Abstract
We are in the midst of a golden age of uncovering defense systems against bacteriophages. Apart from the fundamental interest in these defense systems, and revolutionary applications that have been derived from them (e.g. CRISPR-Cas9 and restriction endonucleases), it is unknown how defense systems contribute to resistance formation against bacteriophages in clinical settings. Bacteriophages are now being reconsidered as therapeutic agents against bacterial infections due the emergence of multidrug resistance. However, bacteriophage resistance through defense systems and other means could hinder the development of successful phage-based therapies. Here, we review the current state of the field of bacteriophage defense, highlight the relevance of bacteriophage defense for potential clinical use of bacteriophages as therapeutic agents and suggest new directions of research.
Keywords: phage resistance, phage therapy, innate immunity, adaptive immunity, bacteria, defense
This review highlights the newest discoveries in the field of bacteriophage defense and discusses the relevance of defense mechanisms for the potential clinical use of bacteriophages as therapeutic agents.
INTRODUCTION
The use of bacteriophages, or phages, as therapeutic agents to treat bacterial infections began immediately after phage discovery in 1917 (Twort 1915; d'Herelle 1917). The initial interest in phages as antibacterial agents faded quickly following the discovery of penicillin two decades later (Wittebole, De Roock and Opal 2014), although phage therapy remained in use in former Soviet republics like Georgia and Russia (Chanishvili 2012). In recent years, Western medicine has started to reconsider the therapeutic use of phages due to the alarming rise in infections caused by multidrug-resistant (MDR) bacteria (Moelling, Broecker and Willy 2018). However, the success of phage therapy might be limited by the development of phage resistance by bacteria, much akin to the resistance developed toward antibiotics. Recently, multiple mechanisms by which bacteria defend against phages have been uncovered (Doron et al. 2018; Gao et al. 2020), some specific for certain species or strains, others more widespread. Unlike antibiotics, phages can adapt and/or deploy anti-defense systems of their own to overcome the defense mechanisms of bacteria (Iida et al. 1987; Atanasiu et al. 2002; Otsuka and Yonesaki 2012; Isaev et al. 2020; Malone et al. 2020; Mendoza et al. 2020; Wiegand et al. 2020).
The evident complexity of phage–bacteria interactions needs to be considered for phage therapy to be implemented successfully (Roach and Debarbieux 2017). It is unknown how defense systems contribute to and impact resistance formation against phages in clinical settings, and this could be a bottleneck in the development of successful phage-based therapies when left without consideration.
Here, we provide an overview of the current state of the field of natural and acquired phage resistance, highlight the relevance of phage defense for potential clinical use of phages as therapeutic agents and suggest new directions of research.
THE MULTISTEP PROCESS OF BACTERIOPHAGE INFECTION
There are multiple families of bacteriophages, each with specific features that influence their process of infection of a bacterial host. For the purposes of this review, we will focus on phages belonging to the order Caudovirales (Fields, Knipe and Howley 1996), which are known as tailed phages and are the most widely used in clinical applications (Wittebole, De Roock and Opal 2014). Tailed phages have double-stranded DNA genomes and a structure made up of an icosahedral head and a tail, which usually incorporates receptor binding proteins (RBPs) such as tail spikes and tail fibers at the distal end (King 2012). These elements are responsible for the first step of infection, i.e. recognition of specific receptors on the surface of bacteria, and subsequent adsorption of the phage (King 2012). Phage receptors on the bacterial surface are typically peptide sequences or polysaccharides present on the bacterial cell wall, as well as protruding structures such as capsules, pili or flagella (Bertozzi Silva, Storms and Sauvageau 2016; Nobrega et al. 2018). Phage attachment to the host surface often occurs first through a reversible interaction with a receptor, which is then followed by an irreversible binding event to the same or a second receptor (Bertozzi Silva, Storms and Sauvageau 2016). Generally, phages recognize receptors with a great degree of specificity, meaning that the host range of a certain phage is often limited at the adsorption stage by the receptors available on the cell surface (de Jonge et al. 2019).
Adsorption of the phage to its native receptor on the cell triggers ejection of the genetic material of the phage into the host cytoplasm. The mechanism of this complex phenomenon is not yet completely understood for many phage types, but in Caudovirales it commonly involves conformational changes of the phage triggered by binding of the phage RBPs to the receptor that result in the opening of the channel required for DNA release from the capsid (González-García et al. 2015; Wang et al. 2019). In some phages, these conformational changes lead also to the ejection of the tape measure protein that reconfigures into a channel through which the genome translocates into the cell cytoplasm (Boulanger et al. 2008; Cumby et al. 2015; Taylor et al. 2016). In phages with short tails (e.g. Podoviridae) proteins ejected together with the genome can work to form a similar channel for genome passage (Leptihn, Gottschalk and Kuhn 2016). The forces behind phage genome ejection into the cell cytoplasm are still unclear, with different models proposed (Molineux and Panja 2013). It seems that thermodynamic and compressing pressures cause the initial release of DNA (Smith et al. 2001), with complete ejection being achieved by further hydrodynamic forces and/or bacterial proteins involved in transcription of the initial segment of the phage genome (Kemp, Gupta and Molineux 2004; Choi et al. 2008; Panja and Molineux 2010).
If the infecting phage is obligately virulent, which is preferred for phage therapy applications (Drulis-Kawa et al. 2013; Petrovic Fabijan et al. 2020a), the infection follows a lytic cycle once the phage genome is inside the cell. In this case, the phage hijacks the cellular machinery of the bacteria, shutting off the expression of host genes and achieving the replication of its genome and the expression of its own genes (De Smet et al. 2017). For this purpose, some phages rely on the bacterial RNA polymerase (Hinton 2010), while others encode and/or co-inject their own (Drobysheva et al. 2021). The lytic cycle culminates with the expression of late genes, which encode structural proteins and proteins necessary for bacterial host lysis. This ultimately leads to the production of more viral particles that will in the end burst out of the host cell (Hobbs and Abedon 2016). However, if the infecting phage is temperate, it may also follow a lysogenic cycle. In this case, the viral genome persists within the host cell, either introduced in the bacterial chromosome as a prophage or in the bacterial cytoplasm as a plasmid. The lysis–lysogeny decision may depend on peptide-based communication between the viruses (Erez et al. 2017) or on host repressor genes that form part of a quorum-sensing system (Silpe and Bassler 2019).
MECHANISMS OF PHAGE RESISTANCE
Bacteria evade phage infections in different ways. Here, we classify different resistance mechanisms in three main categories:
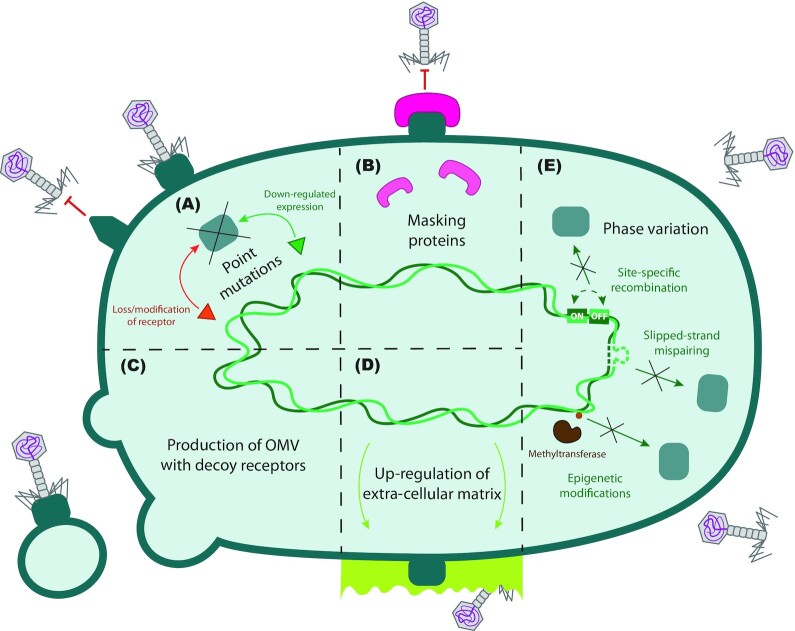
Receptor adaptations: random mutations or phenotypical variations in bacteria that result in decreased phage adsorption (Fig. 1).
Host defense systems: molecular pathways that have specifically evolved in bacteria to prevent or suppress phage infections (Fig. 2).
Phage-derived phage defense systems: molecular pathways encoded by phages to compete with other phages to the benefit of the host (Fig. 3).
Figure 1.
Host adaptations leading to phage resistance. (A) Point mutations can lead to a loss or modification of the phage receptors (green rectangles), or to downregulation of their expression. (B) Receptor masking proteins like TraT of Escherichia coli (pink) can bind to the surface-exposed regions of phage receptors, making them unavailable for the phages. (C)Outer-membrane vesicles (OMVs) presenting phage receptors act as decoys to prevent the phages from encountering the bacteria. (D) An increase in the production of extracellular matrix (light green) leads to phage receptors being physically hidden. (E) Phase variation occurs through three mechanisms: site-specific recombination, slipped-strand mispairing and epigenetic modifications. It can regulate the bacterial phenotype, including the expression of surface proteins like phage receptors.
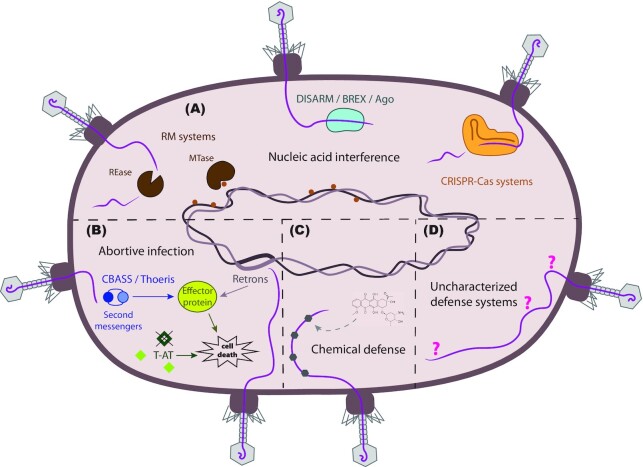
Figure 2.
Host phage defense systems. (A) Multiple defense systems act via nucleic acid interference. R-M systems are generally composed of an MTase that methylates endogenous DNA to distinguish it from exogenous DNA, and of an REase that cleaves the exogenous, non-methylated DNA. DISARM interacts with phage DNA to prevent its circularization, thereby blocking its replication or lysogeny. BREX or Ago systems interact with phage DNA and prevent it from replicating without necessarily cleaving it. CRISPR-Cas systems are known as the adaptive immune system of bacteria. The CRISPR array contains sequences of foreign origin that can be transcribed and processed to act as a guide for the Cas endonuclease, which recognizes and cleaves said sequences upon reentry into the bacteria. (B) Abortive infection comprises a series of mechanisms that lead to bacterial cell suicide. An example in which this can happen is through an imbalance in the concentration of toxins and antitoxins in a cell. Another example is through the action of effector proteins that might get activated directly, like in the case of retrons, or via second messengers, like in the case of CBASS or Thoeris. These effector proteins can lead to cell death in several ways, for instance through inner membrane degradation (CBASS) or through NAD depletion (Thoeris). (C) Bacteria can produce secondary metabolites such as daunorubicin (depicted) that intercalate phage DNA and prevent it from circularizing and replicating. (D) Analysis of genetic defense islands has recently led to the discovery of a series of defense systems that are yet to be fully characterized. These include: Hachiman, Shedu, Gabija, Septu, Lamassu, Zorya, Kiwa, Druantia, Wadjet, RADAR, DRTs, AVAST and pVips, among others.
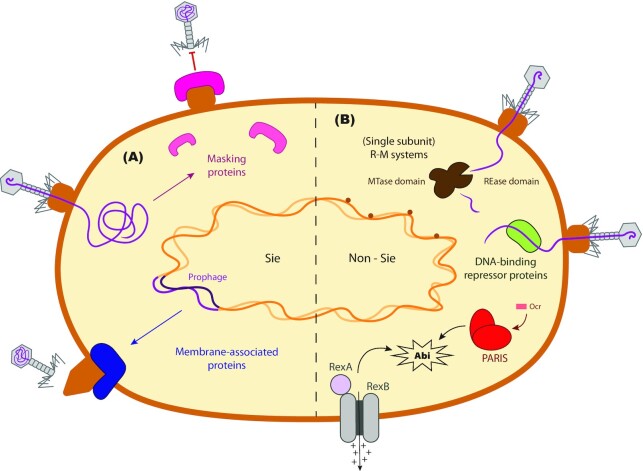
Figure 3.
Phage-derived defense systems. (A) Superinfection exclusion systems (Sie) are encoded by phages to prevent other phages from infecting their host. Some phages like T5 produce proteins that mask their receptor and make it inaccessible. Other phages, especially prophages, encode membrane-associated proteins that interact with the phage receptor, blocking the DNA entry channel, triggering a conformational change or inhibiting the invading phage's enzymes. (B) Prophages like Panchino of Mycobacteriumsmegmatis can confer resistance to their hosts through the expression of R-M systems or DNA-binding repressor proteins that target the DNA of newly infecting phages. Other prophage-encoded systems, like RexA-RexB or the newly characterized PARIS, can trigger an Abi response upon sensing an invasion by a new phage.
Receptor adaptations leading to phage resistance
In their natural environments, bacteria are subjected to constant selective pressure, which has driven bacteria and phages into an arms race to evolve defense systems and to counter them. The arms race is characterized by high mutation rates and horizontal gene transfer, and leads to rapid evolution of genetic traits and genetic diversity (Takeuchi et al. 2012; Puigbò et al. 2014; Hampton, Watson and Fineran 2020). Mutations that cause cell surface alterations can result in blockage of phage adsorption, and are therefore directly beneficial to the host.
Bacteria can pose a barrier to phage adsorption by decreasing the availability of the receptors to which phages bind. The acquisition of point mutations in their genome (Fig. 1A) is probably the simplest way by which bacteria can become fully resistant to phages. In fact, mutations in the receptor genes or their regulation have been a common way to identify the receptor of a phage (Nobrega et al. 2018; Kortright, Chan and Turner 2020). These mutations occur often upon phage challenges, and can lead to a loss or decrease in the gene expression of certain receptors, or to modifications of their structure (Chapman-McQuiston and Wu 2008a,b). For example, E. coli mutates tolC and lipopolysaccharide (LPS) genes to resist infection by phage U136B (Burmeister et al. 2020). Similarly, Acinetobacter baumannii mutates genes involved in the biosynthesis of capsular polysaccharides to avoid infection by phages øFG02 and øCO01 (Gordillo Altamirano et al. 2021). In Listeria monocytogenes, loss or deficiency of wall teichoic acid rhamnosylation leads to resistance to a wide range of phages (Trudelle et al. 2019), and results also in serovar diversification (Eugster et al. 2015). Other proteins involved in phage adsorption and DNA injection, like the phage infection protein from Enterococcus faecalis (PIPEF), can also mutate as a response to phage challenges (Duerkop et al. 2016).
Bacteria may also block phage adhesion by producing proteins that mask or block the phage receptors on the cell surface (Fig. 1B). An example of this is F plasmid-encoded protein TraT, which localizes at the cell outer membrane and binds surface-exposed regions of the outer membrane protein OmpA in E. coli (Riede and Eschbach 1986). This makes this common phage receptor inaccessible for phage binding. Masking molecules, such as lipoproteins, that bind phage receptors can also be produced by bacteria under stress conditions and are released during bacterial lysis (Decker et al. 1994). Some bacteria also produce and release OMVs (Fig. 1C) that act as cell decoys that capture and inactivate phages (Manning and Kuehn 2011). Another mechanism that can prevent phages from reaching their receptors is upregulating the production of extracellular matrix typically consisting of polysaccharides, proteins, lipids and extracellular DNA (Fig. 1D), in a way that protects the embedded bacteria or subsequent biofilm against phage adsorption (Hanlon et al. 2001; Testa et al. 2019). In Lactococcus lactis, plasmids encoding exopolysaccharide biosynthesis genes can also confer protection against phages (Forde and Fitzgerald 2003).
In addition to this, reversible changes in the regulation of gene expression, a phenomenon known as phase variation (Fig. 1E), can lead to a decrease in receptor availability (Gencay et al. 2018). These changes can be mediated by site-specific recombination (Dybvig 1993), in which inversion of a DNA segment in the promoter or regulatory region of a gene causes its expression to be turned on or off. This is exemplified by the development of flagella in Salmonella spp. and fimbriae in E. coli (Abraham et al. 1985; Heichman and Johnson 1990; Choi et al. 2013). Other receptors, such as the outer membrane protein Opc of Neisseria meningitidis and the subunits of Bordetella pertussis fimbriae (Willems et al. 1990; Sarkari et al. 1994; Zhou, Aertsen and Michiels 2014), are regulated by slipped strand mispairing, i.e. programmed mutations that occur in defined regions during DNA recombination. Epigenetic modifications, such as altered methylation patterns on DNA sequences (Casadesús and Low 2006), also regulate expression of phage receptors, such as the O-antigen chains of LPSs in Salmonella enterica (Cota et al. 2015).
All of these alterations act directly on phage receptors and decrease the chances of phage adsorption. However, modifications of surface elements can come with a fitness trade-off for the host bacteria, in terms of reduced virulence or survival ability of the host (Alseth et al. 2019; Mangalea and Duerkop 2020), limiting the possibility to alter the receptor itself. Due to this, more specific defense systems that target phages within the host cell are also necessary, especially in the context of a complex microbial community (Alseth et al. 2019; Broniewski et al. 2020).
Host phage defense systems
Bacteria have evolved defense systems dedicated to defense against mobile genetic elements such as phages. Many of them are clustered in regions of the genome known as defense islands (Koonin, Makarova and Wolf 2017), offering an opportunity for discovering new defense systems by analyzing the genetic regions in the proximity of other known defense systems. Such strategy has resulted in a significant and fast expansion of the known arsenal bacteria use to defend against phage infection. We will cover a number of different phage defense systems that have been identified, including those acting on viral nucleic acids and those causing abortive infection of the host.
Nucleic acid interference (Fig. 2A)
The ability to interfere with viral nucleic acids is a common strategy that hosts employ to limit phage invasion and propagation.
One of the most widespread and longest known examples of phage defense systems are those called Restriction-Modification (R-M) systems that act on phages with DNA genomes (Luria and Human 1952; Luria 1953). In R-M systems, a methyltransferase (MTase) methylates endogenous DNA at specific sites, protecting it from cleavage by the restriction endonuclease (REase) that recognizes the foreign, unmodified DNA and cleaves it within, close to, or at a distance from the recognition site (Tock and Dryden 2005).
There are four classical types of R-M systems (I-IV), classified according to the characteristics of their specific components (Tock and Dryden 2005). The type I R-M system consists of a protein complex of three subunits with distinct activities, the M (MTase), R (REase) and S (specificity) subunits. The S subunit dictates the target sequence specificity of both methylation and restriction by the protein complex. The abundant type II R-M system features a MTase and REase that work independently as separate proteins. These have been the major source for hundreds of commercially available restriction endonucleases used for molecular cloning. The type III R-M system also expresses the two independent MTase and REase proteins, but these exert their function as a complex. The type IV R-M system does not contain an MTase, and is thought to have evolved in response to some phages evading type I-III R-M systems by modifying their genome to evade restriction. Type IV systems overcome this counterattack by restricting the phage's modified DNA, while the bacterial DNA remains unmethylated (Stewart et al. 2000; Loenen and Raleigh 2014). It is interesting to note that MTases tend to be more conserved than REases, since the latter undergo rapid evolution to keep up with mutations in phage genomes (Gupta, Capalash and Sharma 2012).
R-M systems typically put epigenetic marks on the nucleobases. However, similar systems have been described that modify the sugar-phosphate backbone by introducing a phosphorothioate (substitution of a non-bridging oxygen with a sulfur) (Xu et al. 2010). The Dnd system works through the double-stranded phosphorothioation of endogenous DNA by proteins DndABCDE and restriction of foreign, unmodified DNA by DndFGH (Xu et al. 2010). Ssp proteins SspABCD also modify the host genome through phosphorothioation, but of only one of the two DNA strands (Xiong et al. 2020). This activity couples with that of SspE, which requires sensing of SspABCD to introduce nicks into foreign DNA, or with that of SspFGH, which indiscriminately damages non-phosphorothioated DNA, inhibiting its replication (Xiong et al. 2020; Wang et al. 2021).
Our knowledge on R-M-related defense systems is continuously expanding as more systems are being discovered through analysis of bacterial genomes. An example is the DISARM (defense island system associated with restriction–modification) systems (Ofir et al. 2018), which include MTases (adenine MTase DrmMI and/or cytosine MTase DrmMII) and proteins with domains of predicted helicase (DrmA, DrmD) and phospholipase D/nuclease (DrmC) activities or of unknown function (DrmB, DrmE). Although the exact mechanism of action of DISARM is not yet understood, it is clear that it involves methylation of the host DNA to distinguish self from nonself, and that it prevents phage DNA circularization, thereby blocking DNA replication and lysogeny at an early stage of the infection. It is also postulated that DISARM might collaborate with different R-M elements, achieving a synergistic effect against phage infection (Ofir et al. 2018). The bacteriophage exclusion (BREX) defense system also targets phage DNA upon entrance in the host cell (Goldfarb et al. 2015). Similar to R-M systems, BREX methylates host DNA to differentiate it from exogenous DNA. However, BREX does not appear to degrade non-methylated phage DNA, and instead seems to hamper replication of the phage DNA without cleavage (Goldfarb et al. 2015). Methylated or glycosylated phage DNA is not sensitive to BREX, but deletion of the methylase gene of this system does not have deleterious effects on the bacteria (Gordeeva et al. 2019).
In some bacteria, foreign DNA can also be intercepted by proteins of the Argonaute (Ago) family. These proteins are also present in eukaryotic cells, where they mediate the degradation of exogenous RNA using small interfering- or microRNAs (siRNA, miRNA) as guides to recognize their targets. While this process is not as well studied in prokaryotic cells as it is in eukaryotes, prokaryotic Ago proteins (pAgo) have been found in Thermus thermophilus (TtAgo) and in Rhodobacter sphaeroides (RsAgo) (Willkomm, Makarova and Grohmann 2018; Wu et al. 2020). TtAgo bases its mechanism on DNA-DNA interference rather than the RNA–RNA interference of eukaryotic Ago (Swarts et al. 2014). This protein also has an endonuclease (slicer) domain that allows it to cleave both single-stranded DNA and negatively supercoiled double-stranded DNA, normally of plasmid origin. Although it is unclear how the DNA guides used by TtAgo are formed, it appears that the activity of the protein itself is necessary for their production. RsAgo, in contrast, uses small RNA molecules as guides to target foreign DNA molecules (Miyoshi et al. 2016). Of note, RsAgo lacks the slicer domain, meaning that DNA interference is caused simply by binding the target rather than by cleaving it.
A particular form of nucleic acid interference, CRISPR-Cas (clustered regularly interspaced short palindromic repeats and associated proteins) systems constitute the only form of adaptive immunity described in prokaryotes so far (Mojica et al. 2005; Brouns et al. 2008). They are present in many bacterial genomes, and occasionally in plasmids (Millen et al. 2012). A CRISPR locus in a bacterial genome is composed of a CRISPR array and a cas gene operon. The CRISPR array contains repeats and sequences of foreign origin called spacers, which form the immunological memory of the defense system. The cas operon contains all genes coding for Cas proteins that form the machinery required for immunity. Immunity is achieved via a three-stage process that involves adaptation, expression and interference (Jackson et al. 2017; Hille et al. 2018; Koonin and Makarova 2019). During the adaptation stage, parts of the foreign genetic material are captured and integrated into the CRISPR array as a new spacer (Al-Attar et al. 2011; McGinn and Marraffini 2019). In DNA targeting CRISPR systems, functional spacers are derived from invader sequences that are flanked by a protospacer adjacent motif (PAM), a short nucleotide sequence that ensures the targeting of foreign invaders rather than the genomic CRISPR locus (Gleditzsch et al. 2018). At the expression stage, the CRISPR array serves as a template to transcribe a long precursor CRISPR RNA (crRNA) that is further processed into smaller mature crRNAs. Each crRNA is then loaded into Cas proteins to form an effector complex. At the stage of interference, this effector complex patrols the cell, screening for complementary sequences that are flanked by a PAM. Upon PAM recognition, the foreign genetic material is cleaved by the Cas proteins, and the infection is contained.
While sharing the general stages described above, CRISPR-Cas systems are characterized by mechanistic variability and are currently classified in two classes, six types and 33 subtypes (Makarova et al. 2020b). Class 1 systems, which include types I, III and IV, are characterized by the presence of a multi-subunit Cas complex that is involved in the recognition of invader DNA (type I, IV) or RNA (type III) during the interference stage. Class 2 systems, which include types II, V and VI, employ a single-subunit effector protein for recognition and cleavage of the foreign DNA (types II, V) or RNA (type VI) sequence. Of the six types described so far, type II is the best-known due to its applications for genome editing technology (Doudna and Charpentier 2014).
In summary, bacteria explore a diverse set of strategies that directly block or cleave phage nucleic acids to survive phage predation.
Abortive infection (Fig. 2B)
Abortive infection (Abi) is a commonly used phage defense strategy in which the cells sacrifice themselves before the phage completes its replication cycle to protect the rest of the population (Lopatina, Tal and Sorek 2020). Many of the Abi systems rely on a toxin–antitoxin (T–AT) mechanism, in which the balance between a stable toxin and an unstable antitoxin determines the fate of the cell (Fineran et al. 2009; Page and Peti 2016). Infection by a phage triggers repression of the antitoxin promoter or termination of its transcription (Dy et al. 2014). The result is that the toxin prevails, causing death of the bacterium. Some of these systems, like ToxIN of Pectobacterium atrosepticum, are encoded by plasmids (Fineran et al. 2009).
Other common strategies that lead to abortive infection are characterized by the specific depletion of critical cellular resources upon viral infection, including enzymatic cofactors and nucleotides (Snyder 1995). Examples in E. coli include protease Lit, which is activated by the Gol peptide of the T4 major capsid protein and cleaves translation elongation factor Tu to arrest translation (Levitz et al. 1990). Other Abi systems trigger not an individual response but a set of events. For example, exclusion of T7 by the F plasmid-encoded PifA system occurs via reduced synthesis of macromolecules, partially impaired DNA ejection and alteration of membrane permeability (Cheng, Wang and Molineux 2004). In Lactococcus spp., Abi systems that can target phage gene replication and expression are constitutively expressed but are toxic to the cell when overexpressed (Chopin, Chopin and Bidnenko 2005). Notably, most of these lactococcal defense systems are encoded by plasmids (Mills et al. 2006).
Cell suicide upon detection of invading, cytosolic DNA occurs in eukaryotic cells as well. It is mediated by the production of cyclic GMP-AMP, which activates the cGAS-STING pathway (Sun et al. 2013), and causes an upregulation of transcription of inflammatory genes. A similar pathway was found in Vibrio cholerae biotype El Tor, where production of cyclic GMP-AMP (cGAMP) activates a phospholipase that degrades the inner membrane leading to cell death (Cohen et al. 2019). Introduction of the operon encoding this pathway into defective V. cholerae and E. coli strains conferred resistance to a variety of phages, suggesting an important role of this system in antiphage defense. The system, called cyclic-oligonucleotide-based antiphage signaling system (CBASS), has since been found in a broad range of organisms belonging to all major bacterial phyla and at least one archaeal phylum (Millman et al. 2020b). It is thought to be an ancestor of the eukaryotic cGAS-STING pathway.
Mutations in enzymes involved in protein maturation can also be used to prevent the spread of phage infection. In Streptococcus thermophilus, a mutation in the methionine aminopeptidase that impairs its catalytic activity was seen to confer resistance to a broad range of phages, seemingly by hampering virion assembly (Labrie et al. 2019). While not exactly considered an Abi mechanism, this process comes at the cost of impairing bacterial growth for at least several of the strains studied.
In Vibrio cholerae, a parasitic phage satellite known as phage-inducible chromosomal island-like element (PLE) defends the bacterial population from phage attack by functioning akin to an Abi system. PLE are found integrated in the V. choleraechromosomes and are excised upon infection by ICP1 phages (Seed et al. 2013; O'Hara et al. 2017; McKitterick et al. 2019). Using both PLE- and phage-encoded products (McKitterick et al. 2019; Barth et al. 2020), PLE replicates and hijacks the structural components of the phage to encapsidate its own genome (O'Hara et al. 2017), and uses protein LidI to disrupt the mechanism of lysis inhibition that would normally give ICP1 phages more time to produce new virions (Hays and Seed 2020). Via a combination of structural hijacking and accelerated cell bursting, PLE prevent phage spreading and efficiently protect the bacterial population while transducing their own genome to other cells.
Recently described Thoeris seems to operate via an Abi mechanism as well (Ka et al. 2020). It presents a protein with a toll-interleukin receptor (TIR) domain which, upon phage infection, produces an isomer of cyclic ADP-ribose (Ofir et al. 2021). This molecule acts as a second messenger and activates a protein with catalytic NADase activity, leading to NAD depletion in the infected host. As a result of this, the bacterium presumably dies before phage progeny can mature. TIR domains appear to be specific toward certain phages, and multiple TIR proteins can be present within the same host.
Retrons, bacterial genetic elements composed of a reverse transcriptase (RT) and a noncoding RNA (ncRNA), have also been shown to protect against phage infection via abortive infection (Gao et al. 2020; Millman et al. 2020a). Effector proteins of multiple functions were found associated with the retrons, such as ribosyltransferases, two-transmembrane domain (2TM) genes, and genes with ATPase or HNH endonuclease domains, suggesting a diversity of mechanisms by which abortive infection may be achieved. Characterization of retron Ec48 associated with a 2TM domain gene demonstrates that it acts by sensing inhibition of DNA-repair enzyme RecBCD by proteins of the infecting phage, leading to abortive infection and cell death (Millman et al. 2020a).
More recently, dCTP deaminase and dGTPase proteins have been found to protect bacterial cells from phage infection by degrading deoxynucleotides dCTP and dGTP, efficiently eliminating these from the nucleotide pool (Severin et al. 2021; Tal et al. 2021). Depletion of these deoxynucleotides during phage infection halts phage replication and likely leads to cell death (Tal et al. 2021). While abortive infection responses can be encoded by some of the defense systems listed above, some CRISPR-Cas systems have been found to use this strategy as well. Most well-known systems are the type III CRISPR-Cas systems that produce small signal molecules upon target RNA detection (Athukoralage and White 2021). This molecule then activates unspecific nucleases and other potentially damaging activities in the cell, aborting an infection (Makarova et al. 2020a). Likewise, some type I CRISPR-Cas systems can function with an Abi mechanism. In P. atrosepticum, expression of a type I-F CRISPR-Cas system reduces phage progeny while hampering the survival of infected cells (Watson et al. 2019).
In summary, many abortive infection-like strategies have been identified in which cells typically detect infection and initiate a self-damaging response that hampers the virus in its infection process, saving the remaining population of cells.
Chemical defense (Fig. 2C)
It is well documented that bacteria produce secondary metabolites, among which are compounds with antimicrobial activity (Davies 2013). Recently, a panel of bioactive compounds was tested to assess whether they could confer protection to E. coli against lysis by phage Lambda (Kronheim et al. 2018). Several compounds were identified that allow bacteria to proliferate in spite of the phage challenge. Most are DNA-intercalating agents, four of which produced by Streptomyces spp.: daunorubicin, doxorubicin, epirubicin and idarubicin. These compounds inhibit double-stranded DNA phages targeting Streptomyces coelicolor, E. coli and Pseudomonas aeruginosa. DNA intercalation is thought to prevent the circularization of the phage linear DNA inside the bacterial cytoplasm, or its interaction with proteins involved in replication and transcription.
Uncharacterized defense systems (Fig. 2D)
Bioinformatic analysis of genes in defense system clusters has led to the identification of multiple new defense systems in recent years. One such approach identified several putative defense systems based on the requirement that each putative system must contain at least one annotated protein domain enriched in defense islands. Some of these were experimentally confirmed to grant protection against at least one phage: Thoeris (now classified as an Abi system), Hachiman, Shedu, Gabija, Septu, Lamassu, Zorya, Kiwa and Druantia (Doron et al. 2018). Zorya contains components that resemble parts of the flagellar motor, and is more abundant in Gram-negative species, especially Proteobacteria. Its proposed mechanism of action leads to cell death through membrane depolarization. Another defense system, Wadjet, does not seem to confer phage resistance but seems to target foreign plasmids by a still unknown mechanism (Doron et al. 2018).
Additional defense system candidates were identified using an approach independent of domain annotations (Gao et al. 2020). These candidates incorporate enzymatic activities not previously thought to be implicated in antiviral defense. Among them is the phage restriction by an adenosine deaminase acting on RNA (RADAR) system. RADAR edits RNA transcripts by catalyzing the deamination of adenosine into inosine, seemingly blocking the early stages of the phage infection cycle. Another candidate system identified in this study is the RT family defense-associated reverse transcriptases (DRT). DRTs are not linked to mobile elements, unlike most RTs found in prokaryotes, and they seem to alter phage gene expression in various ways. Antiphage activity was likewise detected in a group of nucleoside triphosphatases (NTPases) of the STAND (signal transduction ATPases with numerous associated domains) superfamily, which were given the name antiviral ATPases/NTPases of the STAND superfamily (AVAST). Members of this superfamily found in eukaryotes are often involved in programmed cell death, so it was postulated that the AVAST system may constitute an Abi mechanism. Additionally, the study found some other proteins and systems that provided protection against T7-like phages, of which the mechanisms of action need to be further investigated.
Another example of an antiphage defense mechanism that has recently started to be characterized is prokaryotic viperins (pVips) (Bernheim et al. 2021). In animals, viperins are interferon-induced proteins that block the replication of several viruses (Helbig and Beard 2014). In a similar way, pVips produce modified ribonucleotides that inhibit viral polymerase-dependent transcription, thereby protecting against infection by phage T7 (Bernheim et al. 2021). pVips with antiphage activity were identified by analyzing prokaryotic homologues of human viperin that are encoded in defense islands.
Finally, some novel defense systems that are still uncharacterized have been identified in T4- and T2-like prophages. These are briefly discussed in the following section.
Phage-derived phage defense systems
Interestingly, phages provide bacteria with defense systems against infection by the same or closely related phage, in a phenomenon known as superinfection exclusion (Sie) (Fig. 3A). Some phages produce proteins to mask the cell surface receptors, blocking new infections. This strategy also protects the newly formed phages from being inactivated as a consequence of binding to receptors coming from remains of lysed bacteria. This behavior is observed for example in phage T5, which produces lipoprotein Llp that conceals its own receptor, outer membrane protein FhuA (Pedruzzi, Rosenbusch and Locher 1998). Other phages, mostly prophages (Van Houte, Buckling and Westra 2016), use membrane-anchored or membrane-associated proteins to target and block the entry of phage DNA into the bacterial cytoplasm (Labrie, Samson and Moineau 2010). Such proteins act by inhibiting the formation of the channel through which DNA travels across the cell membrane, by inhibiting the phage lysozyme that degrades the peptidoglycan of the bacterial cell wall, or by changing the conformation of the proteins surrounding the ejection site to prevent translocation (Bondy-Denomy et al. 2016).
Furthermore, prophages can mediate resistance through non-Sie-like mechanisms as well (Fig. 3B). The RexA-RexB system, an Abi system expressed by λ-lysogenic E. coli, works by reducing the membrane potential of the cell, leading to a decrease in ATP production that ultimately results in cell death (Parma et al. 1992). Another example is the phage Panchino of M. smegmatis, which provides lysogens with a single subunit R-M system able to recognize a broad range of phages (Dedrick et al. 2017). Genes encoding repressor proteins that bind phage DNA may also be found in prophages (Pope et al. 2011). They are thought to have a role in protecting the viability of the lysogenized bacteria, counteracting accidental prophage transcription events. Prophage-mediated phenotypic changes in bacteria are sometimes encoded in genetic elements called morons, which are flanked by a promoter and a transcriptional terminator and can be transcribed autonomously, independent of prophage activation (Juhala et al. 2000).
As with host defense systems, the discovery of phage-derived defense systems is ongoing. Analysis of Enterobacteria P4- and P2-like prophages recently led to the discovery of genetic hotspots that encode a variety of bacterial immune mechanisms (Rousset et al. 2021). Among these is the phage anti-restriction-induced system (PARIS). This system triggers an Abi response upon sensing a phage-encoded anti-restriction protein, Ocr, which inhibits R-M systems and BREX (Rousset et al. 2021). The mechanisms of action of PARIS and the other systems identified in this study remain to be further uncovered.
In summary, once inside the host, phages themselves can provide the bacteria with mechanisms of protection against further phage infection that favor both the bacteria and the phage.
PHAGE COUNTERATTACK STRATEGIES
While the mechanisms of phage resistance exhibited by bacteria seem overwhelmingly varied, phages have also developed a broad array of opposing strategies. Just as bacterial defenses target every step in the process of phage infection, every barrier imposed by bacteria has to withstand a phage counterattack (Stern and Sorek 2011).
In response to variations in the bacterial cell surface receptors, phages are able to change their tropism through mutations in their RBPs. In fact, genes encoding RBPs and other proteins related to host recognition are reported to incorporate mutations at a very high frequency. This is often mediated by the activity of diversity-generating retroelements (DGRs) (Guo et al. 2014). These are regions that are subjected to targeted mutation by means of the exchange of two variable repeats by an error-prone reverse transcriptase (Paul et al. 2017). This type of directed mutagenesis is template dependent and affects determined adenine-specific sites, while a conserved scaffold sequence is retained to ensure stability. This process was first described for the specificity switch of the major tropism determinant protein in Bordetella spp. phages. Since then, more phages have been identified that benefit from these systems (Benler et al. 2018).
To overcome the barrier imposed by capsules and extracellular layers, some phages became able to bind to these structures (Bertozzi Silva, Storms and Sauvageau 2016), and to degrade them using depolymerases. These enzymes may be either expressed as part of tail spike or tail fiber proteins or released in a soluble form following lysis of infected bacteria (Latka et al. 2017). A recent review offers an overview of the diversity of phage depolymerases (Knecht, Veljkovic and Fieseler 2020).
Phages have also developed forms of escaping targeting by R-M systems. They can (i) mutate to remove restriction sites from their genome (palindrome avoidance) and therefore avoid recognition by REases (Rocha, Danchin and Viari 2001; Rusinov et al. 2018); (ii) modify the sequences recognized by REases (e.g. the glucosyl-hydroxymethylcytosine of T4 that is used instead of the regular cytosine; Labrie, Samson and Moineau 2010); (iii) change the distance and orientation of restriction sites to avoid restriction by REases that need to recognize two sequences at a determined distance from each other and in a specific orientation (Golovenko et al. 2009); (iv) occlude the restriction sites with proteins (e.g. DarA and DarB of P1 phages) that are ejected together with the phage genome (Iida et al. 1987); (v) sequester REases with proteins that mimic the structure of a DNA double helix (e.g. Ocr from T7) (Zavil'gelskiĭ and Kotova 2014); and (vi) acquire genes encoding an MTase that modifies the phage genome (Hill, Miller and Klaenhammer 1991), or stimulate the activity of the host MTase for the same purpose (Loenen and Murray 1986).
CRISPR-Cas systems can also be evaded by phages in multiple ways (Malone, Birkholz and Fineran 2021). Phages can acquire point mutations or deletions in the PAM sequences or in positions of the protospacer region close to the PAM sequences (i.e. the seed region of the protospacer) (Tao, Wu and Rao 2018). Alternatively, some phages use anti-CRISPR (Acr) proteins, first described in phages of P. aeruginosa (Bondy-Denomy et al. 2012). In general, Acrs work by either preventing recruitment of the crRNA-Cas complex to the target DNA by binding the complex or occluding the PAM sequence, or by inhibiting the endonuclease domain so that cleavage cannot take place (Stanley and Maxwell 2018). Glucosylation of phage genetic material has also been shown to protect phages against some CRISPR-Cas systems (Vlot et al. 2018). A different strategy is employed by the jumbo Serratia phage PCH45 and Pseudomonas phage ϕKZ, which form a protein shell that encloses phage DNA in a nucleus-like compartment, physically shielding it from the CRISPR-Cas complexes (Malone et al. 2020; Mendoza et al. 2020). Of note, this mechanism does not protect the phage from RNA-targeting CRISPR-Cas systems, as the transcribed mRNA is not contained within the nucleus-like compartment during translation.
Abi mechanisms can also be outsmarted by phages. Phages can avoid toxin–antitoxin mechanisms by inhibiting the protease that degrades the antitoxin, or by expressing their own antitoxin analogue (Blower et al. 2012; Otsuka and Yonesaki 2012; Sberro et al. 2013). Furthermore, mutations in genes involved in the metabolism of nucleic acids also prove to be effective in avoiding toxin–antitoxin systems of some bacteria like Lactococcus spp. (Samson, Bélanger and Moineau 2013). Mutations in phage genes encoding peptides that activate Abi-associated enzymes, such as the Lit activator Gol peptide in the major head protein of T4, can also result in hindering of the Abi mechanism (Bingham et al. 2000). Phages ICP1 that infect V. cholerae overcome PLE-mediated Abi by using either a phage-encoded CRISPR-Cas system that targets the PLE genome during infection (Seed et al. 2013), or an endonuclease that binds and cleaves the PLE origins of replication (Barth, Nguyen and Seed 2021). It is expected that phages possess countermeasures against the more newly described defense systems as well. The Ocr protein of phage T7, known to inhibit R-M systems, was recently found to also inactivate the BREX system by binding the methyltransferase BrxX (Isaev et al. 2020). The discovery of other new phage counterattack strategies is likely just a matter of time, as interest in bacterial defense mechanisms and phage anti-defenses continues to grow.
PHAGE RESISTANCE MECHANISMS IN A CLINICAL CONTEXT
The number of phage therapy case studies and clinical trials performed in humans has significantly increased in these past years, as the problem of antibiotic resistance aggravates. The efficacy of phage therapy in these studies is quite variable, ranging from negative outcomes to the resolution of severe infections in human patients (Table 1; Table S1, Supporting Information). Interestingly, while phage resistance has been shown to develop quickly in vitro, studies in humans have described both the presence (Zhvania et al. 2017) and the absence (Khawaldeh et al. 2011) of phage resistance in vivo. As a consequence of such variable results, there is a lack of consensus in the scientific and medical community about the potential of phages as therapeutic agents.
Table 1.
Case studies and clinical trials of phage therapy in humans, with associated safety, and clinical and phage-resistance outcomes.
| Clinical study/case reporta | Safety | Clinical outcomeb | Phage resistancec | Mechanism of resistance | Ref. |
|---|---|---|---|---|---|
| Tibia bone infection with MDR A. baumannii and Klebsiella pneumoniae treated with phages and antibiotics | Well tolerated | Successful | Not found in the patient; found in in vitro experiments with isolates | Mutations in surface adhesin and glycosyl-transferase of the EpsG family (speculated) | (Nir-Paz et al. 2019) |
| Necrotizing pancreatitis patient with an MDR A. baumannii infection treated with phages and antibiotics | Well tolerated | Successful | A resistant bacterial isolate was found and used to select a new phage | Loss of bacterial capsule and increased extracellular polysaccharide production | (Schooley et al. 2017) |
| Cystic fibrosis patient with MDR P. aeruginosa infection treated with phages and antibiotics | Well tolerated | Successful | One resistant isolate was identified | Not described | (Law et al. 2019) |
| Patient with K. pneumoniae urinary tract infection treated with non-active antibiotics and phages | Well tolerated | Successful | Bacteria became resistant to two rounds of phage cocktails but were ultimately sensitive to the combination of a third cocktail and previously inactive antibiotics | Not yet elucidated | (Bao et al. 2020) |
| Patient with periprosthetic joint infection and MDR P. aeruginosa chronic osteomyelitis treated with phages and antibiotics | Well tolerated | Successful | Not identified | Not applicable | (Tkhilaishvili et al. 2020) |
| Patient with MDR P. aeruginosa urinary tract infection treated with phages and antibiotics | Well tolerated | Successful | Not identified | Not applicable | (Khawaldeh et al. 2011) |
| Aortic graft infection with P. aeruginosa treated with a phage targeting an efflux pump protein and antibiotics | Well tolerated | Clinical improvement | Yes, as expected. Explored to cause sensitization of bacteria to antibiotic. | Mutations in receptor protein (M of mexAB and mexXY efflux system) | (Chan et al. 2018) |
| Cystic fibrosis patient with Mycobacterium abscessus and P. aeruginosa infection treated with engineered phages | Well tolerated | Clinical improvement | Resistance detected in vitro for two of the three phages administered | Not described | (Dedrick et al. 2019) |
| Netherton syndrome patient with MDR Staphylococcus aureus infection and allergy to multiple antibiotics treated with phage cocktails topically and orally | Well tolerated | Clinical improvement | Resistance to a phage cocktail was identified after 3 months of treatment | Not described | (Zhvania et al. 2017) |
| Thirteen patients with S. aureus bacteremia treated with phages and antibiotics | Well tolerated | Clinical improvement in eight of the patients | Changes in phage susceptibility were detected in vivo in three patients | Single nucleotide polymorphisms were detected in isolates recovered from one of the patients | (Petrovic Fabijan et al.2020b) |
| Three lung transplant patients with MDR P. aeruginosa and Burkholderia dolosa infections treated with phages and antibiotics | Well tolerated | Clinical improvement in two of three patients | Resistant isolates were identified post-therapy in one of the patients who had a successful outcome | Not described | (Aslam et al. 2019) |
| Patient with bronchiectasis and M. abscessus infection treated with a phage cocktail and antibiotics | Well tolerated | Failure due to antiphage immune response | Resistant isolates appeared to one out of three phages in the cocktail | Not described | (Dedrick et al. 2021) |
| Eight cardiothoracic surgery patients treated with antibiotics, phages and fibrin glue | Well tolerated except for an increase in inflammation | Successful in five of the patients; two patients showed improvement but died of unrelated complications; one patient did not experience sufficient bacterial clearance and died of sepsis | Bacterial isolates appeared to have mutated in one of the patients | Not described | (Rubalskii et al. 2020) |
| Ten patients with diverse MDR bacterial infections treated with intravenous phage cocktails and antibiotics | Well tolerated except by one patient, who developed fever, wheezing and shortness of breath after first dose of the cocktail (subsequent doses were well tolerated) | Successful for seven out of ten patients | Resistance developed in three of the patients, but was overcome through administration of phages specific for the resistant isolates (two of these cases still had a successful outcome) | Not described | (Aslam et al. 2020) |
| Phases I–II clinical trial to assess the safety and efficacy of a phage cocktail to treat burn wounds infected with P. aeruginosa in 26 patients (13 treated, 13 control) | Fewer adverse events were observed in the phage-treated group than in the group treated with the standard of care | Trial was stopped because of insufficient efficacy (patients were being exposed to 10 000-fold lower doses of phages than originally intended) | Intermediately susceptible and resistant isolates were found | Not described | (Jault et al. 2019) |
| Report of phage therapy performed on 153 patients with different infections | Diverse | Diverse | Resistance and changes in the phage typing profile were detected in multiple cases | Not described | (Międzybrodzki et al. 2012) |
Only clinical studies/case reports for which phage resistance was investigated are shown. For a full list of clinical studies and case reports with phages, refer to Table S1 (Supporting Information).
Successful: patient was healed. Clinical improvement: infection and/or associated complications were reduced but not completely resolved.
Not described: resistance was not addressed. Not identified: resistance was investigated but not found.
The human immune response to bacterial infection and the specific phage-resistance mechanisms developed by the bacteria are likely behind the distinct outcomes. The development of an immune response, particularly involving neutrophils, has been shown essential for the success of phage therapy by preventing the outgrowth of phage-resistant mutants (Roach et al. 2017). Multiple studies have demonstrated that phage-resistant phenotypes often associate with decreased pathogenicity, with the strain becoming more susceptible to the human immune defenses (Sumrall et al. 2019). Receptor adaptations such as mutations in bacterial capsule, LPS and other surface components are examples of phage-resistance mechanisms that result in increased immune susceptibility (Cai et al. 2019). Importantly, these surface modifications also often associate with increased antibiotic susceptibility. This effect occurs, for example, in cases where the phage interacts with bacterial structures that function as drug efflux pumps (Chan et al. 2016; Gurney et al. 2020). By mutating the efflux pump to achieve phage resistance, bacteria lose the ability to pump out the antibiotics, thus gaining antibiotic susceptibility as a trade-off (for a review of mechanisms of phage-antibiotic synergism, see e.g. Tagliaferri, Jansen and Horz 2019). Such interactions have been exploited in therapeutic contexts (León and Bastías 2015; Chaudhry et al. 2017; Oechslin et al. 2017; Chan et al. 2018). However, phage-resistance mutations have also been shown to pleiotropically confer increased antibiotic resistance (Burmeister et al. 2020), and other mechanisms of phage resistance (e.g. CRISPR-Cas, R-M systems) may lead to a phage-resistance phenotype that does not render the bacteria more susceptible to the immune system or to antibiotics. In such cases, resistance to phages may develop in vivo even in the presence of a strong immune response.
Unfortunately, the mechanisms underlying phage resistance are seldom, if ever, investigated in human clinical studies and trials, and represent a clear knowledge gap. Most studies look at the safety and/or clinical outcome of phage therapy, and very few have documented the development of phage resistance (Table 1), let alone the mechanisms behind it. There are studies, however, that indicate that addressing and tackling phage resistance can lead to improved treatment outcomes. One such study found phage-resistant clones in a patient suffering from a multidrug-resistant A. baumannii infection after eight days of treatment with intravenous phage therapy (Schooley et al. 2017). Phage-resistance was associated with loss of bacterial capsule and increased extracellular polysaccharide production, and was overcome via an iterative process of phage cocktail formulation that resulted in the resolution of the infection. Of relevance, the phage-resistant phenotype was associated with increased antibiotic sensitivity, suggesting a fitness cost of phage-resistant mutations in vivo. In another study, the association between phage-resistance and increased antibiotic susceptibility was exploited to treat a patient with a chronic multidrug-resistant P. aeruginosa infection of an aortic graft (Chan et al. 2018). The treatment consisted of a combination of the antibiotic ceftazidime and phage OMKO1, which binds to an outer membrane protein that is part of multidrug efflux systems of P. aeruginosa. This combination explored the capacity of the phage to kill the original strain and the ability of ceftazidime to kill any emerging phage-resistant variants with mutations in the multidrug efflux system, to achieve resolution of the infection.
Characterizing the mechanisms of resistance to phages that target pathogens of interest will inform about the relevance that each phage defense system has in a clinical context, in terms of frequency with which they occur in pathogens and their association with virulence and antibiotic susceptibility of the pathogen. Furthermore, certain natively present defense systems like R-M may affect and reduce the choice of phages available to use in a therapeutic setting. Another issue to consider is that the development of resistance (as well as treatment efficacy) may significantly differ when using single- or multi-phage treatment approaches, and may also vary with the timing and order of phage administration (Wright et al. 2019). The more widespread use of bacteriophages for therapeutic purposes could lead to selection for phage-resistant phenotypes that arise through horizontal gene transfer of phage defense systems. Understanding the complexity of interactions and mechanisms leading to phage resistance will aid the development of phage-based treatments with better clinical outcomes and to engineered phages that may overcome host defense systems.
CONCLUDING REMARKS
In this review, we have provided an overview of the current knowledge of mechanisms behind existing and developing phage resistance, and highlighted the potential knowledge gaps and clinical importance of phage resistance for phage therapeutic strategies. While our understanding of the mechanisms behind phage resistance has expanded in recent years, many defense systems remain uncharacterized or yet undiscovered. As such, the complete picture of phage resistance development remains elusive, especially in the context of the human body.
For phage therapy to move forward, it is imperative that clinical studies and trials also assess the development of resistance in a systematic manner, in which both the emergence of phage resistance and the mechanisms behind it are included in the investigation. Sequencing technologies and genome analysis of both bacterial strains and phages may allow for the identification of defense and anti-defense systems in clinical isolates. Such data will prove invaluable for isolating and selecting candidate phages, as well as for predicting the outcome of the therapeutic intervention.
Improved understanding of how defense systems affect phage therapy, combined with an increased knowledge of the anti-defense strategies employed by phages to counteract bacterial defenses, will greatly contribute to the development of more effective phage-based therapeutic approaches.
ACKNOWLEDGMENTS
The present work is part of the research programme of the Netherlands Centre for One Health (NCOH).
Supplementary Material
Contributor Information
Julia E Egido, Medical Microbiology, University Medical Center Utrecht, Utrecht University, Heidelberglaan 100, 3584 CX Utrecht, Netherlands.
Ana Rita Costa, Department of Bionanoscience, Delft University of Technology, van der Maasweg 9, 2629 HZ Delft, Netherlands; Kavli Institute of Nanoscience, van der Maasweg 9, 2629 HZ Delft, Netherlands; Fagenbank, van der Maasweg 9, 2629 HZ Delft, Netherlands.
Cristian Aparicio-Maldonado, Department of Bionanoscience, Delft University of Technology, van der Maasweg 9, 2629 HZ Delft, Netherlands; Kavli Institute of Nanoscience, van der Maasweg 9, 2629 HZ Delft, Netherlands.
Pieter-Jan Haas, Medical Microbiology, University Medical Center Utrecht, Utrecht University, Heidelberglaan 100, 3584 CX Utrecht, Netherlands.
Stan J J Brouns, Department of Bionanoscience, Delft University of Technology, van der Maasweg 9, 2629 HZ Delft, Netherlands; Kavli Institute of Nanoscience, van der Maasweg 9, 2629 HZ Delft, Netherlands; Fagenbank, van der Maasweg 9, 2629 HZ Delft, Netherlands.
FUNDING
This work was supported by donations from the public and the University Fund from the Delft University of Technology to Fagenbank; the Netherlands Organisation for Scientific Research [NWA Startimpulse 17.366 to ARC and Vici VI.C.182.027 to SJJB]; and the European Research Council under the European Union's Horizon 2020 research and innovation programme [grant agreement no. 101003229 to SJJB].
Conflict of interest
None declared.
REFERENCES
- Abraham JM, Freitag CS, Clements JRet al. . An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Attar S, Westra ER, van der Oost Jet al. . Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–89. [DOI] [PubMed] [Google Scholar]
- Alseth EO, Pursey E, Luján AMet al. . Bacterial biodiversity drives the evolution of CRISPR-based phage resistance. Nature. 2019;574:549–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam S, Courtwright AM, Koval Cet al. . Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am J Transplant. 2019;19:2631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam S, Lampley E, Wooten Det al. . Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect Dis. 2020;7:ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Su TJ, Sturrock SSet al. . Interaction of the ocr gene 0.3 protein of bacteriophage T7 with EcoKl restriction/modification enzyme. Nucleic Acids Res. 2002;30:3936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, White MF. Cyclic oligoadenylate signalling and regulation by ring nucleases during type III CRISPR defence. RNA. 2021;27:855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Wu N, Zeng Yet al. . Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg Microbes Infect. 2020;9:771–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ZK, Nguyen MH, Seed KD. A chimeric nuclease substitutes a phage CRISPR-Cas system to provide sequence-specific immunity against subviral parasites. Elife. 2021;10:2021.02.21.432181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ZK, Silvas TV, Angermeyer Aet al. . Genome replication dynamics of a bacteriophage and its satellite reveal strategies for parasitism and viral restriction. Nucleic Acids Res. 2020;48:249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benler S, Cobián-Güemes AG, McNair Ket al. . A diversity-generating retroelement encoded by a globally ubiquitous Bacteroides phage. Microbiome. 2018;6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim A, Millman A, Ofir Get al. . Prokaryotic viperins produce diverse antiviral molecules. Nature. 2021;589:120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi Silva J, Storms Z, Sauvageau D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. 2016;363:fnw002. [DOI] [PubMed] [Google Scholar]
- Bingham R, Ekunwe SIN, Falk Set al. . The major head protein of bacteriophage T4 binds specifically to elongation factor Tu. J Biol Chem. 2000;275:23219–26. [DOI] [PubMed] [Google Scholar]
- Blower TR, Evans TJ, Przybilski Ret al. . Viral evasion of a bacterial suicide system by RNA-based molecular mimicry enables infectious altruism. PLoS Genet. 2012;8:e1003023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KLet al. . Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2012;493:429–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Qian J, Westra ERet al. . Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10:2854–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P, Jacquot P, Plançon Let al. . Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem. 2008;283:13556–64. [DOI] [PubMed] [Google Scholar]
- Broniewski JM, Meaden S, Paterson Set al. . The effect of phage genetic diversity on bacterial resistance evolution. ISME J. 2020;14:828–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren Met al. . Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister AR, Fortier A, Roush Cet al. . Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc Natl Acad Sci USA. 2020;117:11207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Wang G, Le Set al. . Three capsular polysaccharide synthesis-related glucosyltransferases, GT-1, GT-2 and WcaJ, are associated with virulence and phage sensitivity of Klebsiella pneumoniae. Front Microbiol. 2019;10. DOI: 10.3389/fmicb.2019.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan BK, Sistrom M, Wertz JEet al. . Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan BK, Turner PE, Kim Set al. . Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health. 2018;2018:60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanishvili N. Phage therapy—history from Twort and d'Herelle through Soviet experience to current approaches. Adv Virus Res. 2012;83:3–40. [DOI] [PubMed] [Google Scholar]
- Chapman-McQuiston E, Wu XL.. Stochastic receptor expression allows sensitive bacteria to evade phage attack. Part I: experiments. Biophys J. 2008a;94:4525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-McQuiston E, Wu XL. Stochastic receptor expression allows sensitive bacteria to evade phage attack. Part II: theoretical analyses. Biophys J. 2008b;94:4537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry WN, Concepción-Acevedo J, Park Tet al. . Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One. 2017;12:e0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wang W, Molineux IJ. F exclusion of bacteriophage T7 occurs at the cell membrane. Virology. 2004;326:340–52. [DOI] [PubMed] [Google Scholar]
- Choi KH, McPartland J, Kaganman Iet al. . Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J Mol Biol. 2008;378:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Shin H, Lee J-Het al. . Identification and characterization of a novel flagellum-dependent Salmonella-infecting bacteriophage, iEPS5. Appl Environ Microbiol. 2013;79:4829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M-C, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–9. [DOI] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman Aet al. . Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature. 2019;574:691–5. [DOI] [PubMed] [Google Scholar]
- Cota I, Sánchez-Romero MA, Hernández SBet al. . Epigenetic control of Salmonella enterica O-antigen chain length: a tradeoff between virulence and bacteriophage resistance. PLoS Genet. 2015;11:e1005667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumby N, Reimer K, Mengin-Lecreulx Det al. . The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coliphage HK97. Mol Microbiol. 2015;96:437–47. [DOI] [PubMed] [Google Scholar]
- d'Herelle F. Sur un microbe invisible antagoniste des bacilles dysentériques. Comptes rendus Acad Sci Paris. 1917;165:373–5. [Google Scholar]
- Davies J. Specialized microbial metabolites: functions and origins. J Antibiot (Tokyo). 2013;66:361–4. [DOI] [PubMed] [Google Scholar]
- de Jonge PA, Nobrega FL, Brouns SJJet al. . Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019;27:51–63. [DOI] [PubMed] [Google Scholar]
- De Smet J, Hendrix H, Blasdel BGet al. . Pseudomonas predators: understanding and exploiting phage–host interactions. Nat Rev Microbiol. 2017;15:517–30. [DOI] [PubMed] [Google Scholar]
- Decker K, Krauel V, Meesmann Aet al. . Lytic conversion of Escherichia coli by bacteriophage T5: blocking of the FhuA receptor protein by a lipoprotein expressed early during infection. Mol Microbiol. 1994;12:321–32. [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Freeman KG, Nguyen JAet al. . Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat Med. 2021;27:1357–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Guerrero-Bustamante CA, Garlena RAet al. . Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25:730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Jacobs-Sera D, Bustamante CAGet al. . Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol. 2017;2:16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir Get al. . Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359. DOI: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- Drobysheva AV, Panafidina SA, Kolesnik MVet al. . Structure and function of virion RNA polymerase of a crAss-like phage. Nature. 2021;589:306–9. [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska Bet al. . Learning from bacteriophages: advantages and limitations of phage and phage-encoded protein applications. Curr Protein Pept Sci. 2013;13:699–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Huo W, Bhardwaj Pet al. . Molecular basis for lytic bacteriophage resistance in Enterococci. mBio. 2016;7. DOI: 10.1128/MBIO.01304-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy RL, Przybilski R, Semeijn Ket al. . A widespread bacteriophage abortive infection system functions through a type IV toxin–antitoxin mechanism. Nucleic Acids Res. 2014;42:4590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybvig K. DNA rearrangements and phenotypic switching in prokaryotes. Mol Microbiol. 1993;10:465–71. [DOI] [PubMed] [Google Scholar]
- Erez Z, Steinberger-Levy I, Shamir Met al. . Communication between viruses guides lysis–lysogeny decisions. Nature. 2017;541:488–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster MR, Morax LS, Hüls VJet al. . Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol Microbiol. 2015;97:33–46. [DOI] [PubMed] [Google Scholar]
- Fields BN, Knipe DM, David Met al. . Virology. Lippincott-Raven Publishers, 1996. [Google Scholar]
- Fineran PC, Blower TR, Foulds IJet al. . The phage abortive infection system, ToxIN, functions as a protein-RNA toxin–antitoxin pair. Proc Natl Acad Sci USA. 2009;106:894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde A, Fitzgerald GF. Molecular organization of exopolysaccharide (EPS) encoding genes on the lactococcal bacteriophage adsorption blocking plasmid, pCI658. Plasmid. 2003;49:130–42. [DOI] [PubMed] [Google Scholar]
- Gao L, Altae-Tran H, Böhning Fet al. . Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science. 2020;369:1077–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencay YE, Sørensen MCH, Wenzel CQet al. . Phase variable expression of a single phage receptor in Campylobacter jejuni NCTC12662 influences sensitivity toward several diverse CPS-dependent phages. Front Microbiol. 2018;9:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleditzsch D, Pausch P, Müller-Esparza Het al. . PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2018:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T, Sberro H, Weinstock Eet al. . BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34:169–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovenko D, Manakova E, Tamulaitiene Get al. . Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009;37:6613–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García VA, Pulido-Cid M, Garcia-Doval Cet al. . Conformational changes leading to T7 DNA delivery upon interaction with the bacterial receptor. J Biol Chem. 2015;290:10038–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeeva J, Morozova N, Sierro Net al. . BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. 2019;47:253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordillo Altamirano F, Forsyth JH, Patwa Ret al. . Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol. 2021;6:157–61. [DOI] [PubMed] [Google Scholar]
- Guo H, Arambula D, Ghosh Pet al. . Diversity-generating retroelements in phage and bacterial genomes. Microbiol Spectr. 2014;2. DOI: 10.1128/microbiolspec.MDNA3-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Capalash N, Sharma P. Restriction endonucleases: natural and directed evolution. Appl Microbiol Biotechnol. 2012;94:583–99. [DOI] [PubMed] [Google Scholar]
- Gurney J, Pradier L, Griffin JSet al. . Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol Med Public Health. 2020;2020:148–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ, Fineran PC.. The arms race between bacteria and their phage foes. Nature. 2020;577:327–36. [DOI] [PubMed] [Google Scholar]
- Hanlon GW, Denyer SP, Olliff CJet al. . Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2001;67:2746–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SG, Seed KD. Dominant vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite. eLife. 2020;9. DOI: 10.7554/eLife.53200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichman KA, Johnson RC. The Hin invertasome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–7. [DOI] [PubMed] [Google Scholar]
- Helbig KJ, Beard MR. The role of viperin in the innate antiviral response. J Mol Biol. 2014;426:1210–9. [DOI] [PubMed] [Google Scholar]
- Hill C, Miller LA, Klaenhammer TR. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991;173:4363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille F, Richter H, Wong SPet al. . The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239–59. [DOI] [PubMed] [Google Scholar]
- Hinton DM. Transcriptional control in the prereplicative phase of T4 development. Virol J. 2010;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs Z, Abedon ST. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett. 2016;363:fnw047. [DOI] [PubMed] [Google Scholar]
- Iida S, Streiff MB, Bickle TAet al. . Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA− phages. Virology. 1987;157:156–66. [DOI] [PubMed] [Google Scholar]
- Isaev A, Drobiazko A, Sierro Net al. . Phage T7 DNA mimic protein Ocr is a potent inhibitor of BREX defence. Nucleic Acids Res. 2020;48:5397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RDet al. . CRISPR-Cas: adapting to change. Science. 2017;356. DOI: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- Jault P, Leclerc T, Jennes Set al. . Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis. 2019;19:35–45. [DOI] [PubMed] [Google Scholar]
- Juhala RJ, Ford ME, Duda RLet al. . Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. [DOI] [PubMed] [Google Scholar]
- Ka D, Oh H, Park Eet al. . Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+ degradation. Nat Commun. 2020;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp P, Gupta M, Molineux IJ. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol Microbiol. 2004;53:1251–65. [DOI] [PubMed] [Google Scholar]
- Khawaldeh A, Morales S, Dillon Bet al. . Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J Med Microbiol. 2011;60:1697–700. [DOI] [PubMed] [Google Scholar]
- King A. Caudovirales. Virus Taxonomy. Elsevier, 2012, 39–45. [Google Scholar]
- Knecht LE, Veljkovic M, Fieseler L. Diversity and function of phage encoded depolymerases. Front Microbiol. 2020;10:2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E V, Makarova KS, Wolf YI.. Evolutionary genomics of defense systems in archaea and bacteria. Annu Rev Microbiol. 2017;71:233–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc Lond B Biol Sci. 2019;374. DOI: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortright KE, Chan BK, Turner PE. High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc Natl Acad Sci. 2020;117:18670–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronheim S, Daniel-Ivad M, Duan Zet al. . A chemical defence against phage infection. Nature. 2018;564:283–6. [DOI] [PubMed] [Google Scholar]
- Labrie SJ, Mosterd C, Loignon Set al. . A mutation in the methionine aminopeptidase gene provides phage resistance in Streptococcus thermophilus. Sci Rep. 2019;9. DOI: 10.1038/s41598-019-49975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. [DOI] [PubMed] [Google Scholar]
- Latka A, Maciejewska B, Majkowska-Skrobek Get al. . Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl Microbiol Biotechnol. 2017;101:3103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law N, Logan C, Yung Get al. . Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection. 2019;47:665–8. [DOI] [PubMed] [Google Scholar]
- León M, Bastías R. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol. 2015;06:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptihn S, Gottschalk J, Kuhn A. T7 ejectosome assembly: a story unfolds. Bacteriophage. 2016;6:e1128513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz R, Chapman D, Amitsur Met al. . The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA, Murray NE. Modification enhancement by the restriction alleviation protein (Ral) of bacteriophage lambda. J Mol Biol. 1986;190:11–22. [DOI] [PubMed] [Google Scholar]
- Loenen WAM, Raleigh EA. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014;42:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatina A, Tal N, Sorek R. Abortive infection: bacterial suicide as an antiviral immune strategy. Ann Rev Virol. 2020;7:371–84. [DOI] [PubMed] [Google Scholar]
- Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. J Bacteriol. 1952;64:557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE. Host-induced modifications of viruses. Cold Spring Harb Symp Quant Biol. 1953;18:237–44. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Timinskas A, Wolf YIet al. . Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res. 2020a;48:8828–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo Jet al. . Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020b;18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone LM, Birkholz N, Fineran PC. Conquering CRISPR: how phages overcome bacterial adaptive immunity. Curr Opin Biotechnol. 2021;68:30–6. [DOI] [PubMed] [Google Scholar]
- Malone LM, Warring SL, Jackson SAet al. . A jumbo phage that forms a nucleus-like structure evades CRISPR–Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol. 2020;5:48–55. [DOI] [PubMed] [Google Scholar]
- Mangalea MR, Duerkop BA. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect Immun. 2020;88. DOI: 10.1128/IAI.00926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinn J, Marraffini LA. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat Rev Microbiol. 2019;17:7–12. [DOI] [PubMed] [Google Scholar]
- McKitterick AC, LeGault KN, Angermeyer Aet al. . Competition between mobile genetic elements drives optimization of a phage-encoded CRISPR-Cas system: insights from a natural arms race. Philos Trans R Soc Lond B Biol Sci. 2019;374. DOI: 10.1098/rstb.2018.0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza SD, Nieweglowska ES, Govindarajan Set al. . A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature. 2020;577:244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Międzybrodzki R, Borysowski J, Weber-Dąbrowska Bet al. . Clinical aspects of phage therapy. Adv Virus Res. 2012;83:73–121. [DOI] [PubMed] [Google Scholar]
- Millen AM, Horvath P, Boyaval Pet al. . Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS One. 2012;7. DOI: 10.1371/journal.pone.0051663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman A, Bernheim A, Stokar-Avihail Aet al. . Bacterial retrons function in anti-phage defense. Cell. 2020a;183:1551–61. [DOI] [PubMed] [Google Scholar]
- Millman A, Melamed S, Amitai Get al. . Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat Microbiol. 2020b:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S, McAuliffe OE, Coffey Aet al. . Plasmids of lactococci-genetic accessories or genetic necessities?. FEMS Microbiol Rev. 2006;30:243–73. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Ito K, Murakami Ret al. . Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat Commun. 2016;7:11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelling K, Broecker F, Willy C. A wake-up call: we need phage therapy now. Viruses. 2018;10. DOI: 10.3390/v10120688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez Jet al. . Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. [DOI] [PubMed] [Google Scholar]
- Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11:194–204. [DOI] [PubMed] [Google Scholar]
- Nir-Paz R, Gelman D, Khouri Aet al. . Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin Infect Dis. 2019;69:2015–8. [DOI] [PubMed] [Google Scholar]
- Nobrega FL, Vlot M, de Jonge PAet al. . Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018;16:760–73. [DOI] [PubMed] [Google Scholar]
- O'Hara BJ, Barth ZK, McKitterick ACet al. . A highly specific phage defense system is a conserved feature of the Vibrio cholerae mobilome. PLoS Genet. 2017;13:e1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oechslin F, Piccardi P, Mancini Set al. . Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis. 2017;215:703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Herbst E, Baroz Met al. . Antiviral activity of bacterial TIR domains via signaling molecules that trigger cell death. bioRxiv. 2021. DOI: 10.1101/2021.01.06.425286. [DOI] [PubMed] [Google Scholar]
- Ofir G, Melamed S, Sberro Het al. . DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol. 2018;3:90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Yonesaki T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol Microbiol. 2012;83:669–81. [DOI] [PubMed] [Google Scholar]
- Page R, Peti W. Toxin–antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol. 2016;12:208–14. [DOI] [PubMed] [Google Scholar]
- Panja D, Molineux IJ. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys Biol. 2010;7. DOI: 10.1088/1478-3975/7/4/045006. [DOI] [PubMed] [Google Scholar]
- Parma DH, Snyder M, Sobolevski Set al. . The Rex system of bacteriophage lambda: tolerance and altruistic cell death. Genes Dev. 1992;6:497–510. [DOI] [PubMed] [Google Scholar]
- Paul BG, Burstein D, Castelle CJet al. . Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat Microbiol. 2017;2:17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Rosenbusch JP, Locher KP. Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5-encoded lipoprotein. FEMS Microbiol Lett. 1998;168:119–25. [DOI] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Khalid A, Maddocks Set al. . Phage therapy for severe bacterial infections: a narrative review. Med J Aust. 2020a;212:279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic Fabijan A, Lin RCY, Ho Jet al. . Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat Microbiol. 2020b;5:465–72. [DOI] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DAet al. . Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One. 2011;6:e16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P, Lobkovsky AE, Kristensen DMet al. . Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Med. 2014;12. DOI: 10.1186/s12915-014-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede I, Eschbach ML. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986;205:241–5. [DOI] [PubMed] [Google Scholar]
- Roach DR, Debarbieux L. Phage therapy: awakening a sleeping giant. Emerg Top Life Sci. 2017;1:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach DR, Leung CY, Henry Met al. . Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. 2017;22:38–47. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–58. [DOI] [PubMed] [Google Scholar]
- Rousset F, Dowding J, Bernheim Aet al. . Prophage-encoded hotspots of bacterial immune systems. bioRxiv. 2021. DOI: 10.1101/2021.01.21.427644. [Google Scholar]
- Rubalskii E, Ruemke S, Salmoukas Cet al. . Bacteriophage therapy for critical infections related to cardiothoracic surgery. Antibiotics. 2020;9. DOI: 10.3390/antibiotics9050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov IS, Ershova AS, Karyagina ASet al. . Avoidance of recognition sites of restriction-modification systems is a widespread but not universal anti-restriction strategy of prokaryotic viruses. BMC Genomics. 2018;19:885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson JE, Bélanger M, Moineau S. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J Bacteriol. 2013;195:3947–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari J, Pandit N, Moxon ERet al. . Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol. 1994;13:207–17. [DOI] [PubMed] [Google Scholar]
- Sberro H, Leavitt A, Kiro Ret al. . Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol Cell. 2013;50:136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley RT, Biswas B, Gill JJet al. . Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61. DOI: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed KD, Lazinski DW, Calderwood SBet al. . A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin GB, Hsueh BY, Elg CAet al. . A broadly conserved deoxycytidine deaminase protects bacteria from phage infection. bioRxiv. 2021. DOI: 10.1101/2021.03.31.437871. [Google Scholar]
- Silpe JE, Bassler BL. A host-produced quorum-sensing autoinducer controls a phage lysis–lysogeny decision. Cell. 2019;176:268–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SBet al. . The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. [DOI] [PubMed] [Google Scholar]
- Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents?. Mol Microbiol. 1995;15:415–20. [DOI] [PubMed] [Google Scholar]
- Stanley SY, Maxwell KL. Phage-encoded anti-CRISPR defenses. Annu Rev Genet. 2018;52:445–64. [DOI] [PubMed] [Google Scholar]
- Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Panne D, Bickle TAet al. . Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J Mol Biol. 2000;298:611–22. [DOI] [PubMed] [Google Scholar]
- Sumrall ET, Shen Y, Keller APet al. . Phage resistance at the cost of virulence: listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathog. 2019;15:e1008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du Fet al. . Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ERet al. . DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliaferri TL, Jansen M, Horz H-P. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol. 2019;0:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Wolf YI, Makarova KSet al. . Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal N, Millman A, Stokar-Avihail Aet al. . Antiviral defense via nucleotide depletion in bacteria. bioRxiv. 2021. DOI: 10.1101/2021.04.26.441389. [Google Scholar]
- Tao P, Wu X, Rao V. Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci Adv. 2018;4:4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NMI, Prokhorov NS, Guerrero-Ferreira RCet al. . Structure of the T4 baseplate and its function in triggering sheath contraction. Nature. 2016;533:346–52. [DOI] [PubMed] [Google Scholar]
- Testa S, Berger S, Piccardi Pet al. . Spatial structure affects phage efficacy in infecting dual-strain biofilms of Pseudomonas aeruginosa. Commun Biol. 2019;2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkhilaishvili T, Winkler T, Müller Met al. . Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2020;64. DOI: 10.1128/AAC.00924-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–72. [DOI] [PubMed] [Google Scholar]
- Trudelle DM, Bryan DW, Hudson LKet al. . Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019;84:103239. [DOI] [PubMed] [Google Scholar]
- Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet North Am Ed. 1915;186:1241–3. [Google Scholar]
- Van Houte S, Buckling A, Westra ER. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev. 2016;80:745–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot M, Houkes J, Lochs SJAet al. . Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR–Cas effector complexes. Nucleic Acids Res. 2018;46:873–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tu J, Liu Jet al. . Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat Microbiol. 2019;4:1049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wan M, Huang Ret al. . SspABCD-SspFGH constitutes a new type of DNA phosphorothioate-based bacterial defense system. mBio. 2021;12. DOI: 10.1128/mBio.00613-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BNJ, Vercoe RB, Salmond GPCet al. . Type I-F CRISPR-Cas resistance against virulent phages results in abortive infection and provides population-level immunity. Nat Commun. 2019;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand T, Karambelkar S, Bondy-Denomy Jet al. . Structures and strategies of anti-CRISPR-mediated immune suppression. Annu Rev Microbiol. 2020;74:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems R, Paul A, van der Heide HGet al. . Fimbrial phase variation in Bordetella pertussis: a novel mechanism for transcriptional regulation. EMBO J. 1990;9:2803–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willkomm S, Makarova KS, Grohmann D. DNA silencing by prokaryotic Argonaute proteins adds a new layer of defense against invading nucleic acids. FEMS Microbiol Rev. 2018;42:376–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittebole X, De Roock S, Opal SM. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence. 2014;5:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RCT, Friman VP, Smith MCMet al. . Resistance evolution against phage combinations depends on the timing and order of exposure. mBio. 2019;10. DOI: 10.1128/mBio.01652-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang J, Cho WCet al. . Argonaute proteins: structural features, functions and emerging roles. J Adv Res. 2020;24:317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Wu G, Wei Yet al. . SspABCD–SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat Microbiol. 2020;5:917–28. [DOI] [PubMed] [Google Scholar]
- Xu T, Yao F, Zhou Xet al. . A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavil'gelskiĭ GB, Kotova VI. Antirestriction activity of T7 Ocr protein in monomeric and dimeric forms. Mol Biol (Mosk). 2014;48:176–84. [PubMed] [Google Scholar]
- Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2014;38:119–41. [DOI] [PubMed] [Google Scholar]
- Zhvania P, Hoyle NS, Nadareishvili Let al. . Phage therapy in a 16-year-old boy with Netherton syndrome. Front Med. 2017;4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.