Abstract
Kinesin is a molecular motor that moves along microtubules. Testis-enriched kinesin KIF9 (Kinesin family member 9) is localized in the mouse sperm flagellum and is important for normal sperm motility and male fertility; however, it is unclear if the motor domain of KIF9 is involved in these processes. In this study, we substituted threonine of the ATP binding motif in the KIF9 motor domain to asparagine (T100N) in mice using the CRISPR/Cas9 system, which is known to impair kinesin motor activity. T100N mutant mice exhibit reduced sperm motility and male fertility consistent with Kif9 knockout mice. Further, KIF9 was depleted in the spermatozoa of T100N mutant mice although the amounts of KIF9 were comparable between wild-type and T100N mutant testes. These results indicate that the motor domain of KIF9 is essential for its localization in the sperm flagellum.
Keywords: axoneme, genome-editing, kinesin, male fertility, sperm motility
Introduction
Fertilization is the union of two gametes, spermatozoa and eggs. To fertilize eggs, spermatozoa need to travel a long distance in the female reproductive tract and penetrate the zona pellucida, an extracellular matrix that surrounds the eggs. Sperm motility plays critical roles in these processes and impaired sperm motility could lead to male infertility, a condition called asthenozoospermia [1]. The motility apparatus of spermatozoa is the axoneme, which is the ‘9+2’ microtubule arrangement that consists of a central pair (CP) of two singlet microtubules surrounded by nine outer microtubule doublets [2, 3]. Outer and inner dynein arms are attached to the outer microtubule doublets and slide neighboring doublets to induce axoneme bending. In addition, the axoneme contains the radial spokes that extend from the outer microtubule doublets toward the CP and the nexin-dynein regulatory complex that links the outer microtubule doublet to the adjacent doublet.
Kinesin is a molecular motor that moves along microtubules. Forty‐five kinesins have been identified in humans, which compose the kinesin superfamily of proteins (KIFs) [4]. KIF9 is evolutionarily conserved and is localized in the flagellum [5]. In unicellular flagellates, Chlamydomonas, KLP1 (Chlamydomonas ortholog of KIF9) is localized in the CP of the axoneme and regulates flagellar motility [6, 7]. In mice, KIF9 is testis-enriched and deletion of Kif9 results in male subfertility due to impaired sperm flagellar motility [5]. Although KIF9 function in regulating flagellar motility seems to be conserved evolutionarily, it is still unclear if the motor activity of KIF9 is involved in the regulation of motility.
Previously, it has been time-consuming and costly to introduce amino acid substitutions using homologous recombination in embryonic stem cells and generating chimeric mice; however, the emergence of genome editing technology makes it possible to substitute amino acids in a short period of time with relatively low costs [8,9,10]. In this study, we substituted the critical amino acid of the KIF9 motor domain in mice using the CRISPR/Cas9 system.
Materials and Methods
Animals
All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University (#Biken-AP-H30-01). Mice were purchased from CLEA Japan (Tokyo, Japan) or Japan SLC (Shizuoka, Japan).
Egg collection for genome editing
CARD HyperOva (0.1 ml, Kyudo, Saga, Japan) was injected into B6D2F1 female mice, followed by human chorionic gonadotropin (hCG) (five units, ASKA Pharmaceutical, Tokyo, Japan) injection 48 h later. Fertilized eggs were collected from the oviduct of superovulated females that were mated with B6D2F1 males.
Amino acid substitution of KIF9 in mice
Electroporation was performed as described previously [10, 11]. Oligonucleotide and crRNA/tracrRNA/Cas9 ribonucleoproteins (tracrRNA: #TRACRRNA05N-5NMOL, Sigma-Aldrich, MO, USA , and CAS9 protein: #A36497, Thermo Fisher Scientific, MA, USA) were electroporated into the fertilized eggs using a super electroporator NEPA21 (NEPA GENE, Chiba, Japan). The oligonucleotide and gRNA sequences were (5’-) CATATTTTGAAATGTTTGCAGGTACCATCATGTGTTACGGGCAGACAGGAGCTGGAAAAAACTACACCATGACAGGGGCAACGGAGAATTACAAGCACCGCGGAATTCTCCC (−3’) (oligonucleotide) and (5’-) TGTCATGGTGTATGTCTTGC (−3’) (gRNA). The eggs were cultivated in potassium simplex optimization medium (KSOM) [12] and the two-cell-embryos were transferred into pseudopregnant ICR females the next day. Genotyping was performed with PCR and subsequent AluI digestion or Sanger sequencing. The primer sequences used for genotyping were (5’-) GCATGTTGGGGATATGAGG (−3’) (Fw) and (5’-) CAGTTCACTTGAGGGCTGG (−3’) (Rv). Kif9T100N/WT mice (B6D2-Kif9em3 (T100N) Osb) are being processed for deposition to the RIKEN BioResource Research Center (#RBRC11496), Japan and Center for Animal Resources and Development (CARD) (#3140), Kumamoto University, Japan.
Fertility of male mice
Sexually mature wild-type (WT) or knockout (KO) male mice were caged individually with three six-week-old female mice for two months. The numbers of pups and copulation plugs were counted every morning.
Analysis of sperm motility and morphology
Spermatozoa from the cauda epididymis were suspended in a 100 µL drop of TYH medium [13] and incubated at 37°C under 5% CO2. For observing sperm morphology, spermatozoa were placed on MAS coated glass slides (Matsunami Glass, Osaka, Japan) and observed using an Olympus BX53 microscope (Tokyo, Japan). For analyzing sperm motility, spermatozoa were collected from the top of the drop after 10 or 120 min incubation and were analyzed using the CEROS II sperm analysis system (software version 1.5; Hamilton Thorne Biosciences, Beverly, MA, USA). For analyzing sperm flagellar waveforms, spermatozoa were observed with an Olympus BX-53 microscope equipped with a high-speed camera (HAS-L1, Ditect, Tokyo, Japan) [5, 14]. The motility was videotaped at 200 frames per second or 50 frames per second. Waveforms were traced using the sperm motion analyzing software (BohBohsoft, Tokyo, Japan) [15].
Immunoblot analysis
Testis and spermatozoa were homogenized in a lysis buffer containing 6 M urea, 2 M thiourea, and 2% sodium deoxycholate, and then centrifuged at 15,000 g for 15 min to collect supernatant samples. Samples were subjected to SDS‐PAGE followed by western blotting. Blots were blocked with 10% skim milk, incubated with primary antibodies overnight at 4°C, and incubated with secondary antibodies conjugated to horseradish peroxidase (1:10,000, #805‐035‐180, #115‐036‐062, or #111-036-045, Jackson ImmunoResearch, West Grove, PA, USA) for 120 min at room temperature. Primary antibodies used: goat anti‐KIF9 1:100 (#SC99958, Santa Cruz Biotechnology, Dallas, TX, USA); mouse anti‐acetylated tubulin 1:1,000 (#T7451, Sigma‐Aldrich); rabbit anti‐DNAH8 1:500 (#ab121989, Abcam, Cambridge, UK); rabbit anti‐DNAH17 1:500 (#HPA024354, Atlas Antibodies, Bromma, Sweden); rabbit anti‐DNAI2 1:1,000 (#17533-1-AP, Proteintech, Rosemont, IL, USA); rabbit anti‐DNALI1 1:1,000 (#17601-1-AP, Proteintech); rabbit anti-GAS8 1:500 (#HPA041311, Atlas Antibodies); rabbit anti‐RSPH9 1:500 (#HPA031703, Atlas Antibodies). Immunoreactive proteins were detected by an ECL western blotting detection kit (GE Healthcare, Little Chalfont, UK).
Immunofluorescence analysis
Immunofluorescence was conducted as described previously [11, 16]. Germ cells including spermatids were squeezed out from the seminiferous tubules onto glass slides and air-dried at 37°C. The samples were fixed with 4% paraformaldehyde in PBS for 15 min. After washing with PBS three times for 5 min each, the samples were blocked with 5% BSA and 10% goat serum in PBS for 1 h at room temperature. The slides were incubated with rabbit anti‐KIF9 antibody (1:50, #HPA022033, Atlas Antibodies) and mouse anti-acetylated tubulin antibody (1:500, #T7451, Sigma‐Aldrich) overnight at 4°C and washed with PBS three times for 10 min each. After incubation with Alexa Fluor 488 and Alexa Fluor 546-conjugated secondary antibody (1:200, #A11070 and #A11018, Thermo Fisher Scientific) at room temperature for 90 min, the slides were washed with PBS three times for 10 min each. The slides were then incubated with Hoechst 33342 (1:5,000) (#H3570, Thermo Fisher Scientific) for 15 min, washed with PBS three times for 10 min each, and observed with an Olympus BX53 microscope.
Statistical analysis
Two-tailed Student’s t test was used for statistical analyses. Differences were considered significant at P<0.05 (*) or highly significant at P<0.01 (**) and P<0.001 (***). Error bars are standard deviation. At least 3 mice of each genotype were used for each experiment.
Results
Generation of Kif9 T100N mutant mice
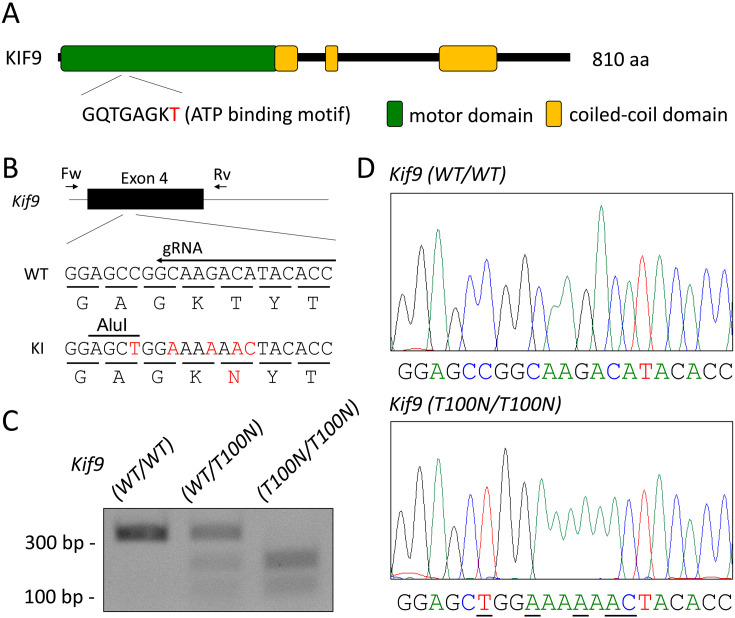
KIF9 possesses a motor domain at the N-terminus, which contains an ATP binding motif (Fig. 1A). To analyze the function of the motor domain, we substituted threonine of the ATP binding motif to asparagine in mice using the CRISPR/Cas9 system. This mutation generates rigor kinesin that can bind to microtubules but cannot move [17, 18]. We electroporated a crRNA/tracrRNA/Cas9 ribonucleoprotein and a single-stranded oligonucleotide containing the T100N mutation into fertilized eggs. Several silent mutations were introduced to prevent the mutated region from being recut by the ribonucleoprotein and to generate the sequence recognized by the restriction enzyme AluI (Fig. 1B). Of the 90 eggs that were electroporated, 86 two-cell embryos were transplanted into the oviducts of three pseudopregnant female mice. Thirty pups were born and 6 pups had the T100N mutation. By subsequent mating, we obtained mice with the T100N mutation, which was confirmed by PCR and AluI digestion (Fig. 1C) as well as Sangar sequencing analysis (Fig. 1D). No overt abnormalities were found in Kif9T100N/T100N mice consistent with Kif9 KO mice [5].
Fig. 1.
Generation of Kif9 T100N mutant mice. (A) The functional domain of KIF9. There is an ATP binding motif in the motor domain. (B) CRISPR/Cas9 targeting scheme. The gRNA was designed to target Exon 4. Primers (Fw, Rv) used for genotyping are shown. Mutated nucleotides and the substituted amino acid are shown in red. (C) Genotyping of Kif9T100N/T100N mice with PCR and subsequent AluI digestion. (D) Wave pattern sequence of Kif9T100N/T100N mice. Mutated nucleotides are underlined.
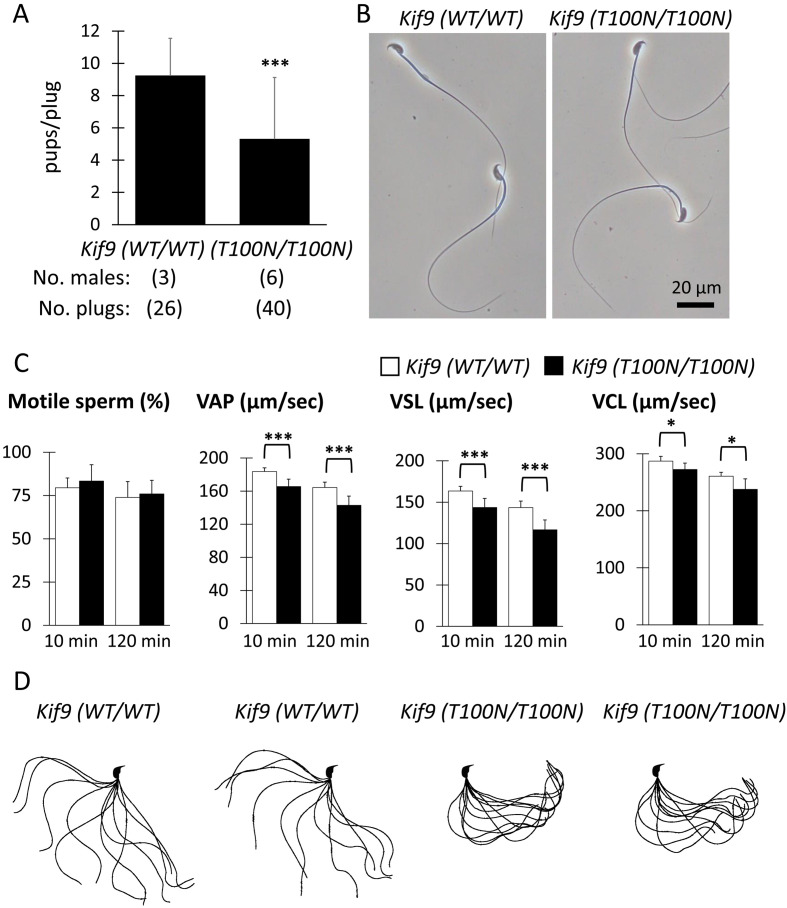
Kif9T100N/T100N mice exhibit impaired male fertility and sperm motility
To examine male fertility, Kif9WT/WT or Kif9T100N/T100N male mice were mated with WT females for two months. The average number of pups obtained per plug of Kif9T100N/T100N males was significantly lower than that of Kif9WT/WT males (Fig. 2A). To examine the cause of impaired male fertility, we observed the morphology of spermatozoa obtained from the cauda epididymis, but no abnormalities were found in Kif9T100N/T100N mice (Fig. 2B). We then analyzed sperm motility using a computer assisted sperm analysis system. Although no differences were observed in the percentage of motile spermatozoa, there were significant differences in the velocity parameters such as average path velocity (VAP), straight-line velocity (VSL), and curvilinear velocity (VCL) both 10 and 120 min after incubating spermatozoa in a capacitating medium (Fig. 2C). Further analysis of sperm flagellar motility using a high-speed camera showed that the majority of spermatozoa from Kif9T100N/T100N mice bent only to the side of the hook (pro-hook stall) (Fig. 2D) (the number of pro-hook stall=65, the number of anti-hook stall=0, the number of spermatozoa without stall=8 out of 73 spermatozoa examined, number of males=3), which caused the circular motion of spermatozoa in Kif9T100N/T100N mice (Supplementary Videos 1 and 2). The spermatozoa with pro-hook stall were also observed in Kif9 KO mice [5]. These results indicate that sperm motility and male fertility are impaired in Kif9T100N/T100N mice consistent with Kif9 KO mice.
Fig. 2.
Kif9T100N/T100N mice exhibit impaired male fertility and sperm motility. (A) Number of pups born per plug detected. (B) Observation of spermatozoa obtained from the cauda epididymis. No overt abnormalities were found in Kif9T100N/T100N mice. (C) Percentage of motile spermatozoa, VAP (average path velocity), VSL (straight-line velocity), and VCL (curvilinear velocity) were analyzed. n=5 males each for Kif9WT/WT and Kif9T100N/T100N mice. (D) Flagellar waveforms were analyzed 120 min after incubation. The motility was videotaped at 200 frames per second. Single frames throughout one beating cycle were superimposed.
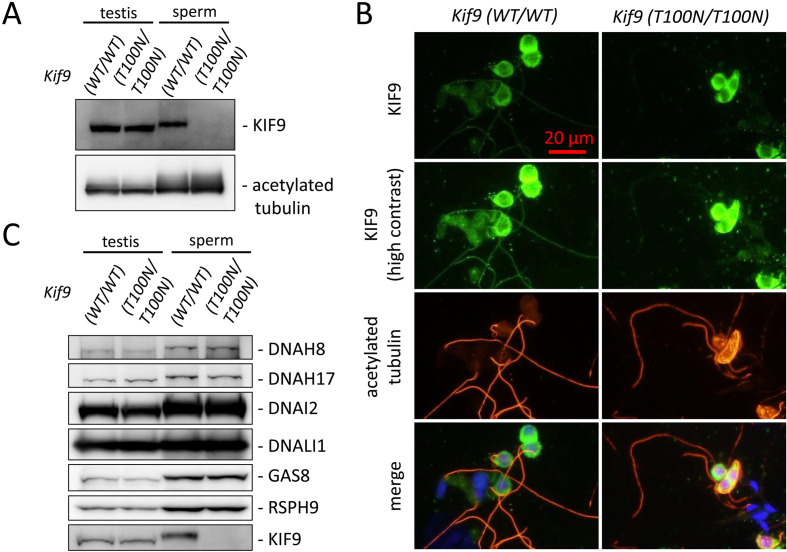
KIF9 with the T100N mutation cannot be localized in the sperm flagellum
Because the amino acid substitution could lead to decreased amounts of targeted protein [19], we examined the amount KIF9 with immunoblotting. In the testis, no clear differences were observed in the amount of KIF9, indicating that a comparable amount of KIF9 was produced in Kif9WT/WT and Kif9T100N/T100N testis (Fig. 3A). In contrast, KIF9 was hardly detected in the spermatozoa of Kif9T100N/T100N mice (Fig. 3A), suggesting that KIF9 with the T100N mutation cannot be localized in the sperm flagellum. To confirm this idea, we analyzed KIF9 localization with immunofluorescence in developing spermatids that were squeezed out of the seminiferous tubules of the testis. Although we detected KIF9 signal in the cytoplasm, no KIF9 signal was found in the sperm flagella that were stained with anti-acetylated tubulin antibody in Kif9T100N/T100N mice (Fig. 3B). These results indicate that the T100N mutation impairs KIF9 transport into the flagellum. We then analyzed if other axonemal proteins can be transported into the flagellum in Kif9T100N/T100N mice. In contrast to KIF9, the amounts of DNAH8, DNAH17, DNAI2 (outer dynein arm) [20,21,22], DNALI1 (inner dynein arm) [22], GAS8 (nexin-dynein regulatory complex) [16], and RSPH9 (radial spoke) [11] in spermatozoa were comparable between Kif9WT/WT and Kif9T100N/T100N mice (Fig. 3C).
Fig. 3.
KIF9 is depleted in the spermatozoa of Kif9T100N/T100N mice. (A) Protein expression of KIF9 in testis and cauda epididymal spermatozoa. Acetylated tubulin is a loading control. (B) Immunofluorescence analysis of spermatids from Kif9WT/WT and Kif9T100N/T100N mice labeled with antibodies against KIF9 (green) and acetylated tubulin (red). Hoechst staining (blue) indicates the nucleus. (C) Western blotting analysis of axonemal proteins. The amount of analyzed proteins in the testis and spermatozoa were comparable between Kif9WT/WT and Kif9T100N/T100N mice.
Discussion
Intraflagellar transport (IFT) is a bidirectional movement of cargos along microtubules that is essential for cilia and flagellum formation. In mouse testes, deletion of IFT components results in impaired flagellar formation and short sperm tails [23, 24]. Kinesin is responsible for the anterograde transport of IFT, which moves cargos from the flagellar base to the tip [25]. In this study, we reveal that mutation of the KIF9 motor domain impaired its localization in the flagellum (Fig. 3A). In developing spermatids, KIF9 with the T100N mutation was detected in the cytoplasm, but not in the elongating axoneme (Fig. 3B), suggesting that the mutated KIF9 cannot be transported into the flagellum. One possibility for this defect is that KIF9 may use its own motor activity to move along microtubules in an elongating flagellum. However, it should be noted that the involvement of KIF9 in transporting molecules during spermiogenesis may be partial and other kinesins may play more critical roles because phenotypes of Kif9 KO mice were milder than those of other IFT mutants that exhibit short flagella [5, 23, 24]. This idea is supported by the fact that the amount of other axonemal proteins in the spermatozoa did not decrease in Kif9T100N/T100N mice (Fig. 3C). Another possibility is that the motor activity of KIF9 may be important to move to the flagellar base and other kinesins transport KIF9 into the elongating axoneme rather than KIF9 using its own motor activity to move along elongating axoneme. KIF3A is known to be involved in IFT [25], which deletion results in severe impairments in sperm flagellar formation in mice [26], and may be involved in transporting KIF9 into the flagellum.
As consistent with Kif9 KO mice, Kif9T100N/T100N mice exhibited impaired sperm motility associated with asymmetric flagellar bending and reduced male fertility. These results indicates that the motor domain of KIF9 is important for normal sperm motility and male fertility. It was shown that KIF9 can be localized to the CP of the axoneme and interacts with HYDIN, a component of the CP, that is thought to function in switching the bending direction by regulating dynein arm activity [5, 27]. Because KIF9 with the T100N mutation cannot be localized in the flagellum, it remains to be determined whether KIF9 in the CP utilizes its motor activity to regulate HYDIN and sperm motility in addition to regulating its localization in the flagellum.
In this study, we reveal that the motor domain of KIF9 is essential for the localization of KIF9 in the flagellum by substituting a critical amino acid using the CRISPR/Cas9 system. Further analysis to understand how KIF9 works in regulating flagellar motility may help us to understand the etiology of asthenozoospermia. Because there is no culture system that produces fully functional spermatozoa in vitro, amino acid substitutions on an organismal level using the CRISPR/CAS9 system is a powerful tool to understand the function of specific domains in sperm motility and male fertility.
Supplementary
Acknowledgments
The authors would like to thank Dr. Julio M. Castaneda for critical reading of the manuscript and Eri Hosoyamada for technical assistance. This research was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP17H04987 to H.M. and JP19H05750, JP21H04753, JP21H05033 to M.I.); Takeda Science Foundation grant to H.M. and M.I.; and the Japan Agency for Medical Research and Development (AMED) grant (JP21gm5010001 to M.I.); and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01HD087157 and R01HD088412 to M.I.); and the Bill & Melinda Gates Foundation (Grand Challenges Explorations grant INV-001902 to M.I.).
References
- 1.Curi SM, Ariagno JI, Chenlo PH, Mendeluk GR, Pugliese MN, Sardi Segovia LM, et al. Asthenozoospermia: analysis of a large population. Arch Androl. 2003; 49: 343–349. doi: 10.1080/01485010390219656 [DOI] [PubMed] [Google Scholar]
- 2.Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod. 2011; 17: 524–538. doi: 10.1093/molehr/gar034 [DOI] [PubMed] [Google Scholar]
- 3.Miyata H, Morohoshi A, Ikawa M. Analysis of the sperm flagellar axoneme using gene-modified mice. Exp Anim. 2020; 69: 374–381. doi: 10.1538/expanim.20-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirokawa N, Tanaka Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp Cell Res. 2015; 334: 16–25. doi: 10.1016/j.yexcr.2015.02.016 [DOI] [PubMed] [Google Scholar]
- 5.Miyata H, Shimada K, Morohoshi A, Oura S, Matsumura T, Xu Z, et al. Testis-enriched kinesin KIF9 is important for progressive motility in mouse spermatozoa. FASEB J. 2020; 34: 5389–5400. doi: 10.1096/fj.201902755R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein M, Beech PL, Katz SG, Rosenbaum JL. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J Cell Biol. 1994; 125: 1313–1326. doi: 10.1083/jcb.125.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yokoyama R, O’toole E, Ghosh S, Mitchell DR. Regulation of flagellar dynein activity by a central pair kinesin. Proc Natl Acad Sci USA. 2004; 101: 17398–17403. doi: 10.1073/pnas.0406817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153: 910–918. doi: 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbasi F, Miyata H, Ikawa M. Revolutionizing male fertility factor research in mice by using the genome editing tool CRISPR/Cas9. Reprod Med Biol. 2017; 17: 3–10. doi: 10.1002/rmb2.12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyata H, Abbasi F, Visconti PE, Ikawa M. CRISPR/CAS9-mediated amino acid substitution reveals phosphorylation residues of RSPH6A are not essential for male fertility in mice. Biol Reprod. 2020; 103: 912–914. doi: 10.1093/biolre/ioaa161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbasi F, Miyata H, Shimada K, Morohoshi A, Nozawa K, Matsumura T, et al. RSPH6A is required for sperm flagellum formation and male fertility in mice. J Cell Sci. 2018; 131: jcs221648. doi: 10.1242/jcs.221648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev. 1995; 41: 232–238. doi: 10.1002/mrd.1080410214 [DOI] [PubMed] [Google Scholar]
- 13.Muro Y, Hasuwa H, Isotani A, Miyata H, Yamagata K, Ikawa M, et al. Behavior of Mouse Spermatozoa in the Female Reproductive Tract from Soon after Mating to the Beginning of Fertilization. Biol Reprod. 2016; 94: 80. doi: 10.1095/biolreprod.115.135368 [DOI] [PubMed] [Google Scholar]
- 14.Miyata H, Satouh Y, Mashiko D, Muto M, Nozawa K, Shiba K, et al. Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science. 2015; 350: 442–445. doi: 10.1126/science.aad0836 [DOI] [PubMed] [Google Scholar]
- 15.Baba SA, Mogami Y. An approach to digital image-analysis of bending shapes of eukaryotic flagella and cilia. Cell Motil Cytoskeleton. 1985; 5: 475–489. doi: 10.1002/cm.970050605 [DOI] [Google Scholar]
- 16.Morohoshi A, Miyata H, Shimada K, Nozawa K, Matsumura T, Yanase R, et al. Nexin-Dynein regulatory complex component DRC7 but not FBXL13 is required for sperm flagellum formation and male fertility in mice. PLoS Genet. 2020; 16: e1008585. doi: 10.1371/journal.pgen.1008585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995; 131: 1039–1053. doi: 10.1083/jcb.131.4.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piddini E, Schmid JA, de Martin R, Dotti CG. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001; 20: 4076–4087. doi: 10.1093/emboj/20.15.4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nozawa K, Satouh Y, Fujimoto T, Oji A, Ikawa M. Sperm-borne phospholipase C zeta-1 ensures monospermic fertilization in mice. Sci Rep. 2018; 8: 1315. doi: 10.1038/s41598-018-19497-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitfield M, Thomas L, Bequignon E, Schmitt A, Stouvenel L, Montantin G, et al. Mutations in DNAH17, Encoding a Sperm-Specific Axonemal Outer Dynein Arm Heavy Chain, Cause Isolated Male Infertility Due to Asthenozoospermia. Am J Hum Genet. 2019; 105: 198–212. doi: 10.1016/j.ajhg.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C, Miyata H, Gao Y, Sha Y, Tang S, Xu Z, et al. Bi-allelic DNAH8 Variants Lead to Multiple Morphological Abnormalities of the Sperm Flagella and Primary Male Infertility. Am J Hum Genet. 2020; 107: 330–341. doi: 10.1016/j.ajhg.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aprea I, Raidt J, Höben IM, Loges NT, Nöthe-Menchen T, Pennekamp P, et al. Defects in the cytoplasmic assembly of axonemal dynein arms cause morphological abnormalities and dysmotility in sperm cells leading to male infertility. PLoS Genet. 2021; 17: e1009306. doi: 10.1371/journal.pgen.1009306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.San Agustin JT, Pazour GJ, Witman GB. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell. 2015; 26: 4358–4372. doi: 10.1091/mbc.E15-08-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Li W, Zhang Y, Zhang L, Teves ME, Liu H, et al. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol Biol Cell. 2016; 27: 3705–3716. doi: 10.1091/mbc.e16-05-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morthorst SK, Christensen ST, Pedersen LB. Regulation of ciliary membrane protein trafficking and signalling by kinesin motor proteins. FEBS J. 2018; 285: 4535–4564. doi: 10.1111/febs.14583 [DOI] [PubMed] [Google Scholar]
- 26.Lehti MS, Kotaja N, Sironen A. KIF3A is essential for sperm tail formation and manchette function. Mol Cell Endocrinol. 2013; 377: 44–55. doi: 10.1016/j.mce.2013.06.030 [DOI] [PubMed] [Google Scholar]
- 27.Lechtreck KF, Witman GB. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J Cell Biol. 2007; 176: 473–482. doi: 10.1083/jcb.200611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.