Abstract
The oral bacteria Actinomyces naeslundii and Actinomyces viscosus are known to contribute to the initiation and progression of human dental caries, especially root caries. We report that both A. naeslundii and A. viscosus react with a component in the Gardenia jasminoides extract to produce a distinct green product. This green color reaction was found to be dependent on the bacterial β-glucosidase. The reaction is specific for cariogenic actinomyces, and it can detect as few as 104 cells of A. naeslundii and A. viscosus per ml.
Actinomyces naeslundii and Actinomyces viscosus are associated with dental caries (13, 25, 27). Either species alone induces periodontal diseases and dental caries in experimental animals (15). In particular, these two bacteria have been implicated as causes of human root surface caries (4, 6, 16, 24, 25). Although A. naeslundii and A. viscosus could be distinguished by their fibril types or catalase activities, genetic analysis of human isolates showed that these two species have similar DNAs (5). More importantly, these two actinomyces are almost identical in all other physiological traits (12, 14), and both of them are cariogenic.
Previous studies have suggested a possible association between oral A. naeslundii and A. viscosus and dental caries (4, 24, 25). Thus, simple and reliable methods for the enumeration of these cariogenic actinomyces will be useful tools for caries diagnosis and risk assessment. Selective medium (9, 29) and antibodies (10, 21, 26) have been used to detect cariogenic actinomyces. Although each of these techniques has its unique positive features, they have their limitations as well. For example, the selective media used for cariogenic actinomyces are only partially selective, and antibody-based methods are relatively complex. The study described here introduces a new, color-based enumeration method for the detection of cariogenic actinomyces.
In the process of a large-scale screening for bioactive compounds in medicinal herbs, we observed that the crude extract of Gardenia jasminoides was able to induce green color development in cultures of A. naeslundii. In this experiment, the G. jasminoides extract was prepared by a water extraction method followed by ethanol precipitation (19) and was stocked at a final concentration of 1 g of dried herb/ml of water. The experiment was performed by the following procedures. First, A. naeslundii strain ATCC 12104 was grown at 37°C under anaerobic conditions (10% H2, 10% CO2, 80% N2) in brain heart infusion (BHI) broth medium (Becton Dickinson, Sparks, Md.). Twenty microliters of each of the overnight cultures (optical density at 600 nm [OD600], 0.6 to 0.8) was inoculated into 2 ml of BHI broth medium containing various dilutions of the G. jasminoides extracts and was allowed to grow and develop a green color reaction at 37°C. The minimum concentration of G. jasminoides extract required for the green color reaction was 0.5 mg of dried herb/ml. Most experiments described below were performed with 10 mg of dried herb/ml. The green color product has a maximum absorption at 605 nm. The green color first appeared at 10 h (OD605, ≈0.1), and it could easily be recognized by visual examination within 24 h (OD605, >0.15).
To test whether this green color reaction was specific for A. naeslundii, we used the same experimental conditions to test a group of other oral bacteria, including Actinobacillus actinomycetemcomitans ATCC 33384, Fusobacterium nucleatum ATCC 25586, Porphyromonas gingivalis ATCC 33277, Streptococcus mutans ATCC 25175, Streptococcus sanguis ATCC 10556, and Streptococcus sobrinus ATCC 6715, as well as actinomyces, including Actinomyces bovis ATCC 13683, Actinomyces denticolens ATCC 43322, Actinomyces gerencseriae ATCC 23860, Actinomyces israelii ATCC 12102, Actinomyces meyeri ATCC 35568, Actinomyces odontolyticus ATCC 17929, and Actinomyces viscosus ATCC 19246. The results showed that the green color reaction was specific for cariogenic A. naeslundii and A. viscosus strains among the bacterial strains tested.
To test the sensitivity of this green color reaction, A. naeslundii at initial cell concentrations of 108, 107, 106, 105, 104, and 103 cells/ml was inoculated into BHI medium containing 10 mg of dried herb/ml and examined for a green color reaction at different time points (Table 1). As shown in Table 1, the minimum cell density required for A. naeslundii to produce a distinct green color within 48 h is 104 cells/ml (Table 1). Similar results were observed when A. viscosus was tested (data not show). It is of interest that previous studies suggested that the cell density of A. naeslundii and A. viscosus in saliva is between 105 and 107 cells/ml (4, 22), which falls within the detectable range of the green color test.
TABLE 1.
Cell density of A. naeslundii and the color reaction
| Initial cell density (cells/ml) | Color development at the following incubation times (h)a:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 2 | 6 | 10 | 24 | 48 | |
| 108 | − | + | ++ | ++ | ++ | ++ |
| 107 | − | − | + | ++ | ++ | ++ |
| 106 | − | − | − | + | ++ | ++ |
| 105 | − | − | − | − | + | ++ |
| 104 | − | − | − | − | − | ++ |
| 103 | − | − | − | − | − | − |
The results of the green color development were defined by −, +, and ++, in which − indicates an OD605 <0.05, + indicates a faint green color (OD605, >0.05), and ++ indicates that the color could clearly be detected by visual examination (OD605, >0.15).
To characterize the active component in A. naeslundii for the color reaction, 108 cells in 1 ml of BHI medium was treated by boiling, sonication, or boiling plus sonication. The G. jasminoides extract was then added to the treated cultures. The mixtures, including a positive control consisting of untreated cells, were incubated at 37°C. Six hours later, a distinct green color was observed in the positive control, as well as in the sonicated mixture. However, no green color was observed for the heat-treated mixtures even after 48 h. This result implies that living cells are not required to perform the green color reaction and that the active component produced by A. naeslundii is heat labile. Since most of the proteins are insensitive to sonication but are easily denatured by heating, it is possible that the active component in A. naeslundii is a heat-labile protein.
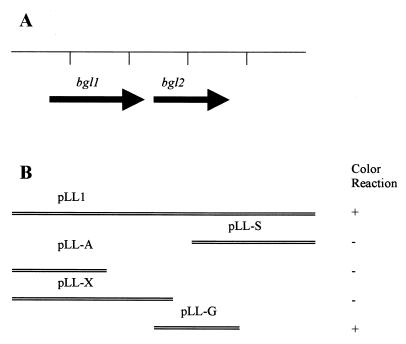
Escherichia coli strain TOP10 (Invitrogen Corp., Carlsbad, Calif.) was negative for the green color test. Thus, we reasoned that this strain could be used as a host to clone the A. naeslundii gene encoding the active protein for the green color reaction. Genomic DNA of A. naeslundii ATCC 12104 was purified with the Easy-DNA kit (Invitrogen) and was then subjected to BamHI digestion. The digested DNA fragments were inserted into the BamHI cloning site in the pZErO-2 vector (Invitrogen) and were then transformed into TOP10 cells. Transformants grown on plates with BHI medium, kanamycin (100 μg/ml), and G. jasminoides extract were screened for green color development. Three green E. coli colonies were identified. Spectrum analysis of the green product produced by these recombinant E. coli colonies showed that they had profiles identical to that produced by A. naeslundii. The plasmid DNAs were isolated from these colonies, and the DNAs from all three of them contained an identical A. naeslundii genomic fragment. One of these plasmids, named pLL1 (Fig. 1), was chosen for further study.
FIG. 1.
Map of the cloned A. naeslundii insert in pLL1. (A) Identified ORFs. (B) Subclones and their ability to produce the green color reaction in E. coli.
The 5-kb BamHI fragment of A. naeslundii genomic DNA in pLL1 was sequenced at the University of California, Los Angeles. Two complete open reading frames (ORFs) were identified. BLAST sequence analyses (1) show that both ORFs share homology with genes involved with the use of β-glucosides. Therefore, these two ORFs were designated bgl1 and bgl2, respectively, in the present study (Fig. 1A). The predicted protein product of bgl1 has 33% identity to the Clostridium longisporum phosphotransferase system enzyme II (PTS II), a protein involved in the transport of β-glucosidic sugars into the cell with concomitant phosphorylation of the sugar (8). Downstream of bgl1 is bgl2, which encodes a predicted protein of 488 amino acids with a calculated molecular mass of 54 kDa (Fig. 2). The bgl2 open reading frame has 63% identity to the phospho-β-glucosidase of C. longisporum (8) and 58% identity to the phospho-β-glucosidase of Bacillus subtilis (18) (Fig. 2).
FIG. 2.
Amino acid sequence of A. naeslundii Bgl2 (A) and its alignment with β-glucosidase proteins from B. subtilis (B) and C. longisporum (C). Matched amino acid residues in these proteins are highlighted in boldface type.
Subclones of pLL1 were constructed and subjected to the functional assay to determine which gene was required for the green color reaction. TOP10 cells harboring pLL-X, pLL-S, and pLL-A (Fig. 1B) did not produce the green color, whereas TOP10/pLL-G cells (Fig. 1B) did produce the green color. These results indicate that bgl2, but not bgl1, is important for the green color reaction.
To verify the predicted β-glucosidase activity of the Bgl2 protein, a group of known glucosides (Table 2) was used to test the bgl2 gene product for glucosidase activity. Cell lysates made from TOP10/pLL-G served as the source of Bgl2, and TOP10/pLL-X cell lysates were used as a negative control. Briefly, 108 cells grown in BHI medium with kanamycin (100 μg/ml) were sonicated and then challenged with various glucosides, each at a final concentration of 4 mM, at 37°C for 30 min. The overall catalytic activity of the mixture against a specific substrate was determined by the released nitrophenol with a maximum absorption at 410 nm (20). The OD410 obtained from TOP10/pLL-G was then compared with that obtained from TOP10/pLL-X to determine the activity exhibited by the Bgl2 protein. We found that β-d-glucosides could be efficiently hydrolyzed by the cellular lysates containing the Bgl2 protein, whereas no effect was observed for the lysates containing the BglI protein (Table 2). No significant catalytic activity was observed when β-d-fucoside, β-d-galactoside, or β-d-xyloside was used to challenge the Bgl2 protein (Table 2). Similar results were observed when A. naeslundii was assayed (data not show). On the basis of these findings, it is likely that Bgl2 is a β-glucosidase and that the green color reaction is dependent upon a functional β-glucosidase.
TABLE 2.
Enzymatic activity of Bgl2
| Compound | A410a
|
|
|---|---|---|
| TOP10/pLL-X | TOP10/pLL-G | |
| p-Nitrophenyl-β-d-glucopyranoside | 0.0575 | 0.7817 |
| o-Nitrophenyl-β-d-glucopyranoside | 0.0272 | 0.4256 |
| p-Nitrophenyl-β-d-galactopyranoside | 0.0514 | 0.1094 |
| o-Nitrophenyl-β-d-galactopyranoside | 0.0532 | 0.0905 |
| p-Nitrophenyl-β-d-fucopyranoside | 0.0321 | 0.0665 |
| p-Nitrophenyl-β-d-xylopyranoside | 0.0250 | 0.0746 |
The enzymatic activity was determined by measuring the released free nitrophenol in the reaction mixture at 410 nm. Four millimolar of the tested glucoside in BHI medium was used as a blank for each assay. Similar results were observed when the experiment was repeated three times.
β-Glucosidase has been found in a variety of organisms and has significant implications in medical and environmental applications (2, 3, 7, 11, 17, 20, 23). In the present study, we cloned a β-glucosidase from cariogenic A. naeslundii and demonstrated that its activity is critical for the generation of a distinct green color product from the G. jasminoides extract. G. jasminoides is a popular medicinal plant with cholagogic and hemostatic functions (28). Further understanding of the green color reaction could be approached by knockout of the bgl2 gene in A. naeslundii and chemical characterization of the chromogenic compound in G. jasminoides.
One unique feature provided by the method for detection of cariogenic actinomyces reported here is that the cell density was determined by the total enzymatic activity instead of by enumeration of CFU. It thus gets around a common concern when selective media are used to enumerate clustered bacteria, like A. naeslundii and A. viscosus. The distinct green color detectable by visual examination also provides a simple and easy way to positively identify the cariogenic actinomyces. The diagnostic implication of this assay needs to be further explored.
Nucleotide sequence accession number.
The 5-kb BamHI fragment of A. naeslundii genomic DNA in pLL1 was registered with GenBank and given accession number AY029505.
Acknowledgments
This study was supported in part by Public Health Service grant DE07296 from the National Institute of Dental Research to Li Chen.
We express appreciation and gratitude to Sharon Hunt-Gerardo, Renate Lux, and Michael Kempf for suggestions and comments on the manuscript. We also thank Fang Gu, Hong Sun, Xiaoyuan Ma, Casey Chen, and Larry Wolinsky for helpful discussions.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E A, Lamed R. The cellulose paradox: pollutant par excellence and/or a reclaimable natural resource? Biodegradation. 1992;3:171–188. doi: 10.1007/BF00129082. [DOI] [PubMed] [Google Scholar]
- 3.Bender I B, Bender A L. Dental observations in Gaucher's disease: review of the literature and two case reports with 13- and 60-year follow-ups. Oral Surg Oral Med Oral Pathol Oral Radiol Endodont. 1996;82:650–659. doi: 10.1016/s1079-2104(96)80440-9. [DOI] [PubMed] [Google Scholar]
- 4.Bowden G. Does assessment of microbial composition of plaque/saliva allow for diagnosis of disease activity of individuals? Community Dent Oral Epidemiol. 1997;25:76–81. doi: 10.1111/j.1600-0528.1997.tb00902.x. [DOI] [PubMed] [Google Scholar]
- 5.Bowden G, Johnson J, Schachatele C. Characterization of actinomyces with genomic DNA fingerprints and rRNA gene probes. J Dent Res. 1993;72:1171–1179. doi: 10.1177/00220345930720080201. [DOI] [PubMed] [Google Scholar]
- 6.Bowden G, Nolette N, Ryding H, Cleghorn B M. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. doi: 10.1177/00220345990780120601. [DOI] [PubMed] [Google Scholar]
- 7.Brady R O. Gaucher's disease: past, present and future. Baillieres Clin Haematol. 1997;10:621–634. doi: 10.1016/s0950-3536(97)80031-5. [DOI] [PubMed] [Google Scholar]
- 8.Brown G D, Thomson J A. Isolation and characterisation of an aryl-β-d-glucoside uptake and utilisation system (abg) from the gram-positive ruminal Clostridium species C. longisporum. Mol Gen Genet. 1998;257:213–218. doi: 10.1007/s004380050641. [DOI] [PubMed] [Google Scholar]
- 9.Bruilsford S R, Lynch E, Beighton D. The isolation of Actinomyces naeslundii from sound root surface and root caries lesions. Caries Res. 1998;32:100–106. doi: 10.1159/000016438. [DOI] [PubMed] [Google Scholar]
- 10.Cisar J O, Barsumain E L, Carl S H, Vatter A E, Sandberg A L, Siraganian R P. Detection and characterization of a lectin on Actinomyces viscosus T14V by monoclonal antibodies. J Immunol. 1981;127:1318–1322. [PubMed] [Google Scholar]
- 11.Esen A. β-Glucosidase: biochemistry and molecular biology. Coordinating ed., A. Esen. American Chemical Society Symposium Series 533. Washington, D.C.: American Chemical Society; 1992. [Google Scholar]
- 12.Fillery E D, Bowden G H, Hardie J M. A comparison of strains of bacteria designated Actinomyces viscosus and Actinomyces naeslundii. Caries Res. 1978;12:299–312. doi: 10.1159/000260349. [DOI] [PubMed] [Google Scholar]
- 13.Frank R M, Guillo B L H. Caries dentaires chez le rat gnotobiote inocule avec A. viscosus et A. naeslundii. Arch Oral Biol. 1972;17:1249–1253. doi: 10.1016/0003-9969(72)90157-4. [DOI] [PubMed] [Google Scholar]
- 14.Gerencser M A, Slack J M. Identification of human strains of Actinomyces viscosus. Appl Microbiol. 1969;18:80–87. doi: 10.1128/am.18.1.80-87.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan H V, Keyes P H. Aerobic gram positive filamentous bacteria as etiologic agents of experimental periodontal disease in hamsters. Arch Oral Biol. 1964;9:401–414. doi: 10.1016/0003-9969(64)90025-1. [DOI] [PubMed] [Google Scholar]
- 16.Jordan H V, Hammond B F. Filamentous bacteria isolated from human root surface caries. Arch Oral Biol. 1972;17:1337–1342. doi: 10.1016/0003-9969(72)90166-5. [DOI] [PubMed] [Google Scholar]
- 17.Landick R, Turnbrough L C, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella, cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. p. 1278. [Google Scholar]
- 18.Le Coq D, Lindner C, Kruger S, Steinmetz M, Stulke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long Z X. The Chinese materia medica. Beijing, China: Academy Press (Xue Yuan); 1998. [Google Scholar]
- 20.Lucas R, Robles A, Cienfuegos G A, Galvez A. β-Glucosidase from Chalara paradoxa CH32: purification and properties. J Agric Food Chem. 2000;48:3698–3703. doi: 10.1021/jf0002591. [DOI] [PubMed] [Google Scholar]
- 21.Nesbitt W E, Beem K P, Stroup S, Swift R, McArther W P, Clark W B. Inhibition of adherence of Actinomyces naeslundii (Actinomyces viscosus) T14V-J1 to saliva-treated hydroxyapatite by a monoclonal antibody to type I fimbriae. Oral Microbiol Immunol. 1996;11:51–58. doi: 10.1111/j.1399-302x.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 22.Schaeken M J, Creugers T J, Van der Hoeven J S. Relationship between dental plaque indices and bacteria in dental plaque and those in saliva. J Dent Res. 1987;66:1499–1502. doi: 10.1177/00220345870660091701. [DOI] [PubMed] [Google Scholar]
- 23.Schnetz K, Toloczyki C, Rak B. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schupbach P, Osterwalder V, Guggenheim B. Human root caries: microbiota in plaque covering sound, caries and arrested carious root surfaces. Caries Res. 1995;29:382–395. doi: 10.1159/000262097. [DOI] [PubMed] [Google Scholar]
- 25.Shu M, Wong L, Miller J H, Sissons C H. Development of multi-species consortia biofilms of oral bacteria as an enamel and root caries model system. Arch Oral Biol. 2000;45:27–40. doi: 10.1016/s0003-9969(99)00111-9. [DOI] [PubMed] [Google Scholar]
- 26.Thurnheer T, Guggenheim B, Gumr R. Characterization of monoclonal antibodies for rapid identification of Actinomyces naeslundii in clinical samples. FEMS Microbiol Lett. 1997;150:255–262. doi: 10.1111/j.1574-6968.1997.tb10378.x. [DOI] [PubMed] [Google Scholar]
- 27.Van der Hoeven J S, Mikx F H M, Konig K G, Plasschaert A J M. Plaque formation and caries in gnotobiotic and SPF rats with A. viscosus. Caries Res. 1974;8:211–223. doi: 10.1159/000260110. [DOI] [PubMed] [Google Scholar]
- 28.Yao Q, Zhao G, Zhu Y, Pan Y, Hu J, Zhang Q. Screening studies on anti-inflammatory function of traditional Chinese herb Gardenia jasminoides Ellis and its possibility in treating soft tissue injuries in animals. Chung Kuo Chung Yao Tsa Chih. 1991;16:489–513. [PubMed] [Google Scholar]
- 29.Zylber L J, Jordan H V. Development of a selective medium for detection and enumeration of Actinomyces viscosus and Actinomyces naeslundii in dental plaque. J Clin Microbiol. 1982;15:253–259. doi: 10.1128/jcm.15.2.253-259.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]