Abstract
Ribosomal proteins (RP) are thought to primarily facilitate biogenesis of the ribosome and its ability to synthesize protein. However, here we show that ribosomal protein L22 like 1 (Rpl22l1) regulates hematopoiesis without affecting ribosome biogenesis or bulk protein synthesis. Conditional loss of murine Rpl22l1 using stage or lineage-restricted Cre drivers impairs development of several hematopoietic lineages. Specifically, Tie2-Cre mediated ablation of Rpl22l1 in hemogenic endothelium impairs the emergence of embryonic hematopoietic stem cells. Ablation of Rpl22l1 in late fetal liver progenitors impairs the development of B lineage progenitors at the pre-B stage and development of T cells at the CD44-CD25+ double-negative stage. In vivo labeling with O-propargyl puromycin revealed that protein synthesis at the stages of arrest was not altered indicating the ribosome biogenesis and function were not generally compromised. The developmental arrest was associated with p53 activation, suggesting that the arrest may be p53-dependent. Indeed, development of both B and T lymphocytes was rescued by p53-deficiency. P53 induction was not accompanied by DNA-damage as indicated by phospho-γH2AX induction or endoplasmic reticulum stress, as measured by phosphorylation of EIF2α, thereby excluding the known likely p53-inducers as causal. Finally, the developmental arrest of T cells was not rescued by elimination of the Rpl22l1 paralog, Rpl22, as we had previously found for the emergence of hematopoietic stem cells. This indicates that Rpl22 and Rpl22l1 play distinct and essential roles in supporting B and T cell development.
Introduction
Ribosomal proteins (RP) play key roles in the assembly of the ribosome and its essential function of translating mRNAs into proteins (1). Inherited defects in RP genes or ribosome assembly factors cause a collection of diseases termed ribosomopathies (2–4). These diseases typically share three developmental anomalies, alterations in body plan, hematopoietic defects, and increased cancer risk, particularly myeloid cancers (5), which are widely thought to result from impairment of ribosome assembly or function (6). However, variations in the spectrum of phenotypes have been observed in ribosomopathies, such as aspleny in patients with RPSA insufficiency (7), raising the possibility that, in addition to supporting ribosome assembly and function, RP may also play distinct regulatory roles important for normal development. These regulatory roles have been proposed to involve either altering ribosome function or extraribosomal activities, performed while the RP is physically separate from the ribosome (1, 8–12).
The ability of RP to regulate biological processes is also, in some cases, influenced by highly homologous RP paralogs. RP paralogs are frequently observed in lower eukaryotes, with ~75% of yeast RP genes having paralogs (13). While many of these paralogous pairs appear to be functionally redundant, others appear to perform somewhat distinct functions(13). Importantly, while RP paralogs are restricted to lower eukaryotes, a handful are conserved in vertebrates(4, 14, 15). Many of those conserved in vertebrates exhibit tissue restricted expression patterns, like the RPS4 paralog, RPS4Y, which is restricted to testis and prostate (14). The conservation into vertebrates and the tissue restriction of certain paralogs suggests an important function. RP paralogs have been proposed to “pimp” or confer unique functions on ribosomes (16); however, the developmental roles of paralagous RP pairs remains largely unexplored, particularly in vertebrates.
The RP, Rpl22 and its highly homologous paralog Rpl22-Like1 (Rpl22l1) are one such paralogous pair that is conserved in vertebrates (17). We have determined that Rpl22 is not required for ribosome biogenesis or global protein synthesis, and yet plays critical roles in the development of both B and T lymphocytes (18–20). Indeed, the germline ablation of the murine Rpl22 locus does not affect mouse viability, gross development, or fertility(18). Instead, it causes a remarkably selective arrest in B and T cell development that occurs at the pre-B and pre-T cell stages, respectively (18–20). The arrest in both B and T cells is p53-dependent and rescued by p53 deficiency (18–20). Surprisingly, while Rpl22 expression is ubiquitous, germline ablation of the Rpl22 locus in mice results in a remarkably selective activation of p53 in B and T cell precursors (18–20). The basis for p53 induction in developing T cells is the activation of excessive endoplasmic reticulum (ER) stress (21).
The role of the paralog of Rpl22, Rpl22l1, is less well understood, but we have begun to provide some insights. We determined that Rpl22l1 is required for embryonic development because it controls the splicing of smad2 pre-mRNA during gastrulation in zebrafish and mice (22). The control of smad2 pre-mRNA splicing is controlled in an antagonistic manner with Rpl22, where both Rpl22 and Rpl22l1 directly bind smad2 but have opposing effects. Rpl22 acts to disrupt splicing while Rpl22l1 opposes splice disruption and promotes normal smad2 pre-mRNA splicing (22). Importantly, Rpl22 and Rpl22l1 also antagonistically regulate the emergence of embryonic hematopoietic stem cells (HSC). They do so through opposing effects on the translation of smad1 mRNA, an essential activator of the transcription factor Runx1, which is required for embryonic HSC emergence (22–24). Thus, Rpl22l1 and Rpl22 do not appear to be functionally redundant, but instead control gastrulation and HSC emergence through antagonistic regulation of common targets. The role of Rpl22l1 in later hematopoietic processes has not been assessed.
Because ablation of Rpl22l1 in zebrafish blocks embryonic HSC emergence, it precludes analysis of Rpl22l1 function at later stages of hematopoiesis. To circumvent his impediment, we conditionally ablated the Rpl22l1 locus in mice using the Vav-Cre driver, which is activated after HSC emergence (25). Using this approach, we determined that Rpl22l1 plays a critical role in more distal stages of hematopoiesis, specifically attenuating the development of B and T lymphocytes. Like Rpl22, the developmental arrests of both B and T cells were not associated with a failure to produce the pre-receptor complex or with changes in protein synthesis, but were p53 dependent, as they were both rescued by p53-deficiency. Finally, the developmental arrests were not rescued by Rpl22 indicating that Rpl22 and Rpl22l1 were not functioning antagonistically in regulating T and B cell development. Taken together, these observations indicate that Rpl22 and Rpl22l1 both play distinct, independent, critical roles in supporting lymphocyte development that do not involve alterations in global protein synthesis.
Materials and Methods
Mice
All mouse strains were housed in the Laboratory Animal Facility at Fox Chase, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and handled under the auspices of protocols approved by the Institutional Animal Care and Use Committee. Rpl22−/−, a germline gene-trap knockout mouse, as well as Rpl22l1fl/fl, vav-cre, pTa-cre, Tie2-cre and p53−/− mice have been previously described (18, 22, 25–28).
Flow Cytometry and Immunofluorescence
Single-cell suspensions were prepared from bone marrow, thymus, spleen, and peritoneal fluid and stained, as indicated, with optimal amounts of the following fluorochrome-conjugated antibodies: anti-CD4 (GK1.5), anti-CD5 (53–7.3), anti-CD8 (53–6.7), anti-CD19 (ID3), anti-CD23 (B3B4), anti-CD25 (PC61), anti-CD43 (S7), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD69 (H1.2F3), anti-B220 (RA3–6B2), anti-AA4.1 (AA4.1), anti-Thy1.2 (53–2.1), anti-IgM (11/41), anti-TCRδ (GL3), anti-CD24 (M1/69), anti-CD73 (TY/11.8), anti-cKit (2B8), anti-Sca1 (D7), and CD150 (TC15–12F12.2), anti-CD48 (HM48–1), and anti-TCRβ (H57–597). The antibodies were purchased from BD Biosciences, eBioscience, BioLegend, or Tonbo Biosciences. Dead cells were excluded from analyses using propidium iodide (PI). Data were analyzed on an LSRII flow cytometer (BD Pharmigen). Cell populations were purified by flow cytometry using a FACSAria Cell Sorter (BD Biosciences). To assess the effect of Rpl22l1 loss on the emergence of embryonic HSC, day 11.5 embryos were fixed and The Rpl22l1 gene was conditionally ablated in hemogenic endothelium by Tie2-Cre, following which HSC emergence in Rpl22l1f/f and Tie2-Cre Tg Rpl22l1f/f embryos was evaluated by immunofluorescence using anti-CD31 to mark vasculature and anti-Runx1 to mark HSC, as described (27, 29). For phospho-γH2AX flow cytometry, thymocytes were stained with Ghost Dye Violet 450 (Tonbo biosciences; 13–0863-T100) and appropriate antibodies, before fixation in cold 70% ethanol. Fixed cells were stained with anti-phospho-γH2AX (Ser139; BioLegend, 2F3) and analyzed by flow cytometry as above. Thymocytes irradiated with 10Gy of g-irradiation served as a positive control. Intracellular TCRβ staining was performed as previously described(18). For immunofluorescence, the same populations of thymocytes were fixed in 4% PFA for 10, permeabilized with 1% Triton X-100, and blocked with 5% goat serum in PBS for 2 hours, before staining overnight with anti-phospho-γH2AX (Ser139; Sigma, JBW301). Bound antibody was visualized with FITC-goat anti mouse (Jackson immunoResearch) for 1 hour, before viewing on a Leica SP8 microscope.
In Vivo Protein Synthesis
Analysis of in vivo protein synthesis was performed as previously described (30). Mice were injected intraperitoneally with O-propargyl-puromycin at 50 mg/kg in PBS. After 1 hour, mice were euthanized and single cell suspensions were generated from bone marrow and thymus. Cells were stained with GhostDye450 or GhostDye710 (Tonbo Biosciences) per manufacturer’s instructions and surfaced stained with optimal amounts of fluorochrome-conjugated antibodies. Cells were subsequently fixed and permeabilized using BD Fixation/Permeabilization Solution (BD Biosciences), and O-propargyl-puromycin incorporation was visualized using the Click-iT Cell Reaction Buffer Kit (Invitrogen) per manufacturer’s instructions.
Retroviral Transduction and OP9-DL1 Cultures
Retroviral particles were produced by transient calcium phosphate transfection of Phoenix cells. MLP and MLP-shp53 vectors have been previously described (18). DN thymocytes were magnetically isolated by depletion using anti-CD4 and anti-CD8 microbeads (Miltenyi Biotec) and underwent spin transduction with the indicated retrovirus. Transduced cells were plated on OP9-DL1 stromal cells, cultured for 4 days in the presence of Flt3L (5 ng/ml) and IL7 (5 ng/ml), and analyzed by flow cytometry.
Quantitative Real-Time PCR
Total RNA was isolated from cells using the RNeasy system (Qiagen). DNA was removed using on-column DNase treatment (Qiagen), and cDNA was synthesized using Superscript IV reverse transcriptase and oligo-dT (Invitrogen). Quantitative real-time PCR was performed on the Prism 7700 thermocycler (Applied Biosystems) using TaqMan real-time PCR primer/probe sets specific for puma/Bbc3 (Mm00519268_m1), noxa/Pmaip1 (Mm00451763_m1), and GAPDH/Gapdh (Mm99999915_g1).
Immunoblot Analysis
Cells were lysed in RIPA buffer (20 mM HEPES pH7, 150 mM NaCl, 1% deoxycholate, 1% Nonidet P-40, 0.1% SDS, 1 mM Na2VO4, 2 mM EDTA, and a complete protease inhibitor mixture; Roche). Samples were resolved on NuPage Novex Bis-Tris gels (Invitrogen) and blotted with the following antibodies: anti-p53 (IMX25; Leica Microsystems), anti-Rpl22l1, anti-EIF2α (5324S, Cell Signaling), anti-phospho-EIF2α (9721S, Cell Signaling), anti-calnexin(31), and anti-GAPDH (6C5; Millipore) (32).
RNA-Seq Analysis
RNA-Seq analysis was performed on RNA extracted from electronically sorted DN3 thymocytes from wild type and Rpl22l1-deficient mice. Briefly, the mRNA-seq library was prepared from the total RNA using poly(A) selection (Truseq RNA Sample Preparation Kit V2, Illumina). RNA concentration was quantified using a NanoDrop, and RNA integrity was measured using a BioAnalyzer chip (Aligent), followed by 100 bp paired end sequencing on a HiSeq2500 (Illumina) according to manufacturer protocols. mRNA-seq reads were mapped to the latest mouse genome assembly (mm10) using STAR alignment algorithm (33). HOMER was used to calculate the RPKM and raw read counts, and differential expression was determined using edgeR R package (34, 35). The raw RNA-Seq data have been deposited at the Gene Expression Omnibus database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183054.
Analysis of ribosomal RNA (rRNA) processing.
The effect of Like1 loss of rRNA processing was assessed in thymic lymphoma cells transduced with either control pMLSII retroviral vector or pMLSII expressing the following miR-30 based shRNA: L1sh-1: 5’-CTCACAGTTGTTTCTGAGAAA; L1sh-2: 5’-CCGGGAGAAGGTTAAAGTCAA. Total RNA was extracted from cells 48hr post transduction, following which 2.5μg RNA were loaded on the RNA gel for northern analysis. The conditions and probes used for Northern hybridizations were as described (36).
Results
Rpl22l1-deficiency attenuates the emergence of fetal hematopoietic stem cells.
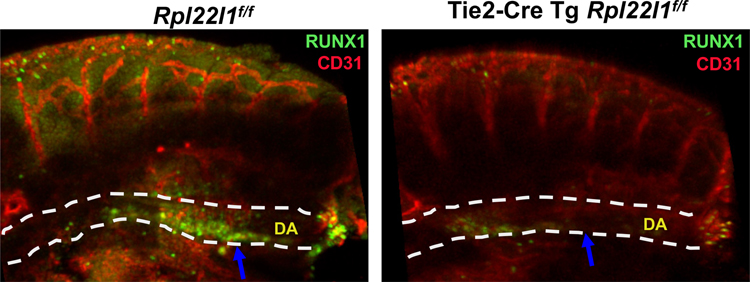
We have previously shown that knockdown of rpl22l1 in zebrafish attenuates the emergence of embryonic HSC (24). To determine if Rpl22l1 also plays an essential role in the emergence of mammalian HSC from the aorta-gonad-mesonephros (AGM), we ablated the Rpl22l1 gene (Rpl22l1cKO) in hemogenic endothelium using Tie2-Cre (27). Emerging HSC were identified by immunofluorescent staining for the transcription factor RUNX1, which is essential for HSC emergence (23). Importantly, ablation of Rpl22l1 in hemogenic endothelium attenuated the emergence of RUNX1-expressing HSC (Figure 1), indicating that Rpl22l1 plays a critical role in HSC emergence in mammals as we had previously reported in zebrafish (24).
Figure 1.
Rpl22l1 loss attenuates embryonic HSC emergence. The Rpl22l1 gene was conditionally ablated in hemogenic endothelium by Tie2-Cre, following which HSC emergence in Rpl22l1f/f and Tie2-Cre Tg Rpl22l1f/f embryos was evaluated by immunofluorescence using anti-CD31 (red) to mark vasculature and anti-Runx1 (green) to mark HSC. DA, dorsal aorta indicated by dashed white lines.
Rpl22l1-deficiency reduces splenic lymphocyte cellularity
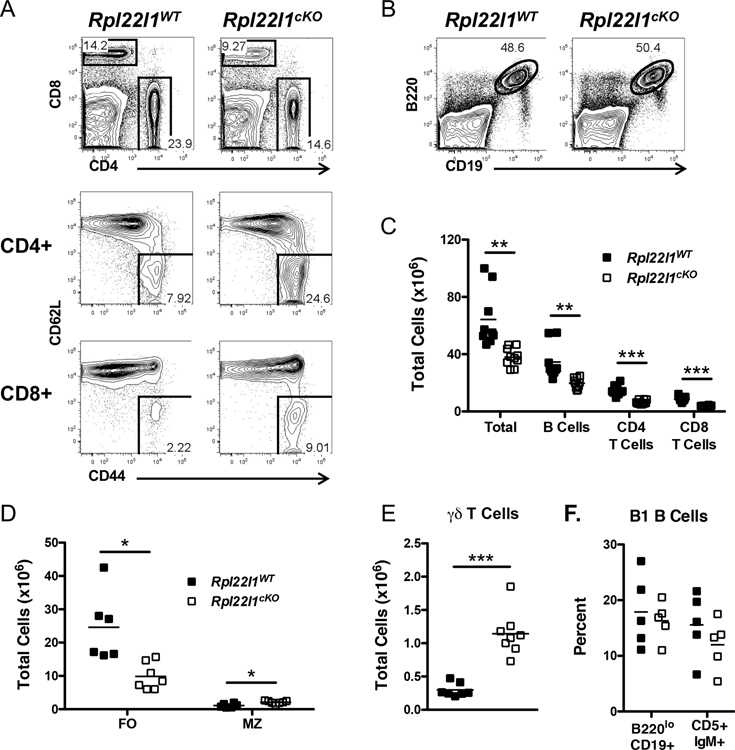
To determine if Rpl22l1 plays a role in later stages of hematopoiesis, we employed Vav-1 Cre to ablate the Rpl22l1 gene (Rpl22l1cKO), since Vav1-Cre induces deletion after HSC emergence (23). Ablation of the Rpl22l1 gene using Vav1-Cre did not alter the frequency or number of early hematopoietic stem and progenitor cells (HSPC) in the bone barrow (Figure S1A–D), suggesting that Rpl22l1 is dispensable for the development and maintenance of adult HSPC; however, Rpl22l1 deficiency did alter the frequency of lymphoid populations in peripheral lymphoid tissues, including the spleen (Figure 2). Indeed, Rpl22l1-deficiency reduced the frequency of T lymphocytes in the spleen and increased the fraction of CD44+CD62L-memory phenotype T cells (Figure 2A). In contrast, the frequency of B lymphocytes was not reduced, but their absolute numbers were reduced as a result of the overall decrease in splenic cellularity (Figure 2B,C). The decrease in B cells numbers was restricted to the follicular subset (FO), since marginal zone B cells were not reduced (Figure 2D). Interestingly, the number of γδ T cells was actually increased (Figure 2E), while innate like B1 B cells were not altered (Figure 2F). Collectively, these results suggest that Rpl22l1 is dispensable for generation and maintenance of adult HSPC, but is selectively required for either the development or survival of some B and T lymphocyte subsets.
Figure 2.
Reduction in peripheral lymphocyte subsets in the absence of Rpl22l1. (A) Splenocytes from Rpl22l1WT (Rpl22l1+/+vav-cre) and Rpl22l1cKO (Rpl22l1fl/fl vav-cre) mice were analyzed for surface expression of CD4, CD8, CD44, and CD62L. The top panels represent CD4 versus CD8 expression. The middle panels represent CD44 and CD62L expression on CD4+ gated cells. The bottom panels represent CD44 and CD62L expression of CD8+ gated cells. (B) Splenocytes from Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of B220 and CD19. (C) Absolute number of total cells, B cells (B220+ CD19+), CD4+ T cells (CD4+ CD8−), and CD8+ T cells (CD4− CD8+) from the spleens of Rpl22l1WT and Rpl22l1cKO mice. (D) Absolute number of follicular B cells (FO; B220+ AA4.1− IgM+ CD23+) and marginal zone B cells (MZ; B220+ AA4.1− IgMhigh CD23−) in the spleens of Rpl22l1WT and Rpl22l1cKO mice. (E) Absolute number of γδ T cells (TCRδ+) in the spleens of Rpl22l1WT and Rpl22l1cKO mice. (F) Peritoneal fluid from Rpl22l1WT and Rpl22l1cKO mice was analyzed for surface expression of B220 and CD19 to identify B1a and B1b cells (B220lo CD19+) or CD5 and IgM to identify CD5+ IgM+ B1a cells. Viable cells were identified as PI−. n = 6–10 mice per genotype (A-E) or 5 mice per genotype (F). *p < 0.01, **p < 0.005, ***p < 0.0001.
Rpl2l1 loss impairs traversal of the β-selection checkpoint
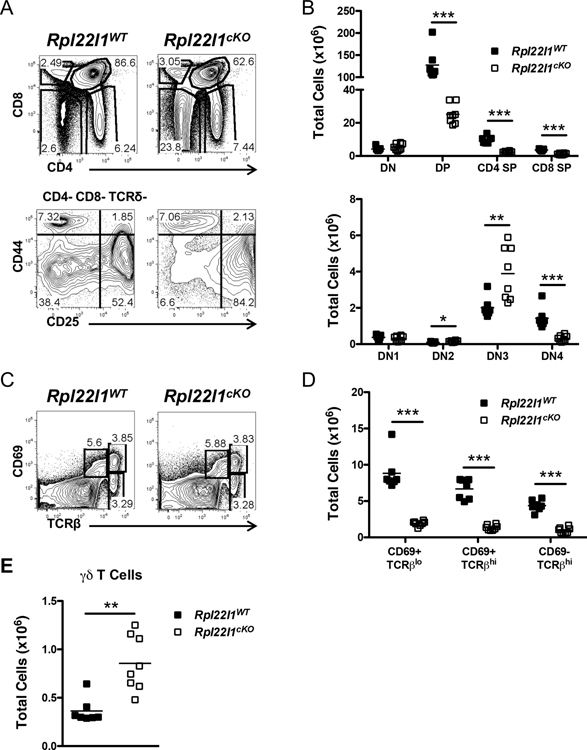
Since the frequency and number of peripheral T cells was reduced, we wished to determine if this was a consequence of impaired development in the thymus. To assess this possibility, we performed flow cytometry on single cell suspensions of thymocytes from Rpl22l1+/+ and Rpl22l1cKO mice. Analysis of thymic subpopopulations separated based on expression of CD4 and CD8 revealed that thymocyte development was impaired between the immature CD4-CD8- double negative (DN) and CD4+CD8+ double positive (DP) stage, since both the percentage and number of DP were diminished in Rpl22l1cKO mice (Figure 3A,B). The arrest appears to result from impaired traversal of the β-selection checkpoint with an accumulation of CD44-CD25+ (DN3) thymocytes and a decrease in CD44-CD25- (DN4) cells (Figure 3A,B). Rpl22l1-deficiency does not appear to affect more distal phases of development. The proportion of more mature CD4+ and CD8+ single positive (SP) thymocytes was not altered (Figure 3A,B); however, their absolute numbers were decreased, presumably as a consequence of the reduced production of pre-selection DP thymocytes, from which mature SP thymocytes are selected (Figure 3A,B). Indeed, the proportion of selection intermediates defined by changes in CD69 and TCRβ expression was unchanged, but there absolute number was reduced (Figure 3C,D). As was observed in the periphery, the number of γδ T cells were increased in the thymus (Figure 3E), suggesting that the increase in the periphery does not result from homeostatic expansion but instead from increased thymic output. A similar defect in T cell development was observed when Rpl22l1 was deleted using pTα-Cre, indicating that Rpl22l1 plays an essential and cell intrinsic role in T cell development, and that the defect in T cell development was not an indirect effect of alterations in earlier, pre-thymic progenitors (Figure S1E). Taken together, these data indicate that the reduced numbers of peripheral αβ T cells and the increase in γδ T cells are a consequence of alterations in thymic development.
Figure 3.
Altered thymopoiesis in the absence of Rpl22l1. (A) Thymocytes from Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of Thy1.2, CD4, CD8, TCRδ, CD44, and CD25. The top panels represent CD4 versus CD8 expression after gating on Thy1.2+ cells. The bottom panels represent CD44 versus CD25 expression after gating on CD4− CD8− TCRδ− cells. (B) Absolute number of double negative (DN; Thy1.2+ CD4− CD8−), double positive (DP; Thy1.2+ CD4+ CD8+), CD4 single positive (CD4 SP; Thy1.2+ CD4+ CD8−), and CD8 single positive (CD8 SP; Thy1.2+ CD4− CD8+) thymocytes (top panel) or DN1 (Thy1.2+ CD4− CD8− TCRδ− CD44+ CD25−), DN2 (Thy1.2+ CD4− CD8− TCRδ− CD44+ CD25+), DN3 (Thy1.2+ CD4− CD8− TCRδ− CD44− CD25+), and DN4 (Thy1.2+ CD4− CD8− TCRδ− CD44− CD25−) thymocytes (bottom panel) from Rpl22l1WT and Rpl22l1cKO mice. (C) Thymocytes from Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of CD69 and TCRβ. (D) Absolute number of CD69+ TCRβlo, CD69+ TCRβhi, and CD69− TCRβhi thymocytes from Rpl22l1WT and Rpl22l1cKO mice. (E) Absolute number of γδ T cells (Thy1.2+ CD4− CD8− TCRδ+) in thymi from Rpl22l1WT and Rpl22l1cKO mice. Viable cells were identified as PI−. n = 7–8 mice per genotype. *p < 0.05, **p < 0.01, ***p < 0.0005.
Development of B lymphocytes beyond the pre-B cell stage is attenuated by Rpl22l1 loss
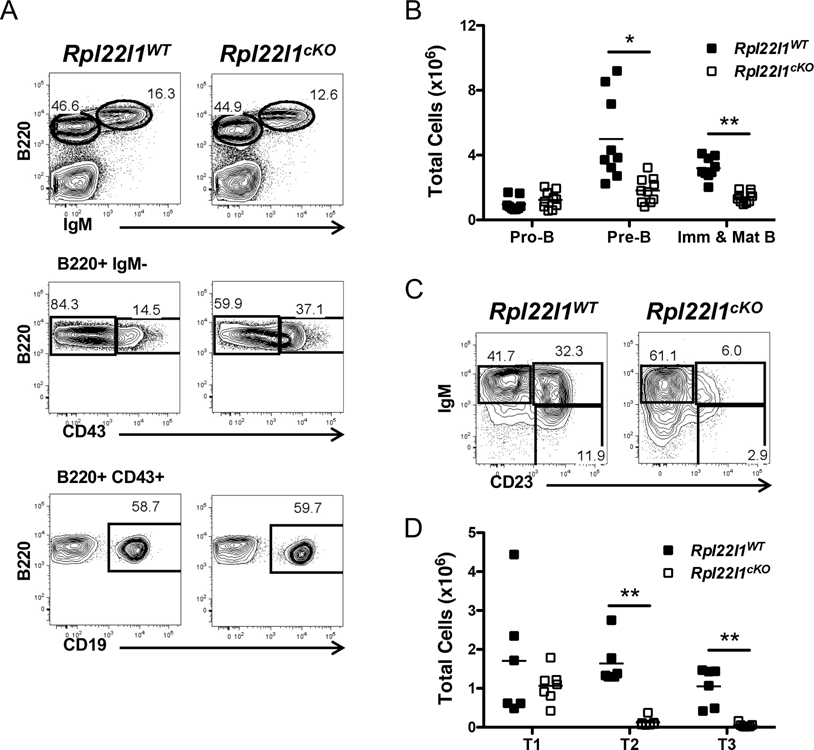
While the proportions of B lymphocytes were unchanged, their absolute number in the spleen was reduced (Figure 2C). Thus, we wished to determine if this resulted from a defect in their development in the bone marrow. The number of B220+CD43+CD19+ pro-B cells was unchanged (Figure 4A,B); however, all distal developmental intermediates were reduced beginning with B220+CD43low pre-B cells (Figure 4A,B) and continuing through immature, mature, and transitional B cell subsets, T1 (IgM+CD23−), T2 (IgM+CD23+), and T3 (IgMlowCD23+) (Figure 4C,D). The transition from B220+CD43+ to B220CD43low pre-B cells is analogous to the DN3 to DN4 transition in thymocyte development. Thus, the developmental arrest points are similar in the bone marrow and thymus. However, unlike thymocyte development where the developmental intermediates distal to the DP stage were not changed in frequency, development of B cells through the transitional intermediates was also impaired (Figure 4C,D).
Figure 4.
B cell development is disrupted in the absence of Rpl22l1. (A) Bone marrow from Rpl22l1WT and Rpl22l1cKO mice was analyzed for surface expression of Ly6C, NK1.1, B220, IgM, CD43, and CD19. The top panels represent B220 versus IgM expression after gating out Ly6C+ and NK1.1+ cells. The middle panels represent B220 versus CD43 expression on B220+ IgM− cells. The bottom panels represent B220 versus CD19 expression of B220+ CD43+ cells. (B) Absolute number of pro-B cells (Ly6C− NK1.1− B220+ IgM− CD43+ CD19+), pre-B cells (Ly6C− NK1.1− B220+ IgM− CD43−), and immature and mature B cells (Ly6C− NK1.1− B220+ IgM+) in the bone marrow from Rpl22l1WT and Rpl22l1cKO mice. (C) Splenocytes from Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of B220, AA4.1, IgM, and CD23. Panels represent IgM versus CD23 expression after gating on B220+ AA4.1+ cells. (D) Absolute number of T1 (B220+ AA4.1+ IgMhigh CD23−), T2 (B220+ AA4.1+ IgMhigh CD23+), and T3 (B220+ AA4.1+ IgMlow CD23+) transitional B cell subsets in the spleens of Rpl22l1WT and Rpl22l1cKO mice. Viable cells were identified as PI−. n = 9–10 mice per genotype (A-B) or 6–7 mice per genotype (C-D). *p < 0.05, **p < 0.0005.
Developmental arrest in Rpl22l1-deficient mice is not associated with impaired protein synthesis
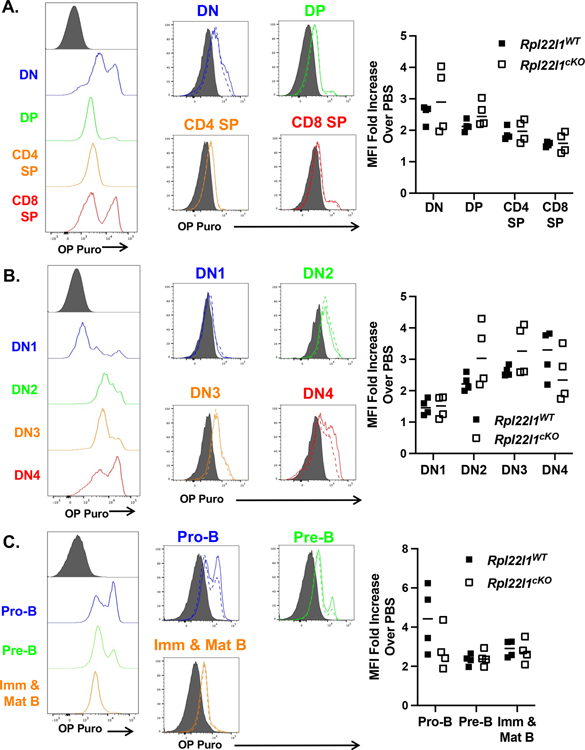
Mutations in numerous RP have previously been linked to defects in hematopoiesis that are associated with impaired ribosome biogenesis and diminished capacity for protein synthesis (8, 30, 37). We have previously shown in cell lines that the elimination of Rpl22l1 does not block protein synthesis in vitro (24). However, the dispensability of Rpl22l1 for protein synthesis in cell lines does not preclude a selective requirement for Rpl22l1 in particular cell lineages or at different stages of development. Consequently, we sought to assess the effect of Rpl22l1-deficiency on protein synthesis in developing primary lymphoid cells in vivo. To do so, we measured protein synthesis in developing lymphocytes in situ, by monitoring O-propargyl-puromycin (OPP) incorporation into nascent polypeptides using flow cytometry (30). Interestingly, we found that during development in the thymus, increased incorporation of OPP was associated with the proliferative stages of development including the DN subpopulations, particularly CD44+CD25+ (DN2), a proportion of DN3 thymocytes, and most DN4 (Figure 5A,B). It should be noted that Rpl22l1-deficiency did not alter the incorporation of OPP in any of these subpopulations (Figure 5A,B; solid v dashed lines). Analysis of B cells developing in the bone marrow revealed that pro-B cells and a small population of pre-B cells exhibited elevated OPP incorporation (Figure 5C); however, as for developing T cells, Rpl22l1-deficiency did not diminish OPP incorporation. Rpl22l1-deficiency did appear to reduce the frequency of OP-puro high pro-B cells (Figure 5C), but this reduction was not statistically significant (p=0.16). Taken together, these data demonstrate that the impairment of B and T cell development caused by Rpl22l1-deficiency was not associated with detectable reductions in protein synthesis.
Figure 5.
Protein synthesis is not impaired in lymphocyte progenitors in the absence of Rpl22l1. Rpl22l1WT and Rpl22l1cKO mice were injected with O-propargyl-puromycin (OP Puro) and analyzed 1 hour later for incorporation of OP Puro in nascent polypeptides. (A-B) Thymocytes from OP Puro-injected Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of Thy1.2, CD4, CD8, TCRδ, CD44, and CD25. (A) The left panel represents OP Puro incorporation in double negative (DN), double positive (DP), CD4 single positive (CD4+ SP), and CD8 single positive (CD8+ SP) thymocytes, as described in Figure 2B, in wild-type mice. The right panel represents OP Puro incorporation in DN, DP, CD4 SP, and CD8 SP thymocytes from Rpl22l1WT (solid line) and Rpl22l1cKO (dotted line) mice. (B) The left panel represents OP Puro incorporation in DN1, DN2, DN3, and DN4 thymocytes, as described in Figure 2B, in wild-type mice. The right panel represents OP Puro incorporation in DN1, DN2, DN3, and DN4 thymocytes from Rpl22l1WT (solid line) and Rpl22l1cKO (dotted line) mice. (C) Bone marrow from OP Puro-injected Rpl22l1WT and Rpl22l1cKO mice were analyzed for surface expression of B220, IgM, CD43, and CD19. The left panel represents OP Puro incorporation in pro-B cells (B220+ IgM− CD43+ CD19+), pre-B cells (B220+ IgM− CD43−) and immature & mature B cells (B220+ IgM+) in wild-type mice. The right panel represents OP Puro incorporation in pro-B cells, pre-B cells, and immature & mature B cells from Rpl22l1WT (solid line) and Rpl22l1cKO (dotted line) mice. Grey histograms represent background staining in PBS-injected wild-type mice. n = 4 mice per genotype. (A-C) Scattergrams of the fold-change in mean fluorescence intensity (MFI) in OP-Puro treated over PBS injected for each mouse are depicted at the right. Each mouse is a symbol. There were no statistically significant changes in Rpl22l1-deficient cell populations, including pro-B cells (p = 0.16).
Rpl22l1 loss is linked to p53 induction
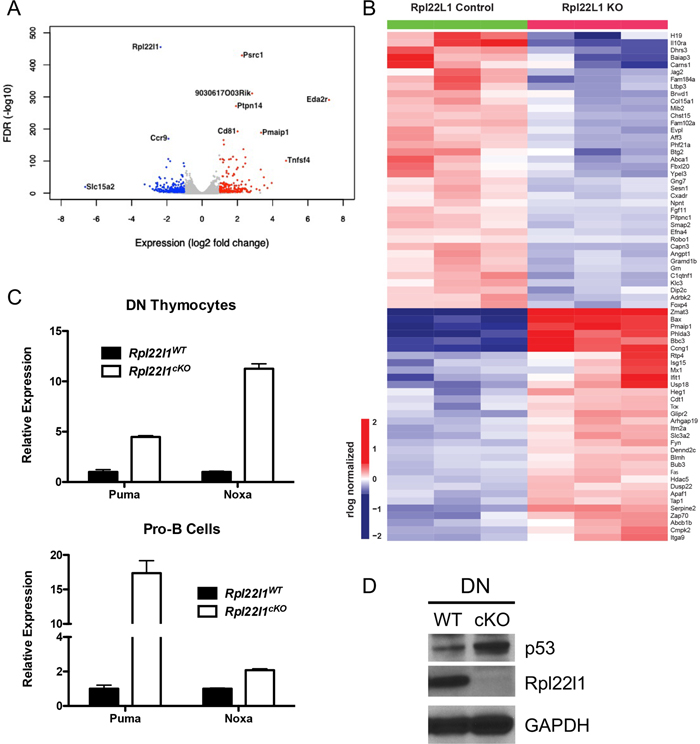
Since Rpl22l1-deficiency did not alter global protein synthesis, to gain insight into the molecular basis for the developmental arrest of T cell precursors, we performed RNA-Seq analysis on immature DN3 thymocytes from Rpl22l1+/+ and Rpl22l1cKO mice. The whole transcriptome analysis identified 781 genes that were differentially expressed in DN3 from Rpl22l1cKO (Figure 6A and S2; Table S1). KEGG and Metascape pathway analysis revealed a number of pathways that exhibited significant enrichment, including T cell activation, innate immune response, lipid metabolism, and cell growth and survival, but the most highly enriched was the p53 pathway (Figure S2). The differentially expressed p53 gene targets comprising the p53 signature include PUMA (Pmaip1) and Noxa (Bbc3) and are displayed in a heat map (Figure 6B). The induction of p53 targets PUMA and Noxa in DN3 from Rpl22l1cKO mice were verified by qPCR in both DN thymocytes and Pro-B cells, the populations in which the developmental arrest begins to manifest (Figure 6C). Consistent with the induction of p53 targets, we observed that p53 protein levels are also induced in DN thymocytes from Rpl22l1cKO mice (Figure 6D). Taken together, these data indicate that the developmental arrest of lymphoid cells in Rpl22l1cKO mice is associated with activation of p53.
Figure 6.
Increased p53 expression in the absence of Rpl22l1 in T and B cell progenitors. (A) Volcano plot of differentially expressed genes in Rpl22l1fl/fl pTa-cre DN3 thymocytes compared to Rpl22l1fl/fl DN3 thymocytes (biological triplicates). Red dots represent genes significantly upregulated (log2 fold change ≥ 1, FDR ≤ 0.05) and blue dots represent genes significantly downregulated (log2 fold change ≤ 1, FDR ≤ 0.05). (B) Heat map of the log2 RPKM values of the p53 signaling pathway genes in Rpl22l1fl/fl (L1WT) and Rpl22l1fl/fl pTa-cre (L1KO) DN3 thymocytes. (C) Quantitative real-time PCR analysis on DN thymocytes (top panel) or sorted pro-B cells (bottom panel) from Rpl22l1WT and Rpl22l1cKO mice. mRNA levels were normalized to the expression of GAPDH. Data are representative of two independent experiments. (D) Detergent extracts from DN thymocytes from Rpl22l1WT and Rpl22l1cKO mice were immunoblotted with anti-p53, anti-Rpl22l1, or GAPDH antibodies. Data are representative of two independent experiments.
Rpl22l1 loss arrests lymphoid development in a p53-dependent manner
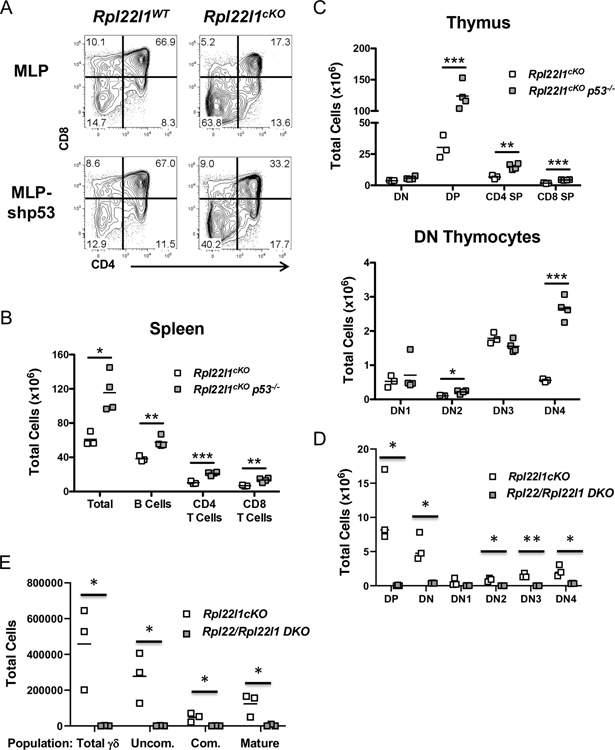
To determine if p53 induction is responsible for the impaired lymphoid development observed in Rpl22l1cKO mice, we performed a loss-of-function experiment using in vitro cultures of adult DN3 thymocytes on OP9-DL1 stroma (38). The development of DN thymocytes from Rpl22l1cKO mice to the DP stage upon culture on OP9-DL1 stroma in vitro was impaired (Figure 7A). Importantly, transduction of the DN thymocytes from Rpl22l1+/+ mice with a p53 shRNA (MLP-shp53) did not alter their development to the DP stage (Figures S3A and 7A); however, transduction of DN from Rpl22l1cKO mice with p53 shRNA markedly improved their development to the DP stage, although it did not fully rescue development (Figure 7A). To assess the role of p53 more broadly in the developmental impairment of lymphocytes in Rpl22l1cKO mice, we crossed them to p53-deficient mice. Importantly, analysis of the spleens of Rpl22l1cKOp53−/− mice revealed a rescue of total splenic cellularity in general, and the numbers of B and T cells specifically (Figure 7B). Moreover, analysis of the thymic subsets of the Rpl22l1cKOp53−/− mice revealed that development to the DP stage and beyond was normalized, as was traversal of the β-selection checkpoint to the DN4 stage, where development in Rpl22l1cKO mice was arrested (Figure 7C). The induction of p53 did not appear to be a consequence of defective ribosome biogenesis, since Rpl22l1 knockdown did not impair rRNA processing, a process that is defective when ribosome biogenesis is impaired (Figure S3B,C). Another possible cause of p53 induction was excessive endoplasmic reticulum (ER) stress, which we had previously determined to be responsible for p53 induction in Rpl22-deficient thymocytes; however, unlike p53 induction in Rpl22-deficient mice, p53 induction in Rpl22l1cKO mice was not associated with significant activation of the unfolded protein response in purified DN3 thymocytes (Figure S3D). Rpl22l-deficiency did not induce DNA-damage as assessed by phospho-γH2AX staining, suggesting that another potential inducer of p53, DNA-damage, was not responsible for p53 induction in Rpl22l1cKO DN3 thymocytes (Figure S3E,F). Because innate immune signaling pathways were activated by Rpl22l1-deficiency, and have been implicated in p53 induction (39, 40), we asked if Rpl22l1-deficient thymocytes were hyperresponsive to interferon (IFN) stimulation; however, they exhibited no difference in IFN induced STAT1-phosphorylation (Figure S3G). Finally, Rpl22l1-deficiency impairs traversal of the pre-receptor checkpoint by both B and T lymphoid progenitors, raising the possibility that VDJ recombination might be impaired; however, intracellular TCRβ staining revealed that the fraction of TCRβ-expressing DN3 cells was unaffected (Figure S3H). Taken together, these data demonstrate that the impairment of lymphoid development in Rpl22l1cKO mice is dependent upon p53 induction and that p53 induction does not appear to result from activation of its known inducers.
Figure 7.
p53 deficiency rescues lymphocyte development in the absence of Rpl22l1. (A) DN thymocytes from Rpl22l1WT and Rpl22l1cKO mice were transduced with empty MLP or MLP-shp53 retroviral vectors and cultured for 4 days on OP9-DL1 cells. Histograms represent CD4 versus CD8 expression on Thy1.2+ GFP+ cells. Data are representative of 4 independent experiments. (B) Absolute number of total cells, B cells (B220+ CD19+), CD4+ T cells (CD4+ CD8−), and CD8+ T cells (CD4− CD8+) from the spleens of Rpl22l1cKO and Rpl22l1cKO p53−/− mice. (C) Absolute number of double negative (DN; Thy1.2+ CD4− CD8−), double positive (DP; Thy1.2+ CD4+ CD8+), CD4 single positive (CD4 SP; Thy1.2+ CD4+ CD8−), and CD8 single positive (CD8 SP; Thy1.2+ CD4− CD8+) thymocytes (top panel) or DN1 (Thy1.2+ CD4− CD8− TCRδ− CD44+ CD25−), DN2 (Thy1.2+ CD4− CD8− TCRδ− CD44+ CD25+), DN3 (Thy1.2+ CD4− CD8− TCRδ− CD44− CD25+), and DN4 (Thy1.2+ CD4− CD8− TCRδ− CD44− CD25−) thymocytes (bottom panel) from Rpl22l1cKO and Rpl22l1cKO p53−/− mice. n = 3–4 mice per genotype. *p < 0.05, **p < 0.01, ***p < 0.001. (D) Absolute number of DN, DP, DN1, DN2, DN3, and DN4 thymocytes from Rpl22l1cKO and Rpl22−/−Rpl22l1cKOmice were depicted graphically as scattergrams. n = 3–4 mice per genotype. *p < 0.05, **p < 0.01. (E) Absolute number of total γδ T cells (γδ TCR+) as well as their developmental intermediates (Uncom., γδ TCR+ CD24+ CD73−; Com., γδ TCR+ CD24+ CD73+, and Mat., γδ TCR+ CD24− CD73+) were depicted graphically as scattergrams. n = 3–4 mice per genotype. *p < 0.05, **p < 0.01.
We had previously reported that that the emergence of embryonic HSC was regulated by the antagonistic balance of Rpl22l1 and Rpl22(24). Indeed, the arrest of emergence of embryonic upon Rpl22l1 knockdown was rescued by the knockdown of Rpl22, indicating that the arrest was caused by Rpl22-mediated repression of HSC emergence in the absence of Rpl22l1(24). To determine if the arrest in T cell development in Rpl22l1cKO mice was also dependent upon Rpl22 function, we generated Rpl22l1cKO Rpl22−/− double deficient mice (Figure S3I and 7D). Interestingly, further elimination of Rpl22 in Rpl22l1cKO mice failed to rescue development, and instead produced an even more profound arrest in T cell development (Figure S3I and 7D). Importantly, this included γδ T cells (Figure 7E) whose development is spared by deletion of either Rpl22 or Rpl22l1 alone (Figure 3E, S3I and 7E)(18). Finally, the block in development of T cells in the double-deficient mice is associated with p53 induction and is likely to be p53-dependent (Figure S3J). Taken together, these findings suggest that Rpl22 and Rpl22l1 each play independent, essential, but not antagonistic, roles in facilitating the development of lymphocytes by repressing the induction of p53.
Discussion
Previous analysis of the role of Rpl22l1 in zebrafish hematopoiesis revealed that Rpl22l1 was critically important for the emergence of embryonic HSC from the AGM (24); however, this early block in hematopoiesis precluded analysis of more distal blood lineages. Here, we report our use of conditional ablation of the Rpl22l1 gene in mice to assess the role of Rpl22l1 in adult hematopoiesis. We found that Rpl22l1 is not essential for the maintenance of adult HSC, but it is critical for B cell development, as Rpl22l1-deficiency attenuates B cell development at the pre-B stage, at which immunoglobulin M heavy chain rearrangement occurs. Moreover, Rpl22l1-deficiency arrests the development of T cells at the equivalent stage, where TCRβ locus rearrangement occurs; however, the arrest was not caused by impaired V(D)J recombination. Instead, the arrest in B and T cell development is associated with p53 induction and is p53-dependent as development is rescued by p53-deficiency. Interestingly, Rpl22l1-loss does impair ribosome biogenesis or global protein synthesis, as has been observed upon loss of other ribosomal proteins (8, 30). Moreover, Rpl22l1-deficiency does not exacerbate endoplasmic reticulum (ER) stress, result in DNA-damage, or exacerbate IFN-responsiveness, three other well-known inducers of p53 (39–42). Consequently, the developmental arrest is not linked to alternations in any of the known inducers of p53.
Rpl22l1 is a highly homologous (i.e., 73% identical) paralog of Rpl22. Unlike yeast, where 75% of RP have paralogs, most RP in plants, flies, and vertebrates are encoded by a single functional gene (6, 16). Nevertheless, a handful of RP paralogs have been preserved in mammals, and such paralogs often exhibit tissue specificity in their expression. The paralog of Rpl3, Rpl3l, is selectively expressed in muscle tissue, while that of Rpl10, Rpl10l, is restricted to the testis (6, 15). The physiologic relevance of the tissue-restricted expression of RP paralogs remains largely unknown, but has been suggested to enable greater control of gene expression in higher eukaryotes and there is some evidence to support this (6, 16, 43). Interestingly, our previous analysis in zebrafish indicated that Rpl22l1 and Rpl22 not only performed distinct functions but were actually antagonistic to one another (22, 24). The elimination of Rpl22l1 expression prior to morphogenesis blocked gastrulation, while its elimination after the maternal to zygotic transcriptional switch blocked embryonic HSC emergence (22, 24). Importantly, both of those developmental defects were rescued by knockdown of Rpl22, indicating that the developmental anomalies in zebrafish embryos lacking Rpl22l1 were Rpl22-dependent. Further gain and loss of function analysis revealed that Rpl22 and Rpl22l1 functions were antagonistic and focused on regulation of a common target, smad1 mRNA translation in the case of HSC emergence, and smad2 pre-mRNA splicing in the case of gastrulation (22, 24). Regulation of these targets is mediated by direct binding to a consensus stem-loop motif (22, 24, 44). Our analysis using conditional ablation of Rpl22l1 in mice revealed that Rpl22l1 plays important roles in both B and T cell development at essentially the same stages were we previously determined that Rpl22 function was critical (18–20). The finding that the functions of both Rpl22 and Rpl22l1 were crucial in supporting these developmental transitions suggested that the functions or Rpl22 and Rpl22l1 at these stages were not antagonistic. We directly tested that possibility in T cell progenitors and found that Rpl22/Rpl22l1 double-deficiency did not rescue development as was previously seen in gastrulation and HSC emergence, and instead created a more profound block in T cell development. Indeed, development beyond the DN stage was completely blocked. Curiously, the loss Rpl22 in Rpl22l1cKO mice allowed traversal of the β checkpoint and arrest at the DN4 stage, but without proliferation, since the absolute number of DN4 cells was markedly reduced. Arrest at the DN4 stage is quite rare, with a similar arrest in Pdpk1−/− mice representing one of the few examples (45). Moreover, the development of γδ T lineage progenitors, which was spared in both Rpl22 and Rpl22l1 singly deficient mice, was blocked by Rpl22/Rpl22l1-double deficiency. Rpl22/Rpl22l1-double-deficient thymocytes also exhibited p53 induction. Consequently, Rpl22-Rpl22l1 antagonism appears to be context-dependent and is not evident at later stages of hematopoiesis where p53-dependent arrest occurs.
The arrest of B and T cell development in Rpl22l1-deficient mice is p53-mediated, since p53 is induced and its genetic ablation rescues both B and T cell development. p53 activation is frequently observed upon genetic ablation of RP-encoding genes. Loss of ribosomal proteins Rps19 and RPs14 in Diamond-Blackfan anemia and 5q-syndrome, respectively, led to the activation of the p53 pathway (5, 37, 46, 47). Likewise, conditional ablation of the Rps6 gene in T cell progenitors induces a p53-dependent block in T cell development (48). p53 induction in these contexts typically results from impaired ribosome biogenesis and general reductions in protein synthesis (8, 37, 48). Disruption of ribosome biogenesis activates p53 because RP, including Rpl5, Rpl11, Rpl23 and Rps7, interact with MDM2 and interfere with its ability to promotes p53 degradation, thereby resulting in p53 accumulation (49–51). Our analysis revealed that Rpl22l1-deficiency does not impair ribosome biogenesis or attenuate protein synthesis in developing progenitors in vivo. Therefore, Rpl22l1 loss does not induce p53 because of generalized effects on the ribosome. It remains possible that p53 induction results from a more subtle effect on the ribosome, but we think this is unlikely because the majority of Rpl22l1 protein appears to reside outside of the ribosome (32). p53 induction did not result from defects in V(D)J recombination, as evidenced by the unimpeded generation of TCRβ+ DN3 thymocytes in Rpl22l1-deficient mice. Moreover, p53 loss fails to produce the robust rescue of the development of pre-TCR deficient thymocytes, that we observed in p53/Rpl22l1-double deficient mice (52).
Because defects in ribosome biogenesis or global protein synthesis were not responsible for p53 induction, we sought to identify the cause. To do so, we performed RNA-Seq, which revealed that nearly 800 genes were differentially-regulated by Rpl22l1-deficiency and the changes were enriched for pathways related to DNA replication, DNA-damage responses, and cell division but this was largely due to the upregulation of p53-targets. Nevertheless, we examined whether DNA-damage might be causal to p53 induction using phopho-γH2AX staining; however, phopho-γH2AX staining was not increased in Rpl22l1-deficient thymic progenitors, suggesting that DNA-damage was not responsible for p53-induction. Our previous analysis of the basis for p53 induction in Rpl22-deficient thymocytes revealed that it was caused by exaggerated ER stress (21). Rpl22-deficiency activated all three ER stress pathways (PERK/ATF4, Ire1a/XBP-1, and ATF6); however, attenuation of the PERK pathway was sufficient to diminish p53 induction and rescue development of Rpl22-deficient thymocytes (21). Importantly, the phosphorylation of EIF2α, the primary target of PERK, was not increased by Rpl22l1-deficiency, suggesting that exaggerated ER stress was also not responsible. Finally, the genes that were differentially-expressed in Rpl22l1-deficient thymocytes clustered in several pathways related to IFN signaling. IFN signaling has been implicated as an activator of p53 (40, 53); however, we observed no excessive activation of IFN-signaling in Rpl22l1-deficient thymocytes. Consequently, p53 induction in Rpl22l1-deficient T cell progenitors appears to be unrelated to any of the known activators of p53 signaling.
Together, our data demonstrate that Rpl22l1 plays a critical role, that is distinct from that of its paralog Rpl22, in supporting the development of B and T lymphocytes. The role of Rpl22l1 in these progenitors is to suppress p53 induction as lymphocytes traverse their pre-receptor checkpoints. These transitions involve an abrupt change between quiescence and rapid proliferation, which produces numerous stresses. The role of Rpl22l1 in supporting these transitions remains unclear as its loss does not diminish global protein synthesis or exacerbate any of the known activators of p53 signaling. It also differs substantially from earlier developmental transitions (gastrulation and HSC emergence), which are governed by the antagonistic balance of Rpl22 and Rpl22l1, since Rpl22-loss did not rescue the arrests caused by Rpl22l1-deficiency. Because Rpl22l1 is primarily extraribosomal (32), its regulation of early lymphoid development likely involves its extaribosomal control of key mRNA targets. Future efforts will be directed at identification of those targets as they will provide new insights into the regulation of p53 as a developmental checkpoint.
Supplementary Material
Key Points:
Rpl22l1 plays a key role in regulating selected stages of hematopoietic development
The function of Rpl22l1 is essential and distinct from its paralog, Rpl22
Rpl22l1 regulates development without affecting ribosome biogenesis of function
Acknowledgements
We thank the following Fox Chase Cancer Center core facilities for their vital service: Bioinformatic and Biostatistics, Cell Culture, Genomics, Imaging, and Laboratory Animal/Zebrafish.
This work was supported by the National Institutes of Health (NIH) grants P30CA006927, P01AI102853, R37AI110985, and the Bishop Fund. SPF was supported by the institutional postdoctoral training grant T32 CA009035.
Footnotes
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Genuth NR, and Barna M. 2018. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nature reviews. Genetics 19: 431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raiser DM, Narla A, and Ebert BL. 2014. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leuk Lymphoma 55: 491–500. [DOI] [PubMed] [Google Scholar]
- 3.Mills EW, and Green R. 2017. Ribosomopathies: There’s strength in numbers. Science 358. [DOI] [PubMed] [Google Scholar]
- 4.Warner JR 2015. Twenty years of ribosome assembly and ribosomopathies. RNA. 21: 758–759. doi: 710.1261/rna.050435.050115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narla A, and Ebert BL. 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue S, and Barna M. 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 13: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, Sackstein P, Puel A, Picard C, Abel L, Quintana-Murci L, Faust SN, Williams AP, Baretto R, Duddridge M, Kini U, Pollard AJ, Gaud C, Frange P, Orbach D, Emile JF, Stephan JL, Sorensen R, Plebani A, Hammarstrom L, Conley ME, Selleri L, and Casanova JL. 2013. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 340: 976–978. doi: 910.1126/science.1234864. Epub 1232013 Apr 1234811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, Abdulhay NJ, Specht H, Keshishian H, Mani DR, Jovanovic M, Ellis SR, Fulco CP, Engreitz JM, Schutz S, Lian J, Gripp KW, Weinberg OK, Pinkus GS, Gehrke L, Regev A, Lander ES, Gazda HT, Lee WY, Panse VG, Carr SA, and Sankaran VG. 2018. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173: 90–103.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, and Barna M. 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, Fujii K, Kovary KM, Genuth NR, Rost HL, Teruel MN, and Barna M. 2017. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol Cell 67: 71–83.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner JR, and McIntosh KB. 2009. How common are extraribosomal functions of ribosomal proteins? Mol Cell 34: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X, Liao WJ, Liao JM, Liao P, and Lu H. 2015. Ribosomal proteins: functions beyond the ribosome. Journal of molecular cell biology 7: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komili S, Farny NG, Roth FP, and Silver PA. 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes AM, Miguel RN, Sargent CA, Ellis PJ, Amorim A, and Affara NA. 2010. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol 11: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong QW, Li J, Ng SR, Lim SG, Yang H, and Vardy LA. 2014. RPL39L is an example of a recently evolved ribosomal protein paralog that shows highly specific tissue expression patterns and is upregulated in ESCs and HCC tumors. RNA Biol 11: 33–41. doi: 10.4161/rna.27427. Epub 22013 Dec 27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerst JE 2018. Pimp My Ribosome: Ribosomal Protein Paralogs Specify Translational Control. Trends Genet. 34: 832–845. doi: 810.1016/j.tig.2018.1008.1004. Epub 2018 Sep 1015. [DOI] [PubMed] [Google Scholar]
- 17.Abbas AK, Williams ME, Burstein HJ, Chang TL, Bossu P, and Lichtman AH. 1991. Activation and functions of CD4+ T-cell subsets. Immunol Rev 123: 5–22. [DOI] [PubMed] [Google Scholar]
- 18.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, and Wiest DL. 2007. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26: 759–772. [DOI] [PubMed] [Google Scholar]
- 19.Fahl SP, Harris B, Coffey F, and Wiest DL. 2015. Rpl22 Loss Impairs the Development of B Lymphocytes by Activating a p53-Dependent Checkpoint. J Immunol 194: 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stadanlick JE, Zhang Z, Lee SY, Hemann M, Biery M, Carleton MO, Zambetti GP, Anderson SJ, Oravecz T, and Wiest DL. 2011. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J Immunol 187: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solanki NR, Stadanlick JE, Zhang Y, Duc AC, Lee SY, Lauritsen JP, Zhang Z, and Wiest DL. 2016. Rpl22 Loss Selectively Impairs alphabeta T Cell Development by Dysregulating Endoplasmic Reticulum Stress Signaling. J Immunol 197: 2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, O’Leary MN, Peri S, Wang M, Zha J, Melov S, Kappes DJ, Feng Q, Rhodes J, Amieux PS, Morris DR, Kennedy BK, and Wiest DL. 2017. Ribosomal Proteins Rpl22 and Rpl22l1 Control Morphogenesis by Regulating Pre-mRNA Splicing. Cell reports 18: 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, and Speck NA. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, and Wiest DL. 2013. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell 24: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, and Print CG. 2002. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis (New York, N.Y. : 2000) 34: 251–256. [DOI] [PubMed] [Google Scholar]
- 26.Luche H, Nageswara Rao T., Kumar S, Tasdogan A, Beckel F, Blum C, Martins VC, Rodewald HR, and Fehling HJ. 2013. In vivo fate mapping identifies pre-TCRalpha expression as an intra- and extrathymic, but not prethymic, marker of T lymphopoiesis. J Exp Med 210: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, and Yanagisawa M. 2001. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol 230: 230–242. [DOI] [PubMed] [Google Scholar]
- 28.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, and Weinberg RA. 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7. [DOI] [PubMed] [Google Scholar]
- 29.Yokomizo T, Yamada-Inagawa T, Yzaguirre AD, Chen MJ, Speck NA, and Dzierzak E. 2012. Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat Protoc 7: 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Signer RA, Magee JA, Salic A, and Morrison SJ. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiest DL, Bhandoola A, Punt J, Kreibich G, McKean D, and Singer A. 1997. Incomplete endoplasmic reticulum (ER) retention in immature thymocytes as revealed by surface expression of “ER-resident” molecular chaperones. Proceedings of the National Academy of Sciences of the United States of America 94: 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Leary MN, Schreiber KH, Zhang Y, Duc AC, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, Martin DB, Kaeberlein M, Ladiges WC, Fink PJ, Mackay VL, Wiest DL, and Kennedy BK. 2013. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet 9: e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, and Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Anikin L, and Pestov DG. 2014. Two orthogonal cleavages separate subunit RNAs in mouse ribosome biogenesis. Nucleic Acids Res 42: 11180–11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider RK, Schenone M, Ferreira MV, Kramann R, Joyce CE, Hartigan C, Beier F, Brummendorf TH, Germing U, Platzbecker U, Busche G, Knuchel R, Chen MC, Waters CS, Chen E, Chu LP, Novina CD, Lindsley RC, Carr SA, and Ebert BL. 2016. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med 22: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, and Zuniga-Pflucker JC. 2004. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol 5: 410–417. [DOI] [PubMed] [Google Scholar]
- 39.Porta C, Hadj-Slimane R, Nejmeddine M, Pampin M, Tovey MG, Espert L, Alvarez S, and Chelbi-Alix MK. 2005. Interferons alpha and gamma induce p53-dependent and p53-independent apoptosis, respectively. Oncogene. 24: 605–615. doi: 610.1038/sj.onc.1208204. [DOI] [PubMed] [Google Scholar]
- 40.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, and Taniguchi T. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 424: 516–523. doi: 510.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 41.Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S, and Fahraeus R. 2010. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol Cell 38: 78–88. [DOI] [PubMed] [Google Scholar]
- 42.Boutelle AM, and Attardi LD. 2021. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 31: 298–310. doi: 210.1016/j.tcb.2020.1012.1011. Epub 2021 Jan 1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segev N, and Gerst JE. 2018. Specialized ribosomes and specific ribosomal protein paralogs control translation of mitochondrial proteins. J Cell Biol. 217: 117–126. doi: 110.1083/jcb.201706059. Epub 201702017 Nov 201706058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobbelstein M, and Shenk T. 1995. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol 69: 8027–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hinton HJ, Alessi DR, and Cantrell DA. 2004. The serine kinase phosphoinositide-dependent kinase 1 (PDK1) regulates T cell development. Nat Immunol 5: 539–545. [DOI] [PubMed] [Google Scholar]
- 46.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, and Ebert BL. 2011. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117: 2567–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, and Zon LI. 2012. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol 40: 228–237 e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, and Volarevic S. 2005. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev 19: 3070–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhat KP, Itahana K, Jin A, and Zhang Y. 2004. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. Embo J 23: 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin A, Itahana K, O’Keefe K, and Zhang Y. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol 24: 7669–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deisenroth C, and Zhang Y. 2010. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 29: 4253–4260. [DOI] [PubMed] [Google Scholar]
- 52.Jiang D, Lenardo MJ, and Zuniga-Pflucker C. 1996. p53 prevents maturation to the CD4+CD8+ stage of thymocyte differentiation in the absence of T cell receptor rearrangement. Journal of Experimental Medicine 183: 1923–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muñoz-Fontela C, Macip S, Martínez-Sobrido L, Brown L, Ashour J, García-Sastre A, Lee SW, and Aaronson SA. 2008. Transcriptional role of p53 in interferon-mediated antiviral immunity. J Exp Med. 205: 1929–1938. doi: 1910.1084/jem.20080383. Epub 20082008 Jul 20080328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.