Abstract
Pectin methylesterase inhibitor (PMEI) inhibits pectin methylesterase (PME) activity at post-translation level, which plays core roles in vegetative and reproductive processes and various stress responses of plants. However, the roles of PMEIs in tea plant are still undiscovered. Herein, a total of 51 CsPMEIs genes were identified from tea plant genome. CsPMEI1-4 transcripts were varied in different tea plant tissues and regulated by various treatments, including biotic and abiotic stresses, sugar treatments, cold acclimation and bud dormancy. Overexpression of CsPMEI4 slightly decreased cold tolerance of transgenic Arabidopsis associated with lower electrolyte leakage, soluble sugars contents and transcripts of many cold-induced genes as compared to wild type plants. Under long-day and short-day conditions, CsPMEI2/4 promoted early flowering phenotypes in transgenic Arabidopsis along with higher expression levels of many flowering-related genes. Moreover, overexpression of CsPMEI2/4 decreased PME activity, but increased sugars contents (sucrose, glucose, and fructose) in transgenic Arabidopsis as compared with wild type plants under short-day condition. These results indicate that CsPMEIs are widely involved in tea plant vegetative and reproductive processes, and also in various stress responses. Moreover, CsPMEI4 negatively regulated cold response, meanwhile, CsPMEI2/4 promoted early flowering of transgenic Arabidopsis via the autonomous pathway. Collectively, these results open new perspectives on the roles of PMEIs in tea plant.
Keywords: Camellia sinensis, Arabidopsis, pectin methylesterase inhibitor, expression patterns, biotic and abiotic stresses, overexpression, cold, flowering
Introduction
Pectin is one of three major polysaccharides in plant primary cell walls, which plays vital roles in different plant tissues and developmental stages, and also in responses to biotic and abiotic stresses. Galacturonic acid (GalA) is the main component of pectin, which can be subdivided into five classes, including apiogalacturonan (AP), homogalacturonan (HG), rhamnogalacturonan I (RG-I), rhamnogalacturonan II (RG-II), and xylogalacturonan (XGA) (Wormit and Usadel, 2018). Among them, HG is the backbone of pectin that constitutes about 65% of total pectin, and its properties could be affected by acetylating at C2–C3 atom and methylesterifing at C6 atom of GalA, respectively (Wolf et al., 2009). Within cell walls, the highly methylesterified HG could be de-methylesterified (DM) by pectin methylesterases (PMEs, E.C. 3.1.1.11) to produce negatively charged carboxyl groups and release methanol and protons (Cantarel et al., 2009). Meanwhile, PMEs activities are regulated by a type of endogenous inhibitors called pectin methylesterases inhibitors (PMEIs) (Juge, 2006).
It is now clear that PMEIs are encoded by a large multigene family both in monocotyledons and dicotyledons, and a conserved PMEI domain (PF04043) was contained in PMEI protein. In plant, the first PMEI protein (AcPMEI) was identified and purified from kiwi fruit, which has been verified to effectively repress PME activity through formation of a 1:1 non-covalent complex (Balestrieri, 1990; Di Matteo et al., 2005). Crystallographic structural analysis showed that AcPMEI is almost all helical mainly consisting of four long helices (α1–4) arranged in an anti-parallel way to form a classical up-and-down four-helical bundle (Di Matteo et al., 2005). The amino acid sequence of PMEI contains an N-terminal signal peptide and four highly conserved cysteine (Cys, C) residues. The four C residues are mainly involved in the formation of two disulfide bridges (S–S), which is critical for the stabilization of four-helical bundle structure (Wormit and Usadel, 2018). However, these structural properties are also found in a type of invertase inhibitor (INH) that shared a structural superimposition with PMEI though their sequence identity is only 20–30%. The main differences between PMEI and INH lie in the N-terminal region and the loops connecting the bundle helix. In particular, an amino acid inserted into helix α2 of INH that partially distorts the helix (Di Matteo et al., 2005). With the development of biological technology, lots of PMEI genes have been identified, and their functions have been explored in different plant species recently. At present, 78 AtPMEIs in Arabidopsis (Müller et al., 2013), 83 LuPMEIs in flax (Pinzon-Latorre and Deyholos, 2014), 49 OsPMEIs in rice (Nguyen et al., 2016), 95 BoPMEIs in Brassica oleracea (Liu et al., 2018a), 100 BcPMEIs in Brassica campestris (Liu et al., 2018b), 55 SbPMEIs in Sorghum bicolor (Ren et al., 2019), and 42 PbrPMEIs in Pyrus bretschneideri (Zhu et al., 2021) etc. have been identified and characterized in succession.
Until now, many research findings have revealed that PMEIs play key roles in plant vegetative and reproductive processes. With the help of mutation and overexpression techniques, the functions of many PMEIs have been extensively explored. In Arabidopsis, there have been more than 12 AtPMEIs verified to mediate plant growth and development. Among them, AtPMEI1 (At1g48020) and AtPMEI2 (At3g17220) inhibited plant-derived PME activity rather than microbial PME activity, which shared 38% sequence identity with AcPMEI from kiwi fruit (Raiola et al., 2004). Similarly, a functional PMEI of Vitis vinifera, VvPMEI1, also typically inhibited the plant PME activity but not microbial PME activity (Lionetti et al., 2015). In addition, the recombinant OsPMEI28 protein showed high inhibitory activity against PME protein, overexpression of OsPMEI28 caused dwarf phenotypes and reduced culm diameter in transgenic rice lines (Nguyen et al., 2017). Similarly, overexpression of AtPMEI3 resulted in hyper-methyl-esterification of HG and effected the formation of flower primordia (Lionetti et al., 2007). The expression of AtPMEI6 showed a spatio-temporal pattern in seed coat epidermal cells, and overexpression of AtPMEI6 inhibited the endogenous PME activity and decreased the total methylesterification of mucilage fractions and demucilaged seeds in transgenic plants (Saez-Aguayo et al., 2013). GUS staining results showed that the promoter of a flower-specific gene, SlPMEI, expressed specifically in mature pollen of tomato and reproductive organs of transgenic Arabidopsis (Raiola et al., 2004).
In addition to mediate plant growth and development, PMEIs have also been reported to participate in defending pathogen infection and responding to environment stresses. In Arabidopsis, the expressions of three PMEIs genes, AtPMEI10/11/12, were induced by Botrytis cinerea infection mainly through jasmonic acid and ethylene signaling. Moreover, the reverse genetic approach results found that the PME activity was increased but the DM of pectin was decreased in pmei10/11/12 mutants, respectively, which finally increased lesion formation during B. cinerea infection. These results indicated that AtPMEI10/11/12 served as mediators to maintain cell wall integrity in plant immunity (Lionetti et al., 2017).
Tea plant (Camellia sinensis) is a type of evergreen economic plant, which is appropriate to growth at normal temperature, high humidity and acid soil (pH 5.5) environments. However, with the frequency of extreme climate in recent years, tea plants usually cannot overwinter safely or grow normally when suffered from drought, chilling, freezing, or cold spell damages. Therefore, more and more studies have been focused on the molecular mechanisms of tea plant stress responses. At present, many functional genes involved in vegetative (Xia et al., 2021), reproductive (Jing et al., 2020), nutrient uptake (Arkorful et al., 2020), secondary metabolism (Yu et al., 2021; Zhao et al., 2021), and stress response (Qian et al., 2018; Wang et al., 2018; Yao et al., 2020) have been extensively explored. However, the functions of PMEIs in tea plant still remain unknown. Herein, we performed a genome-wide identification and characterization analysis of CsPMEIs, analyzed the tissue-specific profiles and spatio-temporal patterns of four CsPMEIs (CsPMEI1-4), and finally explored the functions of CsPMEI2/4 in transgenic Arabidopsis, respectively. Our study opened the door for the functional study of pectin in tea plant, and these results also provided a firm foundation for deeply exploring the functions of PMEs and PMEIs in tea plant.
Materials and Methods
Plant Materials and Multiple Treatments
For tissue-specific analysis, the apical buds, the first leaves, the second leaves, the third leaves, young fruits, mature fruits, young stems, flowers, and roots were sampled from 4-year-old clonal potted seedlings of the ‘LongJing43’ cultivar during flowering season (October). Each tissue contains three independent biological replicates, and all tissues were frozen in liquid nitrogen and then stored at −80°C until used.
For various abiotic stress treatments, 1-year-old clonal hydroponic seedlings of the ‘LongJing43’ cultivar with the similar growth potential were used for 4°C, PEG-6000 [10% (w/v)], 150 mmol⋅L–1 NaCl and 100 μmol⋅L–1 ABA treatments. The detailed treatment methods were carried out as described by Qian et al. (2016). The nutrient solution formulation was shown in Supplementary Table 1. Five-year-old clonal seedlings of the ‘LongJing43’ cultivar at one bud and five leaf stages were used for Colletotrichum fructicola (C. fructicola) infection. The detailed infection method was performed as described by Wang et al. (2016). 15-year-old of four-tea cultivars, including two cold-resistant cultivars (‘ZheNong113’ and ‘LongJing43’) and two cold-susceptible cultivars (‘ZheNong12’ and ‘DaMianBai’) as demonstrated by Wang et al. (2019), were used for cold acclimation (CA) analysis in 2018–2019. The sampling methods were performed according to the methods stated by Qian et al. (2018). 15-year-old clonal tea plants of the ‘LongJing43’ cultivar that planted in field were used for bud dormancy analysis. The sampling methods were implemented as described by Hao et al. (2018). The above mentioned treatments were, respectively, performed three independent biological replicates, and all samples were quickly frozen in liquid nitrogen and then stored at −80°C until used.
2-year-old clonal potted cuttings of the ‘LongJing43’ cultivar at two different growth stages, including one bud and one leaf (OBOL) stage and one bud and three leaves (OBTL) stage, were used for cold treatment. Before proceeding, the cuttings were moved into the greenhouse for normal culture. The growth conditions were as follows: lighting time, 14 h/10 h (light/dark); temperature, 23°C; and humidity, 75%. When the cuttings reached the standards, the temperature of greenhouse was plummeted to 4°C without changing lighting time and humidity. The cuttings with OBOL were continued for 3 days at 4°C, and the samples (OBOL) were collected at 0, 1, 3, 6, 12, 24, 48, and 72 h, respectively. Similarly, the cuttings with OBTL were continued for 8 days at 4°C, and the samples (OBTL) were collected at 0, 1, 2, 3, 4, 5, 6, 7, and 8 days, respectively. Subsequently, the cuttings were returned to 25°C for 2 days, and the samples (OBTL) were collected at 1 and 2 days. All samples were immediately frozen in liquid nitrogen and then stored at −80°C until used. Each sampling time point was performed five biological replicates, and each biological replicate contains 10 pots of tea plants and each pot contained three cuttings.
One-year-old clonal hydroponic cuttings of the ‘LongJing43’ cultivar were used for sugar treatment. 3% sucrose (Suc), 3% glucose (Glu), 3% fructose (Fru), and 3% mannose (Man) treatments were, respectively, performed as described by Qian et al. (2018). Each sampling time point was performed three independent biological replicates, and each biological replicate contains four pots of tea plants and each pot contains eight cuttings. The nutrient solution formulation was shown in Supplementary Table 1.
The wild-type (WT) Arabidopsis thaliana (Columbia-0 ecotype) was used to construct overexpression (OE) lines of the opening reading frame of CsPMEI2 and CsPMEI4. The seeds of the homozygous OE lines and WT were surface sterilized and vernalized for 2 days, and then sown on 1/2 MS medium (1.5% Suc, 0.8% Agar, pH 5.7) for 2 weeks. Thereafter, the seedlings were transplanted on the seedling block in the growth chamber under short-day (SD) (8 h light at 22°C/16 h dark at 20°C) or long-day (LD) (16 h light at 22°C/8 h dark at 20°C) conditions. The light intensity was 100 μmol/m2s both under SD and LD conditions. The OE lines of blank vector were omitted in this study as a same phenotype detected in WT as demonstrated by Qian et al. (2018). Under LD condition, 4-week-old seedlings of both WT and CsPMEI2/4-OE lines were sampled for analyzing the expressions of flowering-related genes. Under SD condition, 10-week-old seedlings of both WT and CsPMEI2/4-OE lines were sampled for measuring PME activities, sugars (sucrose, glucose, and fructose) contents, and flowering-related genes expressions.
Genome-Wide Identification of CsPMEIs
The hidden Markov models (HMM) file of PMEI domain (PF04043) downloaded from Pfam database1 was used to obtain CsPMEIs similar sequences from the tea plant protein database of ‘ShuChaZao’ cultivar (Wei et al., 2018) by using HMMER 3.0 software with the default parameters. Subsequently, all of these sequences were performed alignment analysis to delete the sequence that contains only one or two conserved Cys residues. The redundant sequences were submitted to the SMART server2 and the Conserved Domain Database of NCBI3 for verifying whether they had conserved the PMEI domain. Finally, the sequences that contain only one conserved PMEI domain were retained for further analyses.
Bioinformatics Analysis of CsPEMIs in Tea Plant
The bioinformatics of CsPEMIs, including ORF lengths, amino acid numbers, molecular weights, signal peptides, transmembrane regions, and subcellular locations were performed as described by Wang et al. (2021). For phylogenetic analysis, 262 PMEI domain contained proteins (Supplementary Table 2), including 78 AtPEMIs from Arabidopsis, 55 SbPEMIs from Sorghum bicolor, 55 PbrPEMIs from pear, 2 AcPEMIs from kiwi fruit, 51 CsPEMIs from tea plant and 21 INHs, were used to generate phylogenetic tree by MEGA 7.0 software. The parameters were set as follows: maximum likelihood method, 1,000 bootstrap replications, Jones–Taylor–Thornton (JTT) model, and partial deletion. Thereafter, the generated tree was uploaded to the ITOL website4 for further beautification. The exon–intron structures of CsPMEIs were predicted by using GSDS 2.0 website5. The cis-acting elements of 2,000-bp promoter sequences of CsPMEIs were predicted by using PlantCARE website6. Multiple amino acids sequences alignment analysis was carried out by using Clustlx2.0 software, and then output the result with the help of GENEDOC software.
Cloning the Full-Length of CsPMEIs
Before the tea plant genome sequenced, 20 expressed sequence tags (EST) that annotated as PMEI domain containing proteins were identified from the transcriptome data of the tea plant under CA condition (Wang et al., 2013). After assembled by Seqman software, a total of 4 contigs were obtained and served as templates to design RT-PCR primers for TA cloning. The TA cloning method was performed as described by Qian et al. (2016). The amplified and purified PCR products were inserted into the pEASY-Blunt Zero vector (TransGen Biotech, Beijing, China), then transferred into Trans5α chemically competent cell (TransGen Biotech, Beijing, China) and sequenced finally. All RT-PCR primers were listed in Supplementary Table 3.
Vector Construction, Plant Transformation, and Cold Treatment of Transgenic Plants
The vector construction and plant transformation methods were carried out as stated by Qian et al. (2018) with some modification. The Gateway technology was used to construct overexpression vectors (Landy, 1989). Simply, the ORF of CsPMEI2 and CsPMEI4 without stop codons were firstly cloned into the entry vector (pENTR/D-TOPO) (Invitrogen, CA, United States) following the instruction of manufacturer, respectively. After verified by sequencing, the ORF of CsPMEI2 and CsPMEI4 were transferred into the destination vector (pH7FWG2) by using LR Clonase II enzyme mix kit (Invitrogen, Carlsbad, CA, United States) (Ishimaru et al., 2005), respectively. The plasmids of recombined vectors of CsPMEI2 and CsPMEI4 were separately mobilized into Agrobacterium tumefaciens strain GV3101, and finally transformed into Arabidopsis via Agrobacterium-mediated floral infiltration (Clough and Bent, 2010). The positive CsPMEI2-OE lines and CsPMEI4-OE lines were obtained by hygromycin B screening, and the transcript abundances of CsPMEI2 and CsPMEI4 in each OE-line were quantified by qRT-PCR. Finally, three OE-lines with different transcript abundances of CsPMEI2 and CsPMEI4 were, respectively, used to proceed further experiments. The primers used for vector construction were listed in Supplementary Table 3.
Cold and ABA Treatments of Transgenic Plants
For cold treatment, the seeds of WT plants and three CsPMEI4-OE lines (OE4-2, OE4-7, and OE4-11) were sterilized and vernalized firstly, and then they were sown onto 1/2 MS medium for germinating in the growth chamber with photoperiod (10 h light at 22°C/14 h dark at 20°C) and 100 μmol/m2s. Two weeks later, the seedlings with four rosette leaves were transplanted on the seedling blocks and grown in the growth chamber for another 2 weeks. For cold treatment, both WT plants and CsPMEI4-OE plants were treated at 4°C for 7 days without changing the light time and intensity. The rosette leaves were collected to measure total soluble sugar (TSS) and the expressions of cold-responsive genes. To detect electrolyte leakage, 4-week-old seedlings of both WT plants and CsPMEI4-OE plants were exposed to −6°C for 8 h without changing the light time and intensity. The control seedlings were grown normally in the growth chamber. Each treatment contained three independent biological replicates, and each replicate contained six seedlings of each OE lines and WT plants, respectively.
ABA treatment was performed as described by Jing et al. (2020). Two-week-old seedlings of CsPMEI2/4-OE lines and WT plants were sprayed with a 20 μM ABA solution twice a week until flowering under LD condition. Meanwhile, the controls were sprayed with distilled water. The phenotypes, flowering time, and leaf number were recorded. Each treatment contained three independent biological replicates, and each replicate contained fifteen seedlings of each OE lines and WT plants, respectively.
Flowering Time, Leaf Number, and Plant Height Measurements
The methods used for evaluating the flowering time and other phenotypes were conducted as described by Jing et al. (2020). The WT and OE-lines were cultured in seedling blocks under LD and SD conditions, respectively. The time from seed sterilization to floral bud formation was recorded as flowering time, and the total number of rosette leaves were counted at the same time. The plant heights of WT and OE-lines were measured once the plants stop flowering. The sizes of the leaves, flowers and seeds of WT and OE-lines were also surveyed as described by Wang et al. (2012). Three independent biological replicates were performed, and each replicate contained fifteen seedlings of each OE lines and WT plants, respectively.
Electrolyte Leakage and Sugar Contents Measurements
The electrolyte leakage (EL) was assayed as described by Qian et al. (2018). For detecting soluble sugars contents, 0.1 g fresh sample was extracted with 1.0 mL extraction buffer in a pre-cooled mortar on ice box, and then transferred into 1.5 mL microcentrifuge tube to water bath at 80°C for 10 min. After centrifuged at 25°C, 4,000 × g for 10 min, the supernatants were decolorized. Thereafter, 1.0 mL extraction buffer was added and centrifuged at 25°C, 4,000 × g for 10 min again. The supernatant was used for total soluble sugar (TSS), Suc, Glc, and Fru measurements according to the instructions of the corresponding sugar measurement kits (Suzhou Comin Biotechnology, Suzhou, China), respectively.
Pectin Methylesterase Activity Measurements
Pectin methylesterase activity was assayed by using NaOH indirect titration method as the manufacture’s introduction (Suzhou Comin Biotechnology, Suzhou, China). 1 g sample was thoroughly ground in an ice bath with 2 mL pre-cooled extraction buffer. After centrifuged at 4°C, 12,000 × g for 10 min, the supernatant was used.
Quantitative Real-Time RT-PCR Analysis
Total RNA isolation, first-strand cDNA synthesis and qRT-PCR analysis of all samples were conducted as demonstrated by Qian et al. (2018). The qRT-PCR reaction system was mixed as follows: 5.0 μL SYBR Premix Ex Taq, 1 μL cDNA, 0.8 μL forward/reverse primers, and 3.2 μL distilled water. The qRT-PCR program was run as follows: 95°C, 15 s for predegeneration; then 94°C, 5 s and 58°C, 30 s for amplification with 40 cycles; finally, a melting curve was added. Two housekeeping genes, CsPTB of tea plant (Hao et al., 2014) and AtEF of Arabidopsis (Yuan et al., 2008), were, respectively, used to quantify the relative expression levels of the target genes based on 2–Δ Ct or 2–Δ Δ Ct method (Livak and Schmittgen, 2001). Each cDNA was performed three parallel technical repeats, and the calculated results were visualized as the mean values ± standard error (±SE). The qRT-PCR primers were shown in Supplementary Table 4.
Statistical Analysis
Statistical differences between WT and CsPMEI2/4-OE lines under different conditions were analyzed by one-way Analysis of Variance (ANOVA) followed by Tukey’s HSD test and/or Duncan’s test.
Results
Identification, Cloning, Bioinformatics Analysis of CsPMEIs in Tea Plant
Based on PMEI domain (PF04043), 51 CsPMEIs and 2 CsVIF/CIFs were identified from tea plant genome. The ORF lengths of CsPMEIs were varied from 420 to 924 bp, the encoded amino acids were ranged from 139 to 307 aa, the molecular weights (MW) were changed from 15.48 to 34.12 kD, and the theoretical isoelectric points (pIs) were ranged from 3.93 to 9.76 pIs. More than 35 CsPMEIs were predicted to be stable proteins that are located in cytoplasm. 46 CsPMEIs were predicted to contain N-terminal signal peptides and 10 CsPMEIs were predicted to contain transmembrane helices (TMHs) (Table 1). Meanwhile, four CsPMEIs genes, named as CsPMEI1-4 (accession number: KU884479, KU884480, KU884481, and KU884482), were cloned based on the ESTs sequences as found in the previous transcriptome data. Alignment analysis results found that CsPMEI1-4 shared more than 99% identities with XP_028079167, XP_028079378, XP_028097250, and XP_028127647, respectively. Amino acid alignment analysis result showed that all of the identified CsPMEIs and CsVIF/CIFs contained four conserved and representative Cys residues except for CsPMEI5 (Supplementary Figure 1).
TABLE 1.
Basic information of CsPMEIs.
| Gene name | Accession number | ORF (bp) | AA | MW (kDa) | pI | Instability index | Loc | SignalP | TMHs |
| CsPMEI1 | KU884479 | 594 | 197 | 21.70 | 4.31 | Stable | Nucleus | Yes | No |
| CsPMEI2 | KU884480 | 591 | 196 | 21.52 | 3.93 | Stable | Cytoplasm | Yes | No |
| CsPMEI3 | KU884481 | 630 | 209 | 22.55 | 9.16 | Stable | Nucleus | Yes | 1 |
| CsPMEI4 | KU884482 | 639 | 212 | 22.86 | 7.67 | Stable | Cytoplasm | Yes | No |
| CsPMEI5 | XP_028119293.1 | 816 | 271 | 28.74 | 4.5 | Unstable | Nucleus | Yes | No |
| CsPMEI6 | XP_028063476.1 | 504 | 167 | 18.27 | 5.79 | Stable | Cytoplasm | Yes | No |
| CsPMEI7 | XP_028078455.1 | 561 | 186 | 20.39 | 4.37 | Stable | Nucleus | Yes | No |
| CsPMEI8 | XP_028080229.1 | 552 | 183 | 19.8 | 9.1 | Stable | Cytoplasm | Yes | No |
| CsPMEI9 | XP_028096826.1 | 798 | 265 | 29.6 | 5.34 | Stable | Cytoplasm | No | 1 |
| CsPMEI10 | XP_028103971.1 | 585 | 194 | 20.53 | 6.27 | Stable | Nucleus | Yes | No |
| CsPMEI11 | XP_028105374.1 | 636 | 211 | 23 | 9.32 | Unstable | Nucleus | Yes | No |
| CsPMEI12 | XP_028106458.1 | 615 | 204 | 22.25 | 9.73 | Stable | Nucleus | Yes | No |
| CsPMEI13 | XP_028108659.1 | 639 | 212 | 23.28 | 8.51 | Unstable | Nucleus | Yes | No |
| CsPMEI14 | XP_028108660.1 | 630 | 209 | 22.68 | 9.24 | Stable | Cytoplasm | Yes | 1 |
| CsPMEI15 | XP_028108661.1 | 639 | 212 | 23.14 | 8.71 | Unstable | Nucleus | Yes | No |
| CsPMEI16 | XP_028112715.1 | 564 | 187 | 21.02 | 9.12 | Stable | Nucleus | Yes | 1 |
| CsPMEI17 | XP_028117331.1 | 651 | 216 | 23.89 | 4.58 | Unstable | Cytoplasm | Yes | No |
| CsPMEI18 | XP_028118374.1 | 546 | 181 | 20.92 | 4.65 | Stable | Nucleus | Yes | No |
| CsPMEI19 | XP_028118383.1 | 546 | 181 | 20.7 | 4.64 | Stable | Cytoplasm | Yes | No |
| CsPMEI20 | XP_028118385.1 | 546 | 181 | 20.96 | 4.64 | Stable | Cytoplasm | Yes | No |
| CsPMEI21 | XP_028118830.1 | 582 | 193 | 21.11 | 6.42 | Stable | Nucleus | Yes | No |
| CsPMEI22 | XP_028118831.1 | 630 | 209 | 22.65 | 9.36 | Stable | Cytoplasm | Yes | No |
| CsPMEI23 | XP_028118832.1 | 642 | 213 | 23.24 | 8.88 | Stable | Cytoplasm | Yes | No |
| CsPMEI24 | XP_028118833.1 | 618 | 205 | 22.21 | 8.29 | Stable | Cytoplasm | Yes | No |
| CsPMEI25 | XP_028118856.1 | 588 | 195 | 20.96 | 9.25 | Stable | Cytoplasm | Yes | No |
| CsPMEI26 | XP_028122572.1 | 504 | 167 | 18.18 | 5.28 | Stable | Cytoplasm | Yes | No |
| CsPMEI27 | XP_028123747.1 | 630 | 209 | 22.68 | 9.76 | Stable | Cytoplasm | Yes | No |
| CsPMEI28 | XP_028123748.1 | 621 | 206 | 21.9 | 6.09 | Stable | Cytoplasm | Yes | No |
| CsPMEI29 | XP_028123765.1 | 555 | 184 | 19.46 | 5.8 | Stable | Chloroplast | No | No |
| CsPMEI30 | XP_028051078.1 | 630 | 209 | 22.7 | 8.92 | Stable | Chloroplast | Yes | No |
| CsPMEI31 | XP_028051784.1 | 582 | 193 | 20.86 | 9.41 | Stable | Cytoplasm | Yes | No |
| CsPMEI32 | XP_028055798.1 | 504 | 167 | 18.17 | 5.63 | Stable | Cytoplasm | Yes | No |
| CsPMEI33 | XP_028056276.1 | 609 | 202 | 22.01 | 9.47 | Unstable | Cytoplasm | Yes | No |
| CsPMEI34 | XP_028061043.1 | 585 | 194 | 21.09 | 5.59 | Stable | Cytoplasm | Yes | No |
| CsPMEI35 | XP_028061148.1 | 627 | 208 | 22.14 | 8.85 | Stable | Cytoplasm | Yes | 1 |
| CsPMEI36 | XP_028063328.1 | 420 | 139 | 15.48 | 5.04 | Stable | Cytoplasm | No | No |
| CsPMEI37 | XP_028063329.1 | 558 | 185 | 20.33 | 4.54 | Stable | Cytoplasm | Yes | 1 |
| CsPMEI38 | XP_028063330.1 | 552 | 183 | 20.36 | 5.27 | Stable | Cytoplasm | Yes | 1 |
| CsPMEI39 | XP_028063858.1 | 573 | 190 | 21.3 | 6.23 | Unstable | Cytoplasm | Yes | 1 |
| CsPMEI40 | XP_028069427.1 | 606 | 201 | 21.49 | 4.52 | Stable | Cytoplasm | Yes | No |
| CsPMEI41 | XP_028069450.1 | 621 | 206 | 21.96 | 6.09 | Stable | Cytoplasm | Yes | No |
| CsPMEI42 | XP_028073478.1 | 567 | 188 | 20.52 | 9.14 | Stable | Cytoplasm | No | No |
| CsPMEI43 | XP_028073497.1 | 540 | 179 | 19.67 | 7.66 | Stable | Cytoplasm | Yes | No |
| CsPMEI44 | XP_028074709.1 | 516 | 171 | 18.5 | 6.29 | Stable | Cytoplasm | Yes | No |
| CsPMEI45 | XP_028079349.1 | 504 | 167 | 18.27 | 5.13 | Stable | Cytoplasm | Yes | No |
| CsPMEI46 | XP_028079849.1 | 564 | 187 | 21.58 | 8.82 | Unstable | Cytoplasm | Yes | 1 |
| CsPMEI47 | XP_028081340.1 | 762 | 253 | 27.12 | 5.11 | Stable | Cytoplasm | No | 1 |
| CsPMEI48 | XP_028082183.1 | 606 | 201 | 21.98 | 9.33 | Stable | Cytoplasm | Yes | No |
| CsPMEI49 | XP_028082302.1 | 621 | 206 | 21.93 | 6.09 | Stable | Cytoplasm | Yes | No |
| CsPMEI50 | XP_028082397.1 | 540 | 179 | 19.6 | 8.32 | Stable | Cytoplasm | Yes | No |
| CsPMEI51 | XP_028090516.1 | 924 | 307 | 34.12 | 8.92 | Unstable | Cytoplasm | Yes | No |
| CsVIF/CIF1 | XP_028053538.1 | 525 | 174 | 18.71 | 9.23 | Stable | Cytoplasm | Yes | No |
| CsVIF/CIF2 | XP_028100701.1 | 522 | 173 | 18.48 | 9.57 | Stable | Cytoplasm | Yes | No |
ORF, opening reading fame; AA, the numbers of amino acid residues; MW, molecule weight; pI, theoretical isoelectric point; Loc, subcellular location; SignalP, signal peptide; TMHs, transmembrane helices.
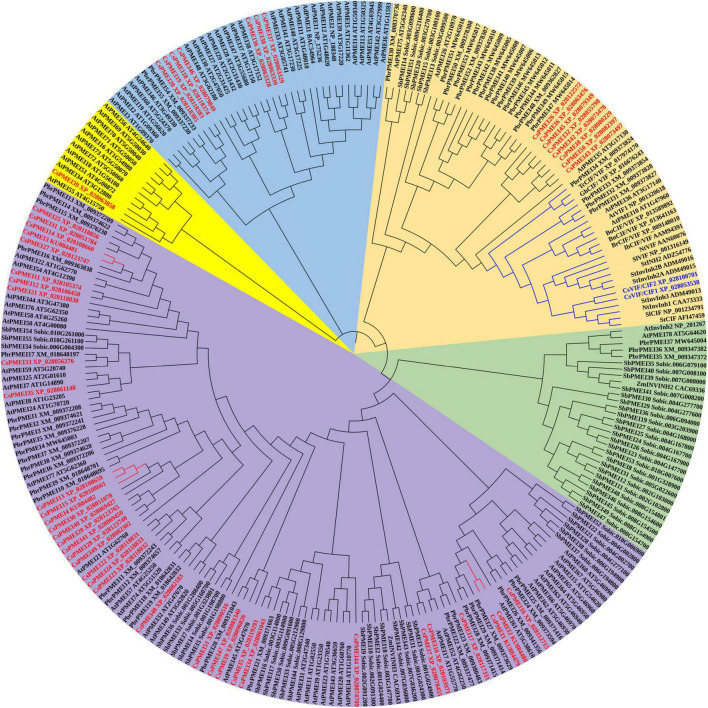
Phylogenetic analysis result showed that 262 PMEI domain contained proteins were clustered into five clades. However, CsPMEIs and CsVIF/CIFs were only grouped into four clades, among them, 35 of CsPMEIs were clustered into Clade I. In addition, we found that CsVIF/CIFs showed closet relationship with lots of known VIF/CIFs, but most of CsPMEIs showed closet relationship with PbrPMEIs (Figure 1). Moreover, the DNA structure of each CsPMEI contains only one exon except for CsPMEI34 and CsPMEI42 (Supplementary Figure 2A). cis-acting elements prediction results showed that CsPMEIs could be regulated by MYB and MYC transcription factors, auxins (GA, MeJ, SA, and ABA etc.) and multiple stresses (drought, cold, and anaerobic etc.) (Supplementary Figure 2B). To verify this conclusion, the expressions of four genes, CsPMEI1-4, were detected under different conditions. Meanwhile, the functions of CsPMEI2 and CsPMEI4 were further explored by means of overexpression technology in Arabidopsis.
FIGURE 1.
Phylogenetic analysis of CsPMEIs and known PMEIs in Arabidopsis, Sorghum bicolor, pear, and kiwi fruit. A total of 262 PMEI domain contained proteins that were used to construct phylogenetic tree by using MEGA 7.0 software. CsPMEIs are highlighted with red color, and different PMEIs subfamilies were covered with different colors.
Expression Profiles of CsPMEIs in Various Tissues, and in Responding to Various Abiotic Stresses in Tea Plant Leaves and Roots
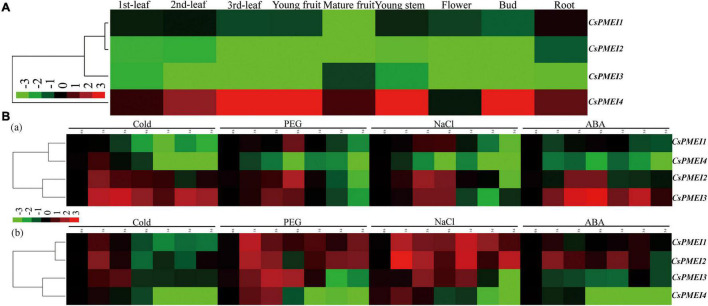
The tissue-specific of CsPMEI1-4 were analyzed in nine different tea plant tissues. As shown in Figure 2A, CsPMEI1-4 showed diverse transcription abundances in various tissues. Among them, CsPMEI1/2 were highly expressed in tender leaves and root, CsPMEI3 was highly expressed in mature fruit. However, CsPMEI4 presented the highest expressions in each detected tissue than the other three CsPMEIs, especially in young fruit, young stem and bud, suggesting that CsPMEIs, especially CsPMEI4, widely involved in the vegetative and reproductive processes of tea plant.
FIGURE 2.
Expression profiles of CsPMEIs in tea plant. (A) Tissue-specifc expression patterns of CsPMEIs in different tea plant tissues. The relative expression levels were calculated by using 2– Δ Ct method. The colorbar was displayed on the lower-left of the heat map, red and green colors represent higher and lower expression levels, respectively. (B) Temporal-spatial expression patterns of CsPMEIs both in tea plant mature leaves and roots under various abiotic stresses. (a) The expression patterns of CsPMEIs in mature leaves; (b) The expression patterns of CsPMEIs in roots. Samples at 0 h were used as control, and the final results were calculated with 2– Δ Δ Ct method. The colorbar was presented on the left side of the heat map, red and green colors represent higher and lower expression levels, respectively. Data are shown as the means ± SE (n = 3).
Within 5 days of different abiotic stresses treatments, the expressions of CsPMEIs showed diverse changes both in tea plant mature leaves and roots at different processing time points (Figure 2B). In mature leaves, CsPMEI1 transcripts were declined by cold treatment, but induced by drought and NaCl treatments; CsPMEI2/3 transcripts were highly induced by different abiotic stresses within 9 h of treatments, then CsPMEI2 was reduced by different abiotic stresses until the 5 days of time point; besides, CsPMEI4 transcripts were significantly decreased within 5 days of different abiotic stresses. In roots, CsPMEI1 transcripts were highly upregulated by drought and NaCl treatments, but downregulated by cold treatment; CsPMEI2 transcripts were highly induced by drought, NaCl and ABA stresses within 5 days of different abiotic stresses, except for a few time points; CsPMEI3 transcripts were increased within 3 h of cold stress and 1 day of drought and NaCl stresses, respectively, but decreased within 5 days of ABA treatment; the mRNA level of CsPMEI4 was reduced by various abiotic stresses after 3 h of treatments, even though it was induced by drought and NaCl within 3 h of treatments. The above results indicated that CsPMEI1-4 play important roles in dealing with various abiotic stresses in tea plant.
Differential Expressions of CsPMEIs Under Biotic, Cold and Sugars Treatments
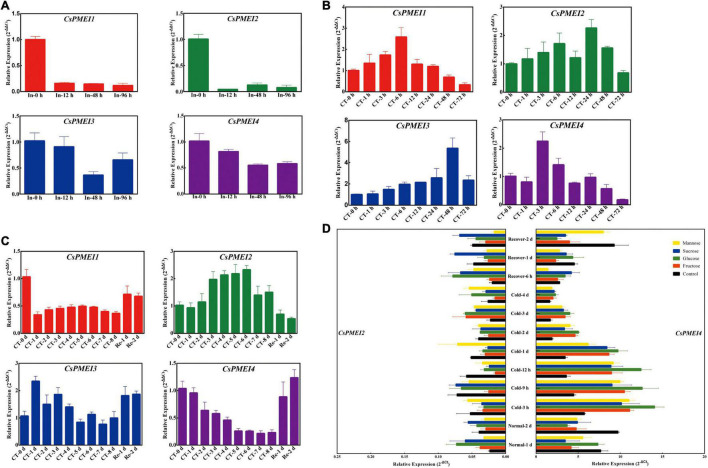
Here, we detected the expression patterns of CsPMEI1-4 under C. fructicola infection condition. As Figure 3A shown, CsPMEI1-4 transcripts were reduced by C. fructicola, especially CsPMEI1/2 were remarkably downregulated by C. fructicola infection.
FIGURE 3.
Temporal-spatial expression patterns of CsPMEIs in response to biotic, cold and various sguar treatments. (A) Expression profiles of CsPMEIs under C. fructicola infection condition. Samples at 0 h were used as control, and the final results were calculated with 2– Δ Δ Ct method. ‘In’ represents infection treatment. Data are shown as the means ± SE (n = 3). (B) Expression profiles of CsPMEIs at one bud and one leaf stage of tea plant under cold condition. Samples at 0 h were used as control, and the final results were calculated with 2– Δ Δ Ct method. ‘CT’ represents cold treatment. Data are shown as the means ± SE (n = 3). (C) Expression profiles of CsPMEIs at one bud and three leaves stage of tea plant under cold condition. Samples at 0 h were used as control, and the final results were calculated with 2– Δ Δ Ct method. ‘CT’ represents cold treatment. Data are shown as the means ± SE (n = 3).(D) Expression analysis of CsPMEIs in tea plant leaves under different sugar and temperature conditions. All results were calculated by using the 2– Δ Ct method. Data are shown as the means ± SE (n = 3).
Besides, a time-course expressions of CsPMEI1-4 were separately performed in OBOL and OBTL tissues under 4°C condition. As Figures 3B,C shown, the expression patterns of CsPMEI1-4 were varied in OBOL and OBTL. At OBOL stage, CsPMEI1/3 were continuously up-regulated with 6 h and 2 days of cold treatment (CT), respectively; CsPMEI2 was also induced by CT within 2 days; CsPMEI4 showed a highest expression at 3 h, and then gradually decreased until 3 days. At OBTL stage, CsPMEI1/4 were gradually downregulated within 8 days of CT, and then recovered to normal expression levels following the temperature increased; in contrast, CsPMEI2/3 were, respectively, upregulated within 8 and 4 days of CT, and also recovered to normal expression levels under normal temperature condition. These results indicated that CsPMEI1-4 had different levels and timings of expressions in different tissues of tea plant under CT condition.
Furthermore, we found exogenous sugars, including Suc, Glc, Fru, and Man independently induced the expression of CsPMEI2 under normal temperature (NT) condition, while CsPMEI2 was reduced within 2 days of CT treatment, except for Man that enhanced the expression of CsPMEI2 under CT treatment. In contrast, the expression of CsPMEI4 were decreased under NT condition, but remarkably induced by exogenous sugars within 4 days of CT. Specifically, CsPMEI4 transcripts were increased more than twofold within 1 days of CT (Figure 3D). These results demonstrated that the cold response of CsPMEI2/4 in tea plant could be mediated by sugar signaling pathway.
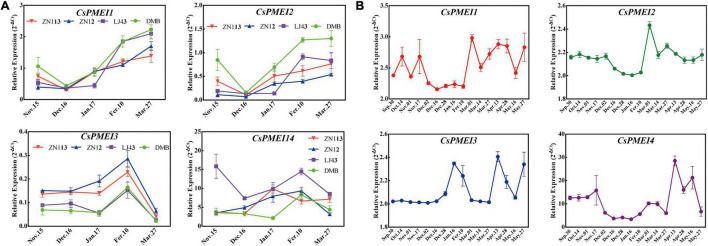
Differential Expressions of CsPMEIs During Cold Acclimation and Bud Dormancy Periods
Four-tea cultivars with different cold-resistance were used for exploring the expression patterns of CsPMEI1-4 during CA periods. As Figure 4A shown, CsPMEIs showed similar expressions patterns among these four-tea cultivars during CA periods. Briefly, CsPMEIs transcripts were decreased during CA periods (from 15 November to 16 December), except for CsPMEI3. Subsequently, the expressions of CsPMEI1/2 were increased from 16 December to 27 March. However, CsPMEI3/4 transcripts were firstly increased from 16 December to 10 February, and then decreased with increasing temperature.
FIGURE 4.
Expression analysis of CsPMEIs during CA and bud dormancy periods. (A) Expression analysis of CsPMEIs in the mature leaves of four-tea cultivars during CA periods. ‘DMB,’ ‘LJ43,’ ‘ZN12,’ and ‘ZN113’ mean ‘DaMianBai,’ ‘LongJing43,’ ‘ZheNong12,’ and ‘ZheNong113’ tea plant cultivar, respectively. The relative expression levels were calculated by using 2– Δ Ct method. Data are shown as the means ± SE (n = 3). (B) Expression analysis of CsPMEIs in axillary buds of ‘LJ43’ cultivar during bud dormancy period. The relative expression levels were calculated by using 2– Δ Ct method. Data are shown as the means ± SE (n = 3).
We also detected the expressions of CsPMEI1-4 in axillary buds during bud dormancy period. As Figure 4B shown, CsPMEI1/2/4 transcripts were decreased throughout the bud dormancy period, but increased with the dormancy released and at bud sprouting stages. However, the expression of CsPMEI3 was not changed during para- and endo-dormancy periods (from 30 September to 16 December), but increased during eco-dormancy (after 28 December) and bud sprouting stages (after 27 March). In a word, CsPMEI1-4 involved in the bud dormancy and release of axillary buds in tea plant, but the comprehensive molecular mechanisms need to be further explored.
Overexpression Analysis of CsPMEI2 and CsPMEI4 in Transgenic Arabidopsis
CsPMEI4 Decreased the Cold Resistance of Transgenic Arabidopsis
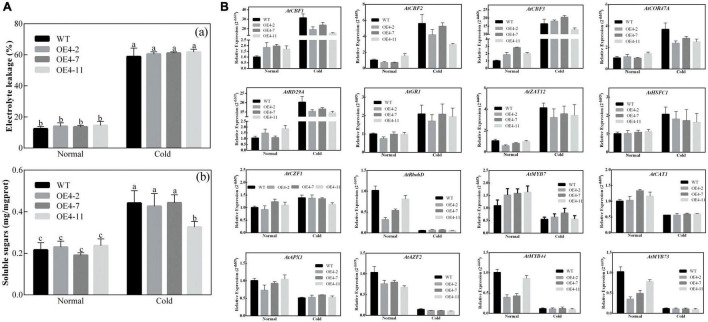
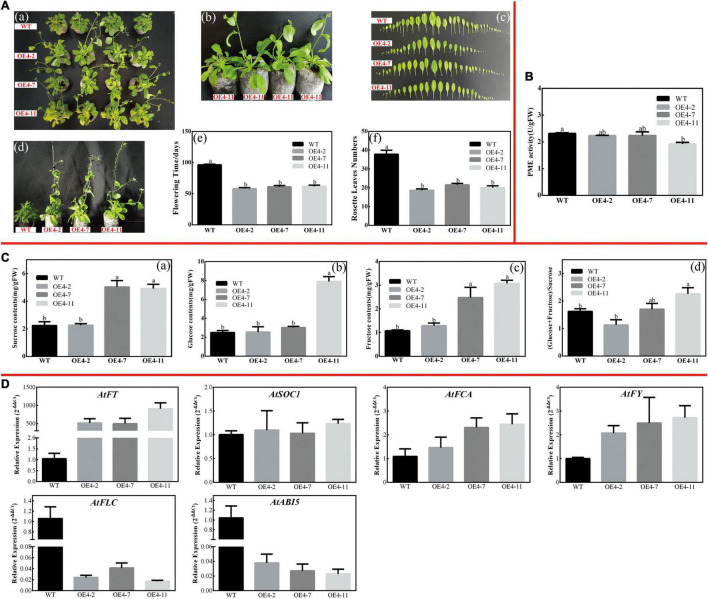
As described in Figures 2, 3, CsPMEI4 was significantly repressed by CT in tea plant. Here, we further explored the cold resistance of CsPMEI4-OE lines under CT conditions. Three independent CsPMEI4-OE lines (OE4-2, OE4-7, and OE4-11) with different transcription abundances were separately selected to perform the following experiments (Figure 5A). Unfortunately, the phenotypes had not shown obvious difference between CsPMEI4-OE lines and WT plants under CT condition (data not shown). However, CsPMEI4-OE lines showed higher EL, but lower TSS contents than the WT plants even though no significant difference detected between them (Figure 6A). Besides, the expression levels of many cold-induced genes (AtCBF1-3, AtCOR47A, AtRD29A, AtGR1, AtZAT12, AtHSFC1, and AtCZF1) in CsPMEI4-OE lines were lower than that in WT plants under CT condition. In addition, the transcript abundances of four cold-repressors (AtAZF and AtMYB7/44/73), two antioxidant-related genes (AtCAT1 and AtAPX1), and one ROS-producing enzyme NADPH oxidase gene (AtRbohD) were decreased both in CsPMEI4-OE lines and WT plants under CT condition (Figure 6B). These results suggested that overexpression of CsPMEI4 slightly reduced the cold-resistance of transgenic Arabidopsis.
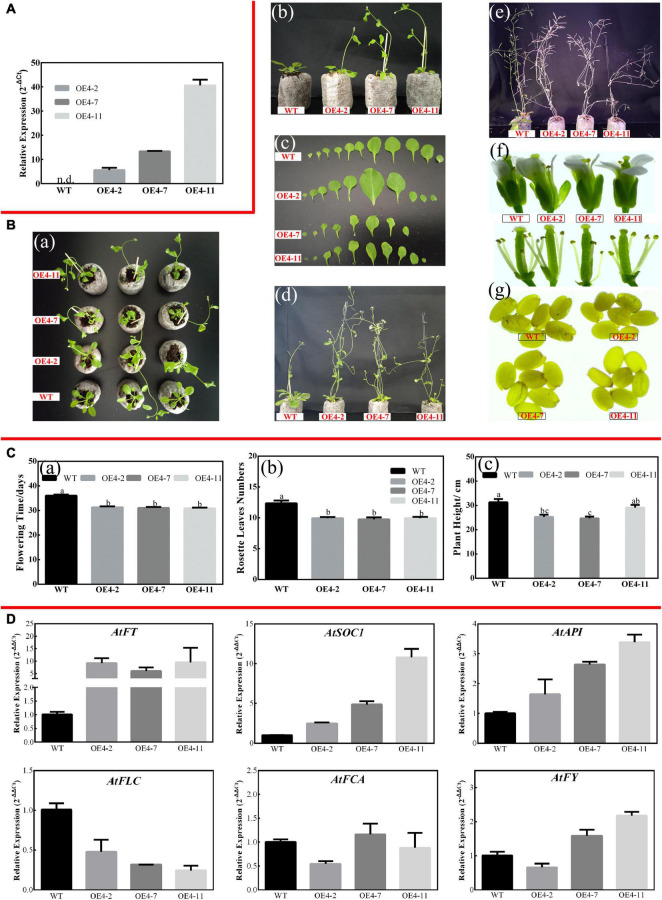
FIGURE 5.
Overexpression of CsPMEI4 promotes early flowering in transgenic Arabidopsis under LD condition. (A) The expression of CsPMEI4 in three CsPMEI4-OE lines and WT plants. The relative expression levels were calculated by using 2– Δ Ct method. Data are shown as the means ± SE (n = 3). (B) Early flowering phenotypes of CsPMEI4-OE lines grown under LD condition. (a) Early-flowering phenotype; (b) phenotypes of CsPMEI4-OE lines and WT plants after 30 d of growth under LD condition; (c) leaf size; (d) phenotype of CsPMEI4-OE lines and WT plants after 40 days of growth under LD condition; (e) phenotype of CsPMEI4-OE lines and WT plants at senescence stage; (f) flowers sizes of WT and CsPMEI4-OE lines; (g) seed size of WT and CsPMEI4-OE lines. (C) Flowering time, rosette leaves numbers, and plant height of WT and CsPMEI4-OE lines at the just bolting stage. (a) Flowering time; (b) rosette leaves numbers; (c) plant height. (D) Expression analysis of flowering-related genes. The final results were calculated with 2– Δ Δ Ct method. Data are shown as the means ± SE (n = 3).
FIGURE 6.
Analysis of EL, TSS contents and cold-related genes transcripts in CsPMEI4-OE lines and WT plants. (A) EL and TSS contents of CsPMEI4-OE lines and WT plants under different temperature conditions. (a) Electrolyte leakage, EL; (b) total soluble sugar content. Data are shown as the means ± SE (n = 3). (B) Expression analysis of cold-related genes in CsPMEI4-OE lines and WT plants under cold condition. The final results were calculated with 2– Δ Δ Ct method. Data are shown as the means ± SE (n = 3).
CsPMEI2 and CsPMEI4 Promote Early Flowering in Transgenic Arabidopsis Both Under Long-Day and Short-Day Conditions
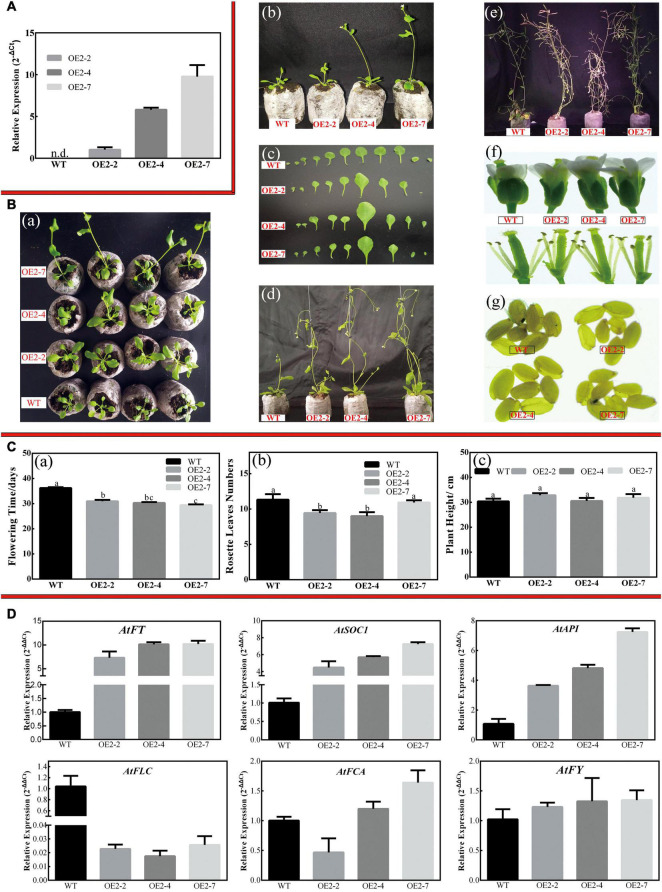
Apart from influencing cold tolerance of transgenic Arabidopsis, an obvious difference was observed on the flowering time between CsPMEI2/4-OE lines and WT plants.
Three independent CsPMEI2-OE lines (OE2-2, OE2-4, and OE2-7) and CsPMEI4-OE lines (OE4-2, OE4-7, and OE4-11) with different transcription abundances were separately selected to perform the following experiments (Figures 5A, 7A). Under LD condition, both CsPMEI2-OE lines and CsPMEI4-OE lines present early flowering than the WT plants (Figures 5B, 7B). Concretely, the WT plants require c. 36 days from seeds sterilization to floral bud formation, while CsPMEI2-OE lines and CsPMEI4-OE lines require c. 30 and 31 days, respectively (Figures 5C, 7C). It is now clear that early-flowering plants usually possess less leaves during flowering periods (Xiong et al., 2019). The similar phenotypes were also detected in CsPMEI2/4-OE lines, where we found that both CsPMEI2-OE lines and CsPMEI4-OE lines formed c. 9 rosette leaves, while the WT plants formed c. 12 rosette leaves as the first flower opened (Figures 5C, 7C). In addition, both CsPMEI2-OE lines and CsPMEI4-OE lines showed earlier senescence phenotypes than the WT plants (Figures 5B, 7B). As compared to WT plants, the plant heights of CsPMEI2-OE lines were taller, while CsPMEI4-OE lines were shorter (Figures 5C, 7C). Besides, the flower sizes of CsPMEI2-OE lines are longer and slender, and the pistils are smaller than the WT plants, while the flower structures (numbers of sepals, petals, and stamens) and the sizes of seeds have not shown significant distances among them (Figure 7B). However, the flower sizes, the flower structures (numbers of sepals, petals, stamens, and pistils) and the sizes of seeds of CsPMEI4-OE lines are similar to the WT plants (Figure 5B).
FIGURE 7.
Overexpression of CsPMEI2 promotes early flowering in transgenic Arabidopsis under LD condition. (A) The expression of CsPMEI2 in three CsPMEI2-OE lines and WT plants. The relative expression levels were calculated by using 2– Δ Ct method. Data are shown as the means ± SE (n = 3). (B) Early flowering phenotypes of CsPMEI2-OE lines grown under LD condition. (a) Early-flowering phenotype; (b) phenotypes of CsPMEI2-OE lines and WT plants after 29 days of growth under LD condition; (c) leaf size; (d) phenotypes of CsPMEI2-OE lines and WT plants after 38 days of growth under LD condition; (e) phenotypes of CsPMEI2-OE lines and WT plants at senescence stage; (f) flowers of WT and CsPMEI2-OE lines; (g) seed size of WT and CsPMEI2-OE lines. (C) Flowering time, rosette leaves, and plant height of WT and CsPMEI2-OE lines at the just bolting stage. (a) Flowering time; (b) rosette leaves numbers; (c) plant height. (D) Expression analysis of flowering-related genes. The final results were calculated with 2– Δ Δ Ct method. Data are shown as the means ± SE (n = 3).
Similarly, CsPMEI2/4-OE lines also promote early flowering in transgenic Arabidopsis under SD condition as compared to WT plants. Concretely, CsPMEI2-OE lines and CsPMEI4-OE lines, respectively, need c. 38 days and c. 58 days from seeds sterilization to the floral bud formation, while WT plants need more than 3 months to bloom. Correspondingly, CsPMEI2-OE lines and CsPMEI4-OE lines formed c. 15 and c. 20 rosette leaves, while the WT plants formed c. 38 rosette leaves as the first flower bloomed (Figures 8A, 9A). These results indicate that CsPMEI2 and CsPMEI4 mediating flowering control may be independent of photoperiod pathway.
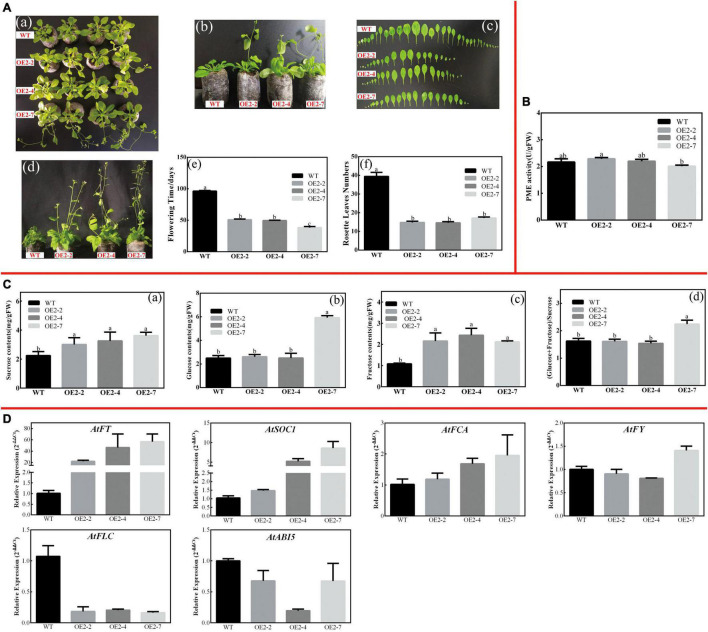
FIGURE 8.
Overexpression of CsPMEI2 promotes early flowering in transgenic Arabidopsis under SD condition. (A) Early flowering phenotypes of CsPMEI2-OE lines grown under SD condition. (a) Early-flowering phenotype; (b) phenotypes of CsPMEI2-OE lines and WT plants after 38 d of growth under SD condition; (c) leaf size; (d) phenotypes of CsPMEI2-OE lines and WT plants after 96 days of growth under SD condition; (e) flowering time; (f) rosette leaves numbers. (B) PME activities of WT and CsPMEI2-OE lines under SD condition. (C) Sugar contents of WT and CsPMEI2-OE lines under SD condition. (a) Suc contents; (b) Glu contents; (c) Fru contents; (d) the ratio of hexose/Suc. (D) Expression analysis of flowering-related genes. The final results were calculated with 2– Δ Δ Ct method. Data are shown as the means ± SE (n = 3).
FIGURE 9.
Overexpression of CsPMEI4 promotes early flowering in transgenic Arabidopsis under SD condition. (A) Early flowering phenotypes of CsPMEI4-OE lines grown under SD condition. (a) Early-flowering phenotype; (b) phenotypes of CsPMEI4-OE lines and WT plants after 58 d of growth under SD condition; (c) leaf size; (d) phenotypes of CsPMEI4-OE lines and WT plants after 96 days of growth under SD condition; (e) flowering time; (f) rosette leaves numbers. (B) PME activities of WT and CsPMEI4-OE lines under SD condition. (C) Sugar contents of WT and CsPMEI4-OE lines under SD condition. (a) Suc contents; (b) Glu contents; (c) Fru contents; (d) the ratio of hexose/Suc. (D) Expression analysis of flowering-related genes. The final results were calculated with 2– Δ Δ Ct method. Data are shown as the means ± SE (n = 3).
CsPMEI2 and CsPMEI4 Affect PME Activities and Sugar Contents in Transgenic Arabidopsis Under Short-Day Condition
Under SD condition, the seedlings were sampled to validate the effects of CsPMEI2 and CsPMEI4 on PME activities of transgenic Arabidopsis. As Figures 8B, 9B show, the PME activities seem to be not significantly inhibited by the exogenous overexpression of CsPMEI2 or CsPMEI4, except for the highest expressions OE-lines (OE2-7 and OE4-11). In addition, the sugars contents, including Suc, Glu, and Fru were increased both in CsPMEI2-OE and CsPMEI4-OE lines. Particularly, Suc and Fru contents in OE lines were significantly higher than that in WT plants, and the ratios of (Glu + Fru)/Suc were also higher both in CsPMEI2-OE and CsPMEI4-OE lines than the WT plants, except for OE4-2 (Figures 8C, 9C). These results indicate that CsPMEI2 and CsPMEI4 mediated flowering was associated with the changes of sugars contents.
CsPMEI2 and CsPMEI4 Affect Multiple Flowering-Relate Genes Transcripts in Transgenic Arabidopsis
To explore the molecular mechanisms of early flowering phenotype of CsPMEI2-OE lines and CsPMEI4-OE lines, we further conducted the expression levels of many flowering responsive genes.
Under LD condition, the expressions of two floral integrators, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), were remarkably increased, while a key flowering repressor gene, FLOWERING LOCUS C (FLC), was significantly inhibited both in CsPMEI2-OE lines and CsPMEI4-OE lines as compared to WT plants. In addition, three floral meristem identity genes, APETALA1 (AP1), FLOWERING CONTROL LOCAL A (FCA), and FLOWERING LOCUS T (FY) were also upregulated both in CsPMEI2-OE lines and CsPMEI4-OE lines (Figures 5D, 7D). Similarly, under SD condition, the expressions of FT, SOC1, FCA, and FY were upregulated both in CsPMEI2-OE lines and CsPMEI4-OE lines, especially for FT gene, which is induced at least c. 20-fold and c. 400-fold in CsPMEI2-OE lines and CsPMEI4-OE lines, respectively. Meanwhile, the mRNA levels of FLC and an ABA stimulated positive factors, ABSCISIC ACID-INSENSITIVE 5 (ABI5) were decreased both in CsPMEI2-OE lines and CsPMEI4-OE lines (Figures 8D, 9D). These results suggested that CsPMEI2 and CsPMEI4 promoting early flowering may partially depend on ABA-dependent pathway. This inference was supported by ABA treatments, where we found the flowering times of CsPMEI2-OE and CsPMEI4-OE lines were delayed by the application of exogenous ABA, even though the development of all plants retarded by ABA treatment (Supplementary Figure 3).
.
Discussion
CsPMEIs Exist in Large Multigene Family Exhibiting Diverse Expression Patterns in Tea Plant
At post-translation level, the activity of PME is antagonized by PMEI. As a type of small molecular protein, PMEI is encoded by large multigene family in various plant species, such as Arabidopsis (Müller et al., 2013), B. campestris (Liu et al., 2018b), rice (Nguyen et al., 2016), pear (Zhu et al., 2021), etc. It is now clear that both PMEI and INH share moderate sequence homology, in particular the presence of four conserved Cys residues that form two disulfide bridges (S–S) (Camardella et al., 2000; Hothorn et al., 2004). However, PMEI and INH are selectively targeted toward PMEs and plant acid INVs (CWIN and VIN), respectively. For INH, a conserved amino acid motif ‘PKF’ (Proline, Lysine, and Phenylalanine) has been demonstrated as core motif that directly targets the active site of the CWIN/VIN (Hothorn et al., 2010). Herein, 53 genes with intact and conserved PMEI domains were identified from ‘ShuChaZao’ tea plant cultivar genome. Bioinformatics analysis results showed that most of CsPMEIs contain one exon on DNA level encoding a type of 21 KD proteins with an N-signal peptide. Phylogenetic analysis result showed that CsPMEIs were clustered into four clades, which are similar to the phylogenetic results of PbrPMEIs (Zhu et al., 2021) and BcPMEIs (Liu et al., 2018b). In addition, we found 2 CsVIF/CIFs showed closet relationships with many known INHs, and only these two CsVIF/CIFs contain the conserved ‘PKF’ motif that directly targets the INV active site (Supplementary Figure 1). These two genes have been cloned and sequenced recently, and their functions will be further explored in future. Besides, phylogenetic analysis result showed that PMEIs and INHs were clearly clustered into different subgroups, most of known INHs, including CsVIF/CIFs, were clustered into subgroup I, while CsPMEI2/4 were clustered into subgroup III (Supplementary Figure 4A). This result was further verified by the amino acid alignment result, where we found all members of subgroup I contain the conserved ‘PKF’ motif, but PMEIs do not contain this motif (Supplementary Figure 4B). Collectively, we considered that 51 CsPMEIs and 2 CsVIF/CIFs genes were identified from ‘ShuChaZao’ tea plant cultivar genome, and CsPMEI2/4 belonged to PMEI family members.
In recent years, lots of experiments have validated that PMEIs function in various biological processes, including seed germination (Müller et al., 2013), pollen growth (Zhu et al., 2021), fertility (Andres-Robin et al., 2020), organ formation and separation, fruit ripening (Srivastava et al., 2012), biotic and abiotic stress response (An et al., 2008; Volpi et al., 2011; Lionetti et al., 2017). Recently, many PMEIs genes involved in abiotic stresses also have been identified and explored in plants, while their molecular regulation mechanisms are rarely studied. Microarray data analysis results showed that many AtPMEIs were differentially expressed in response to different abiotic stresses (Liu et al., 2018b). Similarly, the meta-transcriptional analysis results found that lots of OsPMEIs were transcriptionally up- or down-regulated by abiotic and biotic stresses (Nguyen et al., 2016). Exogenous overexpression of CaPMEI enhanced drought tolerance and alleviated the sensitive to mannitol-induced osmotic stress in transgenic Arabidopsis seedlings as compared to WT plants (An et al., 2008). Herein, CsPMEI1-4 exist transcriptional diversity and functional division. In various tea plant tissues, CsPMEI4 transcripts were remarkably accumulated in each detected tissue, suggesting that it may play an important role in response to various biological processes in tea plant. Besides, CsPMEI1-4 were up- or down-regulated by various abiotic stresses in a certain period of treatment time, suggesting that CsPMEIs possess functional specialization, and a balance mechanism of pectin regulation exists in tea plant in response to abiotic stresses. This assumption was also supported by the results of Table 1, which indicated that the PME activity of tea plant was co-regulated by CsPMEI superfamily genes at the post-translational level, rather than by several CsPMEI genes.
Previous studies found that pectin contents, PME activity and the low-methylated pectin contents increased in the leaves of winter oil-seed rape during CA period, while a contrary tendency exhibited as the de-acclimation proceeded (Solecka et al., 2008). Similarly, Baldwin et al. (2014) found that the levels of arabinose, galactose, galacturonic acid and xylose residues changed in Pisum sativum under CA condition. Moreover, the degree of DM increased after 10–20 days of CA and 2 days of frost treatment as compared to non-CA plants. Meanwhile, the PME activity increased after 10–20 days of CA, but decreased after 2 days of frost treatment in frost-tolerant genotype ‘Champagne’ as compared to non-CA ‘Champagne,’ which indicated that methylesterification of pectins contributes to improving frost-tolerance of pea during CA period (Baldwin et al., 2014). In this study, we found that the expressions of CsPMEI1-2 declined in tea plant leaves during CA period, while increased with the process of de-acclimation. Meanwhile, CsPMEI1-4 transcripts decreased throughout the bud dormancy period, but increased with the breaking dormancy and the bud sprouting. These results inferred that the PME activity, pectin contents and the methyl-esterification degree of pectins may be dynamically changed by temperature both in tea plant leaves and axillary buds. However, this assumption still needs to be further verified due to PME activity was inhibited by a large superfamily members of CsPMEIs as showed in Table 1. Therefore, the expression analysis of all CsPMEIs, the assay of PME activity and pectins contents should be further performed in order to extensively explore the specific regulation mechanism of CsPMEIs involved in freezing tolerance of tea plant.
In addition to responding to abiotic stress, PMEIs transcripts are also affected by hormones. As Srivastava et al. (2012) demonstrated that the expression of a ripening related gene, MaPMEI, relies on ethylene-dependent pathway indirectly during banana ripening. Similarly, the transcription levels of a wheat PMEI gene, TaPMEI, were regulated by SA, ABA, and MeJA in leaves, stem and root (Hong et al., 2010). Meanwhile, the expression levels of CaPMEI1 were not only induced by abiotic stresses, but also by ABA, SA, ethylene and MeJA at various time intervals, suggesting that CaPMEI1 may be mediated the early active defense responses to bacterial pathogen infection and exogenous hormones treatment (An et al., 2008). Herein, we found the transcription abundance of CsPMEI2/3 were strongly induced after 9 h of ABA treatment, while CsPMEI1/4 were significantly declined during ABA treatment period, suggesting that CsPMEI2/3 may be involved in hormone-related signaling pathways associated with defense-responsive.
At present, lots of evidences have verified that PMEIs are involved in plant defense against pathogen infection. CaPMEI1 exhibited basal disease resistance against a variety of plant pathogenic fungi, overexpression of CaPMEI1 enhanced the anti-fungal ability in transgenic Arabidopsis, which indicated that the increased PMEI activity may cause the decrease accessibility of fungal pectin degrading-enzymes and thus enhance disease resistance of transgenic Arabidopsis (An et al., 2008). Similar results were also found in AtPMEI-1 or AtPMEI-2 overexpressed Arabidopsis lines after infected by B. cinerea (Lionetti et al., 2007). However, it has been reported that increasing PME activity also improved the ability of pathogen immunity. During pattern-triggered immunity and after inoculation with necrotrophic fungus and bacterial hemibiotroph in Arabidopsis, the PME activity increased, but the degree of pectin methylesterification decreased. Further research found that pathogen-induced PME activity was dependent on JA signaling pathway. Mutating the selected pme gene resulted in high sensitivity to pathogen, but the total PME activity was not influenced in pme mutants, suggesting that PME enhancing the immunity responsive was not determined by total PME activity, but by some specific effect of PMEs, such as methyl-esterification degree of pectins (Bethke et al., 2014). For tea plant, lots of differentially expressed metabolites (e.g., gluconic acid, fatty acid, amino acid, organic acid, etc.) have been identified after being inoculated with C. camelliae. Meanwhile, the contents of JA and IAA were significantly increased, accompanied by the transcription accumulation of a pathogenesis-related protein 4 (PR4) in JA signaling pathway, which indicated that tea plant-Colletotrichum interaction may be mainly mediated by JA signaling pathway (Lu et al., 2020). Herein, all four CsPMEIs transcripts were significantly inhibited by C. fructicola infection in tea plant, suggesting that the low transcription abundance of CsPMEIs may result in the decrease of PMEI activity, and thus increasing PME activity may change the methyl-esterification degree of pectins depending on JA signaling pathway.
Exogenous Overexpression of CsPMEIs Affects Cold-Resistance and Flowering Time in Transgenic Arabidopsis
It has been reported that the PME activity, pectins content and the methyl-esterification degree of pectins were dynamically changed under cold conditions (Solecka et al., 2008; Baldwin et al., 2014). In present study, there are no obvious phenotypic differences observed between CsPMEI4-OE lines and WT plants under cold condition. However, the higher EL, but the lower TSS contents and the lower expressions of many cold-induced genes were found in CsPMEI4-OE lines as compared with WT plants, suggesting that CsPMEI4 may play negative roles in responding to cold stress in transgenic Arabidopsis. A similar result was also obtained by Chen et al. (2018), where they found overexpression of CbPMEI from Chorispora bungeana or PMEI13 (AT5G62360) from Arabidopsis decreased PME activity and the content of low-DM pectins in transgenic Arabidopsis. Furthermore, both CbPMEI-OE and PMEI13-OE lines are sensitive to freezing stress, while tolerant to salt stress. Expression analysis results showed that CbPMEI and PMEI13 mediated freezing tolerance may be independent on CBF pathway due to CBFs and CORs transcripts were not consistently declined in transgenic lines (Chen et al., 2018). However, the molecular and genetic mechanisms of PMEIs involved in abiotic stress tolerance still remain poorly understood, how the increased PMEI activity affects abiotic resistance needs to be further explored.
It is now clear that PMEIs are involved in pollen tube growth through inhibiting PME activity and hence affect cell wall stability (Rockel et al., 2008). A PMEI gene, BoPMEI, has been demonstrated as a pollen-specific gene, which is critical to pollen tube growth. Antisense expression of BoPMEIl in Arabidopsis suppressed the transcriptions of an orthologous gene, Atlgl0770, which resulted in partial male sterility and decreased seed set (Zhang et al., 2010). In B. campestris, many BcPMEIs transcripts were highly expressed in inflorescences, in particular 10 BcPMEIs transcripts were specifically expressed during flower development periods (Liu et al., 2018b). Moreover, mature transcripts of two PMEI genes, Tdpmei2.1 and Tdpmei2.2, were predominantly detected in floral organs of durum wheat, which indicated that Tdpmei2.1 and Tdpmei2.2 play key roles in flower development, in particular in anther and pollen development (Rocchi et al., 2012). Since many studies have demonstrated that PMEIs are involved in the reproductive process of plant, the roles in promoting flowering have not been reported. In the present study, both CsPMEI2-OE and CsPMEI4-OE lines exhibited early flowering phenotypes as compared to WT plants both under LD and SD conditions. Meanwhile, we found the PME activities were slightly decreased under SD condition. It is well known that the inhibition of PME activity leads to an increase in methylesterification of cell wall HGs, but the changes to cell walls were spatially regulated (Müller et al., 2013). Here, we failed to detect the methylesterification contents as the technology restriction. Instead, we found the degradation productions contents of pectins, including Suc, Glu and Fru were significantly increased both in CsPMEI2-OE and CsPMEI4-OE lines, suggesting that overexpression of CsPMEI2/4 induced the changes of carbohydrate metabolism, and hence affected reproductive process. However, the specific regulation mechanisms need to be further explored. Actually, a similar result was also found by Müller et al. (2013), where they demonstrated that overexpression of AtPMEI5 in Arabidopsis resulted in lower PME activity and higher contents of neutral sugars (i.e., Glu, fucose, and rhamnose) in cell wall, but the levels of uronic acids were not significantly changed as compared with WT plants.
In terms of flowering, at least four major pathways, including autonomous, gibberellin, photoperiod, and vernalization pathways, have been identified to participate in floral promotion (Wang et al., 2012; Jing et al., 2020). During flowering period, two flowering pathway integrators (FT and SOC1), which are commonly regulated by these four pathways, will activate two floral meristem identity genes (AP1 and LFY) to promote the formation of floral meristems (Wang et al., 2012). However, such integrators are negatively regulated by a central upstream regulator, FLC, which encodes a MADS box transcription factor and mediates the autonomous or vernalization pathways (Michaels and Amasino, 2001). The transcription accumulation of genes involved in the autonomous pathway (e.g., FCA and FY) would suppress the expression of FLC through chromatin or RNA modification (Wang et al., 2012). As expected, the transcriptions of FT, SOC1, and AP1 and some genes involved in the autonomous pathway, including FCA and FY, were accumulated, while the expression of FLC was declined both in CsPMEI2-OE and CsPMEI4-OE lines as compared to WT plants under LD and SD condition. A similar phenotype was also found in CsUGT85A53-OE transgenic Arabidopsis. The expressions of FT, SOC1, AP1, LFY, FY, and FCA were increased, while the mRNA level of FLC was repressed in CsUGT85A53-OE lines. Further research found that overexpression of CsUGT85A53 increased the DNA methylation levels of FLC, but decreased the free ABA contents via ABA glucosylation in transgenic Arabidopsis. However, the early flowering phenotype was restored by the application of exogenous ABA (Jing et al., 2020). In this study, we found the expression of an activator of FLC gene, ABI5, was also declined both in CsPMEI2-OE and CsPMEI4-OE lines. However, exogenous ABA treatment did not completely inhibit early flowering phenomenon of CsPMEI2-OE and CsPMEI4-OE lines, suggesting that the early flowering phenomenon in transgenic Arabidopsis may be partially dependent on ABA signaling pathway, and the changes of carbohydrate metabolism may also contribute to this phenomenon. Still, the detailed molecular mechanism of the roles of CsPMEI2/4 in flowering needs to be deeply explored.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WQ conceived and designed the experiments. WQ and HW wrote the original draft. HW and BL performed the experiments. YW, NL, LW, and XH sampled the materials. SH, BL, and WQ analyzed the results and performed figures. ZD and YY reviewed and edited the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (31800588), the Sub Project of Shandong Agricultural Elite Variety Project (2321401), and the School Fund Project of Qingdao Agricultural University (1118025).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.807514/full#supplementary-material
Conserved domains analysis of CsPMEIs and CsVIF/CIFs. Four conserved and representative Cys (C) residues were contained in the red boxes, respectively.
The exon–intron structures, and cis-acting elements of CsPMEIs. (A) Exon–intron structure of CsPMEIs. The coding sequence and the corresponding genomic sequence of each CsPMEI were compared by using GSDS2.0. Blue boxes represent untranslated upstream/downstream regions, green double sided wedges represent exons, and black lines indicate introns. (B) The cis-acting regulatory elements of CsPMEIs. 2,000-bp upstream non-coding region sequences of each CsPMEI gene, except for CsPMEI3/20, were used to predict cis-acting elements, and different colored blocks represent different elements.
Phenotypes, flowering time and rosette leaves numbers of CsPMEI2/4-OE lines under ABA treatment condition. (A) Phenotypes, flowering time and rosette leaves numbers of CsPMEI2-OE lines under ABA treatment condition. (a) Phenotypes of CsPMEI2-OE lines and WT plants after 31 days of growth under ABA treatment condition; (b) phenotypes of CsPMEI2-OE lines and WT plants after 41 days of growth under ABA treatment condition; (c) phenotypes of CsPMEI2-OE lines and WT plants after 51 days of growth under ABA treatment condition; (d) flowering time; (e) rosette leaves numbers. (B) Phenotypes, flowering time and rosette leaves numbers of CsPMEI4-OE lines under ABA treatment condition. (a) Phenotypes of CsPMEI4-OE lines and WT plants after 35 days of growth under ABA treatment condition; (b) phenotypes of CsPMEI4-OE lines and WT plants after 41 days of growth under ABA treatment condition; (c) phenotypes of CsPMEI4-OE lines and WT plants after 51 days of growth under ABA treatment condition; (d) flowering time; (e) rosette leaves numbers.
Phylogenetic and conserved domains analysis of INHs and PMEIs. (A) A total of 30 INHs and PMEIs were used to construct phylogenetic tree by using MEGA 7.0 software. CsVIF/CIFs highlighted with red squares and CsPMEIs are highlighted with red triangle, and different subgroups were covered with different colors. (B) Conserved domains analysis of INHs and PMEIs. The conserved ‘PKF’ motif was contained in the blue box, and the four conserved and representative Cys (C) residues were contained in the red boxes, respectively.
Nutrient solution formulation.
All sequences used to construct phylogenetic tree.
Primer sequences used in ORF of CsPMEI2/4 cloning and vector construction.
Primers information used in qRT-PCR detection.
References
- An S., Sohn K., Choi H., Hwang I., Lee S., Hwang B. (2008). Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228 61–78. 10.1007/s00425-008-0719-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Robin A., Reymond M. C., Brunoud G., Martin-Magniette M. L., Moneger F., Scutt C. P. (2020). Immediate targets of ETTIN suggest a key role for pectin methylesterase inhibitors in the control of Arabidopsis gynecium development. Plant Signal Behav. 15:1771937. 10.1080/15592324.2020.1771937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkorful E., Hu S., Zou Z., Yu Y., Chen X., Li X. (2020). Metabolomic analyses provide new insights into signaling mechanisms for nutrient uptake by lateral roots of pruned tea plant (Camellia sinensis). J. Agric. Food Chem. 68 7890–7903. 10.1021/acs.jafc.0c02053 [DOI] [PubMed] [Google Scholar]
- Baldwin L., Domon J. M., Klimek J. F., Fournet F., Sellier H., Gillet F., et al. (2014). Structural alteration of cell wall pectins accompanies pea development in response to cold. Phytochemistry 104 37–47. 10.1016/j.phytochem.2014.04.011 [DOI] [PubMed] [Google Scholar]
- Balestrieri C. (1990). A glycoprotein inhibitor of pectin methylesterase in kiwi fruit. Eur. J. Biochem. 193 183–187. 10.1111/j.1432-1033.1990.tb19321.x [DOI] [PubMed] [Google Scholar]
- Bethke G., Grundman R., Sreekanta S., Truman W., Katagiri F., Glazebrook J. (2014). Arabidopsis pectin methylesterases contribute to immunity against Pseudomonas syringae. Plant physiol. 164 1093–1107. 10.1104/pp.113.227637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camardella L., Carratore V., Ciardiello M., Servillo L., Balestrieri C., Giovane A. (2000). Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 267 4561–4565. 10.1046/j.1432-1327.2000.01510.x [DOI] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37 D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Chen X., Zhang Q., Zhang Y., Ou X., An L., et al. (2018). A cold-induced pectin methyl-esterase inhibitor gene contributes negatively to freezing tolerance but positively to salt tolerance in Arabidopsis. J. Plant Physiol. 222 67–78. 10.1016/j.jplph.2018.01.003 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (2010). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Di Matteo A., Giovane A., Raiola A., Camardella L., Bonivento D., De Lorenzo G., et al. (2005). Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17 849–858. 10.1105/tpc.104.028886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X. Y., Horvath D., Chao W., Yang Y. J., Wang X. C., Xiao B. (2014). Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L.) O. Kuntze). Int. J. Mol. Sci. 15 22155–22172. 10.3390/ijms151222155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X. Y., Tang H., Wang B., Wang L., Cao H. L., Wang Y. C., et al. (2018). Gene characterization and expression analysis reveal the importance of auxin signaling in bud dormancy regulation in tea plant. J. Plant Growth Regul. 38 225–240. 10.1007/s00344-018-9834-7 [DOI] [Google Scholar]
- Hong M., Kim D., Lee T., Jeon W., Seo Y. (2010). Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes Genet. Syst. 85 97–106. 10.1266/ggs.85.97 [DOI] [PubMed] [Google Scholar]
- Hothorn M., Van den Ende W., Lammens W., Rybin V., Scheffzek K. (2010). Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc. Natl. Acad. Sci. U.S.A. 107 17427–17432. 10.1073/pnas.1004481107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Wolf S., Aloy P., Greiner S., Scheffzek K. (2004). Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16 3437–3447. 10.2307/3872361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru T., Hirose T., Matsuda T., Akitoshi G., Kazunari T., Haruto S., et al. (2005). Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): comparison of caryopses located at different positions in a panicle. Plant Cell Physiol. 46 620–628. 10.1093/pcp/pci066 [DOI] [PubMed] [Google Scholar]
- Jing T., Zhang N., Gao T., Wu Y., Zhao M., Jin J., et al. (2020). UGT85A53 promotes flowering via mediating abscisic acid glucosylation and FLC transcription in Camellia sinensis. J. Exp. Bot. 71 7018–7029. 10.1093/jxb/eraa373 [DOI] [PubMed] [Google Scholar]
- Juge N. (2006). Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11 359–367. 10.1016/j.tplants.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Landy A. (1989). Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58 913–949. 10.1146/annurev.bi.58.070189.004405 [DOI] [PubMed] [Google Scholar]
- Lionetti V., Fabri E., De Caroli M., Hansen A. R., Willats W. G., Piro G., et al. (2017). Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to botrytis. Plant Physiol. 173 1844–1863. 10.1104/pp.16.01185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Raiola A., Camardella L., Giovane A., Obel N., Pauly M., et al. (2007). Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 143 1871–1880. 10.1104/pp.106.090803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V., Raiola A., Mattei B., Bellincampi D. (2015). The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS One 10:e0133810. 10.1371/journal.pone.0133810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Yu H., Xiong X., Yu Y., Yue X., Liu J., et al. (2018a). Genome-wide identification and characterization of pectin methylesterase inhibitor genes in Brassica oleracea. Int. J. Mol. Sci. 19:338. 10.3390/ijms19113338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Yu H., Xiong X., Yue X., Yu Y., Huang L., et al. (2018b). Genome-wide identification, molecular evolution, and expression profiling analysis of pectin methylesterase inhibitor genes in Brassica campestris ssp. chinensis. Int. J. Mol. Sci. 19:338. 10.3390/ijms19051338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. 10.1006/meth.2001 [DOI] [PubMed] [Google Scholar]
- Lu Q., Wang Y., Xiong F., Hao X., Zhang X., Li N., et al. (2020). Integrated transcriptomic and metabolomic analyses reveal the effects of callose deposition and multihormone signal transduction pathways on the tea plant-Colletotrichum camelliae interaction. Sci. Rep. 10:12858. 10.1038/s41598-020-69729-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S., Amasino R. M. (2001). Loss of Flowering locus c activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–941. 10.2307/3871350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K., Levesque-Tremblay G., Fernandes A., Wormit A., Bartels S., Usadel B., et al. (2013). Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal. Behav. 8:e26464. 10.4161/psb.26464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. P., Jeong H. Y., Jeon S. H., Kim D., Lee C. (2017). Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels. J. Plant Physiol. 208 17–25. 10.1016/j.jplph.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Nguyen H. P., Jeong H. Y., Kim H., Kim Y. C., Lee C. (2016). Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol. Biochem. 101 105–112. 10.1016/j.plaphy.2016.01.021 [DOI] [PubMed] [Google Scholar]
- Pinzon-Latorre D., Deyholos M. K. (2014). Pectinmethylesterases (PME) and pectinmethylesterase inhibitors (PMEI) enriched during phloem fiber development in flax (Linum usitatissimum). PLoS One 9:e105386. 10.1371/journal.pone.0105386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W. J., Xiao B., Wang L., Hao X. Y., Yue C., Cao H. L., et al. (2018). CsINV5, a tea vacuolar invertase gene enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 18:228. 10.1186/s12870-018-1456-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W. J., Yue C., Wang Y. C., Cao H. L., Li N. N., Wang L., et al. (2016). Identification of the invertase gene family (INVs) in tea plant and their expression analysis under abiotic stress. Plant Cell Rep. 35 2269–2283. 10.1007/s00299-016-2033-8 [DOI] [PubMed] [Google Scholar]
- Raiola A., Camardella L., Giovane A., Mattei B., De Lorenzo G., Cervone F., et al. (2004). Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors 1. FEBS Lett. 557 199–203. 10.1016/s0014-5793(03)01491-1 [DOI] [PubMed] [Google Scholar]
- Ren A., Ahmed R. I., Chen H., Han L., Sun J., Ding A., et al. (2019). Genome-wide identification, characterization and expression patterns of the pectin methylesterase inhibitor genes in Sorghum bicolor. Genes (Basel) 10:755. 10.3390/genes10100755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi V., Janni M., Bellincampi D., Giardina T., D’Ovidio R. (2012). Intron retention regulates the expression of pectin methyl esterase inhibitor (Pmei) genes during wheat growth and development. Plant Biol. 14 365–373. 10.1111/j.1438-8677.2011.00508.x [DOI] [PubMed] [Google Scholar]
- Rockel N., Wolf S., Kost B., Rausch T., Greiner S. (2008). Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 53 133–143. 10.1111/j.1365-313X.2007.03325.x [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S., Ralet M. C., Berger A., Botran L., Ropartz D., Marion-Poll A., et al. (2013). PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25 308–323. 10.1105/tpc.112.106575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecka D., Zebrowski J., Kacperska A. (2008). Are pectins involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann. Bot. 101 521–530. 10.1093/aob/mcm329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Gupta S. M., Sane A. P., Nath P. (2012). Isolation and characterization of ripening related pectin methylesterase inhibitor gene from banana fruit. Physiol. Mol. Biol. Plants 18 191–195. 10.1007/s12298-012-0102-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi C., Janni M., Lionetti V., Bellincampi D., Favaron F., D’Ovidio R. (2011). The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant Microbe In. 24 1012–1019. 10.1094/mpmi-01-11-0021 [DOI] [PubMed] [Google Scholar]
- Wang B., Jin S. H., Hu H. Q., Sun Y. G., Wang Y. W., Han P., et al. (2012). UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol. 194 666–675. 10.1111/j.1469-8137.2012.04107.x [DOI] [PubMed] [Google Scholar]
- Wang H., Ding Z., Gou M., Hu J., Wang Y., Wang L., et al. (2021). Genome-wide identification, characterization, and expression analysis of tea plant autophagy-related genes (CsARGs) demonstrates that they play diverse roles during development and under abiotic stress. BMC Genomics 22:121. 10.1186/s12864-021-07419-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yao L., Hao X., Li N., Qian W., Yue C., et al. (2018). Tea plant SWEET transporters: expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 96 577–592. 10.1007/s11103-018-0716-y [DOI] [PubMed] [Google Scholar]
- Wang L. Y., Hao X. Y., Li N. N., Wang Y. C., Ding C. Q., Lei L., et al. (2019). Transcriptional and physiological analyses reveal the association of ROS metabolism with cold tolerance in tea plant. Environ. Exp. Bot. 160:14. 10.1016/j.envexpbot.2018.11.011 [DOI] [Google Scholar]
- Wang X. C., Zhao Q. Y., Ma C. L., Zhang Z. H., Cao H. L., Kong Y. M., et al. (2013). Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:415. 10.1186/1471-2164-14-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C., Qian W. J., Li N. N., Hao X. Y., Wang L., Xiao B., et al. (2016). Metabolic changes of caffeine in tea plant (Camellia sinensis (L.) O. kuntze) as defense response to Colletotrichum fructicola. J. Agric. Food. Chem. 64 6685–6693. 10.1021/acs.jafc.6b02044 [DOI] [PubMed] [Google Scholar]
- Wei C., Yang H., Wang S., Zhao J., Liu C., Gao L., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U.S.A. 115 E4151–E4158. 10.1073/pnas.1719622115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Mouille G., Pelloux J. (2009). Homogalacturonan methyl-esterification and plant development. Mol. Plant 2 851–860. 10.1093/mp/ssp066 [DOI] [PubMed] [Google Scholar]
- Wormit A., Usadel B. (2018). The multifaceted role of pectin methylesterase inhibitors (PMEIs). Int. J. Mol. Sci. 19:878. 10.3390/ijms19102878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Mi X., Jin L., Guo R., Zhu J., Xie H., et al. (2021). CsLAZY1 mediates shoot gravitropism and branch angle in tea plants (Camellia sinensis). BMC Plant Biol. 21:243. 10.1186/s12870-021-03044-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Ren J. J., Yu Q., Wang Y. Y., Lu C. C., Kong L. J., et al. (2019). AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. 223 277–292. 10.1111/nph.15756 [DOI] [PubMed] [Google Scholar]
- Yao L. N., Ding C. Q., Hao X. Y., Zeng J. M., Yang Y. J., Wang X. C., et al. (2020). CsSWEET1a and CsSWEET17 mediate growth and freezing tolerance by promoting sugar transport across the plasma membrane. Plant Cell Physiol. 61 1669–1682. 10.1093/pcp/pcaa091 [DOI] [PubMed] [Google Scholar]
- Yu S., Li P., Zhao X., Tan M., Ahmad M. Z., Xu Y., et al. (2021). CsTCPs regulate shoot tip development and catechin biosynthesis in tea plant (Camellia sinensis). Hortic Res. 8:104. 10.1038/s41438-021-00538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Jie L., Zhao W., Du J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18 385–397. 10.1038/cr.2008.26 [DOI] [PubMed] [Google Scholar]
- Zhang G. Y., Feng J., Wu J., Wang X. (2010). BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen Tube growth. Planta 231 1323–1334. 10.1007/s00425-010-1136-7 [DOI] [PubMed] [Google Scholar]
- Zhao X., Zeng X., Lin N., Yu S., Fernie A. R., Zhao J. (2021). CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator-repressor network. Hortic Res. 8:110. 10.1038/s41438-021-00545-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Tang C., Li Q., Qiao X., Li X., Cai Y., et al. (2021). Characterization of the pectin methylesterase inhibitor gene family in rosaceae and role of PbrPMEI23/39/41 in methylesterified pectin distribution in pear pollen tube. Planta 253:118. 10.1007/s00425-021-03638-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved domains analysis of CsPMEIs and CsVIF/CIFs. Four conserved and representative Cys (C) residues were contained in the red boxes, respectively.
The exon–intron structures, and cis-acting elements of CsPMEIs. (A) Exon–intron structure of CsPMEIs. The coding sequence and the corresponding genomic sequence of each CsPMEI were compared by using GSDS2.0. Blue boxes represent untranslated upstream/downstream regions, green double sided wedges represent exons, and black lines indicate introns. (B) The cis-acting regulatory elements of CsPMEIs. 2,000-bp upstream non-coding region sequences of each CsPMEI gene, except for CsPMEI3/20, were used to predict cis-acting elements, and different colored blocks represent different elements.
Phenotypes, flowering time and rosette leaves numbers of CsPMEI2/4-OE lines under ABA treatment condition. (A) Phenotypes, flowering time and rosette leaves numbers of CsPMEI2-OE lines under ABA treatment condition. (a) Phenotypes of CsPMEI2-OE lines and WT plants after 31 days of growth under ABA treatment condition; (b) phenotypes of CsPMEI2-OE lines and WT plants after 41 days of growth under ABA treatment condition; (c) phenotypes of CsPMEI2-OE lines and WT plants after 51 days of growth under ABA treatment condition; (d) flowering time; (e) rosette leaves numbers. (B) Phenotypes, flowering time and rosette leaves numbers of CsPMEI4-OE lines under ABA treatment condition. (a) Phenotypes of CsPMEI4-OE lines and WT plants after 35 days of growth under ABA treatment condition; (b) phenotypes of CsPMEI4-OE lines and WT plants after 41 days of growth under ABA treatment condition; (c) phenotypes of CsPMEI4-OE lines and WT plants after 51 days of growth under ABA treatment condition; (d) flowering time; (e) rosette leaves numbers.
Phylogenetic and conserved domains analysis of INHs and PMEIs. (A) A total of 30 INHs and PMEIs were used to construct phylogenetic tree by using MEGA 7.0 software. CsVIF/CIFs highlighted with red squares and CsPMEIs are highlighted with red triangle, and different subgroups were covered with different colors. (B) Conserved domains analysis of INHs and PMEIs. The conserved ‘PKF’ motif was contained in the blue box, and the four conserved and representative Cys (C) residues were contained in the red boxes, respectively.
Nutrient solution formulation.
All sequences used to construct phylogenetic tree.
Primer sequences used in ORF of CsPMEI2/4 cloning and vector construction.
Primers information used in qRT-PCR detection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.