Abstract
The hallmark autoantibodies in rheumatoid arthritis are characterized by variable domain glycans (VDGs). Their abundant occurrence results from the selective introduction of N-linked glycosylation sites during somatic hypermutation, and their presence is predictive for disease development. However, the functional consequences of VDGs on autoreactive B cells remain elusive. Combining crystallography, glycobiology, and functional B cell assays allowed us to dissect key characteristics of VDGs on human B cell biology. Crystal structures showed that VDGs are positioned in the vicinity of the antigen-binding pocket, and dynamic modeling combined with binding assays elucidated their impact on binding. We found that VDG-expressing B cell receptors stay longer on the B cell surface and that VDGs enhance B cell activation. These results provide a rationale on how the acquisition of VDGs might contribute to the breach of tolerance of autoreactive B cells in a major human autoimmune disease.
Variable domain immunoglobulin glycans affect antigen binding and enhance B cell activation.
INTRODUCTION
A pathogenic role of B cells in autoimmunity is evidenced by the effective treatment of multiple autoimmune diseases, such as rheumatoid arthritis (RA), multiple sclerosis, antineutrophil cytoplasmic antibody (ANCA)–associated vasculitis, and systemic lupus erythematosus with B cell–targeted therapies (1). Together with the observation that disease-specific autoantibody responses often characterize autoimmune diseases, these findings point to the notion that autoreactive B cells are involved in disease pathogenesis. Autoreactive B cells could contribute to disease via diverse mechanisms, such as antigen presentation to T cells, secretion of cytokines, or the production of pathogenic antibodies (2). To mediate these functions, an autoreactive B cell response needs to be induced, which involves the breach of tolerance mechanisms that generally prevent the activation of pathogenic B cells. However, the mechanisms leading to the induction and survival of autoreactive B cells that contribute to autoimmune diseases remain unclear. In mouse models, it has been shown that B cells harboring autoreactive B cell receptors (BCRs) are regulated in the bone marrow and peripheral tissues by clonal deletion, anergy induction (short-lived inactivated cells), or receptor editing (3–7). This multistep process eliminates high-affinity and potentially autoreactive clones (8). During immune responses against non-self or modified proteins carried by unwanted intruders, some initially autoreactive B cells might somatically mutate to be more efficient defense cells, thereby potentially losing affinity to self-antigens (9, 10). This might include a role for variable domain glycans (VDGs) that could regulate self/non–self-discrimination of B cells as has been described in a BCR-transgenic mouse model (11). Notably, the survival of autoreactive B cells involves costimulatory receptors and negative regulators, such as the sialic acid–binding lectin CD22, which will, together with the BCR, determine the outcome of the activation, the timing of T cell help, and the accessibility of antigens (12, 13). At present, although we have some glimpses through the study of autoreactive B cell biology in mice, the processes controlling human autoreactive B cells are largely unexplored.
The hallmark disease-specific autoantibodies in RA, a common human autoimmune disease affecting synovial joints and cartilage, are anti–citrullinated protein antibodies (ACPAs) (14). ACPAs specifically recognize a citrulline side chain and show limited interactions with the side chains of surrounding amino acids on the protein surface, explaining their promiscuity in their protein or peptide specificity (15). Recently, we have found that more than 90% of ACPAs are glycosylated in their variable domains (16, 17). The VDGs displayed on ACPAs are acquired through the introduction of N-linked glycosylation sites following somatic hypermutation (18). VDGs are mainly complex-type carbohydrates containing a high percentage of sialic acids (16). Intriguingly, the presence of VDGs in ACPA-positive healthy subjects is associated with the transition toward RA (19). This observation, together with the selective introduction of N-linked glycosylation sites and the finding that this introduction has taken place in more than 90% of ACPAs in patients with RA (16, 18), underlines the importance of this glycan tag and suggests that VDGs play a regulatory role for ACPA-expressing B cells. Intriguingly, variable domain glycosylation of antibodies is a characteristic not only of RA but also for some other human responses, such as ANCA-associated vasculitis, Sjögren’s syndrome, anti-Hinge, and antidrug antibodies (20–22).
By combining crystallography, glycobiology, and antigen-binding studies, we show that ACPA VDGs can mask binding toward potential self-antigens. Functionally, we demonstrate that BCR VDGs enhance B cell activation and decrease BCR downmodulation. This occurs independently of the negative regulator CD22. Together, the data demonstrate an important role of VDGs in changing the threshold of B cells to respond to self-antigens and to be activated. The generation of B cells with receptors modified by VDGs could lead to a loss of control of the self-reactive response and may play a role in the pathogenesis of RA.
RESULTS
Generation of patient-derived monoclonal antibodies with and without VDGs
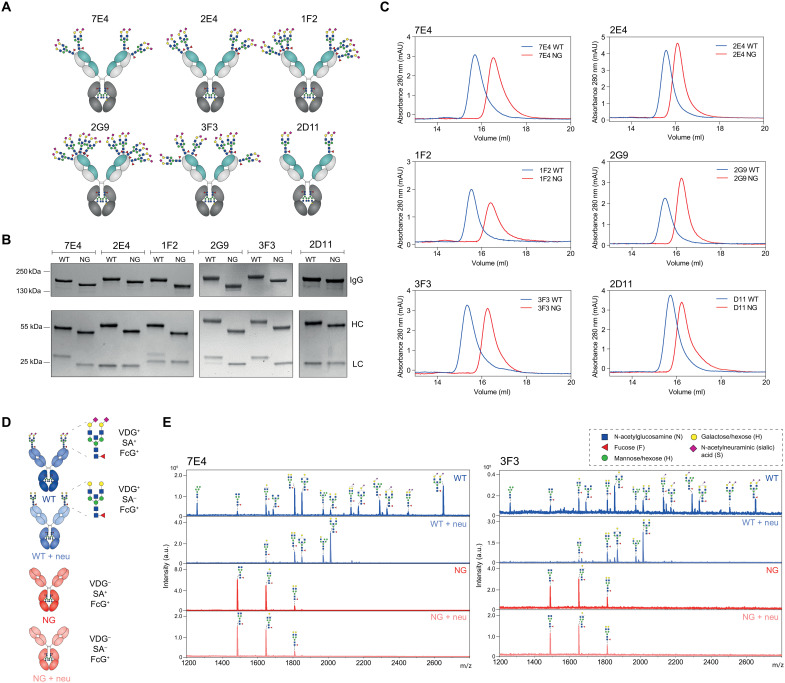
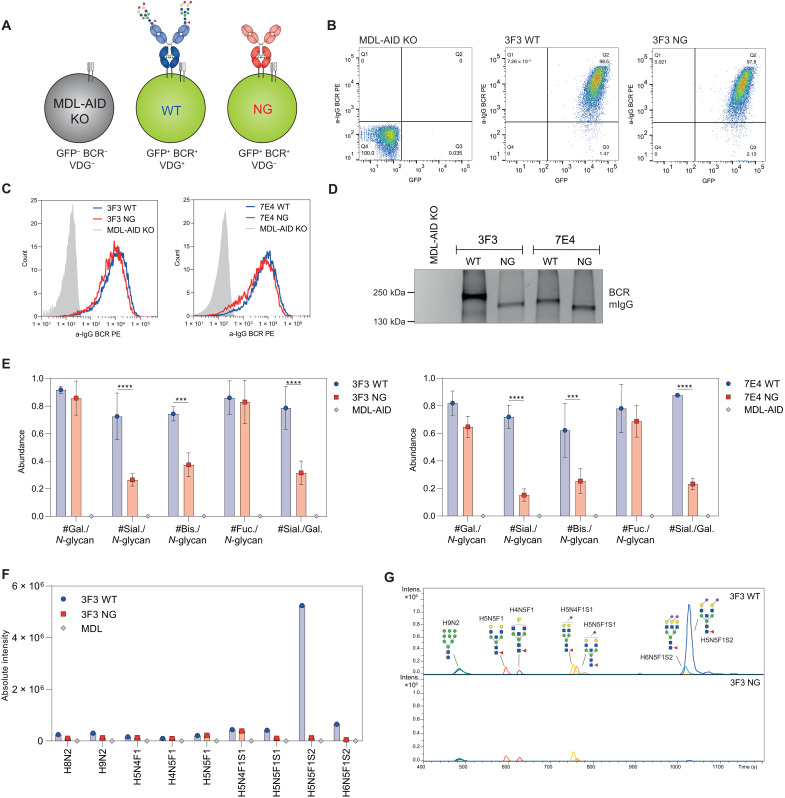
To investigate the putative effects of autoantibody VDG on antigen binding, we generated six monoclonal antibodies (mAbs) based on full-length immunoglobulin G (IgG) BCR sequences isolated from ACPA-positive patients with RA (Fig. 1A and Table 1) (23, 24). We produced the IgG1 mAbs as wild-type (WT) variants including their respective N-linked glycosylation sites, which are required for the acquisition of VDGs. Next, nonglycosylated (NG) mAbs were generated by mutating the N-linked glycosylation sites in the variable domains back to the respective germline sequence (Table 1). Both mAb variants express the typical asparagine(N)297-linked Fc-glycans. The recombinant proteins were expressed in human embryonic kidney cells with transiently overexpressed glycosyltransferases (i.e., for glyco-engineering) (25) to achieve glyco-variants similar to those found on ACPA-IgG isolated from patients with RA (16). We analyzed the mAb integrity and the occupancy of the N-linked glycosylation sites by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and size exclusion chromatography (SEC). Our results confirmed an increased apparent molecular weight of ACPA-IgG due to the presence of VDGs (17), as evidenced by a shift of the WT versus the NG variants in gel electrophoresis and SEC (Fig. 1, B and C). Furthermore, our data indicated that all N-linked glycosylation sites were fully occupied by VDGs, except for the glycosylation sites in the light chain (LC) of the mAbs 1F2 and 2G9, which appeared to be partially occupied (Fig. 1B).
Fig. 1. Generation of patient-derived IgG1 monoclonal anti–citrullinated protein (auto)antibodies (ACPA) with (WT) and without (NG) VDGs.
(A) Schematic representation of the generated ACPA-IgG (7E4, 2E4, 1F2, 2G9, 3F3, and 2D11) carrying various amounts of N-linked glycans in the variable regions of their HC (turquoise) and LC (light gray). (B) Size shift between WT and NG ACPA under nonreduced (IgG) and reduced (HC and LC) conditions on a 4 to 15% gradient SDS protein gel (Bio-Rad). The size was determined using the PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific). (C) Size shift between WT and NG ACPA-IgG identified via analytical size exclusion chromatography (SEC). ACPA-IgG+ fractions were determined via the absorbance at 280 nm. mAU, milli absorbance unit (D) Schematic depiction of the four ACPA variants (WT, NG, WT + neu, and NG + neu) analyzed: variable domain glycosylated (VDG+), Fc-glycosylated (FcG+), sialylated (SA+), and nonsialylated (SA−) after neuraminidase (neu) treatment. (E) MALDI-TOF MS analysis of N-linked variable domain and Fc-glycans from WT, WT + neu, NG, and NG + neu ACPA-IgG 7E4 and 3F3. Glycan structures of the most abundant N-linked glycan peaks are depicted. Blue square, N-acetylglucosamine (N); green circle, mannose/hexose (H); yellow circle, galactose/hexose (H); red triangl,: fucose (F); purple diamond, α 2,6-linked N-acetylneuraminic (sialic) acid (S). a.u., arbitrary units.
Table 1. ACPA-IgG BCR variable region sequence details.
ACPA-IgG BCR sequences isolated from single B cells of three patients with RA(23, 24). Immunoglobulin (IG) heavy and κ (K) or λ (L) LC CDR3 amino acid (AA) sequences are depicted, and the amount of nucleotide (nt) mutations is compared to germline. The N-linked glycan motifs presented in the variable heavy (VH) or light (VL) chain are visualized together with their respective location. The germline motifs are depicted on the basis of the IMGT database.
| ACPA-IgG | IGH-CDR3 AA |
IGH nt
mutations |
IGK/L-CDR3 AA |
IGK/L nt
mutations |
N-linked glycan motifs VH* |
N-linked glycan motifs VL*. |
Germline motifs VH† |
Germline motifs VL†. |
| 7E4 (24) | CVRIRGGSSNW | 28 | CAAWNGRLSAFVF | 28 | NES (CDR1) | NVT (FR1) | SES (CDR1) | KVT (FR1) |
| 2E4 (23) | CARGSFLERPESVPFHPW | 71 | CLQYHAEPYTF | 61 | NIT (FR3) and NST (FR3) |
x | TMT (FR3) and STA (FR3) |
x |
| 1F2 (23) | CVRGGSLGIFGGSVGYW | 44 | CQSYYRGDWVL | 46 | NFS (CDR1) and NLT (CDR2-FR3) |
NLT (FR1) | TFT (CDR1) and NPT (CDR2-FR3) |
NFM (FR1) |
| 2G9 (23) | CVRWGEDRTEGLW | 61 | CMQRLRFPLTF | 31 | NGS (CDR1), NTS (FR3), and NMT (FR3) |
NIS (FR3) | GGS (CDR1), NPS (FR3), and KLS (FR3) |
KIS (FR3) |
| 3F3 (23) | CARGTYLPVDESAAFDVW | 56 | CQQYYEAPYTF | 37 | NMT (FR3) and NTS (FR3) |
NLT (FR3) | TMT (FR3) and STA (FR3) |
TLT (FR3) |
| 2D11 (23) | CARRGGKDNVWGDW | 21 | CQQYNDWPVTF | 11 | NFT (CDR1) | x | SFT (CDR1) | x |
*Amino acid sequence and location.
†Determined by IMGT/V-QUEST.
We analyzed the variable domain and Fc glycan profile of the mAb variants (Fig. 1D) in detail using mass spectrometry (MS) [matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF)] (Fig. 1E and fig. S1). The exemplarily illustrated results of 7E4 and 3F3 (Fig. 1E) showed a highly sialylated glycan profile for the WT variant with a dominant complex type, bisected, and α-2,6–linked sialic acid comprising biantennary glycan peak H5N5F1S2 [mass/charge ratio (m/z) = 2651] (Fig. 1E), similar to the most prominent VDG found on ACPA-IgG directly isolated from patients with RA (16). As we wished to determine the effect of negatively charged sialic acids (specifically N-acetylneuraminic acids) on antigen-binding functionalities, we treated the WT and NG mAb variants with neuraminidase (+neu). As depicted in Fig. 1E, this resulted in an effective cleavage of sialic acids. Likewise, the NG variant showed no sialylated glycan peaks but only the three most abundant Fc-glycan forms comprising different amounts of galactose (H3N4F1, H4N4F1, and H5N4F1) (Fig. 1E). Together, these results demonstrate the successful generation of patient-derived IgG mAbs with engineered VDGs presenting the expected size and glycan composition as well as their NG counterparts.
Spatial orientation of VDG and antigen-binding pockets
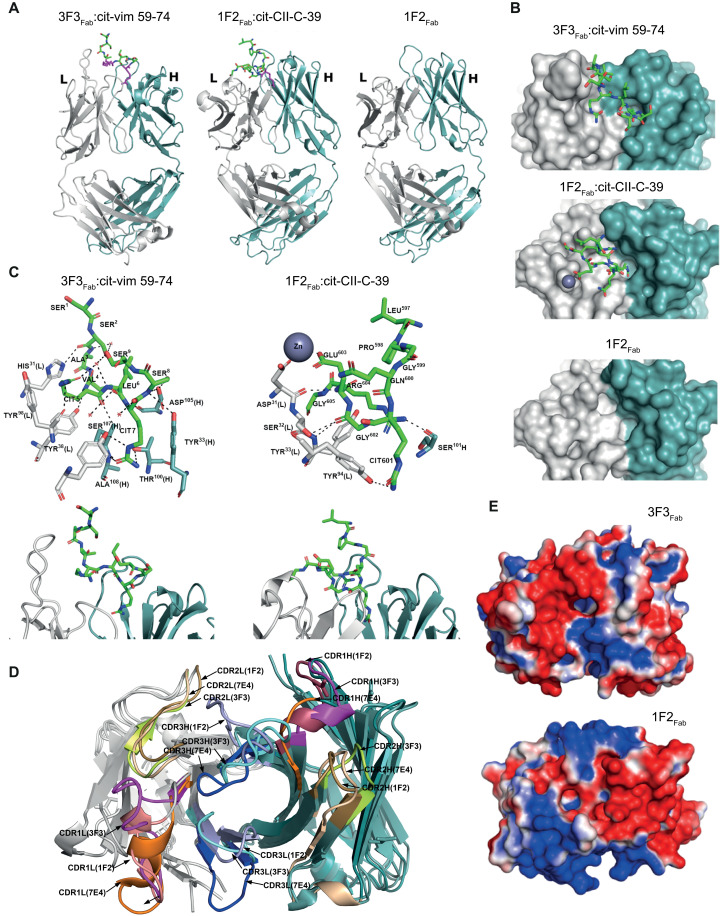
Crystal structures of two ACPA-IgG Fab fragments (3F3Fab and 1F2Fab) were generated to identify the structural composition of the antigen-binding pockets and the spatial orientation of the VDGs. The ACPA Fab fragments were complexed with distinct citrullinated peptides {3F3Fab:cit-vim 59-74 [Protein Data Bank (PDB): 6YXK] and 1F2Fab:cit-CII-C-39 (PDB: 6YXM)} selected on the basis of high reactivities within in vitro binding multiplex assays [as described in (15)] (Fig. 2A). In addition, 1F2Fab was crystallized in an unbound state (PDB: 6YXN) (Fig. 2A). The crystallographic data collection and refinement parameters are summarized in table S3. The results showed that 3F3Fab and 1F2Fab adopted characteristic Ig folds with well-defined binding grooves indicated by electron densities (Fig. 2B). The core binding regions of the citrullinated peptides are well defined, except for the terminal parts, which extend beyond the binding pocket (Fig. 2B). Furthermore, the complexed citrullinated (cit) peptides are visible in the antigen-binding sites, and residues SSAV(cit)L(cit)SS of cit-vim 59-74 and LPGQ(cit)GERG of cit-CII-C-39 could be modeled, while the other peptide residues were invisible (Fig. 2C). The buried molecular surface areas of the Fab fragment and its respective peptide upon binding are 503 Å for 1F2Fab [178-Å heavy chain (HC) and 325-Å LC] and 622 Å for cit-CII-C-39 as well as 659 Å for 3F3Fab (323-Å HC and 336-Å LC) and 789 Å for cit-vim 59-74, respectively. The results indicate that the LC, especially for 1F2Fab:cit-CII-C-39, contributed considerably to antigen binding. Superposition of the Apo-form of 1F2 (unligated) and its peptide-bound complex gave a backbone atom root mean square deviation (RMSD) of 0.4 Å, indicating no major changes in Ig folding upon antigen binding. When analyzing the structures of the unbound state, we found two deep pockets located within the antigen-binding region (Fig. 2B), similar to pockets found in a third ACPA, 7E4Fab, that has been described previously (PDB: 5OCY) (15). Noteworthy, these pockets, composed of polar and hydrophobic residues from HC and LC, are sufficient to accommodate a citrulline side chain (Fig. 2C). The flanking residues around the citrulline were, in both cases, partially embedded in the binding groove, while the terminal residues of the peptides were directed toward the solvent (Fig. 2C). The two peptides adopt a β-sheet–like conformation atop the complementary-determining regions (CDRs). To determine the structural basis required to accommodate citrullinated peptides, we performed a structural comparison between the determined ACPA 3F3Fab and 1F2Fab crystal structures and 7E4Fab. The superposition of the three Fab structures revealed distinct structural features of paratopes formed by the CDRs including different conformations, suggesting heterogenous antigen-binding regions among different ACPAs complexed with various peptides (Fig. 2D). Moreover, there was a major difference in the electrostatic potentials that might determine the recognition of flanking residues of citrulline (Fig. 2E). Noteworthy, the binding pocket of 3F3Fab is positively charged, different from the negative or neutral antigen-binding pocket characteristics of the other ACPA Fab fragments described (Fig. 2E).
Fig. 2. Crystal structures of ACPA Fab fragments and their peptide-binding modes.
(A) Crystal structures of ACPA-IgG Fab domains complexed with their respective peptides (3F3Fab:cit-vim 59-74 and 1F2Fab:cit-CII-C-39) and the unbound (APO) form of 1F2Fab in a cartoon representation. The peptides bound to the Fab are shown as sticks with carbon (green), oxygen (red), and nitrogen (blue) atoms. Citrulline residues are colored in magenta. (B) Top view of the ACPA paratope and the bound peptides for 3F3Fab:cit-vim 59-74 and 1F2Fab:cit-CII-C-39 and for 1F2Fab in an unbound state. The peptides are shown as sticks with carbon (green), oxygen (red), and nitrogen (blue) atoms. (C) Detailed paratope/epitope interactions between Fab fragments and their bound ligands shown for 3F3Fab:cit-vim 59-74 and 1F2Fab:cit-CII-C-39. Fab HC and LC are depicted as cartoon or sticks. All residues involved in the polar/hydrophobic interactions to the ligands are marked. Water molecules are depicted as red spheres, hydrogen bonds are depicted as black dashed lines, and the zinc ion is depicted as a dark sphere. Citrullinated residues are indicated as CIT. (D) Superposition of the CDR loops (CDR1, CDR2, and CDR3) of 3F3Fab, 1F2Fab, and 7E4Fab. (E) Electrostatic surface potentials of 3F3Fab and 1F2Fab are depicted. Positively charged regions are colored in red and negatively charged regions in blue. LC is depicted in light gray and HC in steel blue.
Hence, all ACPA Fab fragments exhibit an open-ended polar/hydrophobic binding groove, fitting the binding of the citrulline side chain, which enables them to promiscuously accommodate different peptide variants, while excluding others due to steric repulsion with other amino acid side chains. This and the fact that interactions are solely formed between the terminal nitrogen and oxygen atom of the citrulline side chain could explain the low affinity toward many of their antigens and the broad cross-reactivity toward various antigens and posttranslational modifications harboring these atoms (acetylated and carbamylated antigens) (23).
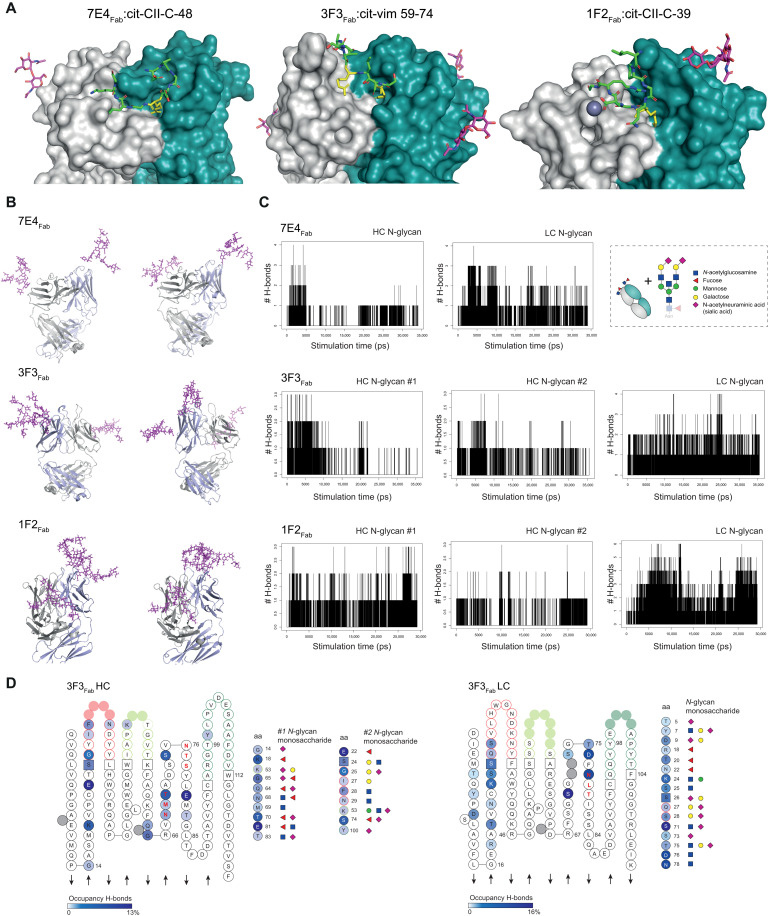
Next, we characterized the spatial localization of the VDG as this could provide insights into the potential impact of ACPA VDGS on antigen binding at the molecular level. Although N-linked glycans are highly flexible structures, the starting monosaccharides (core domain) were cocrystallized with the respective antigen-binding region of the generated monoclonal ACPA 7E4Fab:cit-CII-C-48 (PDB: 6ZJG), 3F3Fab:cit-vim 59-74, and 1F2Fab:cit-CII-C-39 (Fig. 3A). The data illustrate that the glycans are positioned outside but in close vicinity to the peptide-binding domain. To examine whether VDGS could potentially reach the antigen-binding pocket and thereby interfere with antigenic interactions, we used the crystals as a basic framework and modeled the complete disialylated glycan compositions on top of these structures. Fab crystal structures with modeled VDG were subsequently used for molecular dynamics (MD) simulations (Fig. 3B and fig. S2A). The number of hydrogen bonds formed during simulation time was analyzed for every Fab domain and the respective HC and LC N-glycans. The results showed that up to six hydrogen bonds were formed between the LC N-glycan and the mAb structure. The HC glycan formed fewer interactions over time, probably because it is more embedded into the protein structure and therefore more rigid (Fig. 3C). These results suggest a possibly higher impact of the LC glycan on antigen binding. We next visualized the amino acids and N-glycan monosaccharides involved in the hydrogen bond interactions as donors or acceptors within the respective three-dimensional (3D) variable domain structures based on IMGT (the international ImMunoGeneTics information system; Fig. 3D and fig. S2 based on tables S4 to S6). The results show that hydrogen bonds between the core fucose or N-acetylglucosamine and the antibody structure were mainly formed between amino acids within or close to the N-linked glycan sites. Notably, N-glycan galactoses or terminal sialic acids also interacted with amino acids in the CDR1/CDR2 or even in the CDR3 (Fig. 3D and fig. S2C). The negatively charged sialic acids of the 3F3 HC N-glycan interacted, for instance, with Y100, an amino acid located in the positively charged binding pocket. As amino acids located around Y100 in the HC are mainly involved in citrulline binding, an interaction with the N-linked glycan will likely affect antigen recognition (Fig. 2D). Together with the structural composition of the antigen-binding pockets, these results show that carbohydrates are positioned in close vicinity to the antigen-binding pocket and are likely capable of disrupting antibody-antigen interactions by forming hydrogen bond interactions with the CDR structures directly involved in binding.
Fig. 3. Fab structures crystallized with the starting (core) monosaccharides of the VDG and MD simulations to predict VDG-antibody interactions.
(A) Fab structures of 7E4Fab:cit-CII-C-48, 3F3Fab:cit-vim 59-74, and 1F2Fab:cit-CII-C-39 crystallized with the first two N-acetylglucosamines (GlcNAcs) of the VDG. LC is depicted in light gray and HC in steel blue. The peptides bound to the respective Fab are depicted as sticks with citrulline residues (yellow), carbon (green), oxygen (red), and nitrogen (blue) atoms. The GlcNAcs are presented as sticks in magenta. (B) MD simulation, two time points, of 7E4Fab, 3F3Fab, and 1F2Fab crystal structures modeled with full-length disialylated VDG represented as sticks in magenta. (C) Hydrogen bond (H-bond) interactions between the VDG (HC or LC) and the antibody structure are visualized over simulation time. (D) ACPA 3F3Fab HC and LC variable gene amino acid (aa) sequence is depicted on the basis of IMGT (2D). Acceptors/donors of H-bond interactions between the antibody structure and the HC (#1 and #2)/LC N-glycans are visualized. High occupancy is depicted in dark blue (HC: 13%, LC:16%), and low occupancy is depicted in light blue (0.01 to 0.2%). Amino acids and their respective interaction partners (N-glycan monosaccharides) are shown. Blue square, GlcNAc; green circle, mannose; yellow circle, galactose; red triangle, fucose; purple diamond, α 2,6-linked N-acetylneuraminic acid (sialic acid).
Disialylated complex-type VDG modulates binding to citrullinated antigens
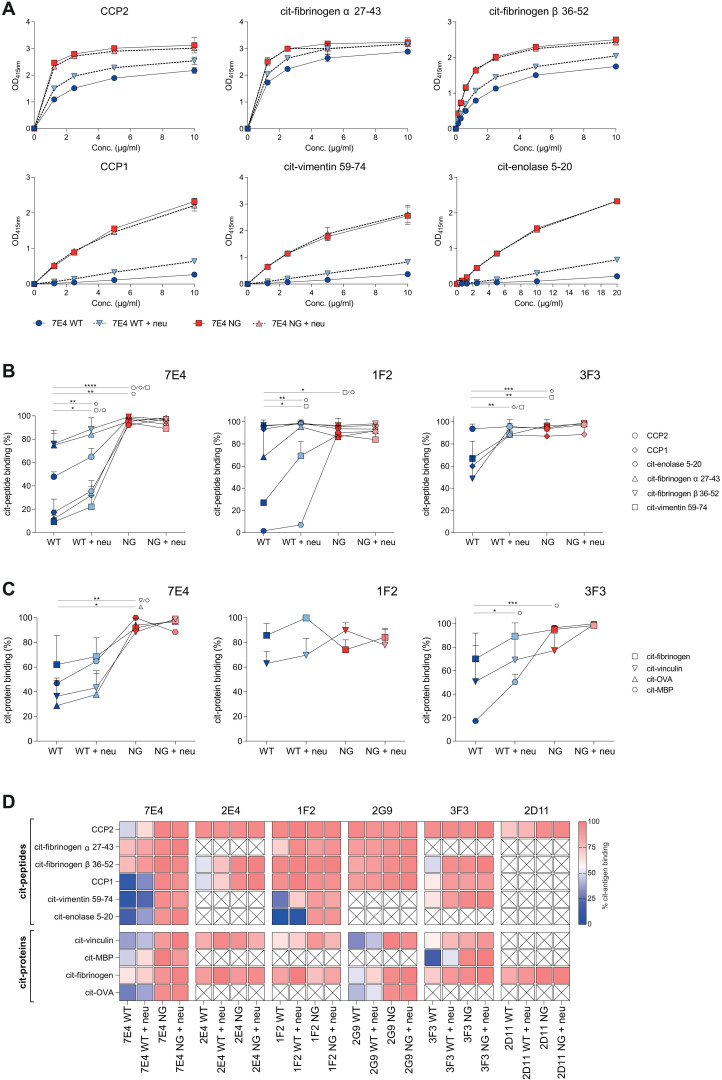
As the data described above indicate that VDGs can modulate antigen binding, we next sought to validate the impact of carbohydrates attached to the variable domain on binding using the different monoclonal ACPA-IgG variants. As shown by crystallography, the minor amino acid substitutions (germline back mutation) used to generate non–variable domain glycosylated variants did not change CDR conformations, and hence, a potential impact on antigen-binding ability can be ruled out (fig. S2B). We identified the effects of various glycan compositions (WT, WT + neu, NG, and NG + neu) (Fig. 1D) within six different mAbs (Table 1) on multiple antigenic interactions. Because of their heterogenous binding behavior, different antigens were tested among different ACPA mAbs. First, binding toward several citrullinated peptides [cyclic citrullinated peptide 2 (CCP2), cit-fibrinogen α 27-43, cit-fibrinogen β 36-52, CCP1, cit-vimentin 59-74, and cit-enolase 5-20] was investigated, and our results showed an overall reduced binding capacity of the VDGs-expressing WT variants (Fig. 4 and figs. S3 and S4). Cleaving off negatively charged sialic acids increased binding to most antigens. The total absence of VDG, as in the NG variants, resulted in considerably higher binding capacities (up to 90%) (Fig. 4). In particular, VDGs affected the binding of ACPAs toward antigens recognized with low-equilibrium KD (equilibrium dissociation constant) values, such as citrullinated enolase for the mAbs 7E4 and 1F2 or CCP1 for the mAb 3F3 (Fig. 4A, fig. S4, and table S2). Interactions with citrullinated antigens that show high maximal binding rates already at low concentrations, as observed for the CCP2 or citrullinated fibrinogen, were less affected by the presence of the carbohydrates (Fig. 4A and fig. S4). Although no direct affinities could be calculated from the biphasic binding curves (table S1), surface plasmon resonance (SPR) measurements showed a similar interference of the 7E4 VDGs with citrullinated antigens. The lack of sialic acids resulted in higher response units, which raised even further, when the entire glycan structure was absent (fig. S5). Similar observations as made using citrullinated peptides as the antigenic target for ACPAs were obtained when analyzing binding toward citrullinated proteins (native autoantigens) such as citrullinated fibrinogen, vinculin, ovalbumin (OVA), and myelin basic protein (MBP). Alike the peptide-binding results, the absence of VDGs raised the binding to most protein targets (Fig. 4, C and D, and fig. S4), although 1F2 binding remained similar, an observation also made for some peptide antigens (Fig. 4B).
Fig. 4. Impact of disialylated ACPA VDG on citrullinated (auto)antigen binding.
(A) ELISA titration binding curves of the ACPA 7E4 (0 to 10 μg/ml) variants (WT, WT + neu, NG, and NG + neu) toward citrullinated peptides (CCP2, CCP1, cit-fibrinogen α 27-43, cit-fibrinogen β 36-52, cit-vimentin 59-74, and cit-enolase 5-20). Binding to the arginine control peptide was subtracted. Reactivity was determined via the optical density (OD) at 415 nm. Each data point represents the mean of two technical replicates, and each binding experiment was repeated two to three times. (B) Relative binding of the ACPA mAb 7E4, 1F2, and 3F3 (2 to 40 μg/ml) variants (WT, WT + neu, NG, and NG + neu) toward citrullinated peptides. N = 2 to 6. Unpaired two-tailed t tests assuming the same SD. 7E4 WT-NG: ****P(CCP2, CCP1, cit-vimentin 59-74) < 0.0001 and **P(cit-enolase 5-20) = 0.0005; 7E4 WT-WT + neu: **P(CCP1) = 0.029, *P(CCP2) = 0.0227, and *P(cit-enolase 5-20) = 0.0419; 1F2 WT-NG: *P(cit-vimentin 59-74) = 0.0104 and *P(cit-enolase 5-20) = 0.0101; 1F2WT-WT + neu: **P(cit-enolase 5-20) = 0.0096 and *P(cit-vimentin 59-74) = 0.0449; 3F3 WT-NG: ***P(CCP1) = 0.0009 and **P(cit-vimentin 59-74) = 0.0010; 3F3WT-WT + neu: **P(CCP1) = 0.0017 and **P(cit-vimentin 59-74) = 0.0082. (C) Relative binding of the ACPA mAb 7E4, 1F2, and 3F3 (5 to 20 μg/ml) variants (WT, WT + neu, NG, and NG + neu) toward citrullinated proteins. N = 2. Unpaired two-tailed t tests assuming the same SD. 7E4 WT-NG: **P(cit-vinculin) = 0.0022, **P(cit-MBP) = 0.0014, and *P(cit-OVA) = 0.0032; 3F3 WT-NG: ***P(cit-MBP) = 0.0003; 3F3 WT-WT + neu: *P(cit-MBP) = 0.0213. (D) Heatmap of relative binding (0%, blue; 100%, red) of all monoclonal ACPA (7E4, 2E4, 1F2, 2G9, 3F3, and 2D11) toward citrullinated peptides and proteins. Nondetermined reactivities are illustrated with a cross. N = 2 to 3.
Thus, the enzyme-linked immunosorbent assay (ELISA)–based binding assays of six different patient-derived monoclonal autoantibodies showed an overall negative impact of VDGs and sialic acids on binding to several potential (auto)antigens (Fig. 4D and figs. S3 and S4). Thus, these data are in line with our prediction from the crystal structure analyses and indicate that VDGs can interact with and mask antigen-binding pockets of ACPAs.
VDGs expressed by autoreactive BCRs masks antigen binding on the B cell surface
The data described above provide novel insights into the potential influence of autoantibody VDGs on antigen binding. To address the question of whether VDGs also affects autoantigen binding on the B cell surface, we investigated the involvement of VDGs on antigen binding at the cellular level. To this end, the human B cell line Ramos, in which the endogenous IGHM, IGHD, and IGLC and activation-induced cytidine deaminase were knocked out (MDL-AID KO) (23), was used to express citrullinated protein–directed BCRs. Two Ramos B cell transfectants carrying variable domain glycosylated 3F3 and 7E4 membrane IgG (mIgG) BCRs (23) and their NG counterparts were generated (Fig. 5A). NG variants were produced as mentioned above by mutating the N-linked glycan sites back to the respective germline amino acid sequences. Transduced B cells were sorted on the basis of green fluorescent protein (GFP) and mIgG expression to ensure an identical amount of WT and NG BCRs on the surface of both cell lines (Fig. 5, B and C). The human Ramos B cells transduced with patient-derived autoreactive BCRs were highly GFP and mIgG BCR positive, while the nontransduced control MDL-AID KO cell line showed no BCR and GFP expression (Fig. 5B). The occupancy of the N-linked glycan sites and thus the expression of VDGs on the WT mIgGs were verified by gel electrophoresis after B cell lysis and BCR capturing (Fig. 5D). A size shift was present between the WT and NG BCR for both cell lines. This mass shift was higher when comparing the 3F3 to the 7E4 BCR variants due to six N-linked glycans attached to the 3F3 BCR HC and LC compared to four glycans attached to the 7E4 mIgG (Fig. 5D). Furthermore, we analyzed the specific IgG BCR VDG composition after an in-gel total glycan release (PNGase F treatment) followed by liquid chromatography–MS (LC-MS). Although this method does not allow a site-specific glycan analysis, the results obtained show that WT BCRs isolated from human Ramos B cells harbor fully processed complex-type H5N5F1S2 glycans as opposed to their NG counterparts (Fig. 5, F and G, and fig. S6, A and B). Likewise, we observed more bisecting N-acetylglucosamine and sialic acid moieties on the VDG (WT) BCR compared to the non-VDG (NG) BCR, which only contained Fc-glycans, while galactosylation and fucosylation were comparable between the variants (Fig. 5E). We confirmed the expression of complex-type glycans on surface IgG BCRs by cell surface biotinylating and Western blot analysis of the captured biotinylated BCRs. To analyze the nature of surface IgG–specific glycans, we treated the biotinylated BCR with EndoH, an enzyme that is unable to cleave complex-type N-glycans and only able to remove mannosylated glycans, or PNGase F, which cleaves all N-glycan structures as also elegantly described in previous publications (26). The results showed a size shift toward the NG BCR after PNGase F treatment and almost no susceptibility of the BCR VDG to EndoH treatment (fig. S6C). Thus, these data indicate that surface IgG BCRs on Ramos B cells predominantly express fully processed complex-type H5N5F1S2 glycans in their variable domains, such as the VDG found on isolated ACPA-IgG from patients, and no high-mannose structures.
Fig. 5. Generation of human Ramos B cell lines carrying disialylated VDG BCRs.
(A) Schematic depiction of generated human Ramos B cell transfectants with mIgG VDG+ BCRs (WT) and VDG− BCRs (NG). The untransduced MDL-AID KO cell line shows no GFP and endogenous BCR expression (GFP−, BCR−, VDG−). (B) GFP and BCR surface expression of the MDL-AID KO control cell line and Ramos cells expressing 3F3 mIgG with (WT) and without (NG) VDG. (C) Histograms depicting an identical WT and NG mIgG BCR expression of the 3F3 and 7E4 Ramos B cell lines and no BCR expression of the MDL-AID KO cell line. (D) Size shift between VDG+ and VDG− 3F3 and 7E4 BCRs visualized on a 4 to 15% SDS–polyacrylamide gel (Bio-Rad). No mIgG BCR protein band was detected for the MDL-AID KO cell line. The size was determined using the PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific). (E) Bar graphs of the relative abundance of galactosylation, sialylation, bisecting GlcNAc, and fucosylation per N-glycan and sialylation per galactosylation on the Ramos 3F3/7E4 WT, NG, and MDL IgG BCR after passing quality control (QC) settings. N = 3 (biological replicates). Multiple nonpaired t tests (Bonferroni-Dunn method): ****P < 0.0001, ***(3F3)P = 0.0008, and ***(7E4)P = 0.0004. (F) Absolute intensity of glycan traits expressed on the MDL and 3F3 WT and NG BCR after passing QC settings. (G) LC chromatogram of glycan traits expressed on the 3F3 WT and NG BCR after passing QC settings.
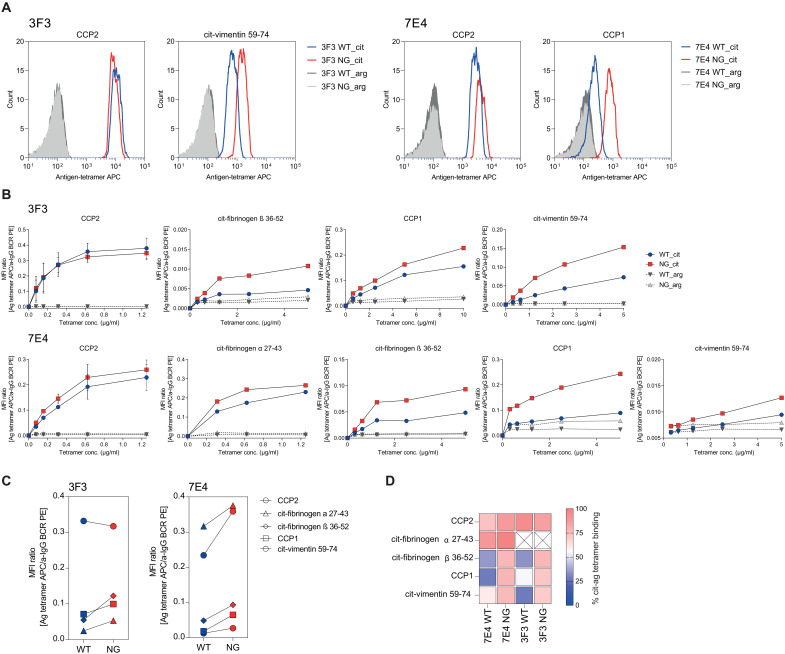
Next, we used the autoreactive 7E4 and 3F3 BCR–expressing cell lines to identify the impact of mIgG H5N5F1S2 VDG on autoantigen binding by flow cytometry analyses using labeled citrullinated peptide tetramers (27). The results obtained are consistent with the ELISA observations and indicate a reduction in antigen binding for B cells that express VDG BCRs (Fig. 6). Similarly to the mAb data, binding of low-affinity antigens by a given ACPA, such as CCP1 for 7E4 or cit-vimentin 59-74 for 3F3, was more affected compared to binding of antigens that display higher-equilibrium KD values, such as cit-fibrinogen α 27-43 for 7E4 and CCP2 for 3F3 (Fig. 6 and table S3).
Fig. 6. Binding of Ramos B cell lines carrying a VDG+ or VDG− BCR to citrullinated antigens.
(A) Histograms of Ramos 3F3 and 7E4 B cells carrying VDG+ or VDG− BCRs and their binding to CCP2/cit-vimentin 59-74 or CCP2/CCP1 strep.–APC–labeled tetramers, respectively. Binding to the arginine control peptides is shown in dark (WT) and light (NG) gray. (B) Binding titration curves of 7E4/3F3 WT and NG BCRs to citrullinated peptide-strep. tetramers (CCP2, CCP1, cit-fibrinogen α 27-43, cit-fibrinogen β 36-52, and cit-vimentin 59-74) and their respective control peptides. N = 2. The y axis depicts the MFI ratio between antigen binding and mIgG BCR expression. (C) Binding of 3F3/7E4 WT and NG Ramos B cells to citrullinated peptide-strep. tetramers (1 to 5 μg/ml). The y axis depicts the MFI ratio between antigen binding and mIgG BCR expression. (D) Relative binding of 7E4/3F3 WT and NG BCRs toward five citrullinated peptides (0%, blue; 100%, red).
Overall, our results demonstrate the first human B cell model that can be used to study the impact of BCR glycans on autoantigen recognition, B cell functions, or B cell fate. Our findings show that VDGs are not only able to modulate antigen interactions on secreted autoantibodies but also capable of affecting the interplay between BCRs and their autoantigens.
Functional impact of sialylated BCR VDG on B cell activation
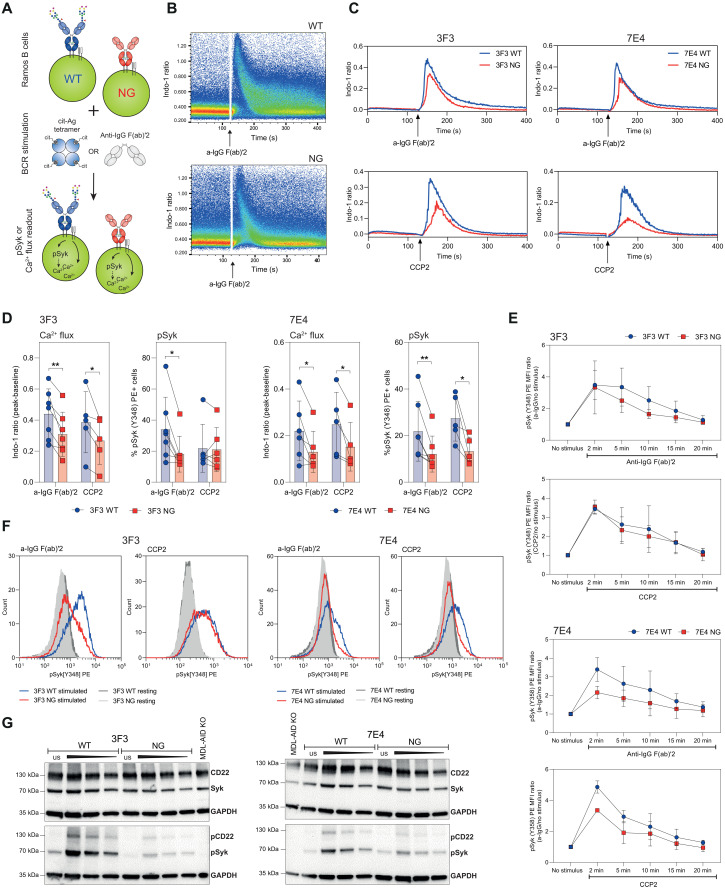
Next, we wished to delineate whether disialylated VDGs expressed on mIgG BCRs influence B cell activation independently of the “masking” effect on antigen binding. For this, we performed calcium flux experiments and investigated the phosphorylation of the spleen tyrosine kinase (Syk), a central kinase in BCR signal initiation and amplification (23, 28). Ramos WT and NG B cell lines, displaying an identical BCR surface expression (Fig. 5C), were either stimulated with anti-IgG F(ab)′2 or with a citrullinated antigen (Fig. 7A). For these experiments, the high-affinity antigen CCP2-streptavidin (strep.) tetramer was used, which exhibits similar binding strengths to both WT and NG BCRs (Fig. 6B). Both stimuli triggered calcium flux within a few seconds, as measured by the ratio of Indo-1 in a calcium-bound and unbound state (Fig. 7, B and C). No calcium flux was detected after stimulating the cells with the noncitrullinated, arginine-containing control tetramers (CArgP2-strep.) (fig. S7B). Likewise, activation with antigens that are recognized with lower affinity, such as cit-fibrinogen β 36-52, did not result in sufficient B cell activation in this assay (fig. S7B).
Fig. 7. Impact of mIgG BCR VDGs on human Ramos B cell activation.
(A) Ramos B cells (WT and NG) were stimulated with antigen (CCP2-strep.) or anti-IgG F(ab)′2. B cell activation was analyzed via Ca2+ release or the phosphorylation of Syk. (B) Ca2+ flux (Ca2+-bound Indo-1/unbound Indo-1) of 3F3 WT and NG Ramos B cells after stimulation with a-IgG F(ab)′2. (C) Ca2+ flux overlays of 7E4/3F3 WT and NG Ramos B cells after stimulation [CCP2-strep. or a-IgG F(ab)′2]. (D) Paired analysis of Ca2+ flux and pSyk(Y348) expression after 5 min of stimulation [a-IgG F(ab)′2 or CCP2-strep.] for 3F3/7E4 WT and NG Ramos B cells. Paired two-tailed t test. N = 5 to 7. 3F3 Ca2+ flux: **P = 0.005 and *P = 0.0316; 3F3 pSyk: *P = 0.0109; 7E4 Ca2+ flux: *P = 0.0309 and *P = 0.0441; 7E4 pSyk: **P = 0.0089 and *P = 0.0276. (E) pSyk (Y348) time-point analysis of 3F3/7E4 WT and NG Ramos B cells after adding no stimulus or 2, 5, 10, 15, and 20 min of stimulation [a-IgG F(ab)′2 or CCP2-strep.]. pSyk(Y348) MFI ratio (stimulated/unstimulated cells) is depicted. (F) pSyk (Y348) histograms of unstimulated or CCP2-strep.– or a-IgG F(ab)′2–stimulated 3F3/7E4 WT and NG Ramos B cells. (G) Western blot analyses of unstimulated (us) or 5-min a-IgG F(ab)′2–stimulated 3F3/7E4 WT and NG Ramos B cells. CD22, Syk, pCD22 (Y822), and pSyk(Y352) expressions are shown. Cell lysates of 1 million (unstimulated and stimulated first slot), 0.5 million (stimulated second slot), and 0.25 million (stimulated third slot) cells were blotted. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control, and 1 million MDL-AID KO cells were added as an additional control.
Intriguingly, when stimulated with either anti-IgG F(ab)′2 or the citrullinated CCP2 antigen, both WT cell lines (3F3 and 7E4) exhibited higher calcium flux peaks compared to the cells expressing NG BCRs (Fig. 7, C and D). B cells expressing VDG BCRs displayed not only a higher maximal calcium flux but also a faster calcium release, as indicated by the slope of the curve (Fig. 7C and fig. S7A). In addition, we analyzed the phosphorylation of the BCR signal-transducing kinase Syk in these cell lines by phospho-flow and Western blotting. Notably, and in line with the calcium release, both WT cell lines showed a higher pSyk level than the NG BCR–carrying cells when stimulated with either anti-IgG F(ab)′2 or CCP2-strep. (Fig. 7, D, F, and G, and fig. S7D). In addition, depending on the stimulus given, our data indicated not only a higher activation signal but also a prolonged Syk phosphorylation after stimulations for different time periods (2 to 20 min) (Fig. 7E). Thus, these data indicate that human B cell lines expressing a variable domain glycosylated autoreactive BCR depict a higher activation after stimulation, potentially explaining why autoreactive B cells with VDG-modified BCRs have a selection advantage in RA.
Effect of VDG on BCR downmodulation and antigen internalization
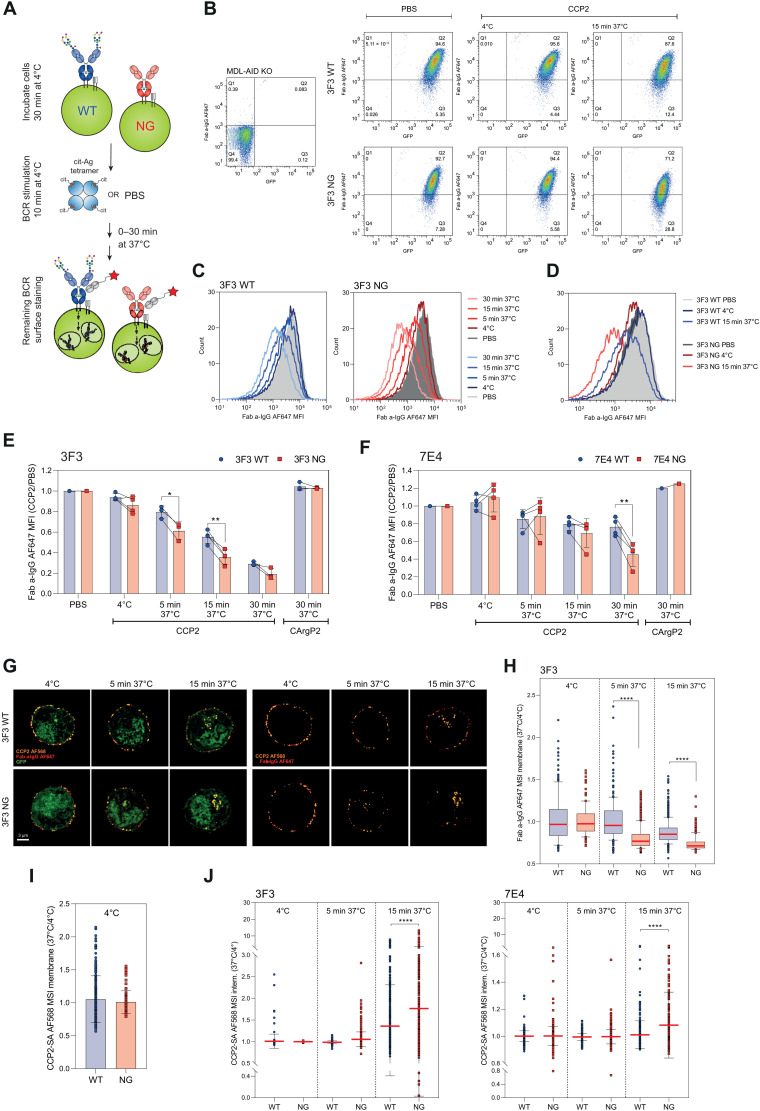
To determine whether variable domain glycosylation also regulates BCR downmodulation after antigen exposure, we stimulated the Ramos B cell lines for several minutes with the CCP2-strep. or the CArgP2-strep. control tetramer at 4°C. We then let the cells incubate at 37°C to allow internalization and measured mIgG BCR downmodulation using a Fab anti-human IgG detection antibody (Fig. 8A). The transduced and GFP-positive Ramos cells showed no changes in cell surface BCR expression after phosphate-buffered saline (PBS) treatment or when incubated on ice. In contrast, downmodulation could be readily detected after 15 min of incubation at 37°C (Fig. 8B), indicating rapid antigen-induced BCR internalization. We identified a time-dependent decrease in BCR surface expression for both the WT and the NG B cell line (Fig. 8C). Notably, the signal of surface-bound BCRs after antigenic stimulation was significantly decreased for the B cell lines carrying NG BCRs compared to their variable domain glycosylated counterparts (Fig. 8, D to F), despite the equivalent binding intensity of CCP2 at baseline. Loss of cell surface BCR expression was faster for the 3F3-expressing cell lines as most of the mIgG was lost within 30 min, whereas no downmodulation could be detected after stimulating the cells with the control peptide (Fig. 8, E and F).
Fig. 8. Effect of BCR VDGs on antigen internalization and BCR downmodulation.
(A) Ramos B cells were incubated at 4°C, stimulated with antigen or PBS, and incubated at 4° or 37°C to allow BCR downmodulation. The remaining surface BCRs were stained (Fab anti-human IgG-AF647). (B) GFP and mIgG expression of MDL-AID KO, 3F3 WT, and NG cells after PBS or CCP2-strep. treatment (4°C or 15 min at 37°C). (C) Histograms (mIgG) of CCP2-strep.–stimulated 3F3 WT or NG Ramos B cells (4°C or 5, 15, and 30 min at 37°C). (D) Histogram (mIgG) overlay of 3F3 WT and NG Ramos B cells after PBS or CCP2-strep. treatment (4°C or 15 min at 37°C). (E) BCR downmodulation of 3F3 WT and NG (F) 7E4WT and NG after PBS, CCP2-strep., or CArgP2-strep. stimulation (4°C or 5, 15, and 30 min at 37°C). Paired two-tailed t test. N = 3 to 4. 3F3: *(5 min)P = 0.0146 and **P = 0.0054; 7E4: **P = 0.0054. (G) Spinning disk confocal microscopy of GFP+ 3F3 WT and NG cells after CCP2-strep.-AF568 stimulation (incubation at 4°C or 5 and 15 min at 37°C), 2% PFA fixation, and a-IgG-AF647 surface staining. (H) 3F3 WT and NG BCR expression after CCP2-strep. stimulation (4°C or 5 and 15 min at 37°C). N = 114, 213, 474, 224, 495, and 595 cell slices, respectively. Ordinary one-way ANOVA, ****P < 0.0001. MSI, mean signal intensity. (I) CCP2-strep. binding of 3F3 WT and NG at 4°C. (J) CCP2-strep. internalization of 3F3/7E4 WT and NG BCRs at 4°C or after 5 and 15 min of incubation at 37°C. N(3F3) = 114, 213, 474, 224, 495, and 595 cell slices, respectively. N(7E4) = 619, 459, 645, 433, 738, and 302 cell slices, respectively. Ordinary one-way ANOVA, ****P < 0.0001.
Furthermore, confocal microscopy confirmed the flow cytometry–based BCR downmodulation data. We assessed antigen internalization by stimulating the cells with AF568-labeled CCP2-strep. tetramers. BCR modulation was evaluated by staining the remaining BCRs on the fixed cell surface after antigenic stimulation. mIgG BCRs were detected using a Fab anti-human IgG antibody coupled to AF647. All antigen-bound mIgG BCRs were still located on the B cell surface after incubation on ice. Incubating the stimulated B cells at 37°C for several minutes induced BCR clustering followed by antigen internalization and a reduced amount of surface mIgGs (Fig. 8G). The quantified results showed that especially NG BCRs were markedly downmodulated after antigenic stimulation and incubation for 5 min at 37°C, while WT BCRs were still stably expressed on the B cell surface (Fig. 8H). Despite a similar binding toward the CCP2-strep. tetramer (Fig. 8I), more antigen-bound BCRs were internalized, when no VDGs were present on the 3F3 and 7E4 mIgGs (Fig. 8J and fig. S7D). These results, together with the flow cytometry–based BCR expression data, indicate that human BCRs harboring VDGs stay longer on the B cell surface in an antigen-bound manner.
The role of the sialic acid–binding lectin CD22 on BCR VDG–mediated effects
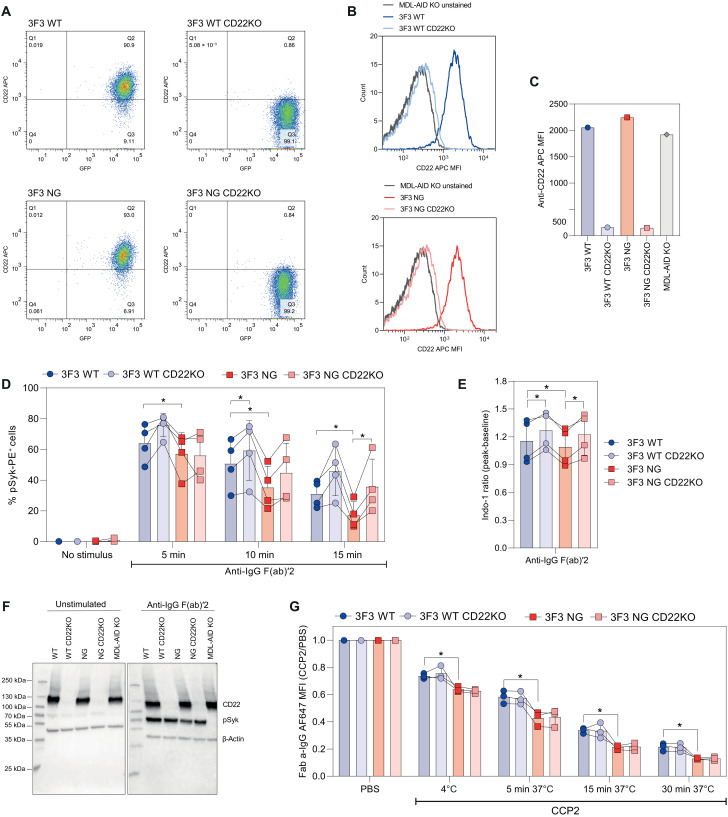
To identify whether interactions with the α 2-6–linked sialic acid–binding lectin (Siglec) CD22 can explain the effect of VDG on BCR signaling and cell surface expression, we performed CRISPR-Cas9 KO in the 3F3 WT– and NG BCR–expressing Ramos B cell lines. The KO was validated by flow cytometry, showing that the surface expression of CD22 was clearly absent in both the 3F3 WT and NG CD22KO cell lines compared to their non-KO counterparts (Fig. 9, A to C). Furthermore, we verified the KO of the CD22 target gene by bulk sequencing. Both KO cell lines showed a gene KO efficiency of more than 90% as evidenced by TIDE (tracking of indels by decomposition) analysis (fig. S8A). Mostly nucleotide deletions (>70%) were observed and, to a lower extent, +1 nucleotide insertions (<20%) with the highest frequency of guanine insertions (>85%) (fig. S8, A and B). Both CD22KO cell lines, expressing a VDG (WT) or NG BCR, as well as their CD22+ counterparts, were assessed in functional assays. Flow cytometry–based phospho-flow depicted, in line with our previous experiments, an increased pSyk expression of stimulated 3F3 cells carrying a WT compared to a NG BCR (Fig. 9D). Furthermore, we identified an increased expression of pSyk in stimulated CD22KO cell lines compared to their CD22-expressing counterparts (Fig. 9D). The increased signaling capacity after the CD22KO was similar for the WT and NG variants, indicating that VDG-mediated differences in BCR signaling are independent of CD22. These results were confirmed by calcium flux (Fig. 9E) and Western blot analyses, which showed that the absence of CD22 did not affect an increased pSyk expression in the presence of the VDG (Fig. 9F). In addition, we could not observe an impact of the Siglec CD22 on BCR downmodulation, as evidenced by the BCR surface expression data of the CD22KO cell lines after activation with a citrullinated antigen (Fig. 9G). However, similar to our previously presented results, the NG BCR was downmodulated quicker compared to its VDG counterpart (Fig. 9G). Together, these data point out that the highly sialylated VDG alters BCR downmodulation and signaling via a mechanism that does not involve the sialic acid–binding lectin CD22.
Fig. 9. Impact of CD22 CRISPR-Cas9 KO on BCR VDG–mediated effects.
(A) GFP and CD22 (anti-CD22 APC) expression of 3F3 WT, WT CD22KO, NG, and NG CD22KO Ramos B cells. (B) Histogram overlay (CD22) of unstained cells, 3F3 WT, and WT CD22KO or 3F3NG and NG CD22KO Ramos B cells. (C) CD22 expression of 3F3 WT, WT CD22KO, NG, NG CD22KO, and MDL-AID KO Ramos B cells. (D) Percentage of pSyk(Y348)-positive cells of 3F3 WT, WT CD22KO, NG, and NG CD22KO Ramos B cells after 5, 10, and 15 min of a-IgG F(ab)′2 stimulation. Paired two-tailed t test. N = 4. 5 min: *(3F3 WT versus NG) P = 0.0318; 10 min: *(3F3 WT versus NG) P = 0.0106 and *(3F3 WT versus WT CD22KO) P = 0.0298; 15 min: *(3F3 WT versus NG) P = 0.0166 and *(3F3NG versus NG CD22KO) P = 0.0315. (E) Ca2+ flux of 3F3 WT, WT CD22KO, NG, and NG CD22KO Ramos B cells after a-IgG F(ab)′2 stimulation. Paired two-tailed t test. N = 4. *(3F3 WT versus WT CD22KO) P = 0.0177, *(3F3 WT versus NG) P = 0.0342, and *(3F3NG versus NG CD22KO) P = 0.0171. (F) Western blot analyses of 3F3 WT, WT CD22KO, NG, and NG CD22KO Ramos B cell lysates after 5 min of a-IgG F(ab)′2 stimulation. CD22 and pSyk(Y352) expression are shown. β-Actin was used as a loading control, and MDL-AID KO cell lysates were used as an additional control. (G) BCR downmodulation of 3F3 WT, WT CD22KO, NG, and NG CD22KO Ramos B cells after PBS or CCP2-strep. stimulation and incubation at 4°C or for 5, 15, or 30 min at 37°C. Paired two-tailed t test. N = 3. *(4°C) P = 0.0474, *(5 min, 37°C) P = 0.0132, *(15 min, 37°C) P = 0.0229, and *(30 min, 37°C) P = 0.0463.
DISCUSSION
We here investigated the impact of VDGs on human B cell biology using citrullinated protein–directed BCR sequences isolated from ACPA-positive patients with RA. These molecules display naturally occurring N-linked glycan sites within the variable regions that are introduced during somatic hypermutation (SHM) (18) and allow the incorporation of VDGs. More than 90% of ACPA-IgG harbor VDGs, which makes them a hallmark feature of the RA-specific ACPAs immune response (16). Although readily found on ACPAs from patients with RA, VDGs are not abundantly present in healthy individuals (19). Furthermore, it has become evident that the transition to ACPA-positive disease is associated with a rise in VDG levels before disease (19), which is intimately connected to an increase in autoantibody levels and a “maturation” of the ACPA response as defined by an increased citrullinated antigen recognition profile and isotype usage. Notably, this maturation is not coupled to an increase in avidity as the ACPA response displays only limited avidity maturation across different (pre)disease stages (29). Together, these observations are intriguing as they imply that affinity maturation does not drive the selection of autoreactive ACPA-expressing memory B cells but rather their propensity to introduce glycosylation sites into their variable domains that potentially support their expansion and/or survival (30). BCR analyses of ACPA-expressing B cells from patients with RA indicate that the introduction of glycosylation sites is a consequence of a selective process not explained by the number of SHM. In line with this concept, the most prominent genetic risk factor for RA, the human leukocyte antigen class II region, does not associate with the presence of ACPA as such but mainly with ACPA-expressing VDG (31, 32). Hence, the acquisition of VDG represents an important event connected to the expansion of the B cell response occurring before RA onset that is also predictive for disease development in healthy first-degree family members of patients with RA (19). Furthermore, accumulating evidence suggests that N-linked VDGs are also at play in several other human autoimmune diseases including ANCA-associated vasculitis and Sjögren’s syndrome (20, 21).
The data presented in this manuscript support a concept in which VDGs act as a “threshold” for human autoreactive B cell activation, by showing that citrullinated protein–directed B cells increase BCR signaling upon the presence of VDGs. The introduction of VDGs during SHM is thus likely involved in the selection and activation process of autoreactive B cells in RA and possibly other autoimmune diseases. Next to their ability to increase B cell activation, VDGs are able to delay BCR downmodulation after antigenic stimulation and “mask” binding to especially “low-affinity” antigens.
Whether or how these functional events are interconnected and contribute in setting the threshold for autoreactive B cell activation and survival is unclear. However, it is conceivable that VDGs reduce the breadth of self-antigen recognition in the GC and thereby promote B cell survival. In case of a ubiquitous expression of self-antigens in the GC environment, self-reactive B cells might be eliminated as demonstrated in mouse studies (33). A differential impact of VDGs on antigen binding, although not self-antigens, has been reported for high-affinity anti-adalimumab and anti-infliximab clones (22) as well as for follicular lymphoma–derived VDGs artificially introduced into anti-NIP or anti-HEL BCRs and expressed in a murine B cell–derived reporter cell line (34). Our crystallographic data indicate that naturally occurring VDGs can interact with the antibody paratope and thereby compete with antigens for binding. This might provide a mechanism for autoreactive B cells to escape elimination through recognition of (abundantly) expressed autoantigens while allowing activation via cross-reactivity to (higher-affinity) foreign epitopes. Structural data together with modeling studies demonstrate that VDGs undergo hydrogen bond formations with CDR domains and can interfere with antigen binding. Our results also show that ACPAs display an open-ended binding groove and interact specifically with the modified (citrullinated) residue and, to a minor extent, with backbone structures surrounding the modification. This might explain the overall low affinity of ACPA and indicates that already marginal disturbances, by, e.g., carbohydrates, can interrupt binding.
One limitation of our study is that NG mAb variants, used for the binding assays, were generated by mutating N-linked glycan sites back into the predicted germline sequence. This prediction is based on the highest identity with the respective germline sequence but might cause altered antibody functions. However, the crystal structures of, e.g., 7E4Fab showed no structural differences in CDR conformations when expressed with or without VDGs, indicating no potential impact of the germline backmutation on antigen-binding ability. In line with the crystallographic results, our ELISA data illustrate that VDGs of autoantibodies in RA can diminish binding to several citrullinated peptides and proteins. The interactions between VDGs and the binding pockets can be more easily disrupted by high-affinity antigens, whereas low-affinity antigens are more strongly affected by the masking carbohydrates. We assessed the impact of VDG on antigen binding not only with germline reverted variants but also confirmed them with neuraminidase-treated mAbs harboring glycan traits without the terminal sialic acids. Furthermore, the results obtained for the mAbs 7E4 and 3F3 were in line with the binding data of the variable domain glycosylated versus non–variable domain glycosylated mIgG BCRs 7E4 and 3F3 expressed on human Ramos B cells.
Although BCR surface expression levels and glycan composition might differ from the in vivo situation, we show that the Ramos model B cell lines are able to express disialylated and bisected VDG structures, similar to glycan traits observed on secreted autoantibodies from patients with RA. Despite the limitation of testing only two recombinant BCRs, our data provide the first and consistent experimental evidence that VDGs are able to boost BCR signaling, as evidenced by an increase in Syk phosphorylation and calcium release, which presumably provides an advantage to these autoreactive B cells. Enhanced variable domain glycosylation is also thought to provide a survival advantage in follicular lymphoma B cells, although this has, so far, not been investigated experimentally (35, 36). A limitation of our study is that we could not confirm our BCR signaling results with citrullinated protein–reactive B cells directly obtained from patients as these cells represent a rare population in peripheral blood of patients with RA (approximately 0.05% of CD19+ B cells). Moreover, at present, it is not feasible to separate these citrullinated protein–reactive B cells in subsets of cells that do or do not express VDG.
Currently, it is unclear how VDGs affect BCR triggering and thereby set the threshold for activation. It has been suggested that sialylated VDGs interact with sialic acid–binding lectins, such as CD22 (37), potentially leading to an altered colocalization of the IgG BCR and the Siglec in a resting or activated state. Our CD22KO data show, however, no impact of CD22 on the VDG-mediated effects on BCR downmodulation and signaling. Furthermore, interactions with other Siglecs or (soluble-secreted) lectins, such as galectin 9, may be involved in altered signaling, similar to the role of galectin 9 described in IgM BCR signaling (38). Moreover, VDGs might alter the molecular organization of B cell surface structures by forming distinct clusters that are absent on B cells expressing BCRs without VDGs. A diverse BCR organization on B cells expressing variable domain glycosylated versus non–variable domain glycosylated BCRs could explain the significant differences in B cell activation after BCR cross-linking as could differences in BCR internalization, degradation, and modulation (39). The presence of VDG modulates the dynamics of the BCR surface expression after antigenic stimulation. We found a decreased BCR downmodulation and uptake after antigen exposure for variable domain glycosylated BCRs compared to NG BCRs. Thus, BCRs containing N-linked glycans show a decreased ability to be downmodulated from the cell surface and are consequently retained longer on the B cell surface after triggering with antigen. The prolonged surface expression might modulate BCR signaling strength and duration, as observed by our B cell activation studies similar to previous studies describing increased BCR signaling after inhibition of BCR uptake pathways (40). Further studies are needed to identify whether signal attenuation and BCR internalization are directly linked and, if so, how VDG affects BCR uptake. In this respect, it will be important to delineate the temporal dynamics and spatial organization of variable domain glycosylated mIgG BCRs at the nanoscale level also in relation to its association with lipid rafts, the actin cytoskeleton, and clathrin-coated pits.
Together, we show that the presence of N-linked glycans on the variable domain affects fundamental mechanisms involved in the functionality of B cells (antigen binding, BCR signaling, and downmodulation) and thus might critically determine the outcome of the B cell responses. In this respect, the abundance of VDG on the hallmark autoantibody response in a prominent human autoimmune disease is noteworthy. Thus, we provide experimental evidence for the selective and abundant presence of VDGs on autoreactive B cells in RA that likely conveys a signaling advantage, potentially explaining the outgrowth of these B cells and the increase of autoantibody levels toward disease onset. Our findings are highly relevant for a better understanding of B cell–driven autoimmune diseases as they provide a rationale on how the acquisition of VDGs might contribute to the escape of critical immune checkpoints in humans and thus the breach of B cell tolerance.
MATERIALS AND METHODS
Blood samples from patients diagnosed with RA
Peripheral blood samples from anti–citrullinated protein (auto)antibody–positive patients with RA visiting the outpatient clinic of the Rheumatology Department at the Leiden University Medical Center (LUMC) were included in this study. All patients fulfilled the EULAR/ACR 2010 criteria for the classification of RA (41). None of the patients were previously treated with B cell depletion therapies. Blood samples were taken upon obtaining written informed consent before inclusion and with approval from the local ethics committee of the LUMC, The Netherlands.
Recombinant monoclonal (auto)antibody production
Monoclonal (auto)antibodies were generated on the basis of full-length BCR sequences from APCA+ patients with RA (23, 24). CCP2-strep. and CArgP2-strep. tetramers were used to isolate autoreactive B cells as described previously (27). Single-sorted B cells were cultured on irradiated CD40 L cells and a cytokine mixture in IMDM (Gibco) medium for 10 to 12 days. RNA isolation, complementary DNA (cDNA) synthesis, ARTISAN polymerase chain reaction (PCR), and BCR sequencing were performed as described earlier (42). The 7E4 sequence was provided by T.R., Sanquin, The Netherlands (24). For the generation of NG variants, the N-linked glycosylation sites (N-X-S/T, X ≠ P) were specifically back-mutated into the germline sequence (based on IMGT) at the respective position (Table 1). WT sequences, including the N-glycan sites, and NG sequences were codon-optimized and the HC/LC variable genes together with 5′-Bam HI and 3′-Xho I restriction sites, the Kozak sequence, and the IGHV1-18*01 leader sequence ordered from GeneArt (Life Technologies). The constructs were ligated into a pcDNA3.1 (+) expression vector (Invitrogen) carrying the IGHG1 or the IGLC3/IGKC constant regions (UniProt), respectively, flanking the 3′-Xho I site. The recombinant mAbs were produced in Freestyle 293-F cells (Gibco) as previously stated (23). Glyco-engineering was performed by adding d-galactose substrate to the medium before transfection (Sigma-Aldrich, G0750-5G). Furthermore, to generate complex-type N-glycans on the antibody variable domains, the recombinant IgGs were coexpressed with 1% β-1,4-N-acetylglucosaminyltransferase III (GnTIII), 2.5% α2,6-sialyltransferase 1 (ST6galT), and 1% β-1,4-galactosyltransferase 1 (B4GalT1). The supernatants were harvested 5 to 6 days after transfection.
IgG purification
IgG1 antibodies were purified using a 1-ml HiTrap Protein G HP affinity column (GE Healthcare) followed by a direct buffer exchange using a 53-ml HiPrep 26/10 Desalting column (GE Healthcare) according to the manufacturer’s instructions. The recombinant mAbs were concentrated with Amicon Ultra-15 50K filter devices (Merck) to a final concentration of 1 mg/ml and used for further experiments.
mAb variable domain glycosylation analysis
Human monoclonal (auto)antibodies were analyzed for levels of variable domain glycosylation via SEC, gel electrophoresis, and MALDI-TOF MS analysis. SEC was performed using a Superose 6 Increase 10/300 GL column (GE Healthcare) according to the manufacturer’s instructions. The recombinant proteins were monitored by ultraviolet (UV) absorption at 280 nm. For SDS-PAGE, 1.5 μg of the mAbs was diluted in 4× Laemmli buffer (Bio-Rad) with (reduced) or without (nonreduced) 2% β-mercaptoethanol (Sigma-Aldrich) and incubated for 5 min at 95°C. Sample (10 μl) and 3 μl of PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific) were loaded on 4 to 15% SDS–polyacrylamide gels (Bio-Rad). For protein detection, gels were stained with Coomassie Brilliant Blue G-250 Dye (Thermo Fisher Scientific). MS analysis of released N-glycans was performed as previously described (43). In brief, the IgG samples (10 μg in 5 μl) were reduced in 10 μl of 2% SDS and incubated with 0.5 U of PNGase F (Roche Diagnostics, Germany), in 10 μl of 1:1 5× PBS/4% NP-40, overnight at 37°C. The total released glycan mixture was subjected to sialic acid stabilization by adding 100 μl of ethylation reagent [0.5 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.5 M 1-hydroxybenzotriazole hydrate] and incubating for 1 hour at 37°C. For glycan purification, the samples were brought to 85% acetonitrile (ACN) (Biosolve, Valkenswaard, The Netherlands) and purified with 15-μl cotton hydrophilic interaction LC (HILIC) tips using 85% ACN and 85% ACN + 1% trifluoroacetic acid for washing. Released and purified N-glycans were eluted from the HILIC tips using 10 μl of MQ. Three microliters of the purified and ethyl-esterified glycans was spotted on a MALDI target (MTP AnchorChip 800/384 TF, Bruker Daltonics) together with 1 μl of super-DHB (5 mg/ml) in 50% ACN and 1 mM NaOH. Spots were dried at room temperature (RT) and analyzed on an UltrafleXtreme (Bruker Daltonics) operated under flexControl 3.3 (Build 108, Bruker Daltonics). A mass spectrum from m/z 1000 to 3000 was recorded, combining 10,000 shots in a random walk pattern at 1000 Hz. The instrument was calibrated with a peptide calibration standard (Bruker Daltonics). MS data were analyzed using flexControl 3.3, and glycan peaks above S/N of nine were included into the analysis.
Fab fragment generation, crystallization, and structure determination
For Fab fragment preparation, chimeric 3F3 and 1F2 antibodies were produced on the basis of the human variable regions and mouse constant (Fc) domains, expressed in Expi393F cells (Life Technologies), purified as previously described (15) and dialyzed against PBS. The 3F3 and 1F2 Fab fragments were prepared using the ImmunoPure Fab Preparation Kit (Pierce) following the manufacturer’s instructions. Cleavage was evaluated by SDS-PAGE. Fab fragments were further purified by SEC on a HiLoad 16/600 Superdex 200 column. The peptides cit-vimentin 59-74, cit-CII-C-39, and cit-CII-48 used for cocrystallization are described in previous studies (15, 23). The crystals used for data collection were grown as follows: For 3F3Fab:cit-vimentin 59-74, the sitting drop consisted of 1 μl of 3F3Fab (10 mg/ml) mixed with cit-vimentin 59-74 in twofold molar excess in 20 mM tris (pH 7.5), 20 mM NaCl, and 0.5 μl of reservoir solution [0.2 M ammonium chloride (pH 6.3) and 20% (w/v) PEG 3350]. For 1F2Fab:cit-CII-C-39, the sitting drop consisted of 1 μl of 1F2Fab solution (10 mg/ml) in 20 mM tris (pH 7.4), 20 mM NaCl mixed with cit-CII-C-39 in twofold molar excess, and 0.5 μl of reservoir solution [20% (w/v) PEG 6000, 0.1 M Hepes 7.0 (pH 7.0), and 0.01 M zinc chloride]. For 1F2Fab, the sitting drop was set up from 1 μl of 1F2Fab (10 mg/ml) in 20 mM tris (pH 7.4), 20 mM NaCl, and 1 μl of reservoir solution [0.2 M sodium acetate and 20% (w/v) PEG 3350]. For 7E4Fab-cit-CII-48, the sitting drop consisted of 1 μl of 7E4Fab solution (10 mg/ml) in 20 mM tris (pH 7.4), 20 mM NaCl mixed with cit-CII-C-48 in twofold molar excess, and 1 μl of reservoir solution [20% (w/v) PEG 3350, 0.1 M bis-tris propane (pH 6.5), and 0.2 M potassium thiocyanate]. The complexes were incubated overnight before crystallization. All crystals were cryoprotected by briefly soaking in the corresponding reservoir solution containing 30% (v/v) ethylene glycol or glycerol before flash-freezing in liquid nitrogen. Diffraction data were collected at the beamlines and with the statistics given in table S3. The images were processed using XIA2 (44) and scaled by AIMLESS (45) from the CCP4 program suite (46). The structures of unbound Fab fragments or Fab-peptide complexes were solved by molecular replacement using PHASER (47). Iterative cycles of the manual model building were performed using COOT (48) and TLS as well as restrained refinement with Phenix or REFMAC5 (49) until R factors converged. Refinement statistics are given in table S3. About 5% of the reflections were randomly selected and set aside for unbiased cross-validation (calculation of Rfree). The “Protein Interfaces, Surfaces, and Assemblies” service PISA at the European Bioinformatics Institute (www.ebi.ac.uk/pdbe/prot_int/pistart.html) (50) was used to analyze molecular surfaces. Structure comparisons and RMSD calculations were performed with SSM (51) as implemented in PyMOL or COOT. Figures were prepared with PyMOL. The crystallographic coordinates and structure factors have been deposited in the PDB with the accession codes 6YXK, 6YXM, 6YXN, and 6ZJG.
MD simulations
The crystal structures of 7E4Fab, 3F3Fab, and 1F2Fab were used as initial structures for the MD simulation. Quick MD Simulator integrated with a module of Glycan Reader at the CHARMM-GUI website (www.charmm-gui.org) was used for preparing the input files for simulation (52). The N-glycans were predicted by NetNGlyc1.0 Server (www.cbs.dtu.dk/services/NetNGlyc/) (53). For simulation, we adopted the default setting from CHARMM-GUI for preparing the input files. Briefly, the TIP3P model was used for explicit water molecules. A distance of 15 Å between the protein atoms and the cubic system boundary was introduced, and 150 mM NaCl was added to the system. The CHARMM36 force field was used for proteins and carbohydrates. All calculations were performed at 300 K. The particle mesh Ewald algorithm was applied to calculate electrostatic forces. Each system was equilibrated in constant particle number, volume, and temperature condition with restraints using CHARMM36. An additional short period of constant particle number, pressure, and temperature equilibration was also applied without restraints for each system. The simulation was performed in Amber 18 (54). All analyses were performed in Visual Molecular Dynamics (55).
Protein citrullination and citrullinated peptide synthesis
Vinculin, fibrinogen, MBP, and OVA (Sigma-Aldrich) protein modification (citrullination) was performed as previously described (56). Four linear N-terminal biotinylated peptides fibrinogen α 27-42, fibrinogen β 36-52, vimentin 59-74, and enolase 5-20 and two cyclic N-terminal biotinylated peptides CCP1 and CCP2 (patent EP2071335) were synthesized including the native arginine and the altered citrullinated residue(s) at the same positions within the peptide sequence (23). The integrity of the synthesized peptides after purification was examined by ultraperformance LC on an Acquity instrument (Waters), and the exact mass was measured via MS on a Microflex instrument (Bruker) and crosschecked with the calculated masses.
Antigen-binding assays
To identify the impact of VDGs on antigen binding, ELISAs were performed as described earlier (23). Briefly, 1 to 10 μg/ml of biotinylated citrulline or arginine-containing peptides were coupled to precoated streptavidin plates (Microcoat, #65001) for 1 hour at RT. mAbs were added in PBS/1% bovine serum albumin (BSA)/0.05% Tween (PBT) (Sigma-Aldrich) and incubated for 1 hour at 37°C. For protein ELISAs, citrullinated or native proteins (10 μg/ml) were directly coupled to Nunc Maxisorp plates (Thermo Fisher Scientific) and incubated overnight on ice at 4°C. Following blocking with PBS/2% BSA for 6 hours, mAbs were added in PBT and incubated overnight on ice at 4°C. Antibody interactions were detected using a horseradish peroxidase (HRP)–conjugated monoclonal mouse anti-human IgG (Fc) CH2 antibody (Bio-Rad, MK1A6) to prevent VDG influences on secondary antibody binding. ELISA readout was performed using ABTS and H2O2. For SPR measurements, biotinylated citrullinated peptides were mounted on a streptavidin iSPR chip (Ssenc BV), and mAb analyte binding was assessed. An SPR imaging system for multiplexing 96 biomolecular interactions (IBISMX96, IBIS Technologies, Enschede, The Netherlands) was used.
Human Ramos B cell transfectants
Human Ramos B cell transfectants carrying 7E4 and 3F3 mIgG BCRs were generated as described earlier (23). In brief, HC and LC containing single vector constructs were created with the In-Fusion HD Cloning Kit (Clontech) using the pMIG-IRES-GFP-2AP vector backbone including the IGHG1 transmembrane domain. Variable gene sequences were cloned with and without N-linked glycan sites to generate WT and NG constructs. Inserts were verified by Sanger sequencing performed on Applied Biosystems 96-capillary (ABI3730) systems (LGTC facility, Macrogen). Retroviral transductions were performed as previously described (57). Briefly, Phoenix-ECO (American Type Culture Collection, CRL-3212) cells were transfected with PolyJet DNA transfection reagent (SignaGen Laboratories). Retrovirus-containing supernatants were collected 72 hours after transfection and used for the transduction of MDL-AID (IGHM, IGHD, IGLC, and AID) KO lymphoma Ramos B cells carrying slc7a1. Ramos cell lines were cultured in RPMI 1640/GlutaMAX/10% fetal calf serum (FCS)/10 mM Hepes medium (Thermo Fisher Scientific) with penicillin/streptomycin (100 U/ml; P/S) (Lonza).
Ramos BCR glycan analysis
For BCR glycan analysis, 20 million human Ramos B cells were lysed in PBS + 1% Triton X-100 for 60 min at 37°C, followed by total IgG BCR capturing using CaptureSelect FcXL Affinity Matrix (Thermo Fisher Scientific) and an overnight incubation at 4°C. Laemmli buffer (4×) was added to the IgG BCR/FcXL bead slurry, boiled for 5 min at 95°C, and loaded on a 4 to 15% SDS gel (Bio-Rad). Proteins were detected with Coomassie Brilliant Blue G-250 Dye (Thermo Fisher Scientific). The visualized IgG BCR bands were extracted from the gel, to exclude glycosylated contaminants, and transferred to Eppendorf tubes. The gel pieces were washed first with 25 mM ammonium bicarbonate (ABC), followed by ACN and, for protein reduction, incubated for 30 min at 56°C in 10 mM dithiothreitol (DTT)/25 mM ABC. After the reduction step, the gel pieces were washed with ACN and, for alkylation, incubated for 30 min at RT in the dark in 55 mM iodoacetamide/25 mM ABC to block reactive cysteine groups. After washing steps with 25 mM ABC and ACN, the gel bands were dried in a centrifugal vacuum concentrator for 5 min. N-linked glycans were released with 30 U of PNGase F (Roche Diagnostics, Germany) in 2% NP-40/2.5× PBS by an overnight incubation at 37°C. Released glycans were subsequently labeled with 2-aminobenzoic acid (Sigma-Aldrich) and 2-picoline borane (Sigma-Aldrich) and purified with 15-μl cotton HILIC tips as described above. Glycans were separated on an Ultimate 3000 UHPLC system (Dionex/Thermo Fisher Scientific, Breda, The Netherlands) coupled to a MaXis Impact HD quadrupole-TOF mass spectrometer (MaXis HD, Bruker Daltonics, Bremen Germany) equipped with a CaptiveSpray NanoBooster source (Bruker Daltonics, Bremen, Germany) as previously described (58). In short, 20% of the released, purified, and labeled glycans were loaded onto a C18 trap column (Acclaim PepMap 100; 100 μm by 2 cm, particle size of 5 μm, pore size of 100 Å; Dionex/Thermo Fisher Scientific) and washed for 2 min with 15 μl/min of 0.1% formic acid (FA)/1% ACN. Glycans were subsequently separated on a C18 analytical column (Acclaim PepMap 100; 75 μm by 15 cm, particle size of 3 μm, pore size of 100 Å; Dionex/Thermo Fisher Scientific), and elution was performed at a flow rate of 700 nl/min with buffer A [0.1% FA (v/v)] and buffer B [95% ACN/0.1% FA (v/v)]. A gradient of 1 to 70% buffer B in 70 min was applied (t = 0 min, B = 1%; t = 5 min, B = 1%; t = 30 min, B = 50%; t = 31 min, B = 70%; t = 35 min, B = 70%; t = 36 min, B = 1%; t = 70 min, B = 1%). The CaptiveSpray NanoBooster was operated with ACN-enriched gas (0.2 bar) and dry gas (3 liters/min) at 180°C and a capillary voltage of 1150 V. Mass spectra were acquired within a mass range of m/z 550 to 1800. LC-MS data were first examined manually using DataAnalysis (Bruker Daltonics). Data processing, including peak integration, was performed using LaCyTools v1.1.0. Glycans with a S/N above nine, a mass accuracy of ±20, and an isotopic peak quality of 0.2 were included into the analysis (59).
Cell surface biotinylating and surface IgG Ramos BCR glycan analysis
Ramos cell surface biotinylating was performed according to the manufacturer’s instructions of the Pierce Cell Surface Protein Biotinylating and Isolation Kit (#A44390). In brief, 24 million cells were harvested and biotinylated using Sulfo-NHS-SS-Biotin for 10 min at RT. The label was removed by washing with tris-buffered saline following cell lysis for 30 min on ice. Biotinylated surface proteins were captured using NeutrAvidin Agarose and eluted in elution buffer containing 10 mM DTT. Subsequently, 2% SDS was added, and the samples were incubated for 30 min at 60°C. The denatured, reduced, and biotinylated surface proteins were treated with 2 U of EndoH (Roche Diagnostics, Germany) in 50 mM sodium acetate buffer (pH 5.5) or 2 U of PNGase F (Roche Diagnostics, Germany) in 1:1 5× PBS/4% NP-40 overnight at 37°C. The presence of high-mannose (EndoH treatment) or complex-type (PNGase F treatment) N-glycans on the surface IgG BCRs was identified via Western blot analysis as described below.
Assembly of ribonucleoprotein complexes
Assembly of ribonucleoproteins (RNPs) was performed in a similar way as described in the CRISPR genome editing user guide from Integrated DNA Technologies (IDT). Briefly, preannealed CD22 exon 1–specific complexes of CRISPR-targeting RNA (crRNA) and transactivating crRNA were designed via IDT. The single-guide RNA (sgRNA) Hs.Cas9.CD22.1.AB (sequence: 5′-TGTCATTGAGGTGCACCGGG-3′; position: 35332832; on-target score: 52; off-target score: 76) was diluted to 44 μM in IDTE buffer (IDT). Alt-R S.p. Cas9 Nuclease V3 (IDT) was diluted to 36 μM in resuspension buffer R (IDT). sgRNA (90 pmol) was combined with 30 pmol of Cas9 to form the RNP complex.
Ramos cell electroporation with RNP complexes
Two million 3F3 WT and NG mIgG–expressing Ramos B cells were used per transfection reaction and washed with PBS to remove culture medium FCS. Ramos cell pellet was homogenously resuspended with 100 μl of 11 μM RNP and 11 μM Cas9 Electroporation Enhancer (IDT) diluted in resuspension buffer R (Invitrogen). Electrolyte buffer (3 ml; Invitrogen) was added to the transfection tube of the Neon transfection machine (Invitrogen). Cell/RNP mixture was pipetted into the Neon tip and transferred to the transfection tube. Electroporation was performed using the following settings: V = 1350, W = 30 ms, and P = 1. Electroporated cells were transferred into a 24-well plate and supplemented with 1 ml of prewarmed medium. Fluorescence-activated cell sorting (FACS) or flow cytometry validation experiments were performed after 1 week of culturing using an anti-CD22 allophycocyanin (APC)–labeled antibody (BD, #562850) on a BD 4 laser FACSAria or a BD LSRII flow cytometry instrument, respectively.
Genomic DNA isolation, bulk sequencing, and TIDE analysis
Genomic DNA was isolated from 5 million 3F3 WT/NG Ramos B cells with and without CD22 CRISPR-Cas9 KO using the Isolate II Genomic DNA Kit (Bioline, BIO-52066) according to the manufacturer’s instructions. A stretch of 500 to 1500 base pairs of the isolated genomic DNA (50 ng) was PCR-amplified enclosing the designed editing site using the predesigned forward and reverse primers (10 μM) and myTaq mix (Bioline, BIO-25041). PCR gel bands were purified using the PCR Clean-up Gel Extraction Kit (Takara), and 25 to 50 ng/μl were prepared for Sanger sequencing performed on an Applied Biosystems 96-capillary (ABI3730) system (LGTC facility, Macrogen). Sequences were analyzed using SnapGene software V5.0.4. The efficiency of the CD22KO was quantified using TIDE analysis version 3.3.0 (alignment window: 100 to 217, decomposition window: 242 to 481, indel size: 10, and p threshold: 0.001) (60).
Flow cytometer experiments
Binding of the generated 7E4/3F3 WT and NG mIgG– and GFP-expressing B cell lines toward citrullinated antigens was analyzed by flow cytometry. Ramos B cells were stained with APC-labeled cit-peptide SA-tetramers (0 to 5 μg/ml) (CCP2, CCP1, cit-fibrinogen α 27-43, cit-fibrinogen β 38-52, and cit-vimentin 59-74) and goat anti-human IgG-Fc phycoerythrin (PE) (0.5 μg/ml; #12-4998-82) in staining solution (PBS/0.5% BSA/0.02% NaN3) via incubation for 30 min on ice. Tetramer formation was performed as previously described (27). Unmodified peptides including arginine residues were measured as negative controls in the same concentration range (0 to 5 μg/ml) to analyze antigen specificity.
B cell activation was measured via calcium release of the generated B cell lines after stimulation. Therefore, 1 million mIgG- and GFP-expressing B cells were collected and stained with 200 μl of calcium indicator loading dye medium, 2 μM Indo-1 AM (#ab142778), and 0.05% pluronic acid (#P6866) in 1 ml of stimulation medium [RPMI 1640/P/S (100 U/ml)/GlutaMAX/10 mM Hepes/2% FCS] for 35 min at 37°C in the dark. After washing with stimulation medium only, B cells were incubated with 500 μl of stimulation medium plus 2 mM calcium and incubated on ice, in the dark until usage. Fifteen minutes before the analysis, B cells were prewarmed in a water bath at 37°C to decrease baseline activation upon measurement. The analysis was performed on a BD LSRII flow cytometry or a Cytek Aurora 5L instrument including a UV laser and acquiring 500 cells/s at a high speed. After 2 min of baseline measurement, 100 μl of stimulus [CCP2/CArgP2 (5 μg/ml) or anti-human IgG F(ab)′2 (80 μg/ml)] was added to the samples and mixed adequately, and the measurement continued for another 5 min until the signal reached almost baseline again. The calcium flux was measured as the ratio of calcium-bound Indo-1 to unbound Indo-1. MDL-AID KO B cells, without an endogenous BCR, were used as negative controls in all experiments. The maximal calcium flux (peak-baseline signal) and its speed (slope of the curve) were analyzed.
BCR signaling was analyzed via the intracellular expression of pSyk after activation. Therefore, 1 million mIgG- and GFP-expressing B cells were collected and stimulated with CCP2-strep. tetramer (5 μg/ml) or 15 μg of anti-human IgG F(ab)′2 for 0, 2, 5, 10, 15, or 20 min at 37°C in stimulation medium [RPMI/P/S (100 U/ml)/GlutaMAX/10 mM Hepes/1% FCS]. Afterward, cells were fixed (BioLegend Fixation Buffer, #420801) and permeabilized (True-Phos Perm Buffer, #425401). After washing, the intracellular expression of phosphorylated Syk was analyzed with a mouse anti-human pSyk(Y348)-PE mAb (#moch1ct, eBioscience) diluted 1:20 in staining solution. The rate of pSyk expression in Ramos cells was calculated as the percentage of pSyk(Y348)+ cells or the pSyk median fluorescence intensity (MFI) ratio between stimulated and unstimulated cells. Gating was based on the MDL-AID KO control cell line stimulated with CCP2 or anti-IgG F(ab)′2, respectively.
To assess BCR modulation, 0.2 million mIgG- and GFP-expressing Ramos B cells were first incubated for 30 min on ice followed by a 15-min stimulation at 4°C with either control PBS or CCP2-/CArgP2-strep. tetramers (5 μg/ml) in PBS/2% FCS. The stimulated B cells were then incubated for 0, 5, 15, or 30 min at 37°C to allow BCR downmodulation. The remaining surface BCRs were stained with a AF647 NHS (N-hydroxysuccinimide) ester (Thermo Fisher Scientific, #A20006)–labeled Fab goat anti-human IgG (Jackson ImmunoResearch, #109007003) diluted 1:2000 in staining solution. Stained cells were analyzed on a BD LSRII flow cytometry instrument. Data were analyzed with FlowJo_V10.
Western blot analysis
CD22, Syk, pCD22, pSyk, and glyceraldehyde-3-phosphate dehydrogenase expression of Ramos cells before and after stimulation were analyzed via Western blot analyses. Therefore, 4 million Ramos cells were lysed in 20 μl of NP-40 cell lysis buffer (Thermo Fisher Scientific) including 1× Protease/Phosphatase Inhibitor Cocktail (100×; Cell Signaling Technology) via incubation for 30 min on ice. Before cell lysis, cells were either stimulated with CCP2 or anti-human IgG F(ab)′2 (20 μg/ml) or incubated with stimulation medium [RPMI 1640/P/S (100 U/ml)/GlutaMAX/10 mM Hepes/2% FCS] only for 5 min at 37°C. The supernatant of the lysed cells was stored at 20°C for further usage. Lysates (10 μl; 1 million cells) were subjected to 4 to 15% SDS–polyacrylamide gels (Bio-Rad) together with 4× Laemmli buffer (Bio-Rad) after incubation for 5 min at 95°C. Subsequently, immunoblotting was performed on a nitrocellulose membrane (Bio-Rad). Blots were incubated in PTE (3% skim milk powder/PBS/0.05% Tween) for 2 hours at RT. Following washing with PBS/0.05% Tween (PT), the blots were incubated at 4°C overnight with 1:20,000 mouse anti-human GADPH (#MAD374, Bio-Rad) or 1:3000 rabbit anti-human β-actin (D6A8, #8457S), 1:15,000 rabbit anti-human CD22 (#ab207727, BioLegend), and 1:300 rabbit anti-human Syk (#2712S, Cell Signaling Technology) or 1:200 rabbit anti-human pCD22 (Y822, #ab32123, BioLegend) and 1:1000 rabbit anti-human pSyk (p-Zap-70, Y319, #2717S, Cell Signaling Technology), respectively, diluted in 5 ml of PTE. After washing with PT, blots were incubated for 1 hour at RT with 5 ml of PTE containing 1:1000 goat anti-rabbit Ig HRP (#P0448, Dako) and 1:5000 goat anti-mouse Ig HRP (#P0447, Dako) secondary antibodies. Blots were washed, and bound antibodies were visualized using enhanced chemiluminescence (GE Healthcare, RPN2109). The readout was performed on a Bio-Rad Chemidoc Touch Imaging system. For biotinylated surface IgG BCR detection after EndoH or PNGase F treatment, samples were mixed with 4× Laemmli buffer, incubated for 5 min at 95°C, and loaded on 4 to 15% gels (Bio-Rad). Western blot analysis was performed as described above using a goat anti-human IgG (#31410, Invitrogen) HRP-labeled antibody diluted 1:1000 in PTE.
Spinning disk confocal microscopy
BCR modulation and antigen internalization were identified using spinning disk confocal microscopy. One million Ramos B cells were incubated for 30 min on ice followed by a 10-min stimulation at 4°C with CCP2-strep. AF568 tetramers (5 μg/ml) in PBS/2% FCS. The stimulated B cells were then incubated for 0, 5, or 15 min at 37°C to allow BCR downmodulation and antigen internalization. After fixation with 2% paraformaldehyde (PFA), the cells were stained for 15 min with an AF647 NHS ester (Thermo Fisher Scientific, #A20006)–labeled Fab goat anti-human IgG (Jackson ImmunoResearch, #109007003) antibody diluted 1:2000 in staining solution to detect the remaining BCRs on the fixed cell surface. After washing, the cells were settled to poly-d-lysine–precoated coverslips (Ibidi) for at least 40 min. Imaging was performed on an Andor Dragonfly 500 spinning disk confocal microscope at the Light and Electron Microscopy Facility of the LUMC, The Netherlands. Z-stacks of 20 slices per cell were acquired for three laser lines (488, 561, and 637), and the mean signal intensity on the B cell membrane or inside the cell was assessed using Fiji/ImageJ.
Statistical analysis
Antigen-binding data of mAbs were analyzed using two-tailed unpaired t tests assuming that all samples show the same scatter (SD). LC-MS mIgG BCR glycan data were analyzed using multiple t tests assuming that all samples show the same SD and corrected for multiple comparisons using the Bonferroni-Dunn method. Paired two-tailed t tests were performed for B cell activation and BCR downmodulation assays with a number of three to five pairs. Ordinary one-way analysis of variance (ANOVA) tests were performed for parametric nonmatched confocal microscopy data. Significant differences are denoted as follows: not significant (ns; P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, or ****P<0.0001.
Acknowledgments
We thank J.-W. Drijfhout (LUMC, Leiden, The Netherlands) for providing the citrullinated peptides, J. Eken (LUMC, Leiden, The Netherlands) for expert assistance with the CRISPR-Cas9 KOs, the Protein Science Facility at Karolinska Institute for providing crystallization infrastructure, and the whole group of Affinity Proteomics at SciLifeLab Stockholm for efforts, and we thankfully acknowledge access to beamlines at the Diamond Light Source and MAX IV Laboratory as well as the staff for assistance with crystal testing and data collection. Furthermore, we thank the LUMC Flow cytometry Core Facility (FCF) and the Light and Electron Microscopy Facility for help with cell sorting and imaging.
Funding: This work was supported by the ReumaNederland 17-1-402 (to R.E.M.T.), the IMI-funded project RTCure 777357 (to T.W.J.H.) , ZonMw TOP 91214031 (to R.E.M.T.), Target-to-B LSHM18055-SGF (to R.E.M.T.), NWO-ZonMW clinical fellowship 90714509 (to H.U.S.), NWO-ZonMW VENI grant 91617107 (to H.U.S.), ZonMW Enabling Technology Hotels grant 435002030 (to H.U.S.), Dutch Arthritis Foundation 15-2-402 and 18-1-205 (to H.U.S.), Excellence Initiative of the German Federal and State Governments EXC 294 (to M.R.), DFG through TRR130-P02 (to M.R.), RO1 grant A031503 (to M.R.), Knut and Alice Wallenberg Foundation 2019.0059 (to R.H.), The Swedish Research Council 2017-06014 (to R.H.), National Science Foundation of China, 1R15AI154248-01A1 (KR-K210) (to R.H.), and European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X grant agreement no. 283570 (to R.H.).
Author contributions: All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Conceptualization: T.K., C.G., L.H., T.W.J.H., H.U.S., M.R., R.H., and R.E.M.T. Methodology: T.K., C.G., L.M.V., L.H., L.M.S., M.C., G.J.M.P., and M.W. Software: C.G., L.M.V., and LH. Investigation: T.K., C.G., L.H., J.C.K., L.M.S., Y.H., K.A.v.S., R.D.V., A.S.B.K., S.R., G.S.-R., and B.X. Visualization: T.K., C.G., and L.H. Supervision: M.C., R.H., T.W.J.H., H.U.S., and R.E.M.T. Writing—original draft: T.K. and R.E.M.T. Writing—review and editing: C.G., L.H., J.C.K., L.M.S., M.C., Y.H., K.A.v.S., R.D.V., A.S.B.K., S.R., G.S.-R., L.M.V., L.H., B.X., G.J.M.P., M.W., T.R., T.W.J.H., H.U.S., M.R., and R.H.
Competing interests: C.G., B.X., T.R., and R.H. are named as coinventors of a patent application (PCT/EP2018/082236) regarding the arthritis-protective effects of ACPA. H.U.S., T.W.J.H., and R.E.M.T. are mentioned as inventors on a patent on ACPA-IgG V-domain glycosylation. G.J.M.P. is mentioned as inventor on the CCP2-related patent EP2071335. The other authors declare that they have no competing interests.
Data and materials availability: We confirm that the data supporting the findings of this study are available within the article and/or the Supplementary Materials. The CCP2 peptide can be provided by G.J.M.P. (Radboud University, The Netherlands) or J.-W. Drijfhout (LUMC, The Netherlands) pending scientific review and a completed material transfer agreement. Requests for the CCP2 peptide should be submitted to G.Pruijn@ncmls.ru.nl. The 7E4 sequence and antibodies can be provided by R.H. (Karolinska Institutet, Sweden) or T.R. (Sanquin Research, The Netherlands) under the protection of a completed material transfer agreement. Requests for the 7E4 sequence should be submitted to Rikard.Holmdahl@ki.se.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S6
REFERENCES AND NOTES
- 1.Barnas J. L., Looney R. J., Anolik J. H., B cell targeted therapies in autoimmune disease. Curr. Opin. Immunol. 61, 92–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampe C. S., B cells in autoimmune diseases. Scientifica 2012, 1–18 (2012). [Google Scholar]
- 3.Erikson J., Radic M. Z., Camper S. A., Hardy R. R., Carmack C., Weigert M., Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature 349, 331–334 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Goodnow C. C., Crosbie J., Adelstein S., Lavoie T. B., Smith-Gill S. J., Brink R. A., Pritchard-Briscoe H., Wotherspoon J. S., Loblay R. H., Raphael K., Trent R. J., Basten A., Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 334, 676–682 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Hartley S. B., Crosbie J., Brink R., Kantor A. B., Basten A., Goodnow C. C., Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature 353, 765–769 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Pelanda R., Schwers S., Sonoda E., Torres R. M., Nemazee D., Rajewsky K., Receptor editing in a transgenic mouse model: Site, efficiency, and role in B cell tolerance and antibody diversification. Immunity 7, 765–775 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Tiegs S. L., Russell D. M., Nemazee D., Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177, 1009–1020 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wardemann H., Yurasov S., Schaefer A., Young J. W., Meffre E., Nussenzweig M. C., Predominant autoantibody production by early human B cell precursors. Science 301, 1374–1377 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Diaz M., Klinman N. R., Relative roles of somatic and Darwinian evolution in shaping the antibody response. Immunol. Res. 21, 89–102 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Burnett D. L., Langley D. B., Schofield P., Hermes J. R., Chan T. D., Jackson J., Bourne K., Reed J. H., Patterson K., Porebski B. T., Brink R., Christ D., Goodnow C. C., Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination. Science 360, 223–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabouri Z., Schofield P., Horikawa K., Spierings E., Kipling D., Randall K. L., Langley D., Roome B., Vazquez-Lombardi R., Rouet R., Hermes J., Chan T. D., Brink R., Dunn-Walters D. K., Christ D., Goodnow C. C., Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc. Natl. Acad. Sci. U.S.A. 111, E2567–E2575 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bretscher P., Cohn M., A theory of self-nonself discrimination. Science 169, 1042–1049 (1970). [DOI] [PubMed] [Google Scholar]
- 13.Reijm S., Kissel T., Toes R. E. M., Checkpoints controlling the induction of B cell mediated autoimmunity in human autoimmune diseases. Eur. J. Immunol. 50, 1885–1894 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Schellekens G. A., de Jong B. A., van den Hoogen F. H., van de Putte L. B., van Venrooij W. J., Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J. Clin. Invest. 101, 273–281 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge C., Xu B., Liang B., Lonnblom E., Lundstrom S. L., Zubarev R. A., Ayoglu B., Nilsson P., Skogh T., Kastbom A., Malmstrom V., Klareskog L., Toes R. E. M., Rispens T., Dobritzsch D., Holmdahl R., Structural basis of cross-reactivity of anti-citrullinated protein antibodies. Arthritis Rheumatol. 71, 210–221 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Hafkenscheid L., Bondt A., Scherer H. U., Huizinga T. W., Wuhrer M., Toes R. E., Rombouts Y., Structural analysis of variable domain glycosylation of anti-citrullinated protein antibodies in rheumatoid arthritis reveals the presence of highly sialylated glycans. Mol. Cell. Proteomics 16, 278–287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rombouts Y., Willemze A., van Beers J. J., Shi J., Kerkman P. F., van Toorn L., Janssen G. M., Zaldumbide A., Hoeben R. C., Pruijn G. J., Deelder A. M., Wolbink G., Rispens T., van Veelen P. A., Huizinga T. W., Wuhrer M., Trouw L. A., Scherer H. U., Toes R. E., Extensive glycosylation of ACPA-IgG variable domains modulates binding to citrullinated antigens in rheumatoid arthritis. Ann. Rheum. Dis. 75, 578–585 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Vergroesen R. D., Slot L. M., Hafkenscheid L., Koning M. T., van der Voort E. I. H., Grooff C. A., Zervakis G., Veelken H., Huizinga T. W. J., Rispens T., Scherer H. U., Toes R. E. M., B-cell receptor sequencing of anti-citrullinated protein antibody (ACPA) IgG-expressing B cells indicates a selective advantage for the introduction of N−glycosylation sites during somatic hypermutation. Ann. Rheum. Dis. 77, 956–958 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hafkenscheid L., de Moel E., Smolik I., Tanner S., Meng X., Jansen B. C., Bondt A., Wuhrer M., Huizinga T. W. J., Toes R. E. M., El-Gabalawy H., Scherer H. U., N-linked glycans in the variable domain of IgG anti-citrullinated protein antibodies predict the development of rheumatoid arthritis. Arthritis Rheumatol. 71, 1626–1633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vletter E. M., Koning M. T., Scherer H. U., Veelken H., Toes R. E. M., A comparison of immunoglobulin variable region N-linked glycosylation in healthy donors, autoimmune disease and lymphoma. Front. Immunol. 11, 241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biermann M. H., Griffante G., Podolska M. J., Boeltz S., Sturmer J., Munoz L. E., Bilyy R., Herrmann M., Sweet but dangerous - the role of immunoglobulin G glycosylation in autoimmunity and inflammation. Lupus 25, 934–942 (2016). [DOI] [PubMed] [Google Scholar]
- 22.van de Bovenkamp F. S., Derksen N. I. L., Ooijevaar-de Heer P., van Schie K. A., Kruithof S., Berkowska M. A., van der Schoot C. E., IJspeert H., van der Burg M., Gils A., Hafkenscheid L., Toes R. E. M., Rombouts Y., Plomp R., Wuhrer M., van Ham S. M., Vidarsson G., Rispens T., Adaptive antibody diversification through N-linked glycosylation of the immunoglobulin variable region. Proc. Natl. Acad. Sci. U.S.A. 115, 1901–1906 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kissel T., Reijm S., Slot L. M., Cavallari M., Wortel C. M., Vergroesen R. D., Stoeken-Rijsbergen G., Kwekkeboom J. C., Kampstra A., Levarht E., Drijfhout J. W., Bang H., Bonger K. M., Janssen G., van Veelen P. A., Huizinga T., Scherer H. U., Reth M., Toes R., Antibodies and B cells recognising citrullinated proteins display a broad cross-reactivity towards other post-translational modifications. Ann. Rheum. Dis. 79, 472–480 (2020). [DOI] [PubMed] [Google Scholar]
- 24.van de Stadt L. A., van Schouwenburg P. A., Bryde S., Kruithof S., van Schaardenburg D., Hamann D., Wolbink G., Rispens T., Monoclonal anti-citrullinated protein antibodies selected on citrullinated fibrinogen have distinct targets with different cross-reactivity patterns. Rheumatology (Oxford) 52, 631–635 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Dekkers G., Plomp R., Koeleman C. A., Visser R., von Horsten H. H., Sandig V., Rispens T., Wuhrer M., Vidarsson G., Multi-level glyco-engineering techniques to generate IgG with defined Fc-glycans. Sci. Rep. 6, 36964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiodin G., Allen J. D., Bryant D., Rock P., Martino E. A., Valle-Argos B., Duriez P. J., Watanabe Y., Henderson I., Blachly J. S., McCann K. J., Strefford J. C., Packham G., Geijtenbeek T. B., Figdor C., Wright G., Staudt L. M., Burack R., Bowden T., Crispin M., Stevenson F. K., Forconi F., Insertion of atypical glycans into the tumor antigen-binding site identifies DLBCLs with distinct origin and behavior. Blood 138, 1570–1582 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkman P. F., Fabre E., van der Voort E. I., Zaldumbide A., Rombouts Y., Rispens T., Wolbink G., Hoeben R. C., Spits H., Baeten D. L., Huizinga T. W., Toes R. E., Scherer H. U., Identification and characterisation of citrullinated antigen-specific B cells in peripheral blood of patients with rheumatoid arthritis. Ann. Rheum. Dis. 75, 1170–1176 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Klasener K., Maity P. C., Hobeika E., Yang J., Reth M., B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. eLife 3, e02069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suwannalai P., van de Stadt L. A., Radner H., Steiner G., El-Gabalawy H. S., Zijde C. M., van Tol M. J., van Schaardenburg D., Huizinga T. W., Toes R. E., Trouw L. A., Avidity maturation of anti-citrullinated protein antibodies in rheumatoid arthritis. Arthritis Rheum. 64, 1323–1328 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Volkov M., van Schie K. A., van der Woude D., Autoantibodies and B cells: The ABC of rheumatoid arthritis pathophysiology. Immunol. Rev. 294, 148–163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kissel T., van Schie K. A., Hafkenscheid L., Lundquist A., Kokkonen H., Wuhrer M., Huizinga T. W., Scherer H. U., Toes R., Rantapaa-Dahlqvist S., On the presence of HLA-SE alleles and ACPA-IgG variable domain glycosylation in the phase preceding the development of rheumatoid arthritis. Ann. Rheum. Dis. 78, 1616–1620 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Kissel T., van Wesemael T. J., Lundquist A., Kokkonen H., Kawakami A., Tamai M., van Schaardenburg D., Wuhrer M., Huizinga T. W., Scherer H. U., van der Woude D., Rantapaa-Dahlqvist S., Toes R. E. M., Genetic predisposition (HLA-SE) is associated with ACPA-IgG variable domain glycosylation in the predisease phase of RA. Ann. Rheum. Dis. annrheumdis-2021-220841 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Chan T. D., Wood K., Hermes J. R., Butt D., Jolly C. J., Basten A., Brink R., Elimination of germinal-center-derived self-reactive B cells is governed by the location and concentration of self-antigen. Immunity 37, 893–904 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Schneider D., Duhren-von Minden M., Alkhatib A., Setz C., van Bergen C. A., Benkisser-Petersen M., Wilhelm I., Villringer S., Krysov S., Packham G., Zirlik K., Romer W., Buske C., Stevenson F. K., Veelken H., Jumaa H., Lectins from opportunistic bacteria interact with acquired variable-region glycans of surface immunoglobulin in follicular lymphoma. Blood 125, 3287–3296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu D., McCarthy H., Ottensmeier C. H., Johnson P., Hamblin T. J., Stevenson F. K., Acquisition of potential N-glycosylation sites in the immunoglobulin variable region by somatic mutation is a distinctive feature of follicular lymphoma. Blood 99, 2562–2568 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Radcliffe C. M., Arnold J. N., Suter D. M., Wormald M. R., Harvey D. J., Royle L., Mimura Y., Kimura Y., Sim R. B., Inoges S., Rodriguez-Calvillo M., Zabalegui N., de Cerio A. L., Potter K. N., Mockridge C. I., Dwek R. A., Bendandi M., Rudd P. M., Stevenson F. K., Human follicular lymphoma cells contain oligomannose glycans in the antigen-binding site of the B-cell receptor. J. Biol. Chem. 282, 7405–7415 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Peaker C. J., Neuberger M. S., Association of CD22 with the B cell antigen receptor. Eur. J. Immunol. 23, 1358–1363 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Cao A., Alluqmani N., Buhari F. H. M., Wasim L., Smith L. K., Quaile A. T., Shannon M., Hakim Z., Furmli H., Owen D. M., Savchenko A., Treanor B., Galectin-9 binds IgM-BCR to regulate B cell signaling. Nat. Commun. 9, 3288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou P., Araujo E., Zhao T., Zhang M., Massenburg D., Veselits M., Doyle C., Dinner A. R., Clark M. R., B cell antigen receptor signaling and internalization are mutually exclusive events. PLoS Biol. 4, e200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoddart A., Jackson A. P., Brodsky F. M., Plasticity of B cell receptor internalization upon conditional depletion of clathrin. Mol. Biol. Cell 16, 2339–2348 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aletaha D., Neogi T., Silman A. J., Funovits J., Felson D. T., Bingham C. O. III, Birnbaum N. S., Burmester G. R., Bykerk V. P., Cohen M. D., Combe B., Costenbader K. H., Dougados M., Emery P., Ferraccioli G., Hazes J. M., Hobbs K., Huizinga T. W., Kavanaugh A., Kay J., Kvien T. K., Laing T., Mease P., Menard H. A., Moreland L. W., Naden R. L., Pincus T., Smolen J. S., Stanislawska-Biernat E., Symmons D., Tak P. P., Upchurch K. S., Vencovsky J., Wolfe F., Hawker G., 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 69, 1580–1588 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Koning M. T., Kielbasa S. M., Boersma V., Buermans H. P. J., van der Zeeuw S. A. J., van Bergen C. A. M., Cleven A. H. G., Kluin P. M., Griffioen M., Navarrete M. A., Veelken H., ARTISAN PCR: Rapid identification of full-length immunoglobulin rearrangements without primer binding bias. Br. J. Haematol. 178, 983–986 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Reiding K. R., Lonardi E., Hipgrave Ederveen A. L., Wuhrer M., Ethyl esterification for MALDI-MS analysis of protein glycosylation. Methods Mol. Biol. 1394, 151–162 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Sauter N. K., Grosse-Kunstleve R. W., Adams P. D., Robust indexing for automatic data collection. J. Appl. Cryst. 37, 399–409 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4 , The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 47.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H., Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Case D. A., Cheatham T. E. III, Darden T., Gohlke H., Luo R., Merz K. M. Jr., Onufriev A., Simmerling C., Wang B., Woods R. J., The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Kampstra A. S. B., Dekkers J. S., Volkov M., Dorjee A. L., Hafkenscheid L., Kempers A. C., van Delft M., Kissel T., Reijm S., Janssen G. M. C., van Veelen P. A., Bang H., Huizinga T. W. J., Trouw L. A., van der Woude D., Toes R. E. M., Different classes of anti-modified protein antibodies are induced on exposure to antigens expressing only one type of modification. Ann. Rheum. Dis. 78, 908–916 (2019). [DOI] [PubMed] [Google Scholar]
- 57.He X., Klasener K., Iype J. M., Becker M., Maity P. C., Cavallari M., Nielsen P. J., Yang J., Reth M., Continuous signaling of CD79b and CD19 is required for the fitness of Burkitt lymphoma B cells. EMBO J. 37, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plomp R., de Haan N., Bondt A., Murli J., Dotz V., Wuhrer M., Comparative glycomics of immunoglobulin A and G from saliva and plasma reveals biomarker potential. Front. Immunol. 9, 2436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen B. C., Falck D., de Haan N., Hipgrave Ederveen A. L., Razdorov G., Lauc G., Wuhrer M., LaCyTools: A targeted liquid chromatography-mass spectrometry data processing package for relative quantitation of glycopeptides. J. Proteome Res. 15, 2198–2210 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Brinkman E. K., Kousholt A. N., Harmsen T., Leemans C., Chen T., Jonkers J., van Steensel B., Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 46, e58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S6