Summary
Inhibiting the histone 3 lysine 79 (H3K79) methyltransferase, disruptor of telomeric silencing 1-like (DOT1L), increases the efficiency of reprogramming somatic cells to induced pluripotent stem cells (iPSCs). Here, we find that, despite the enrichment of H3K79 methylation on thousands of actively transcribed genes in somatic cells, DOT1L inhibition (DOT1Li) does not immediately cause the shutdown of the somatic transcriptional profile to enable transition to pluripotency. Contrary to the prevalent view, DOT1Li promotes iPSC generation beyond the mesenchymal to epithelial transition and even from already epithelial cell types. DOT1Li is most potent at the midpoint of reprogramming in part by repressing Nfix that persists at late stages of reprogramming. Importantly, regulation of single genes cannot substitute for DOT1Li, demonstrating that H3K79 methylation has pleiotropic effects in maintaining cell identity.
Keywords: DOT1L, H3K79 methylation, reprogramming, iPSCs induced pluripotent stem cells, NFIX
Highlights
-
•
DOT1L is a barrier of reprogramming, especially at the mid-point
-
•
DOT1L inhibition increases pluripotency beyond MET
-
•
DOT1L inhibition does not immediately suppress somatic expression
-
•
Single factors cannot replace the pleiotropic effects of DOT1L inhibition
In this article, Wille and Sridharan utilize somatic cell reprogramming to uncover that the enigmatic histone modification H3K79me is a barrier to pluripotency acquisition beyond the promotion of an epithelial state. Inhibition of the H3K79 methyltransferase, DOT1L, has pleiotropic effects in maintaining cell identity that cannot be recapitulated by modulating individual genes.
Introduction
Differential gene expression allows for functional diversity that translates to tissue specialization in multicellular organisms. In response to signaling and spatial cues, transcription factors engage with the epigenome to culminate in gene expression patterns that establish cell identity (Kelsey et al., 2017). The abundance of specific epigenetic modifications changes dynamically during development, right from the formation of the zygote (Dang-Nguyen and Torres-Padilla, 2015; Smith and Meissner, 2013). Fertilization of an oocyte triggers a decrease in histone 3 lysine (K) 79 dimethylation (H3K79me2) independent of genome replication (Ooga et al., 2008). H3K79me2 levels remain low during the pre-implantation phase until the blastocyst stage (Ooga et al., 2008). H3K79me2 is the most differential histone modification between somatic cells and embryonic stem cells (ESCs) that are derived from the blastocyst (Sridharan et al., 2013).
Disruptor of telomeric silencing 1-like (DOT1L) is the sole methyltransferase that performs all levels (me1, me2, and me3) of H3K79 methylation (called H3K79me henceforth; Black et al., 2012). Dot1l knockout (KO) mice are embryonic lethal between days 9.5 and 13.5, demonstrating the importance of H3K79me in development (Feng et al., 2010; Jones et al., 2008). Dot1l KO results in disorganized yolk sacs containing primitive erythrocytes. DOT1L is also necessary for development of the heart (Nguyen and Zhang, 2011), cerebral cortex (Franz et al., 2019), and chondrocytes (Castaño Betancourt et al., 2012) and normal CD8+ T cell differentiation (Kwesi-Maliepaard et al., 2020). H3K79me is not required for pluripotency, as ESCs continue to self-renew in the absence of DOT1L or DOT1L catalytic activity (Barry et al., 2009; Cao et al., 2020; Jones et al., 2008). We and others have shown that DOT1L is a barrier for transcription-factor-mediated reprogramming to induced pluripotent stem cells (iPSCs) from mouse neural stem cells and human fibroblasts (Jackson et al., 2016; Onder et al., 2012).
Collectively, these phenotypes provide evidence for the importance of DOT1L in cell fate determination; however, the function of H3K79me in mammals has remained elusive. H3K79me2 is enriched on the bodies of rapidly elongating genes (Duffy et al., 2018; Veloso et al., 2014), implicating the modification as a positive regulator of transcription. In acute leukemia, fusion proteins between mixed-lineage leukemia (MLL) and numerous DOT1L-associated proteins (ENL, AF9, and AF10) frequently drive oncogenesis (Mohan et al., 2010). MLL target genes, such as the HOXA cluster, are upregulated concurrent with the corresponding locus becoming H3K79 methylated as DOT1L is mislocalized via the MLL-DOT1L-interacting fusion protein. In contrast, H3K79me enrichment directly downregulates the expression of the epithelial sodium channel gene in mouse cells (Zhang et al., 2006), confounding the role of DOT1L in transcription.
Here, we use the dynamic system of somatic cell reprogramming from mouse embryonic fibroblasts (MEFs) to iPSCs to investigate DOT1L function in maintaining cell identity. We find that DOT1L inhibition enhances MEF pluripotency acquisition throughout the process but acts most potently at mid-reprogramming. This dramatic increase in reprogramming is accompanied by few steady-state transcriptional changes. Previous studies have reported that DOT1L inhibition (DOT1Li) enhances reprogramming by facilitating the mesenchymal to epithelial transition (MET) that occurs when reprogramming fibroblasts (Apostolou and Hochedlinger, 2013; Onder et al., 2012). However, with orthogonal experiments, such as expressing Cdh1 during reprogramming, testing cells after MET, and reprogramming keratinocytes that are already epithelial, we conclusively demonstrate that DOT1Li enhances pluripotency far beyond the transition to the epithelial identity. Using a small interfering RNA (siRNA) screen, we identify Nfix, which remains highly expressed throughout reprogramming, as a regulator that collaborates with DOT1L to reduce iPSC formation. Therefore, DOT1Li increases pluripotency acquisition in part by preventing upregulation of reprogramming-associated genes, which contributes to the continued requirement of DOT1Li into the mid-phase of reprogramming. DOT1Li does not immediately switch off the somatic transcriptome to favor pluripotency and functions through pleiotropic effects.
Results
DOT1L enzymatic activity is a barrier for reprogramming
We used a secondary reprogramming system to assess how DOT1L affects MEF pluripotency acquisition. MEFs isolated from mice that contain Oct4, Sox2, Klf4, and c-Myc (OSKM) (Sridharan et al., 2013) under a doxycycline-inducible promoter were reprogrammed with Dot1l knockdown using siRNA. With depletion of just half of the Dot1l transcript (Figure S1A), reprogramming was increased 2-fold (Figure 1A). Because DOT1L protein can function independent of catalytic activity (Cao et al., 2020), we next inhibited DOT1L using two extremely specific small molecules with no reported off-target effects (Figure 1B; Daigle et al., 2013; Kaniskan et al., 2018; Yu et al., 2012). SGC0946, hereafter called DOT1Li, disrupts DOT1L interaction with S-adenosyl methionine (SAM) by inducing a conformational change and is 10-fold more potent than the first-generation DOT1Li inhibitor, EPZ004777 (Yu et al., 2012). Both EPZ004777 (Nassa et al., 2019) and SGC0946 (Wu et al., 2021) were recently shown to reduce DOT1L occupancy at target genes. EPZ5676, known as pinometostat, is a highly selective DOT1L inhibitor that also prevents SAM interaction and has entered clinical trials (Campbell et al., 2017). Both chemical inhibitors decreased H3K79me2 with similar kinetics compared with control (Figure 1B) and comparably increased reprogramming of MEFs as measured by NANOG expression (Figures 1C, S1B, and S1C). Importantly, DOT1Li produced at least 9-fold more bona fide pluripotent colonies that maintained NANOG expression after removal of transgene expression by withdrawal of doxycycline (Figures 1C, S1B, and S1C, right). Similar concordance of increased reprogramming efficiency from human somatic cells has been reported when using RNAi-mediated depletion and catalytic inhibition with EPZ00477 (Onder et al., 2012).
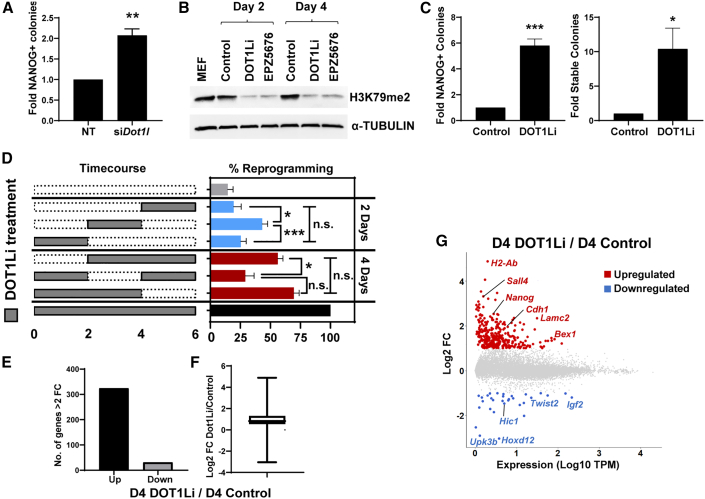
Figure 1.
Loss of H3K79me during reprogramming results in few steady-state transcriptional changes
(A) Reprogramming efficiency of MEFs treated with control non-targeting (NT) siRNA or depleted for Dot1l (siDot1l). Error bars represent the SEM of three independent experiment replicates, each composed of two or three technical replicates. Colonies obtained in NT were set to 1. ∗∗p < 0.01 by unpaired t test.
(B) Immunoblot of H3K79me2 and TUBULIN loading control of reprogramming cells on day 2 or 4 treated with control DMSO, SGC0946 (DOT1Li), or EPZ5676.
(C) NANOG+ colonies obtained on days 6 to 7 after induction of OSKM in MEFs (left) and stable (NANOG+/DPPA4+) colonies 2–4 days post-doxycycline (dox) and drug removal (right) with control or DOT1Li. Colonies obtained in control treatment were set to 1. Error bars represent the SEM of three independent experiment replicates, each composed of two or three technical replicates. ∗∗∗p < 0.001 and ∗p < 0.05 by unpaired t test.
(D) (Left) Scheme of exposure to DOT1Li (gray boxes) or control (dotted boxes). (Right) Number of NANOG+ colonies on day 6 is shown. Colonies obtained in day 0–6 continuous DOT1Li treatment were set to 100% (black bar). Error bars represent the SEM of three independent experiment replicates, each consisting of two or three technical replicates. Each duration of DOT1Li treatment, 2 days (blue bars) or 4 days (red bars), was assessed by one-way ANOVA. Significance was calculated post hoc with the Tukey test: ∗∗∗p < 0.001, ∗p < 0.05, and not significant (n.s.), p > 0.05.
(E) Number of genes upregulated or downregulated by a more than 2-fold change (FC) with a posterior probability of differential expression greater than 0.95 determined by EBSeq in day 4 DOT1Li versus day 4 control.
(F) Boxplot of log2 FC of all genes with a posterior probability of differential expression greater than 0.95 in day 4 DOT1Li relative to day 4 control.
(G) Log10 of the averaged transcripts per million (TPM) of day 4 DOT1Li and 4 control versus log2 FC of all genes. More than 2-fold upregulated is indicated in red and downregulated indicated in blue.
See also Figure S1.
To determine the temporal requirement for DOT1Li to enhance reprogramming, we performed a 6-day time course (Figure 1D, left). When reprogramming populations were exposed to DOT1Li in 2-day intervals, the greatest increase in reprogramming was observed with treatment between days 2 and 4 (Figure 1D, blue bars). Removing DOT1Li between days 2 and 4 resulted in fewer NANOG-positive colonies compared with 4 days of continuous DOT1Li treatment either early or late in reprogramming (Figure 1D, red bars). The greatest number of NANOG+ colonies formed when DOT1L was inhibited throughout reprogramming. Thus, DOT1L activity is a barrier to reprogramming throughout the conversion process but has the greatest effect in the intermediate phase.
Loss of H3K79me during reprogramming results in few steady-state transcriptional changes
To elucidate how loss of H3K79me enhances iPSC generation, the starting MEFs, reprogramming populations on days 2 and 4, and ESCs were transcriptionally profiled using RNA sequencing (RNA-seq). Independent experiment replicate samples collected on day 4 were treated continuously with DOT1Li, treated “early” from day 0 to 2, treated “mid” from day 2 to 4, or treated continuously with vehicle control (Figures S1D and S1E) to capture how DOT1Li specifically increases mid reprogramming (Figure 1D). Continuous DOT1Li for 4 days resulted in the most transcriptional alterations compared with early or mid treatment (Figures 1E, S1G, and S1H). Three hundred fifty-two genes were differentially expressed (Figure 1E; experimental procedures), with 10-fold more genes upregulated than downregulated. The majority of differentially expressed (DE) genes had a modest change in expression (Figure 1F). This low-magnitude change in expression occurred in genes with low levels of absolute expression (Figure 1G). Mid treatment from day 2 to 4 with DOT1Li yielded the next highest number of changes, with 125 upregulated genes and 2 downregulated genes (Figure S1H), followed by day 2 DOT1Li (51 upregulated and 1 downregulated; Figure S1F). Removal of DOT1Li after 2 days (early treatment) resulted in only nine DE genes (Figure S1G). Accordingly, early treatment clustered closest with day 4 control-treated replicates (Figure S1D). At every time point, a far greater number of upregulated than downregulated genes were DE.
H3K79me2 is enriched on numerous genes, yet few change their steady-state mRNA levels
DOT1Li could promote iPSC generation by erasing the somatic program, enhancing pluripotent gene expression, or a combination of the two. To distinguish between these possibilities, significantly DE genes from all time points (n = 438; “DOT1Li-DE”) were compared with the change in gene expression in ESCs versus MEFs. While about 50% of DOT1Li-downregulated genes were also decreased in ESCs, far fewer of the DOT1Li-upregulated genes were upregulated in ESCs, suggesting that most of the steady-state transcriptional change is aberrant to that observed in ESCs (Figure 2A).
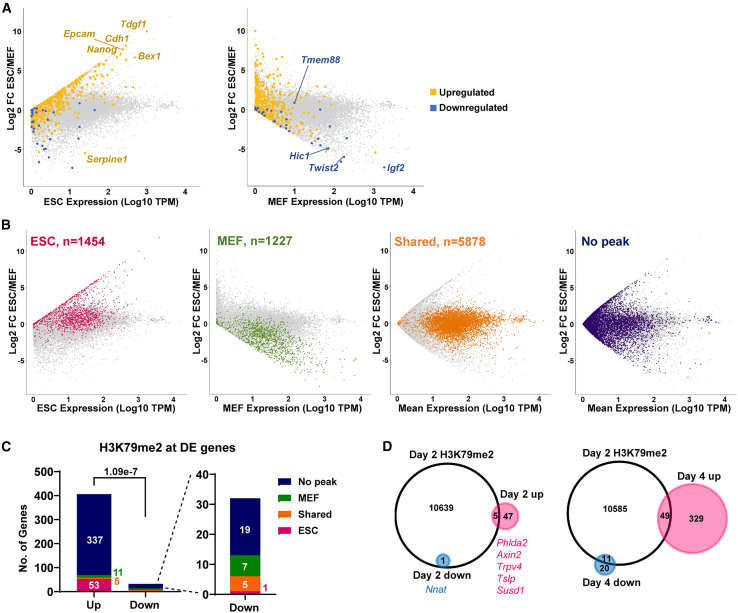
Figure 2.
H3K79me2 is enriched on numerous genes, yet few change transcriptionally in steady-state mRNA levels
(A) Genes upregulated (gold) or downregulated (blue) by DOT1Li treatment at any time point relative to the matched control (DOT1Li-DE list) plotted on expression calculated as log10 of ESC TPM (left) or MEF TPM (right) versus log2 FC in ESCs relative to MEFs.
(B) Genes with H3K79me2 called peaks in ESCs (red), in MEFs (green), shared in both cell types (orange), or with no peak (purple) were plotted onto the indicated expression calculated as log10 TPM versus log2 FC in ESCs relative to MEFs. H3K79me2 location data were analyzed from GEO: GSE90895 (Chronis et al., 2017).
(C) Bar graph of H3K79me2 called peaks at DOT1Li-DE genes. Significance calculated by Fisher's exact test is shown.
(D) Overlap of genes with an H3K79me2 peak on day 2 of reprogramming (Chronis et al., 2017) with DE genes at the indicated time points.
See also Figure S2.
We then asked whether the transcriptional changes were correlated with the presence of H3K79 methylation. Chromatin immunoprecipitation sequencing (ChIP-seq) of H3K79me2 in MEFs derived from the same reprogrammable mouse line and ESCs was analyzed (Figure 2B; Chronis et al., 2017). As expected, highly expressed genes were enriched for H3K79me2 in both MEFs and ESCs. Genes with “shared peaks” were highly expressed and predominantly housekeeping genes (Figure S2A). In addition, many low-expressed genes did not have a significant H3K79me2 peak, yet a subset of these genes were DE in ESCs versus MEFs (Figure 2B, “no peak”).
The majority of DE genes did not have an H3K79me2 peak (Figures 2C, S2B, and S2C). Thirteen percent of upregulated genes had a peak in ESCs, while only 2.7% had a peak in MEFs, and 1.2% had a shared peak (Figure 2C). Only 22% of downregulated genes had a peak in MEFs, 16% had a shared peak, and 1 gene had a peak in ESCs (Figure 2C). To capture whether transient gain in H3K79me2 during reprogramming affects gene expression, DE genes at day 2 or 4 of reprogramming overlapped with genes modified by H3K79me2 48 h after OSKM induction. Very few DE genes contained an H3K79me2 peak during reprogramming, and the majority were upregulated (Figure 2D).
DOT1L inhibition leads to transcriptional changes not observed in MEFs or ESCs
To ascertain the functional role of DOT1Li during reprogramming irrespective of time point, all DOT1Li-DE genes were clustered and visualized on a heatmap relative to their expression in MEFs (Figure 3A). Genes in clusters I–III were changed by DOT1Li to resemble ESC-like expression and included transcription factors involved in structure morphogenesis, epithelium, and proliferation genes (Figures 3A, 3B, and S3A). In contrast to genes in clusters I–III, the majority of genes (clusters IV and V) do not resemble the transcriptional profile of ESCs relative to MEFs (Figure 3A). Genes in cluster IV (n = 60) are a mixed population of aberrantly upregulated genes, or their downregulation is prevented by DOT1Li (Figures 3A and S3A) and they are not functionally related. Cluster V (n = 160) contains genes that are upregulated by DOT1Li treatment yet are largely unchanged in ESCs relative to MEFs (Figures 3A and S3A) that function in transmembrane signaling (Figure 3B).
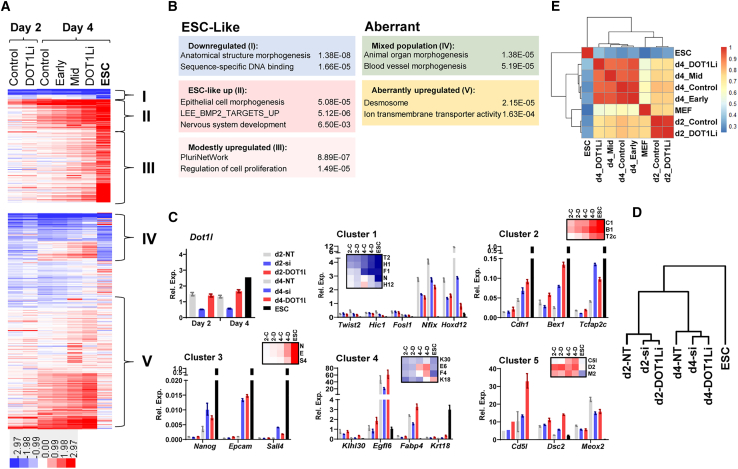
Figure 3.
DOT1L inhibition leads to transcriptional changes not observed in MEFs or ESCs
(A) k-means clustering of DOT1Li-DE genes organized based on transcriptional change in ESCs relative to MEFs. Heatmap intensity indicates log2 FC TPM relative to MEF for each sample.
(B) Representative gene ontology categories for each cluster. The most enriched categories containing unique sets of genes are displayed (HOMER).
(C) qPCR validation of select genes from every cluster in non-targeting control (NT) treated with control DMSO, siDot1l (si), and DOT1Li. Normalized expression in MEFs was set to 1 for Dot1l and clusters 1, 4, and 5. Normalized expression in ESCs was set to 1 for clusters 2 and 3. All samples shown are from day 2 (left) to day 4 (right). Insets: zoomed-in RNA-seq heatmaps of assessed genes in control (C), DOT1Li (D), and ESCs at day 2 (2) and day 4 (4). Genes are abbreviated as first initial and number.
(D) Hierarchical clustering of relative expression of tested genes (C).
(E) Spearman correlation of DOT1Li-DE gene TPM across all samples.
See also Figure S3.
As an independent validation of these trends in transcriptional profile, we verified the expression changes in an siRNA-mediated depletion of Dot1l over a 6-day time course. Samples were collected on day 2 and day 4 and showed a greater than 2-fold decrease in Dot1l expression (Figure 3C). We selected at least three genes in each cluster that represented varying levels of change on day 2 and day 4 by DOT1Li (Figure 3C, inset). The results obtained from the genetic knockdown cluster with DOT1L catalytic inhibition (Figure 3D). Another study using the first-generation EPZ004777 inhibitor and shDOT1L in breast cancer lines found a large overlap in gene-expression changes between the two methods of depleting DOT1L function (Nassa et al., 2019). However, it remains possible that some of the transcriptional alterations that we observe are due to off-target effects of the small-molecule DOT1Li.
Although the changes in clusters I–III (Figure 3A) position DOT1Li-treated cells closer to pluripotency acquisition than control-treated cells, Spearman correlation of DOT1Li-DE gene expression shows that all time points more closely resemble MEFs than ESCs (Figure 3E). Both day 2 samples cluster together and are more correlated to day 4 control compared with other day 4 samples, suggesting day 4 control treatment is kinetically behind the other DOT1Li treatments. Because there are few transcriptional alterations, all day 4 samples cluster closely together (Figure 3E). Of the day 4 samples, day 4 early (d0–2) and day 4 control treatments are most highly correlated, explaining why a removal of DOT1Li on day 2 of reprogramming barely increases NANOG+ colony formation relative to control treatment (Figure 1D).
To determine how the clusters may be regulated, we performed motif analysis and found that cluster III and, to a lesser extent, cluster II were enriched for E-boxes that can be bound by a cadre of proteins, depending on the motif context (Figure S3B). Mesenchymal E-box binding genes that repress expression are not DE by DOT1Li, with the exception of Twist2, which is 2-fold downregulated on day 4 with constant DOT1Li inhibition (Figure S3C). Of note, DOT1Li inhibition also activates Mycn (2.7-fold in day 4 DOT1Li) that binds E-boxes to promote gene activity. Thus, the balance of these two proteins may be responsible for the modest increase in expression of cluster III, which is much more significantly enriched for E-boxes compared with cluster II (Figure S3B). Cluster IV is enriched for TCF7 motifs, a transcriptional activator, which is upregulated by DOT1Li treatment. Finally, cluster V is enriched for HOXD12 and HIC1 motifs, both transcriptional regulators downregulated by DOT1Li treatment.
Thus, DOT1Li may promote development of reprogramming intermediates expressing lineage genes (Polo et al., 2012) due to subtle changes in transcription factor levels. However, as the two clusters that most resemble ESCs (I and II) are devoid of motifs bound significantly by any of the DOT1Li DE genes, it is unlikely that DOT1L regulates pluripotency acquisition through such intermediates.
Inhibition of DOT1L enhances reprogramming of epithelial cells
To investigate whether individual genes in the upregulated cohort could replace DOT1Li, we first examined the epithelial genes. MET is an early phase of reprogramming when starting with MEFs (Apostolou and Hochedlinger, 2013). Cluster II genes (Figure 3A) that were most similar in their expression changes to ESCs (Figure S3A) are enriched for epithelial genes and E-box motifs (Figure S3B). We introduced Cdh1, which has previously been shown to be required for pluripotency (Chen et al., 2010; Hawkins et al., 2012), into reprogrammable MEFs using a lentivirus to achieve expression comparable to that in ESCs (Figure 4A). Although CDH1 was detected at the cell surface (Figure 4A), it did not enhance MEF reprogramming (Figure 4B). It is important to note that exogenous Cdh1 expression was incapable of causing mesenchymal gene downregulation (Figure S4A). This result supports the notion that mesenchymal gene downregulation and Cdh1 upregulation are regulated independent of each other from our co-expression analysis of single-cell (sc) RNA-seq data, where we found that reprogramming cells can simultaneously express genes from both programs (Tran et al., 2019). Given that expression of Cdh1 alone could not replace DOT1Li function (Figure 4B), we assessed how reduction of H3K79me affects reprogramming of cells that are already epithelial. Reprogramming MEFs were sorted on day 3.5 post-OSKM induction for cell-surface CDH1 expression (Figure 4C) to a level similar to that of ESCs (Figure S4B). Reprogramming of the CDH1− and CDH1+ sorted cells was then continued in the presence of DOT1Li. Interestingly, reprogramming of both populations of cells was increased to similar extents (6.8-fold) when exposed to DOT1Li (Figure 4C), indicating a response beyond the upregulation of the epithelial program.
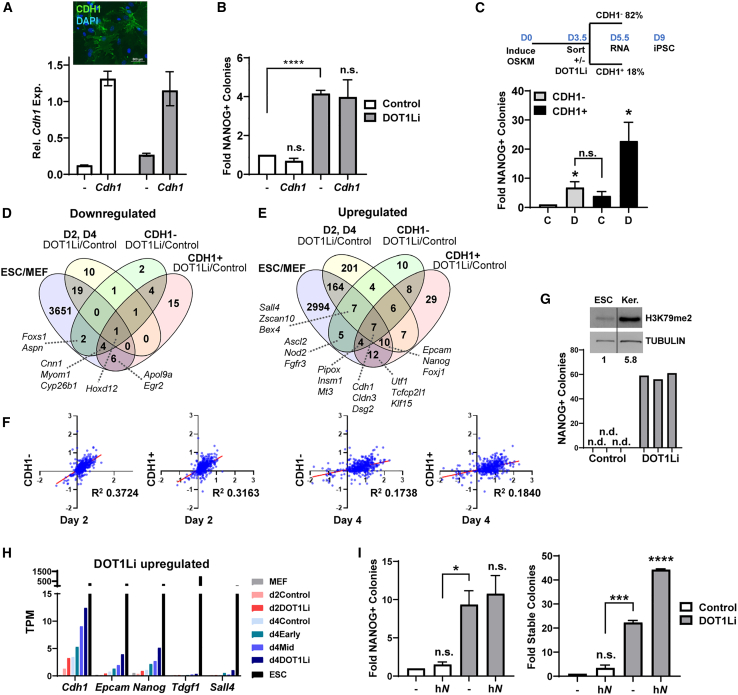
Figure 4.
Inhibition of DOT1L enhances reprogramming of epithelial cells
(A) (Top) CDH1 immunofluorescence on day 4 of reprogramming of Cdh1-transduced cells. Scale bar represents 500 μm. (Bottom) Relative Cdh1 expression measured on day 3 of reprogramming is shown. ESCs are set to 1. Cells were treated with control (white bars) or DOT1Li (gray bars).
(B) Fold NANOG+ colonies of empty-vector- (–) or Cdh1-transduced cells, treated with control (white bars) or DOT1Li (gray bars). Empty-vector-transduced cells treated with control are set to 1. Error bars represent the SEM of three independent experiment replicates, each consisting of two technical replicates. ∗∗∗∗p < 0.0001 and n.s., p > 0.05, by unpaired t test.
(C) Reprogramming scheme (top): cells were grown for 3 days in KnockOut serum replacement (KSR)-containing medium to accelerate CDH1 expression. On day 3.5, cells were sorted with flow cytometry for CDH1 surface expression, followed by DOT1Li or control treatment. RNA-seq samples were collected at day 5.5, and reprogramming was evaluated on days 8 to 9. (Bottom) NANOG+ colony formation of CDH1-sorted cells treated with DOT1Li (D) or control (C) is shown. Error bars represent the SEM of three independent experiment replicates, each consisting of two or three technical replicates. ∗p < 0.05 and n.s., p > 0.05, by unpaired t test.
(D) Overlap of 1.5-FC DOT1Li downregulated genes in CDH1− and CDH1+ cells with the downregulated genes from all time points of the DOT1Li time course (2-FC; Figure 3A) and genes downregulated in ESCs relative to MEFs (2-FC). A selection of genes is displayed.
(E) Overlap of 1.5-FC DOT1Li upregulated genes in CDH1− and CDH1+ cells with the upregulated genes from all time points of the DOT1Li time course (2-FC; Figure 3A) and genes upregulated in ESCs relative to MEFs (2-FC). A selection of genes is displayed.
(F) Scatterplot of log2 FC TPM of DOT1Li versus matched control, of the indicated time course (Figure 1) x axis, and CDH1 sort (Figure 4) y axis RNA-seq samples at DOT1Li-DE genes.
(G) NANOG+ colonies on day 8 of keratinocyte reprogramming, treated with DOT1Li or control. Bars represent technical replicates. n.d., not detected. Inset: immunoblot of H3K79me2 and TUBULIN loading control in ESCs and keratinocytes (Ker.) is shown.
(H) Expression (TPM) bar graph of DOT1Li upregulated reprogramming factors.
(I) Cells transduced with vector control (–) or human NANOG (hN), treated with control (white bars) or DOT1Li (gray bars). (Left) NANOG+ colonies during reprogramming are shown. (Right) Stable colonies post-dox removal are shown. Colonies obtained in vector control are set to 1. Error bars represent the SEM of three independent experiment replicates, each consisting of two or three technical replicates. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05, and n.s., p > 0.05, by unpaired t test.
See also Figure S4.
To identify a transcriptional response after epithelial upregulation, we profiled the gene expression of CDH1− and CDH1+ populations with DOT1Li. Independent experiment replicate transcriptional data from these sorted populations were highly reproducible (Figure S4C) and clustered with one another rather than with MEFs or ESCs (Figure S4D). Similar to the results from the unsorted time course (Figures 1 and 3), more genes were upregulated than downregulated by DOT1Li in both CDH1− and CDH1+ populations. Hoxd12 was the only commonly downregulated gene in CDH1−, CDH1+, DOT1Li-DE days 2 and 4, and ESCs relative to MEFs (Figure 4D). Upregulated genes that were altered in the same direction as their expression in ESCs (Figure 4E) were enriched for the functional categories of stem cell population maintenance and epithelial cell development. The changes mediated by DOT1Li in unsorted populations were positively correlated with those in CDH1-sorted populations (Figure 4F). Interestingly, the changes in CDH1− cells more closely resemble day 2 of the unsorted time course, whereas the CDH1+ cells are comparatively more correlated to day 4 alterations (Figure 4F). Indeed, irrespective of the status of CDH1 expression in the sorted population, addition of DOT1Li further enhances the expression of pluripotent genes, such as Nanog, albeit to a much lower extent than in ESCs (Figure S4E).
CDH1 is an important, but not the sole, determinant of epithelial identity. Given that DOT1Li increased reprogramming from MEFs that overexpressed or were induced to express CDH1, we next investigated the effect of DOT1L on an epithelial cell type. Keratinocytes are epithelial and, despite not having to undergo MET (Li et al., 2010; Samavarchi-Tehrani et al., 2010), still reprogram poorly compared with MEFs (Nefzger et al., 2017). We isolated keratinocytes from reprogrammable mice and confirmed that they express similar levels of CDH1 on their surface compared with ESCs (Figure S4F). Because our mass spectrometry comparison of global histone modification levels was from MEFs (Sridharan et al., 2013), we performed an immunoblot for H3K79me2. Keratinocytes had over 5-fold more H3K79me2 globally than ESCs (Figure 4G, inset), confirming that high levels of this modification are a common feature of somatic cells. DOT1Li applied during keratinocyte reprogramming increased both the efficiency and the kinetics with the appearance of tens of NANOG+ colonies by day 8 of reprogramming (Figure 4G). No colonies were obtained in the control at this time point, likely due to their lower reprogramming efficiency compared with MEFs (Nefzger et al., 2017). Thus, DOT1Li promotes pluripotency beyond the upregulation of epithelial identity.
We next assessed whether DOT1Li increases reprogramming through upregulation of pluripotency factors. In our previous analysis of single-cell transcriptomic analysis, we found that the co-expression of a quartet of genes, Nanog, Sall4, Tdgf1, and Epcam, within the same cell predicted a more homogeneous transition to an iPSC state (Tran et al., 2019). DOT1Li upregulated the same genes but to a much smaller magnitude than that in ESCs (Figure 4H). We introduced exogenous human NANOG (Figures S4G and S4H) and found that it enhanced transgene independence in the presence of DOT1Li (Figure 4I). Thus, DOT1Li does not increase reprogramming through Nanog upregulation but collaborates with NANOG specifically to maintain pluripotent colonies.
To identify whether any other DOT1Li DE genes regulate pluripotency acquisition, they were overlapped with genes identified in a large short hairpin RNA (shRNA) screen starting with the same OSKM-inducible secondary MEF system (Borkent et al., 2016). Among the genes that affected reprogramming more than 2-fold from the screen (∼2,300 genes), only 10 overlapped. However, seven barrier genes were upregulated and three enhancers were downregulated by DOT1Li, in the opposite direction of pluripotency promotion (Figure S4I). Thus, the DOT1Li-mediated increase in reprogramming is unlikely to be replaced by upregulating single-gene expression. In addition, DOT1Li treatment does not increase cellular proliferation (Figure S4J) or cause changes in the cell cycle (Figures S4K and S4L), indicating that it does not enhance pluripotency by increased cell number.
Inhibition of DOT1L enhances reprogramming beyond modulation of single genes
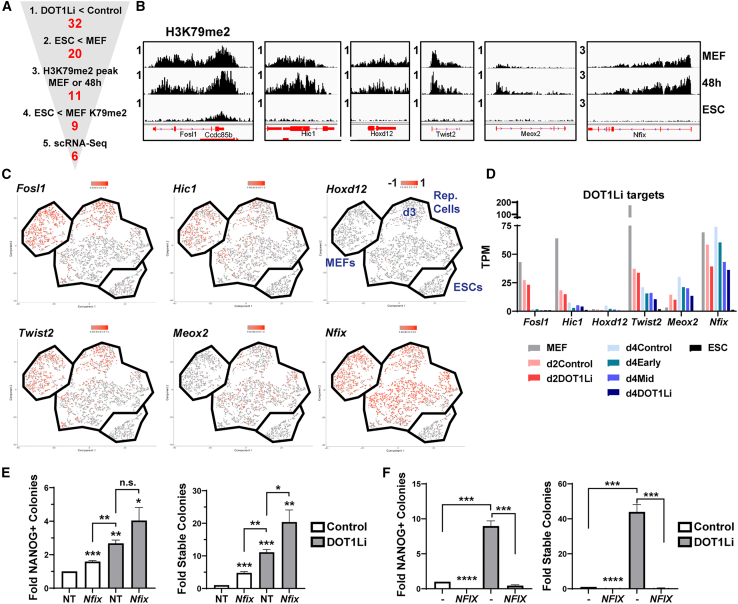
Because DOT1Li did not change the entire transcriptional landscape either away from a somatic program or toward a pluripotency program, we used a candidate approach to screen DOT1Li-DE genes for their contribution to pluripotency acquisition. As a higher percentage of downregulated genes were modified by H3K79me2 (Figures 2C and S3D), and DOT1L has previously been reported as a barrier to reprogramming by maintaining fibroblast gene transcription (Onder et al., 2012), we comprehensively ranked DOT1Li directly downregulated genes (Figure 5A, step 1). Among the DOT1Li genes that were downregulated in ESCs versus MEFs (step 2), we filtered those that had a significant H3K79me2 peak in MEFs or 48-h post-OSKM induction (step 3) that is reduced in ESCs (step 4; Figure 5B). A change in gene expression levels between MEFs and ESCs could occur in every single cell of the starting population or could reflect a population average. We have previously performed scRNA-seq of MEFs, ESCs, and a time course of reprogramming (Tran et al., 2019). Using these data, we then chose genes that were expressed in the entire population of starting MEFs or reprogramming cells (step 5; Figure 5C), narrowing to six potential targets that could be critical for the mechanism of DOT1Li (Figure 5D). Notably, all six targets were also downregulated by siRNA-mediated Dot1l depletion (Figure 3C).
Figure 5.
Inhibition of DOT1L enhances reprogramming beyond modulation of single genes
(A) Schematic to identify DOT1L direct target genes. Genes were filtered for (1) at least 2-FC downregulation in expression by DOT1Li treatment at any time point, (2) at least 2-FC downregulation in expression in ESC versus MEFs, (3) a significant gene body H3K79me2 peak (p-4) with 3-fold enrichment over input in MEFs or 48 h OSKM (B), (4) a 2-fold reduction in H3K79me2 gene body reads in ESCs, and (5) expression in a substantial portion of MEFs or reprogramming cells by scRNA-seq (C).
(B) Integrative Genomics Viewer (IGV) tracks of H3K79me2 signal in MEFs, 48 h OSKM, and ESCs of potential DOT1L direct targets.
(C) Cells expressing the DOT1L target gene are labeled in red on the scRNA-seq t-distributed stochastic neighbor embedding (t-SNE). MEFs, reprogramming cells (days 3, 6, 9, and 12), and ESCs are indicated and labeled. Day 3 of reprogramming is indicated, but the other days are mixed, as reprogramming becomes more heterogeneous as it proceeds (Tran et al., 2019).
(D) Expression (TPM) bar graph of candidate DOT1Li downregulated genes.
(E) NANOG+ colonies on days 6 to 7 of reprogramming (left) and stable colonies post-dox removal (right) of cells transfected with NT and siRNA against Nfix, treated with control (white bars) or DOT1Li (gray bars). Control-treated NT was set to 1. Error bars represent the SEM of three independent experiment replicates, each consisting of two or three technical replicates. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, and n.s., p > 0.05, by unpaired t test.
(F) NANOG+ colonies at day 6 of reprogramming (left) and stable colonies post-dox removal (right) of cells transduced with empty vector control (−) or NFIX, treated with control (white bars) or DOT1Li (gray bars). Error bars represent the SEM of three independent experiment replicates, each consisting of two or three technical replicates. ∗∗∗∗p < 0.0001 and ∗∗∗p < 0.001 by unpaired t test.
See also Figure S5.
Each target was reduced individually using siRNA-mediated knockdown in the presence or absence of DOT1Li. Depletion of Fosl1 resulted in cell death with three different siRNAs (data not shown) and was not analyzed further. For each of the other five targets, siRNA-mediated depletion was robust and resulted in a similar fold change compared with that obtained by DOT1Li (Figure S5A). When siRNA for each target was combined with DOT1Li, mRNA levels were further reduced (Figure S5A). Hic1, Hoxd12, and Twist2 depletion did not enhance reprogramming in the presence or absence of DOT1Li (Figures S5B and S5C). Meox2 depletion alone or combined with DOT1Li slightly decreased NANOG+ colony formation (Figure S5B) but did not affect bona fide colonies that remained after doxycycline withdrawal (Figure S5C). Depletion of Nfix alone increased the occurrence of NANOG+ colonies 2-fold compared with a 3-fold increase with DOT1Li (Figure 5E, left). However, Nfix depletion did not lead to a robust formation of bona fide colonies, as DOT1Li treatment increased transgene-independent colonies 11-fold, whereas only a 4.8-fold increase was observed with siNfix (Figure 5E, right). When combined with DOT1Li, Nfix depletion enhanced transgene-independent colony formation (Figure 5E, right). Thus, Nfix depletion acts in conjunction with DOT1Li to promote reprogramming.
To interrogate whether these genes are barriers of the process, we overexpressed MEOX2 and NFIX individually in MEFs prior to the induction of reprogramming (Figure S5D). Exogenous MEOX2 expression inhibited reprogramming on its own but did not affect DOT1Li-mediated reprogramming (Figure S5E, left). Surprisingly, MEOX2 increased stable colony formation in the absence of DOT1Li (Figure S5E, right). These contrary effects of Meox2 depletion and overexpression on reprogramming may be related to its heterogeneous expression in the reprogramming population (Figure 5C) and the increase in proliferation in MEFs overexpressing MEOX2 (data not shown). Ectopic expression of NFIX during reprogramming reduced DOT1Li-mediated NANOG acquisition below control levels and prevented stable colony formation (Figure 5F), suggesting it is a potent reprogramming barrier independent of DOT1L activity. Thus, the downregulation of Nfix acts in an additive manner to increase pluripotency by contributing to the DOT1Li phenotype.
By comparing the results of the depletion experiments (Figures 5E, S5B, and S5C) with the pattern of expression from single-cell analysis (Figure 5C), we find that DOT1L acts in a collaborative manner with Nfix that is still expressed at later stages of reprogramming. Hic1, Fosl1, and Twist2 are downregulated in most cells by day 3 of reprogramming, and Meox2 is upregulated in only a portion of reprogramming cells (Figures 5C and 5D). Taken together with the temporal effectiveness of DOT1Li at the mid stages of reprogramming (Figure 1D), DOT1Li sets the stage for enhancing reprogramming efficiency along with the downregulation of persistently expressed genes such as Nfix.
Discussion
DOT1L is crucial for mammalian development, yet its role in cell-fate determination is still unknown. Here, we find that DOT1L is a barrier to pluripotency acquisition of MEFs throughout reprogramming but acts most strongly during mid-reprogramming (Figure 1). Stages of reprogramming when starting from MEFs include an early inactivation of the somatic program, an important component of which is the mesenchymal genes (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Inhibition of DOT1L was reported to enhance human fibroblast reprogramming by facilitating MET (Onder et al., 2012). The reverse process, epithelial to mesenchymal transition (EMT), is prevented in other systems, such as renal (Liu et al., 2019) and breast cancer cell lines (Cho et al., 2015), by DOT1Li. We demonstrate that DOT1L control of cell identity extends far beyond epithelial transitions. Although inhibition of DOT1L increases Cdh1 expression in our mouse reprogramming studies, it also enhances reprogramming of keratinocytes that do not have to undergo MET and MEF reprogramming post-MET (Figure 4). This corroborates our recent discovery that the epithelial and mesenchymal programs are independently regulated in the presence of DOT1Li during reprogramming using scRNA-seq (Tran et al., 2019).
In the course of assessing DOT1L transcriptional regulation, we identified contributions of the reprogramming barrier Nfix that maintain cellular identity with DOT1L (Figure 5). NFIX is a transcription factor important for neural (Pekarik and Belmonte, 2008) and muscle development (Pistocchi et al., 2013). In addition, NFIX is required to maintain murine hair follicle stem cell enhancer function (Adam et al., 2020), and its depletion increases the number of pluripotent colonies (Yang et al., 2011). From our previous analysis of reprogramming with scRNA-seq, we ordered single cells in a trajectory (Tran et al., 2019). We found a major branchpoint where cells stalled and did not complete the transition to iPSCs (Tran et al., 2019). Nfix was found to be a branchpoint gene such that downregulation was required to continue in the reprogramming trajectory, further suggesting that it may have a specific role in mid-reprogramming. Of the identified DOT1L targets, Nfix continues to be expressed in cells later in reprogramming and gains H3K79me2 after OSKM induction (Figures 5B and 5C), suggesting DOT1Li may facilitate its downregulation by preventing the gain in H3K79me2 enrichment at later reprogramming time points. This result is in contrast to previous findings that DOT1Li enhances pluripotency acquisition by downregulation of the established transcriptional program (Onder et al., 2012). It is important to note that depletion of Nfix does not reach the reprogramming efficiency of DOT1Li, and the effect is additive with DOT1Li (Figure 5E). Thus, downregulation of reprogramming-associated factors may contribute but is not solely causal of the DOT1L reprogramming phenotype.
We find that modulation of DOT1L-DE genes does not substitute for DOT1L catalytic inhibition (Figures 4 and 5), suggesting that H3K79me may have a role in reprogramming beyond transcriptional regulation of single genes. During development, only four genes were DE in Dot1l KO mouse c-kit+ cells sorted from embryonic day 10.5 (E10.5) yolk sacs, where profound phenotypic alterations in vascular morphology and erythrocyte maturation were observed (Feng et al., 2010). These small transcriptional changes in key factors like Nanog during reprogramming or Gata2 during erythrocyte maturation suggest that loss of H3K79me may alter global epigenetic profiles rather than local expression profiles. For example, loss of H3K79me2 allowed for spread of H3K27me3 on downregulated genes in leukemia cells (Deshpande et al., 2014). H3K79me2/3 are enriched at certain intronic enhancers in leukemia cell lines and regulate future deposition of H3K27ac (Godfrey et al., 2019), a modification associated with enhancer activity. In neural differentiation, DOT1Li decreased accessibility of a subset of enhancers (Ferrari et al., 2020). In support of further epigenetic alterations, both upregulated and downregulated genes can be decorated by H3K79me2 before DOT1L depletion in development of the cerebral cortex (Franz et al., 2019). Thus, H3K79me may affect histone modification, depending on chromatin context.
We observed that many more genes are upregulated rather than downregulated in their steady-state expression by DOT1Li during reprogramming (Figures 1 and 4). Many of the upregulated genes are expressed in other lineages and are not modified by H3K79me2 in MEFs or ESCs (Figures 3 and S3D). Alternatively, these genes may be regulated indirectly by the binding of DOT1Li-DE transcription factors, such as HOXD12 or HIC1. The aberrant expression of lineage-specific genes may promote reprogramming, a notion that can be further investigated by single-cell transcriptional analysis. Regardless, the boost to reprogramming cells by DOT1Li seems to outweigh the burden of spurious lineage gene expression. Taken together, our results demonstrate that DOT1L activity functions beyond steady-state alterations to the somatic transcriptome; rather it collaborates with reprogramming-associated factors to safeguard cell identity.
Experimental procedures
Cell isolation and culture
Male and female MEFs were isolated on E13.5 from embryos that were homozygous for the Oct4-2A-Klf4-2A-IRES-Sox2-2A-c-Myc (OKSM) transgene at the Col1a1 locus and either heterozygous or homozygous for the reverse tetracycline transactivator (rtTA) allele at the Rosa26 locus, as previously described (Tran et al., 2019). MEFs were grown in DMEM, 10% fetal bovine serum (FBS), 1× non-essential amino acids, 1× GlutaMAX, 1× penicillin/streptomycin, and 2-mercaptoethanol (4 μL/500 mL). Feeder MEFs were maintained and isolated as above from DR4 mice genetically resistant to geneticin (G418), puromycin, hygromycin, and 6-thioguanine. Feeder cells were irradiated with 9,000 rad after three passages. ESCs V6.5 were grown on feeder MEFs in KO DMEM, 15% FBS, 1× non-essential amino acids, 1× GlutaMAX, 1× penicillin/streptomycin, 2-mercaptoethanol (4 μL/525 mL), and leukemia inhibitory factor. Keratinocytes were isolated from reprogrammable mice 4 days postnatal as previously described (Li et al., 2017) and cultured in EpiLife medium with 60 μM calcium (Thermo Fisher Scientific MEPI500CA) with the EpiLife defined growth supplement (Thermo Fisher Scientific S0125). 293T cells were acquired from ATCC and grown in DMEM and 10% FBS. Mice were maintained according to the UW-Madison institutional animal care and use committee (IACUC)-approved protocol.
A detailed description of all materials and methods is provided in the supplemental information.
Data and code availability
The accession number for all RNA-seq datasets reported in this paper is National Center for Biotechnology Information Gene Expression Omnibus (GEO): GSE160580.
Author contributions
C.K.W. performed experiments and bioinformatic analysis, C.K.W. and R.S. wrote the manuscript and acquired funding, and R.S. conceived and directed the project.
Conflict of interests
The authors declare no competing interests.
Acknowledgments
This work was supported by a UW-Madison Stem Cell and Regenerative Medicine Center postdoctoral award to C.K.W., a UW-Madison Fall competition, and a Shaw Scientist award to the R.S. lab. We thank Dr. Roice Wille for R script design, Stefan Pietrzak and Brenton Halvorson for scRNA-seq analysis, Dr. Jason Tchieu for the NFIX vector, and Dr. Konstantinos Chronis and the R.S. lab members for critical reading of the manuscript.
Published: January 6, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.12.004.
Supplemental information
References
- Adam R.C., Yang H., Ge Y., Infarinato N.R., Gur-Cohen S., Miao Y., Wang P., Zhao Y., Lu C.P., Kim J.E., et al. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nat. Cell Biol. 2020;22:640–650. doi: 10.1038/s41556-020-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry E.R., Krueger W., Jakuba C.M., Veilleux E., Ambrosi D.J., Nelson C.E., Rasmussen T.P. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells. 2009;27:1538–1547. doi: 10.1002/stem.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkent M., Bennett B.D., Lackford B., Bar-Nur O., Brumbaugh J., Wang L., Du Y., Fargo D.C., Apostolou E., Cheloufi S., et al. A serial shRNA screen for roadblocks to reprogramming identifies the protein modifier SUMO2. Stem Cell Rep. 2016;6:704–716. doi: 10.1016/j.stemcr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.T., Haladyna J.N., Drubin D.A., Thomson T.M., Maria M.J., Yamauchi T., Waters N.J., Olhava E.J., Pollock R.M., Smith J.J., et al. Mechanisms of Pinometostat (EPZ-5676) treatment-emergent resistance in MLL-rearranged leukemia. Mol. Cancer Ther. 2017;16:1669–1679. doi: 10.1158/1535-7163.MCT-16-0693. [DOI] [PubMed] [Google Scholar]
- Cao K., Ugarenko M., Ozark P.A., Wang J., Marshall S.A., Rendleman E.J., Liang K., Wang L., Zou L., Smith E.R., et al. DOT1L-controlled cell-fate determination and transcription elongation are independent of H3K79 methylation. Proc. Natl. Acad. Sci. U S A. 2020;117:27365–27373. doi: 10.1073/pnas.2001075117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño Betancourt M.C., Cailotto F., Kerkhof H.J., Cornelis F.M.F., Doherty S.A., Hart D.J., Hofman A., Luyten F.P., Maciewicz R.A., Mangino M., et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc. Natl. Acad. Sci. U S A. 2012;109:8218–8223. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Cho M.-H., Park J.-H., Choi H.-J., Park M.-K., Won H.-Y., Park Y.-J., Lee C.H., Oh S.-H., Song Y.-S., Kim H.S., et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat. Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle S.R., Olhava E.J., Therkelsen C.A., Basavapathruni A., Jin L., Boriack-Sjodin P.A., Allain C.J., Klaus C.R., Raimondi A., Scott M.P., et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang-Nguyen T.Q., Torres-Padilla M.-E. How cells build totipotency and pluripotency: nuclear, chromatin and transcriptional architecture. Curr. Opin. Cell Biol. 2015;34:9–15. doi: 10.1016/j.ceb.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Deshpande A.J., Deshpande A., Sinha A.U., Chen L., Chang J., Cihan A., Fazio M., Chen C., Zhu N., Koche R., et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26:896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy E.E., Canzio D., Maniatis T., Simon M.D. Solid phase chemistry to covalently and reversibly capture thiolated RNA. Nucleic Acids Res. 2018;46:6996–7005. doi: 10.1093/nar/gky556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yang Y., Ortega M.M., Copeland J.N., Zhang M., Jacob J.B., Fields T.A., Vivian J.L., Fields P.E. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F., Arrigoni L., Franz H., Izzo A., Butenko L., Trompouki E., Vogel T., Manke T. DOT1L-mediated murine neuronal differentiation associates with H3K79me2 accumulation and preserves SOX2-enhancer accessibility. Nat. Commun. 2020;11:5200. doi: 10.1038/s41467-020-19001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz H., Villarreal A., Heidrich S., Videm P., Kilpert F., Mestres I., Calegari F., Backofen R., Manke T., Vogel T. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic Acids Res. 2019;47:168–183. doi: 10.1093/nar/gky953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey L., Crump N.T., Thorne R., Lau I.-J., Repapi E., Dimou D., Smith A.L., Harman J.R., Telenius J.M., Oudelaar A.M., et al. DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat. Commun. 2019;10:2803. doi: 10.1038/s41467-019-10844-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins K., Mohamet L., Ritson S., Merry C.L.R., Ward C.M. E-cadherin and, in its absence, N-cadherin promotes Nanog expression in mouse embryonic stem cells via STAT3 phosphorylation. Stem Cells. 2012;30:1842–1851. doi: 10.1002/stem.1148. [DOI] [PubMed] [Google Scholar]
- Jackson S.A., Olufs Z.P.G., Tran K.A., Zaidan N.Z., Sridharan R. Alternative routes to induced pluripotent stem cells revealed by reprogramming of the neural lineage. Stem Cell Rep. 2016;6:302–311. doi: 10.1016/j.stemcr.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G.A., Kadam S., Zhai H., Valdez R., et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. Plos Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniskan H.Ü., Martini M.L., Jin J. Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 2018;118:989–1068. doi: 10.1021/acs.chemrev.6b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G., Stegle O., Reik W. Single-cell epigenomics: recording the past and predicting the future. Science. 2017;358:69–75. doi: 10.1126/science.aan6826. [DOI] [PubMed] [Google Scholar]
- Kwesi-Maliepaard E.M., Aslam M.A., Alemdehy M.F., van den Brand T., McLean C., Vlaming H., van Welsem T., Korthout T., Lancini C., Hendriks S., et al. The histone methyltransferase DOT1L prevents antigen-independent differentiation and safeguards epigenetic identity of CD8+ T cells. Proc. Natl. Acad. Sci. U S A. 2020;117:20706–20716. doi: 10.1073/pnas.1920372117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Adase C.A., Zhang L.-J. Isolation and culture of primary mouse keratinocytes from neonatal and adult mouse skin. J. Vis. Exp. 2017;125:e56027. doi: 10.3791/56027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liu L., Zou J., Guan Y., Zhang Y., Zhang W., Zhou X., Xiong C., Tolbert E., Zhao T.C., Bayliss G., et al. Blocking the histone lysine 79 methyltransferase DOT1L alleviates renal fibrosis through inhibition of renal fibroblast activation and epithelial-mesenchymal transition. FASEB J. 2019;33:11941–11958. doi: 10.1096/fj.201801861R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M., Lin C., Guest E., Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat. Rev. Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Nassa G., Salvati A., Tarallo R., Gigantino V., Alexandrova E., Memoli D., Sellitto A., Rizzo F., Malanga D., Mirante T., et al. Inhibition of histone methyltransferase DOT1L silences ERα gene and blocks proliferation of antiestrogen-resistant breast cancer cells. Sci. Adv. 2019;5:eaav5590. doi: 10.1126/sciadv.aav5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefzger C.M., Rossello F.J., Chen J., Liu X., Knaupp A.S., Firas J., Paynter J.M., Pflueger J., Buckberry S., Lim S.M., et al. Cell type of origin dictates the route to pluripotency. Cell Rep. 2017;21:2649–2660. doi: 10.1016/j.celrep.2017.11.029. [DOI] [PubMed] [Google Scholar]
- Nguyen A.T., Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B., et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooga M., Inoue A., Kageyama S., Akiyama T., Nagata M., Aoki F. Changes in H3K79 methylation during preimplantation development in mice. Biol. Reprod. 2008;78:413–424. doi: 10.1095/biolreprod.107.063453. [DOI] [PubMed] [Google Scholar]
- Pekarik V., Belmonte J.C.I. NFIX--one gene, two knockouts, multiple effects. J. Biol. 2008;7:29. doi: 10.1186/jbiol94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistocchi A., Gaudenzi G., Foglia E., Monteverde S., Moreno-Fortuny A., Pianca A., Cossu G., Cotelli F., Messina G. Conserved and divergent functions of Nfix in skeletal muscle development during vertebrate evolution. Development. 2013;140:1528–1536. doi: 10.1242/dev.076315. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J., et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.-K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M., et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K.A., Pietrzak S.J., Zaidan N.Z., Siahpirani A.F., McCalla S.G., Zhou A.S., Iyer G., Roy S., Sridharan R. Defining reprogramming checkpoints from single-cell analyses of induced pluripotency. Cell Rep. 2019;27:1726–1741.e5. doi: 10.1016/j.celrep.2019.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A., Kirkconnell K.S., Magnuson B., Biewen B., Paulsen M.T., Wilson T.E., Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Zhi J., Tian T., Cihan A., Cevher M.A., Liu Z., David Y., Muir T.W., Roeder R.G., Yu M. DOT1L complex regulates transcriptional initiation in human erythroleukemic cells. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2106148118. e2106148118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-S., Lopez C.G., Rana T.M. Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem Cells. 2011;29:1528–1536. doi: 10.1002/stem.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Chory E.J., Wernimont A.K., Tempel W., Scopton A., Federation A., Marineau J.J., Qi J., Barsyte-Lovejoy D., Yi J., et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- Zhang W., Xia X., Reisenauer M.R., Hemenway C.S., Kone B.C. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCα in an aldosterone-sensitive manner. J. Biol. Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for all RNA-seq datasets reported in this paper is National Center for Biotechnology Information Gene Expression Omnibus (GEO): GSE160580.