Summary
Taste bud cells are renewed throughout life in a process requiring innervation. Recently, we reported that R-spondin substitutes for neuronal input for taste cell regeneration. R-spondin amplifies WNT signaling by interacting with stem-cell-expressed E3 ubiquitin ligases RNF43/ZNRF3 (negative regulators of WNT signaling) and G-protein-coupled receptors LGR4/5/6 (positive regulators of WNT signaling). Therefore, we hypothesized that RNF43/ZNRF3 may serve as a brake, controlled by gustatory neuron-produced R-spondin, for regulating taste tissue homeostasis. Here, we show that mice deficient for Rnf43/Znrf3 in KRT5-expressing epithelial stem/progenitor cells (RZ dKO) exhibited taste cell hyperplasia; in stark contrast, epithelial tissue on the tongue degenerated. WNT signaling blockade substantially reversed all these effects in RZ dKO mice. Furthermore, innervation becomes dispensable for taste cell renewal in RZ dKO mice. We thus demonstrate important but distinct functions of RNF43/ZNRF3 in regulating taste versus lingual epithelial tissue homeostasis.
Keywords: taste stem cells, RNF43, ZNRF3, WNT, R-spondin, LGR5
Graphical abstract
Highlights
-
•
Rnf43/Znrf3 deficiency in taste stem cells leads to taste cell hyperplasia
-
•
Lingual epithelial tissue degenerated in Rnf43/Znrf3-deficient mice
-
•
Innervation is dispensable for taste cell renewal in Rnf43/Znrf3-deficient mice
-
•
Wnt signaling blockade prevents phenotypic changes in Rnf43/Znrf3-deficient mice
Genetic knockout of Rnf43/Znrf3 leads to both overgrowth in taste cells and undergrowth in epithelial tissue on the tongue.
Introduction
The sense of taste enables humans and other species to detect nutritious substances or potentially harmful/toxic substances (Bachmanov and Beauchamp, 2007; Chaudhari and Roper, 2010). Taste sensation begins when substances are detected by a variety of taste receptor cells clustered in taste buds, comprising different taste fields in the oral cavity: in anterior tongue, fungiform papillae typically house single taste buds (e.g., rodents), whereas, in posterior tongue, the circumvallate papillae and foliate papillae contain many dozens of taste buds each. Once taste receptor cells are activated by taste stimuli, these cells send signals to the brain to generate taste sensation or perception (Bachmanov and Beauchamp, 2007; Chaudhari and Roper, 2010).
Despite the sensory nature of taste bud cells, they are epithelial cells, not nerve cells, and, like other epithelial cells, taste cells turn over throughout life (Beidler and Smallman, 1965), with an average life span of about 1–2 weeks (Barlow and Klein, 2015; Perea-Martinez et al., 2013). This rapid turnover requires continuous generation of new taste cells by taste stem/progenitor cells, and interruption of this process can lead to an altered sense of taste (Barlow and Klein, 2015).
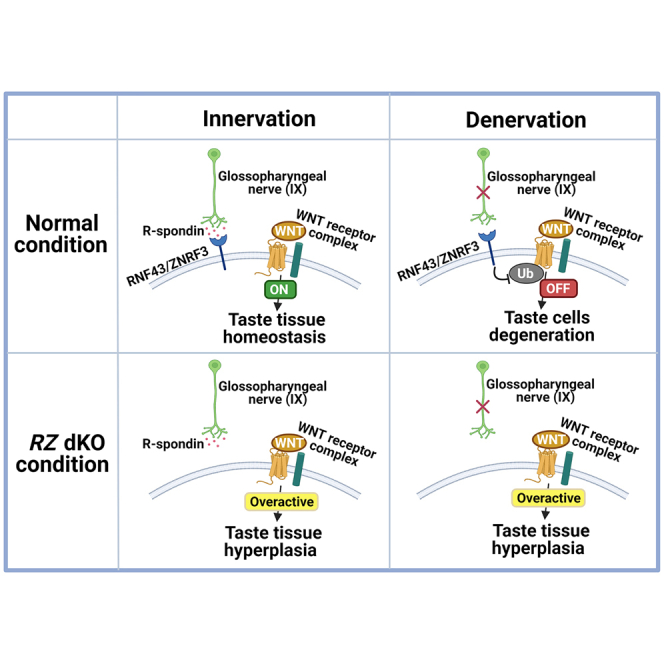
Taste tissue homeostasis is regulated, in part, by gustatory (taste-related) neurons. A well-known phenomenon described more than a century ago is that cutting the gustatory nerve leads to degeneration of taste buds; once the nerve has healed, taste buds regenerate (Cheal and Oakley, 1977; Guth, 1958; Olmsted, 1921; Vintschgau and Hönigschmied, 1876). Previously, we and others demonstrated that the protein LGR5 uniquely marks adult taste stem/progenitor cells (Aihara et al., 2015; Ren et al., 2014, 2017; Takeda et al., 2013; Yee et al., 2013). Recently, we showed that R-spondin, the ligand for the LGR4/5/6 receptors, as well as for the RNF43/ZNRF3 receptors, can substitute for neuronal input for taste cell regeneration, suggesting that one or more members of the R-spondin family (e.g., RSPO2) may be the gustatory-neuron-derived factor that regulates LGR5-expressing taste stem/progenitor cells to maintain taste tissue homeostasis (Lin et al., 2021).
R-spondin interacts both with LGR4/5/6 and with the two E3 ligases RNF43/ZNRF3 to regulate stem cell activity (de Lau et al., 2011, 2014; Hao et al., 2012; Koo et al., 2012). In intestinal epithelial stem cells, RNF43/ZNRF3 act as negative regulators of WNT signaling, whereas LGRs act as positive regulators (de Lau et al., 2014). Loss of Rnf43/Znrf3 in intestinal epithelial tissue leads to overgrowth of gut epithelial tissue (hyperplasia) (Koo et al., 2012), whereas loss of Lgr4/5 leads to abolishment of the crypt stem cell compartment (de Lau et al., 2011). This ternary interaction among LGRs, RNF43/ZNRF3, and R-spondin regulates WNT signaling and thus either promotes or retards stem cell proliferation and differentiation (de Lau et al., 2014).
Despite these recent advances in understanding the R-spondin–LGR4/5/6–RNF43/ZNRF3 pathway, its role in taste stem cell regulation and other tissues is as yet undefined. Here we show (1) that Rnf43/Znrf3 is expressed in the lingual and taste stem/progenitor cell compartments (i.e., basal cells of lingual and taste epithelium where stem/progenitor cells reside); (2) that ablation of Rnf43/Znrf3 in lingual epithelium, including taste stem/progenitor cells, leads to taste cell hyperplasia but lingual epithelial cell degeneration; and (3) that the effects of RNF43/ZNRF3 on both lingual and taste epithelium appear to be mediated by WNT signaling, but in opposite directions. Importantly, taste cell hyperplasia in Rnf43/Znrf3 double-knockout (RZ dKO) mice resembled the effect of exogenous R-spondin we previously showed (Lin et al., 2021): regeneration of taste cells occurred in the RZ dKO mice even in the absence of nerve input (i.e., after nerve transection).
We thus propose the following model: in the oral cavity, RNF43/ZNRF3 regulates activity of stem/progenitor cells through WNT signaling, in a tissue-dependent pattern. In taste epithelium, gustatory-neuron-derived R-spondin interacts with taste stem/progenitor-cell-expressed RNF43/ZNRF3 to regulate taste stem cell activity: RNF43/ZNRF3 acts as a brake for adult taste stem/progenitor cells; R-spondin releases the RNF43/ZNRF3 brake to allow stem cells to become active to generate mature taste cells. In contrast, in lingual epithelial tissues, RNF43/ZNRF3 appears to be required for maintaining lingual epithelial tissue, serving a promoting rather than braking function.
Results
Rnf43 and Znrf3 are expressed in the taste and lingual basal cell compartments, where stem/progenitor cells are typically located
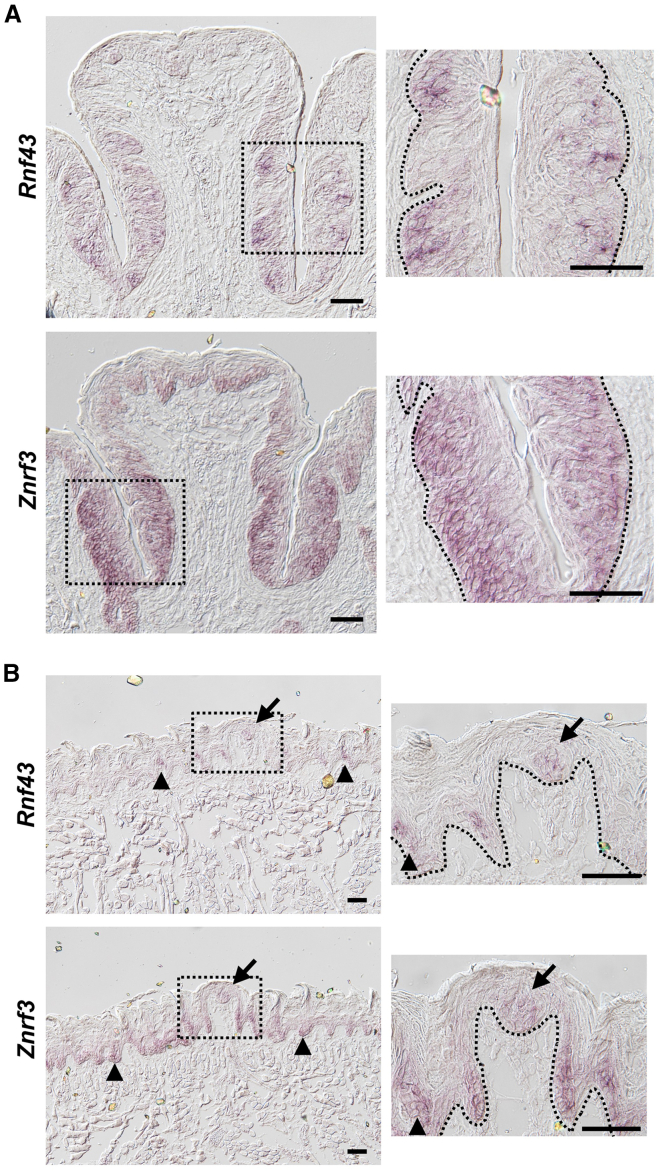
Previous transcriptome analysis of taste tissue demonstrated the presence of both Rnf43 and Znrf3 in taste tissue (Hevezi et al., 2009). In other tissues, Rnf43/Znrf3 are often found in the stem/progenitor cell compartment and appear to serve redundant functions (Koo et al., 2012). To determine whether Rnf43 and Znrf3 are expressed in taste stem/progenitor cells, we performed in situ hybridization using antisense riboprobes. We found that both Rnf43 and Znrf3 are expressed in the basal cell compartment underneath taste buds in the circumvallate papilla; Rnf43 distribution appears to be more restricted in the basal cell compartment (e.g., only underlying taste buds) (Figure 1A). Nevertheless, some apparent signals were detected even within taste buds outside the basal cell compartment for both Rnf43 and Znrf3 (Figure 1A). In anterior tongue, we detected Rnf43 and Znrf3 in the basal cell compartment underneath taste buds in the fungiform papillae (arrow), as well as in the lingual epithelial layer (i.e., in filiform papillae, which do not contain taste buds; arrowhead) (Figure 1B). Again, Rnf43 distribution appears to be more restricted in the basal cell compartment than Znrf3. Furthermore, both Rnf43 and Znrf3 in situ signals were detected within taste buds as well. Given their expression pattern and their established function in stem cells in other tissues, both molecules may function in both taste and lingual stem/progenitor cells.
Figure 1.
Expression of Rnf43 and Znrf3 in the circumvallate papillae, fungiform papillae, and surrounding lingual epithelium
(A and B) In situ hybridization of Rnf43 and Znrf3 with antisense riboprobes in circumvallate papilla in posterior tongue (A) and in fungiform papillae and the surrounding lingual epithelium (e.g., filiform papillae) in anterior tongue (B). Dotted lines in insets demarcate epithelial-mesenchymal boundaries. Arrows, fungiform papillae; arrowheads, filiform papillae. Scale bars, 50 μm.
Ablation of Rnf43/Znrf3 leads to a wrinkled and thinner dorsal lingual surface with reduced numbers of proliferating lingual cells
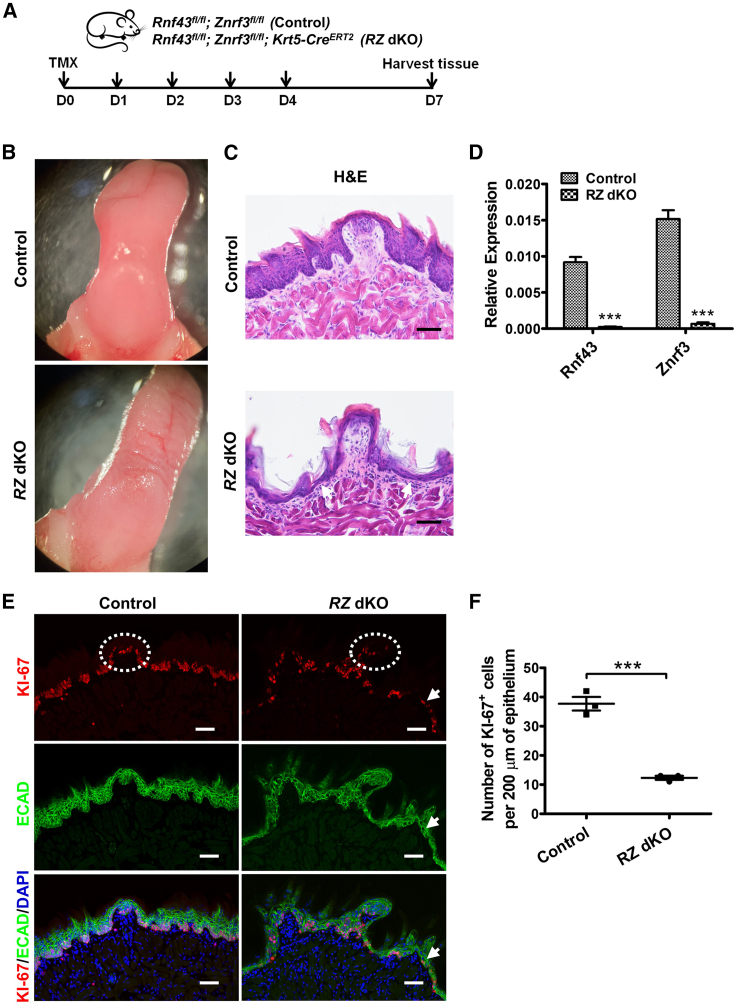
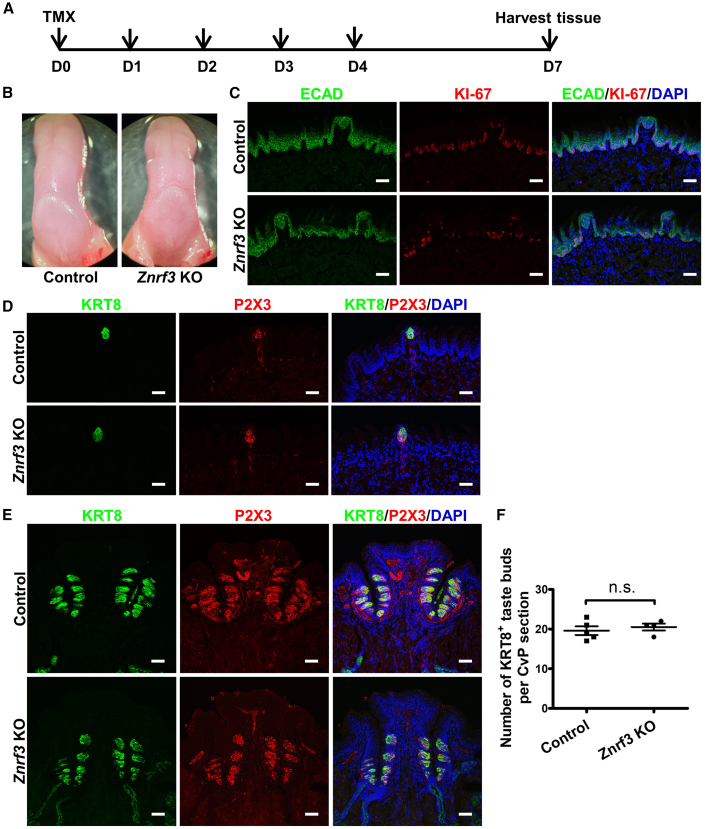
To determine if RNF43/ZNRF3 act as regulators for taste tissue homeostasis, as demonstrated in intestinal epithelium, we designed a strategy to specifically ablate these two molecules in KRT5-expressing epithelial tissue. This strategy was used previously for investigating taste tissue maintenance (Ohmoto et al., 2020). To generate the RZ dKO mice, we injected Krt5CreERT2/+;Rnf43fl/fl;Znrf3fl/fl mice with tamoxifen for five consecutive days and analyzed taste tissues at day 7 (Figure 2A). After day 7, mice showed significant body weight loss and lethality, preventing us from further analysis. The gross morphology of the tongue differed between RZ dKO and control mice (Rnf43fl/fl;Znrf3fl/fl mice, also treated with tamoxifen) (Figure 2B). As typically observed in normal wild-type mice, in control mice the dorsal surface of the anterior tongue was smooth (Figures 2B and 2C). However, in the RZ dKO mice, the dorsal surface was uneven, with shrinkage in the anterior field (Figures 2B and 2C). In contrast, the ventral surface appeared to be normal, suggesting that ablation of Rnf43/Znrf3 led to dorsal lingual epithelial tissue atrophy. We confirmed the ablation of these two genes by quantitative RT-PCR (qRT-PCR) (Figure 2D).
Figure 2.
Ablation of Rnf43/Znrf3 leads to a wrinkled tongue and thinner tongue epithelium
(A) Schematic illustration of the experimental design. Control (Rnf43fl/fl;Znrf3fl/fl) and Rnf43fl/fl;Znrf3fl/fl;Krt5-CreERT2 mice were treated with tamoxifen for five continuous days (D0–D4), and tongue tissue was collected at day 7. TMX, tamoxifen.
(B) Representative bright-field images of tongues collected from control (n = 3) and RZ dKO (n = 3) mice.
(C) Representative images of hematoxylin and eosin (H&E) staining of anterior tongue sections from control and RZ dKO mice. Note the thinner lingual epithelium (arrows) in RZ dKO mice. n = 3 for each group. Scale bars, 50 μm.
(D) qRT-PCR analysis of the expression of Rnf43 and Znrf3 in tongue epithelium in control and RZ dKO mice. Expression levels of Rnf43 and Znrf3 were normalized to Gapdh. Data are presented as mean ± SEM. ∗∗∗p < 0.0001. n = 3 for control group and n = 4 for RZ dKO group.
(E) Immunofluorescence staining of sections of anterior tongues of control and RZ dKO mice for KI-67 (red) and E-cadherin (ECAD, green). We frequently noted a single layer of sparse KI-67+ cells in the dorsal lingual epithelium in RZ dKO sections (arrow), but not in sections from control mice. Dashed circles show fungiform papillae. Cell nuclei were counterstained with DAPI (blue). n = 3 for each group. Scale bars, 50 μm.
(F) Tabulation of KI-67+ cells in the dorsal lingual epithelium. Fewer KI-67+ cells are present in the dorsal lingual epithelium of RZ dKO mice than of control mice. Data are presented as mean ± SEM. ∗∗∗p = 0.0005. n = 3 for each group. See also Figures S1.
We further analyzed the anterior tongue using the cell proliferation marker KI-67 and epithelial cell marker E-cadherin (ECAD). Control mice had two to three layers of KI-67+ cells at the basal part of the dorsal tongue epithelium, and multiple layers of ECAD+ cells (Figure 2E). In contrast, in RZ dKO mice, only a single layer of sparsely distributed KI-67+ cells and a thin layer of ECAD+ cells were detected in a large portion of the dorsal lingual epithelium, which had filiform (nontaste) papillae but was devoid of fungiform (taste) papillae (Figure 2E, arrow). We found significantly fewer KI-67+ cells in the dorsal lingual epithelium of RZ dKO mice than in control mice (Figure 2F). These results suggest that ablating Rnf43/Znrf3 can affect dorsal lingual epithelial stem cell proliferation. In addition, the epithelium became much thinner, possibly because stem cells became quiescent in the absence of Rnf43/Znrf3 (Figures 2C, 2E, and S1). In contrast to the dorsal lingual epithelium, the distribution pattern of KI-67+ cells appeared to be comparable between control and RZ dKO mice in fungiform papillae (Figure 2E, circle). Although the configuration of fungiform taste buds makes quantifying the number of KI-67+ cells difficult, RNF43/ZNRF3 appears to have distinct effects on lingual epithelium stem cells and taste stem cells on the dorsal tongue. At the same time, we noted thinner dorsal lingual epithelium, suggesting that lingual epithelium degenerated in the dKO mice, presumably due to slower or arrested proliferation/differentiation of dorsal lingual stem cells.
Conditional ablation of Rnf43/Znrf3 leads to taste tissue hyperplasia in the fungiform papillae and in the circumvallate papilla
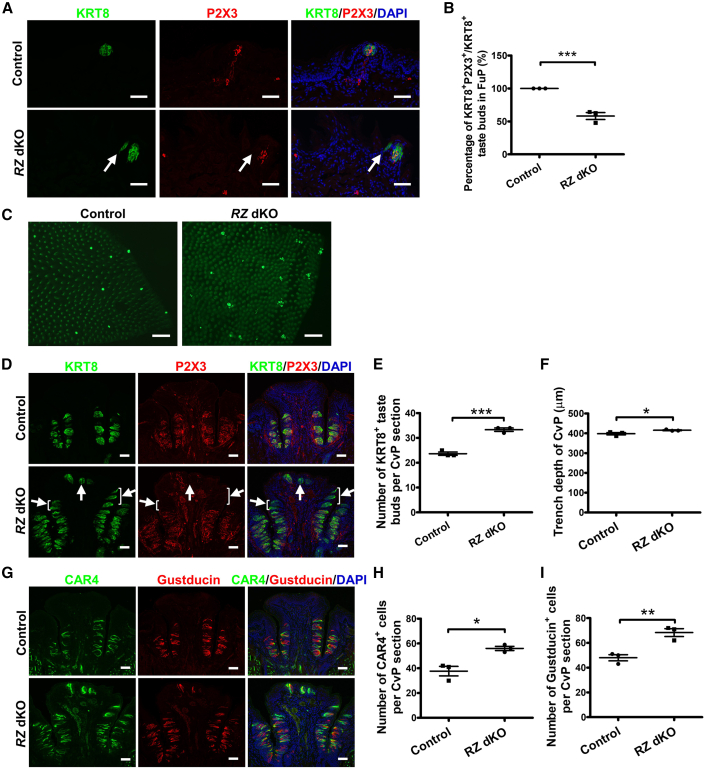
To determine if loss of Rnf43/Znrf3 affects taste tissue homeostasis, we immunostained thin sections from RZ dKO mice and control mice using antibodies against the taste cell markers KRT8, α-Gustducin, and CAR4, along with the taste neuron marker P2X3 to label gustatory nerve terminals. In control mice, as expected, KRT8+ taste buds in fungiform papillae were all innervated by P2X3+ terminals (Figure 3A). In contrast, small KRT8+ taste buds that were not innervated by P2X3+ nerve fibers (ectopically expressed taste bud cells) were frequently found near fungiform taste buds in dKO mice but never in control mice (Figures 3A and 3B).
Figure 3.
Deletion of Rnf43/Znrf3 leads to taste tissue hyperplasia in the fungiform papillae and circumvallate papilla
(A) Immunofluorescence staining for KRT8 (green) and P2X3 (red) in fungiform papillae. Ectopic (uninnervated) taste bud cells (arrow) are present near fungiform taste buds in dKO mice. Cell nuclei were counterstained with DAPI (blue). n = 3 for each group. Scale bars, 50 μm.
(B) Percentage of KRT8+ taste buds that are also positive for P2X3 (i.e., innervated) in fungiform papillae (FuP). Data are presented as mean ± SEM. ∗∗∗p = 0.0001. n = 3 for each group. Each point represents a single mouse.
(C) Representative images of KRT8 immunostaining of whole-mount tongue epithelium in the tip of the tongue. Unlike the single KRT8+ bright spot (i.e., KRT8+ taste bud) in each fungiform papilla in control mice (n = 3) shown in (A), here a few small scattered spots are visible in some fungiform papillae in RZ dKO mice (n = 3). Scale bars, 200 μm.
(D) Representative images of KRT8 and P2X3 immunostaining of circumvallate papilla sections from control and RZ dKO mice. KRT8+ ectopic taste buds (arrows) are present in the upper cleft and dorsum of the circumvallate papilla in RZ dKO mice. Cell nuclei were counterstained with DAPI (blue). n = 3 for each group. Scale bars, 50 μm.
(E) Number of KRT8+ taste buds in the circumvallate papilla from control and RZ dKO mice. Data are presented as mean ± SEM. ∗∗∗p = 0.0005. n = 3 for each group. Each point represents a single mouse. CvP, circumvallate papilla.
(F) Analysis of the depth of trench of the circumvallate papilla in control and RZ dKO mice. Note the small variation of the depth of trench of the circumvallate papilla within the group. Data are presented as mean ± SEM. ∗p = 0.049. n = 3 for each group. CvP, circumvallate papilla.
(G) Immunofluorescence staining for CAR4 (green) and α-Gustducin (red) of circumvallate papilla sections. Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(H and I) Numbers of CAR4+ cells (H) and α-Gustducin+ cells (I) show proportionate increases in type II and III taste cell with loss of Rnf43 and Znrf3 in circumvallate papilla. Data are presented as mean ± SEM. ∗p = 0.0122, ∗∗p = 0.0074. n = 3 for each group. Each point represents a single mouse. See also Figures S2.
Because fungiform taste buds are scattered throughout the dorsal surface of the tongue, sectioning them may not fully identify the distribution of ectopically expressed taste bud cells. We therefore adopted a different strategy to visualize fungiform taste buds and ectopic taste buds in a whole-mount preparation, using peeled tongue epithelium, which was previously described in Lu et al. (2018). After immunostaining the epithelium using the anti-KRT8 antibody, we immediately noted satellite ectopic cells near prototypical fungiform taste buds, consistent with results we obtained from thin-section staining, but in striking contrast to the pattern in control mice (Figures 3C and S2). The generation of ectopic taste buds near fungiform papillae in RZ dKO mice suggests that RNF43/ZNRF3 act as negative regulators for taste tissue homeostasis and that deleting them promotes generation of additional taste buds and taste bud cells (e.g., satellite ectopic taste buds without gustatory innervation).
Next, we examined the circumvallate papillae in the posterior tongue. We observed numerous taste buds in the lateral trench wall, upper cleft region, and the dorsal surface of the circumvallate papillae in dKO mice, with a distribution pattern sharply distinct from control mice (Figure 3D). KRT8+ taste cells, with a typical elongated shape, cluster with each other, which makes counting cells difficult. We therefore counted and compared taste buds in RZ dKO and control mice. In each section examined, the number of taste buds in RZ dKO mice was ∼40% more than that in control mice (Figure 3E). However, the depth of the trench appears to be only slightly greater (4.4%) in RZ dKO than in control mice (control group, 398.2 ± 5.865 μm; RZ dKO group, 415.7 ± 2.085 μm; Figure 3F). The increased number of taste buds and taste bud cells is consistent with the presence of satellite taste buds we observed in anterior tongue. In addition, some taste buds in the dorsal surface and upper cleft of the circumvallate papillae appeared to be ectopic, as they had no P2X3+ innervation.
Similar to KRT8+ cells, α-Gustuducin+ and CAR4+ cells were also increased in RZ dKO mice compared with control mice (Figures 3G–3I). However, we observed no apparent difference in the distribution pattern of KI-67+ cells in the circumvallate papillae between RZ dKO mice and control mice (Figure S3).
More importantly, we previously demonstrated that exogenous R-spondin can substitute for neuronal input for taste cell generation (Lin et al., 2021). In the present study, the pattern of taste buds we observed in RZ dKO mice was strikingly similar to what we previously observed in mice receiving R-spondin, with or without glossopharyngeal nerve transection, including the presence of ectopic taste buds in the dorsal surface and upper cleft region of the circumvallate papillae (Lin et al., 2021). Thus, either overexpression of R-spondin or knockout of Rnf43/Znrf3, the receptor for R-spondin with inhibitory effects, can lead to a nearly identical phenotype, suggesting that neuron-derived R-spondin and its stem/progenitor-cell-expressed RNF43/ZNRF3 receptors interact to govern taste tissue homeostasis.
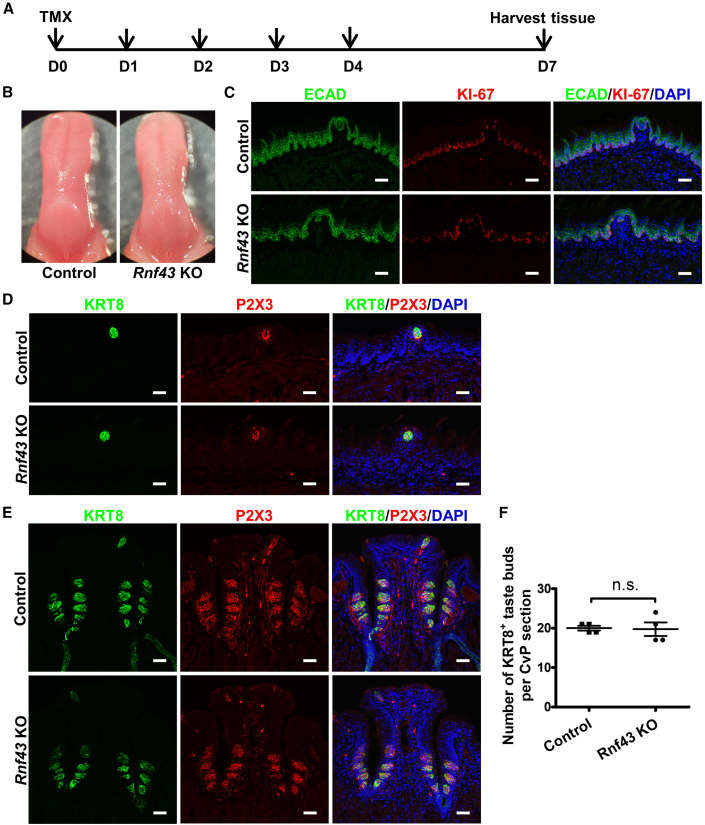
To determine if deletion of either Rnf43 or Znrf3 would produce a phenotype similar to deletion of both Rnf43/Znrf3, we generated Krt5CreERT2/+;Rnf43fl/fl and Krt5CreERT2/+;Znrf3fl/fl mice and examined taste tissues and lingual epithelial tissues upon tamoxifen injection (dosing scheme was the same as that for RZ dKO mice). Unlike in RZ dKO mice, in single-knockout mice no apparent taste tissue hyperplasia occurred. Rnf43fl/fl or Znrf3fl/fl mice treated with tamoxifen were used as controls. However, mild degeneration of the dorsal lingual epithelium was noted in Rnf43 or Znrf3 single-knockout mice (Figures 4 and 5). This set of data suggests that RNF43 and ZNRF3 have redundant roles in regulating taste tissue homeostasis and that they have similar but nonredundant roles in regulating homeostasis of dorsal lingual epithelial tissue (e.g., filiform papillae).
Figure 4.
Mice deficient for Rnf43 show no detectable changes in taste tissues and mild changes in the dorsal lingual epithelium
(A) Schematic illustration of the experimental design.
(B) Representative bright-field images of tongues collected from control and single Rnf43 KO mice.
(C) Immunofluorescence staining of anterior tongue sections of control and single Rnf43 KO mice for E-cadherin (ECAD, green) and KI-67 (red). Note that KI-67+ cells are fewer in Rnf43 KO mice than control mice. Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(D and E) Immunofluorescence staining for KRT8 (green) and P2X3 (red) of anterior tongue sections (D) and circumvallate papilla sections (E) from control and single Rnf43 KO mice. There are no apparent changes in the KRT8 and P2X3 staining patterns. Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(F) Number of KRT8+ taste buds in the circumvallate papilla from control and single Rnf43 KO mice. Data are presented as mean ± SEM. n.s., not significant. n = 4 for each group. Each point represents a single mouse. CvP, circumvallate papilla.
Figure 5.
Mice deficient for Znrf3 show no detectable changes in taste tissues and mild changes in the dorsal lingual epithelium
(A) Schematic illustration of the experimental design.
(B) Representative bright-field images of tongues collected from control and single Znrf3 KO mice.
(C) Immunofluorescence staining of anterior tongue sections of control and single Znrf3 KO mice for E-cadherin (ECAD, green) and KI-67 (red). Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(D and E) Immunofluorescence staining for KRT8 (green) and P2X3 (red) of anterior tongue sections (D) and circumvallate papilla sections (E) from control and single Znrf3 KO mice. Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(F) Number of KRT8+ taste buds in the circumvallate papilla from control and single Znrf3 KO mice. Data are presented as mean ± SEM. n.s., not significant. n = 5 for control group and n = 4 for Znrf3 KO group. Each point represents a single mouse. CvP, circumvallate papilla.
To further determine if the observed atrophic phenotype in the dorsal lingual epithelium of RZ dKO mice was due to potential exhaustion of stem/progenitor cells from a rapid increase in proliferation of stem/progenitor cells at the early phase (Gaillard et al., 2015), we examined taste tissues and dorsal lingual epithelial tissue (e.g., filiform papillae) 2 days after the first of the two tamoxifen injections (the second was performed 1 day after the first). No apparent changes were detected in the lingual epithelium (e.g., KI-67 staining) or in taste organs (Figure S4). Therefore, it seems that stem/progenitor cell exhaustion at the early phase in RZ dKO mice is unlikely.
Taste cell hyperplasia remains in RZ dKO mice in the absence of nerve input
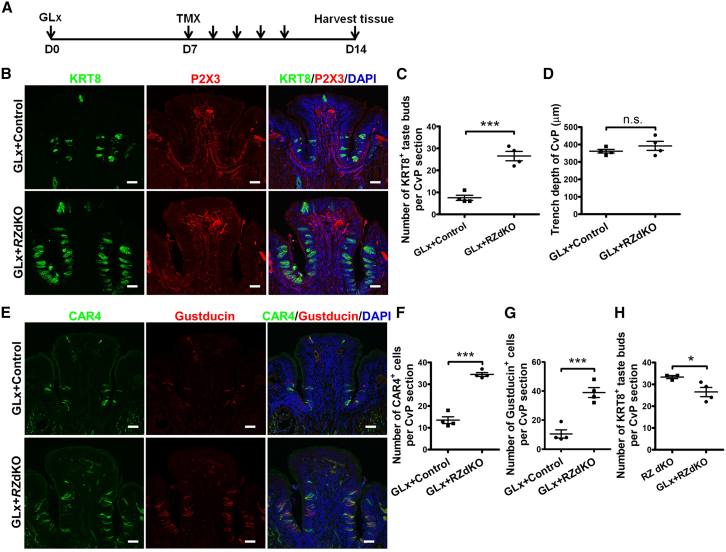
We reasoned that, if the principal activity of neuron-derived R-spondin is mediated by taste stem/progenitor-cell-expressed RNF43/ZNRF3, then removing the brake function of RNF43/ZNRF3 should render neuronal input unnecessary for taste tissue maintenance. To this end, we first performed bilateral glossopharyngeal nerve transection (GLx) of control (Rnf43fl/fl;Znrf3fl/fl) and Krt5CreERT2/+;Rnf43fl/fl;Znrf3fl/fl mice. Seven days after GLx, we induced ablation of Rnf43 and Znrf3 by injecting Krt5CreERT2/+;Rnf43fl/fl;Znrf3fl/fl mice with tamoxifen for five consecutive days, and 2 days later we harvested tissue (Figure 6A). As before, control mice received tamoxifen injection as well. Circumvallate papilla sections were then examined using antibodies against the taste cell markers KRT8, α-Gustducin, and CAR4 and the taste nerve marker P2X3. As predicted, after GLx, KRT8+ taste bud cells were present in RZ dKO mice but reduced dramatically in control mice (Figure 6B), demonstrating that taste bud cells regenerated in RZ dKO mice despite denervation. Counting the number of KRT8+ taste buds showed more taste buds in GLx RZ dKO mice than in GLx control mice (Figure 6C). It appeared that the trench was slightly deeper in GLx RZ dKO mice than in GLx control mice, albeit not significantly (control group, 361.8 ± 10.61 μm; RZ dKO group, 392.0 ± 25.97 μm; Figure 6D). Similarly, significantly more α-Gustducin+ and CAR4+ cells were present in GLx RZ dKO mice than in GLx control mice (Figures 6E–6G). These results further support the idea that neuronal input with R-spondin regulates taste tissue homeostasis by releasing the brake function of RNF43/ZNRF3. Interestingly, when we compared the number of taste buds in GLx RZ dKO mice (from Figure 6C) and in RZ dKO mice (from Figure 3E), we noted that there were more taste buds in RZ dKO than in GLx RZ dKO mice (p = 0.044; Figure 6H), which suggests that nerve-supplied R-spondin may interact with taste stem-cell-expressed LGR4/5/6 as well, to further promote taste cell generation in the absence of Rnf43 and Znrf3.
Figure 6.
Rnf43/Znrf3 dKO promotes maintenance of taste bud after glossopharyngeal nerve transection (GLx)
(A) Schematic illustration of the experimental design. Bilateral GLx was performed at day 0 (D0), followed by tamoxifen induction of conditional deletion of Rnf43/Znrf3 at day 7. Tissues were collected at day 14. TMX, tamoxifen.
(B) Representative images of KRT8 and P2X3 immunostaining of circumvallate papilla sections from control and RZ dKO mice. Only a few residual KRT8+ taste cells are present in control mice; in contrast, numerous KRT8+ taste cells have regenerated in RZ dKO mice. Little or no P2X3 staining is present in taste tissues in either control or RZ dKO mice, although P2X3 signal is detected in the mesenchymal core. n = 4 for each group. Scale bars, 50 μm.
(C) Number of KRT8+ taste buds in the circumvallate papilla from control and RZ dKO GLx mice. Data are presented as mean ± SEM. ∗∗∗p = 0.0002. n = 4 for each group. Each point represents a single mouse. CvP, circumvallate papilla.
(D) Analysis of the depth of trench of the circumvallate papilla in GLx + Control and GLx + RZ dKO mice. Data are presented as mean ± SEM. n.s., not significant. n = 4 for each group.
(E) Immunofluorescence staining for CAR4 (green) and α-Gustducin (red) of circumvallate papilla sections. Few residual CAR4+ or α-Gustducin+ cells are present in control mice after GLx, compared with multiple CAR4+ or α-Gustducin+ cells in RZ dKO mice after GLx. Cell nuclei were counterstained with DAPI (blue). Scale bars, 50 μm.
(F and G) Numbers of CAR4+ taste cells (F) and α-Gustducin+ taste cells (G) in circumvallate papilla in RZ dKO and control mice after GLx. Data are presented as mean ± SEM. ∗∗∗p < 0.0001 in (F) and p = 0.0007 in (G). n = 4 for each group. Each point represents a single mouse.
(H) Number of KRT8+ taste buds in the circumvallate papilla from RZ dKO and GLx RZ dKO mice. Data are presented as mean ± SEM. ∗p = 0.0437. n = 3 for RZ dKO group and n = 4 for GLx RZ dKO group. Each point represents a single mouse. CvP, circumvallate papilla.
WNT signaling blockade prevents taste cell hyperplasia and lingual epithelium degeneration in RZ dKO mice
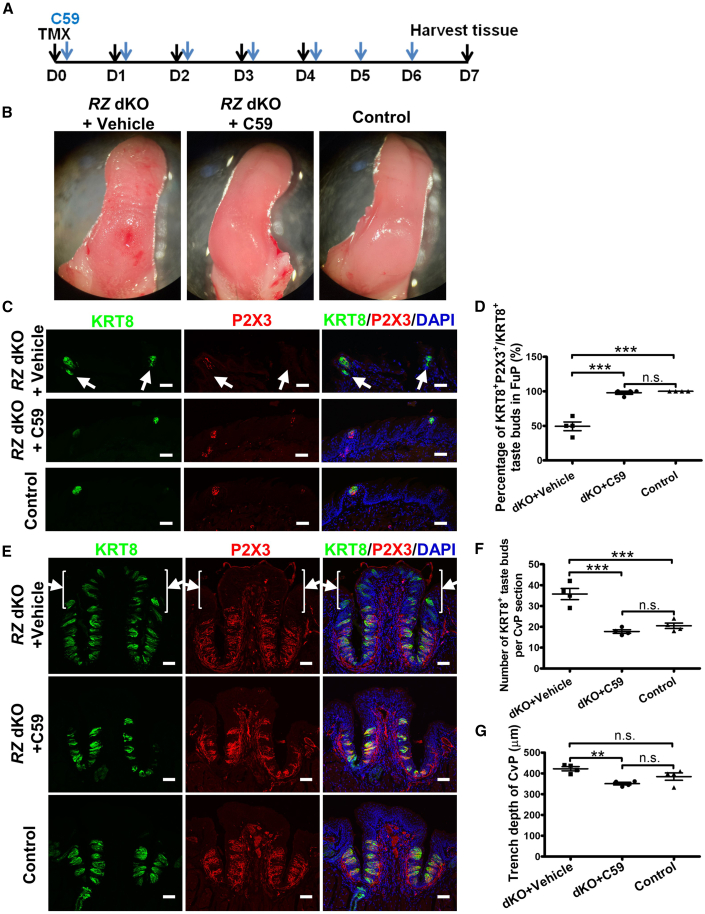
Mechanistically, the R-spondin-LGR4/5/6-RNF43/ZNRF3 pathway is known to augment WNT signaling (Hao et al., 2012; Koo et al., 2012, 2015), thus generally promoting stem cell activity. Because we noted the apparent opposite effects of ablation of Rnf43/Znrf3—promoting taste bud and taste cell expansion yet inhibiting dorsal lingual epithelium (e.g., filiform) renewal—we asked whether the WNT signaling pathway is involved in mediating the effect of RNF43/ZNRF3 in both tissues. If so, we would expect that blocking WNT signaling would inhibit both taste cell hyperplasia and lingual epithelium degeneration in RZ dKO mice. To inhibit WNT signaling, we treated RZ dKO mice (injected with tamoxifen once a day for five consecutive days) by gavaging C59 (or vehicle solution as control) on a daily basis for 7 days (Figure 7A) (Koo et al., 2015). C59 inhibits porcupine, a membrane-bound O-acyltransferase necessary for WNT activation (e.g., palmitoylation, secretion, and biological activity) (Takada et al., 2006). In RZ dKO mice treated with vehicle, the dorsal lingual surface displayed a profound phenotypic change, with a highly wrinkled and thinner surface. In contrast, in dKO mice treated with C59, the dorsal lingual surface showed normal gross morphology, comparable with control mice (Rnf43fl/fl;Znrf3fl/fl, receiving tamoxifen and vehicle control) (Figure 7B).
Figure 7.
WNT signaling blockade prevents taste tissue hyperplasia and lingual epithelium degeneration in RZ dKO mice
(A) Schematic illustration of the experiment design. Rnf43fl/fl;Znrf3fl/fl and Rnf43fl/fl;Znrf3fl/fl;Krt5-CreERT2 mice were treated with tamoxifen for five continuous days and C59 for seven continuous days, and tongue tissue was harvested at day 7. TMX, tamoxifen.
(B) Representative bright-field images of the tongue (dorsal surface) from RZ dKO mice with or without C59 treatment and from control mice. The dorsal surface of RZ dKO mice that received C59 treatment reverted to a normal, smooth appearance. n = 4 for each group.
(C) Immunofluorescence staining of KRT8 (green) and P2X3 (red) of anterior tongue sections. Ectopic taste bud cells (arrows) are present in dKO mice treated with vehicle but not in dKO mice treated with C59. Cell nuclei were counterstained with DAPI (blue). n = 4 for each group. Scale bars, 50 μm.
(D) Percentage of KRT8+ taste buds that are also positive for P2X3 (i.e., innervated) in fungiform papillae (FuP). Data are presented as mean ± SEM. ∗∗∗p < 0.0001. n.s., not significant. n = 4 for each group. Each point represents a single mouse.
(E) Immunofluorescence staining of KRT8 (green) and P2X3 (red) of circumvallate papilla sections. Ectopic taste bud cells (arrows) are present in the upper cleft and dorsum of the circumvallate papilla in RZ dKO mice treated with vehicle but not in dKO mice treated with C59. Cell nuclei were counterstained with DAPI (blue). n = 4 for each group. Scale bars, 50 μm.
(F) Number of KRT8+ taste buds in the circumvallate papilla in RZ dKO mice with or without C59 treatment and in control mice. Data are presented as mean ± SEM. ∗∗∗p < 0.001. n.s., not significant. n = 4 for each group. Each point represents a single mouse. CvP, circumvallate papilla.
(G) Analysis of the depth of trench of the circumvallate papilla in RZ dKO mice receiving vehicle or C59 or in control mice. Data are presented as mean ± SEM. ∗∗p = 0.0083. n.s., not significant. n = 4 for each group.
See also Figures S5–S7.
Using the antibodies against taste cell markers (KRT8, α-Gustducin, and CAR4) and taste nerves (P2X3), we found that C59 reversed the phenotypic changes in taste tissue in RZ dKO mice. For instance, KRT8+ ectopic taste buds (lacking P2X3 innervation) were eliminated, and taste hyperplasia was subdued in fungiform papillae (Figures 7C and 7D) and in circumvallate papillae (Figures 7E and 7F). Meanwhile, the depth of the trench was significantly decreased in RZ dKO + C59 mice compared with RZ dKO + vehicle mice, but no significant difference was noted between RZ dKO + vehicle mice and control mice (RZ dKO + vehicle group, 422.2 ± 10.04 μm; RZ dKO + C59 group, 350.6 ± 6.238 μm; control group, 384.5 ± 17.62 μm; Figure 7G). However, there was a trend that the trench was slightly deeper in RZ dKO mice than in control mice (also see Figure 3F). Moreover, we noted taste bud cell (e.g., α-Gustducin+, CAR4+) degeneration in circumvallate papillae in C59-treated RZ dKO mice (Figure S5), although to a lesser extent than observed after GLx nerve transection (see Figures 6E–6G). Furthermore, proliferation of lingual epithelial stem cells was also partly rescued in anterior tongue as demonstrated by KI-67 staining (Figure S6). These results suggest that, in both dorsal lingual epithelium and taste epithelium, the activity of RNF43/ZNRF3 is mediated by WNT signaling, despite our observations of the exact opposite effects of RNF43/ZNRF3 in the two tissues.
To corroborate our finding that RNF43/ZNRF3 modulates WNT signaling, we also performed immunostaining of lymphoid enhancer-binding factor 1 (LEF1), a key mediator of the WNT signaling pathway (Iwatsuki et al., 2007; Liu and Millar, 2010). As expected, increased expression of LEF1 was detected in basal cells in the circumvallate papillae in RZ dKO mice, but decreased expression of LEF1 was found in basal cells in the dorsal lingual epithelium (e.g., filiform papillae) (Figure S7), consistent with the idea that RNF43/ZNRF3 modulates WNT signaling in a context-dependent manner in taste and lingual epithelia. C59 treatment blunted the increase of LEF1 expression in the circumvallate papillae and the decrease of LEF1 expression partially in the dorsal lingual epithelium (Figure S7).
Discussion
In the present work, using in situ hybridization, we demonstrated the presence of Rnf43/Znrf3 in basal cells in lingual epithelial tissue and taste tissue. Using a tissue-specific knockout mouse, we demonstrated that RNF43/ZNRF3 act as negative regulators to maintain taste tissue homeostasis and positive regulators to maintain lingual epithelial tissue homeostasis. Furthermore, we demonstrated that gustatory neurons regulate taste tissue homeostasis by removing the inhibitory effect of RNF43/ZNRF3 that keeps taste stem/progenitor cell quiescent, via neuron-supplied R-spondin. Mechanistically, RNF43/ZNRF3 appear to regulate WNT signaling in both taste and lingual epithelial tissues.
RNF43/ZNRF3 as negative regulators for taste tissue homeostasis
Three pieces of evidence support RNF43/ZNRF3 as negative regulators for taste tissue homeostasis. First, epithelial-specific deletion of Rnf43/Znrf3 in KRT5+ (marks basal cells in epithelial tissues including taste tissue) cells led to taste cell hyperplasia in circumvallate papillae and fungiform papillae, as evidenced by increased numbers of taste buds and taste cells (e.g., CAR4+ and α-Gustducin+ cells). Importantly, de novo generation of taste buds and taste cells in RZ dKO mice occurred ectopically, without apparent innervation as determined by P2X3 staining, such as those observed in the dorsal region and upper cleft of the circumvallate papillae and satellite ectopic taste buds in fungiform papillae. However, we cannot completely exclude the possibility that existing taste cells may persist longer and thus may potentially contribute to more taste cells observed in RZ dKO mice. Second, neuronal input was not necessary for taste tissue renewal in RZ dKO mice. In normal mice, taste tissue homeostasis requires neuronal input and gustatory nerve transection leads to degeneration of taste cells and taste buds, which do not regenerate until taste fields are reinnervated. In contrast, in RZ dKO mice, taste cells in the circumvallate papillae regenerated even after GLx. Third, the pattern of taste bud distribution (e.g., ectopic taste buds in the dorsal surface and upper cleft of the circumvallate papillae) is virtually identical in RZ dKO mice (present results) and in mice receiving adenovirus encoding recombinant R-spondin, with or without nerve transection (Lin et al., 2021). In the gut system, R-spondin interacts with RNF43/ZNRF3 to neutralize their E3 ligase activity (Park et al., 2018; Yan et al., 2017). Our data suggest this is also the case in the taste system, where neuron-produced R-spondin (e.g., RSPO2) releases the braking effect exerted by RNF43/ZNRF3 to allow taste stem/progenitor cells to become active and generate taste cells and taste buds. In normal mice, taste buds are generated only in areas that receive projections from gustatory nerve fibers, the source of R-spondin. In contrast, in RZ dKO mice, which lack the braking exerted by RNF43/ZNRF3, taste buds are generated even in areas without gustatory nerve projections.
Taste stem/progenitor cells
Previously, we and others have shown that LGR5 marks taste stem/progenitor cells in posterior tongue (Takeda et al., 2013; Yee et al., 2013). In anterior tongue, transcripts for Lgr4, Lgr5, and Lgr6 can be detected, and LGR6-expressing cells give rise to taste cells as well (Ren et al., 2014). In RZ dKO mice, we noted taste cell hyperplasia and that multiple taste bud cells were generated ectopically. Previously we had noted a similar pattern of taste bud cell distribution in mice receiving adenovirus encoding recombinant R-spondin, with or without nerve transection (Lin et al., 2021). The broad distribution of taste buds in these mice suggests that LGR5-negative taste/progenitor cells are present, that these cells express the E3 ligase RNF43 or ZNRF3, and that these cells are typically quiescent because of a lack of innervation.
One caveat is that we analyzed RZ dKO mice 7 days after the first injection of tamoxifen. Whether taste cell hyperplasia was long-lasting cannot be addressed using our current models, due to lethality of dKO mice (the exact cause of lethality is unknown but it could be partially due to degeneration of lingual epithelial cells) and the short-term nature of effects created by the adenovirus-based method (Wei et al., 2008). It would be intriguing to test the long-term effect of taste tissue hyperplasia, by using either another, more specific Cre driver that is selectively expressed in taste tissues, to potentially avoid lethality of ablating Rnf43/Znrf3, or an AAV-based protein delivery method to provide systemic delivery of recombinant R-spondin (Lahde et al., 2021).
WNT signaling
Our data suggest that the interaction of R-spondin and RNF43/ZNRF3 plays an important role in regulating taste stem/progenitor cell activity. Whether R-spondin also regulates the activity of LGR4/5/6 to augment WNT signaling, similar to the R-spondin–LGRs–RNF43/ZNRF3 pathway in the gut, remains to be tested. However, our comparison of the number of taste buds in the circumvallate papilla between RZ dKO and GLx RZ dKO mice suggests that, aside from interacting with RNF43/ZNRF3, R-spondin may also interact with LGR4/5/6 to promote taste cell generation, as there were more taste buds in RZ dKO than in GLx RZ dKO mice. Regardless, taste cell hyperplasia induced by deleting Rnf43/Znrf3 is apparently dependent on WNT signaling, which has been shown to play a key role in taste tissue homeostasis (Gaillard et al., 2015, 2017). We showed that inhibiting WNT signaling with C59 can abolish taste cell hyperplasia in RZ dKO mice. C59 inhibits porcupine, which is required for WNT palmitoylation, secretion, and biological activity. Therefore, as in the case of intestinal stem cells, RNF43/ZNRF3 is likely to modulate WNT signaling in taste stem/progenitor cells. A number of receptors for WNT (Frizzled receptors) are expressed in taste tissue (Hevezi et al., 2009; Ohmoto et al., 2020), as are a number of WNT molecules (Xu et al., 2017). The exact WNT-FZD pair involved in regulating taste stem/progenitor cells via autocrine or paracrine signaling is largely unknown. However, WNT10A has been shown to regulate taste and nontaste homeostasis in humans and mice (Xu et al., 2017). It is also plausible that the system has built-in redundancy, such that all these pairs can regulate taste stem/progenitor cell activity; thus, simply removing one FZD or WNT may not be sufficient to detect any phenotypic changes in taste tissue homeostasis. Nevertheless, C59 inhibits porcupine, thereby inhibiting WNT signaling triggered by all WNTs. As a consequence, it not only blocks the effect observed in RZ dKO but also appears to suppress normal WNT activity, reducing numbers of taste cells by slowing down stem cell activity. We speculate that longer treatment of mice with C59 (beyond the 7 days in this study) may lead to more severe degeneration of taste buds.
RNF43/ZNRF3 is required for maintaining dorsal lingual epithelial tissue (e.g., filiform papillae) homeostasis
RNF43 and ZNRF3 are known as negative regulators of the WNT signaling pathway for intestinal stem cells (Koo et al., 2012), liver zonation (Planas-Paz et al., 2016), adrenal homeostasis (Basham et al., 2019), and now taste stem cells (present results). Remarkably, we found that, in dorsal lingual epithelium, ablating Rnf43/Znrf3 led not to lingual epithelial cell hyperplasia but, instead, to degeneration of lingual epithelial tissue. The dorsal lingual epithelium is covered with filiform (nontaste) papillae, whereas the ventral lingual epithelium is not. Therefore, it appears that filiform papilla renewal requires RNF43/ZNRF3. C59 can partially ameliorate degeneration of dorsal lingual epithelial tissue induced by loss of Rnf43/Znrf3, suggesting activating WNT signaling may lead to degeneration of lingual epithelium. However, we cannot rule out the possibility that in this context, or even in taste tissues, C59 targets other molecules instead of porcupine. Interestingly, the expression of LEF1, an indicator of WNT signaling (Iwatsuki et al., 2007; Liu and Millar, 2010), was decreased in the dorsal lingual epithelium in the absence of Rnf43/Znrf3, which argues against the idea that activating WNT signaling leads to degeneration of lingual epithelium, instead suggesting that activation of WNT signaling in the dorsal lingual epithelium (i.e., filiform papillae) requires the presence of RNF43/ZNRF3. Paradoxically, C59 can partially reverse the decreased expression of LEF1 after ablating Rnf43/Znrf3. However, as described above, this could be mediated by the blockade of either porcupine or other molecules. Regardless, given the strong phenotype we observed in this study, further study is warranted to understand the molecular basis of the context-dependent effects of RNF43/ZNRF3 on WNT signaling in dorsal lingual epithelium (e.g., filiform papillae) and in taste tissues. Our ongoing proteomic work may lead to identification of potentially distinct target molecules (e.g., different WNT signaling elements) of the two E3 ligases RNF43/ZNRF3 in the dorsal lingual epithelium and in taste tissues.
Experimental procedures
Mice
The Rnf43fl/fl;Znrf3fl/fl mice were supplied by Drs. Hans Clevers and Bon-Kyoung Koo and maintained at the Monell Center. B6N.129S6(Cg)-Krt5tm1.1(cre/ERT2)Blh/J (Krt5CreERT2/+; stock no. 029155) mice and C57BL/6 (stock no. 000664) mice were purchased from Jackson Laboratory. All transgenic mice were maintained on the C57BL/6 genetic background. Mice between 8 and 10 weeks old of both sexes were used for analyses. All experiments were performed under National Institutes of Health guidelines for the care and use of animals in research and approved by the Institutional Animal Care and Use Committee of the Monell Chemical Senses Center.
Tamoxifen and C59 administration
Conditional Rnf43/Znrf3 double-knockout (RZ dKO) mice were generated by crossing Rnf43fl/fl;Znrf3fl/fl mice with Krt5-CreERT2 mice. The Cre enzyme was induced by intraperitoneal injection of tamoxifen (100 mg/kg body weight; Sigma Aldrich, no. T5648) for five consecutive days, unless specified otherwise. Tamoxifen was dissolved in corn oil at a concentration of 20 mg/mL by shaking overnight at 37°C. The porcupine inhibitor C59 (50 mg/kg body weight; Cayman Chemical, no. 16644) was mixed with 0.5% methylcellulose and 0.1% Tween 80 and then administrated by oral gavage for seven consecutive days.
Glossopharyngeal nerve transection
Mice were anesthetized with continuous 2% isoflurane and placed on isothermal heating pads. GLx was performed following procedures described previously (Lin et al., 2021). Details are provided in the supplemental experimental procedures.
Tissue preparation
Mice were euthanized on the indicated days. Tongues were dissected, fixed in 4% (wt/vol) paraformaldehyde for 2 h, cryoprotected with 30% sucrose overnight, and embedded in the frozen O.C.T. compound (Sakura, no. 4583). Cryosections (10 μm) were prepared using a Leica CM3050 S cryostat (Leica Biosystems) and mounted on tissue-adhesive-coated glass slides (Electron Microscopy Sciences, no. 71869-40).
In situ hybridization
In situ hybridization was carried out as described previously (Lin et al., 2021). Additional details are provided in the supplemental experimental procedures.
Tongue epithelium peeling
The procedure was performed following the description previously (Lu et al., 2018). Additional details are provided in the supplemental experimental procedures.
Immunostaining and imaging
Immunostaining was performed essentially as described previously (Lin et al., 2021). Details are provided in the supplemental experimental procedures.
Cell counting
Taste buds were identified as onion-like structures with KRT8+ immunostaining. Type II or III taste cells were identified as visible elongated cell profiles with a clear nucleus with α-Gustducin+ or CAR4+ immunostaining. Details are provided in the supplemental experimental procedures.
RNA isolation and quantitative PCR
Total RNA was extracted from peeled tongue epithelium using the PureLink RNA Micro Kit (Thermo Fisher, no. 12183-016) plus TRIzol (Life Technologies, no. 15596-026) according to the manufacturers’ directions. Details are provided in the supplemental experimental procedures.
Statistical analyses
Data are shown as the mean ± SEM. GraphPad Prism 5 software was used for the graphs and statistical analyses. Student's t test was used to compare difference between two groups, and one-way ANOVA followed by Tukey's test was used to determine differences among three groups. Chi-square test was used to compare difference of taste bud innervation in fungiform papillae.
Author contributions
C.L. and P.J. designed the experiments. C.L., X.L., J.Y., R.X., M.Z., and I.M. performed the experiments. C.L., X.L., R.X., I.M., and P.J. analyzed data and interpreted results of experiments. Y.V.Z., H.W., R.F.M., B.-K.K., and H.C. contributed reagents. P.J. conceived of the work and wrote the manuscript with C.L.
Conflicts of interest
The authors declare no competing interests, with the exception of H.C., who is the inventor on several patents related to organoid technology; cofounder of Surrozen, D1Med, and Xilis; member of the board of directors of Roche/Genentech; and Scientific Advisory Board (SAB) member of Volastra, Decibel, and Merus; his full disclosure is given at https://www.uu.nl/staff/JCClevers/.
Acknowledgments
This work was supported by NIH grants DC010842 (to P.J.), DC018627 (to P.J.), DC015491 (to I.M.), and G20 OD020296 (to Danielle R. Reed). Imaging was performed at the Cell and Developmental Biology Core at the University of Pennsylvania and at the Monell Histology and Cellular Localization Core, which was supported in part by NIH National Institute on Deafness and Other Communication Disorders Core Grant DC011735 (to R.F.M.). The graphical abstract was created with BioRender.com.
Published: January 6, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.12.002.
Supplemental information
References
- Aihara E., Mahe M.M., Schumacher M.A., Matthis A.L., Feng R., Ren W., Noah T.K., Matsu-ura T., Moore S.R., Hong C.I., et al. Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid. Sci. Rep. 2015;5:17185. doi: 10.1038/srep17185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmanov A.A., Beauchamp G.K. Taste receptor genes. Annu. Rev. Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow L.A., Klein O.D. Developing and regenerating a sense of taste. Curr. Top. Dev. Biol. 2015;111:401–419. doi: 10.1016/bs.ctdb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham K.J., Rodriguez S., Turcu A.F., Lerario A.M., Logan C.Y., Rysztak M.R., Gomez-Sanchez C.E., Breault D.T., Koo B.K., Clevers H., et al. A ZNRF3-dependent Wnt/beta-catenin signaling gradient is required for adrenal homeostasis. Genes Dev. 2019;33:209–220. doi: 10.1101/gad.317412.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beidler L.M., Smallman R.L. Renewal of cells within taste buds. J. Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari N., Roper S.D. The cell biology of taste. J. Cell Biol. 2010;190:285–296. doi: 10.1083/jcb.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheal M., Oakley B. Regeneration of fungiform taste buds: temporal and spatial characteristics. J. Comp. Neurol. 1977;172:609–625. doi: 10.1002/cne.901720405. [DOI] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M., et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- de Lau W., Peng W.C., Gros P., Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard D., Bowles S.G., Salcedo E., Xu M., Millar S.E., Barlow L.A. beta-catenin is required for taste bud cell renewal and behavioral taste perception in adult mice. PLoS Genet. 2017;13:e1006990. doi: 10.1371/journal.pgen.1006990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard D., Xu M., Liu F., Millar S.E., Barlow L.A. Beta-catenin signaling biases multipotent lingual epithelial progenitors to differentiate and acquire specific taste cell fates. PLoS Genet. 2015;11:e1005208. doi: 10.1371/journal.pgen.1005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth L. Taste buds on the cat's circumvallate papilla after reinnervation by glossopharyngeal, vagus, and hypoglossal nerves. Anatomical Rec. 1958;130:25–37. doi: 10.1002/ar.1091300104. [DOI] [PubMed] [Google Scholar]
- Hao H.X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H., et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Hevezi P., Moyer B.D., Lu M., Gao N., White E., Echeverri F., Kalabat D., Soto H., Laita B., Li C., et al. Genome-wide analysis of gene expression in primate taste buds reveals links to diverse processes. PLoS One. 2009;4:e6395. doi: 10.1371/journal.pone.0006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki K., Liu H.X., Gronder A., Singer M.A., Lane T.F., Grosschedl R., Mistretta C.M., Margolskee R.F. Wnt signaling interacts with Shh to regulate taste papilla development. Proc. Natl. Acad. Sci. U S A. 2007;104:2253–2258. doi: 10.1073/pnas.0607399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.K., Spit M., Jordens I., Low T.Y., Stange D.E., van de Wetering M., van Es J.H., Mohammed S., Heck A.J., Maurice M.M., Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Koo B.K., van Es J.H., van den Born M., Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc. Natl. Acad. Sci. U S A. 2015;112:7548–7550. doi: 10.1073/pnas.1508113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahde M., Heino S., Hogstrom J., Kaijalainen S., Anisimov A., Flanagan D., Kallio P., Leppanen V.M., Ristimaki A., Ritvos O., et al. Expression of R-spondin 1 in Apc(min/+) mice suppresses growth of intestinal adenomas by altering Wnt and transforming growth factor beta signaling. Gastroenterology. 2021;160:245–259. doi: 10.1053/j.gastro.2020.09.011. [DOI] [PubMed] [Google Scholar]
- Lin X., Lu C., Ohmoto M., Choma K., Margolskee R.F., Matsumoto I., Jiang P. R-spondin substitutes for neuronal input for taste cell regeneration in adult mice. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2001833118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Millar S.E. Wnt/beta-catenin signaling in oral tissue development and disease. J. Dent Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W.-J., Mann R.K., Nguyen A., Bi T., Silverstein M., Tang J.Y., Chen X., Beachy P.A. Neuronal delivery of Hedgehog directs spatial patterning of taste organ regeneration. Proc. Natl. Acad. Sci. U S A. 2018;115:E200–E209. doi: 10.1073/pnas.1719109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmoto M., Lei W., Yamashita J., Hirota J., Jiang P., Matsumoto I. SOX2 regulates homeostasis of taste bud cells and lingual epithelial cells in posterior tongue. PLoS One. 2020;15:e0240848. doi: 10.1371/journal.pone.0240848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted J.M.D. Effects of cutting the lingual nerve of the dog. J. Comp. Neurol. 1921;33:149–154. doi: 10.1002/cne.900330204. [DOI] [Google Scholar]
- Park S., Cui J., Yu W., Wu L., Carmon K.S., Liu Q.J. Differential activities and mechanisms of the four R-spondins in potentiating Wnt/beta-catenin signaling. J. Biol. Chem. 2018;293:9759–9769. doi: 10.1074/jbc.RA118.002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Martinez I., Nagai T., Chaudhari N. Functional cell types in taste buds have distinct longevities. PLoS ONE. 2013;8:e53399. doi: 10.1371/journal.pone.0053399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Paz L., Orsini V., Boulter L., Calabrese D., Pikiolek M., Nigsch F., Xie Y., Roma G., Donovan A., Marti P., et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell. Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- Ren W., Aihara E., Lei W., Gheewala N., Uchiyama H., Margolskee R.F., Iwatsuki K., Jiang P. Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Sci. Rep. 2017;7:4004. doi: 10.1038/s41598-017-04099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Lewandowski B.C., Watson J., Aihara E., Iwatsuki K., Bachmanov A.A., Margolskee R.F., Jiang P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. U S A. 2014;111:16401–16406. doi: 10.1073/pnas.1409064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev. Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Takeda N., Jain R., Li D., Li L., Lu M.M., Epstein J.A. Lgr5 identifies progenitor cells capable of taste bud regeneration after injury. PLoS One. 2013;8:e66314. doi: 10.1371/journal.pone.0066314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintschgau M.V., Hönigschmied J. Nervus Glossopharyngeus und Schmeckbecker. Arch. Ges. Physiol. 1876;14:443–448. [Google Scholar]
- Wei K., Kuhnert F., Kuo C.J. Recombinant adenovirus as a methodology for exploration of physiologic functions of growth factor pathways. J. Mol. Med. (Berl) 2008;86:161–169. doi: 10.1007/s00109-007-0261-7. [DOI] [PubMed] [Google Scholar]
- Xu M., Horrell J., Snitow M., Cui J., Gochnauer H., Syrett C.M., Kallish S., Seykora J.T., Liu F., Gaillard D., et al. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat. Commun. 2017;8:15397. doi: 10.1038/ncomms15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee K.K., Li Y., Redding K.M., Iwatsuki K., Margolskee R.F., Jiang P. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells. 2013;31:992–1000. doi: 10.1002/stem.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.