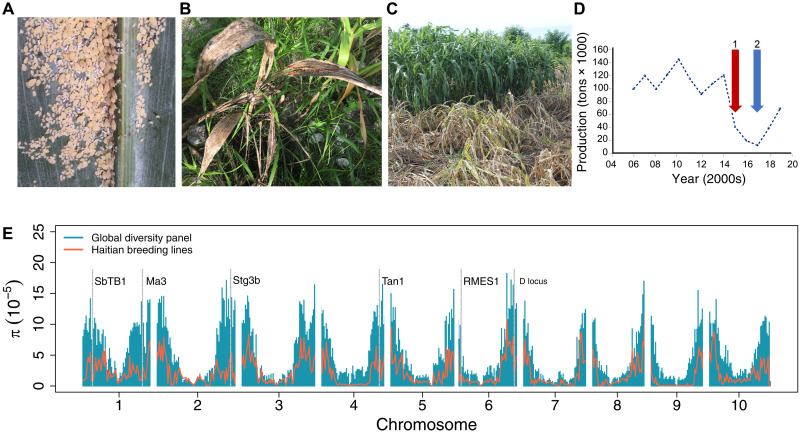
Sorghum [Sorghum bicolor L. (Moench)] is among the world’s most important staple crops for smallholder farmers in semiarid regions, as well as a commercial grain and forage crop in industrialized nations (18). Since 2013, an aggressive biotype of the sugarcane aphid (SCA; Melanaphis sacchari) has become a major threat to global sorghum production, with widespread and substantial yield loss (Fig. 1A) (19, 20). The M. sacchari superclone has been rapidly expanding (21), putting >90% of the sorghum-producing areas of North America at risk and threatening to end sorghum cultivation in some areas (19). In Haiti, a Caribbean nation with one of the world’s highest rates of food insecurity, sorghum is among the most important staple crops (22). However, heavy infestations by M. sacchari since 2015 have caused the loss of over 70% of sorghum production in the country and prevented production of most local landraces (Fig. 1B) (23). Shortly before the SCA outbreak, a new Haitian breeding population (HBP) had been launched by Chibas, a public crop improvement program now a part of University of Quisqueya. This program uses global admixed germplasm, rapid-cycling intercrossing (two generations of crossing per year using nuclear male sterility), and selection under smallholder conditions (e.g., no insecticidal treatment) for development of pureline varieties (Fig. 1C) (24). Selecting from a small number of breeding lines that survived SCA infestation, a new SCA-resistant sorghum variety, Papèpichon, was developed and distributed nationally (Fig. 1D) (22), and intercrossing and advancement of resistant breeding lines have continued.
Fig. 1. Evolutionary rescue following a continental outbreak of a sorghum pest.
(A) Infestation of SCA, M. sacchari, on a commercial hybrid in the U.S. sorghum-growing production region (Kansas). (B) SCA infestation on a traditional sorghum variety on a smallholder farm in Haiti (brown plant in foreground; green leaves in background are maize and wild grasses). (C) Reaction of susceptible (brown plants; foreground) and resistant (green plants; background) sorghum breeding lines under natural SCA infestation during breeding trials in Haiti. (D) Estimates of annual sorghum production in Haiti (2006–2019) indicating the start of the SCA outbreak (1, red arrow) and the start of national distribution of SCA-resistant variety, Papèpichon (2, blue arrow). (E) Genome-wide nucleotide diversity (π) in the HBP (red line) compared to a global diversity panel (GDP; blue bars). Nucleotide diversity was calculated for a nonoverlapping sliding window of 1 Mbp across the genome. The gray vertical dashed lines indicate the position of a priori candidate genes for breeding targets of the Haiti program, which colocalized with genomic regions of reduced π (see file S3 for details). Photo credit: (A and B) Geoffrey Morris, Colorado State University; (C) Gael Pressoir, University of Quisqueya.
Here, we used a retrospective genomic analysis of the Haitian sorghum breeding population that was subjected to strong selection under SCA infestation to understand the genetic basis of the evolutionary rescue following the SCA outbreak, as well as the origins of the SCA resistance alleles. We find that the rapid adaptation of the HBP to the SCA outbreak was due to selection for a globally rare Ethiopian allele at the RMES1 SCA resistance locus, which is shared across programs in Africa, Asia, and the Americas because of >50 years of global germplasm exchange before the SCA outbreak. Furthermore, we developed a convenient low-cost molecular marker based on the evolutionary genome scan and validated it in eight commercial and public sorghum breeding programs, demonstrating the value of leveraging global germplasm exchange and evolutionary population genomics to improve crop resilience.
RESULTS
Genome-wide polymorphism and nucleotide diversity
To understand the evolutionary rescue of sorghum following the SCA outbreak (Fig. 1, A to D), we conducted a retrospective genomic analysis of the HBP in comparison to a global diversity panel (GDP). Genotyping-by-sequencing of 296 HBP and 767 GDP (fig. S1 and file S1) sorghum lines generated 159,683 polymorphic single-nucleotide polymorphisms (SNPs) with an average SNP density of 75 and 229 per Mb in the HBP and GDP, respectively (fig. S2). The GDP had a higher proportion of low-frequency minor alleles (<5% MAF) compared to the HBP (fig. S3). The average inbreeding coefficient (FIS) is high in both the HBP and GDP, at 0.7 and 0.9, respectively (table S1). The effect of selection on genetic diversity in the HBP was assessed on the basis of genome-wide nucleotide diversity (π) in the HBP in comparison to (i) the GDP and (ii) a major public program in the United States [Texas A&M pre-breeding lines (TAM-PBLs), N = 35]. Average nucleotide diversity (π) in the HBP was 2.3 × 10−5, moderately lower than π in the GDP (5.8 × 10−5) and TAM-PBL (4.8 × 10−5) (Fig. 1E and table S2). In the HBP, 31% of 1-Mb windows have negative average Tajima’s D values, indicating an excess of rare variants, while in the GDP predominantly positive values of Tajima’s D were observed, indicating an excess of common variants (fig. S4).
Contributions of global sorghum diversity to the HBP
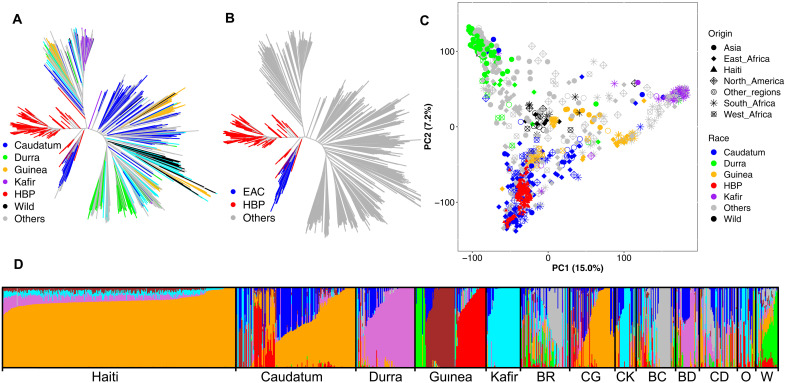
The genetic ancestry of the HBP from global germplasm was inferred on the basis of population structure analyses. In a neighbor-joining analysis, the HBP clusters with caudatum accessions (Fig. 2A), specifically caudatums from East Africa (Fig. 2B). Similarly, in principal coordinate analysis, the HBP clusters with East African caudatum accessions (Fig. 2C). To estimate ancestry coefficients for HBP lines, we used Bayesian model–based clustering in ADMIXTURE, projecting HBP lines onto ancestral populations and allele frequencies defined using only GDP (with HBP lines omitted). With the GDP, the lowest cross-validation error was observed at K = 8 (fig. S5) and accessions clustered by ecogeographic region and botanical type, as expected. ADMIXTURE projection analysis suggests that the HBP is admixed, largely consisting of caudatum haplotypes (>80% of the genome) with a remaining small percentage being contributed by durra and guinea sorghums (Fig. 2D).
Fig. 2. Population structure of the HBP in relation to global sorghum diversity reflects its derivation from East African germplasm.
Genetic relatedness of the HBP to the global diversity assessed by neighboring joining method, color-coded by botanical type (A) or highlighting the close relationship between the HBP and East African caudatum (EAC) germplasm (B). (C) Scatterplot of the first two principal components (PC) of genome-wide SNP variation, demonstrating the clustering of HBP within EAC germplasm. (D) Bayesian hierarchical clustering of the HBP and GDP with the probability of membership (Q) in each of K = 8 ancestral populations. The Q value bar plots are arranged by botanical types to reflect the relationship of the HBP to the GDP. Note that color-coding of the bar plots in (D) is arbitrary and does not reflect the color code in (A) to (C). BR, bicolor; CG, caudatum-guinea; CK, caudatum-kafir; BC, bicolor-caudatum; BD, bicolor-durra; CD, caudatum durra; O, others (includes botanical types containing less than 10 individuals); W, wild.
Evidence of a selective sweep in the HBP at RMES1
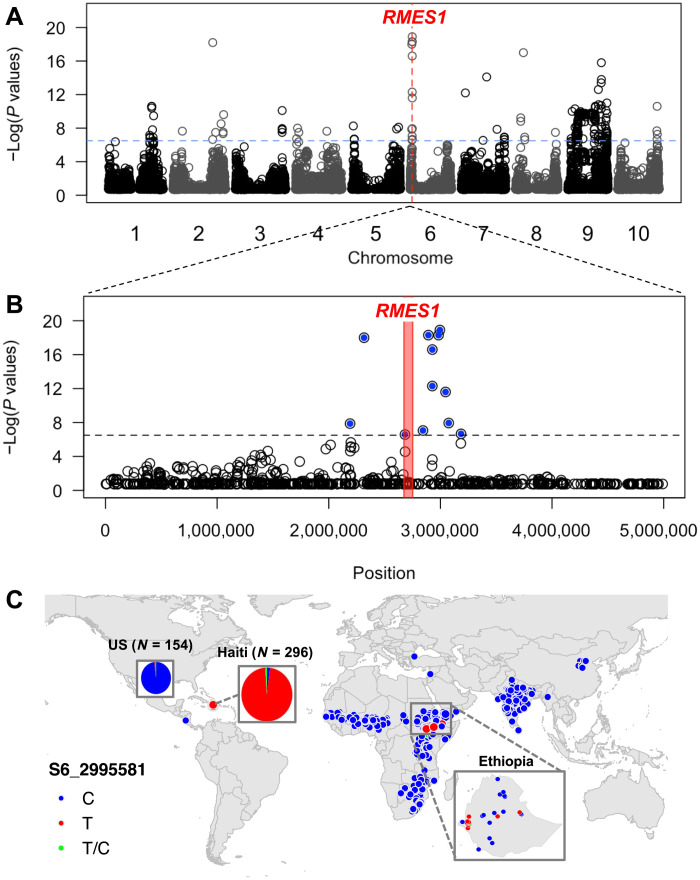
To identify genome regions implicated in the evolutionary rescue of the HBP, genome-wide scans for outlier loci were performed on the basis of an FST test. Overall, the HBP is moderately differentiated from the GDP, with an average genome-wide FST of 0.16 (Fig. 3A and file S2). On the basis of a Bonferroni-adjusted P < 0.01, FST analysis identified 171 outlier genomic regions, which are candidate selective sweep regions. Several genomic regions with FST outliers and low diversity (Fig. 1E) colocalized with candidate genes for traits under selection by the Chibas breeding program, including photoperiodic flowering, inflorescence architecture, stay-green, stem sugar content, and SCA resistance (file S3). The most extreme FST outliers were observed on chromosome 6, precisely colocalizing with RMES1, a locus previously shown to underlie SCA resistance in a Chinese sorghum line of unknown pedigree (Fig. 3, A and B) (25). To characterize the prevalence of the putative selected haplotype and identify its geographic origin, we mapped the allelic distribution of the highest FST SNP S6_2995581 in global georeferenced sorghum landraces (Fig. 3C) and compared these distributions to the allele frequency in U.S. and Haitian breeding germplasm (Fig. 3C, left inset). Globally, the allele is rare (<2%), found only in Ethiopian caudatum landraces and a few breeding lines from West Africa and the United States. However, the sweep-associated allele is common (~40%) in Ethiopian caudatum accessions (Fig. 3C and table S3). The high local frequency of the sweep-associated allele in Ethiopia suggests a likely origin of the SCA resistance allele in the Ethiopian highlands (Fig. 3C, right inset).
Fig. 3. Genome scan for selection identifies the major aphid resistance allele at RMES1 originating in Ethiopia.
(A) Genome-wide scan for selection in the HBP using fixation index (FST) with the −log(FST P value) (y axis) plotted against position on the chromosome (x axis). (B) Detailed view (5 Mb) of top FST peak on chromosome 6 that colocalizes with the RMES1 locus. The ~130-kb region from 2,667,082 to 2,796,847 base pairs (bp) corresponding to the published RMES1 interval is denoted with the red bar. (C) Global allele distribution of the SNP that showed the highest FST value (S6_2995581), which colocalized with the RMES1 locus. Allelic state for georeferenced global germplasm is denoted with points. Allele frequencies in the U.S. (C = 151, T = 2, T/C = 1) and Haiti (C = 6, T = 287, T/C = 3) breeding germplasm, denoted in pie charts with area proportional to the number of accessions, show that the allele is almost fixed in Haitian breeding germplasm and rare in U.S. breeding germplasm.
Comparative genomic analysis to identify candidate causative variants
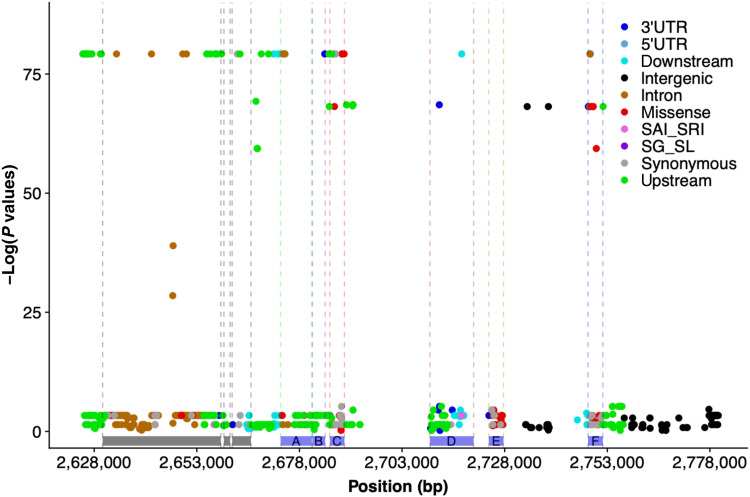
To identify candidate causative variants for the RMES1 locus, we used whole-genome resequencing and de novo genome sequencing of sorghum accessions with known SCA reactions. The RMES1 interval previously defined on the basis of biparental linkage mapping (25) includes seven gene models (Sobic.006G017000, Sobic.006G017100, Sobic.006G017200, Sobic.006G017332, Sobic.006G017266, Sobic.006G017400, and Sobic.006G017500) that were candidates for the causative gene. Comparative genomic analyses based on local multiple sequence alignment of de novo genome sequence of the resistant accession (PI 276837, the Ethiopian progenitor of SCA-resistant line SC170) and three sorghum reference genomes of SCA-susceptible lines (BTx623, Tx430, and BTx642) were used to identify potential causative variants. No sequence variants were identified in the exons of three of the seven genes (Sobic.006G017000, Sobic.006G017100, and Sobic.006G017266). A total of 35, 32, and 29 nonsynonymous SNPs were detected in the exons of Sobic.006G017200, Sobic.006G017400, and Sobic.006G017500, when comparing the sequences of the resistant PI 276837 and the three susceptible accessions. In addition, three insertion-deletion variants resulting in frameshifts were detected in Sobic.006G017500 (file S4). To further refine the set of candidate causative variants, we performed a localized association analysis for SCA resistance (“resistant” or “susceptible,” based on literature classification) around RMES1 with resequencing data for diverse sorghum accessions (Fig. 4 and file S5) that detected 101 highly significant associations (P > 0.0001). Annotations of the variants within the RMES1 locus indicate that only 10 of 101 associated variants are nonsynonymous (5 of 10 in Sobic.006G017200 and the remaining 5 of 10 in Sobic.006G017500).
Fig. 4. Whole-genome resequencing and local association mapping identifies potential causative variants at RMES1.
Functional annotation and association mapping of nucleotide polymorphisms within the RMES1 locus across a set of 13 diverse sorghum accessions with known SCA resistance or susceptibility. The −log of P values of local marker-trait association scan plotted against the chromosomal positions at the RMES1 locus on chromosome 6. Variants are color-coded by annotation generated by the SnpEff program. Blue bars represent the seven annotated genes within the RMES1 interval (A, Sobic.006G017000; B, Sobic.006G017100; C, Sobic.006G017200; D, Sobic.006G017332 and Sobic.006G017266; E, Sobic.006G017400; F, Sobic.006G017500.v3.1). Gray bars indicate genes outside the RMES1 interval as originally defined (25). 3′UTR, 3′ untranslated region variant; SAI_SRI, splice acceptor/intron or splice region intron variants; SG_SL, stop gained or stop loss variant.
Development and validation of a molecular marker based on the selective sweep
Next, we sought to test the hypothesis that the genome region identified by the FST scan underlies variation for SCA resistance in other global sorghum germplasm. Therefore, we developed a KASP [kompetitive allele-specific polymerase chain reaction (PCR)] marker based on the SNPs at the RMES1 locus identified in the FST scan. Of the candidate SNPs (table S3 and files S2 and S6), SNP 06_02892438 was determined to have the best combination of linkage, linkage disequilibrium, and technical KASP functionality of the SNPs. Alternative SNPs were also developed into markers (file S6), and while the markers are often used as technical checks, testing has confirmed the priority of the marker based on SNP 06_02892438 (Sbv3.1_06_02892438R). Initial validation of the Sbv3.1_06_02892438R KASP marker using DNA samples from known resistant lines (SC110, Tx2783, and IRAT204), susceptible lines (BTx623 and BTx642) (26), and multiple F2 families segregating for SCA resistance demonstrated that the KASP marker Sbv3.1_06_02892438R was in complete agreement with historical phenotypes of inbred lines and segregated within F2 populations (file S7).
A recombinant family derived from a cross between Tx430 (susceptible U.S. breeding line) and IRAT204 (a variety developed in Senegal that is SCA resistant) was used to further validate the utility and predictiveness of the KASP marker for marker-assisted selection. A total of 50 segregating lines (F3 and F4), together with resistant (IRAT204 and SC110) and susceptible controls (RTx430), were genotyped with the KASP marker Sbv3.1_06_02892438R. The resistant controls and 23 F3 and F4 lines were homozygous for the resistant allele (+/+). The susceptible control and 9 F3 and F4 lines were homozygous for the susceptible allele (−/−), while the remaining 18 F3 and F4 lines were heterozygous (+/−). A subset of 23 homozygous F3 and F4 lines (either +/+ or −/−), along with three resistant and three susceptible control lines, were tested for SCA reaction in a free-choice flat screen assay in the greenhouse, scoring aphid damage rating, leaf greenness [soil plant analysis development (SPAD)], and seedling height (Fig. 5, A and B, and file S7). The SCA reaction phenotypes match the KASP marker genotypes, demonstrating the reliability and predictability of using KASP markers in marker-assisted selection for SCA resistance breeding.
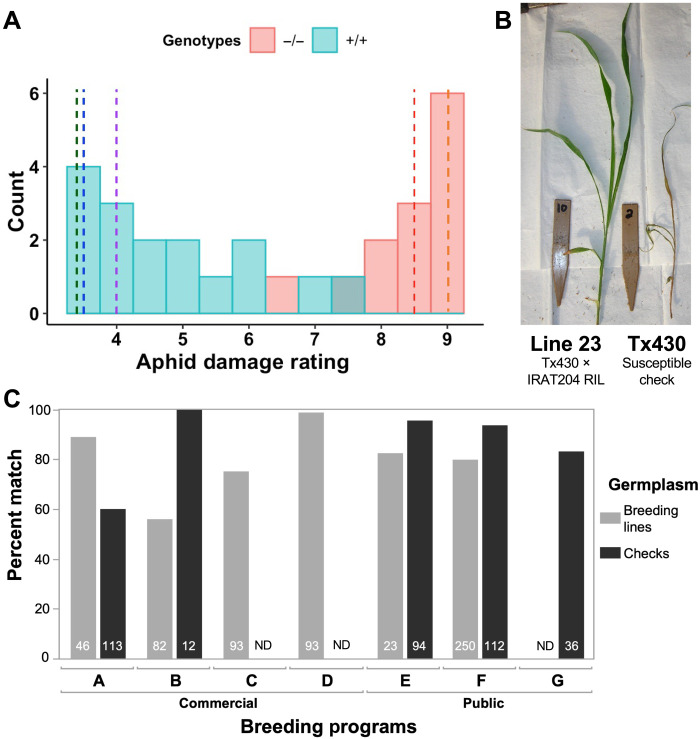
Fig. 5. Multiprogram evaluation of a molecular marker developed on the basis of the selective sweep validates its global utility.
(A) The KASP marker predicts SCA resistance in independent U.S. × Senegal breeding lines. The histogram represents the aphid damage ratings of F3 and F4 lines from a Tx430 × IRAT204 recombinant family (N = 23) and controls (N = 6) under infestation by M. sacchari at the seedling stage in a choice greenhouse assay. Cyan bars represent the aphid damage ratings for lines carrying the +/+ genotypes at the SNP 06_02892438, while the red bars represent aphid damage ratings of the lines carrying the −/− genotypes (including the controls). The blue dashed lines represent the average aphid damage rating of the resistance controls Tx2783, IRAT204, and SC110 (green, blue, and purple dashed lines, respectively), while red dashed line represents the average damage rating of susceptible controls, KS585 (orange) and Tx7000 and Tx430 (red). (B) Representative SCA reaction from the choice greenhouse assay for an F4 line carrying the +/+ genotype (left) versus the susceptible parent Tx430 (right). (C) Evaluation of the same marker in eight U.S. breeding programs. Percent match of KASP marker genotyping prediction with breeder-provided SCA resistance classification for four commercial breeding programs and three public breeding programs. The number of genotypes evaluated in each program is noted at the bottom of each bar. ND, not determined. Photo credit: Scott Armstrong, USDA-ARS.
Multiprogram validation and deployment in commercial and public breeding programs
To further validate the utility of the SCA resistance KASP markers, we tested them with four U.S. commercial seed company breeding programs and three U.S. public sector breeding programs, representing a large fraction of the U.S. sorghum breeding community (Fig. 5C). (The programs are anonymized to avoid disclosing proprietary information.) Under the hypothesis that (i) RMES1 underlies SCA resistance in U.S. breeding programs and (ii) the KASP marker (Sbv3.1_06_02892438R) tags the relevant resistant versus susceptible haplotypes, the breeders’ phenotype-based classification of SCA resistance should largely match the KASP marker genotype-based prediction. As expected, the match between the phenotype-based breeder classification and KASP marker genotypes is high, ranging from ~60 to 100%, with most germplasm sets (9 of 12) having >80% matching (Fig. 5C and fig. S6). Less than 0.5% of mismatches (5 of 1100) were observed among technical replicates (independent tissue samples from the same plant), so mismatches are unlikely to be due to KASP genotyping errors. Mismatches may be due to differences among programs of SCA-resistant or SCA-susceptible haplotypes, or errors in the phenotype-based resistance classifications [some of which are based on visual ratings under natural field infestations, which are prone to false positives (27)]. There were also some genotype-phenotype mismatches in public germplasm checks used by commercial and public programs (Fig. 5C). In nearly all cases, further investigation revealed that mismatches were due to unexpected heterogeneity in public germplasm within or among breeding programs (table S4).
DISCUSSION
RMES1 is a major resistance gene underlying evolutionary rescue of sorghum worldwide
Understanding the genetics of evolutionary rescue, including the genetic architecture and molecular basis, could contribute to more resilient conservation and breeding strategies (28). Here, we hypothesized, parsimoniously, that a single Mendelian SCA resistance locus RMES1 could underlie the global evolutionary rescue of sorghum to the new M. sacchari superclone. Previous studies had suggested that a single dominant locus is responsible for SCA resistance in families derived from resistant Chinese grain sorghum variety Henong 16 (H16) and susceptible BTx623 or in families derived from U.S. breeding lines, resistant RTx2783 and susceptible CK60 (25, 29). The H16 resistance was mapped to a ~130-kb region at 2.7 Mb on chromosome 6 (RMES1) (25). Consistent with the RMES1 evolutionary rescue hypothesis, the genome region with the highest FST in the HBP colocalized precisely with RMES1 (Fig. 3). Together, the evolutionary genome scan (Fig. 3) and multiprogram marker validation (Fig. 5) provide evidence that RMES1 is the major SCA resistance locus globally, shared across the Americas, Asia, and Africa. However, our findings do not preclude the hypotheses that different resistance alleles are found at or near RMES1 or that other SCA resistance loci were selected in Haiti and were required for the evolutionary rescue. In particular, other FST scan peaks on chromosomes 2, 7, 8, and 9 (Fig. 3) could correspond to other SCA resistance loci. Given that SCA resistance is fixed in the Haitian program, further population development and quantitative trait locus mapping for SCA resistance will be necessary to test this hypothesis.
Identifying the causal variant underlying SCA resistance would advance our understanding of aphid resistance mechanisms in plants (30) and facilitate development of perfectly predictive molecular markers for SCA resistance breeding (31). Our comparative genomic analysis between the resistant PI 276837 and the three susceptible reference genomes identified four candidate genes with putative functional variants within the RMES1 locus (Sobic.006G017200, Sobic.006G017332, Sobic.006G017400, and Sobic.006G017500; file S4). Three of the four genes in the candidate region encode leucine-rich repeat (LRR) proteins, a gene family involved in immune responses to invading pathogens and insects (32). Given that some LRR genes mediate plant resistance to aphids and other phloem-feeding insects (30), these genes represent promising candidates for the RMES1 causative gene. Functional annotation and sequence comparison between the resistant and susceptible accession identified nonsynonymous variants only in Sobic.006G017200 and Sobic.006G017500 (Fig. 4), suggesting that these nucleotide-binding LRRs are promising candidates for the RMES1 gene. Another nearby candidate is Sobic.006G016900, a cyanoalanine synthase that may regulate cyanide levels in sorghum tissues (33). Fine-mapping and positional cloning will be needed to test these hypotheses and positively identify the causative variant.
Evolutionary rescue of sorghum depended on a half century of global germplasm exchange
In the 20th century, sorghum genebanks and breeding programs exchanged germplasm widely (34, 35). On the basis of pedigree records and morphology, we hypothesized that the HBP originated from global admixed germplasm with a primary contribution of Ethiopian caudatum of the zerazera type (36). Consistent with this hypothesis, HBP genotypes clustered with caudatum sorghum of East Africa (Fig. 2), but admixture analysis identified a contribution from durra and guinea sorghum from West Africa (Fig. 2D). Combining population genomics findings (Figs. 2 and 3) with genebank and pedigree records (37, 38), we can map the history of global germplasm exchange that led to the evolutionary rescue of sorghum in Haiti following the SCA outbreak (Fig. 6A), as well as the spread of the SCA resistance allele from Ethiopia to breeding programs around the world (Fig. 6B). Notably, the evolutionary rescue of sorghum in the Americas (Haiti and United States) involved germplasm and knowledge exchange over a period of >50 years, involving nine countries on three continents.
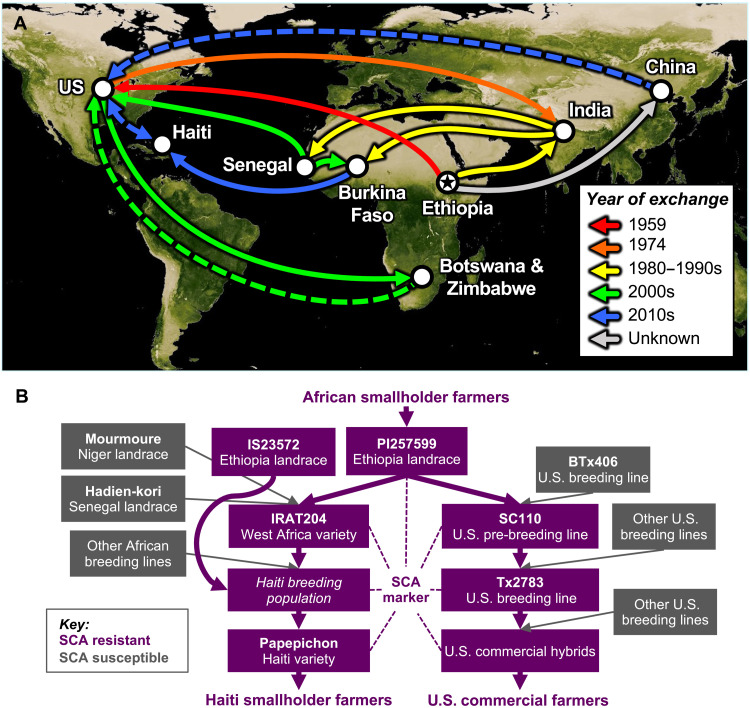
Fig. 6. Evolutionary rescue of sorghum through >50 years of global exchange of germplasm and knowledge.
(A) Germplasm and knowledge exchange inferred from pedigree records and genomic analyses. Germplasm exchange is denoted by solid lines. Knowledge exchange through scientific literature is denoted by dashed lines. The star indicates the inferred origin of the SCA resistance allele in the Ethiopian highlands, with at least two paths to the Americas, via IS 23572 (yellow line) or PI 257599 (red line). (B) Pedigree relationships among global accessions, breeding lines, breeding programs, or varieties, color-coded by inferred SCA resistance or susceptibility. Note that with respect to U.S. commercial hybrids, the diagram is illustrative and is not meant to imply that all U.S. commercial hybrids used Tx2783 as the SCA resistance donor. Some known pedigree information has been omitted from the diagram for clarity. An annotated version of this figure, with detailed evidence for each arrow, is provided as fig. S8. Map: Visible Earth/NASA.
In the case of the SCA outbreak, the global sorghum improvement community was fortunate that the rare SCA resistance allele originated in East African caudatums, since this germplasm is preferred by many sorghum breeders worldwide and widely used by breeding programs in Africa, Asia, and the Americas (34, 39). The SCA resistance allele appears to have been inadvertently spread across sorghum breeding programs across the three continents long before the recent SCA outbreak (Fig. 6). For example, SC110, a converted version of an Ethiopian caudatum landrace (PI 257599/IS 12610) identified as SCA resistant in several world regions (26, 40), is a major contributor to the pedigrees of many SCA-resistant breeding lines in the United States (Fig. 6B) (41). The same progenitor line (IS 12610) was used by breeding programs in West Africa (Fig. 6B) as a parent of IRAT204 (CE151-262; PI 656031), a widely adopted variety (42) and key progenitor of current West African breeding programs (39).
Another potential benefit of germplasm exchange is the maintenance of diversity in breeding programs following strong selection, including evolutionary rescue. Given the strong selection on the HBP during the SCA outbreak, it might be expected that the postselection HBP no longer retains sufficient diversity for future adaptation and genetic gain (7). However, the HBP was founded with diverse admixed global germplasm (Fig. 2) and extensively intercrossed, so it appears to have retained sufficient genetic diversity for future adaptation and crop improvement. While the abundance of windows with negative Tajima’s D is consistent with a recent population bottleneck and/or selective sweep, only a modest reduction in nucleotide diversity is observed throughout the genome of the HBP relative to global accessions, East African caudatums, or a major public pre-breeding program (Fig. 1E and fig. S7). Recombination during intercrossing cycles (before the SCA outbreak) presumably reshuffled the SCA resistance allele onto many backgrounds, suggesting that the intercrossing approach was critical to allow the Haitian program to retain diversity for future genetic gain and adaptation.
Rapid discovery and deployment of a global trait-predictive molecular marker using evolutionary population genomics
Molecular marker development based on phenotype-to-genotype mapping of trait loci (e.g., linkage or association mapping) is limited by availability of suitable mapping populations, phenotyping capacity, and genotyping resources, which can take years to develop (13, 43). For instance, spatial and temporal variability of SCA infestation in field trials limits the effectiveness of field phenotyping (27), while greenhouse assays can be complicated and time-consuming for lower-resourced programs. Thus, an evolutionary genomics approach, which leverages a history of selection by smallholder farmers or plant breeders, could have advantages for marker discovery. Despite wide use of evolutionary genome scans in crops, the hypotheses generated on adaptive loci are rarely, if ever, tested by independent experimental approaches (e.g., with near isogenic lines) (44). To our knowledge, this is the first example where an evolutionary or population genomic scan led directly to molecular breeding technology in use in commercial and public varietal development (Figs. 3 and 5).
Here, we demonstrated the effectiveness of the evolutionary population genomic approach, showing that a marker discovered in a single developing country breeding program (Chibas-Haiti) can link crop improvement efforts across three continents (North America, Africa, and Asia; Figs. 5A and 6) and the commercial and public sector (Fig. 5, A and C). Thus, our findings establish the value of evolutionary population genomics to facilitate and guide global crop improvement. The KASP marker developed and validated in this study can facilitate the rapid conversion of existing farmer-preferred varieties for SCA resistance (e.g., by marker-assisted introgression) (45). While the RMES1 resistance allele is currently conferring effective resistance, a further biotype shift in the aphid could overcome this gene. Several biotype shifts occurred in the 1960s to 1980s for the greenbug aphid Schizaphis graminum (46) and slowed genetic gain in sorghum for many years (47). In programs that have already fixed RMES1 (such as Chibas), the markers developed here could also facilitate identification of additional SCA resistance genes via counterselection of RMES1 allele to reveal unidentified SCA resistance genes after crossing with non-RMES1 resistance sources. These outsourced KASP markers are convenient for breeding programs, since they require no laboratory labor or facilities, and are low cost relative to dedicated field or greenhouse phenotyping capacity, at ~$2 per sample for DNA extraction and marker genotyping (48).
Synergy of long-standing germplasm exchange practices with genomics technologies
In this study, we integrated evolutionary population genomic analyses and historical records on global germplasm exchange to show that the recent evolutionary rescue of sorghum depended on >50 years of germplasm exchange. Germplasm exchange led to global diffusion of a rare SCA resistance allele, sometimes purposely and sometimes inadvertently, from smallholder farmers in Ethiopian highlands across breeding programs in Africa, the Americas, and Asia. Over the past several decades, movement of crop genetic resources through international cooperation of germplasm exchange has provided access to adaptive genetic variation for crop improvement (35, 49). However, germplasm exchange is increasingly restricted because of commercial or institutional interests asserting intellectual property (IP) rights and governments asserting national or local sovereignty over genetic resources (50, 51). While IP rights and sovereignty are important considerations, the question remains how to balance these aims with the benefits of free exchange of global public goods (52, 53). While we are not in a position to resolve these societal trade-offs, our study does highlight the global food security benefits of germplasm exchange and the opportunities that could be lost because of restrictions on exchange.
Together, our findings suggest that genomic technologies will be most powerful when leveraged with global exchange of germplasm and knowledge. No matter how powerful genomic technologies are in terms of accuracy or throughput, their utility will depend on the germplasm assayed, since all genetic mapping approaches require effective recombination and allelic diversity (54, 55). Global germplasm exchange vastly increases both these parameters, providing a “bank” of historical recombinations and allelic variants that can be rapidly leveraged with genomic tools (Figs. 3 and 4). Therefore, our best opportunity to address challenges of global change may be to leverage new genomic technologies with long-standing practices of global germplasm exchange.
MATERIALS AND METHODS
Sorghum breeding and production in Haiti
The Chibas sorghum breeding program was launched in 2013 using admixed global germplasm, including heterogeneous breeding material from West Africa carrying ms3 nuclear male sterility and inbred global accessions. During 2015–2018, the material was selected in breeding nurseries under low-input conditions (approximating local smallholder practices) and extensively intercrossed using the ms3 sterility system. No insecticides were used to limit SCA infestations in breeding nurseries in this period, and natural SCA infestations were intense during this period (e.g., Fig. 1C). Note that selection pressure on sorghum by SCA in Haiti is expected to be greater than in temperate zones (e.g., United States) because the SCA infestation occurs year-round in this tropical environment (19). Annual sorghum production estimates for Haiti are based on Food and Agriculture Organization Corporate Statistical Database (FAOSTAT; 2009–2014 and 2018) (56) and the U.S. Department of Agriculture (USDA) forecast for 2019–2020 (22). FAOSTAT data for 2015–2017 and 2019 were not used because they were based on imputation (“FAO data based on imputation methodology”) that did not account for the known effects of SCA (e.g., “this aphid spread throughout the country and decimated Haiti sorghum production”) (22). Production for the missing years of SCA outbreak was inferred on the basis of 2009 agriculture survey acreage before infestation in each region and assessment of sorghum production levels compared to pre-infestation levels, adjusted to FAOSTAT (1990–2014) production averages for each region.
Plant genetic resources
The HBP (N = 296) are inbred lines derived from a recurrent selection breeding population developed by intercrossing germplasm that survived natural SCA infestation. For genomic DNA extraction, fresh leaf tissue of each accession was collected from 2-week-old seedlings raised in a greenhouse. Tissue was lyophilized for 2 days and then ground using a 96-well plate plant tissue grinder (Retsch Mixer Mill). Genomic DNA was extracted using the BioSprint 96 DNA Plant Kit (QIAGEN), quantified using the Quant-iT PicoGreen dsDNA Assay Kit, and normalized to 10 ng/μl. An additional set of global accessions (GDP, N = 767) was assembled on the basis of published datasets (57, 58), selecting the subset of accessions with georeference information and other passport data such as country of origin and morphological type (fig. S1 and file S1). The GDP accessions were from 52 countries on five continents and all major botanical types including 164 caudatum, 96 guinea, 81 durra, 57 bicolor, and 47 kafir accessions; 288 of other botanical types; and 34 accessions of unknown botanical type. A subset of publicly available pre-breeding lines (N = 35) from Texas A&M University’s sorghum program was identified from the GDP.
Genotyping-by-sequencing
Genotypes for the 296 Haitian breeding lines were generated with genotyping-by-sequencing. Genomic DNA digestion, ligation, and PCR amplification processes were performed according to the methods previously described (57). The libraries were sequenced using the single-end 100-cycle sequencing by Illumina HiSeq2500 (Illumina, San Diego, CA, USA) at the University of Kansas Medical Center, Kansas City, MO, USA. A total of 220 million reads for the HBP were combined with published data for the GDP (57) for SNP calling. TASSEL 5 GBS v2 pipeline (59) was used to perform the SNP calling of the sequence data obtained from Illumina sequencing. Reads were aligned to the BTx623 sorghum reference genome v.3.1 (60) with the Burrows-Wheeler alignment (61). The SNPs were filtered for 20% missingness, and then missing data were imputed using BEAGLE 4.0 (62). Raw sequence reads are available at the Sequence Read Archive under accession PRJNA757369, and genotyping data are available at Dryad (https://doi.org/10.5061/dryad.n02v6wwx0).
Population genomic analyses
Genome-wide nucleotide diversity (π) and Tajima’s D statistics for HBP and GDP were estimated on the basis of a nonoverlapping sliding window of 1 million base pairs (Mbp) across the genome using VCFtools (63). The characterization of the population structure of the HBP was based on a discriminant analysis of principal components in the Adegenet package in R (64). A distance matrix calculated on the basis of a modified Euclidean distance model was used to create a cladogram based on a neighbor-joining algorithm in TASSEL (65). Neighbor-joining analysis was visualized using the APE package in R (66). The population structure of the germplasm panel was further assessed by the Bayesian model–based clustering method implemented in the ADMIXTURE program (67). Pairwise SNP differentiation (FST) between the HBP and the GDP was calculated, and outlier loci were detected on the basis of an inferred distribution of neutral FST using the R package OutFLANK (68).
Whole-genome resequencing
Around the 130-kb mapped interval in BTx623, SNPs from 10 sorghum accessions with known SCA resistance status were examined to search for functional mutations responsible for SCA resistance. Six of the 10 resequenced accessions represent known susceptible lines, which include RTx430 (PI 655996), BTx623 (PI 564163), Tx7000 (PI 655986), Tx2737 (PI 655978), BTx642, and RTx436. The remaining four resequenced accessions represent known resistant lines, which include PI 257599 (SC110 original exotic parent), PI 276837 (SC170 original exotic parent), PI 534157 (SC170), and IS 36563 (IRAT204). These samples were used before publication for this interval analysis with permission from TERRA-REF (T.C.M.), JGI Sorghum Pan-genome project (T.C.M.), BMFG Sorghum Genomic Toolbox (T.C.M. and G.P.M.), JGI Sorghum Diversity project (J. Mullet), and JGI EPICON project (J.P.V.). The reads were mapped to Sorghum bicolor v3.1 using bwa-mem. The bam file was filtered for duplicates using Picard (http://broadinstitute.github.io/picard) and realigned around indels using GATK (69). Multisample SNP calling was done using SAMtools mpileup and VarScan V2.4.0 with a minimum coverage of 8 and a minimum alternate allele frequency of 4. Repeat content of the genome was masked using 24–base pair (bp) kmers. Kmers that occur at a high frequency, up to 5%, were masked. SNPs around 25 bp of the mask were removed for further analysis. An SNP was included for further analysis only when it has coverage in 75% of the samples and a MAF > 0.005. Functional annotation of the variants within the RMES1 locus was performed using SnpEff.
De novo genome sequencing
De novo genome assembly of the resistance sorghum line PI 276837 was used to perform comparative genomic analysis to identify the causative variant for SCA resistance at the RMES1 locus. PI 276837 main assembly consisted of 101.47× of PACBIO coverage with an average read size of 11,931 bp. The genome was assembled using Canu 1.8, a fork of the Celera Assembler designed for high-noise single-molecule sequencing. The resulting sequence was polished using ARROW. The assembled genome resulted in contig N50 sizes ranging from 14 to 19 kb and scaffold N50 sizes ranging from 5 to 65 kb. Sequence variations at RMES1 locus between the de novo sequence of PI 276837 were compared to the reference genomes of BTx623, Tx430, and BTx642.
KASP marker development
SNPs from the FST genomic selection scan were selected for development into markers based on several factors: LOD score (logarithm of the odds ratio for linkage) of the FST analysis, proximity to RMES1 locus, and suitability of the flanking sequence for KASP assay development. KASP assays were developed using the KASP-by-design service from LGC Biosearch Technologies (Middlesex, UK) through a third-party genotyping service provider, Intertek (Alnarp, Sweden), who genotyped the KASP assays using Kraken software (LGC Biosearch Technologies). All genomic DNA extraction and KASP genotyping were performed by Intertek using two 6-mm leaf punches dried with silica beads. Initial technical validation of the KASP marker was performed using known resistant (SC110, IRAT204, and Tx2783) and susceptible (KS585 and Tx7000) sorghum lines. The KASP markers developed for SCA resistance selection (file S6) are publicly available through the third-party genotyping service provided by Intertek. For further information on accessing markers, contact the corresponding author.
Marker validation in public and commercial breeding programs
To test the predictiveness of the marker, a population segregating for SCA resistance was developed by crossing the susceptible Tx430 and resistant IRAT204. F3 and F4 lines of the Tx430 × IRAT204 population were genotyped with the KASP marker together with the susceptible and resistant parents. The same population was evaluated for SCA reaction under artificial inoculation using a free-choice flat-screen trial in the greenhouse. Tx2783 and SC110 were included as known resistant genotypes, along with the known susceptible genotypes, KS 585 and Tx7000 (26, 70). SCAs used for artificial inoculations originated from collections off grain sorghum near Bay City, TX in August of 2013 and have been maintained in an experimental colony, as previously described (27). Free-choice flat-screen assay, data collection (damage rating, SPAD score, and plant height difference), and analysis were conducted as previously described (27).
Validation of the KASP marker across different breeding programs was performed in seven additional breeding programs, four commercial and three public in the United States. The marker testing was done under an agreement that the identity of the programs would remain confidential, so we have anonymized the findings. Each program collected tissue samples from known tolerant and susceptible parental breeding lines, F1’s of the parental lines, and later-generation lines from their SCA tolerance breeding populations; the SCA reaction phenotypes of the late-generation lines may or may not have been known. For the parental breeding lines, both technical replicates (tissue samples from the same plant) and biological replicates (tissue samples from separate plants) were collected to test both the technical function of the markers and the reliability of the germplasm, respectively. In addition, most programs included public sources (e.g., Tx2783) of known SCA tolerance as checks. Tissue samples were sent to Intertek, who extracted DNA and performed the KASP genotyping.
Acknowledgments
Funding: This study is made possible by the support of the American people provided to the Feed the Future Innovation Lab for Collaborative Research on Sorghum and Millet through the U.S. Agency for International Development (USAID) under associate award no. AID-OAA-LA-16-00003, “Feed the Future Innovation Lab for Genomics-Assisted Sorghum Breeding.” The contents are the sole responsibility of the authors and do not necessarily reflect the views of USAID or the U.S. government. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. We thank the Joint Genome Institute and collaborators for prepublication access to the genomes of Tx430 and Tx642 for use in this study. Additional support was provided by the Bill & Melinda Gates Foundation under the “Sorghum Genomics Toolkit” project. Such use does not constitute an official endorsement or approval by the USDA or the Agricultural Research Service of any product or service to the exclusion of others that may be suitable. USDA is an equal opportunity provider and employer. Support for the marker testing program was provided by the Collaborative Sorghum Investment Program, including additional coordination provided by Sarah Sexton-Bowser. We thank the breeding programs participating in the marker testing for their in-kind support.
Author contributions: Conceptualization: G.P. and G.P.M. Investigation: K.T.M., T.F., G.P., N.W., R.W., J.R.C., J.S.A., S.M., C.P., J.P.V., P.G.L., J.S., and G.P.M. Formal analysis: K.T.M., T.F., J.S.A., and S.M. Visualization: K.T.M., T.F., and G.P.M. Supervision: G.P., G.P.M., J.G., and J.S. Funding acquisition: G.P., T.C.M., J.P.V., P.G.L., J.S., and G.P.M. Writing (original draft): K.T.M., T.F., S.M., and G.P.M.
Competing interests: The authors declare that they have no competing interests. The breeding programs that participated in the marker testing did so under the agreement that (i) their data would remain confidential and (ii) anonymized results would be published. The authors provided participating breeding programs complimentary marker genotyping support and data in exchange for their participation in the testing program. After completion of the marker testing, one participating program provided funding to KSU for further marker implementation and development.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper, the Supplementary Materials, Dryad (https://doi.org/10.5061/dryad.n02v6wwx0), or the Sequence Read Archive (PRJNA757369).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Files S1 to S7
REFERENCES AND NOTES
- 1.Palumbi S. R., Humans as the world’s greatest evolutionary force. Science 293, 1786–1790 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Wheeler T., von Braun J., Climate change impacts on global food security. Science 341, 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez A., Ronce O., Ferriere R., Hochberg M. E., Evolutionary rescue: An emerging focus at the intersection between ecology and evolution. Philos. Trans. R. Soc. B Biol. Sci. 368, 20120404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell G., Evolutionary rescue. Annu. Rev. Ecol. Evol. Syst. 48, 605–627 (2017). [Google Scholar]
- 5.Peter B. M., Huerta-Sanchez E., Nielsen R., Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLOS Genet. 8, e1003011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexander H. K., Martin G., Martin O. Y., Bonhoeffer S., Evolutionary rescue: Linking theory for conservation and medicine. Evol. Appl. 7, 1161–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr H. A., Unckless R. L., The population genetics of evolutionary rescue. PLOS Genet. 10, e1004551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner R. S., After the famine: Plant pathology, Phytophthora infestans, and the late blight of potatoes, 1845–1960. Hist. Stud. Phys. Biol. Sci. 35, 341–370 (2005). [Google Scholar]
- 9.Burke J. M., Burger J. C., Chapman M. A., Crop evolution: From genetics to genomics. Curr. Opin. Genet. Dev. 17, 525–532 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Wilson B. A., Pennings P. S., Petrov D. A., Soft selective sweeps in evolutionary rescue. Genetics 205, 1573–1586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pennings P. S., Hermisson J., Soft sweeps III: The signature of positive selection from recurrent mutation. PLOS Genet. 2, e186 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitti J. J., Grossman S. R., Sabeti P. C., Detecting natural selection in genomic data. Annu. Rev. Genet. 47, 97–120 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., Kiani S., Forrest K., Saintenac C., Brown-Guedira G. L., Akhunova A., See D., Bai G., Pumphrey M., Tomar L., Wong D., Kong S., Reynolds M., da Silva M. L., Bockelman H., Talbert L., Anderson J. A., Dreisigacker S., Baenziger S., Carter A., Korzun V., Morrell P. L., Dubcovsky J., Morell M. K., Sorrells M. E., Hayden M. J., Akhunov E., Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 8057–8062 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuber C. W., Moll R. H., Goodman M. M., Schaffer H. E., Weir B. S., Allozyme frequency changes associated with selection for increased grain yield in maize (ZEA MAYS L.). Genetics 95, 225–236 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wisser R. J., Murray S. C., Kolkman J. M., Ceballos H., Nelson R. J., Selection mapping of loci for quantitative disease resistance in a diverse maize population. Genetics 180, 583–599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coque M., Gallais A., Genomic regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor. Appl. Genet. 112, 1205–1220 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Hufford M. B., Mier y Teran J. C. B., Gepts P., Crop biodiversity: An unfinished magnum opus of nature. Annu. Rev. Plant Biol. 70, 727–751 (2019). [DOI] [PubMed] [Google Scholar]
- 18.R. Monk, C. Franks, J. Dahlberg, in Yield Gains in Major US Field Crops (Crop Science Society of America, 2014), pp. 293–310; https://dl.sciencesocieties.org/publications/books/abstracts/cssaspecialpubl/yieldgainsinmaj/293.
- 19.Bowling R. D., Brewer M. J., Kerns D. L., Gordy J., Seiter N., Elliott N. E., Buntin G. D., Way M. O., Royer T. A., Biles S., Maxson E., Sugarcane aphid (Hemiptera: Aphididae): A new pest on sorghum in North America. J. Integr. Pest Manag. 7, 12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordy J. W., Brewer M. J., Bowling R. D., Buntin G. D., Seiter N. J., Kerns D. L., Reay-Jones F. P. F., Way M. O., Development of economic thresholds for sugarcane aphid (Hemiptera: Aphididae) in susceptible grain sorghum hybrids. J. Econ. Entomol. 112, 1251–1259 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Nibouche S., Costet L., Holt J. R., Jacobson A., Pekarcik A., Sadeyen J., Armstrong J. S., Peterson G. C., McLaren N., Medina R. F., Invasion of sorghum in the Americas by a new sugarcane aphid (Melanaphis sacchari) superclone. PLOS ONE 13, e0196124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.USDA-FAS, Grain and Feed Annual Report: Haiti (HA2020-0001) (2020); https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Grain%20and%20Feed%20Annual_Port-au-Prince_Haiti_04-15-2020.
- 23.W. Calvin, thesis, University of Florida, Gainesville, FL (2019). [Google Scholar]
- 24.Muleta K. T., Pressoir G., Morris G. P., Optimizing genomic selection for a sorghum breeding program in Haiti: A simulation study. G3 9, 391–401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F., Zhao S., Han Y., Shao Y., Dong Z., Gao Y., Zhang K., Liu X., Li D., Chang J., Wang D., Efficient and fine mapping of RMES1 conferring resistance to sorghum aphid Melanaphis sacchari. Mol. Breed. 31, 777–784 (2013). [Google Scholar]
- 26.Armstrong J. S., Rooney W. L., Peterson G. C., Villenueva R. T., Brewer M. J., Sekula-Ortiz D., Sugarcane aphid (Hemiptera: Aphididae): Host range and sorghum resistance including cross-resistance from greenbug sources. J. Econ. Entomol. 108, 576–582 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Armstrong J. S., Mbulwe L., Sekula-Ortiz D., Villanueva R. T., Rooney W. L., Resistance to Melanaphis sacchari (Hemiptera: Aphididae) in forage and grain sorghums. J. Econ. Entomol. 110, 259–265 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Burgarella C., Barnaud A., Kane N. A., Jankowski F., Scarcelli N., Billot C., Vigouroux Y., Berthouly-Salazar C., Adaptive introgression: An untapped evolutionary mechanism for crop adaptation. Front. Plant Sci. 10, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tetreault H. M., Grover S., Scully E. D., Gries T., Palmer N. A., Sarath G., Louis J., Sattler S. E., Global responses of resistant and susceptible sorghum (Sorghum bicolor) to sugarcane aphid (Melanaphis sacchari). Front. Plant Sci. 10, 145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y., Zhang C.-X., Chen R., He S. Y., Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 23390–23397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen J. R., Lübberstedt T., Functional markers in plants. Trends Plant Sci. 8, 554–560 (2003). [DOI] [PubMed] [Google Scholar]
- 32.McHale L., Tan X., Koehl P., Michelmore R. W., Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 7, 212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machingura M., Salomon E., Jez J. M., Ebbs S. D., The β-cyanoalanine synthase pathway: Beyond cyanide detoxification. Plant Cell Environ. 39, 2329–2341 (2016). [DOI] [PubMed] [Google Scholar]
- 34.C. W. Smith, R. A. Frederiksen, in Sorghum: Origin, History, Technology, and Production, C. W. Smith, R. A. Frederiksen, Eds. (Wiley, 2000), vol. 2, p. 191. [Google Scholar]
- 35.Klein R. R., Miller F. R., Dugas D. V., Brown P. J., Burrell A. M., Klein P. E., Allelic variants in the PRR37 gene and the human-mediated dispersal and diversification of sorghum. Theor. Appl. Genet. 128, 1669–1683 (2015). [DOI] [PubMed] [Google Scholar]
- 36.J. A. Dahlberg, D. T. Rosenow, in Achieving Sustainable Cultivation of Sorghum Volume 1 (Burleigh Dodds Science Publishing, 2018), pp. 23–86. [Google Scholar]
- 37.Peterson G. C., Johnson J. W., Teetes G. L., Rosenow D. T., Registration of Tx2783 greenbug resistant sorghum germplasm line. Crop. Sci. 24, 390 (1984). [Google Scholar]
- 38.République du Sénégal, Catalogue officiel des espèces et des variétés cultivées au Sénégal [Official catalog of cultivated species and varieties in Senegal] (2012); www.fao.org/pgrfa-gpa-archive/sen/docs/senegal_varietes/Catalogue_%20varietes.htm.
- 39.Faye J. M., Maina F., Akata E. A., Sine B., Diatta C., Mamadou A., Marla S., Bouchet S., Teme N., Rami J.-F., Fonceka D., Cisse N., Morris G. P., A genomics resource for genetics, physiology, and breeding of West African sorghum. Plant Genome 14, e20075 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Mbulwe L., Peterson G. C., Scott-Armstrong J., Rooney W. L., Registration of sorghum germplasm Tx3408 and Tx3409 with tolerance to sugarcane aphid [Melanaphis sacchari (Zehntner)]. J. Plant Regist. 10, 51–56 (2016). [Google Scholar]
- 41.Peterson G. C., Armstrong J. S., Pendleton B. B., Stelter M., Brewer M. J., Registration of RTx3410 through RTx3428 sorghum germplasm resistant to sugarcane aphid [Melanaphis sacchari (Zehntner)]. J. Plant Regist. 12, 391–398 (2018). [Google Scholar]
- 42.J. Ndjeunga, K. Mausch, F. Simtowe, in Crop Improvement, Adoption, and Impact of Improved Varieties in Food Crops in Sub-Saharan Africa, T. S. Walker, J. Alwang, Eds. (CABI, Wallingford, 2015), pp. 123–147; www.cabi.org/cabebooks/ebook/20153367543.
- 43.Nuzhdin S. V., Turner T. L., Promises and limitations of hitchhiking mapping. Curr. Opin. Genet. Dev. 23, 694–699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoban S., Kelley J. L., Lotterhos K. E., Antolin M. F., Bradburd G., Lowry D. B., Poss M. L., Reed L. K., Storfer A., Whitlock M. C., Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yohannes T., Abraha T., Kiambi D., Folkertsma R., Hash C. T., Ngugi K., Mutitu E., Abraha N., Weldetsion M., Mugoya C., Masiga C. W., de Villiers S., Marker-assisted introgression improves Striga resistance in an Eritrean farmer-preferred sorghum variety. Field Crops Res. 173, 22–29 (2015). [Google Scholar]
- 46.Royer T. A., Pendleton B. B., Elliott N. C., Giles K. L., Greenbug (Hemiptera: Aphididae) biology, ecology, and management in wheat and sorghum. J. Integr. Pest Manag. 6, 19 (2015). [Google Scholar]
- 47.F. R. Miller, Y. Kebede, in Genetic Contributions to Yield Gains of Five Major Crop Plants (John Wiley & Sons Ltd., 1984), pp. 1–14; 10.2135/cssaspecpub7.c1. [DOI]
- 48.Thomson M. J., High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2, 195–212 (2014). [Google Scholar]
- 49.Fowler C., Hodgkin T., Plant genetic resources for food and agriculture: Assessing global availability. Annu. Rev. Env. Resour. 29, 143–179 (2004). [Google Scholar]
- 50.P. Heisey, K. Day-Rubenstein, Using crop genetic resources to help agriculture adapt to climate change: Economics and policy (SSRN Scholarly Paper ID 2709190, Social Science Research Network, Rochester, NY, 2015).
- 51.Prathapan K. D., Pethiyagoda R., Bawa K. S., Raven P. H., Rajan P. D.; 172 co-signatories from 35 countries , When the cure kills—CBD limits biodiversity research. Science 360, 1405–1406 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Esquinas-Alcázar J., Protecting crop genetic diversity for food security: Political, ethical and technical challenges. Nat. Rev. Genet. 6, 946–953 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Kotschi J., Horneburg B., The Open Source Seed Licence: A novel approach to safeguarding access to plant germplasm. PLOS Biol. 16, e3000023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernardo R., Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 48, 1649–1664 (2008). [Google Scholar]
- 55.Brachi B., Morris G. P., Borevitz J. O., Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 12, 232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.FAO, FAOSTAT (2021); www.fao.org/faostat/en/.
- 57.Morris G. P., Ramu P., Deshpande S. P., Hash C. T., Shah T., Upadhyaya H. D., Riera-Lizarazu O., Brown P. J., Acharya C. B., Mitchell S. E., Harriman J., Glaubitz J. C., Buckler E. S., Kresovich S., Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. U.S.A. 110, 453–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lasky J. R., Upadhyaya H. D., Ramu P., Deshpande S., Hash C. T., Bonnette J., Juenger T. E., Hyma K., Acharya C., Mitchell S. E., Buckler E. S., Brenton Z., Kresovich S., Morris G. P., Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 1, e1400218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J., Sun Q., Buckler E. S., TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLOS ONE 9, e90346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCormick R. F., Truong S. K., Sreedasyam A., Jenkins J., Shu S., Sims D., Kennedy M., Amirebrahimi M., Weers B. D., McKinley B., Mattison A., Morishige D. T., Grimwood J., Schmutz J., Mullet J. E., The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Browning B. L., Browning S. R., Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jombart T., Devillard S., Balloux F., Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bradbury P. J., Zhang Z., Kroon D. E., Casstevens T. M., Ramdoss Y., Buckler E. S., TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitlock M. C., Lotterhos K. E., Reliable detection of loci responsible for local adaptation: Inference of a null model through trimming the distribution of FST. Am. Nat. 186, S24–S36 (2015). [DOI] [PubMed] [Google Scholar]
- 69.der Auwera G. A. V., Carneiro M. O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K. V., Altshuler D., Gabriel S., DePristo M. A., From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paudyal S., Armstrong J. S., Giles K. L., Payton M. E., Opit G. P., Limaje A., Categories of resistance to sugarcane aphid (Hemiptera: Aphididae) among sorghum genotypes. J. Econ. Entomol. 112, 1932–1940 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4
Files S1 to S7