Abstract
We developed a rapid pulsed-field gel electrophoresis (PFGE) method that required 3 days to complete, an improvement over the standard method that required as many as 8 days. The accuracy and reproducibility of the rapid method were verified by analysis of DNA band sizes of our control group B streptococcus isolate. The rapid method was superior to the standard method, providing more precise molecular sizing and gels of higher image quality. The reproducibility of rapid PFGE substantiated its value and continued use.
Pulsed-field gel electrophoresis (PFGE), a powerful tool for resolving large DNA molecules (4, 6), is used for typing bacterial isolates (2), examination of serotype prevalence and evolutionary divergence (1, 7), and investigation of outbreaks (15, 16, 19). PFGE provides increased discrimination compared to PCR-based methods (20) and has been found to be preferable to restriction enzyme digestion with conventional electrophoresis (8, 12).
A major drawback to PFGE is the length of the procedure. The standard procedure requires up to 8 working days for completion (8). Therefore, the PFGE procedure (13, 17) used for other bacterial genera has been modified to reduce the time required for completion (5, 10, 11, 18). A rapid procedure for group B streptococcus (GBS) isolates would be advantageous for examining large numbers of isolates and responding quickly to nursery clusters of disease in infants. Our rapid method described here, specific for GBS, overcame the disadvantages of the standard PFGE method by reducing the time required from 6 to 8 days to 39 h (3 work days) while improving the results visually and quantitatively.
The 57 GBS isolates examined using the rapid PFGE method were from our laboratory's GBS culture collection. They represented nine serotypes: 8 Ia, 4 Ib, 5 II, 15 III, 12 V, 3 VII, 8 VIII, and 1 each of IV and VI. Isolates were stored at −30°C in Todd-Hewitt broth (Difco, Detroit, Mich.) with 5% defibrinated sheep blood. Serotyping was done by Ouchterlony assay with type-specific antisera generated in our laboratory as previously described (14).
The standard PFGE method was adapted from our previously published method (8) with minor modifications. To form plugs, a cell pellet from 2 ml of growth in Todd-Hewitt broth with yeast extract (Difco) was mixed with 750 μl of 1.0% low-melting-point agarose (ultraPURE; Gibco-BRL, Rockville, Md.). These were incubated initially for 4 to 6 h at 37°C, washed four times with 12 ml of TE buffer (10 mM Tris, 1 mM EDTA, pH 7.6), and electrophoresed for 29 h. This procedure required 89 h over 6 to 8 days with 16 h of hands-on work time.
We modified our adapted standard method implementing features described for the rapid method for Escherichia coli (10). The main modifications were growth on solid medium, treatment of plugs with higher enzyme concentrations, higher temperature for washing and wash solutions, and electrophoresis at a higher temperature. Specifically, the isolates were grown on 5% sheep blood agar plates for 48 h at 37°C. Bacterial cells were suspended in TE buffer (100 mM Tris, 100 mM EDTA, pH 7.6), and the suspension was incubated in 10 μl of lysozyme (10 mg/ml) (Sigma) for 15 min at 37°C. The cell suspension was mixed with 5 μl of proteinase K (20 mg/ml) (Gibco-BRL), 90 μl of mutanolysin (1 mg/ml) (Sigma), and 250 μl of tempered 1.5% low-melting-point agarose to form plugs. These were incubated in 1.5 ml of ES buffer (0.5 M EDTA [pH 9.0], 1% n-lauryl sarcosine) and 75 μl of proteinase K (20 mg/ml) (Gibco-BRL) for 90 min in a 55°C water bath with occasional shaking. The plugs were washed once at 50°C for 15 min with 50°C sterile water and three times with 50°C TE buffer (10 mM Tris and 1 mM EDTA, pH 7.6). These higher temperatures and enzyme concentrations permitted a savings of 45 h. Washed plug slices were digested for 3 h at room temperature (RT) with 50 U of SmaI (Gibco-BRL), 20 μl of React4 restriction enzyme buffer (Gibco-BRL), 2 μl of bovine serum albumin (1 mg/ml) (Sigma), and sterile type I water. Plug slices were placed on the well comb, and tempered agarose was poured in the gel mold. The gel was run at 6.0 V/cm with an initial switch time of 10 s to a final switch time of 45 s at 14°C in 0.5× TBE (Tris-borate-EDTA) running buffer for 22 h, a savings of 7 h. Molecular sizes were determined using the Kodak Digital Science 1D image analysis software. The time required to complete this procedure was 39 h over 3 days, of which 10.5 h were hands-on time.
To verify the reproducibility of each method, the internal control isolate 89-022, a wild-type Ib/α+β isolate from our laboratory, was run in each gel, as space permitted. Molecular sizes of the DNA bands from 89-022 were calculated by the 1D software, using the standard curve determined from the lambda size standard. Bands 2, 3, and 4 were analyzed because these were in the linear area of the standard curve. The standard deviations (SD) from the mean of these three bands were calculated using InStat statistical software (GraphPad Software version 2.03). The accuracy of the rapid PFGE method was examined within an individual gel by assaying the control isolate, 89-022, seven times in a single gel to ensure that lane location was not a factor.
The DNA profiles from the 57 GBS isolates were examined by a modification of the Tenover criteria for bacterial strain typing (22) because we were studying nonoutbreak isolates. Isolates were classified as genetically related if they differed by one to three DNA bands.
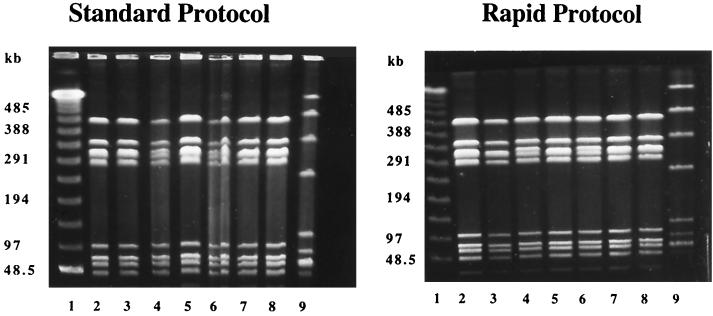
The results from the two methods were examined and compared using 18 rapid-method and seven standard-method gels. Examples of gels run by each method are shown in Fig. 1. In lanes 2 to 8 of both gels were type V isolates expressing the R1 and R4 proteins, demonstrating that DNA band patterns obtained by the two methods were nearly identical. Furthermore, the molecular sizes of the bands were approximately the same (data not shown). However, the rapid method produced DNA profiles that were visually superior, with finer resolution than the standard method. Band distortion, as seen in lane 6 in the standard-method gel, was not observed in the rapid-method gel. In addition, the number of distinct lambda DNA restriction bands available for standard curve determination was higher for the rapid (11 to 12 bands) than for the standard (9 to 10 bands) method.
FIG. 1.
PFGE gel from the standard method and the rapid method stained with ethidium bromide. The standard-method gel (left) was run for 29 h at 10°C, and the rapid-method gel (right) was run for 22 h at 14°C. Both gels were run at 6.0 V/cm and used switching times of 10 to 45 s. Lane 1, lambda ladder. Lanes 2 to 8, GBS type V/R1, R4 isolates. Lane 9, control GBS strain 89-022 (type Ib/α+β).
To analyze the reproducibility of the PFGE methods, we used molecular sizes of bands 2, 3, and 4 from 89-022 (lane 9, standard- and rapid-method gels). The SD of each of these bands was calculated from seven standard- and 14 rapid-method gels. The SD of the second, third, and fourth bands were 4.49 kb (1.7% of the mean band size), 5.91 kb (1.6%), and 7.57 kb (1.6%) respectively, for the rapid method and 10.12 kb (4.0%), 12.98 kb (3.6%), and 23.35 kb (5.0%), respectively, for the standard method. The accuracy of molecular sizes derived from the rapid method was supported by the results from the intragel analysis, which showed minimal variation from lane to lane. The SD for bands 2 to 4 were 2.37, 3.95, and 3.38 kb, respectively.
Using our rapid PFGE method, 57 GBS isolates were studied and their macro-restriction profiles were analyzed for relatedness. We found 21 restriction profiles among the 57 isolates of the designated serotypes (Table 1). However, there were only 18 distinct DNA profiles because each of three profiles of isolates of specific serotypes was similar to a profile of a different serotype. Isolates of serotypes III, V, and VIII demonstrated one to three DNA profiles, while the five isolates of serotype II had different DNA patterns. All serotype V isolates had a single restriction profile, and serotype VIII isolates also showed great homology, as seven of the eight were related to the prototype VIII profile.
TABLE 1.
Macrorestriction analysis of 57 GBS isolates grouped by serotype
| Type | No. of isolates examined | No. of restriction profiles within each serotypea |
|---|---|---|
| Ia | 8 | 4 |
| Ib | 4 | 2 |
| II | 5 | 5 |
| III | 15 | 3 |
| IV | 1 | 1 |
| V | 12 | 1 |
| VI | 1 | 1 |
| VII | 3 | 2 |
| VIII | 8 | 2 |
| Total | 57 | 21 |
Profiles within three band differences were counted as one profile.
Comparison of the results from the rapid and standard methods indicated that the gel image from the rapid method was superior. This likely resulted from the reduction in electrophoresis time, reduced manipulation of the plug slices, and method of gel loading. In contrast to the rapid method, the finished gel from the standard method sat for up to 14 h before it was stained. It was during this time that the DNA likely degraded and started to diffuse into the surrounding gel, leading to less distinct bands with less reproducible molecular sizes. Degeneration of the plug slices was rare in the rapid method due to the ease of the gel loading procedure. It is important to note that while the rapid method produced a higher-quality result, it was sensitive to biochemical alteration or activity of reagents and solutions. This was minimized by careful handling and use of the plugs soon after they were prepared and the use of fresh reagents.
An advantage to the rapid method was the ability to process a large number of isolates in less time than with the standard method. Using the rapid method, we found considerable genetic diversity within capsular polysaccharide types, just as we and others had described previously (3, 7, 8, 12, 21). However, serotype V isolates were an exception, since isolates of this serotype had nearly identical DNA restriction profiles. These profiles were closely related to other serotype V isolates from our laboratory (9) and from other study centers (3, 7).
The reproducibility of this rapid PFGE method was established by inter- and intra-assay statistical analysis of three bands from the control GBS isolate. Comparability of DNA restriction profiles by the rapid method and the standard method suggested that rapid-method profiles were valid. Increased reproducibility, higher image quality, and reduction of the time for completion by as many as 5 days supported replacement of the standard by the rapid PFGE method.
Acknowledgments
This work was supported in part by research contract NO1-AI-75326 from the National Institute for Allergy and Infectious Diseases, National Institutes of Health.
We acknowledge Aurea E. Flores for critical review of the manuscript.
REFERENCES
- 1.Arbeit R D, Arthur M, Dunn R, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg H M, Stephens D S, Modansy M, Erwin M, Elliot J, Facklam R R, Schuchat A, Baughman W, Farley M M. Invasive group B streptococcal disease: the emergence of serotype V. J Infect Dis. 1996;173:365–373. doi: 10.1093/infdis/173.2.365. [DOI] [PubMed] [Google Scholar]
- 4.Carle G F, Frank M, Olson M V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986;232:65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- 5.Chang N, Chui L. A standardized protocol for the rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. Diagn Microbiol Infect Dis. 1998;31:275–279. doi: 10.1016/s0732-8893(98)00007-8. [DOI] [PubMed] [Google Scholar]
- 6.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 7.Elliott J A, Farmer K D, Facklam R R. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J Clin Microbiol. 1998;36:2115–2116. doi: 10.1128/jcm.36.7.2115-2116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fasola E, Livdahl C, Ferrieri P. Molecular analysis of multiple isolates of the major serotypes of group B streptococci. J Clin Microbiol. 1993;31:2616–2620. doi: 10.1128/jcm.31.10.2616-2620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrieri P, Cho D S, Livdahl C, Rubens C E, Flores A E. DNA restriction profiles of nontypeable group B streptococcal clinical isolates. In: Horaud T, Bouvet A, Leclercq R, de Montclos H, Sicard M, editors. Streptococci and the host. Proceedings of the XIIIth Lancefield International Symposium on Streptococci and Streptococcal Diseases (Paris). Adv. Exp. Med. Biol. 418:343–346. 1997. [DOI] [PubMed] [Google Scholar]
- 10.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goering R V, Winters M A. Rapid method for epidemiological evaluation of gram-positive cocci by field inversion gel electrophoresis. J Clin Microbiol. 1992;30:577–580. doi: 10.1128/jcm.30.3.577-580.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordillo M E, Singh K V, Baker C J, Murray B E. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. J Clin Microbiol. 1993;31:1430–1434. doi: 10.1128/jcm.31.6.1430-1434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grothues D, Koopman U, von der Hardt H, Tümmler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26:1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson D R, Ferrieri P. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J Clin Microbiol. 1984;19:506–510. doi: 10.1128/jcm.19.4.506-510.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee R, Peppe J, George H. Pulsed-field gel electrophoresis of genomic digests demonstrates linkages among food, food handlers, and patrons in a food-borne Salmonella javiana outbreak in Massachusetts. J Clin Microbiol. 1998;36:284–285. doi: 10.1128/jcm.36.1.284-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonard R B, Mayer J, Sasser M, Woods M L, Mooney B R, Brinton B G, Newcomb-Gayman P L, Carroll K C. Comparison of MIDI Sherlock system and pulsed-field gel electrophoresis in characterizing strains of methicillin-resistant Staphylococcus aureus from a recent hospital outbreak. J Clin Microbiol. 1995;33:2723–2727. doi: 10.1128/jcm.33.10.2723-2727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 18.Matushek M G, Bonten M J M, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolland K, Marois C, Siquier V, Cattier B, Quentin R. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J Clin Microbiol. 1999;37:1892–1898. doi: 10.1128/jcm.37.6.1892-1898.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover F C, Arbeit R D, Goering R V, Mickelson P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]