Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen and a leading cause of chronic infection in the lungs of individuals with cystic fibrosis. After colonization, P. aeruginosa often undergoes a phenotypic conversion to mucoidy, characterized by overproduction of the alginate exopolysaccharide. This conversion is correlated with poorer patient prognoses. The majority of genes required for alginate synthesis, including the alginate lyase, algL, are located in a single operon. Previous investigations of AlgL have resulted in several divergent hypotheses regarding the protein’s role in alginate production. To address these discrepancies, we determined the structure of AlgL and, using multiple sequence alignments, identified key active site residues involved in alginate binding and catalysis. In vitro enzymatic analysis of active site mutants highlights R249 and Y256 as key residues required for alginate lyase activity. In a genetically engineered P. aeruginosa strain where alginate biosynthesis is under arabinose control, we found that AlgL is required for cell viability and maintaining membrane integrity during alginate production. We demonstrate that AlgL functions as a homeostasis enzyme to clear the periplasmic space of accumulated polymer. Constitutive expression of the AlgU/T sigma factor mitigates the effects of an algL deletion during alginate production, suggesting that an AlgU/T-regulated protein or proteins can compensate for an algL deletion. Together, our study demonstrates the role of AlgL in alginate biosynthesis, explains the discrepancies observed previously across other P. aeruginosa ΔalgL genetic backgrounds, and clarifies the existing divergent data regarding the function of AlgL as an alginate degrading enzyme.
Keywords: Pseudomonas aeruginosa, alginate lyase, biofilm, polysaccharide, bacterial genetics, crystallography, enzyme structure, structure–function
Abbreviations: Carb, carbenicillin; CAZy, Carbohydrate-Active enZYmes; CF, cystic fibrosis; E.C., enzyme commission; eDNA, extracellular DNA; Gen, gentamicin; GulA, L-guluronic acid/guluronate; HRP, horseradish peroxidase; IPTG, isopropyl β-D-1-thiogalactopyranoside; Kan, kanamycin; kcat, turnover number; LB, lysogeny broth; MAP, modified alginate producing; ManA3, mannuronate trisaccharide; ManA, D-mannuronic acid/mannuronate; NSLB, no-salt lysogeny broth; OD, optical density; PL, polysaccharide lyase; polyM, polymannuronate; polyMG, polymannuronate-guluronate; PNAG, poly-N-acetylglucosamine; SAD, single-wavelength anomalous diffraction; SeMet, selenomethionine; TBS, tris-buffered saline; TBST, tris-buffered saline with Tween-20; TEM, transmission electron microscopy; VBMM, Vogel–Bonner minimal medium; VSV-G, vesicular stomatitis virus glycoprotein
Biofilms are highly structured communities of bacterial cells embedded in a self-produced matrix (1, 2). Their ability to adhere to a variety of biotic and abiotic surfaces makes biofilms ubiquitous in natural, industrial, and clinical settings. Biofilms are found in deep-sea vents, freshwater rivers, drinking water distribution systems, food processing equipment, and even the International Space Station and are responsible for the contamination of medical devices such as catheters, prosthetic heart valves, and cardiac pacemakers (3, 4, 5, 6, 7, 8). Biofilms are also responsible for tissue-related infections including diffuse panbronchiolitis, lung infections in individuals with cystic fibrosis (CF), and chronic wound infections (9, 10, 11, 12, 13, 14). Composed of proteins, extracellular DNA (eDNA), and exopolysaccharides, the biofilm matrix confers an advantage to the bacteria by providing protection from antibiotic treatments and the host’s immune response (3, 15). The opportunistic human pathogen Pseudomonas aeruginosa is notorious for establishing chronic infections in the lungs of individuals with CF and tolerating antibiotic treatment through formation of a biofilm.
P. aeruginosa is genetically capable of producing three distinct exopolysaccharides as part of its biofilm matrix: Pel, Psl, and alginate. Each polymer plays an important role in chronic infections (16). For example, Pel binds to eDNA in the stalk of the biofilm near the point of attachment, is associated with P. aeruginosa aggregates in CF sputum, and provides protection from aminoglycoside antibiotics (17, 18, 19). Psl has also been associated with P. aeruginosa aggregates in CF sputum and is important for surface adhesion and biofilm structure (19, 20, 21), while alginate is typically associated with the chronic lung infections suffered by individuals with CF where it blocks cell-mediated phagocytosis and hence aids in evasion of the host immune response (22, 23). Alginate is produced when P. aeruginosa converts to a mucoid state (24). This conversion is typically induced by a mutation in the antisigma factor MucA that is responsible for regulating the sigma factor AlgU/T (25, 26). Alginate overproduction also promotes P. aeruginosa coinfection with Staphylococcus aureus, thus influencing CF patient outcomes as coinfection is associated with decreased lung function (27, 28, 29). Upregulation of genes involved in alginate biosynthesis has also been observed in a P. aeruginosa murine burn wound model, demonstrating the polymer’s importance in biofilm formation outside of the CF lung environment (30).
Except for algC, the genes required for alginate biosynthesis are clustered in a single operon (31) (Fig. S1). Alginate is initially synthesized as an anionic homopolymer that is chemically modified by acetylation and epimerization in the periplasm prior to the export of the polymer (Fig. S1). Proteins involved in this process are hypothesized to form a multiprotein complex that spans the inner and outer membranes. AlgA, AlgC, and AlgD are involved in the production of the alginate precursor molecule GDP-mannuronic acid, which, in response to cyclic di-GMP binding to the PilZ domain of Alg44, is polymerized by Alg8 and Alg44 to form β-1,4-linked D-mannuronic acid (ManA) (31, 32, 33, 34, 35). Once in the periplasm, AlgF, AlgI, AlgJ, and AlgX acetylate the ManA homopolymer at the O2 and/or O3 hydroxyls (31, 36, 37, 38), while AlgG selectively epimerizes nonacetylated ManA residues to L-guluronate (GulA) (31). The polymer is exported from the cell via AlgK and the outer membrane ß-barrel porin AlgE (39, 40).
The alg operon also encodes a periplasmic lyase, AlgL. Characterization of this enzyme in vitro demonstrated that it preferentially degrades nonacetylated polymannuronate (polyM) via a ß-elimination mechanism (41, 42). Despite our detailed understanding of its reaction mechanism, the function of AlgL in alginate biosynthesis remains poorly understood. Several different roles for the enzyme have been proposed. These include the regulation of the length of secreted alginate polymer by cleaving β-1,4-linkages between ManA residues prior to export (43, 44); aiding in biofilm detachment (45); and degradation of alginate that is not exported from the cell to prevent its accumulation within the periplasmic space (46, 47). Studies have also suggested that the enzyme is part of a multiprotein complex with AlgG, AlgX, and AlgK and that it assists in transporting the polymer across the outer membrane (47). This contrasts with more recent studies that suggest AlgL does not associate with other alginate biosynthesis proteins (48). Most notably, AlgL has variably been suggested to be required for alginate production (49), required for P. aeruginosa viability during alginate production (47), and completely dispensable for alginate production and biofilm biomass (48).
In the present study, we employed a multidisciplinary approach to address these discrepancies and determine the role of AlgL. Our structure of wild-type (WT) P. aeruginosa AlgL in complex with ManA, its comparison with other bacterial alginate lyases, and in vitro enzyme kinetic analyses have enabled the identification of active site residues important for alginate binding and catalysis. In a genetically engineered strain where alginate biosynthesis can be controlled using arabinose, we demonstrate that absence of algL or mutation of key catalytic residues is detrimental for P. aeruginosa growth during alginate biosynthesis and results in abnormal cellular morphology. Furthermore, we show that AlgL prevents lethal accumulation of alginate during polymer production suggesting that the enzyme is important for homeostasis of the periplasm. Finally, using a mucA22 P. aeruginosa background, we found that absence of algL is tolerated. Taken together, our data suggest that, when necessary, AlgL functions as a periplasmic homeostasis enzyme during alginate production and that a protein or proteins can compensate for its loss when the entire AlgU/T regulon is upregulated.
Results
AlgL has an (α/α)5 toroid fold
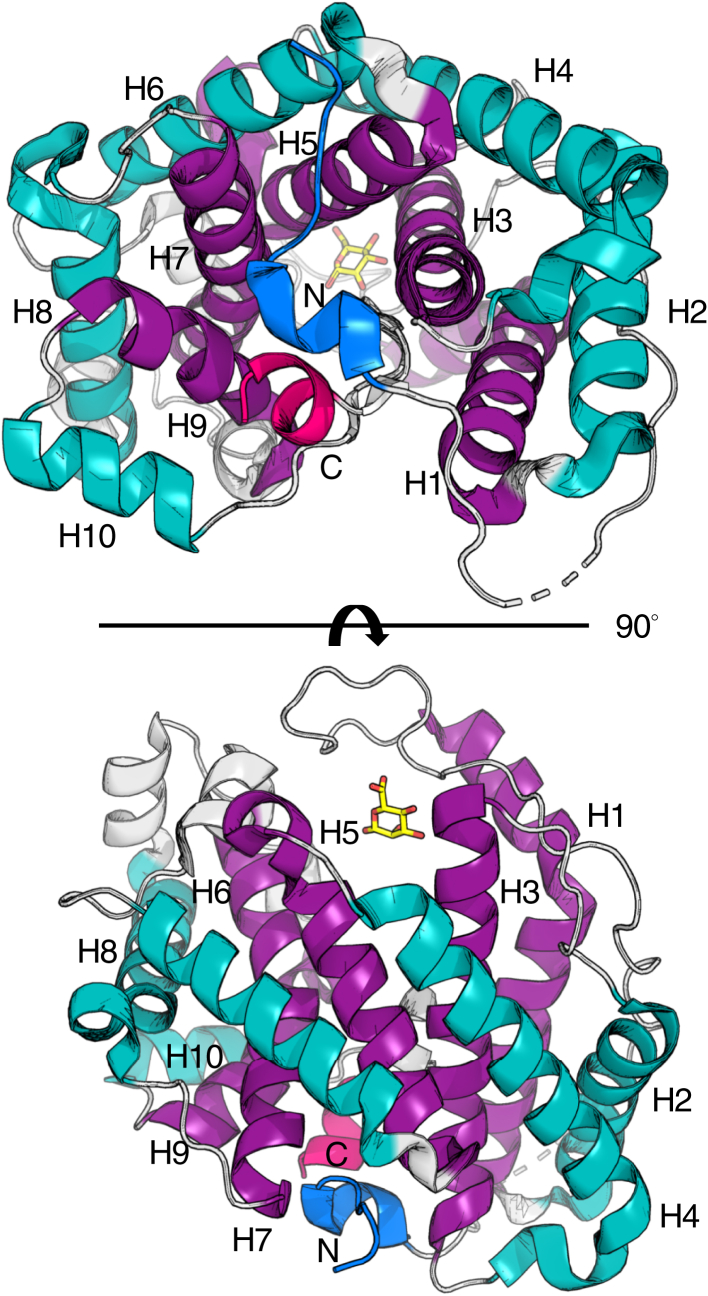
To enable our functional in vivo studies of AlgL and delineate the role of this enzyme in alginate biosynthesis, we first determined its structure, minus its signal sequence (NHis6-AlgL28–362), to 1.65 Å resolution using selenomethionine (SeMet) incorporation and the single-wavelength anomalous dispersion (SAD) technique. AlgL crystallized in space group P212121 with one molecule in the asymmetric unit (Table 1). The final model of AlgL was refined to a final Rwork and Rfree of 17.7% and 19.4%, respectively. The structure reveals that AlgL adopts an (α/α)5 toroid fold with five pairs of antiparallel α-helices (Fig. 1). Examination of the carbohydrate-active enzymes (CAZy; http://www.cazy.org) database reveals that this (α/α)n spatial arrangement has been reported previously in alginate lyases from the polysaccharide lyase (PL) family PL-5 to which AlgL belongs, as well as 12 other PL families (50, 51).
Table 1.
Data collection and refinement statistics
| AlgL SeMet | AlgL | AlgL H202A | AlgL K66A | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.97920 | 1.075 | 1.075 | 1.5406 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Space Group | P212121 | P212121 | P212121 | P21212 |
| Cell Dimensions | ||||
| a, b, c (Å) | 56.0, 59.8, 94.6 | 56.4, 59.6, 102.1 | 56.4, 59.4, 102.1 | 67.5, 58.7, 76.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.1 (2.18–2.10) | 50.0–1.64 (1.70–1.64) | 50.0–1.64 (1.70–1.64) | 20.3–2.50 (2.60–2.50) |
| Total No. of Reflections | 253,565 | 542,308 | 594,045 | 127,670 |
| No. of Unique Reflections | 19,041 | 43,027 | 43,079 | 10,903 |
| Redundancy | 13.4 (10.5) | 13.2 (10.6) | 15.0 (12.8) | 10.3 (7.75) |
| Completeness (%) | 95.2 (71.2) | 99.0 (94.0) | 99.3 (100.0) | |
| Average I/σ (I) | 14.2 (2.5) | 25.8 (3.2) | 22.0 (11.6) | 16.0 (3.3) |
| Rmergea (%) | 16.5 (65.1) | 8.9 (55.6) | 9.4 (26.2) | 9.8 (43.5) |
| Refinementb | ||||
| Rwork/Rfree (%)c | 17.7/19.4 | 16.8/19.3 | 24.6/26.9 | |
| No. atoms | ||||
| Protein | 2601 | 2606 | 2458 | |
| Ligand | 13 | |||
| Solvent | 107 | 161 | 38 | |
| Average B-factors (Å2) | ||||
| Protein | 25.0 | 22.1 | 32.8 | |
| Ligand | 48.2 | |||
| Water | 25.0 | 25.9 | 35.0 | |
| Root mean square deviations | ||||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 | |
| Bond angles (°) | 0.97 | 0.98 | 1.5 | |
| Ramachandran plotd | ||||
| Total favored (%) | 99.0 | 99.0 | 97.0 | |
| Total allowed (%) | 100 | 100 | 100 | |
| Est. coordinate error (Å)e | 0.16 | 0.12 | 0.30 | |
| PDB code | 4OZV | 4OZW | 7SA8 |
Values in parentheses are for the highest-resolution shell.
Rmerge = Σhkl Σi |Ii(hkl) – <I(hkl)>|/Σhkl Σi Ii(hkl), where Ii(hkl) and <I(hkl)> represent the diffraction-intensity values of the individual measurements and the corresponding mean values, respectively.
AlgL and AlgL H202A were refined using PHENIX.REFINE (95); AlgL K66A was refined using REFMAC5 (99).
Rwork = Σ||Fobs| − k|Fcalc||/|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
As calculated using MolProbity (114).
Figure 1.
Structure of AlgL from Pseudomonas aeruginosa reveals an (α/α)5toroid fold with five pairs of antiparallel α-helices. Helices are numbered from H1 to H10. Outer helices are colored in teal, inner helices are colored in purple, the N terminus is colored in blue, and the C terminus is colored in magenta. A single monomer of mannuronate (ManA) (yellow) is observed.
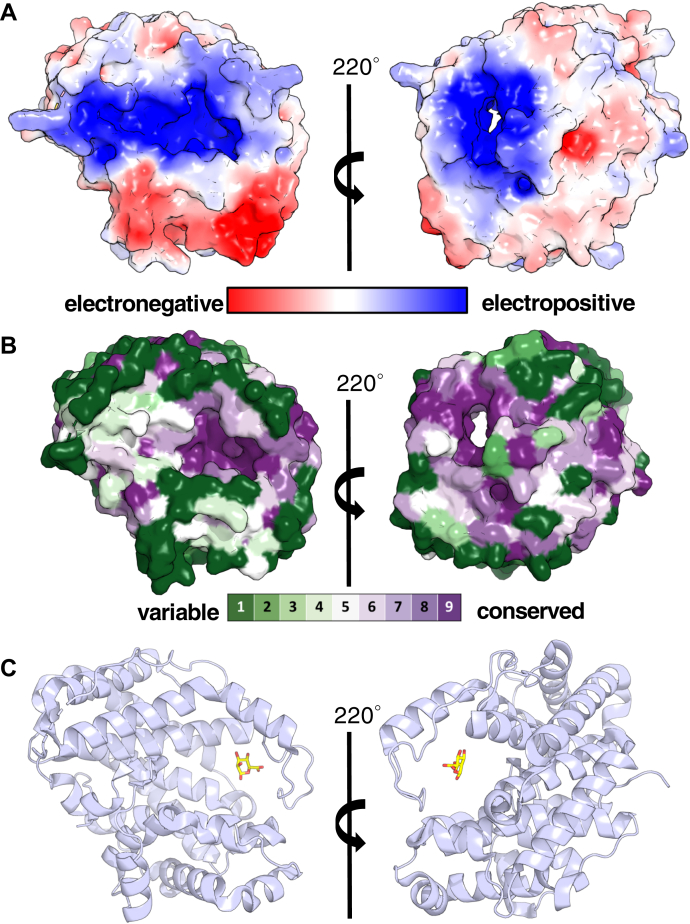
Although we cocrystallized the protein with a mannuronate trisaccharide (ManA3), we were only able to detect electron density for a sugar monomer suggesting perhaps that cleavage of the ManA3 substrate may have occurred in crystallo or that the remaining sugar units/moieties were disordered in the structure (Fig. 1). While this ManA identifies the location of the active site, we anticipate that it will contain multiple sugar-binding sites given that AlgL’s activity increases linearly with the number of residues in the substrate (41). Indeed, examination of the enzyme’s surface electrostatic properties and residue conservation revealed an elongated, highly conserved, and strongly electropositive groove (Fig. 2, A and B). In addition to the pronounced, long substrate-binding groove, we also identified an extended loop that partially closes over the ManA-binding site, known as the lid-loop (Fig. 2C).
Figure 2.
Structure of Pseudomonas aeruginosa AlgL reveals an elongated, highly conserved, and strongly electropositive groove.A, electrostatic surface representation of AlgL calculated by APBS Tools; contoured from +5 (blue) to −5 (red) kT/e (102). B, conservation surface representation of AlgL calculated by the ConSurf server; green indicates residues that are less conserved, and purple indicates residues that are highly conserved (103). C, AlgL in complex with a mannuronate residue (yellow) reveals a partially enclosed structure.
Structural comparisons identify AlgL residues required for alginate binding and catalysis
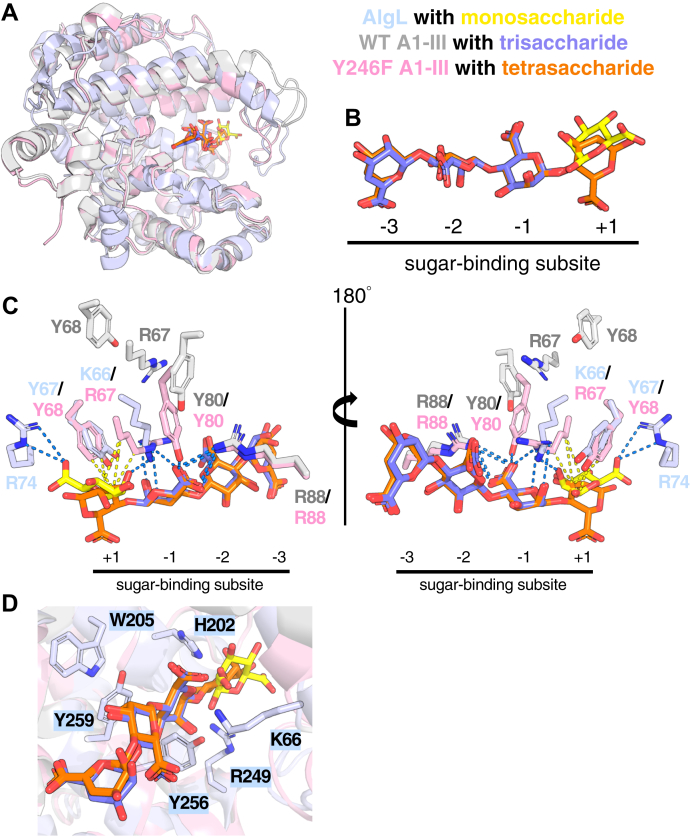
To gain further insight into the structure and function of AlgL and identify key residues involved in substrate binding and catalysis, we compared our structure with the known structures of the PL-5 family member, Sphingomonas sp. A1-III. Of the five available A1-III structures, two are complexed with polyM oligosaccharides. The WT A1-III in complex with a polyM trisaccharide (PDB: 1HV6) (52) and the Y246F mutant in complex with a polyM tetrasaccharide (PDB: 4F13) (53) were therefore compared with our AlgL-ManA structure. Superposition of our AlgL structure with WT and Y246F A1-III resulted in a Cα RMSD of 1.965 Å and 1.895 Å, respectively, highlighting the similarity of the overall tertiary structure of the enzymes (Fig. 3A).
Figure 3.
The polysaccharide family 5 bacterial alginate lyases Pseudomonas aeruginosa AlgL and Sphingomonas sp. A1-III are structurally similar.A, superposition of P. aeruginosa AlgL (light blue) in complex with a monosaccharide of mannuronate (yellow) (PDB: 4OZV), Sphingomonas sp. WT A1-III (grey) in complex with a mannuronate trisaccharide (purple) (PDB: 1HV6) (52), and Sphingomonas sp. Y256F A1-III (pink) in complex with a mannuronate tetrasaccharide (orange) (PDB: 4F13) (53). B, ligands bound and their sugar-binding subsite positions in the P. aeruginosa AlgL and Sphingomonas sp. A1-III structures. C, close-up of the lid-loop region of the P. aeruginosa AlgL and Sphingomonas sp. A1-III active sites. Hydrophobic and hydrogen-bonding interactions are represented by the dashed yellow and blue lines, respectively. D, P. aeruginosa AlgL active site residues (light blue) chosen for downstream in vitro and in vivo mutagenic studies, including the proposed catalytic acid and base Y256, the residue that stabilizes the anionic intermediate H202, the lid-loop residue K66, the highly conserved residues W205 and Y259, and R249 involved in neutralizing the C-5 carboxylate group.
A noticeable difference between the three structures involves the position and orientation of the bound ligands (Fig. 3, A and B). Within the sugar-binding site of WT A1-III, the trisaccharide occupies subsites −3 to −1 (Fig. 3B; (52)), while the tetrasaccharide in the catalytically inactive Y246F A1-III mutant structure occupies subsites −3 to +1. Cleavage of the ligand occurs between subsites −1 and +1 (Fig. 3B; (53)). Both ligands bind in a similar orientation in subsites −3 to −1 (Fig. 3B). Similar to Y246F A1-III, the sugar monomer modeled in our AlgL structure occupies subsite +1 (Fig. 3B). However, this monomer adopts a different orientation; a variation that is most likely attributable to the lack of additional conformational restraints that a longer sugar polymer would provide (Fig. 3B).
Comparison of the three structures reveals a striking difference in the extended loop that partially encloses the ligand, termed the lid-loop: residues 64 to 85 in A1-III, and 63 to 90 in AlgL (Figs. S2 and 3A; (53)). This loop has previously been suggested to undergo an induced-fit motion during alginate binding and catalysis (53). Although the AlgL and A1-III lid-loops differ in size by six residues, both adopt similar conformations (Fig. 3A). The conformation of the lid-loop correlates with the presence of a ligand in the +1 subsite, with the lid-loop in the Y246F A1-III and AlgL structures in a more enclosed conformation, while the loop adopts a more open conformation in the WT A1-III structure (Fig. 3, A and B). A more enclosed orientation of the lid-loop is also observed in the structure of the H192A A1-III mutant in complex with a polyMG tetrasaccharide, which occupies subsites −3 to +1 (53). In our AlgL structure, we found that the Nζ atom of lid-loop residue K66 interacts via a hydrogen bond with the hydroxyl group of the C-2 of ManA (Fig. 3C). In contrast, in the Y246F A1-III structure, the difference in orientation of the ligand results in residues Y68 forming hydrophobic interactions with the +1 ligand (Fig. 3C). In Y246F A1-III, residues R67, Y80, and R88, which are adjacent to the lid-loop, also hydrogen bond to the ligand in the −1 subsite (Fig. 3C; (53)). Mutation of residues in the lid-loop region of A1-III, including R67, Y68, and Y80, result in less catalytically active enzymes in vitro compared with WT A1-III (53), highlighting the importance of this loop in the enzymatic mechanism. R67, Y68, and R80 in A1-III correspond to K66, Y67, and F85 in P. aeruginosa AlgL (Fig. S2).
Examination of the interactions between the protein and ligand in the active site of the WT and Y246F A1-III structures also revealed that Q134, Q138, H245, R306, R312, D314, and R342 directly hydrogen bond to the ligand, while R88, Q134, Y137, W141, N191, R306, and R312 make hydrophobic contacts (52, 53). In the WT A1-III structure, residues W141 and Y249 also form hydrogen bonds with the bound sugar (52). Of the residues that interact with the ligand in Al-III, only residues W141, N191, Y249, and R342 are conserved in AlgL, and these correspond to W146, N201, Y259, and R352, respectively (Fig. S2; (52)). In addition, in A1-III, residue W195, which corresponds to W205 in AlgL, while not directly involved in binding the carbohydrate, is highly conserved across PL-5 alginate lyases (Fig. S2; (52)).
Studies on A1-III have implicated Y246 as both the catalytic acid and base, with H192 stabilizing the anionic intermediate and R239 interacting with the sugar to neutralize the C-5 carboxylate group (52). Mutation of H192 to alanine and Y246 to phenylalanine drastically decreased A1-III enzyme activity in vitro (53). These residues are conserved in AlgL and correspond to Y256, H202, and R249, respectively (Fig. S2).
Mutation of predicted AlgL catalytic site residues abrogates enzymatic activity in vitro
Based on our sequence and structural analysis, we chose six residues for downstream in vitro and in vivo mutagenic studies (Table 2 and Fig. 3D). The residues chosen include the proposed catalytic acid and base Y256, the residue that stabilizes the anionic intermediate H202, the lid-loop residue K66, the highly conserved residues W205 and Y259, and the residue involved in neutralizing the C-5 carboxylate group R249. To determine the importance of these residues in alginate degradation, we first assessed the impact of the point mutations using an in vitro alginate lyase activity assay with a polyM substrate derived from P. aeruginosa. The assay enabled the steady-state kinetic parameters for the enzymatic reaction to be calculated, including the catalytic efficiency (kcat/KM), for the WT and each mutant enzyme (Table 3). As anticipated, WT AlgL is the most catalytically efficient enzyme with a turnover rate of 15.7 ± 0.274 s−1 and catalytic efficiency of 147 ± 10.6 × 103 (s−1 M−1) (Table 3). We were unable to detect any catalytic activity using this assay for mutants H202A, R249A, R249E, and Y256F (Table 3). The conservative mutant R249K retained approximately 1.5% catalytic efficiency compared with the WT enzyme (Table 3). Mutation of residues implicated in substrate binding, such as K66 and Y259, led to the retention of enzymatic activity, although these mutants were significantly less catalytically efficient than the WT. K66A and Y259F retained approximately 1% and 30% catalytic efficiency, respectively, compared with WT (Table 3). Mutation of the highly conserved residue W205 also resulted in an enzyme with ∼2.5% catalytic efficiency relative to WT (Table 3). However, loss of enzyme activity in this case could be attributable to protein instability or misfolding as the melting temperature for W205F was more than 4 °C less than the WT enzyme (Table S1). Overall, our data demonstrate that mutation of AlgL residues implicated in catalysis, with the exception of the conservative mutant R249K, results in loss of in vitro alginate lyase activity, while mutation of residues implicated in substrate binding greatly reduced, but did not abrogate, enzymatic activity.
Table 2.
Proposed role of P. aeruginosa AlgL residues and the point mutants used in this study
| Residue | Proposed function | Point mutants studied |
|---|---|---|
| K66 | Lid-loop residue directly involved in substrate binding | K66A |
| H202 | Neutralizes anionic intermediate of the substrate | H202A |
| W205 | Highly conserved residue indirectly involved in substrate binding | W205F |
| R249 | Neutralizes C-5 carboxylate group on substrate for proton abstraction | R249A, R249E, R249K |
| Y256 | Catalytic acid/base | Y256F |
| Y259 | Directly involved in substrate binding | Y259F |
Table 3.
Pseudomonas aeruginosa AlgL reaction steady-state kinetic parametersa
| AlgL enzymea | kcat/KM (s−1 M−1) | kcat (s−1) | KM (μM) |
|---|---|---|---|
| WT | 147 ± 10.6 × 103 | 15.7 ± 0.274 | 107 ± 5.80 |
| K66A | 1.37 ± 0.141 × 103 | 0.128 ± 0.00303 | 93.5 ± 7.44 |
| H202A | Activity not detected | ||
| W205F | 3.89 ± 0.350 × 103 | 1.92 ± 0.0647 | 494 ± 27.9 |
| R249K | 2.23 ± 0.429 × 103 | 0.322 ± 0.0165 | 144 ± 20.3 |
| R249A | Activity not detected | ||
| R249E | Activity not detected | ||
| Y256F | Activity not detected | ||
| Y259F | 44.5 ± 7.05 × 103 | 15.8 ± 0.864 | 354 ± 36.8 |
Initial velocities were fitted to the Michaelis–Menten equation.
A functional AlgL is required for P. aeruginosa viability during alginate production
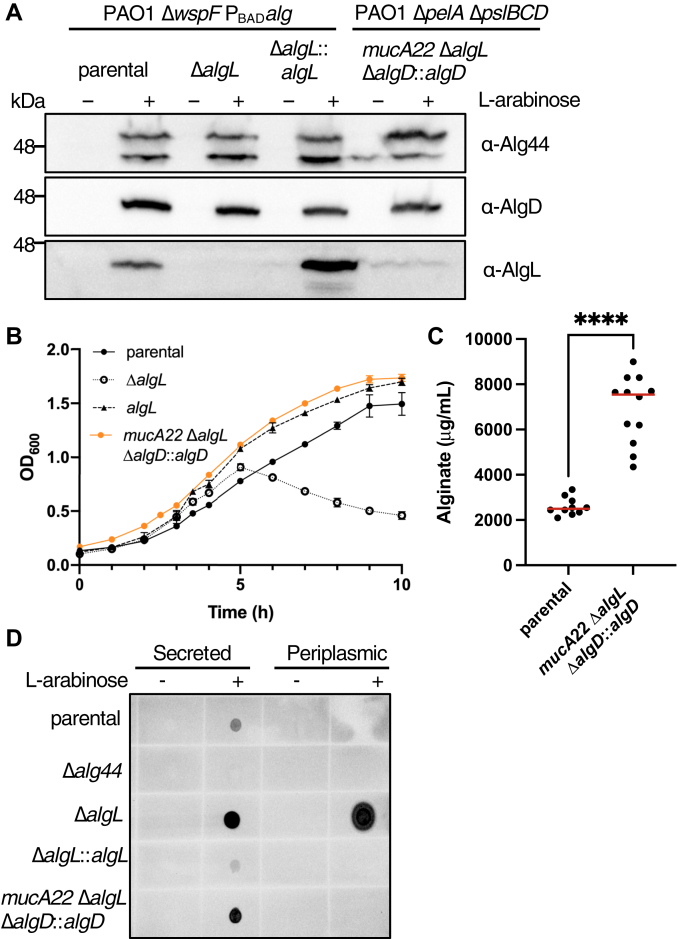
Previous studies reported variable results when alginate biosynthesis is induced in algL deletion mutant strains. Absence of the enzyme was shown to cause cell death in P. aeruginosa FRD1 (47), while no significant loss of biofilm biomass was observed in P. aeruginosa PDO300 (48). To further probe the role of AlgL in vivo and its impact on cell viability during alginate biosynthesis, we first generated an algL deletion in our P. aeruginosa PAO1 ΔwspF PBADalg strain background that allows for induction of alginate expression using L-arabinose in a high c-di-GMP background (28). In this strain, alginate production is isolated from its native AlgU/T system of regulation and can readily be switched on and off with the addition of L-arabinose to the growth medium. Complementation of the ΔalgL strain with either WT algL or active site point mutants was performed using an integrating plasmid at the chromosomal attTn7 site with the complemented gene also under the control of an arabinose-inducible promoter, thus allowing for simultaneous expression with other alginate proteins within the operon. To ensure that the results we observed could be correlated with a particular mutation, we first confirmed using Western blot analysis that each AlgL variant is expressed 1 h after induction with 0.5% (w/v) L-arabinose in the complemented PAO1 ΔwspF PBADalg strains (Fig. S3).
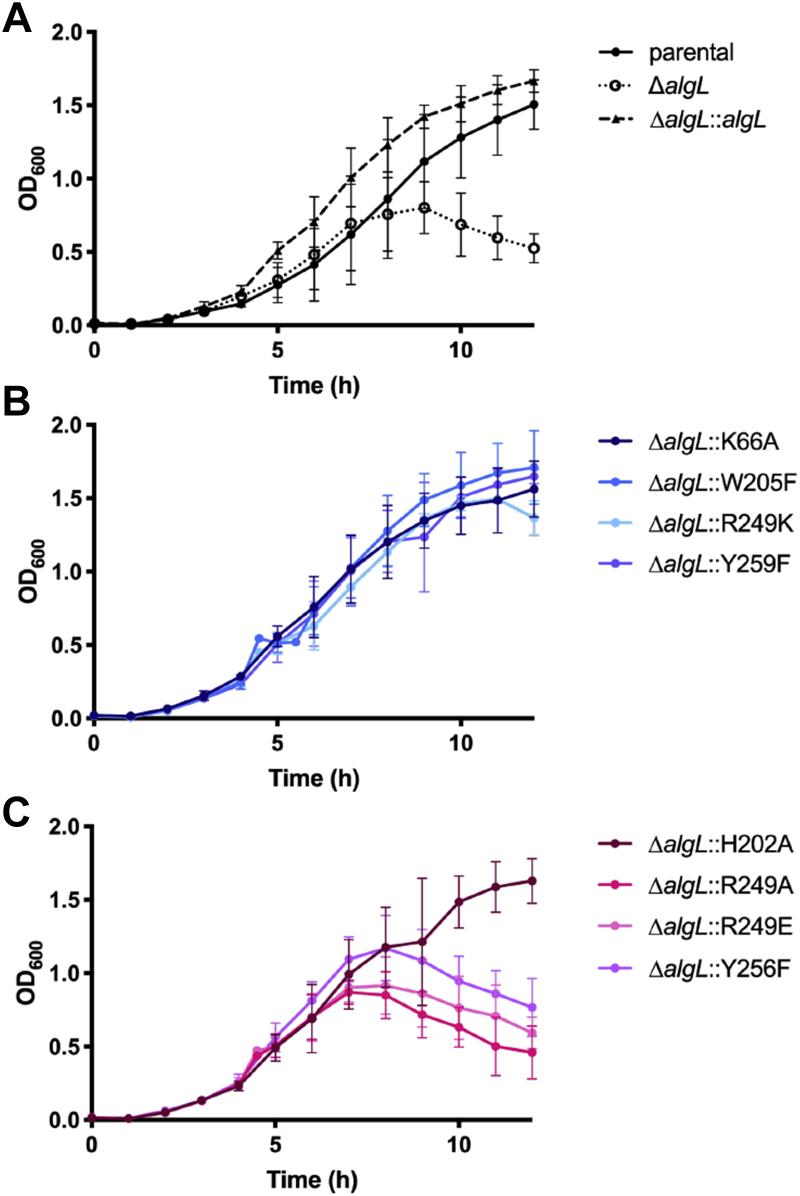
We next investigated whether growth of each strain was compromised in liquid medium (Fig. 4). The optical density of the bacterial culture at 600 nm (OD600) was measured every 1 h for 12 h. When strains reached an approximate OD600 of 0.500, 0.5% (w/v) L-arabinose was added to the medium. Four hours after induction with L-arabinose, a noticeable growth defect was observed in ΔalgL compared with the parental strain (Fig. 4A). Complementation with WT AlgL (ΔalgL::algL) restored P. aeruginosa growth in the presence of L-arabinose, demonstrating that absence of AlgL during alginate production is detrimental to the cell (Fig. 4A). When we examined the growth characteristics of AlgL point mutants that retained alginate lyase enzymatic activity in vitro, we observed that complementation of the deletion strain with the K66A, W205F, R249K, and Y259F variants exhibited similar growth characteristics as ΔalgL::algL (Fig. 4, A and B). The effects on growth in the presence of L-arabinose were more pronounced when we examined the AlgL point mutants whose alginate lyase activity was abrogated in vitro (Fig. 4C). The R249A, R249E, and Y256F strains each displayed a growth defect phenotype (Fig. 4C). Interestingly, although we were unable to detect enzymatic activity in vitro for the H202A variant, the H202A complemented strain did not display the same in vivo growth patterns as the other catalytically inactive mutants (Table 3 and Fig. 4C).
Figure 4.
Growth of Pseudomonas aeruginosa PAO1 ΔwspF PBADalg strains reveals mutation of R249 and Y256 impacts cell viability during alginate production. After reaching an OD600 of approximately 0.5, cells were induced to produce alginate with 0.5% (w/v) L-arabinose. Data points represent averages from three biological replicates with two technical replicates per biological replicate. A, control strains. B, ΔalgL strains complemented with AlgL point mutants that retained alginate lyase enzymatic activity in vitro. C, ΔalgL strains complemented with AlgL point mutants whose alginate lyase enzymatic activity in vitro was compromised.
Taken together, these data suggest that deletion of algL is inhibitory to growth when alginate production is induced with L-arabinose in our genetically engineered strain. In support of this conclusion, we also found that nonconservative mutations of active site residues required for alginate lyase activity, including R249A, R249E, and Y256F, were similarly not tolerated and did not grow under alginate-producing conditions. Mutations that might compromise AlgL’s ability to bind to alginate were found to be tolerated. Thus, our data suggest that the presence of AlgL alone is insufficient to restore viability and that its catalytic activity is required for growth during alginate production in our genetically engineered strain.
Deletion of AlgL or mutation of its catalytic residues results in abnormal cellular morphology during alginate production
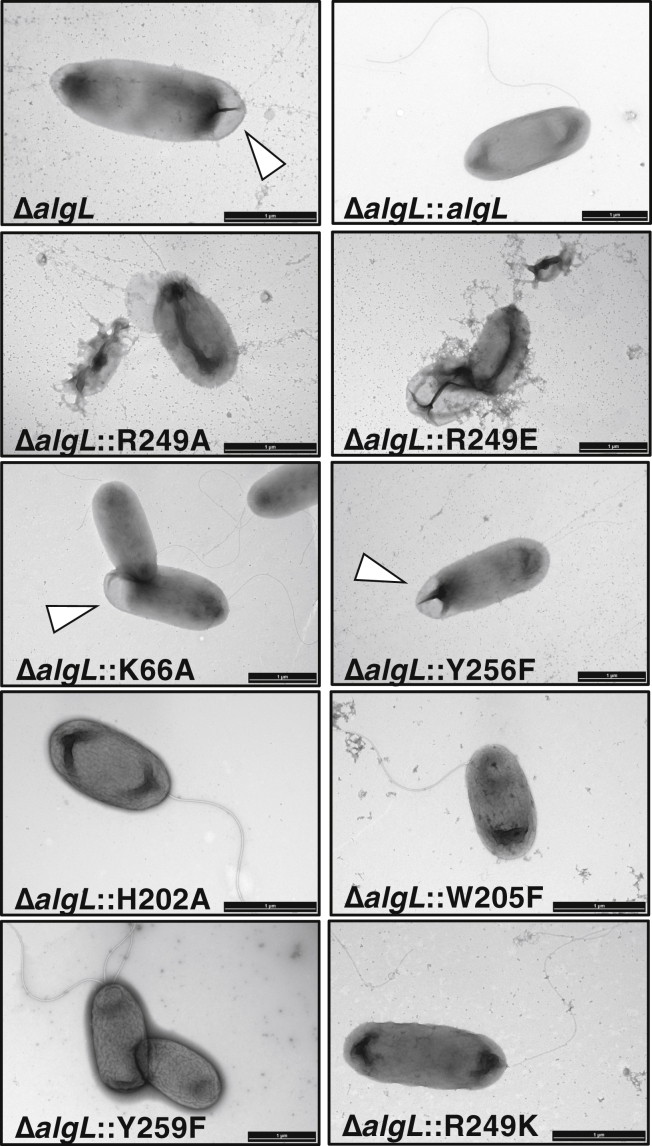
To further investigate the impact of algL mutations on aberrant cell growth, we next sought to assess whether the cell morphology was altered during alginate production. Large zones of separation between membranes after 4 h of alginate production in P. aeruginosa FRD1 ΔalgL and general cell lysis after 6 h have been reported (47). It was hypothesized that these large zones of separation between membranes are indicative of periplasmic alginate accumulation (47). We therefore grew the cells for 4 h post 0.5% (w/v) L-arabinose induction in liquid medium, and transmission electron microscopy (TEM) was used to visualize whole cellular morphologies during alginate production. The micrographs revealed that the ΔalgL strain and strains for which mutation of the catalytic active site residues resulted in aberrant growth phenotypes, i.e., R249A, R249E, and Y256F (Fig. 4), all displayed abnormal cell morphology compared with the ΔalgL::algL strain (Fig. 5). In particular, ΔalgL and Y256F cells have perturbations in the cell membrane (Fig. 5). Cell lysis was observed for R249E and R249A (Fig. 5). Some K66A cells also appear to have membrane perturbations (Figs. S4 and 5). Strains that were observed to grow in alginate-producing conditions, including H202A, W205F, Y259F, and R249K, have cellular morphologies comparable to ΔalgL::algL (Figs. 4B and 5). Thus, taken together with our growth data, the TEM images suggest that algL mutations that impact function are not tolerated under alginate-producing conditions. In particular, the ΔalgL strain displays membrane perturbations that we hypothesize are due to the deleterious periplasmic accumulation of alginate.
Figure 5.
Transmission electron microscopy images of whole Pseudomonas aeruginosa cells after 4 h induction of alginate biosynthesis.White arrows indicate cell membrane perturbations. Scale bar is 1 μm.
Overall, the data suggest that AlgL point mutants that have retained some in vitro enzymatic activity against polyM do not have a deleterious effect in vivo, except for K66A and H202A (Table 3, Figs. 4B and 5). Although K66A retains enzymatic activity and the strain does not demonstrate an in vivo growth defect, some cells were observed to display aberrant cell membrane phenotypes by TEM (Table 3, Figs. 4B, 5 and S4). In contrast, while no enzymatic activity was detected for the H202A variant in our in vitro assay, this variant did not display the same growth or cell morphology phenotypes observed for other catalytically inactive mutants (Table 3, Figs. 4B and 5), but rather behaved like ΔalgL::algL.
The AlgL lid-loop is important for alginate binding and catalysis
To investigate how the H202A and K66A active site mutants might affect enzymatic activity and account for the discrepancies between in vivo phenotypes and in vitro catalytic activities observed, we determined the structures of the AlgL H202A and K66A variants (Table 1).
Comparison of the K66A variant with WT AlgL revealed that the two structures are highly similar with a Cα RMSD of 0.352 Å (Fig. S5). However, residues 66 to 81 within the lid-loop region of K66A AlgL could not be built into the model, suggesting that the lid-loop region in this mutant is flexible (Fig. S5). Similarly, we were unable to observe any corresponding density for the mannuronate substrate soaked into the crystal prior to data collection (Fig. S5). As noted above, K66 interacts with the sugar ligand and appears to contribute to loop enclosure in the WT enzyme (Fig. 3C). Mutation of K66 and the resulting flexibility of the lid-loop region suggest that K66 in AlgL is important for priming the enzyme for catalysis by locking the lid-loop in a closed conformation after substrate binding. This could explain the abnormal cellular morphology observed in vivo in the K66A strain and the reduced enzymatic efficiency of this mutant variant in vitro.
In keeping with its proposed role in neutralizing the anionic substrate intermediate, we found that H202A AlgL was catalytically inactive in our in vitro alginate lyase assay and thus were anticipating that this variant would display similar growth and cellular morphology phenotypes to the other inactive variants (Table 3). However, H202A displays a similar growth pattern and cellular morphology to WT (Figs. 4 and 5). When we compared the structure of the mutant with the WT AlgL enzyme, we found that there were strikingly similar with a Cα RMSD of 0.062 Å (Fig. S6). There were only minor differences in overall structure of the active sites. Examination of the structure reveals that the orientation of the side chain of K66 is altered, with the Nζ oriented toward the ManA ligand in the WT structure and oriented away from the active site in the H202A structure (Fig. S6). Although no ligands were present during the crystallization process, the lid-loop in the H202A structure adopts a more closed conformation similar to that found in the WT AlgL structure and Y246F and H192A A1-III cocrystal structures (Fig. S7A). This conformation appears to be stabilized by hydrogen bonding between R74 and R249 and hydrophobic interactions between K66 and L67 (Fig. S7B). To allow for the interaction, R74 in H202A AlgL is oriented toward subsite +1 within the active site, which is occupied by ManA in the WT structure (Fig. S7B). In WT AlgL, R74 is oriented away from the active site, suggesting that this residue may be important for anchoring the lid-loop in a closed orientation in the absence of a ligand (Fig. S7B). Given the discrepancies observed between in vivo growth phenotypes and enzymatic inactivity in vitro, and the similarity of the WT and H202A structures, we hypothesize that the H202A mutant is catalytically active, but its activity is below the level that can be detected in our current assay. In support of this, mutation of H192, the equivalent catalytic residue in A1-III, resulted in a large decrease but not complete abrogation of the alginate lyase activity (53).
AlgL prevents accumulation of alginate within the periplasmic space
In addition to functioning as an alginate-degrading enzyme, AlgL has also been hypothesized to have a structural role in the biosynthetic complex, interacting with other alginate proteins such as AlgG, AlgK, AlgX, or Alg44 (47). However, protein pull-down and immunoblot assays failed to demonstrate that AlgL is associated with any of these proteins (48). Using a comparable approach, we performed coimmunoprecipitation experiments using AlgL C-terminally tagged with the vesicular stomatitis virus glycoprotein (VSV-G) epitope followed by mass spectrometry analysis and similarly did not enrich for any other alginate proteins or any other protein that was not found in the ΔalgL::algL control sample (WT) (Table S2). Our data reinforce the previous findings and suggest that AlgL does not interact with the alginate secretion complex. These data suggest that AlgL does not need to interact with the rest of the alginate complex to degrade accumulated alginate within the periplasm. To test this hypothesis, we next sought to determine whether the substance we observed accumulating within the cell envelope in our ΔalgL TEM images is alginate. Periplasmic and secreted fractions of the parental, ΔalgL, ΔalgL::algL, and Δalg44 strains grown in liquid culture were extracted and then analyzed via dot blot assays using a commercially available monoclonal alginate antibody. A signal for alginate was detected in the parental, ΔalgL, and ΔalgL::algL secreted fractions when polymer production was induced with 0.5% (w/v) L-arabinose (Fig. 6). As expected, alginate is not detected in the Δalg44 strain, which lacks the c-di-GMP receptor that posttranslationally regulates polymerization (Fig. 6). Alginate was detected in the periplasmic fraction of ΔalgL, demonstrating for the first time that loss of AlgL directly results in periplasmic accumulation of the polymer (Fig. 6). Taken together with the ΔalgL TEM micrographs, these data support the hypothesis that deletion of AlgL results in accumulation of alginate within the periplasmic space during alginate production (Figs. 5 and 6) and suggests that AlgL can function as a periplasmic housekeeping enzyme that maintains cell viability by preventing the accumulation of alginate in the periplasmic space.
Figure 6.
Alginate is retained in the periplasmic space during alginate production in the absence of AlgL. Chemiluminescence detection of a dot blot on secreted and periplasmic fractions from P. aeruginosa strains using a monoclonal alginate-reactive antibody.
Growth defects due to absence of AlgL during alginate biosynthesis are mitigated by constitutive expression of the AlgU/T regulon
Thus far, using our P. aeruginosa PAO1 ΔwspF PBADalg strain, we have demonstrated that AlgL is not part of the alginate secretion complex and that it degrades polymer that accumulates within the periplasmic space. While the genetic background we used in our in vivo studies was crucial for dissecting the role of AlgL, alginate production is normally under the control of the AlgU/T sigma factor. AlgU/T regulates hundreds of genes in PAO1, including genes responsible for mitigating cell wall stress, peptidoglycan biosynthesis, pyoverdine biosynthesis, and lipopolysaccharide biosynthesis (54, 55, 56, 57, 58, 59).
In chronic P. aeruginosa CF lung infections, alginate production can be induced by a truncation mutation in the AlgU/T antisigma factor, MucA (mucA22). This leads to constitutive activation of AlgU/T and thus expression of its vast regulon (25). In our genetically engineered strain, the alginate operon is transcribed in the absence of other genes present in the AlgU/T regulon, which could potentially include genes that can respond to and mitigate the periplasmic stress induced by the accumulation of alginate. We hypothesized therefore that the discrepancies in the current literature regarding the effect of ΔalgL on P. aeruginosa could potentially be attributed to differences in regulation of AlgU/T and the alginate operon in the strains used. To test this hypothesis, we first generated a ΔalgL mutation in a PAO1 mucA22 background that is incapable of producing Pel (ΔpelA), Psl (ΔpslBCD), or alginate (ΔalgD) (Table S3). To complement the alginate production defect, algD was reintroduced at the chromosomal attTn7 site under the control of an arabinose-inducible promoter (Table S3). Thus, proteins within the alginate operon, with the exception of algD, are under the control of their native promoter. We first confirmed that the newly generated PAO1 ΔpelA ΔpslBCD mucA22 ΔalgL ΔalgD::algD (mucA22 ΔalgL ΔalgD::algD) strain was able to express the appropriate alginate proteins by probing for the expression of AlgD, Alg44, and AlgL (Figs. 7A and S8). As expected, Alg44 was expressed in the absence of L-arabinose, while AlgD was only expressed after addition of 0.5% (w/v) L-arabinose to the growth media (Figs. 7A and S8). Although a faint band can be observed at the expected molecular weight of AlgL, colony PCR of the strain confirms that algL is deleted from the genome (Figs. 7A and S9). Therefore, we speculate that this band represents nonspecific binding of the antibody to a protein that is expressed as a consequence of constitutive expression of the AlgU/T regulon. We next examined the growth of our parental, ΔalgL, ΔalgL::algL, and mucA22 ΔalgL ΔalgD::algD strains in liquid culture. When strains reached an approximate OD600 of 0.500, 0.5% (w/v) L-arabinose was added to the media. As anticipated, the ΔalgL strain demonstrated an aberrant growth phenotype and the ΔalgL::algL strain grew similarly to the parental strain (Figs. 4A and 7B). Interestingly, the mucA22 ΔalgL ΔalgD::algD strain also grew similarly to the parental and ΔalgL::algL strains, illustrating that loss of algL is tolerated during alginate production when the alginate operon is under control of AlgU/T and AlgU/T is free to transcribe all of the genes within its regulon (Fig. 7B). Moreover, we found that our mucA22 ΔalgL ΔalgD::algD strain is capable of producing at least as much alginate as the parental strain in the presence of 0.5% (w/v) L-arabinose (Fig. 7C). After 22 h of alginate production, the parental and mucA22 ΔalgL ΔalgD::algD strain reached an average OD600 of 2.156 and 2.125, respectively, and produced approximately 2.5 mg/ml and 6.8 mg/ml alginate, respectively (Fig. 7C). This suggests that tolerance of an algL deletion in mucA22 ΔalgL ΔalgD::algD cannot be attributed to a deficiency in alginate production (Fig. 7C). Consistent with these results, analysis of the secreted and periplasmic fractions demonstrates that alginate is secreted in the mucA22 ΔalgL ΔalgD::algD strain and not retained in the periplasm (Fig. 7D).
Figure 7.
Expression of the AlgU/T regulon in a ΔalgL strain background rescues Pseudomonas aeruginosa lethality during alginate production.A, Western blot analysis of whole cell lysates expressing Alg44, AlgD, or AlgL after 1 h induction with 0% or 0.5% (w/v) L-arabinose using poly-clonal Alg protein specific antibodies. B, growth curves of PAO1 ΔwspF PBADalg (parental), PAO1 ΔwspF PBADalg ΔalgL (ΔalgL), PAO1 ΔwspF PBADalg ΔalgL::algL (ΔalgL::algL), and PAO1 ΔpelA ΔpslBCD mucA22 ΔalgL ΔalgD::algD (mucA22 ΔalgL ΔalgD::algD). After reaching an OD600 of approximately 0.5, cells were induced to produce alginate with 0.5% (w/v) L-arabinose. Values shown represent averages of two biological replicates with two technical replicates for each biological replicate. C, quantification of alginate produced over the course of 22 h by parental and mucA22 ΔalgL ΔalgD::algD with addition of 0.5% (w/v) L-arabinose to the growth media. Values represent all technical replicates across four separate experiments. Red lines represent the median. Statistical analysis was carried out using a Mann–Whitney test: ∗∗∗∗ indicates p < 0.0001. D, chemiluminescence detection of a dot blot on secreted and periplasmic fraction samples from P. aeruginosa strains using a monoclonal alginate-reactive antibody.
Discussion
In this study, by genetically engineering a strain that enables alginate production under the control of arabinose, we have been able to dissect the role of AlgL in P. aeruginosa alginate biosynthesis. We found that this enzyme prevents the lethal periplasmic accumulation of polymer, suggesting that the enzyme has a role in homeostasis. Structural analyses of WT P. aeruginosa AlgL in complex with ManA, coupled with site-directed mutagenesis and in vitro enzymatic assays, enabled the identification of key active site residues involved in alginate binding and catalysis (Figs. 1, 2 and S2; Table 2). We found that mutation of the catalytic site residues R249 and Y256 that abrogate in vitro activity was also detrimental for P. aeruginosa viability during alginate production (Figs. 4 and 5). Importantly, we demonstrated that any detrimental effects due to the loss of AlgL in P. aeruginosa are mitigated by constitutive expression of the AlgU/T regulon (Fig. 7). Collectively, our results define the role of AlgL in alginate biosynthesis and can explain the variations in the in vivo phenotypes observed across different P. aeruginosa ΔalgL strains.
Our analyses show that R249 and Y256 are important for AlgL in vitro catalytic activity and in vivo viability (Table 3 and Fig. 4). PL-5 family alginate lyases are proposed to cleave polyM via a syn β-elimination reaction where the carboxylate group on C-5 is neutralized and a general base abstracts a proton from C-5 resulting in the elimination of the substituent at C-4 (41, 60). As expected, when the proposed catalytic acid/base residue Y256 was mutated to Y256F, we observed a complete loss of catalytic activity in our in vitro alginate lyase assay (Table 3). However, it is still unclear whether Y256 is the only catalytic residue involved in proton transfer during cleavage of ManA residues (53). To cleave ManA substrates in A1-III, R249 is proposed to act by lowering the pKa of Y256 to facilitate the abstraction and donation of protons by Y256 (53). In agreement with this hypothesis, no alginate lyase activity was detected for our R249A and R249E mutants, while R249K retained approximately 1.5% catalytic efficiency compared with the WT enzyme (Table 3). However, AlgL was previously shown to also cleave GulA residues via an anti β-elimination reaction, which could not be mediated by Y256 alone (41). Thus, it was suggested that AlgL could potentially utilize two different catalytic bases for cleavage of each pair of epimeric substrates, as has been reported for chondroitin lyase ABC that cleaves the epimers chondroitin sulfate and dermatan sulfate using a composite active site with two separate histidine residues that act as a general base for each substrate (41, 61). In AlgL, R249 may act as the general base to cleave GulA residues. Although arginine residues are poor catalytic bases because of their relatively high pKa (∼12.5) and thus their poor propensity to be protonated at physiological pH, the role of arginine residues as a general base in proton abstraction has been previously reported in other bacterial enzymes including pectate/pectin lyases and fumarate reductase (62, 63, 64, 65). The pKa of arginine residues can be perturbed to favor a deprotonated state by surrounding the catalytic arginine with positively charged residues, as is the case in AlgL where K248 and R250 are near R249. This mechanism was previously demonstrated in acetoacetate decarboxylase from Clostridium acetobutylicum where the spatial proximity of K116 decreased the pKa of the catalytic base K115 to allow for efficient decarboxylation of substrate (66). Furthermore, the mutants K116C and K116N demonstrated significantly reduced enzymatic activity (∼2% of WT activity) while K116R only demonstrated modestly decreased activity (∼20% of WT activity) (66). Thus, R249 may act to reduce the pKa of the catalytic acid and base residue Y256 to catalyze the syn reaction, while Y256 and R249 function as the catalytic acid and base, respectively, to catalyze the anti-reaction.
Herein, we have demonstrated that AlgL functions as a homeostasis enzyme that degrades alginate polymer that accumulates within the periplasmic space (Figure 4, Figure 5, Figure 6). While this role has been proposed previously, the current study is the first to directly probe the periplasmic contents of AlgL-deficient bacteria during alginate production (Fig. 6). The biosynthetic operons of synthase-dependent exopolysaccharide secretion systems each contain an enzyme that can degrade the polymer being synthesized. To date, while it has been hypothesized that the glycoside hydrolases PelA and BcsZ may be required to clear the periplasmic space of Pel and accumulated glucan chains, respectively, no data have been reported to support these functions (67, 68). Of note, in the synthase-dependent Escherichia coli poly-N-acetylglucosamine (PNAG) system, deletion of the periplasmic bifunctional deacetylase/hydrolase enzyme PgaB did not hinder PNAG synthesis but did prevent its export and resulted in a dramatic expansion of the periplasmic space at the cell poles, the putative site of PNAG biosynthesis (69). Combined with the results of our current study, this adds support to the proposal that glycoside hydrolases/lyases present in synthase-dependent exopolysaccharide secretion systems play a role in bacterial periplasmic homeostasis.
Recognizing the importance of strain backgrounds in studying alginate biosynthesis, we employed the use of our PAO1 mucA22 ΔalgL ΔalgD::algD strain to investigate if deletion of algL is deleterious in the commonly used P. aeruginosa mucA22 background (Fig. 7). Unlike the results obtained for our arabinose-inducible ΔalgL strain, mucA22 ΔalgL ΔalgD::algD did not demonstrate a growth defect and did not retain alginate in the periplasmic space (Figs. 4 and 7). The critical difference between these two strains is that MucA is truncated in mucA22 ΔalgL ΔalgD::algD and therefore unable to regulate AlgU/T (56, 70). Thus, we speculate that there must be one or more genes within the AlgU/T regulon that compensate for the loss of algL in a mucA22 background. This could potentially explain the discrepancies in observed phenotypes across different P. aeruginosa ΔalgL genetic backgrounds such as PDO300, a mucA22 derivative of PAO1 (48, 71). For example, analysis of the P. aeruginosa genome reveals two similar and lesser-known alginate lyases, PA1167 and PA1784, which share 47.0% sequence identity with each other. Their homologue PSPTO_5015 was found to be regulated by AlgU/T in Pseudomonas syringae pv. Tomato DC3000 (42, 72, 73). Although PA1167 and PA1784 do not share any significant sequence similarity with AlgL, they share 35.2% and 30.7% sequence identity, respectively, with the homologous PL-7 family alginate lyase A1-II from Sphingomonas sp. which preferentially degrades polyMG (42, 72). PA1167 was demonstrated to preferentially degrade polyMG in vitro, in contrast to AlgL, which preferentially degrades polyM (42). Given the structure of alginate, it seems ideal for P. aeruginosa to have alginate lyases with different substrate preferences to prevent polymer accumulation in the periplasm. It is also possible that, given the lethal consequence of periplasmic alginate retention, P. aeruginosa has redundant systems to degrade polymer that is not exported to ensure cell viability. In this instance, AlgL is the primary alginate lyase for alginate degradation and the additional alginate lyases, PA1167 and PA1784, may function as a fail-safe. Moreover, other genes within the vast AlgU/T regulon, such as sugar ABC-transporters may function to recycle cleaved alginates back into the cytosol (74). Thus, the genetic differences between widely used P. aeruginosa strains may have nuanced implications on investigating alginate biosynthesis.
Considering the potential significance of a constitutively active AlgU/T in P. aeruginosa (75), we might speculate that studies investigating algL in the mucA22 strain FRD1 may demonstrate similar phenotypes as were reported in our mucA22 ΔalgL ΔalgD::algD strain and in PDO300 (76). However, in FRD1 with the alginate operon isolated and under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter, a different phenotype was observed (47). Although the alginate operon should theoretically be expressed alongside other AlgU/T genes when IPTG is added to the growth medium, aberrant phenotypes such as cell death and membrane separation were observed in a ΔalgL mutant as a consequence of alginate production (47). Further study will be required to reconcile the remaining discrepancies between the mucA22 FRD1 and our mucA22 ΔalgL ΔalgD::algD strains. Similarly, in the mucoid Pf201 strain of Pseudomonas fluorescens, deletion of algL was also found to be detrimental to growth in liquid culture; however, whether the genetic mutation resulting in P. fluorescens Pf201 mucoidy is related to mucA is unclear (46). Mucoidy can arise from mutations in other genes, such as a truncation of rsmE in Pseudomonas putida KT2440 and deletion of kinB in PAO1 (76, 77). Thus, alginate overproduction can occur in the presence of a WT mucA allele, suggesting that there are other mechanisms outside of the AlgU/T regulon, which can induce mucoidy. For example, in a kinB mutant, the sigma factor RpoN is required for mucoidy and regulates 926 genes, including genes involved in alginate biosynthesis, carbohydrate metabolism, iron regulation, motility, and quorum sensing (77, 78). As the additional alginate lyases PA1167 and PA1784 are not present in the KinB-RpoN regulon, AlgL would function as the only mechanism by which accumulated alginate in the periplasm can be degraded (79). Thus, mucoidy that occurs independent of the AlgU/T regulon may result in different consequences if algL is deleted.
In conclusion, we report the WT structure of AlgL in complex with ManA and identify key active site residues important for alginate binding and catalysis. Our in vivo studies demonstrate that AlgL does not associate with other alginate biosynthetic proteins and functions as a periplasmic homeostasis enzyme to clear the periplasm of accumulated alginate to prevent cell lysis. Furthermore, we demonstrate that AlgL is necessary for P. aeruginosa viability during alginate production when the alg operon is expressed in isolation from the AlgU/T regulon, and that a protein or proteins can compensate for its deletion when the AlgU/T regulon is upregulated. Future studies to investigate the gene(s) responsible for suppressing the effect of an algL deletion in a mucA22 genetic background will improve our understanding of alginate biosynthesis in the context of CF lung infections and provide insight into potential therapeutic target development.
Experimental procedures
Bacterial strains, plasmids, and growth conditions
A complete list of the bacterial strains and plasmids used in this study can be found in Tables S3 and S4. All P. aeruginosa strains were derived from PAO1 (80). P. aeruginosa mutant and complemented strains were generated using allelic exchange and mini-Tn7 mutagenesis, as previously described (81, 82).
Lysogeny broth (LB) contained, per liter of ultrapure water: 10.0 g tryptone, 5.0 g yeast extract, and 5.0 g NaCl. Vogel-Bonner minimal medium (VBMM) was prepared as a 10× concentrate, which contained per liter: 2.0 g MgSO4·7 H2O, 20 g citric acid, 100 g K2HPO4, and 35 g NaNH4HPO4·4 H2O, and was adjusted to pH 7.0 and sterilized by filtration. The 10× VBMM solution was diluted to 1× as needed in sterile, ultrapure water. Semisolid media was prepared by adding 1.0% (w/v) noble agar to VBMM, and 1.5% (w/v) agar to LB. Where appropriate, antibiotic selection was added to growth media as follows: for P. aeruginosa, carbenicillin (Carb) at 300 μg/ml, and gentamicin (Gen) at 30 or 60 μg/ml, depending on the application as described below; for E. coli, Gen at 10 μg/ml, Carb at 100 μg/ml, and kanamycin (Kan) at 50 μg/ml.
Basic molecular biology methods
Molecular and microbiological techniques were performed according to standard protocols (83). Genomic DNA isolation, plasmid preparation, and DNA gel extraction were performed using nucleotide purification kits purchased from Bio Basic Inc. All primers were purchased from Sigma Aldrich.
Construction of P. aeruginosa chromosomal mutations
In-frame and unmarked deletion of algL in P. aeruginosa PAO1 ΔwspF PBADalg was generated using two-step allelic exchange (82). Flanking upstream and downstream regions of the algL ORF were amplified and joined by splicing-by-overlap extension PCR (primers listed in Table S4). The upstream forward and downstream reverse primers were tailed with EcoRI and HindIII restriction enzyme sequences, respectively, to enable cloning of the spliced PCR products. The PCR product was gel purified, digested with EcoRI (Thermo Fischer Scientific) and HindIII (Thermo Fischer Scientific) restriction enzymes as per manufacturer’s instructions, and ligated into pEX18Gm using T4 DNA ligase (Thermo Fischer Scientific). The resulting allelic exchange vector, pEX18Gm::ΔalgL, was selected for on LB agar supplemented with 10 μg/ml Gen, identified by colony PCR, and verified by Sanger sequencing using M13 forward and reverse primers (Table S4). Deletions of alg44 and algD were similarly constructed (Table S4).
The deletion allele encoded by pEX18Gm::ΔalgL was introduced into P. aeruginosa PAO1 ΔwspF pBADalg or PAO1 ΔpelA ΔpslBCD mucA22 ΔalgD via biparental mating with the donor strain E. coli SM10 (84). Merodiploids were selected on VBMM supplemented with 60 μg/ml Gen. SacB-mediated counter-selection was carried out by selecting for double crossover mutations on no-salt LB (NSLB) agar supplemented with 15% (w/v) sucrose. Unmarked gene deletions were identified by colony PCR with primers targeting the outside, flanking regions of algL (Table S4). To confirm the deletion, PCR products were gel purified and sent for Sanger sequencing.
Construction of mini-Tn7 vectors
The use of the pUC18-mini-Tn7T-Gm for generating single-copy chromosomal insertions at the attTn7 site in P. aeruginosa was previously reported (81). The vector was modified for arabinose-dependent expression of complemented genes, as was previously reported (28). The araC-PBAD promoter form pJJH187 was amplified using the primer pair miniTn7-pBAD_F and miniTn7-pBAD_R. The latter contains flanking sequence encoding SmaI, NotI, PstI, and NcoI sites to generate an additional multiple cloning site downstream of the araC-PBAD promoter (85) (Table S4). The algL ORF was amplified using the primer pair algL_miniTn7_NcoI and algL_miniTn7_SacI, which encode a synthetic ribosome binding site upstream of the start codon (Table S4). The resultant PCR product was cloned into pUCT18T-miniTn7T-Gm-pBAD using NcoI and SacI restriction enzyme cut sites, selected on LB agar with 10 μg/ml Gen and 100 μg/ml Carb, and confirmed by Sanger sequencing using the miniTn7 Seq_F and miniTn7 Seq_R primers (Table S4). Construction of mini-Tn7 vectors with algD was similarly constructed (Table S4).
Complemented P. aeruginosa strains were generated through incorporation of miniTn7 vectors at the neutral attTn7 site on the P. aeruginosa chromosome via electroporation of miniTn7 vectors and the pTNS2 helper plasmid, as previously described (81). Transposon mutants were selected on LB agar supplemented with 30 μg/ml Gen and confirmed by colony PCR using the miniTn7 Seq_F and miniTn7 Seq_R primers (Table S4).
Growth curve assay
P. aeruginosa strains were grown in 5 ml of modified alginate producing (MAP) defined medium containing 100 mM monosodium glutamate, 7.5 mM monosodium phosphate, 16.8 mM dipotassium phosphate, and 10 mM magnesium sulfate supplemented with 30 μg/ml Gen for 16 h overnight at 37 °C shaking (86). The following morning, 2% (v/v) overnight starter cultures were inoculated into 25 ml MAP medium supplemented with 30 μg/ml Gen. Growth was monitored approximately every hour for 12 h by measuring the OD600 using an Ultrospec 21000 pro (Biochrom). After reaching mid-logarithmic growth phase at an approximate OD600 of 0.5, cultures were subsequently induced with 0.5% (w/v) L-arabinose to induce expression of alginate proteins.
Imaging of whole cells using transmission electron microscopy (TEM)
For each strain, 5 μl of culture was added to a carbon-coated 200-mesh copper grid and then blotted. Grids were washed once by applying 5 μl of water, and then samples were negatively stained with 2% uranyl acetate. Samples were viewed with a Phillips CM-10 transmission electron microscope operating at 80 kV under standard operating conditions, and images were collected using a SIS/Olympus Morada 11-megapixel charge-coupled device camera.
P. aeruginosa AlgL gene expression in E. coli
The nucleotide sequence of P. aeruginosa PAO1 AlgL was obtained from the Pseudomonas Genome Database (87). AlgL28–362 was PCR amplified from genomic DNA, as previously described (88). The primers account for amino acids 28 to 362, thus excluding the N-terminal signal sequence as predicted by SignalP (89). The gene was incorporated into a pET28b vector with a 3′ stop codon for N-terminal His6-tag expression, as previously described (88). For each AlgL protein construct, E. coli Origami 2(DE3) Competent Cells (Novagen) were transformed with the expression vector and grown in LB Miller broth supplemented with 50 μg/ml Kan at 37 °C. Once the bacterial cell culture reached an OD600 of 0.8, protein expression was induced by the addition of IPTG to a final concentration 1 mM. After the cell culture was incubated at 18 °C for 16 h, cells were harvested by centrifugation at 6700g for 30 min at 4 °C. Cell pellets were stored at −20 °C until required for protein purification.
Purification of AlgL protein from E. coli
The pellet from 1 l of E. coli bacterial culture was thawed and resuspended in 50 ml of cold lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM PMSF, 100 mg/ml lysozyme, 100 mg/ml DNase I, with one SIGMAFAST Protease Inhibitor Cocktail EDTA-free tablet). Cells were mechanically lysed by three passes through an Emulsiflex C3 (Avestin Inc) at 15,000 psi. The cell lysate was centrifuged at 20,100g for 40 min at 4 °C to remove cellular debris. The resultant cell lysate was loaded onto Ni-NTA resin at 4 °C equilibrated with Buffer B (20 mM Tris-HCl pH 8.0, 500 mM NaCl) with 10 mM imidazole. The resin was washed with 30 ml of Buffer B with 30 mM imidazole. The His-tagged protein was eluted off the column using 30 ml of Buffer B with 300 mM imidazole and concentrated by centrifugation with a 30 kDa cutoff Vivaspin Turbo centrifugal concentrator (Sartorius). The concentrated protein was further purified by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare) in 50 mM Tris pH 8.0, 150 mM NaCl, and 2% (v/v) glycerol. Finally, the protein was concentrated by centrifugation with a 30 kDa cutoff Vivaspin Turbo centrifugal concentrator (Sartorius) and frozen in aliquots at −80 °C until required. Protein purification was monitored throughout by SDS-PAGE.
Purification of 6His-tagged AlgL protein for structural studies
The cloning, protein expression, and purification of the native NHis6-AlgLPa28–362 were previously described (88). The SetMet-incorporated protein was produced using E. coli B834 Met− competent cells (Novagen) (90). The SeMet-incorporated NHis6-AlgLPa28–362 was purified as described for the native protein (88). Mutants of AlgL were constructed with the pET28b::AlgLPa expression vector as a template using the QuikChange Lightning Site-directed mutagenesis Kit (Stratagene) according to manufacturer’s instructions. Constructs were verified by Sanger Sequencing. All AlgL mutants were expressed and purified as described for the native protein (88).
Crystallization, data, collection, structure determination, and analysis
To determine the structure of the AlgL-alginate complex, native AlgL was cocrystallized in the presence of 8 mM ManA3. The ManA3 was prepared by acid hydrolysis as described previously (91). Native AlgL was crystallized as previously described (89). Crystals of SeMet AlgL were obtained in condition 9 from the Crystal Screen Suite (Hampton Research; 0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 30% (w/v) PEG 4000) using 4.7 mg/ml protein. Crystals were cryoprotected by soaking them for 30 s in crystallization solution supplemented with 20% (v/v) glycerol before vitrification by flash freezing. The cryoprotection solution for native AlgL also contained 8 mM ManA3. Subsequently, the frozen crystals were stored in liquid nitrogen prior to data collection.
H202A AlgL was crystallized as described for the native protein (89). The best crystals of H202A AlgL were obtained in 0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 4.6, 26% (w/v) PEG 4000, 0.01 M taurine. Crystals were cryoprotected by soaking for 30 s in crystallization solution supplemented with 20% (v/v) glycerol. The best crystals of K66A AlgL were obtained in 0.275 M K2SO4, 19% (w/v) PEG 3350, and 0.1 M HEPES pH 6.9. Crystals were cryoprotected by soaking for 10 min with 2 mM mannuronate tetrasaccharide and 20% (v/v) PEG 4000.
For native AlgL and H202A AlgL, data were collected at beam line X29 at the National Synchrotron Light Source (Brookhaven National Laboratory). 360 images of 1° Δφ oscillation on an ADSC Q315 CCD detector with a 200 mm crystal-to-detector distance with an exposure time of 0.4 s per image were collected. The data were processed, integrated, and scaled using the HKL-2000 program suite (92). SeMet SAD data consisting of 360 images of 1° Δφ oscillation on an ADSC Q315 CCD detector with a 300 mm crystal-to-detector distance with an exposure time of 0.5 s per image were also collected, processed, integrated, and scaled using the HKL-2000 program suite (93).
For K66A AlgL, data collection was completed using a D8 Venture X-ray Diffractometer (Bruker AXS) at the Structural & Biophysical Core Facility (The Hospital for Sick Children). In total, 510 total image scans of 1° Δφ oscillation with 60 s exposure times per image were collected using the diffractometer with a Kappa four-circle goniometer and a Photon 100 detector at a crystal-to-detector distance of 75 mm. Data was indexed, integrated, scaled, and merged using the Proteum 2 software (Bruker AXS).
The SeMet-SAD data in conjunction with HKL2MAP (93) were used to locate nine out of 11 selenium sites. Density-modified phases were calculated using SOLVE/RESOLVE (94). The electron density map was interpretable, and the model was built to 75% by PHENIX Autobuild and briefly refined using PHENIX.REFINE (95). The PHENIX AutoMR wizard was used to determine the structure of H202A AlgL by molecular replacement. Additional residues were built using COOT, and the structure was refined with PHENIX.REFINE (95, 96). The native AlgL structure was determined using the PHENIX AutoMR wizard using the H202A AlgL mutant structure as a search model and refined using PHENIX.REFINE. The K66A AlgL structure was solved by molecular replacement with native WT as the starting model using Phaser (97). Translation/Libration/Screw groups used during the refinement were determined automatically using the TLSMD web server (98). The electron density map was of sufficient quality for subsequent manual model building and refinement using COOT and REFMAC5 (99).
Multiple sequence analysis
Bacterial PL-5 family alginate lyases were identified from the CAZy database (http://www.cazy.org/) (50, 51, 100). Sequences of the six PL-5 family alginate lyases with the established enzyme commission (E.C.) number 4.2.2.3 for mannuronate-specific alginate lyase reactions were taken from GenBank (100). The GenBank accession numbers for the sequences are as follows: Azotobacter vinelandii CA (AGK12841.1), Azotobacter chroococcum B3 (ASL27682.1), Cobetia marina N-1 (BAA33966.1), P. syringae 31R1 (SDR80954.1), P. aeruginosa (SIP51704.1), Sphingomonas sp. A1 (BAB03312.1). Sequences were input into Clustal Omega (101) in FASTA format.
Structure analysis tools
The electrostatic surface potentials were calculated using APBS Tools (102). Conservation analysis was performed using the ConSurf server (103). All structural figures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.2, Schrödinger, LLC).
Alginate lyase activity assay
The activities of AlgL and its mutants were determined by monitoring the formation of the product of the lyase reaction, unsaturated uronic acids, in a Synergy Neo2 Multi-Mode Plate Reader (BioTek Instruments). The lyase reaction was performed at room temperature in 200 μl of 100 mM Tris-HCl pH 7.5 and 150 mM NaCl, or a similar buffer with 100 mM MES pH 6.0, containing various concentrations of nonacetylated polyM and AlgL. Reactions were allowed to progress for 10 min, measuring the OD of the solution at 240 nm (OD240) every 10 s. OD240 values were converted to molar concentration using the extinction coefficient of 6150 M−1 cm−1. Kinetic parameters (Km and the turnover number kcat) were calculated from initial velocities fitted to the Michaelis–Menten equation. All activity assays were performed in triplicate. Nonacetylated polyM was prepared from P. aeruginosa strain FRD462 as described previously (104).
Periplasmic extractions, alginate purification and detection
A 5 ml starter culture for each strain was grown in MAP medium supplemented with 30 μg/ml Gen for 16 h overnight at 37 °C shaking. The following morning, 2% (v/v) overnight starter cultures were inoculated into 50 ml MAP medium supplemented with 30 μg/ml Gen. Strains were grown to an OD600 of ∼0.3 and subsequently supplemented with 0.5% (w/v) L-arabinose to induce expression of alginate protein. After 1 h of growth, the cells were collected by centrifugation and resuspended in 1 ml cold shock buffer (0.2 M Tris-HCl pH 8.0, 0.2 g/ml sucrose, 0.1 M EDTA) and incubated on ice for 20 min. The cells were harvested by centrifugation and resuspended in a periplasmic extraction buffer (10 mM Tris-HCl pH 8.0, 5 mM MgSO4, 0.2% (v/v) SDS, and 1% (v/v) Triton X-100) and incubated on ice for 10 min with regular inversion. To collect the periplasmic contents, samples were centrifuged at 2100g and supernatants collected. Three volumes of cold 100% isopropanol were added to the bacterial supernatants and periplasmic extraction supernatants and incubated for 16 h overnight at −20 °C to precipitate the exopolysaccharides. The following morning, precipitated material was collected by centrifugation at 6700g for 20 min at 4 °C. The supernatants were discarded, and tubes were allowed to air dry for 16 h overnight at room temperature. The following morning, the precipitated material was collected with ultra-pure H2O and lyophilized using the VirTis Freeze Dryer Freezemobile 35EL Sentry 2.0 Lyophilizer (SP Scientific) until samples were completely dry. Secreted and periplasmic extractions were resuspended in 1 ml and 500 μl ultra-pure H2O, respectively, prior to blotting 4 μl onto a nitrocellulose membrane. The membrane was blocked using 5% (w/v) skim milk powder dissolved in Tris-buffered saline (TBS) (50 mM Tris:HCl pH 7.5 and 150 mM) with 0.1% (v/v) Tween-20 (TBST) for 1 h at room temperature. Blots were washed twice in TBST, and then the membrane was incubated with a Pseudomonas-reactive alginate monoclonal antibody (QED Bioscience Inc) at a 1:1000 dilution in TBST at 4 °C for 16 h. Blots were washed five times in TBST and then probed with goat α-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Bio-Rad) at 1:3000 dilution in TBST for 1 h at room temperature. Blots were washed four times in TBST and once with TBS. Blots were developed using the Super Signal West Pico chemiluminescent substrate from Pierce (Thermo Scientific). Blots were imaged using the ChemiDoc XRS System (Bio-Rad).
Antibody production
Protein-specific antibodies for Alg44 were produced as described previously (35). AlgL was purified as described in this study, and antibodies were produced as described previously (35). AlgD was expressed and purified as described previously (105, 106), and antibodies were produced as described previously (35).
Western blot analysis
Overnight cell cultures were grown in MAP medium supplemented with 30 μg/ml Gen for 16 h overnight at 37 °C shaking. Cell culture aliquots were normalized to an OD600 of 1.000 and centrifuged at 25,000g for 10 min to isolate cell pellets. Cell pellets were combined with SDS-PAGE sample buffer (4% (w/v) SDS, 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol, and 200 mM dithiothreitol) in a 1:1 ratio and boiled at 95 °C for 20 min prior to loading each sample onto a 12% (v/v) polyacrylamide gel. Protein was transferred to a polyvinylidene fluoride membrane for immunoblotting (Bio-Rad). The membrane was stained with Ponceau S (0.1% (w/v) Ponceau S in 1% (v/v) acetic acid) for 5 min. The membrane was washed with water and imaged using the ChemiDoc XRS System (Bio-Rad). The membrane was blocked using 5% (w/v) skim milk dissolved in TBST for 1 h at room temperature. Blots were washed twice in TBST, and the membrane was then incubated with alginate protein-specific antibodies (QED Bioscience) at a 1:1000 dilution in TBST at 4 °C for 16 h. Blots were washed five times in TBST and then probed with goat α-rabbit HRP-conjugated secondary antibody (Bio-Rad) at 1:3000 dilution in TBST for 1 h at room temperature. Blots were washed four times in TBST and once with TBS. Alginate protein bands were detected using the Super Signal West Pico chemiluminescent substrate from Pierce (Thermo Scientific). Blots were imaged using the Chemidoc XRS System (Bio-Rad).
Purification and quantification of alginate
Twenty-five millliliters of MAP media supplemented with 30 μg/ml Gen, with addition of 0.5% (w/v) L-arabinose, was inoculated with cells from solid media and grown for 22 h at 37 °C shaking. Cells were removed by centrifugation and culture supernatants were collected. To precipitate alginate, 3× volume of cold isopropanol was added to the supernatants and incubated at −20 °C overnight. Precipitated alginates were collected by centrifugation, and excess isopropanol was removed by air drying samples at room temperature overnight. Samples were collected and resuspended in 15 ml ultrapure H2O and then lyophilized to dryness using the VirTis BenchTop Pro Freeze Dryer (SP Scientific Products). Samples were resuspended in 1 ml PBS and incubated with 30 μg/ml each DNase I (Bio-Basic) and RNase A (Bio-Basic) overnight at 37 °C. The following day, samples were incubated with 30 μg/ml proteinase K (Bio-Basic) overnight at 37 °C. Samples were dialyzed against ultrapure H2O overnight using a 3.5 kDa molecular weight cutoff dialysis membrane (FisherBrand). Samples were collected and lyophilized to dryness using the VirTis BenchTop Pro Freeze Dryer (SP Scientific Products). Samples were assayed for alginate concentration using a colorimetric test for uronic acids with alginic acid from Macrocystis pyrifera (Sigma-Aldrich) used as the standard, as was previously described (47, 107).
Data availability
All the data described are located within the manuscript and the supplemental information. The coordinates and structure factors for WT AlgL and the K66A and H202A mutants have been deposited in the PDB, ID codes 4OZV, 7SA8, and 4OZW, respectively.
Supporting information
This article includes supporting information (100, 108, 109, 110, 111, 112, 113).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Crystallization crystallographic data collection utilized the Structural and Biophysical Core Facility at The Hospital for Children supported in part by the Canadian Foundation for Innovation. Beam line X29 at the National Synchrotron Light Source is supported by the U.S. Department of Energy and the National Institutes of Health National Center for Research Resources.
Author contributions
A. A. G. and P. L. H. conceptualization; A. A. G. and P. L. H. formal analysis; P. L. H. funding acquisition; A. A. G., F. W., S. S. Y. W., G. B. W., H. M. J., R. P., S. S. Y. W., A. K. G., A. M. B, and M. C. G. investigation; P. L. H. project administration; C. M. K. and M. R. P. resources; C. M. K., M. R. P., and P. L. H. supervision; A. A. G. and P. L. H. writing—original draft: A. A. G., G. B. W., and P. L. H. writing—review and editing.
Funding and additional information
This work was supported in part by grants from the Canadian Institutes of Health Research (CIHR) to P. L. H. (MOP 43998 and FDN154327), and C. M. K. (PJT 156111). P. L. H. was the recipient of a Tier I Canada Research Chair (2006–2020). This research has been supported by graduate scholarships from Cystic Fibrosis Canada (A. A. G. and G. B. W.); The Hospital for Sick Children Foundation Student Scholarship Program (A. A. G. and S. S. Y. W.) and the Natural Sciences and Engineering Research Council of Canada (G. B. W. and S. S. Y. W.). M. R. P. was supported by the NIH: R01AI134895, R01AI077628, and R01AI143916-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Gerald Hart
Footnotes
Present address for Gregory B. Whitfield: Département de Microbiologie, Infectiologie et Immunologie, Université de Montréal, Montréal, Quebec, Canada.
Present address for Allison K. Guitor: Department of Biochemistry and Biomedical Sciences and the Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, Ontario, Canada.
Present address for Alison M. Berezuk: Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, British Columbia, Canada.
Supporting information
References
- 1.Costerton J.W., Gessey G.G., Cheng K.J. How bacteria stick. Sci. Am. 1978;238:86–95. doi: 10.1038/scientificamerican0178-86. [DOI] [PubMed] [Google Scholar]
- 2.Costerton J.W., Lewandowski Z., DeBeer D., Caldwell D., Korber D., James G. Biofilms, the customized microniche. J. Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Coughlan L.M., Cotter P.D., Hill C., Alvarez-Ordóñez A. New weapons to fight old enemies: Novel strategies for the (bio)control of bacterial biofilms in the food industry. Front. Microbiol. 2016;7:1–21. doi: 10.3389/fmicb.2016.01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donlan R.M., Pipes W.O., Yohe T.L. Biofilm formation on cast iron substrata in water distribution systems. Water Res. 1994;28:1497–1503. [Google Scholar]
- 6.Kim W., Tengra F.K., Young Z., Shong J., Marchand N., Chan H.K., Pangule R.C., Parra M., Dordick J.S., Plawsky J.L., Collins C.H. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0062437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda K.I., Nagahori R., Yamada S., Sugimoto S., Sato C., Sato M., Iwase T., Hashimoto K., Mizunoe Y. The composition and structure of biofilms developed by Propionibacterium acnes isolated from cardiac pacemaker devices. Front. Microbiol. 2018;9:1–12. doi: 10.3389/fmicb.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piper C., Körfer R., Horstkotte D. Prosthetic valve endocarditis. Heart. 2001;85:590–593. doi: 10.1136/heart.85.5.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagihara K., Tomono K., Sawai T., Hirakata Y., Kadota J., Koga H., Tashiro T., Kohno S. Effect of clarithromycin on lymphocytes in chronic respiratory Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 1997;155:337–342. doi: 10.1164/ajrccm.155.1.9001333. [DOI] [PubMed] [Google Scholar]
- 10.Yanagihara K., Tomono K., Sawai T., Kuroki M., Kaneko Y., Ohno H., Higashiyama Y., Miyazaki Y., Hirakata Y., Maesaki S., Kadota J.I., Tashiro T., Kohno S. Combination therapy for chronic Pseudomonas aeruginosa respiratory infection associated with biofilm formation. J. Antimicrob. Chemother. 2000;46:69–72. doi: 10.1093/jac/46.1.69. [DOI] [PubMed] [Google Scholar]
- 11.Nagata T., Mukae H., Kadota J., Hayashi T., Fujii T., Kuroki M., Shirai R., Yanagihara K., Tomono K., Koji T., Kohno S. Effect of erythromycin on chronic respiratory infection caused by Pseudomonas aeruginosa with biofilm formation in an experimental murine model. Antimicrob. Agents Chemother. 2004;48:2251–2259. doi: 10.1128/AAC.48.6.2251-2259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Hengzhuang W., Wu H., Damkiær S., Jochumsen N., Song Z., Givskov M., Høiby N., Molin S. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 2012;65:366–376. doi: 10.1111/j.1574-695X.2012.00936.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakagami G., Sanada H., Sugama J., Morohoshi T., Ikeda T., Ohta Y. Detection of Pseudomonas aeruginosa quorum sensing signals in an infected ischemic wound: An experimental study in rats. Wound Repair Regen. 2008;16:30–36. doi: 10.1111/j.1524-475X.2007.00329.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang H., Wu H., Ciofu O., Song Z., Høibya N. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob. Agents Chemother. 2012;56:2683–2690. doi: 10.1128/AAC.06486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flemming H.C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield G.B., Marmont L.S., Howell P.L. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front. Microbiol. 2015;6:1–21. doi: 10.3389/fmicb.2015.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colvin K.M., Gordon V.D., Murakami K., Borlee B.R., Wozniak D.J., Wong G.C.L., Parsek M.R. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jennings L.K., Storek K.M., Ledvina H.E., Coulon C., Marmont L.S., Sadovskaya I., Secor P.R., Tseng B.S., Scian M., Filloux A., Wozniak D.J., Howell P.L., Parsek M.R. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings L.K., Dreifus J.E., Reichhardt C., Storek K.M., Secor P.R., Wozniak D.J., Hisert K.B., Parsek M.R. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021;34:108782. doi: 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L., Jackson K.D., Landry R.M., Parsek M.R., Wozniak D.J. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrd M.S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A.B., Richardson S.H., Ma L., Ralston B., Parsek M.R., Anderson E.M., Lam J.S., Wozniak D.J. Genetic and biochemical analyses of the Pseudomonas aeruginosa psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in psl and LPS production. Mol. Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mai G.T., Seow W.K., Pier G.B., McCormack J.G., Thong Y.H. Suppression of lymphocyte and neutrophil functions by Pseudomonas aeruginosa mucoid exopolysaccharide (alginate): Reversal by physicochemical, alginase, and specific monoclonal antibody treatments. Infect. Immun. 1993;61:559–564. doi: 10.1128/iai.61.2.559-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pier G.B., Coleman F., Grout M., Franklin M., Ohman D.E. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Govan J.R., Deretic V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D.W., Schurr M.J., Mudd M.H., Govan J.R.W., Holloway B.W., Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher J.C., Yu H., Mudd M.H., Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: Characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limoli D.H., Yang J., Khansaheb M.K., Helfman B., Peng L., Stecenko A.A., Goldberg J.B. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:947–953. doi: 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 28.Limoli D.H., Whitfield G.B., Kitao T., Ivey M.L., Howell P.L., O'Toole G.A., Goldberg B. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio. 2017;8:1–18. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price C.E., Brown D.G., Limoli D.H., Phelan V.V., O'Toole G.A. Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J. Bacteriol. 2020;202:1–17. doi: 10.1128/JB.00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandenburg K.S., Weaver A.J., Karna S.L.R., You T., Chen P., van Stryk S., Qian L., Pineda U., Abercrombie J.J., Leung K.P. formation of Pseudomonas aeruginosa biofilms in full-thickness scald burn wounds in rats. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-019-50003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain S., Ohman D.E. Springer US; Boston, MA: 2004. Alginate Biosynthesis in Pseudomonas. [Google Scholar]
- 32.Merighi M., Lee V.T., Hyodo M., Hayakawa Y., Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 33.Remminghorst U., Rehm B.H.A. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 34.Remminghorst U., Rehm B.H.A. Bacterial alginates: From biosynthesis to applications. Biotechnol. Lett. 2006;28:1701–1712. doi: 10.1007/s10529-006-9156-x. [DOI] [PubMed] [Google Scholar]
- 35.Whitney J.C., Whitfield G.B., Marmont L.S., Yip P., Neculai A.M., Lobsanov Y.D., Robinson H., Ohman D.E., Howell P.L. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J. Biol. Chem. 2015;290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley L.M., Weadge J.T., Baker P., Robinson H., Codée J.D.C., Tipton P.A., Ohman D.E., Howell P.L. Structural and functional characterization of Pseudomonas aeruginosa AlgX: Role of AlgX in alginate acetylation. J. Biol. Chem. 2013;288:22299–22314. doi: 10.1074/jbc.M113.484931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker P., Whitfield G.B., Hill P.J., Little D.J., Pestrak M.J., Robinson H., Wozniak D.J., Howell P.L. Characterization of the Pseudomonas aeruginosa glycoside hydrolase PslG reveals that its levels are critical for Psl polysaccharide biosynthesis and biofilm formation. J. Biol. Chem. 2015;290:28374–28387. doi: 10.1074/jbc.M115.674929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker P., Ricer T., Moynihan P.J., Kitova E.N., Walvoort M.T.C., Little D.J., Whitney J.C., Dawson K., Weadge J.T., Robinson H., Ohman D.E., Codée J.D.C., Klassen J.S., Clarke A.J., Howell P.L. P. aeruginosa SGNH hydrolase-like proteins AlgJ and AlgX have similar topology but separate and distinct roles in alginate acetylation. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keiski C.-L., Harwich M., Jain S., Neculai A.M., Yip P., Robinson H., Whitney J.C., Riley L., Burrows L.L., Ohman D.E., Howell P.L. AlgK is a TPR-containing protein and the periplasmic component of novel exopolysaccharide secretin. Structure. 2010;18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitney J.C., Hay I.D., Li C., Eckford P.D.W., Robinson H., Amaya M.F., Wood L.F., Ohman D.E., Bear C.E., Rehm B.H., Howell P.L. Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrell E.K., Tipton P.A. Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry. 2012;51:255–262. doi: 10.1021/bi301425r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamasaki M., Moriwaki S., Miyake O., Hashimoto W., Murata K., Mikami B. Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: A novel alginate lyase with a β-sandwich fold. J. Biol. Chem. 2004;279:31863–31872. doi: 10.1074/jbc.M402466200. [DOI] [PubMed] [Google Scholar]
- 43.Boyd A., Ghosh M., May T.B., Shinabarger D., Chakrabarty A.M., Keogh R. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene. 1993;131:1–8. doi: 10.1016/0378-1119(93)90662-m. [DOI] [PubMed] [Google Scholar]
- 44.May T.B., Chakrabarty A.M. Pseudomonas aeruginosa: Genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 45.Boyd A., Chakrabarty A.M. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1994;60:2355–2359. doi: 10.1128/aem.60.7.2355-2359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakkevig K., Sletta H., Gimmestad M., Aune R., Ertesvåg H., Degnes K., Christensen B.E., Ellingsen T.E., Valla S. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J. Bacteriol. 2005;187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain S., Ohman D.E. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 2005;73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Moradali M.F., Goudarztalejerdi A., Sims I.M., Rehm B.H.A. Biological function of a polysaccharide degrading enzyme in the periplasm. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep31249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albrecht M.T., Schiller N.L. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:3869–3872. doi: 10.1128/JB.187.11.3869-3872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombard V., Bernard T., Rancurel C., Brumer H., Coutinho P.M., Henrissat B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 2010;432:437–444. doi: 10.1042/BJ20101185. [DOI] [PubMed] [Google Scholar]
- 51.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon H.J., Hashimoto W., Miyake O., Murata K., Mikami B. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J. Mol. Biol. 2001;307:9–16. doi: 10.1006/jmbi.2000.4509. [DOI] [PubMed] [Google Scholar]
- 53.Mikami B., Ban M., Suzuki S., Yoon H.J., Miyake O., Yamasaki M., Ogura K., Maruyama Y., Hashimoto W., Murata K. Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III. Acta Crystallogr. D Biol. Crystallogr. 2012;68:1207–1216. doi: 10.1107/S090744491202495X. [DOI] [PubMed] [Google Scholar]
- 54.Hershberger C.D., Ye R.W., Parsek M.R., Xie Z.D., Chakrabarty A.M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative δ factor (σE) Proc. Natl. Acad. Sci. U. S. A. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H., Schurr M.J., Deretic V. Functional equivalence of Escherichia coli σ(E) and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 1995;177:3259–3268. doi: 10.1128/jb.177.11.3259-3268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schurr M.J., Yu H., Martinez-Salazar J.M., Boucher J.C., Deretic V. Control of AlgU, a member of the σ(E)-like family of stress sigma factors, by the negative regulators mucA and mucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J. Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wood L.F., Ohman D.E. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 2009;72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 58.Wood L.F., Ohman D.E. Identification of genes in the σ22 regulon of Pseudomonas aeruginosa required for cell envelope homeostasis in either the planktonic or the sessile mode of growth. mBio. 2012;3:1–11. doi: 10.1128/mBio.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu H., Boucher J.C., Hibler N.S., Deretic V. Virulence properties of Pseudomonas aeruginosa lacking the extreme- stress sigma factor AlgU (σ(E)) Infect. Immun. 1996;64:2774–2781. doi: 10.1128/iai.64.7.2774-2781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerlt J.A., Gassman P.G. Understanding enzyme-catalyzed proton abstraction from carbon acids: Details of stepwise mechanisms for β-elimination reactions. J. Am. Chem. Soc. 1992;114:5928–5934. [Google Scholar]
- 61.Shaya D., Hahn B.S., Bjerkan T.M., Kim W.S., Park N.Y., Sim J.S., Kim Y.S., Cygler M. Composite active site of chondroitin lyase ABC accepting both epimers of uronic acid. Glycobiology. 2008;18:270–277. doi: 10.1093/glycob/cwn002. [DOI] [PubMed] [Google Scholar]
- 62.Mowat C.G., Moysey R., Miles C.S., Leys D., Doherty M.K., Taylor P., Walkinshaw M.D., Reid G.A., Chapman S.K. Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase. Biochemistry. 2001;40:12292–12298. doi: 10.1021/bi011360h. [DOI] [PubMed] [Google Scholar]
- 63.Guillén Schlippe Y.v., Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch. Biochem. Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 64.Basu S., Roy A., Ghosh A., Bera A., Chattopadhyay D., Chakrabarti K. Arg235 is an essential catalytic residue of Bacillus pumilus DKS1 pectate lyase to degum ramie fibre. Biodegradation. 2011;22:153–161. doi: 10.1007/s10532-010-9384-6. [DOI] [PubMed] [Google Scholar]
- 65.Seyedarabi A., To T.T., Ali S., Hussain S., Fries M., Madsen R., Clausen M.H., Teixteira S., Brocklehurst K., Pickersgill R.W. Structural insights into substrate specificity and the anti β-elimination mechanism of pectate lyase. Biochemistry. 2010;49:539–546. doi: 10.1021/bi901503g. [DOI] [PubMed] [Google Scholar]
- 66.Highbarger L.A., Gerlt J.A., Kenyon G.L. Mechanism of the reaction catalyzed by acetoacetate decarboxylase. Importance of lysine 116 in determining the pKa of active-site lysine 115. Biochemistry. 1996;35:41–46. doi: 10.1021/bi9518306. [DOI] [PubMed] [Google Scholar]
- 67.Colvin K.M., Alnabelseya N., Baker P., Whitney J.C., Lynne Howell P., Parsek M.R. PelA deacetylase activity is required for pel polysaccharide synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2013;195:2329–2339. doi: 10.1128/JB.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazur O., Zimmer J. Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ. J. Biol. Chem. 2011;286:17601–17606. doi: 10.1074/jbc.M111.227660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Itoh Y., Rice J.D., Goller C., Pannuri A., Taylor J., Meisner J., Beveridge T.J., Preston J.F., Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Z.D., Hershberger C.D., Shankar S., Ye R.W., Chakrabarty A.M. Sigma factor-anti-sigma factor interaction in alginate synthesis: Inhibition of AlgT by MucA. J. Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathee K., Ciofu O., Sternberg C., Lindum P.W., Campbell J.I.A., Jensen P., Johnsen A.H., Givskov M., Ohman D.E., Molin S., Høiby N., Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: A mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 72.Markel E., Stodghill P., Bao Z., Myers C.R., Swingle B. AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 2016;198:2330–2344. doi: 10.1128/JB.00276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin D.W., Schurr M.J., Yu H., Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: Relationship to σ(E) and stress response. J. Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaneko A., Uenishi K., Maruyama Y., Mizuno N., Baba S., Kumasaka T., Mikami B., Murata K., Hashimoto W. A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate. J. Biol. Chem. 2017;292:15681–15690. doi: 10.1074/jbc.M117.793992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schofield M.C., Rodriguez D., Kidman A.A., Michaels L.A., Elizabeth A., Jorth P.A., Tseng B.S., Vegas L. Inhibition of AlgU by MucA is required for viability in Pseudomonas aeruginosa. bioRxiv. 2020 doi: 10.1101/2020.08.17.253617. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schofield M.C., Rodriguez D., Kidman A.A., Cassin E.K., Michaels L.A., Campbell E.A., Jorth P.A., Tseng B.S. The anti-sigma factor MucA is required for viability in Pseudomonas aeruginosa. Mol. Microbiol. 2021;116:550–563. doi: 10.1111/mmi.14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim W., Racimo F., Schluter J., Levy S.B., Foster K.R. Importance of positioning for microbial evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1639–E1647. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Damron F.H., Qiu D., Hongwei D.Y. The Pseudomonas aeruginosa sensor kinase kinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 2009;191:2285–2295. doi: 10.1128/JB.01490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Damron F.H., Owings J.P., Okkotsu Y., Varga J.J., Schurr J.R., Goldberg J.B., Schurr M.J., Yu H.D. Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN. J. Bacteriol. 2012;194:1317–1330. doi: 10.1128/JB.06105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S.L., Hufnagle W.O., Kowallk D.J., Lagrou M., Garber R.L., Goltry L., Tolentino E., Westbrock-Wadman S., Yuan Y., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 81.Choi K.H., Schweizer H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 82.Hmelo L.R., Borlee B.R., Almblad H., Love M.E., Randall T.E., Tseng B.S., Lin C., Irie Y., Storek K.M., Yang J.J., Siehnel R.J., Howell P.L., Singh P.K., Tolker-Nielsen T., Parsek M.R., et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Green M.R., Joseph S. Cold Spring Harbor Laboratory Press; New York, NY: 2012. Molecular Cloning: A Laboratory Manual, Fourth. [Google Scholar]
- 84.de Lorenzo V., Timmis K.N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 85.Almblad H., Harrison J.J., Rybtke M., Groizeleau J., Givskov M., Parsek M.R., Tolker-Nielsen T. The cyclic AMP-Vfr signaling pathway in Pseudomonas aeruginosa is inhibited by cyclic Di-GMP. J. Bacteriol. 2015;197:2190–2200. doi: 10.1128/JB.00193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohman D.E., Chakrabarty A.M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolfram F., Arora K., Robinson H., Neculai A.M., Yip P., Howell P.L. Expression, purification, crystallization and preliminary X-ray analysis of Pseudomonas aeruginosa AlgL. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:584–587. doi: 10.1107/S1744309112012808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 90.Lee J.Y., Kwak J.E., Moon J., Eom S.H., Liong E.C., Pedelacq J.D., Berendzen J., Suh S.W. Crystal structure and functional analysis of the SurE protein identify a novel phosphatase family. Nat. Struct. Biol. 2001;8:789–794. doi: 10.1038/nsb0901-789. [DOI] [PubMed] [Google Scholar]
- 91.Campa C., Holtan S., Nilsen N., Bjerkan T.M., Stokke B.T., Skjåk-Bræk G. Biochemical analysis of the processive mechanism for epimerization of alginate by mannuronan C-5 epimerase AlgE4. Biochem. J. 2004;381:155–164. doi: 10.1042/BJ20031265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 93.Pape T., Schneider T.R. HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- 94.Terwilliger T.C., Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adams P.D., Afonine P.v., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 97.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 99.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chitnis C.E., Ohman D.E. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 1990;172:2894–2900. doi: 10.1128/jb.172.6.2894-2900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Naught L.E., Gilbert S., Imhoff R., Snook C., Beamer L., Tipton P. Allosterism and cooperativity in Pseudomonas aeruginosa GDP-mannose dehydrogenase. Biochemistry. 2002;41:9637–9645. doi: 10.1021/bi025862m. [DOI] [PubMed] [Google Scholar]
- 106.Roychoudhury S., May T.B., Gill J.F., Singh S.K., Feingold D.S., Chakrabarty A.M. Purification and characterization of guanosine diphospho-D-mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J. Biol. Chem. 1989;264:9380–9385. [PubMed] [Google Scholar]
- 107.Knutson C., Jeanes A. A new modification of the carbazole analysis: Application to heteropolysaccharides. Anal. Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 108.Borlee B.R., Goldman A.D., Murakami K., Samudrala R., Wozniak D.J., Parsek M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harrison J.J., Almblad H., Yasuhiko I., Wolter D.J., Eggleston H.C., Randall T.E., Kitzman J.O., Stackhouse B., Emerson J.C., Mcnamara S., Larsen T.J., Shendure J., Hoffman L.R., Wozniak D.J., Parsek M.R. Elevated exopolysaccharide levels in Pseudomonas aeruginosa flagellar mutants have implications for biofilm growth and chronic infections. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoang T.T., Karkhoff-Schweizer R.R., Kutchma A.J., Schweizer H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 111.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983;1:784–791. [Google Scholar]
- 112.Tan J., Rouse S.L., Li D., Pye V.E., Vogeley L., Brinth A.R., El Arnaout T., Whitney J.C., Howell P.L., Sansom M.S.P., Caffrey M. A conformational landscape for alginate secretion across the outer membrane of Pseudomonas aeruginosa. Acta. Crystallogr. D. Biol. Crystrallogr. 2014;70:2054–2068. doi: 10.1107/S1399004714001850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolfram F., Kitova E.N., Robinson H., Walvoort M.T.C., Codée J.D.C., Klassen J.S., Howell P.L. Catalytic mechanism and mode of action of the periplasmic alginate epimerase AlgG. J. Biol. Chem. 2014;289:6006–6019. doi: 10.1074/jbc.M113.533158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described are located within the manuscript and the supplemental information. The coordinates and structure factors for WT AlgL and the K66A and H202A mutants have been deposited in the PDB, ID codes 4OZV, 7SA8, and 4OZW, respectively.