Abstract
Taxonomy
Potato virus X is the type‐member of the plant‐infecting Potexvirus genus in the family Alphaflexiviridae.
Physical properties
Potato virus X (PVX) virions are flexuous filaments 460–480 nm in length. Virions are 13 nm in diameter and have a helical pitch of 3.4 nm. The genome is approximately 6.4 kb with a 5′ cap and 3′ poly(A) terminus. PVX contains five open reading frames, four of which are essential for cell‐to‐cell and systemic movement. One protein encodes the viral replicase. Cellular inclusions, known as X‐bodies, occur near the nucleus of virus‐infected cells.
Hosts
The primary host is potato, but it infects a wide range of dicots. Diagnostic hosts include Datura stramonium and Nicotiana tabacum. PVX is transmitted in nature by mechanical contact.
Useful website
Keywords: plant virus, Potato virus X, Potexvirus, RNA virus
Potato virus X, one of the oldest known and well‐studied plant viruses, has contributed much to our current knowledge of plant immunity in potato and tobacco, and is a versatile vector for expressing foreign genes in plants.

1. INTRODUCTION
Potato virus X is the type‐member of the genus Potexvirus in the family Alphaflexiviridae, and is one of the oldest known potato viruses, with the earliest published scientific report in 1938 (Loughnane & Murphy, 1938). Potato virus X (PVX) causes a mild mosaic disease in potato, tomato, tobacco, and other primarily solanaceous plants. PVX is one of the most widely distributed viruses and this is largely attributed to global commerce. PVX is typically transmitted via pollen or true seeds, by contaminated farming equipment, or from plant to plant by contact between healthy and infected foliage or roots (Bercks, 1949). PVX does not have a known invertebrate vector. Transmission by chewing insects has been reported but none are considered to be specific vectors for PVX. PVX was once studied as a critical problem in seed potato production; however, potato seed certification programmes and the incorporation of disease resistance genes into potato breeding programmes have been successful to mitigate the economic impact of PVX (Bragard et al., 2020).
PVX is known to confer severe disease when it occurs in mixed infections with other viruses, especially potyviruses such as potato virus Y (PVY) and potato virus S (PVS), but also with the luteovirus potato leafroll virus (PLRV) (Pacheco et al., 2012; Syller, 2012). Mixed virus infections involving PVX lead to increases in the titres of PVY, PVS, or PLRV as well as more severe foliar symptoms. PVX and PVY co‐infections are a well‐studied synergism that produces rugose mosaic foliar disease, necrosis in some plant varieties, and stunting. PVX can also exacerbate diseases caused by Verticillium dahliae and Colletotrichum armamentarium while also decreasing the susceptibility of potato tubers to Fusarium roseum and Phytophthora infestans (Fernandez de Cubillos & Thurston, 1975; Goodell et al., 1981; Jones & Mullen, 1974; Loebenstein et al., 2001; Müller & Munro, 1951; Pietkiewicz, 1975).
Potato virus X is one of 48 species belonging to the genus Potexvirus, and many of these viruses cause severe diseases in their hosts (Martelli et al., 2008). PVX made the list of top 10 plant viruses in molecular plant pathology, as reported in Molecular Plant Pathology in 2011, because of its scientific development as a model for studying plant–virus interactions (Scholthof et al., 2011). PVX has been at the forefront of important advances in plant biology, such as the antiviral gene silencing machinery, the development of viral vectors to express foreign genes, the use of the green fluorescent protein (GFP) to track plant virus infection, discovering the role of viral movement proteins in plasmodesmata transport, and cloning of the Rx gene in potato via map‐based cloning, as the first antiviral R gene isolated from potato (Batten et al., 2003; Bendahmane et al., 1999; Lacorte et al., 2010; Verchot‐Lubicz et al., 2007).
2. VIRION MORPHOLOGY AND GENOME ORGANIZATION
The PVX genome consists of a single, single‐stranded, positive‐sense RNA molecule that is approximately 6400 bp in length. The RNA genome of PVX, as for all members of Alphaflexiviridae, has a 5′ methyl guanosine cap and 3′ poly(A) tail (Kreuze et al., 2020; Martelli et al., 2008). The genome has five open reading frames encoding the RNA‐dependent RNA polymerase (RdRp), three proteins required for virus cell‐to‐cell movement known as the triple gene block (TGB), and a single coat protein (CP) gene (Figure 1a). The virions are relatively short, 470–580 nm in length, and are flexuous filaments. There are 1300 copies of the CP forming a left‐handed helical virion (Grinzato et al., 2020). On entering a cell, PVX virions unpack through phosphorylation of the exposed N‐terminus of the CP by cellular enzymes. The exposed RNA is translated to produce the viral RdRp, enabling replication and subgenomic RNA synthesis. Replication occurs in structures called X‐bodies that form near the nucleus (Tilsner & Oparka, 2012).
FIGURE 1.
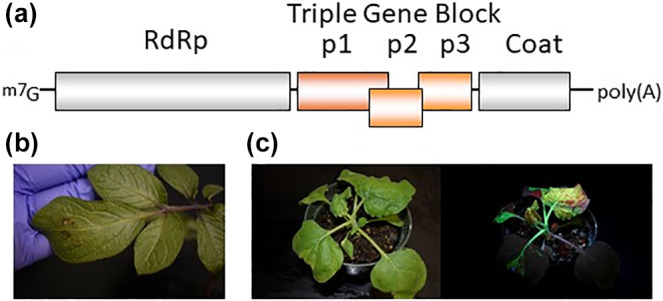
(a) Genome structure of PVX. The line indicates viral genomic RNAs. Labelled boxes represent open reading frames. (b) Solanum venturi leaf inoculated with PVX shows mild symptoms. (c) Nicotiana benthamiana plant inoculated with PVX‐GFP also shows mild symptoms but under an ultraviolet lamp the plant is fully systemically infected
PVX uses two strategies for gene expression. First, the viral RdRp is the sole protein expressed from the genomic RNA while the remaining viral proteins derive from 3′ coterminal subgenomic (sg)RNAs (Huisman et al., 1988; Ravin et al., 2008). The RdRp is an approximately 166 kDa protein and contains a conserved methyltransferase motif, necessary for adding the 5′ methyl guanosine cap, as well as central helicase and C‐terminal RNA polymerase catalytic motifs. The triple gene block proteins are a conserved block of three overlapping open reading frames that encode virus movement proteins expressed from two subgenomic RNAs. Until recently the sgRNA1 was considered a monocistronic mRNA that produces only the 25 kDa triple gene block 1 (TGB1) protein, although it has the coding capacity for other TGB proteins and the CP. A study by Fujimoto et al. (2021) showed that the sgRNA1 can produce all TGB proteins via a mechanism of ribosome leaky scanning. The sgRNA2 encodes the 12 kDa TGB2 and 8 kDa TGB3 proteins from overlapping open reading frames, which also rely on a mechanism of leaky scanning for expression of TGB3 (Verchot et al., 1998). The sgRNA3 encodes the virus CP.
The 5′ and 3′ untranslated regions (UTRs) of the viral genome contains cis‐regulatory elements that control gene expression and replication; these studies were conducted in PVX in the late 1990s and it is possible that with newer technologies we could learn more about the functions of these regions (Park et al., 2008). Thermodynamic predictions identified two stem loop structures, SL1 (106 nucleotides) and SL2 (39 nucleotides), within the 5′ UTR and mutational analysis was used to demonstrate that these are critical for PVX plus‐strand RNA accumulation (Kwon & Kim, 2006). The SL1 contains a terminal tetraloop with a GAAA motif that supports polymerase recognition and the initiation of RNA synthesis. Protein binding studies determined that the SL1 interacts with soluble proteins in tobacco protoplast extracts, which are essential for virus replication. The 3′ UTR also has a stem loop, SL3, adjacent to the poly(A) tail that is important for minus‐strand RNA accumulation (Pillai‐Nair et al., 2003). Within SL3 is a hexanucleotide motif in the loop region (ACAUAA) that is complementary to a conserved 5′ octanucleotide segment (AACUAAAC). There is a 3′ U‐rich element that is also required for minus‐strand RNA accumulation. In addition, sequences upstream of the TGB and CP genes regulate sgRNA accumulation and have core octanucleotide elements similar to the genomic 5′ untranslated leader, and are complementary to the hexanucleotide sequence of the SL3 (Kim & Hemenway, 1999). The short lead of the sgRNA1 also contains elements that drive translation of downstream TGB proteins by leaky scanning (Fujimoto et al., 2021).
3. GENE FUNCTION
PVX encodes a 166 kDa viral replicase derived from open reading frame 1 (ORF1) (Huisman et al., 1988). This protein maintains an N‐terminal methyltransferase‐like domain, NTP‐binding/helicase‐like domain, and C‐terminal RNA‐dependent polymerase. Immunolocalization and cell fractionation studies showed that the viral replication complexes (VRCs) also accumulate along membranes derived from the endoplasmic reticulum (ER) and are associated with TGB2 and TGB3 proteins (Bamunusinghe et al., 2009). VRCs are membrane‐bound bodies that accumulate near the nucleus or near the plasmodesmata and researchers have hypothesized that the neighbouring plasmodesmata specifically provide viral RNA for cell‐to‐cell movement. The recruitment of viral genomes from VRCs to the plasmodesmata entrance is known as “co‐replicational insertion” into plasmodesmata (Linnik et al., 2013; Tilsner et al., 2013). The TGB2 and TGB3 proteins recruit the TGB1 and CP to the transport RNA within the plasmodesmata to neighbouring cells. The viral TGB proteins and CP localize to distinct subdomains of the plasmodesmata. TGB1and CP localize inside plasmodesmata channels while TGB2 and TGB3 reside in caps and appressed cortical ER near the entry and exit of plasmodesmata pores (Tilsner et al., 2013).
The TGB1 is a multifunctional protein that contains a canonical NTPase/helicase domain and has ATPase activity, RNA binding, and RNA helicase activity (Leshchiner et al., 2006; Morozov & Solovyev, 2015). TGB1 increases the size exclusion limit of plasmodesmata to enable intercellular trafficking of viral genomes (Howard et al., 2004; Park et al., 2014). TGB1 binds to the virion CP to promote viral RNA translation (Mukhamedzhanova et al., 2009; Zayakina et al., 2008). TGB1 is also a suppressor of posttranscriptional gene silencing (PTGS) by inactivating SGS3, which is a cofactor for RDR6 and is necessary for synthesis of secondary viral siRNAs that contribute to systemic RNA silencing. TGB1 also suppresses RNA silencing by interacting with AGO1, AGO2, AGO3, and AGO4 and driving their degradation via the proteasome (Voinnet et al., 2000). Inside cells TGB1 aggregates to organize X‐bodies, which are actin‐ and membrane‐rich inclusion bodies (Linnik et al., 2013). TGB1 remodels host actin, ER, and Golgi membranes forming these perinuclear bodies.
The TGB2 and TGB3 proteins are small membrane‐binding proteins that embed into the ER and contribute to virus movement (Krishnamurthy et al., 2002; Mitra et al., 2003; Samuels et al., 2007). TGB2 causes restriction of ER tubules causing membrane curvature (Lazareva et al., 2021). TGB2 has been shown to produce anchored vesicles along the ER on occasion and data show these membrane altering functions are essential for virus cell‐to‐cell movement (Ju et al., 2005). TGB2 and TGB3 colocalize and recruit TGB1 to the ER, building intercellular transport complexes. The structural changes in the ER contribute to the inherent structure of the X‐bodies. The model for cell‐to‐cell movement is a “co‐replication” model in which newly synthesized viral genomes are recognized by the TGB1 and transported into the plasmodesmata for transfer into neighbouring cells (Tilsner et al., 2013; Tilsner & Oparka, 2012). In addition, TGB3 activates the unfolded protein response (UPR), which triggers ER‐to‐nuclear signalling and transcriptional changes via transcription factors known as bZIP60, bZIP28, and bZIP17 (Gayral et al., 2020).
4. PVX GENETIC VARIABILITY AND INNATE IMMUNITY AGAINST PVX
PVX strains differ in severity of the mosaic symptoms and were initially categorized by Cockerham (1955, 1970) as strain groups based on their phenotypic responses to three host resistance genes, namely Nx and Nb, which confer hypersensitive resistance to PVX, and Rx, which confers extreme resistance (Cockerham, 1970; Valkonen et al., 1996). Strain group 1 produces a hypersensitive response (HR) in plants with Nx and Nb genes; strain group 2 overcomes Nx but not Nb hypersensitivity; strain group 3 overcomes Nb but not Nx resistance; and strain group 4 overcomes both genes but is restricted by the Rx gene. Later, monoclonal antibodies were used to differentiate strains and four major serological groups were identified, indicating differences among strains probably occur in the surface epitopes of the viral CP. These differences correlate to some degree with the resistance groups, leading researchers to experimentally confirm a role for the CP in Rx recognition and gene‐for‐gene resistance (Feigelstock et al., 1995). Given that Nx and Nb contribute to resistance against the broadest PVX strains, these genes have been exploited in potato breeding programmes globally.
In the 1990s there were extensive investigations using resistant and susceptible plants and protoplasts to identify the viral determinants of HR resistance in these hosts. These studies used European PVX strains such as Roth1, UK3, and X3, as well as the American strains CP, CP4, and HB. Molecular analysis of PVX strains in Asia has been performed more recently and some isolates appear to be more similar to European isolates, suggesting that PVX was introduced before there were strict quarantine measures (Hajizadeh & Sokhandan‐Bashir, 2017; Komatsu et al., 2005; Yu et al., 2010). Japanese strains of PVX inoculated to Solanum demissium were categorized into the “common group” and the “necrosis group”. The common group causes no symptoms or small necrotic symptoms in the inoculated leaves (Figure 1b) and spreads systemically; the “necrosis group” causes severe necrotic spots on the inoculated leaves and does not spread systemically. Amino acid changes in the viral RdRp are responsible for producing various symptoms associated with the common or necrosis group (Komatsu et al., 2005). Neighbour‐joining analysis comparing large numbers of isolates from Asia, Australia, Europe, and the Americas revealed two distinct lineages: clade I consisted of >70 isolates sharing >93% nucleotide identity that were described as the Eurasia group; and clade II contained fewer isolates sharing >82% identity and known as the Americas group (Fuentes et al., 2021).
A unique form of innate immunity to potexviruses, including PVX, is led by the Jacalin‐type lectin required for potexvirus resistance 1 (JAX1) gene discovered in Arabidopsis. JAX1 is often compared to another lectin resistance gene, RTM1, that inhibits long‐distance movement of tobacco etch virus in Arabidopsis (Yamaji et al., 2012). Unlike RTM1, JAX1 interacts with the viral RdRp and directly inhibits virus replication. A single amino acid substitution of glutamine to histidine at amino acid position 336 in the PVX RdRp is sufficient to overcome JAX1‐mediated resistance (Sugawara et al., 2013). PVX does not normally infect Arabidopsis ecotype Col‐0 but infects other ecotypes such as C24. The basis for Col‐0 resistance is attributed to the specific natural variant of Argonaute 2 (AGO2), an endonuclease incorporated into the RNA‐induced silencing complexes (RISC) found in Col‐0 that affects its antiviral activity. AGO2 alleles largely affect systemic infection of Arabidopsis by PVX (Brosseau et al., 2020). Furthermore, AGO5 acts in concert with AGO2 to more fully compromise PVX infection (Brosseau & Moffett, 2015). Research shows that JAX1‐mediated resistance is epistatic to the effects of AGO2 on PVX susceptibility and that salicylic acid (SA) biosynthesis is not linked to AGO2 regulation of PVX in systemic leaves.
The EU Commission's Panel on Plant Health recently reviewed the threat of non‐EU isolates of PVX and assessed whether such isolates maybe deemed harmful and qualify as potential quarantine pests (Bragard et al., 2020). Given that EU isolates already impact EU potato production, the panel evaluated whether non‐EU isolates may additionally impact the current situation if they are introduced and spread in the EU. The assessment determined that non‐EU strains do not have the potential for adverse environmental or economic impact within EU territory.
5. VIRAL SYNERGISTIC INTERACTIONS PRODUCE SYSTEMIC NECROSIS
Synergistic interactions involving PVX, mainly with potyviruses including PVY, potato virus A (PVA), plum pox virus, or tobacco etch virus, manifest as severe symptoms including systemic necrosis and increased virus titres (Syller, 2012). Early investigations showed that HC‐Pro, the potyvirus silencing suppressor protein, could dramatically increase PVX levels by up to 10‐fold in systemic leaves (Shi et al., 1997). HC‐Pro within this synergistic infection contributes to alterations in miRNA metabolism and function. This synergism between PVX and PVY as well as other potyviruses led to the discovery that HC‐Pro is a viral silencing suppressor protein that limits the antiviral effects of small interfering RNAs (siRNAs). In mixed infections there are changes in the accumulation of microRNAs (miRNAs) miR156, 168, 171, and 398 as well as their mRNA targets and this is suggested to influence symptoms (Moyo et al., 2017). In mixed infections involving PVX and PVA, the HC‐Pro also interferes with S‐adenosyl‐l‐methionine synthetase and S‐adenosyl‐l‐homocysteine hydrolase, which modulate glutathione (GSH) levels (De et al., 2018). GSH is an antioxidant that also contributes to the regulation of PVX sgRNA expression. Shortages in GSH were shown to result in more severe symptoms in mixed infections. Co‐expression of the potyvirus HC‐Pro and potexvirus TGB1 silencing suppressor proteins also triggers ER stress as another path to causing tissue necrosis (Aguilar et al., 2019).
PVX has been important for early characterization of the homology‐dependent PTGS pathway that targets viral RNAs for degradation. PTGS recognizes highly structured regions of viral genomes or replication intermediates that can be digested by the plant dsRNA‐specific Dicer to produce siRNAs. These fundamental studies involved using PVX as an expression vector harbouring cDNAs encoding foreign sequences, including green fluorescent protein (GFP) (Figure 1c), which could be used to monitor siRNA production and mRNA turnover (Batten et al., 2003; Voinnet et al., 2000).
6. CONTROL OR MANAGEMENT
The most effective methods for controlling PVX are through incorporating disease resistance genes into potato varieties and through certified seed programmes that test for and eliminate virus‐contaminated potato seed from future planting stocks (Fuller et al., 2020). Stem cuttings and micropropagation technologies have been used to develop virus‐free planting stocks that are then used to produce clean seeds. Several generations of plants are grown in fields to produce clean seeds for commercial sales (Hutton et al., 2015). Most countries have a national potato certification programme that conducts disease testing to verify these seeds test clean for distribution (Loebenstein et al., 2001).
The most recent diagnostic technology innovations focus on methods to detect multiple viruses of a specific crop. Researchers focused on oligonucleotide‐based microarray and multiplex PCR methods for detecting viruses in potato (Boonham et al., 2003; Bystricka et al., 2005). This and other hybridization approaches using synthetic probes are cheaper and less labour‐intensive while providing a spectrum of diagnostic data.
7. CONCLUSIONS
Potato is one of the most important staple crops globally and infection by various viruses has been recognized to be among the most significant disease problems affecting global production. PVX is one of many viruses that causes reductions in tuber yields. The wide use of virus‐resistant germplasm, as well as the willingness of producers to conform to seed certification standards, has been quite effective for minimizing the risk of PVX disease.
ACKNOWLEDGEMENTS
This work was supported by a grant from NSF (IOS #1759034).
Verchot, J. (2022) Potato virus X: A global potato‐infecting virus and type member of the Potexvirus genus. Molecular Plant Pathology, 23, 315–320. 10.1111/mpp.13163
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed.
REFERENCES
- Aguilar, E. , del Toro, F.J. , Brosseau, C. , Moffett, P. , Canto, T. & Tenllado, F. (2019) Cell death triggered by the P25 protein in Potato virus X‐associated synergisms results from endoplasmic reticulum stress in Nicotiana benthamiana . Molecular Plant Pathology, 20, 194–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamunusinghe, D. , Hemenway, C.L. , Nelson, R.S. , Sanderfoot, A.A. , Ye, C.M. , Silva, M.A.T. et al. (2009) Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology, 393, 272–285. [DOI] [PubMed] [Google Scholar]
- Batten, J.S. , Yoshinari, S. & Hemenway, C. (2003) Potato virus X: a model system for virus replication, movement and gene expression. Molecular Plant Pathology, 4, 125–131. [DOI] [PubMed] [Google Scholar]
- Bendahmane, A. , Kanyuka, K. & Baulcombe, D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. The Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercks, R. (1949) Potato virus X. Nature, 163, 502–503. [DOI] [PubMed] [Google Scholar]
- Boonham, N. , Walsh, K. , Smith, P. , Madagan, K. , Graham, I. & Barker, I. (2003) Detection of potato viruses using microarray technology: towards a generic method for plant viral disease diagnosis. Journal of Virological Methods, 108, 181–187. [DOI] [PubMed] [Google Scholar]
- Bragard, C. , Dehnen‐Schmutz, K. , Gonthier, P. , Jacques, M.A. , Jaques Miret, J.A. , Justesen, A.F. et al. (2020) Pest categorisation of potato virus X (non‐EU isolates). EFSA Journal, 18, e05937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosseau, C. , Bolaji, A. , Roussin‐Léveillée, C. , Zhao, Z. , Biga, S. & Moffett, P. (2020) Natural variation in the Arabidopsis AGO2 gene is associated with susceptibility to potato virus X. New Phytologist, 226, 866–878. [DOI] [PubMed] [Google Scholar]
- Brosseau, C. & Moffett, P. (2015) Functional and genetic analysis identify a role for Arabidopsis ARGONAUTE5 in antiviral RNA silencing. The Plant Cell, 27, 1742–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricka, D. , Lenz, O. , Mraz, I. , Piherova, L. , Kmoch, S. & Sip, M. (2005) Oligonucleotide‐based microarray: a new improvement in microarray detection of plant viruses. Journal of Virological Methods, 128, 176–182. [DOI] [PubMed] [Google Scholar]
- Cockerham, G. (1955) Strains of potato virus X. In: Streutgers, E. (Ed.) Proceedings of the second conference on potato virus diseases, 1954, Lisse‐Wageningen. Wageningen: H. Veenman, pp. 25–29. [Google Scholar]
- Cockerham, G. (1970) Genetical studies on resistance to potato viruses X and Y. Heredity, 25, 309–348. [Google Scholar]
- De, S. , Chavez‐Calvillo, G. , Wahlsten, M. & Mäkinen, K. (2018) Disruption of the methionine cycle and reduced cellular gluthathione levels underlie potex–potyvirus synergism in Nicotiana benthamiana . Molecular Plant Pathology, 19, 1820–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigelstock, D.A. , Tozzini, A.C. & Hopp, H.E. (1995) Coat protein sequence of a resistance‐breaking strain of potato virus X isolated in Argentina. Virus Genes, 10, 289–292. [DOI] [PubMed] [Google Scholar]
- Fernandez de Cubillos, C. & Thurston, H.D. (1975) The effect of viruses on infection by Phytophthora infestans (Mont.) De Bary in potatoes. American Potato Journal, 52, 221–226. [Google Scholar]
- Fuentes, S. , Gibbs, A.J. , Hajizadeh, M. , Perez, A. , Adams, I.P. , Fribourg, C.E. et al. (2021) The phylogeography of potato virus X shows the fingerprints of its human vector. Viruses, 13, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, Y. , Keima, T. , Hashimoto, M. , Hagiwara‐Komoda, Y. , Hosoe, N. , Nishida, S. et al. (2021) Short 5′ UTR enables optimal translation of plant virus tricistronic RNA via leaky scanning. bioRxiv. 10.1101/2021.05.14.444105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller, K.B. , McIntosh, C. & Zidack, N. (2020) Valuing disease prevention in a vegetatively propagated annual crop: benefits from the Montana seed potato certification program. Plant Disease, 104, 2060–2067. [DOI] [PubMed] [Google Scholar]
- Gayral, M. , Arias Gaguancela, O. , Vasquez, E. , Herath, V. , Flores, F.J. , Dickman, M.B. et al. (2020) Multiple ER‐to‐nucleus stress signaling pathways are activated during Plantago asiatica mosaic virus and Turnip mosaic virus infection in Arabidopsis thaliana . The Plant Journal, 103, 1233–1245. [DOI] [PubMed] [Google Scholar]
- Goodell, J.J. , Powelson, M.L. & Allen, T. (1981) Interaction between PVX and Verticillium in potato. Phytopathology, 72, 631–634. [Google Scholar]
- Grinzato, A. , Kandiah, E. , Lico, C. , Betti, C. , Baschieri, S. & Zanotti, G. (2020) Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family. Nature Chemical Biology, 16, 564–569. [DOI] [PubMed] [Google Scholar]
- Hajizadeh, M. & Sokhandan‐Bashir, N. (2017) Population genetic analysis of potato virus X based on the CP gene sequence. Virus Disease, 28, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, A.R. , Heppler, M.L. , Ju, H.‐J. , Krishnamurthy, K. , Payton, M.E. & Verchot‐Lubicz, J. (2004) Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology, 328, 185–197. [DOI] [PubMed] [Google Scholar]
- Huisman, M.J. , Linthorst, H.J. , Bol, J.F. & Cornelissen, J.C. (1988) The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus‐stranded RNA viruses. Journal of General Virology, 69, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Hutton, F. , Spink, J.H. , Griffin, D. , Kildea, S. , Bonner, D. , Doherty, G. et al. (2015) Distribution and incidence of viruses in Irish seed potato crops. Irish Journal of Agricultural and Food Research, 54, 98–106. [Google Scholar]
- Jones, E.D. & Mullen, J.M. (1974) The effect of potato virus X on susceptibility of potato tubers to Fusarium roseum “avenaceum”. American Potato Journal, 51, 209–215. [Google Scholar]
- Ju, H.‐J. , Samuels, T.D. , Wang, Y.‐S. , Blancaflor, E. , Payton, M. , Mitra, R. et al. (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum‐derived vesicles during virus infection. Plant Physiology, 138, 1877–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.H. & Hemenway, C.L. (1999) Long‐distance RNA‐RNA interactions and conserved sequence elements affect potato virus X plus‐strand RNA accumulation. RNA, 5, 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, K. , Kagiwada, S. , Takahashi, S. , Mori, T. , Yamaji, Y. , Hirata, H. et al. (2005) Phylogenetic characteristics, genomic heterogeneity and symptomatic variation of five closely related Japanese strains of Potato virus X . Virus Genes, 31, 99–105. [DOI] [PubMed] [Google Scholar]
- Kreuze, J.F. , Vaira, A.M. , Menzel, W. , Candresse, T. , Zavriev, S.K. , Hammond, J. et al. (2020) ICTV virus taxonomy profile: Alphaflexiviridae . Journal of General Virology, 101, 699–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy, K. , Mitra, R. , Payton, M.E. & Verchot‐Lubicz, J. (2002) Cell‐to‐cell movement of the PVX 12K, 8K, or coat proteins may depend on the host, leaf developmental stage, and the PVX 25K protein. Virology, 300, 269–281. [DOI] [PubMed] [Google Scholar]
- Kwon, S.J. & Kim, K.H. (2006) The SL1 stem‐loop structure at the 5′‐end of Potato virus X RNA is required for efficient binding to host proteins and for viral infectivity. Molecules and Cells, 21, 63–75. [PubMed] [Google Scholar]
- Lacorte, C. , Ribeiro, S.G. , Lohuis, D. , Goldbach, R. & Prins, M. (2010) Potato virus X and Tobacco mosaic virus‐based vectors compatible with the Gateway cloning system. Journal of Virological Methods, 164, 7–13. [DOI] [PubMed] [Google Scholar]
- Lazareva, E.A. , Lezzhov, A.A. , Chergintsev, D.A. , Golyshev, S.A. , Dolja, V.V. , Morozov, S.Y. et al. (2021) Reticulon‐like properties of a plant virus‐encoded movement protein. New Phytologist, 229, 1052–1066. [DOI] [PubMed] [Google Scholar]
- Leshchiner, A.D. , Solovyev, A.G. , Morozov, S.Y. & Kalinina, N.O. (2006) A minimal region in the NTPase/helicase domain of the TGBp1 plant virus movement protein is responsible for ATPase activity and cooperative RNA binding. Journal of General Virology, 87, 3087–3095. [DOI] [PubMed] [Google Scholar]
- Linnik, O. , Liesche, J. , Tilsner, J. & Oparka, K.J. (2013) Unraveling the structure of viral replication complexes at super‐resolution. Frontiers in Plant Science, 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebenstein, G. , Berger, P.H. , Brunt, A.A. & Lawson, R.H. (2001) Virus and virus‐like diseases of potatoes and production of seed‐potatoes. Dordrecht, Netherlands: Springer. [Google Scholar]
- Loughnane, J. & Murphy, P. (1938) Mode of dissemination of potato virus X. Nature, 141, 120–121. [Google Scholar]
- Martelli, G.P. , Adams, M.J. , Kreuze, J.F. & Dolja, V.V. (2008) Family Flexiviridae: a case study in virion and genome plasticity. Annual Review of Phytopathology, 45, 73–100. [DOI] [PubMed] [Google Scholar]
- Mitra, R. , Krishnamurthy, K. , Blancaflor, E. , Payton, M. , Nelson, R.S. & Verchot‐Lubicz, J. (2003) The Potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell‐to‐cell movement. Virology, 312, 35–48. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. & Solovyev, A.G. (2015) Phylogenetic relationship of some “accessory” helicases of plant positive‐stranded RNA viruses: toward understanding the evolution of triple gene block. Frontiers in Microbiology, 6, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo, L. , Ramesh, S.V. , Kappagantu, M. , Mitter, N. , Sathuvalli, V. & Pappu, H.R. (2017) The effects of potato virus Y‐derived virus small interfering RNAs of three biologically distinct strains on potato (Solanum tuberosum) transcriptome. Virology Journal, 14, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhamedzhanova, A.A. , Karpova, O.V. , Rodionova, N.P. & Atabekov, I.G. (2009) Nonspecific activation of translation of encapsidated potexviral RNA with involvement of potato virus X movement protein TGB1. Doklady Biochemistry and Biophysics, 428, 239–241. [DOI] [PubMed] [Google Scholar]
- Müller, K.O. & Munro, J. (1951) The reaction of virus infected potato plants to Phytophthora infestans . Annals of Applied Biology, 38, 765–773. [Google Scholar]
- Pacheco, R. , García‐Marcos, A. , Barajas, D. , Martiáñez, J. & Tenllado, F. (2012) PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana . Virus Research, 165, 231–235. [DOI] [PubMed] [Google Scholar]
- Park, M.‐R. , Jeong, R.‐D. & Kim, K.‐H. (2014) Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Frontiers in Plant Science, 5, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.R. , Kwon, S.J. , Choi, H.S. , Hemenway, C.L. & Kim, K.H. (2008) Mutations that alter a repeated ACCA element located at the 5′ end of the Potato virus X genome affect RNA accumulation. Virology, 378, 133–141. [DOI] [PubMed] [Google Scholar]
- Pietkiewicz, J. (1975) Effect of viruses on the reaction of potato to Phytophthora infestans II. Mechanism of changes in the reaction to Ph. infestans in virus‐infected plants. Journal of Phytopathology, 82, 49–55. [Google Scholar]
- Pillai‐Nair, N. , Kim, K.H. & Hemenway, C. (2003) Cis‐acting regulatory elements in the potato virus X 3′ non‐translated region differentially affect minus‐strand and plus‐strand RNA accumulation. Journal of Molecular Biology, 326, 701–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin, N.V. , Mardanova, E.S. , Kotlyarov, R.Y. , Novikov, V.K. , Atabekov, J.G. & Skryabin, K.G. (2008) Complete sequencing of potato virus X new strain genome and construction of viral vector for production of target proteins in plants. Biochemistry, 73, 44–49. [DOI] [PubMed] [Google Scholar]
- Samuels, T.D. , Ju, H.‐J. , Ye, C.‐M. , Motes, C.M. , Blancaflor, E.B. & Verchot‐Lubicz, J. (2007) Subcellular targeting and interactions among the Potato virus X TGB proteins. Virology, 367, 375–389. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.‐B. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. et al. (2011) Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology, 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X.M. , Miller, H. , Verchot, J. , Carrington, J.C. & Vance, V.B. (1997) Mutations in the region encoding the central domain of helper component‐proteinase (HC‐Pro) eliminate potato virus X/potyviral synergism. Virology, 231, 35–42. [DOI] [PubMed] [Google Scholar]
- Sugawara, K. , Shiraishi, T. , Yoshida, T. , Fujita, N. , Netsu, O. , Yamaji, Y. et al. (2013) A replicase of Potato virus X acts as the resistance‐breaking determinant for JAX1‐mediated resistance. Molecular Plant‐Microbe Interactions, 26, 1106–1112. [DOI] [PubMed] [Google Scholar]
- Syller, J. (2012) Facilitative and antagonistic interactions between plant viruses in mixed infections. Molecular Plant Pathology, 13, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner, J. , Linnik, O. , Louveaux, M. , Roberts, I.M. , Chapman, S.N. & Oparka, K.J. (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. Journal of Cell Biology, 201, 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner, J. & Oparka, K.J. (2012) Missing links? ‐ The connection between replication and movement of plant RNA viruses. Current Opinion in Virology, 2, 699–705. [DOI] [PubMed] [Google Scholar]
- Valkonen, J.P.T. , Jones, R.A.C. , Slack, S.A. & Watanabe, K.N. (1996) Resistance specificities to viruses in potato: standardization of nomenclature. Plant Breeding, 115, 433–438. [Google Scholar]
- Verchot, J. , Angell, S.M. & Baulcombe, D.C. (1998) In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. Journal of Virology, 72, 8316–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Ye, C.‐M. & Bamunusinghe, D. (2007) Molecular biology of potexviruses: recent advances. Journal of General Virology, 88, 1643–1655. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. , Lederer, C. & Baulcombe, D.C. (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana . Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Yamaji, Y. , Maejima, K. , Komatsu, K. , Shiraishi, T. , Okano, Y. , Himeno, M. et al. (2012) Lectin‐mediated resistance impairs plant virus infection at the cellular level. The Plant Cell, 24, 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X.Q. , Jia, J.L. , Zhang, C.L. , Li, X.D. & Wang, Y.J. (2010) Phylogenetic analyses of an isolate obtained from potato in 1985 revealed potato virus X was introduced to China via multiple events. Virus Genes, 40, 447–451. [DOI] [PubMed] [Google Scholar]
- Zayakina, O. , Arkhipenko, M. , Kozlovsky, S. , Nikitin, N. , Smirnov, A. , Susi, P. et al. (2008) Mutagenic analysis of Potato virus X movement protein (TGBp1) and the coat protein (CP): in vitro TGBp1‐CP binding and viral RNA translation activation. Molecular Plant Pathology, 9, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed.


