Abstract
Biprobe identification assays based on real-time PCR were designed for 15 species of coagulase-negative staphylococci (CNS). Three sets of primers and four biprobes were designed from two variable regions of the 16S rRNA gene. An identification scheme was developed based on the pattern of melting peaks observed with the four biprobes that had been tested on 24 type strains. This scheme was then tested on 100 previously identified clinical isolates and 42 blindly tested isolates. For 125 of the 142 clinical isolates there was a perfect correlation between the biprobe identification and the result of the ID 32 Staph phenotypic tests and PCR. For 12 of the other isolates a 300-bp portion of the 16S rRNA gene was sequenced to determine identity. The remaining five isolates could not be fully identified. LightCycler real-time PCR allowed rapid and accurate identification of the important CNS implicated in infection.
The genus Staphylococcus currently includes 38 species (13), and coagulase-negative staphylococci (CNS) are isolated frequently in clinical microbiology laboratories (11, 12, 20). CNS are associated with the normal skin flora and mucous membranes and can be isolated from many other sources (including foodstuffs) such as meat, milk and cheese, soil, sand, water, and air (12). CNS may cause bacteremia, endocarditis, catheter-related infections, central nervous system shunt infections, urinary tract infections, endophthalmitis, and infections of prosthetic joints (9). CNS give rise to significant hospital infections often associated with the use of medical devices and immunocompromised patients (13).
With increasing numbers of CNS infections being recognized, it has become necessary to have fast and reliable identification methods (2, 11). Biochemical methods exist (2, 4, 10, 18), but none can reliably identify the important CNS because of the variable expression of some phenotypic characters. Multilocus enzyme electrophoresis or analysis of cellular fatty acid composition have also been used, but identification remains incomplete (21). Genotypic methods for identification have been developed (5, 6,7, 15, 16, 23) but also have limitations. For example, the individual PCRs designed by Gribaldo et al. (6) to identify eight species of CNS are time-consuming and expensive.
Real-time PCR and biprobe (3) assays have recently been used to distinguish related bacterial species of, for example, the genus Campylobacter (14). The aim of the present study was to establish whether real-time PCR was suitable for the identification of CNS involved in human infection.
The LightCycler (Bio/Gene Ltd., Kimbolton, England) is a real-time PCR machine that allows both rapid PCR cycling and continuous monitoring of product formation (24). The formation of double-stranded PCR products is detected by Sybr Green I. Sybr Green I is an intercalating dye which fluoresces strongly when bound to double-stranded DNA; thus, when PCR products are formed an increase in fluorescence is observed (8, 19). After PCR amplification the LightCycler can monitor melting of the DNA with increasing temperature by measuring the decrease in fluorescence as Sybr Green I is released. For convenience, the negative derivative of fluorescence versus temperature is plotted to give a discrete melting peak. When the melting temperature of the PCR products is analyzed in this way, it is not necessary to visualize the PCR products on agarose gels.
Sequence-specific detection of the amplicons can be achieved by the addition of a short-labeled probe. When a probe is included in the reaction, after the melt cycle two peaks can be observed. One corresponds to a decrease in fluorescence due to the melting of the PCR product, and the other is due to the release of the probe. Biprobes are sequence-specific probes labeled with the fluorophore Cy5. When the probe binds to the complementary sequence in the PCR product, the Cy5 label is excited by the energy transfer from Sybr Green I, resulting in an increase of light emitted by Cy5. This fluorescence is measured at a different wavelength from that emitted by Sybr Green I and so can be distinguished from it. Biprobes have another useful feature: as well as hybridizing to a perfectly matched sequence, they will also bind when there is some mismatch (usually up to five mismatches). When the biprobe binds to a mismatched sequence, a melting temperature lower than that of a perfectly matched sequence can be distinguished.
We designed different biprobes to bind to variable regions of the 16S rRNA gene of staphylococci with various degrees of mismatch. We have applied one to four probes to amplicons from 15 species of staphylococci and confirmed melting peaks for a collection of 24 type strains. Further, the identification results of 142 clinical isolates determined by using the biprobes were compared with the results obtained by biochemical and PCR identification.
MATERIALS AND METHODS
Bacterial strains, culture conditions, phenotypic tests, and PCR.
Type strains used in the study are listed in Table 1 and were obtained from the National Collection of Type Cultures (NCTC) (Central Public Health Laboratory, London, United Kingdom). The 142 clinical isolates were selected from those submitted for identification to the Laboratory of Hospital Infection, Central Public Health Laboratory. All strains were grown overnight on blood agar plates at 37°C. Colony suspensions of the strains were made by suspending several colonies from one plate in 1 ml of water, heating to 99°C for 5 min, and centrifuging at 3,000 × g for 2 min. The supernatant was then used in LightCycler assays. Phenotypic identification of strains was made using ID 32 Staph (bioMérieux) according to the manufacturer's instructions (4). PCR identification was made using the primers and conditions of Gribaldo et al. (6).
TABLE 1.
Type strains used in the study
| Species | Strain |
|---|---|
| S. arlettae | NCTCa 12413 |
| S. aureus | NCTC 8523 |
| S. auricularis | NCTC 12101 |
| S. capitis | NCTC 11045 |
| S. caprae | NCTC 12196 |
| S. chromogenes | NCTC 10530 |
| S. cohnii | NCTC 11041 |
| S. delphini | NCTC 12225 |
| S. epidermidis | NCTC 11047 |
| S. equorum | NCTC 12414 |
| S. gallinarum | NCTC 12195 |
| S. haemolyticus | NCTC 11042 |
| S. hominis | NCTC 11320 |
| S. hyicus | NCTC 10350 |
| S. intermedius | NCTC 11048 |
| S. kloosii | NCTC 12415 |
| S. lentus | NCTC 12102 |
| S. lugdunensis | NCTC 12217 |
| S. saprophyticus | NCTC 7292 |
| S. sciuri | NCTC 12103 |
| S. schleiferi | NCTC 12218 |
| S. simulans | NCTC 11046 |
| S. warneri | NCTC 11044 |
| S. xylosus | NCTC 11043 |
| Stomatococcus mucilaginosus | NCTC 10663 |
| Micrococcus luteus | NCTC 2665 |
NCTC, National Collection of Type Cultures (Central Public Health Laboratory, London, England).
DNA extraction.
DNA was extracted from the type strains using the Wizard Genomic DNA purification kit (Promega, Southampton, United Kingdom) according to the manufacturer's instructions, except that lysis was achieved by resuspending cells in 100 μl of lysostaphin (1 mg/ml in Tris-EDTA [TE] buffer; Sigma) and 50 μl of lysozyme (50 mg/ml in TE buffer; Sigma).
LightCycler primers and biprobes.
The sequences of all primers and biprobes are shown in Table 2. All biprobes (3) were labeled with Cy5 at the 5′ end and with biotin at the 3′ end to prevent them from acting as primers. All primers and probes were synthesized by MWG-Biotech UK Ltd. (Milton Keynes, England). CNS Probe A was used with primers STAR1 (5 pmol/μl) and STAF1 (1 pmol/μl), CNS Probe B was used with STAR1 (1 pmol/μl) and STAF2 (5 pmol/μl), CNS Probe C was used with STBF1 (5 pmol/μl) and STBR1 (1 pmol/μl), and CNS Probe D was used with primers STBF1 (1 pmol/μl) and STBR1 (5 pmol/μl).
TABLE 2.
DNA sequences of primers and biprobes
| Name of primer or probe | Sequence |
|---|---|
| Primers | |
| STAR1 | 5′-GCG GdT CCA TCT ATA AGT GA-3′a |
| STAF1 | 5′-GGG TGA GTA ACA CGT GGA-3′ |
| STAF2 | 5′-GGG TGA GTA ACA CGT GGG-3′ |
| STBF1 | 5′-ATT CGA AGC AAC GCG AAG-3′ |
| STBR1 | 5′-CCA TGC ACC ACC TGT CAC-3′ |
| Probes | |
| CNS Probe A | 5′-GGA TAA TAT ATT GAA CCG CA-3′ |
| CNS Probe B | 5′-TCC GGT TTC CCG AAG TTG TC-3′ |
| CNS Probe C | 5′-GAG CGG TCA AAG GAT GTC AAG-3′ |
| CNS Probe D | 5′-CCT CTG ATC CCT CTA GAA ATA-3′ |
d, A or G.
LightCycler assays.
The LightCycler (Bio/Gene Ltd.) was used for all biprobe assays. PCR mixtures (10 μl) contained 1 μl of colony suspension or 10 ng of DNA, 500 mM Tris-HCl (pH 8.3), MgCl2 (5 mM), bovine serum albumin (1 mg/ml), deoxynucleoside triphosphates (200 μM concentrations of each; Gibco BRL), forward and reverse primers (one at 5 pmol/μl and the other at 1 pmol/μl), platinum Taq polymerase (0.4 U; Gibco BRL), Sybr Green I (Bio/Gene Ltd.) diluted 0.01%, and a biprobe (5 pmol/μl). The cycling parameters were one denaturation cycle at 94°C for 5 s and 40 amplification cycles (temperature transition rate of 20°C/s) of 94°C for 0 s, 55°C for 1 s, 60°C for 15 s, and 74°C for 10 s. Fluorescence readings were taken after annealing at 55°C for 1 s. PCR cycling was followed by melting curve analysis of 40 to 95°C (temperature transition rate of 0.2°C/s) with continuous fluorescence readings. For ease of interpretation, controls for all of the peaks were run alongside the samples being tested. For example, with CNS Probe A, three controls were run, one for each of the three peaks. This allowed easy scoring of the results and corrected for slight variations in the melting temperatures which were observed from run to run.
DNA sequencing.
The primers Epsilon F (AAGAGTTTGATCCTGGCTCAG) and 1510R (GGTTACCTTGTTACGACTT) were used in a PCR to amplify a 1,500-bp amplicon of the 16S rRNA gene. The PCR mixtures (100 μl) contained forward and reverse primers (20 pmol each), deoxynucleoside triphosphates (200 μM concentrations of each), platinum Taq polymerase (0.5 U), MgCl2 (3 mM), 1× PCR buffer (as supplied with platinum Taq polymerase), and 5 μl of a colony suspension. The PCR was performed in a Perkin-Elmer 9600 machine, and cycling consisted of 1 cycle of 95°C for 5 min, 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min followed by final extension at 72°C for 5 min. The PCR products were purified using the Wizard PCR Clean up kit (Promega) and quantified by agarose gel electrophoresis. The PCR products were sequenced according to the manufacturer's instructions for the ABI prism sequencing kit (Perkin-Elmer) using the primers Epsilon F and 320R (TTGACCGTGTCTCAGTTCCA).
RESULTS
Biprobe and primer design.
The 16S rRNA gene sequences of 35 species of Staphylococcus, Stomatococcus mucilaginosus, and Micrococcus luteus were retrieved from the GenBank database. The sequences were aligned using the Lasergene Dnastar computer package. Two variable regions between bp 1 and 250 and between bp 970 and 1080 were identified as being suitable for the design of biprobes. Four biprobes were designed, CNS Probe A and CNS Probe B from the region between bp 1 and 250 and CNS Probe C and CNS Probe D from the region between bp 970 and 1080. CNS Probe A was designed to be a perfect match to Staphylococcus epidermidis, CNS Probe B was a perfect match to Staphylococcus lugdunensis, CNS Probe C had no mismatches with Staphylococcus warneri, Staphylococcus sciuri, S. lugdunensis, Staphylococcus intermedius, and Staphylococcus schleiferi and CNS Probe D was a perfect match with Staphylococcus capitis. All other species had one or more mismatches with the probes. It was predicted that for S. epidermidis it would be necessary to use only one probe (CNS Probe A) to make an accurate identification. Since S. epidermidis is the most commonly found CNS in nosocomial infections the use of only one probe would allow a fast and inexpensive identification. The probes were all designed to have an annealing temperature at least 5°C higher than that of the primers and were also predicted to have minimum secondary structure that might prevent them from binding to the target sequence.
PCR primers were designed for use with the probes and were checked with the computer program OLIGO. For CNS Probe A and CNS Probe B, three primers were designed: STAF1 and STAR1 for CNS Probe A and STAF2 and STAR1 for CNS Probe B. For CNS Probes C and D two primers were designed, STBF1 and STBR1. All primers were designed to amplify an approximately 100-bp amplicon, which is the optimal size for use with a biprobe and for optimal amplification on the LightCycler. For ease of use all of the primers were designed to have an annealing temperature of 55°C, thus allowing the same cycling conditions to be used for all four PCRs.
Biprobe evaluation.
The four CNS biprobes were tested on DNA extracted from the 15 type strains of staphylococcal species of clinical interest. During the cycling step the temperature was held at 55°C to allow the primers to anneal. It was also found that for the biprobe to anneal and hence fluoresce it was also necessary to hold the temperature at 60°C for 15 seconds. In addition, it was necessary to reduce the concentration of the primer closest to the probe; otherwise, the primer prevented the probe from binding.
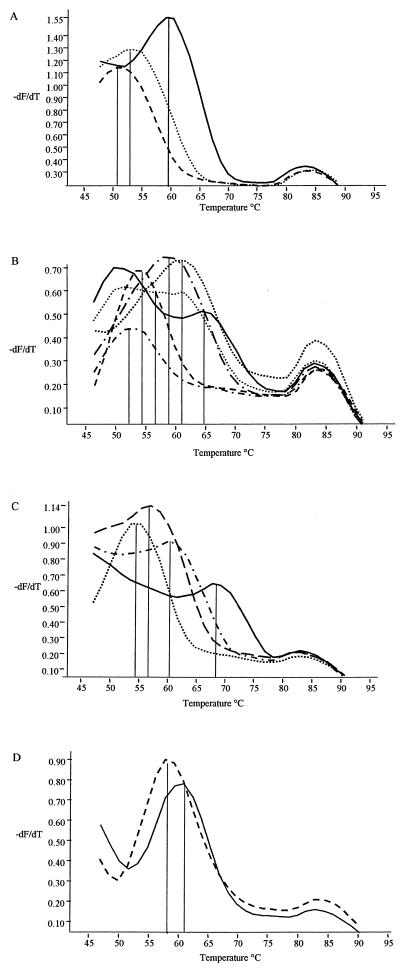
After the melting step CNS Probe A gave three discrete melting peaks (Fig. 1A). The first peak had a melting temperature of 58°C (peak 0) and was seen with S. epidermidis; the second peak had a melting temperature of 52°C (peak 1) and was seen with S. warneri; and the third peak melted at 50°C (peak 2) and was seen with S. sciuri, S. lugdunensis, S. aureus, S. intermedius, and S. chromogenes. Only 7 of the 15 type strains displayed a melting peak with this probe and the remaining 8 gave a negative result. The second probe tested, CNS Probe B, displayed six discrete melting peaks, designated 0 to 5 (Fig. 1B), melting at 64, 60, 56, 58, 53, and 52°C, respectively. CNS Probe B bound to and gave observable melting peaks with all 15 type strains and was found to be the most discriminatory of the four probes. CNS Probe C showed four discrete melting peaks (Fig. 1C) melting at 68, 60, 56, and 54°C, respectively. As with CNS Probe B, each of the 15 type strains showed a melting peak with this probe. The final probe, CNS Probe D, only displayed two discrete melting peaks (Fig. 1D), and these were observed in only three of the type strains tested. Peak 0 had a melting temperature of 60°C and was observed only with S. capitis. Peak 1 was observed with S. caprae and S. epidermidis and had a melting temperature of 58°C.
FIG. 1.
Biprobe peak profiles. (A) Peak profiles obtained with biprobe A. Peak 0 melted at 58°C, peak 1 melted at 52°C, and peak 2 melted at 50°C. (B) Peak profiles obtained with biprobe B. Peak 0 melted at 64°C, peak 1 melted at 60°C, peak 2 melted at 56°C, peak 3 melted at 58°C, peak 4 melted at 53°C, and peak 5 melted at 52°C. (C) Peak profiles obtained with biprobe C. Peak 0 melted at 68°C, peak 1 melted at 60°C, peak 2 melted at 56°C, and peak 3 melted at 54°C. (D) Peak profiles obtained with biprobe D. Peak 0 melted at 60°C, and peak 1 melted at 58°C.
Development of an identification scheme.
The four biprobes were tested on 15 Staphylococcus species of interest, namely, S. epidermidis, S. warneri, S. lugdunensis, S. aureus, S. intermedius, S. chromogenes, S. sciuri, S. hominis, S. haemolyticus, S. capitis, S. caprae, S. schleiferi, S. simulans, S. saprophyticus, and S. cohnii. The results are shown in Table 3. Based on these results it was concluded that CNS Probe A identified S. epidermidis and S. warneri, and therefore it was not necessary to use any of the other three probes for identification. CNS Probe B used alone can identify S. lugdunensis and S. haemolyticus, and the combination of CNS Probe A and CNS Probe B allows identification of S. sciuri, S. lugdunensis, S. aureus, S. hominis, S. haemolyticus, and S. cohnii. CNS Probe C in combination with Probes A and B can be used to identify S. intermedius, S. chromogenes, S. schleiferi, and S. saprophyticus. The final probe, CNS Probe D, was designed to distinguish S. capitis and S. caprae.
TABLE 3.
Biprobe identification scheme for CNSa
| Organism | Biprobe melting peakb obtained with:
|
|||
|---|---|---|---|---|
| CNS Probe A | CNS Probe B | CNS Probe C | CNS Probe D | |
| S. epidermidis | 0 | 3 | 3 | 1 |
| S. warneri | 1 | 1 | 0 | – |
| S. lugdunensis | 2 | 0 | 0 | – |
| S. aureus | 2 | 3 | 2 | – |
| S. intermedius | 2 | 4 | 0 | – |
| S. chromogenes | 2 | 4 | 3 | – |
| S. sciuri | 2 | 5 | 0 | – |
| S. hominis | – | 1 or 2 | 1 | – |
| S. haemolyticus | – | 2 | 2 | – |
| S. capitis | – | 3 | 3 | 0 or 1 |
| S. caprae | – | 3 | 3 | 1 |
| S. schleiferi | – | 4 | 0 | – |
| S. simulans | – | 4 | 2 | – |
| S. saprophyticus | – | 4 | 3 | – |
| S. cohnii | – | 5 | 2 | – |
Results highlighted in bold are not necessary for identification of that species. –, no peak observed.
Designated 0 through 5, as defined in the legend to Fig. 1.
The four probes were also tested on nine other species of staphylococci, namely, S. kloosii, S. arlettae, S. lentus, S. delphini, S. equorum, S. hyicus, S. xylosus, S. auricularis, and S. gallinarum, as well as Stomatococcus mucilaginosus and Micrococcus luteus. These strains were readily available and were used to check for any possible cross-reactions. Each of these species gave a pattern of reactions distinct from that of any of the species required to be identified (results not shown).
Clinical isolate testing.
A total of 142 clinical isolates of staphylococci were tested with all four CNS probes to determine whether reactions observed with the type strains were consistent. Of the 142, 100 were tested as known isolates and 42 were tested as part of a blind study. Where possible, at least 10 isolates of each species were selected, but for six species only a few clinical isolates were available, and for S. caprae none were obtainable. The results are shown in Table 4. All of the S. hominis, S. sciuri, S. cohnii, S. intermedius, and S. simulans strains showed complete agreement between biochemical identification and LightCycler identification. Four additional S. epidermidis isolates, one additional S. aureus isolate, and one additional S. saprophyticus isolate that were not identified by biochemical tests were identified by real-time PCR.
TABLE 4.
Results of testing of 142 clinical isolates by ID 32 Staph and PCR and by real-time PCR
| Organism | Total no. of isolates | No. of isolates identified by:
|
||
|---|---|---|---|---|
| ID 32 Staph and PCR | Real-time PCR | 16S rRNA sequencing | ||
| S. hominis | 17 | 17 | 17 | –b |
| S. sciuri | 3 | 3 | 3 | – |
| S. cohnii | 2 | 2 | 2 | – |
| S. intermedius | 2 | 2 | 2 | – |
| S. simulans | 5 | 5 | 5 | – |
| S. epidermidis | 30 | 26 | 30 | 4 |
| S. aureus | 11 | 10 | 11 | 1 |
| S. saprophyticus | 6 | 5 | 6 | 1 |
| S. warneri | 11 | 13 | 11 | – |
| S. haemolyticus | 24 | 27 | 24 | – |
| S. schleiferi | 4 | 6 | 4 | – |
| S. lugdunensis | 10 | 12 | 10 | – |
| S. capitis | 12 | 14 | 7 | 1 |
| S. arlettae | 1 | 0 | 0 | 1 |
| S. pasteuri | 1 | 0 | 0 | 1 |
| Staphylococcus sp. | 1 | 0 | 0 | 1 |
| Acinetobacter lwoffii | 1 | 0 | 0 | 1 |
| Micrococcus luteus | 1 | 0 | 0 | 1 |
Number of isolates identified using a combination of all tests and using 16S rRNA sequencing to confirm identity when discrepancies occurred between the ID 32 Staph and PCR and the real-time PCR identifications.
–, not performed.
Two of the S. warneri isolates gave unknown profiles, and so a variable region of the 16S rRNA gene was sequenced. Sequencing results indicated that the most likely identification of one isolate was S. pasteuri, and the other isolate was identified as a Staphylococcus species. Three of the isolates previously identified as S. haemolyticus did not yield the expected LightCycler pattern. One of these isolates was identified as S. aureus, one as S. epidermidis, and the other as an unknown strain by the LightCycler. The S. aureus and S. epidermidis identifications were confirmed by DNA sequencing, and sequencing indicated that the likely identification of the unknown isolate was S. arlettae. One of the S. schleiferi isolates was identified as S. saprophyticus, and another gave an unknown profile. The S. saprophyticus identification was confirmed by DNA sequencing, and the other isolate was identified by sequencing as Acinetobacter lwoffii. Of the 12 S. lugdunensis isolates tested, one was identified as S. capitis and another gave an unknown profile. DNA sequencing identified the S. capitis strain as either S. capitis or S. caprae and the unknown strain as Micrococcus luteus. Of the 14 S. capitis isolates tested only 6 were correctly identified, 5 being identified as S. caprae and 3 being identified as S. epidermidis. The three identified as S. epidermidis were sequenced, and this confirmed the LightCycler identification. CNS Probe D is based on the region of bp 1004 to 1025, within which at base 1011 the type strain of S. capitis has a T and S. caprae has a C. After testing on the LightCycler, this region of the sequence was examined more closely and an alignment was formed using three sequences of S. capitis and three sequences of S. caprae obtained from the GenBank database. It was found that in two of the three S. capitis sequences a C was found at position 1011 and not the T that the probe was based on. It is therefore not possible to distinguish S. capitis from S. caprae reliably using this probe. The 16S rRNA gene sequences were examined for further differences, and none that would differentiate S. capitis from S. caprae could be found.
DISCUSSION
We have described a rapid and accurate test for identification of the main species of CNS of clinical interest, as well as S. aureus, S. intermedius, and S. schleiferi, which are or can be coagulase-positive species. It is becoming increasingly important that reference laboratories identify CNS accurately, particularly S. epidermidis (11).
Of the 142 clinical isolates tested by the biprobe assays, 125 showed concordance between the biprobe assay results and those of the traditional PCR and phenotypic identification method. The 12 strains that did not correlate were further characterized by partial sequencing of the 16S rRNA gene, and these results, except in the case of the S. capitis strains, confirmed the biprobe assay result. There were five strains of S. capitis that were identified by the biprobe assays as S. caprae, but further examination of the 16S rRNA gene sequences indicated that it was not possible to distinguish S. caprae from S. capitis on the basis of 16S rRNA sequence. This has been reported also by Takahashi et al. (22), who compared 16S rRNA gene sequences of 38 species of staphylococci, and by Bannerman and Kloos (1), who carried out DNA-DNA hybridization studies. It may be more convenient to report these species as “S. capitis or S. caprae” or to use an alternative test to identify them.
Several methods are currently being used or have been evaluated to identify CNS. The most common is the use of commercially available biochemical tests that allow phenotypic identification, but these are not always reliable and results are often operator dependent (17). For example, phosphatase-negative variants of S. epidermidis can often be misidentified as S. hominis (7, 13). The introduction of molecular identification of CNS has improved the accuracy of results and allows the identification of the more unusual CNS which occasionally cause infection (6). Species-specific PCRs can be used to replace the phenotypic tests or be used in conjunction with them, but this can require the use of several primer sets, which is time-consuming and expensive. Furthermore, primers are not available for all of the species of CNS that can be implicated in infection. Several genotypic tests are available for the identification of S. epidermidis only (7, 16, 23), but with increasing numbers of the other CNS being implicated in infection it is necessary to have available a fuller range of identification tests.
The real-time PCR and biprobe approach to identifying staphylococci to the species level has several advantages. LightCycler tests can be completed in 30 to 40 min, and it is not necessary to open the tubes after amplification. The risk of contamination is therefore reduced. The assay requires easily prepared colony suspensions, and it is not necessary to perform time-consuming DNA extractions. Only three PCRs need to be performed to characterize up to 15 species of CNS. The more widespread use of these assays is constrained by the initial expense of the LightCycler, and only 32 samples can be included in one run compared to 96 samples in most conventional PCR systems. With future generations of real-time PCR machines having the capacity for additional fluorescence channels, there will be the opportunity to include more than one probe in each reaction, thus reducing the number of PCRs required.
The biprobe assays proved to be accurate and robust, and by using a series of control strains in each experiment the assays are easy to interpret. The rapid identification of S. epidermidis using only one PCR and only one probe will significantly improve the turnaround time for identification to the species level of the most common CNS.
ACKNOWLEDGMENT
This work was partially funded by the Biotechnology and Biological Sciences Research Council (BBSRC).
REFERENCES
- 1.Bannerman T L, Kloos W E. Staphylococcus capitis subsp. ureolyticus subsp. nov. from human skin. Int J Syst Bacteriol. 1991;41:144–147. doi: 10.1099/00207713-41-1-144. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman T L, Kleeman K T, Kloos W E. Evaluation of the Vitek Systems Gram-Positive Identification card for species identification of coagulase-negative staphylococci. J Clin Microbiol. 1993;31:1322–1325. doi: 10.1128/jcm.31.5.1322-1325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bio/Gene Ltd. and The Secretary of State for Defense. July 1999. Nucleic acid detection system. Great Britain patent GB2333359A.
- 4.Brun Y, Bes M, Boeufgras J M, Monget D, Fleurette J, Auckenthaler R, Devriese L A, Kocur M, Marples R R, Piemont Y. International collaborative evaluation of the ATB 32 staph gallery for identification of the Staphylococcus species. Zentbl Bakteriol. 1990;273:319–326. doi: 10.1016/s0934-8840(11)80435-4. [DOI] [PubMed] [Google Scholar]
- 5.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gribaldo S, Cookson B, Saunders N, Marples R, Stanley J. Rapid identification by specific PCR of coagulase-negative staphylococcal species important in hospital infection. J Med Microbiol. 1997;46:45–53. doi: 10.1099/00222615-46-1-45. [DOI] [PubMed] [Google Scholar]
- 7.Hedin G. Comparison of genotypic and phenotypic methods for species identification of coagulase-negative staphylococcal isolates from blood cultures. APMIS. 1994;102:855–864. doi: 10.1111/j.1699-0463.1994.tb05245.x. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 9.Huebner J, Goldmann D A. Coagulase-negative staphylococci: role as pathogens. Annu Rev Med. 1999;50:223–236. doi: 10.1146/annurev.med.50.1.223. [DOI] [PubMed] [Google Scholar]
- 10.Ieven M, Verhoeven J, Pattyn S R, Goossens H. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J Clin Microbiol. 1995;33:1060–1063. doi: 10.1128/jcm.33.5.1060-1063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleeman K T, Bannerman T L, Kloos W E. Species distribution of coagulase-negative staphylococcal isolates at a community hospital and implications for selection of staphylococcal identification procedures. J Clin Microbiol. 1993;31:1318–1321. doi: 10.1128/jcm.31.5.1318-1321.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloos W E, Schleifer K H, Gotz F. The genus Staphylococci. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1991. pp. 1369–1420. [Google Scholar]
- 13.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan J M J, Edwards K J, Saunders N A, Stanley J. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J Clin Microbiol. 2001;39:2227–2232. doi: 10.1128/JCM.39.6.2227-2232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller M A, Herwaldt L A. Laboratory, clinical, and epidemiological aspects of coagulase-negative staphylococci. Clin Microbiol Rev. 1988;1:281–299. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renneberg J, Rieneck K, Gutschik E. Evaluation of Staph ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J Clin Microbiol. 1995;33:1150–1153. doi: 10.1128/jcm.33.5.1150-1153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ririe K M, Rasmussen R P, Wittwer C T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 20.Sewell C M, Clarridge J E, Young E J, Guthrie R K. Clinical significance of coagulase-negative staphylococci. J Clin Microbiol. 1982;16:236–239. doi: 10.1128/jcm.16.2.236-239.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoakes L, John M A, Lannigan R, Schieven B C, Ramos M, Harley D, Hussain Z. Gas-liquid chromatography of cellular fatty acids for identification of staphylococci. J Clin Microbiol. 1994;32:1908–1910. doi: 10.1128/jcm.32.8.1908-1910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol. 1999;49:725–728. doi: 10.1099/00207713-49-2-725. [DOI] [PubMed] [Google Scholar]
- 23.Wieser M, Busse H J. Rapid identification of Staphylococcus epidermidis. Int J Syst Evol Microbiol. 2000;50:1087–1093. doi: 10.1099/00207713-50-3-1087. [DOI] [PubMed] [Google Scholar]
- 24.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]