Locatelli et al. (6) reported on the quantitative detection of human herpesvirus 6 (HHV-6) in plasma and cell suspensions by TaqMan-based PCR. We wish to add our experience. We have developed a rapid (<30-min) assay for the simultaneous detection, quantification, and differentiation of HHV-6 variants A and B using a LightCycler (LC; Bio/Gene Ltd., Kimbolton, England). The instrument performs real-time PCR, combining rapid cycling with fluorescence-based identification of PCR products in glass reaction capillaries (1).
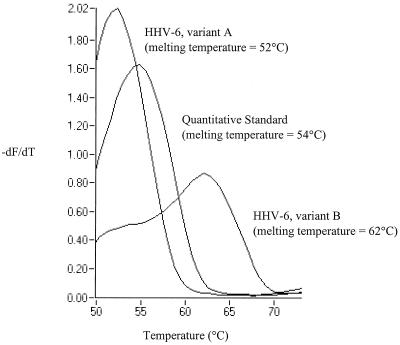
We designed primers and a 5′-Cy5-labeled biprobe (2) to target a 119-bp fragment of the large tegument protein gene. Detection of PCR product occurs when the biprobe hybridizes to the amplicon, leading to an increase in fluorescence resonance energy transfer between the double-stranded DNA fluorophore, SYBR green 1 and Cy5 (2, 5). The biprobe was designed to be complementary to the variant B sequence and to detect a 2-bp polymorphism between the variants (Table 1). Using PCR-directed mutagenesis, a plasmid-cloned quantitative standard (QS) was generated, incorporating a single-nucleotide change in the same region with respect to the variant B sequence, which could also be detected by the biprobe (Table 1). Simultaneous identification of the three targets with the biprobe was achieved by melting curve analysis (1, 5), with variants A and B and the QS yielding probe and product melting temperatures of 52, 62, and 54°C, respectively (Fig. 1).
TABLE 1.
Sequence variation in the large tegument protein of HHV-6 detected by the biprobe
| Target | Sequence | GenBank accession no. | Positions |
|---|---|---|---|
| HHV-6 variant A | -AAT- | X83413 | 46119–46121 |
| HHV-6 variant B | -CAA- | AF157706 | 47239–47241 |
| QS | -GAA- | N/Aa | N/A |
N/A, not applicable.
FIG. 1.
Melting curve graph showing differentiation of HHV-6 variants A and B and the QS. Melting peaks were derived by plotting the negative derivative of fluorescence (−dF/dT) with respect to temperature.
We routinely receive surveillance samples from immunocompromised patients (including severe combined immune deficiency syndrome patients and solid-organ and bone marrow transplant recipients) and have evaluated the LC assay using not only plasma but also EDTA whole-blood, serum, bone marrow, tissue, and respiratory samples, all extracted using a simple commercially available method (QIAamp DNA minikit; Qiagen, West Sussex, United Kingdom). To date, we have tested 129 samples in parallel with an in-house modification of a solid-block-based nested PCR (9). Forty samples have tested positive by both assays. Using the LC assay, all positive samples were identified as variant B, which has been shown to be the predominant variant in transplant recipients (3, 4, 7, 8) and the viral loads (ranging from 5 × 103 to 2.55 × 107 copies/ml) proved to be reproducible (±0.5 log10 units).
Our assay offers rapid detection (<30 min, excluding DNA extraction), quantitation, and typing of HHV-6 DNA in a closed-tube format which minimizes the risk of carryover contamination. The assay is sensitive (detection limit, ≤10 copies), with a 6-log10 unit dynamic range (10 to ≥106 copies), is applicable to a wide range of sample types, and compared well with nested PCR.
HHV-6 is an emerging pathogen and has been associated with severe disease in the immunocompromised (3, 4, 7, 8). We concur with Locatelli et al. (6) that determination of viral load may be a useful indicator of active infection. The availability of quantitative PCR methods means that results are available in a clinically helpful time-frame (2 h by LC versus 2 days by nested PCR), which should assist with implementing timely therapeutic intervention and assessing response to treatment. In addition, use of this LC assay may help to elucidate the epidemiology of the two variants and their role in disease.
Footnotes
Ed. Note: The authors of the published article declined to respond.
REFERENCES
- 1.Bernard P S, Lay M J, Wittwer C T. Integrated amplification and detection of the C677T point mutation in the methylenetetrahydrofolate reductase gene by fluorescence resonance energy transfer and probe melting curves. Anal Biochem. 1998;255:101–107. doi: 10.1006/abio.1997.2427. [DOI] [PubMed] [Google Scholar]
- 2.Bio/Gene Ltd.; The Secretary of State for Defence. Nucleic acid detection system. Great Britain patent GB2333359A. 21 July 1999. [Google Scholar]
- 3.Chan P K S, Peiris J S M, Yuen K Y, Liang R H S, Lau Y L, Chen F E, Lo S K F, Cheung C Y, Chan T K, Ng M H. Human herpesvirus-6 and human herpesvirus-7 infections in bone marrow transplant recipients. J Med Virol. 1997;53:295–305. doi: 10.1002/(sici)1096-9071(199711)53:3<295::aid-jmv20>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 4.Clark D A. Human herpesvirus 6. Rev Med Virol. 2000;10:155–173. doi: 10.1002/(sici)1099-1654(200005/06)10:3<155::aid-rmv277>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Gibson J R, Saunders N A, Burke B, Owen R J. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999;37:3746–3748. doi: 10.1128/jcm.37.11.3746-3748.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Locatelli G, Santoro F, Veglia F, Gobbi A, Lusso P, Malnati M S. Real-time quantitative PCR for human herpesvirus 6 DNA. J Clin Microbiol. 2000;38:4042–4048. doi: 10.1128/jcm.38.11.4042-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh N, Carrigan D R. Human herpesvirus-6 in transplantation: an emerging pathogen. Ann Intern Med. 1996;124:1065–1071. doi: 10.7326/0003-4819-124-12-199606150-00007. [DOI] [PubMed] [Google Scholar]
- 8.Stoeckle M Y. The spectrum of human herpesvirus 6 infection: from roseola infantum to adult disease. Annu Rev Med. 2000;51:423–430. doi: 10.1146/annurev.med.51.1.423. [DOI] [PubMed] [Google Scholar]
- 9.Wakefield A J, Fox J D, Sawyerr A M, Taylor J E, Sweenie C H, Smith M, Emery V C, Hudson M, Tedder R S, Pounder R E. Detection of herpesvirus DNA in the large intestine of patients with ulcerative colitis and Crohn's disease using the nested polymerase chain reaction. J Med Virol. 1992;38:183–190. doi: 10.1002/jmv.1890380306. [DOI] [PubMed] [Google Scholar]