Abstract
The sequence of the citrate synthase gene (gltA) of 13 ehrlichial species (Ehrlichia chaffeensis, Ehrlichia canis, Ehrlichia muris, an Ehrlichia species recently detected from Ixodes ovatus, Cowdria ruminantium, Ehrlichia phagocytophila, Ehrlichia equi, the human granulocytic ehrlichiosis [HGE] agent, Anaplasma marginale, Anaplasma centrale, Ehrlichia sennetsu, Ehrlichia risticii, and Neorickettsia helminthoeca) have been determined by degenerate PCR and the Genome Walker method. The ehrlichial gltA genes are 1,197 bp (E. sennetsu and E. risticii) to 1,254 bp (A. marginale and A. centrale) long, and GC contents of the gene vary from 30.5% (Ehrlichia sp. detected from I. ovatus) to 51.0% (A. centrale). The percent identities of the gltA nucleotide sequences among ehrlichial species were 49.7% (E. risticii versus A. centrale) to 99.8% (HGE agent versus E. equi). The percent identities of deduced amino acid sequences were 44.4% (E. sennetsu versus E. muris) to 99.5% (HGE agent versus E. equi), whereas the homology range of 16S rRNA genes was 83.5% (E. risticii versus the Ehrlichia sp. detected from I. ovatus) to 99.9% (HGE agent, E. equi, and E. phagocytophila). The architecture of the phylogenetic trees constructed by gltA nucleotide sequences or amino acid sequences was similar to that derived from the 16S rRNA gene sequences but showed more-significant bootstrap values. Based upon the alignment analysis of the ehrlichial gltA sequences, two sets of primers were designed to amplify tick-borne Ehrlichia and Neorickettsia genogroup Ehrlichia (N. helminthoeca, E. sennetsu, and E. risticii), respectively. Tick-borne Ehrlichia species were specifically identified by restriction fragment length polymorphism (RFLP) patterns of AcsI and XhoI with the exception of E. muris and the very closely related ehrlichia derived from I. ovatus for which sequence analysis of the PCR product is needed. Similarly, Neorickettsia genogroup Ehrlichia species were specifically identified by RFLP patterns of RcaI digestion. If confirmed this technique will be useful in rapidly identifying Ehrlichia spp.
Ehrlichiae were previously known mainly as important agents of veterinary disease (13). For example, Ehrlichia canis, Ehrlichia equi, Ehrlichia phagocytophila, Ehrlichia platys, Ehrlichia risticii, Cowdria ruminantium, Anaplasma marginale, and Neorickettsia helminthoeca have been known as veterinary pathogens. However, over the last decade, several new Ehrlichia species or strains have been isolated and characterized from human patients and are known as major emerging tick-borne pathogens. The human granulocytic ehrlichiosis (HGE) agent, Ehrlichia chaffeensis, and Ehrlichia ewingii are now included among emerging ehrlichial agents of humans. Diagnostic methods of emerging ehrlichial infection include isolation, serology, and molecular techniques. Isolation is the “gold standard” for diagnosis; however, this method is time-consuming and expensive. Although serology is the most frequently used method for diagnosis, serological cross-reactions occur between closely related ehrlichiae, leading to misinterpretation and misdiagnosis (3, 17). With the recent development of molecular biology methods, specific and sensitive assays such as PCR and sequencing are now used for detection of ehrlichiae.
The 16S rRNA encoding gene sequence is most often used for the identification of Ehrlichia. Using the 16S rRNA, the genus Ehrlichia was found to belong to the alpha-subgroup of Proteobacteria closely related to the genus Rickettsia (5). The Ehrlichia clade also includes the genera Neorickettsia, Cowdria, and Anaplasma and the species Wolbachia pipientis (27). Polyphasic taxonomy had been advocated in order to ensure well-balanced determinations of taxonomic relationships (26), but few genes are available for investigating the genetics of ehrlichiae. A phylogenetic tree derived from nucleotide sequences of the heat shock protein gene (groESL) was the only alternative tree, and it supported the relationships among Ehrlichia species previously determined by comparison of 16S rRNA gene sequence (24). Other determined ehrlichial sequences, i.e., those of the quinolinate synthetase gene (31) and the ankA gene (4, 12, 28), have provided useful information for phylogenetic study of ehrlichiae although a limited number of strains or isolates have been tested. Consequently, studies of additional genes are required to improve the classification, identification, and diagnosis of ehrlichiae and ehrlichial diseases.
The citrate synthase gene (gltA) encodes the first enzyme of the tricarboxylic acid cycle, which is a key regulator of intracellular ATP production in nearly all living cells (29). Sequences of gltA contribute to the phylogenetic analysis and identification of Rickettsia (19) and Bartonella species (2, 9) and exhibit higher variation than the 16S rRNA gene, therefore allowing better discrimination among closely related species. gltA analysis is currently one of the best tools for this purpose and for phylogenetic analysis of these two closely related genera (2, 19).
We determined the gltA sequences of 13 ehrlichial species by combining consensus degenerate PCR and Genome Walker approaches. gltA-based phylogenetic analyses were performed, and consensus primers were developed to amplify partial sequences of gltA of ehrlichial species within a group. A preliminary PCR-restriction fragment length polymorphism (RFLP) assay was developed to allow identification of ehrlichial species.
MATERIALS AND METHODS
Ehrlichia strains and DNA preparation.
All ehrlichial strains included in this study are listed in Table 1. The HGE agent and E. equi were cultured in HL-60 cells, and E. canis, E. chaffeensis, Cowdria ruminantium, E. risticii, E. sennetsu, and N. helminthoeca were cocultured with DH82 cells. E. phagocytophila-infected sheep blood was provided by A. Garcia-Perez, Foundation Hospital Alcoron, Derio, Spain. Anaplasma centrale-infected bovine blood was provided by Y. Terada, National Institute of Animal Health, Tsukuba, Japan (8). Genomic DNA was extracted from these infected cells by using the QIAamp blood kit (Qiagen GmbH, Hilden, Germany) and stored in 200 μl of Tris-EDTA (TE) buffer at −20°C until use. Genomic DNA of a recently discovered Ehrlichia species originally isolated from Ixodes ovatus (22) was extracted from an I. ovatus tick collected from a bear in Yamaguchi Prefecture, Japan (Inokuma et al., unpublished data). This isolate was a strain variant of the newly described Ehrlichia species previously isolated from I. ovatus. DNA extracted from A. marginale, strains South Idaho and Florida, and Ehrlichia muris were kindly provided by G. Palmer, Washington State University, Pullman, and M. Kawahara, Nagoya City Public Health Research Institute, Nagoya, Japan, respectively.
TABLE 1.
Ehrlichial strains studied
| Organism | Strain | Source |
|---|---|---|
| HGE agent | Webster | J. S. Dumler, The Johns Hopkins Medical Institutions, Baltimore, Md. |
| E. equi | MRK | J. E. Madigan, University of California, Davis |
| E. phagocytophila | 1602 | A. Garcia-Perez, Fundation Hospital Alcoron |
| A. marginale | South Idaho | G. Palmer, Washington State University |
| A. marginale | Florida | G. Palmer, Washington State University |
| A. centrale | Aomori | Y. Terada, National Institute of Animal Health |
| E. canis | Oklahoma | J. Dawson, Centers for Disease Control and Prevention, Atlanta, Ga. |
| E. chaffeensis | Arkansas | J. Dawson, Centers for Disease Control and Prevention, Atlanta, Ga. |
| E. muris | M. Kawahara, Nagoya City Public Health Research Institute | |
| Ehrlichia detected from I. ovatus | Yamaguchi | H. Inokuma |
| C. ruminantium | C. E. Yunker, University of Pretoria, Pretoria, Republic of South Africa | |
| E. risticii | ATCC | |
| E. sennetsu | Miyayama | G. Dasch, Naval Medical Research Center, Bethesda, Md. |
| N. helminthoeca | Y. Rikihisa, Ohio State University, Columbus |
PCR amplification of gltA of HGE agent.
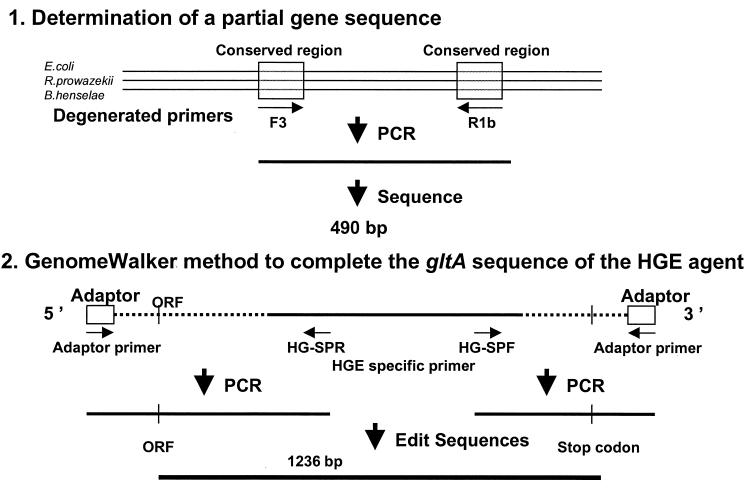
The strategy for determining gltA sequences of HGE is summarized in Fig. 1. A partial sequence of the HGE agent gltA was first determined by using degenerated primers F3 and R1b designed after the alignment of the conserved regions of gltA among Rickettsia prowazekii, Bartonella henselae, and Escherichia coli (Fig. 1; Table 2). For the amplification, the reaction mixture contained 50 pmol of each primer, 1.5 U of Taq DNA polymerase (GibcoBRL, Gaithersburg, Md.), a 20 mM concentration of each deoxynucleoside triphosphate, 10 mM Tris-HCl, 50 mM KCl, 1.6 mM MgCl2, and 5 μl of template DNA in a final volume of 50 μl. The amplifications were performed in a Peltier model PTC-200 thermal cycler (MJ Research, Inc., San Francisco, Calif.) with the following program: initial 5-min denaturation step at 95°C; 35 cycles of denaturation (95°C for 30 s), annealing (50°C for 30s), and extension (72°C for 90 s); and a final 5-min extension step at 72°C. Distilled water and DNA of B. henselae were included as negative and positive controls in each PCR. The amplification products were visualized on a 1% agarose gel after electrophoretic migration. The PCR products were purified for DNA sequencing using the QIAquick PCR purification kit (Qiagen) and sequenced using PCR primers when a single clear band was observed on the ethidium bromide-stained agarose gel. When multiple bands including bands of the expected size were obtained in PCR, the Qiagen gel extraction kit was used to purify the expected bands from the gel. After determination of the partial sequence, the unknown sequences of the 3′- and 5′-ends of the gene were amplified using the Universal Genome Walker Kit (Clontech Laboratories, Palo Alto, Calif.). Briefly, genomic DNA was digested with EcoRV, DraI, PvuII, StuI, and ScaI. DNA fragments were ligated with a Genome Walker adapter, which had one blunt end and one end with a 5′ overhang. The ligation mixture of the adapter and ehrlichial genomic DNA fragments was used as template for PCR. This PCR was performed using an adapter primer supplied by the manufacturer and ehrlichial gltA-specific primers to walk downstream on the DNA sequence (Table 3). For the amplification, 1.5 U of ELONGASE (GibcoBRL) was mixed with 10 pmol of each primer, a 20 mM concentration of each deoxynucleoside triphosphate, 10 mM Tris-HCl, 50 mM KCl, 1.6 mM MgCl2, and 5 μl of template DNA in a final volume of 50 μl. Distilled water and genomic DNA extracted from uninfected host cells (HL-60) was included as a negative control in each PCR. The following program was used for the amplification: an initial 2-min denaturation step at 94°C; 44 cycles of denaturation (94°C for 30 s), annealing (53°C for 60s), and extension (68°C for 60 s); and a final 3-min extension step at 68°C.
FIG. 1.
Strategy for determination of the sequence of the citrate synthase gene (gltA) of the HGE agent. Primers F3 and R1b were determined after alignment of the gltA of E. coli, R. prowazekii, and B. henselae. After determination of the partial sequence, the unknown sequences of both the 3′ and 5′ ends of the gene were amplified by PCR using an adapter primer provided in the Universal Genome Walker kit and the HGE agent-specific primers based on the partial sequence. Assembly of these sequences determines the complete gltA sequence of HGE. ORF, open reading frame.
TABLE 2.
Oligonucleotide primers used for PCR to amplify partial citrate synthase gene
| Primer | Sequence (5′-3′)a | Organisms in which primer is used |
|---|---|---|
| Forward | ||
| F3 | TCT-TCT-CAT-CCT-ATG-GC | HGE agent |
| F4b | CCG-GGT-TTT-ATG-TCT-ACT-GC | HGE agent, E. equi, E. phagocytophila, A. marginale South Idaho, A. marginale Florida, E. chaffeensis, E. muris, C. ruminantium |
| F4e | ACT-GCT-TCK-TGT-SAR-TC | N. helminthoeca |
| F1 | CAT-GAR-CAR-AAT-GCT-TC | E. sennetsu |
| Chaf-F | GGA-TTA-TGG-TWR-AAR-AAG-C | E. canis |
| EH130F | GGW-TTY-ATG-TCY-ACT-GCT-GC | A. centrale |
| SEN330F | AAA-TAT-CCG-TTC-TTT-CCC-AAC-G | E. risticii |
| Reverse | ||
| R1b | CGA-TGA-CCA-AAA-CCC-AT | HGE agent, E. equi, E. phagocytophila, A. marginale South Idaho, A. marginale Florida, E. chaffeensis, E. muris, C. ruminantium, N. helminthoeca, E. sennetsu |
| Chaf-R | TAY-AAC-TGR-CGT-GGR-CG | E. canis |
| HG1085R | ACT-ATA-CCK-GAG-TAA-AAG-TC | A. centrale |
| SEN890R | GCT-TTA-ATA-TGG-CTG-CAC-GAG | E. risticii |
S, G or C; R, A or G; W, A or T; Y, C or T; K, G or T.
TABLE 3.
Oligonucleotide primers and restriction genome libraries used for genome walking of the ehrlichial citrate synthase gene
| Target species | Primer | Sequence (5′-3′)b | Restriction genomic library(ies) |
|---|---|---|---|
| HGE agent | HG-SPF | CGG-ATC-AAT-GCG-ACT-TGC-G | PvuII |
| HG-SPR | TTG-CTG-AGA-TTG-GTT-CAC-C | PvuII | |
| A. marginale South Idaho | MAR-SPR | GCC-ATT-GGG-TGG-GCA-TCA | DraI |
| MAR-SPF1 | ATG-CTT-GAG-GAA-ATT-GGT-CGC-C | DraI | |
| MAR-SPF2a | CCA-GTG-AAC-ATG-TTT-ACC-GCG | DraI | |
| A. centrale | CENT-SPR1 | GAT-GAG-CAA-CGC-TGC-AGA-ACC | ScaI, DraI |
| CENT-SPR2a | CCA-CCT-GTC-GTG-ACA-AGA-TC | ScaI, DraI | |
| CENT-SPF1 | TTG-AGT-GCA-GGT-GCT-GCA-AC | PvuII | |
| CENT-SPF2a | TTT-GAG-CTT-GAA-AGA-GTG-GCC | PvuII | |
| E. chaffeensis | F3R | CAT-AGG-ATG-TGA-ATC-T | DraII |
| CHAF-SPRa | ATT-TTG-CAA-ATC-CTC-AAG-ACC | DraII | |
| F1 | CAT-GAR-CAR-AAT-GCT-TC | EcoRV | |
| CHAF-SPFa | TGT-GAT-TAA-TAT-GTT-AAT-GGC | EcoRV | |
| E. canis | CAN-SPR1 | TAA-CTT-TAT-TTC-CAT-TAG-TAT-CAC | EcoRV, DraI |
| CAN-SPR2a | CGG-TAA-TTT-CAC-TTT-TTG-ACC | EcoRV, DraI | |
| CAN-SPF | GTG-GTA-TGA-GAT-GGT-GGC-AGA-TAA-G | DraI | |
| E. muris | MUR-SPR1 | TAA-ATC-TAC-TAT-GTT-ATG-TCC | EcoRV |
| MUR-SPR2a | TCA-TAA-GTT-AAA-ACT-CCT-GTG-TC | EcoRV | |
| MUR-SPF1 | AAT-GAC-AAT-TGA-AAA-ACC-AAG | DraI | |
| ER-R1Fa | ATG-GGT-TTT-GGT-CAT-AGA-G | DraI | |
| C. ruminantium | R1b | CGA-TGA-CCA-AAA-CCC-AT | DraI |
| CR-SPRa | TAA-ATT-CCT-GAA-GTT-GCT-CAG-CAT | DraI | |
| F1b | GAT-CAT-GAR-CAR-AAT-GCT-TC | PvuII | |
| CR-SPFa | GGA-TTC-CAG-TTA-AAA-TGT-TTA-CG | PvuII | |
| E. sennetsu | R1b | CGA-TGA-CCA-AAA-CCC-AT | DraII, StuI |
| SEN-SPR1a | CTG-TAG-CCG-CAG-AAC-ATG-CC | DraII, StuI | |
| SEN-SPR2 | TTG-CAT-GGA-GCA-GTT-TTT-GC | EcoRV, ScaI | |
| SEN-SPR3a | GCA-TAC-CCT-GGA-TCA-TAA-AAG | EcoRV, ScaI | |
| SEN-SPF1 | GAG-AGG-TAT-AGT-TGA-AAG-CG | EcoRV | |
| SEN-SPF2a | AAG-GGA-AGA-GTA-CTT-TCT-GAG-TC | EcoRV | |
| SEN-SPF4 | GAG-TGA-CAG-TGA-ACA-AAA-AC | ScaI, StuI | |
| SEN-SPF5a | GCA-GAC-CTA-GAC-AAA-TTT-ACG | ScaI, StuI | |
| E. risticii | RIS-SPR | CGA-GCG-AAG-CAA-GTG-CAC-A | EcoRV, ScaI, DraI, PvuII |
| RIS-SPF | GAG-AGG-TAT-CAT-TGA-AAG-CGG | EcoRV, ScaI | |
| N. helmintoeca | NEO-SPR1 | CCA-TGG-GAT-GTG-CAT-C | EcoRV |
| NEO-SPR2a | TTT-TGC-GCG-GAC-AGG-GTC | EcoRV | |
| F1 | CAT-GAR-CAR-AAT-GCT-TC | EcoRV | |
| NEO-SPFa | GAG-AAA-GTC-CTG-CAC-ATG-CTG | EcoRV |
Used in the nested PCR.
R, A or G.
Determination of other ehrlichial gltA sequences.
Additional primers were designed based upon the alignment of the complete gltA sequences of the HGE agent, R. prowazekii, and B. henselae (Table 2). Primers F4b and R1b were used for the amplification of E. equi, E. phagocytophila, E. chaffeensis, E. muris, C. ruminantium, and A. marginale strains South Idaho and Florida; primers F4e and R1b were used for the amplification of N. helminthoeca; and primers F1 and R1b were used for the amplification of E. sennetsu. The optimal annealing temperature (48 to 55° C) was determined for each species by empirical testing. After determination of the sequences of these short fragments (230 to 730 bp), the gltA sequences of E. chaffeensis, E. muris, C. ruminantium, A. marginale strain South Idaho, N. helminthoeca, and E. sennetsu were completed by using the Genome Walker method as described above for the HGE agent. Based upon the complete sequences of ehrlichial gltA described above, new primer sets were designed to amplify partial gltA sequences of E. canis, A. centrale, and E. risticii (Table 2). After sequencing of these partial gltA fragments, the Genome Walker method was used to determine the 3′ and 5′ ends of these three species. As the material of the Ehrlichia species detected from I. ovatus was not abundant enough to perform the Genome Walker method, two primer pairs, CAN-M61F–R1b and F1b-MUR1251R, were used to obtain a partial gltA sequence of the species (Table 4).
TABLE 4.
Oligonucleotide primers used for PCR amplification and sequencing to determine the gltA sequences of various species and to confirm sequences of the gltA coding region in other ehrlichial strains studieda
| Target species | Primer | Sequence (5′-3′)c |
|---|---|---|
| HGE agent, E. equi, E. phagocytophila | HG-M28F | GTA-ATA-AAT-TGT-ATT-ATC-AGA-G |
| HG1257R | AAT-ACG-TGA-GTT-TGA-AAC-CA | |
| HG602Fb | TGG-ATG-ATG-CAC-ATC-GTG | |
| HG700Ra | TAC-GCA-CAG-TGG-AAG-TAG | |
| A. marginale Florida and South Idaho | MAR-M35F | GTC-TGG-TGA-GTT-TGT-TGT-CC |
| MAR-1287R | GCT-TGC-ACA-TCG-CTC-AAT-AA | |
| A. centrale | CENT-M32F | GTG-TCC-AGT-AAA-CTT-GTT-GTC-GG |
| CENT1534R | AAA-GCA-TGG-TGC-GAG-CAT-A | |
| Ehrlichia detected from I. ovatus | CAN-M61F | TTA-TCT-GTT-TAT-GTT-ATA-TAA-GC |
| R1b | CGA-TGA-CCA-AAA-CCC-AT | |
| F1b | GAT-CAT-GAR-CAR-AAT-GCT-TC | |
| MUR1251R | CTA-GAT-TTT-TGT-AAT-ATG-GCC-AG | |
| E. chaffeennsis | CAN-M61F | TTA-TCT-GTT-TAT-GTT-ATA-TAA-GC |
| CHAFF1285R | AAA-CAA-TAA-GCA-ATG-ATA-ATT-CAA | |
| E. muris | CAN-M61F | TTA-TCT-GTT-TAT-GTT-ATA-TAA-GC |
| MUR1278R | AAT-TTG-ATA-ACA-ATA-GCA-TAA-AAA-C | |
| E. canis | CAN-M61F | TTA-TCT-GTT-TAT-GTT-ATA-TAA-GC |
| CAN1317R | CAG-TAC-CTA-TGC-ATA-TCA-ATC-C | |
| C. ruminantium | CR-M31F | ACG-CTT-TGT-TGT-TAT-TGT-ATT-AG |
| CR1278R | CTA-CAA-AAG-GAA-ATA-CCT-TCA-C | |
| E. sennetsu, E. risticii | SEN-M26F | CTC-AAC-TGG-AGA-ATA-TTT-AAA-GAA-G |
| SEN–RIS-1218R | GCG-KAG-AAC-CAC-TAA-MAG-CTG | |
| N. helminthoeca | NH-M91F | CAT-AGA-TTA-ACT-GTG-CTA-C |
| NH1239R | GAT-GAA-ATT-CCA-TCC-TCG-TGT-G |
Specific or degenerated primers shown in Table 3 were also used as supplementary sequence primers if necessary.
Sequence primers.
R, A or G; M, A or C.
DNA sequencing.
The fluorescence-labeled dideoxynucleotide technology was used for DNA sequencing reactions (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). The sequencing fragments were separated using an Applied Biosystems model ABI 310 automated DNA sequencer (Perkin-Elmer), and data were collected with an ABI PRISM 310 Genetic Analyzer package (Perkin-Elmer). The collected sequences were assembled and edited with the AutoAssembler (version 1.4; Perkin-Elmer).
Confirmation of the ehrlichial gltA sequence.
In order to avoid the editorial error of the Genome Walker method, obtained sequences of the citrate synthase coding region, including the open reading frame at the 5′ end and the stop codon at the 3′ end, from each ehrlichial species except for the Ehrlichia sp. detected from I. ovatus were confirmed by PCR with the primers shown in Table 4 and also were sequenced.
Data analysis.
The sequences of ehrlichial gltA and the registered gltA sequence of R. prowazekii and B. henselae deposited in GenBank were analyzed for GC content, level of similarity, and phylogenetic relationships. Pairwise percent identities of the sequences with all gaps omitted were calculated by a program designed by H. Ogata, IGS, CNRS-UMR, France. Multiple alignment analysis, distance matrix calculation, and construction of a phylogenetic tree were performed with the ClustalW program (25), version 1.8 (available from the DNA Data Bank of Japan, Mishima, Japan [http://www.ddbj.nig.ac.jp/htmls/E-mail/clustalw-e.html]). The distance matrices for the aligned sequences with all gaps ignored were calculated using the Kimura two-parameter method (10), and the neighbor-joining method was used for constructing a phylogenetic tree (21). The stability of the tree obtained was estimated by bootstrap analysis for 1,000 replications using the same program. Tree figures were generated using the TreeView program, version 1.61 (15). The same analysis of similarity and phylogenetic relationships was also performed for the deduced amino acid sequences of gltA and the 16S rRNA gene sequences.
Consensus PCR and PCR-RFLP analysis.
Based upon the alignment analysis of the ehrlichial gltA sequences, a pair of primers, EHR-CS136F (5′-TTY-ATG-TCY-ACT-GCT-GCK-TG-3′) and EHR-CS778R (5′-GCN-CCM-CCA-TGM-GCT-GG-3′), was designed in order to specifically amplify partial sequences of the gltA gene of the following tick-borne Ehrlichia species: E. chaffeensis, E. canis, E. muris, the Ehrlichia sp. detected from I. ovatus, C. ruminantium, the HGE agent, E. equi, E. phagocytophila, A. marginale, and A. centrale. Another pair of primers, NEO-CS142F (5′-ATY-ACY-TTC-RTA-GAY-GGT-GA-3′) and NEO-CS730R (5′-CGT-GCA-GTG-GWC-CCC-ATA-A-3′), was designed to specifically amplify N. helminthoeca, E. risticii, and E. sennetsu. The conditions for these two PCRs were the same as those described above with an annealing temperature at 55°C. Amplified products were digested with Acs I (Roche, Mannheim, Germany) and XhoI (Roche) for tick-borne Ehrlichia species and RcaI (Roche) for Neorickettsia genogroup ehrlichial species. Briefly, 7 μl of each PCR product was incubated with 2 μl of each enzyme and 1 μl of 10× buffer supplied by the manufacturer, and this was followed by incubation at 50°C for 1 h. Digestion products were separated by 1.5% agarose gel electrophoresis.
Nucleotide sequence accession numbers.
The herein-determined gltA sequences of the following organisms have been deposited in the GenBank database under the indicated accession numbers: HGE agent, AF304136; E. equi, AF304137; E. phagocytophila, AF304138; A. marginale strain South Idaho, AF304139; A. marginale strain Florida, AF304140; A. centrale, AF304141; E. chaffeensis, AF304142; E. canis, AF304143; E. muris, AF304144; Ehrlichia sp. detected from I. ovatus, AF304145; C. ruminantium, AF304146; E. risticii, AF304147; E. sennetsu, AF304148; and N. helminthoeca, AF304149. The GenBank accession numbers of the gltA sequences of R. prowazekii, B. henselae, and E. coli used in this study were M17149, L38987, and J01619, respectively. The GenBank accession numbers of the following 16S rRNA gene sequences used to calculate percent identities and construct phylogenetic trees are as indicated: HGE agent, U02521; E. equi, M73223; E. phagocytophila, M73224; A. marginale, M60313; A. centrale, AF283007; E. chaffeensis, M73222; E. canis, M73221; E. muris, U15527; Ehrlichia sp. detected from I. ovatus, AF260591; C. ruminantium, AF069758; W. pipientis, AF179630; E. risticii, M21290; E. sennetsu, M73225; N. helminthoeca, U12457; R. prowazekii, M21789; and B. henselae, AJ223779. The GenBank accession numbers of the following heat shock protein-coding genes for most Ehrlichia species, except for A. centrale and N. helminthoeca or for the glutathione synthetase gene for A. centrale, that were used to compare the GC contents with those of gltA are as indicated: HGE agent, AF172163; E. equi, AF173988; E. phagocytophila, U96735; A. marginale, AF165812; A. centrale, M80425; E. chaffeensis, L10917; E. canis, U96731; E. muris, AF210459; Ehrlichia sp. detected from Ixodes ovatus, AB032712; C. ruminantium, U13638; E. risticii, AF206299; E. sennetsu, AF060197; R. prowazekii, Y15783; and B. henselae, U78514.
RESULTS
Determination of ehrlichial gltA sequences.
After determination of the 482-bp partial sequence of the gltA of HGE agent, a 1,236-bp open reading frame extending from the ATG start codon down to the TAA stop codon was determined using the Genome Walker PCR method.
Complete gltA nucleotide sequences of E. chaffeensis, E. muris, C. ruminantium, A. marginale strain South Idaho, N. helminthoeca, and E. sennetsu have been determined, with lengths of 1,251, 1,251, 1,248, 1,254, 1,212, and 1,197 bp, respectively. As the 730-bp partial gltA sequence of E. equi and E. phagocytophila exhibited more than 99.0% similarity with that of HGE, primers HG-M28F and HG1257R, which could amplify the complete gltA sequence of the HGE agent, were used for determining the 1,236-bp gltA sequences of E. equi and E. phagocytophila (Table 4). As the 730-bp partial sequence of A. marginale strain Florida was identical to that of strain South Idaho, primers MAR-M35F and HG1287R, which could amplify the complete gltA sequence of A. marginale strain South Idaho, were used to amplify the 1,254-bp sequences of the gltA of A. marginale strain Florida (Table 4).
The complete gltA nucleotide sequences of E. canis, A. centrale, and E. risticii have been determined in the third step of the strategy. The gltA genes of these organisms have lengths of 1,251, 1,254, and 1,197 bp, respectively. Two primer sets, CAN-M61F–R1b and F1b-MUR1251R, were used to obtain a partial gltA sequence of the Ehrlichia sp. detected from I. ovatus (Table 2) and resulted in a 1,228-bp open reading frame near the 3′ end.
Comparison of gltA sequences.
The GC content of the ehrlichial gltA genes varied from 30.5% for the Ehrlichia sp. detected from I. ovatus to 51.0% for A. centrale (Table 5).The multiple alignment analysis by the ClustalW program demonstrated several gaps in the alignment (data not shown). The percentages of similarity varied from 49.7% (E. risticii versus A. centrale) to 99.8% (the HGE agent versus E. equi) for the nucleotide sequence and from 44.4% (E. sennetsu versus E. muris) to 99.5% (the HGE agent versus E. equi) for the deduced amino acid sequence. The percent identities of the gltA nucleotide sequences between species in the Neorickettsia group (N. helminthoeca, E. sennetsu, and E. risticii) and other ehrlichial species varied from 49.7 to 55.3%; these were lower than those between other ehrlichial species and R. prowazekii (53.8 to 62.2%) or B. henselae (52.7 to 58.5%). The gltA nucleotide and deduced amino acid sequences of the HGE agent, E. equi, and E. phagocytophila were very similar: there were 3 nucleotide and 2 amino acid differences between the HGE agent and E. equi sequences, 9 nucleotide and 4 amino acid differences between the HGE agent and E. phagocytophila, and 10 nucleotide and 4 amino acid differences between E. equi and E. phagocytophila.
TABLE 5.
The length and GC contents of ehrlichial citrate synthase gene
| Organism | Citrate synthase gene
|
%GC contents of other gene registered in GenBank (gene, accession number) | |
|---|---|---|---|
| Length (bp)a | GC content (%) | ||
| HGE agent | 1,236 | 38.3 | 42.4 (heat shock protein gene, AF172163) |
| E. equi | 1,236 | 38.3 | 42.1 (heat shock protein gene, AF173988) |
| E. phagocytophila | 1,236 | 38.2 | 42.1 (heat shock protein gene, U96735) |
| A. marginale | 1,254 | 50.5 | 48.8 (heat shock protein gene, AF165812) |
| A. centrale | 1,254 | 51.0 | 50.4 (glutathione synthetase gene, M80425) |
| E. canis | 1,251 | 32.1 | 33.2 (heat shock protein gene, U96731) |
| E. chaffeensis | 1,251 | 31.3 | 33.2 (heat shock protein gene, L10917) |
| E. muris | 1,251 | 31.2 | 34.0 (heat shock protein gene, AF210459) |
| Ehrlichia detected from I. ovatus | 1,228 b | 30.5 | 33.7 (heat shock protein gene, AB032712) |
| C. ruminantium | 1,248 | 32.6 | 33.0 (heat shock protein gene, U13638) |
| E. risticii | 1,197 | 43.7 | 41.8 (heat shock protein gene, AF206299) |
| E. sennetsu | 1,197 | 43.9 | 42.4 (heat shock protein gene, AF060197) |
| N. helminthoeca | 1,212 | 44.2 | 50.5 (16S rRNA gene, U12457) |
| R. prowazekii | 1,311 | 33.6 | 33.8 (heat shock protein gene, Y15783) |
| B. henselae | 1,296 | 38.6 | 44.3 (heat shock protein gene, U78514) |
From ATG start codon to stop codon.
Partial sequence; sequencing of the 3′ end has not been completed.
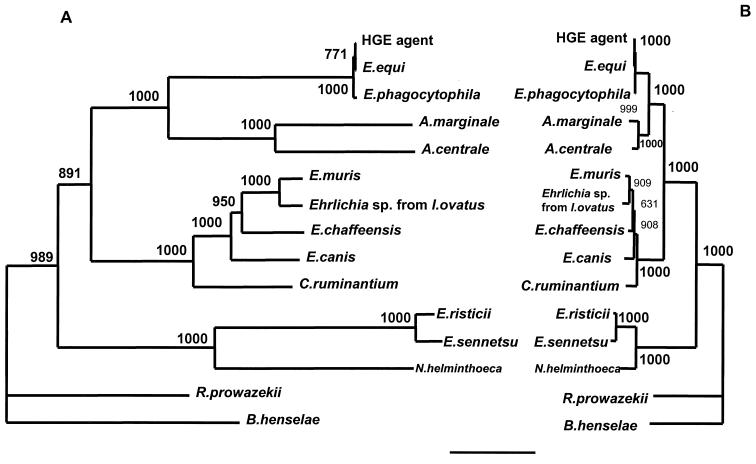
Phylogenetic analyses.
GltA-based phylogenetic reconstruction was compared with 16S rRNA-based analysis (Fig. 2). The phylogenetic tree based on the deduced amino acid sequences of ehrlichial species, R. prowazekii, and B. henselae was similar to the gltA gene tree (data not shown)
FIG. 2.
Phylogenetic relationship of various Ehrlichia spp. based on the nucleotide sequences of citrate synthase gene (A) and 16S rRNA gene (B). The neighbor-joining method was used to construct the phylogenetic tree by using the ClustalW program. The scale bar represents 1% divergence. The numbers at nodes are the proportions of 1,000 bootstrap resamplings that support the topology shown.
The topologies of the gltA-based phylogenetic trees were almost the same as those derived from the 16S rRNA gene sequence analyses. However, the trees constructed by gltA nucleotide sequences or amino acid sequences showed better bootstrap values than the 16S rRNA-based tree. Higher bootstrap values were obtained in the nucleotides of gltA-based trees for the relationships between E. muris and the Ehrlichia species detected from I. ovatus (bootstrap value: 1,000) and between E. chaffeensis and these two species (bootstrap value: 950). However, the bootstrap value between the HGE agent and E. equi was comparatively low (771 and 497 for nucleotide- and amino acid sequence-based trees, respectively).
Consensus PCR and PCR-RFLP analysis.
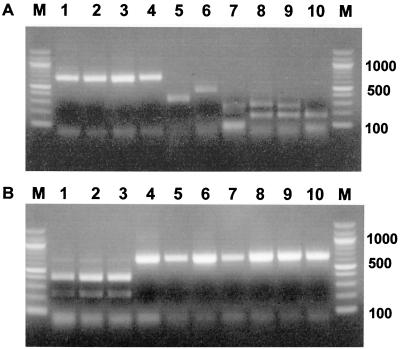
A consensus primer pair, EHR-CS136F and EHR-CS778R, amplified a 643-bp partial sequence of gltA in 10 tick-borne Ehrlichia species. Predicted AcsI RFLP patterns were a single band (no digestion) for the HGE agent, E. equi, E. phagocytophila, and A. marginale; two bands of 312 and 331 bp for A. centrale; three bands of 37, 162, and 44 bp for E. canis; five bands of 14, 84, 87, 199, and 259 bp for E. chaffeensis; four bands of 38, 162, 171, and 272 bp for E. muris; five bands of 14, 38, 162, 171, and 258 bp for the Ehrlichia sp. detected from I. ovatus; and five bands of 37, 75, 167, 171, and 192 bp for C. ruminantium (Fig. 3A). The predicted patterns for XhoI were two bands of 242 and 401 bp for the HGE agent, E. equi, and E. phagocytophila and a unique band for the seven other species (Fig. 3B). Experimental results differed from those predicted, because bands that have molecular size of <100 bp were not easily detected and two or three bands which have similar molecular sizes could not be distinguished in agarose gels. The combination of both AcsI and XhoI digestion identified tick-borne ehrlichial species except those of the E. phagocytophila genogroup (HGE, E. equi, and E. phagocytophila), and E. muris and the Ehrlichia sp. detected from I. ovatus that showed the same RFLP patterns for both AcsI and XhoI digestion.
FIG. 3.
Restriction profiles obtained after AcsI (A) and XhoI (B) digestion of a portion of the citrate synthase gene amplified from 10 tick-borne ehrlichial species by PCR using consensus primers EHR-CS136F–EHR-CS778R. Lanes: M, molecular weight markers (in thousands); 1, HGE agent; 2, E. equi; 3, E. phagocytophila; 4, A. marginale; 5, A. centrale; 6, E. canis; 7, E. chaffeensis; 8, E. muris; 9, Ehrlichia sp. detected in I. ovatus; 10, C. ruminantium.
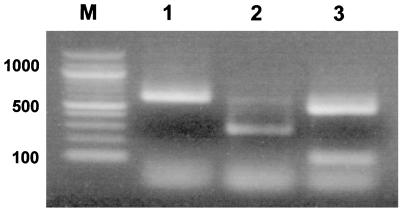
A consensus primer pair, NEO-CS142F and NEO-CS730R, amplified a 596-bp partial sequence in E. sennetsu, E. risticii, and N. helminthoeca. Predicted RcaI RFLP patterns included a unique band for E. sennetsu, two bands of 285 and 304 bp for E. risticii, and two bands of 109 and 487 bp for N. helminthoeca. The result of the RFLP is shown in Fig. 4. Although two bands of 285 and 304 bp for E. risticii were not distinguished in agarose gel, the RFLP profiles of these three species were apparently different from one to another.
FIG. 4.
Restriction profiles obtained after RcaI digestion of a portion of the citrate synthase gene amplified from three species of the Neorickettsia genogroup by PCR using consensus primer NEO-CS142F–NEO-CS730R. Lanes: M, molecular weight markers (in thousands); 1, E. sennetsu; 2, E. risticii; 3, N. helminthoeca.
DISCUSSION
To determine the complete gltA sequence of most ehrlichial species, a combination of consensus PCR amplification, sequencing, and the Genome Walker method was used in this study. To determine the RNA polymerase beta subunit (rpoB) gene of Leptospira biflexa, this strategy was recently evaluated as a convenient means for amplifying unknown sequences on the 3′ and 5′ ends (18). Semipurified genomic DNA of ehrlichial bacteria, including host cell genomic DNA, was used in this study, although the Genome Walker method is, as recommended by the manufacturer, usually performed with purified DNA in order to avoid nonspecific amplification. By using DNA from noninfected host cells as a negative control, nonspecific amplification in the Genome Walker PCR can be identified and controlled. Avoiding the purification steps saves material and time, which are critical issues when dealing with fastidious intracellular organisms such as ehrlichiae.
The length of the gltA sequences varied in ehrlichial species from 1,197 bp (E. sennetsu and E. risticii) to 1,254 bp (A. marginale and A. centrale), encoding proteins with deduced sequences of 398 to 421 amino acid residues. The length of ehrlichial gltA was shorter than that of gltA from closely related genera of Rickettsia (R. prowazekii, 1,311 bp) and Bartonella (B. henselae, 1,296 bp) (14, 30). The level of similarity among ehrlichial gltA was much lower than that of 16S rRNA gene sequences in the same species. The percent identities of the gltA nucleotide and deduced amino acid sequences vary from 49.7 to 99.8% and 44.4 to 99.5%, respectively. In contrast, those of the 16S rRNA gene vary from 83.5 to 99.9%. Percent identities were also found to be lower than those reported for groESL sequences (23), although the differences are small. These findings suggest that ehrlichial gltA sequencing may offer a tool with increasing discriminatory power for both phylogenetic and identification studies because of the greater variation in gltA than in any other gene currently determined for these species. Interestingly, the level of similarity between species in the Neorickettsia genogroup (N. helminthoeca, E. sennetsu, and E. risticii) and other ehrlichial species was lower than that between these species and R. prowazekii or B. henselae. The gltA sequence analysis confirmed that this group of ehrlichiae forms a clade distinct from other tick-borne ehrlichial agents.
GC contents of the gltA gene also shows greater variation from 30.5 to 51.0%. The C. ruminantium genogroup (E. canis, E. chaffeensis, E. muris, Ehrlichia sp. detected from I. ovatus, and C. ruminantium) has lower GC content (30.5 to 32.6%) than the E phagocytophila genogroup (GC: 38.2 to 38.3%) and the Neorickettsia genogroup (GC: 43.7 to 44.2%). A. marginale and A. centrale show the highest percentages (50.5 and 51.0%, respectively). The GC content of other genes, mainly heat shock protein, shows values similar to those of gltA; 30.5 to 34.0% for the heat shock protein gene of C. ruminantium genogroup species, 48.8% for the heat shock protein gene of A. marginale, and 50.4% for the glutathione synthetase gene of A. centrale. The architecture of gltA-based phylogenetic trees was almost the same as the that of the tree derived from the 16S rRNA gene sequences. However, the trees constructed from gltA show more divergence than that from the 16S rRNA gene. The relationships of E. muris, E. chaffeensis, and the recently detected Ehrlichia species originally isolated from I. ovatus were well defined, with higher bootstrap values in the gltA-based tree than for those of the 16S rRNA-based tree. The bootstrap values for all of the nodes were greater than 85% in both nucleotide and deduced amino acid analyses. The only exception was the branching of the HGE agent and E. equi, due to the high sequence homology. These findings suggest that the gltA-based phylogeny of ehrlichial agents can be an additional phylogenetic tool and support the 16S rRNA-based phylogeny.
Although A. marginale, A. centrale, E. phagocytophila, E. equi, and the HGE agent are all tick-borne agents and most often detected in the cells in the peripheral blood that derive from bone marrow precursors in vivo, both A. marginale and A. centrale show biological differences from E. phagocytophila, E. equi, and the HGE agent. Anaplasma species infect predominantly erythrocytes in the ruminant host, while E. phagocytophila genogroup ehrlichiae are most often detected in granulocytes of various mammalian hosts, including humans. In vitro, E. equi and the HGE agent grew in the HL-60 human promyelocytic cell line (6, 7, 11), whereas no mammalian cell system allowed active replication of Anaplasma species. In the present study, both A. marginale and A. centrale show low levels of similarity with the E. phagocytophila genogroup (63.8 to 64.0%). The GC contents of both A. marginale and A. centrale are 50.5 and 51.0%, respectively, while those of the E. phagocytophila genogroup are of 38.2 or 38.3%. These two groups were distant in the phylogenetic tree. These data regarding the level of similarity between the gltA nucleotide sequences, GC content, and the gltA-based trees suggest that the Anaplasma group (A. marginale and A. centrale) forms a clade independent from the E. phagocytophila genogroup.
W. pipientis occupies a position intermediate between tick-borne Ehrlichia and the Neorickettsia clade as shown by 16S rRNA gene analysis (5). The gltA sequence of this species was not been analyzed in the present study, but its sequencing is under way in our laboratory.
The gltA nucleotide sequences of 13 ehrlichial species used in this study demonstrate both very conserved regions that allowed us to amplify DNA fragments by using both B. henselae- and R. prowazekii-derived degenerate primers and highly variable regions that allowed better definition of Ehrlichia species. Consequently, the design of Ehrlichia genus-specific primers has not been successful. However, tick-borne-Ehrlichia-specific or Neorickettsia genogroup-specific primer sets that amplify partial gltA genes of several ehrlichial agents were designed. In the present study, two sets of primers, EHR-CS136F–EHR-CS778R and NEO-CS142F–NEO-CS730R, amplified 10 tick-borne Ehrlichia species and three species among the Neorickettsia genogroup, respectively. This gltA-based group-specific PCR may be useful for epidemiological studies of ehrlichiosis, as previously demonstrated for Rickettsia (1) (2, 20). Although a unique isolate of each species has been tested herein, the conservation of these primer pairs among the different species argues for their conservation within a species suggesting their usefulness. Indeed, two new Ehrlichia genotypes have been recently found in African ticks in our laboratory, and partial gltA sequences of these species were determined by using a consensus primer set to characterize the phylogenetic position in ehrlichiae (16).
The results of RFLP analysis revealed that the combination of consensus PCR and RFLP can identify ehrlichiae at the species level with the exception of the E. phagocytophila genogroup and E. muris or Ehrlichia sp. detected in I. ovatus. It has been suggested that E. phagocytophila, E. equi, and the HGE agent are different strains of the same species and are not convincingly distinguishable by 16S rRNA analysis (4). Moreover, little is known about the newly described ehrlichia isolated from I. ovatus or its phylogenetic relationship with E. muris. RFLP analysis of gltA PCR products offers an effective new tool for identification of ehrlichial species among tick-borne Ehrlichia and Neorickettsia genogroup Ehrlichia.
ACKNOWLEDGMENTS
We thank H. Ogata for analyzing the sequence data and J. S. Dumler for correction of the English version of the manuscript and helpful discussion.
H. Inokuma was supported by a grant from the EGIDE, Paris, France.
REFERENCES
- 1.Billings A N, Teltow G J, Weaver S C, Walker D H. Molecular characterization of a novel Rickettsia species from Ixodes scapularis in Texas. Emerg Infect Dis. 1998;4:305–309. doi: 10.3201/eid0402.980221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birtles R J, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 3.Brouqui P, Dumler J S. Serologic evidence of human monocytic and granulocytic ehrlichiosis in Israel. Emerg Infect Dis. 2000;6:314–315. doi: 10.3201/eid0603.000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chae J S, Foley J E, Dumler J S, Madigan J E. Comparison of the nucleotide sequences of 16S rRNA, 444 Ep-ank, and groESL heat shock operon genes in naturally occurring Ehrlichia equi and human granulocytic ehrlichiosis agent isolates from Northern California. J Clin Microbiol. 2000;38:1364–1369. doi: 10.1128/jcm.38.4.1364-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drancourt M, Raoult D. Taxonomic position of the rickettsiae: current knowledge. FEMS Microbiol Rev. 1994;13:13–24. doi: 10.1111/j.1574-6976.1994.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 6.Goodman J L, Nelson C, Vitale B, Madigan J E, Dumler J S, Kurtti T J, Munderloh U G. Direct cultivation of the causative agent of human granulocytic ehrlichiosis N. Engl J Med. 1996;334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 7.Heimer R, Van Andel A, Wormser G P, Wilson M L. Propagation of granulocytic Ehrlichia spp. from human and equine sources in HL-60 cells induced to differentiate into functional granulocytes. J Clin Microbiol. 1997;35:923–927. doi: 10.1128/jcm.35.4.923-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inokuma H, Terada Y, Kamio T, Raoult D, Brouqui P. Analysis of the 16S rRNA gene sequence of Anaplasma centrale and its phylogenetic relatedness to other ehrlichiae. Clin Diagn Lab Immunol. 2001;8:241–244. doi: 10.1128/CDLI.8.2.241-244.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joblet C, Roux V, Drancourt M, Gouvernet J, Raoult D. Identification of Bartonella (Rochalimaea) species among fastidious gram-negative bacteria on the basis of the partial sequence of the citrate-synthase gene. J Clin Microbiol. 1995;33:1879–1883. doi: 10.1128/jcm.33.7.1879-1883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 11.Klein M B, Miller J S, Nelson C M, Goodman J L. Primary bone marrow progenitors of both granulocytic and monocytic lineages are susceptible to infection with the agent of human granulocytic ehrlichiosis. J Infect Dis. 1997;176:1405–1409. doi: 10.1086/517332. [DOI] [PubMed] [Google Scholar]
- 12.Massung R F, Owens J H, Ross D, Reed K D, Petrovec M, Bjoersdorff A, Coughlin R T, Beltz G A, Murphy C I. Sequence analysis of the ank gene of granulocytic ehrlichiae. J Clin Microbiol. 2000;38:2917–2922. doi: 10.1128/jcm.38.8.2917-2922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDade J E. Ehrlichiosis—a disease of animals and humans. J Infect Dis. 1990;161:609–617. doi: 10.1093/infdis/161.4.609. [DOI] [PubMed] [Google Scholar]
- 14.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 16.Parola, P., H. Inokuma, J. L. Camicas, P. Brouqui, and D. Raoult. Detection and identification of spotted fever group Rickettsia and Ehrlichia in African ticks. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 17.Parola P, Raoult D. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin Infect Dis. 2001;32:897–928. doi: 10.1086/319347. [DOI] [PubMed] [Google Scholar]
- 18.Renesto P, Lorvellec-Guillon K, Drancourt M, Raoult D. rpoB gene analysis as a novel strategy for identification of spirochetes from the genera Borrelia, Treponema, and Leptospira. J Clin Microbiol. 2000;38:2200–2203. doi: 10.1128/jcm.38.6.2200-2203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 20.Rydkina E, Roux V, Rudakov N, Gafarova M, Tarasevich I, Raoult D. New Rickettsiae in ticks collected in territories of the former Soviet Union. Emerg Infect Dis. 1999;5:811–814. doi: 10.3201/eid0506.990612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Med Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Shibata S, Kawahara M, Rikihisa Y, Fujita H, Watanabe Y, Suto C, Ito T. New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J Clin Microbiol. 2000;38:1331–1338. doi: 10.1128/jcm.38.4.1331-1338.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumner J W, Nicholson W L, Massung R F. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J Clin Microbiol. 1997;35:2087–2092. doi: 10.1128/jcm.35.8.2087-2092.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumner J W, Storch G A, Buller R S, Liddell A M, Stockham S L, Rikihisa Y, Messenger S, Paddock C D. PCR amplification and phylogenetic analysis of groESL operon sequences from Ehrlichia ewingii and Ehrlichia muris. J Clin Microbiol. 2000;38:2746–2749. doi: 10.1128/jcm.38.7.2746-2749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson J C, Higgins D G, Gibson T J. CUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position, specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandamme P, Pot B, Gillis M, De Vos A J, Kersters K, Swings J G. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker D H, Dumler J S. Emergence of the ehrlichioses as human health problems. Emerg Infect Dis. 1996;2:18–29. doi: 10.3201/eid0201.960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walls J J, Caturegli P, Bakken J S, Asanovich K M, Dumler J S. Improved sensitivity of PCR for diagnosis of human granulocytic ehrlichiosis using epank1 genes of Ehrlichia phagocytophila-group ehrlichiae. J Clin Microbiol. 2000;38:354–356. doi: 10.1128/jcm.38.1.354-356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiegarg G, Remington S J. Citrate synthase: structure, control, and mechanism. Annu Rev Biophys Chem. 1986;15:97–117. doi: 10.1146/annurev.bb.15.060186.000525. [DOI] [PubMed] [Google Scholar]
- 30.Wood D O, Willamson L R, Winkler H H, Kranse D C. Nucleotide sequence of the Rickettsia prowazekii citrate synthase gene. J Bacteriol. 1987;169:3564–3572. doi: 10.1128/jb.169.8.3564-3572.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X J, Walker D H. Sequence and characterization of an Ehrlichia chaffeensis gene encoding 314 amino acids highly homologous to the NAD A enzyme. FEMS Microbiol Lett. 1997;154:53–58. doi: 10.1111/j.1574-6968.1997.tb12623.x. [DOI] [PubMed] [Google Scholar]