Abstract
The mu-opioid receptor (MOR) mediates the rewarding properties of many psychoactive drugs and is an important target in the treatment of addictions. Functional interactions between the opioid and endocannabinoid systems are established and have been hypothesized to contribute to the effects of cannabis. We investigated associations between three single nucleotide polymorphisms in the MOR gene OPRM1 (rs1799971, rs2281617, and rs510769) and subjective responses to smoked cannabis. Fifty-two regular cannabis users (1-4 days/week) were given a cannabis cigarette (12.5% THC) and rated their subjective responses on visual analog scales at baseline and at multiple time points after smoking. Blood samples were collected for THC quantification. There was a significant impact of the intronic variant rs510769 on subjective cannabis effects and THC blood levels. The influence of this gene variant may thus be mediated by pharmacodynamics and/or pharmacokinetic factors. We provide novel evidence that variability in OPRM1 contributes to individual responses to cannabis and may affect risk of cannabis use disorder. Our findings add to the growing body of literature on the genetic basis of individual responses to cannabis and may have implications for targeting the endogenous opioid system in the treatment of cannabis use disorder.
Keywords: Cannabis, genetics, mu opioid receptor, subjective effects
Introduction
Acute subjective effects of cannabis include euphoria, sedation, cognitive and psychomotor impairment, and sensory distortion; however self-reported experiences vary considerably between individuals [1]. Early positive reactions to cannabis have been associated with progression to heavier use, including cannabis use disorder [2-4]. There is evidence suggesting that individual subjective responses to cannabis are partially heritable and may be mediated by genetic factors [5]. However, further research is required to determine whether there is a genetic contribution to individual differences and identify which genes or genetic polymorphisms are involved.
Δ9-Tetrahydrocannabinol (THC) induces the subjective effects of cannabis primarily by the activation of CB1 receptors (CB1R) in the endocannabinoid system. Endogenous opioid signaling is also thought to contribute to the rewarding properties of cannabis, through functional interactions with the endocannabinoid and dopamine systems [6]. The mu opioid receptor (MOR), encoded by OPRM1, indirectly modulates the rewarding effects of many drug classes, including cannabinoids, through the mesolimbic dopamine system [6]. Early studies provided initial evidence of this, showing that cannabinoid-induced increases in dopamine release are blocked by MOR antagonism [7,8]. Moreover, MORs and CB1Rs co-localize within the reward system [9,10], and there is evidence suggesting that the two receptor subtypes form heterodimers and act synergistically [11].
The implication of the MOR in THC reinforcement is further supported by preclinical research. In rodent studies, the MOR antagonist naloxone fully abolishes conditioned place preference (CPP) to the cannabinoid agonist CP 55,940 [12] and reduces self-administration of the same compound [13]. Similarly, THC CPP is completely eliminated in MOR knockout mice [14]. MOR antagonism by naloxone also blocks THC-induced hyperphagia in rats [15,16]. In squirrel monkeys, pretreatment with the opioid antagonist naltrexone reduced intravenous self-administration of THC on a fixed-ratio schedule [17]. Drug-taking responses were markedly attenuated, but remained significantly elevated compared to those of animals self-administering saline. Under the same experimental conditions, pretreatment with the CB1 antagonist SR141716A completely eliminated THC self-administration and flattened the dose-response curve to vehicle-control levels [18]. The rewarding effects of THC are thus attributed to CB1 activation and are subject to modulation by endogenous opioids.
Cannabinoid-opioid interactions appear to be more complex in humans. Low doses of naltrexone can reduce intoxication to low doses of THC in regular cannabis smokers; however, this effect was not found at a higher dose of THC. Interestingly, the opposite effects were found in non-cannabis smokers, suggesting that chronic THC exposure modifies the interactions between cannabis and endogenous opioids [19]. A subsequent study found that a wide range of therapeutic naltrexone doses increased the subjective effects and self-administration of cannabis in heavy cannabis users [20]. In this population, endogenous opioids may counteract cannabinoid reinforcement rather than mediate subjective reward. However, when administered repeatedly to daily cannabis users on a maintenance schedule, naltrexone reduces cannabis intoxication and self-administration [21]. Although the nature of the relationship is not fully understood, these studies provide evidence for the implication of MOR in cannabinoid reward and abuse liability in humans.
Given the importance of the MOR in THC reward, we investigated the effect of three OPRM1 single nucleotide polymorphisms (SNPs) on subjective responses to cannabis. The most extensively studied OPRM1 variant is rs1799971, which is an A118G (Asn40Asp) substitution in exon 1. The G allele results in reduced MOR expression in vitro [22] and an increased affinity of the receptor for the endogenous opioid substrate beta-endorphin [23]. It has been investigated as a candidate gene for drug addiction, but association studies have reported inconsistent findings. The G allele has been associated with increased risk of alcohol, heroin, and general substance dependence [24-26], but some studies have reported the opposite effect [27], or shown no effect at all [28-30]. We also examined two intronic variants: rs2281617 and rs510769. Both are C/T polymorphisms located in intron 1 and have unclear effects on gene expression and protein function [31]. The rs2281617 T-allele has been associated with lower dietary fat preference and body fat mass [32], as well as reduced energy and stimulation in response to amphetamine [31]. These findings could suggest a loss of function of OPRM1 and reduced subjective reward in minor allele carriers. The rs510769 T-allele has been associated with an increased risk of heroin dependence [33], increased smoking behavior in patients undergoing methadone therapy [34], and decreased subjective responses to amphetamine [31].
To determine whether the rewarding properties of cannabis are modulated by OPRM1, we investigated the impact of SNPs on visual analogue scale (VAS) ratings of subjective drug effects in healthy regular (1-4 days/week) cannabis users.
Methods
This study was conducted in follow-up to a previous study conducted at the Centre for Addiction and Mental Health (CAMH) [35], during which participants had the option of being included in an additional genetic investigation. Those who consented provided a 20 mL blood sample from which DNA was extracted for genotyping. All study procedures were conducted in accordance with the Declaration of Helsinki and approved by the CAMH Research Ethics Board (Protocol #097-2019), and the Health Canada Research Ethics Board (Protocol #2011-0024). All participants provided written informed consent prior to participating in any study procedures.
Participants
Participants included in the study were male and female active cannabis users (using 1-4 days per week) between the ages of 19-25 years. Current cannabis use was confirmed by a positive urine toxicology screen for THC. Those who met criteria for a severe psychiatric disorder, DSM-IV cannabis dependence or any current or lifetime substance dependence (with the exception of nicotine dependence) were excluded. Participants were also excluded if they regularly used medications affecting brain function (e.g., antidepressants, stimulants, benzodiazepines), were pregnant, breastfeeding, or trying to become pregnant.
Out of the 99 participants enrolled in the original study, 70 completed the trial and consented to the genetic analysis. Fifty-two of them were randomized to the active cannabis group and were included in the present analysis.
Study procedure
Participants were required to abstain from alcohol and recreational drugs 48 hours prior to and for the duration of the study. This was verified by alcohol breathalyzer tests and point-of-care urine toxicology prior to each study session. Participants who were randomized to the active cannabis group received one cannabis cigarette with a mass of 750 mg and a potency of 12.5% THC. They were instructed to smoke ad libitum, in an externally ventilated reverse airflow room over a duration of 10 minutes. Total smoking duration was timed, and cannabis cigarettes were weighed before and after smoking. To obtain an estimate of total THC dose for each participant, the potency of the cannabis (0.125) was multiplied by the change in mass of the cigarette. Ratings of subjective drug effects were collected at baseline, 5, 15, 30 minutes and 1, 2, 3, 4, 5, 6, 24, 48 hours after cannabis administration. Participants reported the intensity of drug effects at each time point on a seven-item visual analog scale. The scale assessed ratings of “I feel a drug effect”, “I feel this high”, “I feel the drug’s good effects”, “I feel the drug’s bad effects”, “I like the drug”, “I feel a rush”, and “It feels like cannabis”. A blood sample was drawn at each data collection time point for the measurement of THC concentrations. More details regarding procedures used for blood sample collection and THC quantification can be found in our previous manuscript [35]. It should be noted that THC was measured in whole blood (typically leading to lower values as compared to plasma measurements).
Genotyping
Approximately 650,000 polymorphic sites were genotyped using the Infinium Global Screening Array (Illumina, Inc., San Diego, CA, USA) at the CAMH Biobank and Molecular Core Facility. The array data underwent standard quality control procedures as described previously [36], and genotypes were extracted for the three OPRM1 polymorphisms. The cluster plots for these three polymorphisms are shown in Supplementary Figure 1. As verified in the quality control steps, SNP genotypes did not deviate significantly from Hardy-Weinberg Equilibrium (P>5e-8). For each polymorphism, only two individuals in our sample were homozygous for the minor allele. Therefore, they were pooled with the heterozygous genotype as one group and compared against the homozygous wild type (WT) genotype. Allele frequencies are presented in Table 1.
Table 1.
Allelic frequencies of OPRM1 polymorphisms
| Polymorphism | Location | Alleles (WT/SNP) | Frequency (N) | ||
|---|---|---|---|---|---|
|
| |||||
| WT/WT | WT/SNP | SNP/SNP | |||
| rs1799971 | 154,360,797 | A/G | 29 | 21 | 2 |
| rs2281617 | 154,529,113 | C/T | 30 | 20 | 2 |
| rs510769 | 154,362,019 | C/T | 38 | 12 | 2 |
Data analysis
All data were analyzed using SPSS version 25. Differences in demographic characteristics and cannabis use between genotype groups were analyzed by independent-sample T-tests and Chi square tests. Maximum ratings and area under the curve (AUC) for each VAS item were determined, and differences between groups were compared using independent-sample t-tests.
Results
Participant characteristics
Fifty-two healthy regular cannabis users were included in the study. Demographic characteristics and cannabis use frequency did not differ between genotype groups (Table 2).
Table 2.
Participant characteristics by genotype (Mean (SD))
| rs1799971 | rs2281617 | rs510769 | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| AA (N = 29) | AG + GG (N = 23) | CC (N = 30) | CT + TT (N = 22) | CC (N = 38) | CT + TT (N = 14) | |
| Age | 22.55 (1.70) | 22.13 (2.14) | 22.43 (1.72) | 22.27 (2.16) | 22.21 (2.00) | 22.79 (1.58) |
| Sex (% female) | 24.14 | 39.13 | 36.67 | 22.72 | 34.21 | 21.43 |
| BMI | 25.33 (5.50) | 23.63 (2.89) | 25.54 (5.30) | 23.25 (2.99) | 25.02 (4.98) | 23.48 (3.31) |
| Cannabis use (times per week) | 2.55 (0.93) | 2.52 (0.83) | 2.65 (0.97) | 2.39 (0.74) | 2.57 (0.89) | 2.46 (0.89) |
rs510769 and subjective drug effects
rs510769 C/T and T/T genotypes reported significantly higher maximum VAS ratings of “Effect” t(48.44) = -2.15, P = 0.037, “Good” t(47.59) = -3.28, P = 0.002, “Liking” t(48.11) = -2.17, P = 0.035, “Rush” t(36.98) = -2.85, P = 0.007 and “Feels like cannabis” t(48.99) = -3.55, P = 0.001, compared to the C/C genotype. Maximum ratings of “High” and “Bad” drug effects were not statistically different between the groups (Figure 1A).
Figure 1.
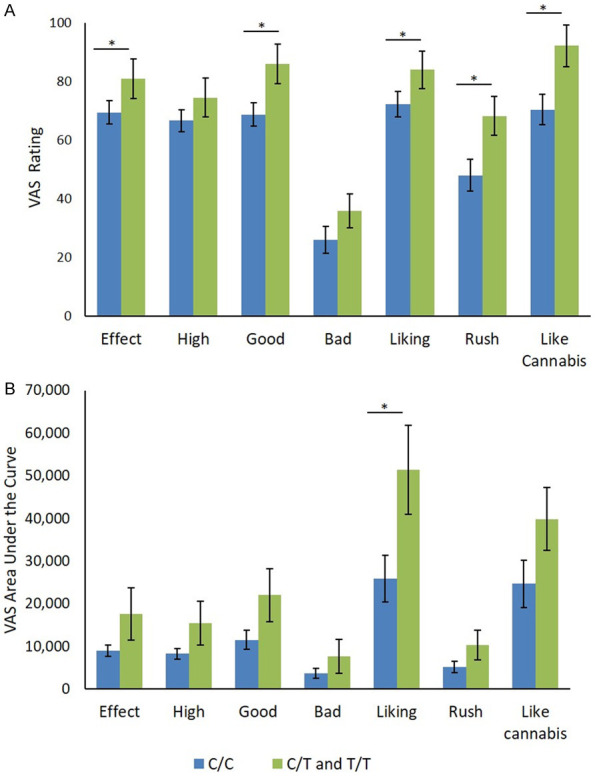
Effects of rs510769 genotype on subjective responses to cannabis measured by visual analog scales. Data expressed as mean ± SEM of (A) maximum VAS ratings by rs510769 genotype (C/C: N = 38; C/T and T/T: N = 13) and (B) area under the curve of VAS ratings over time (C/C: N = 34; C/T and T/T: N = 11). C/C genotype compared to C/T and T/T genotypes using independent sample t-tests. *P<0.05.
Mean area under the curve (AUC) of “Liking” was significantly elevated in rs510769 T-allele carriers compared to C allele homozygotes t(43) = -2.25, P = 0.029. AUC of all other VAS items did not differ significantly between groups (Figure 1B).
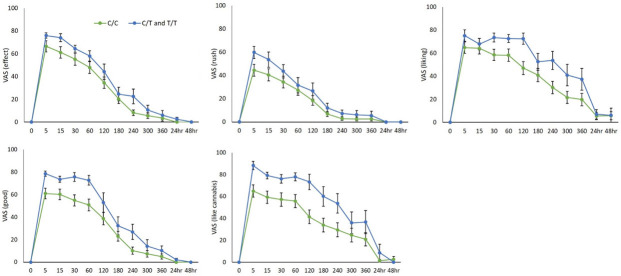
Mean ratings over time of significant VAS items are presented in Figure 2.
Figure 2.
Effects of rs510769 genotype on subjective responses to cannabis over time (minutes or hours) for individual VAS items “Effect”, “Rush”, “Liking”, “Good”, and “Like Cannabis”. Data expressed as mean ± SEM (C/C: n = 38; C/T and T/T: n = 13).
rs510769 and THC pharmacokinetics
The mean estimated THC dose was 81.64 mg (SD = 23.06) in the C/C group and 85.18 mg (SD = 17.70) in the C/T and T/T group. This was not a statistically significant difference (t(46) = -0.50, P = 0.619).
Blood THC pharmacokinetics for both groups over time are presented in Figure 3. Repeated measures ANOVA of blood concentration over time revealed no statistically significant effect of genotype (F(1,42) = 2.35, P = 0.133), or time by genotype interaction (F(1.07,44.96) = 2.59 P = 0.113). THC concentrations peaked at five minutes and decreased over time in both groups. There was no significant difference in the area under the THC concentration/time curve between groups (C/C: M = 22.55, SD = 21.52, C/T and T/T: M = 36.05, SD = 27.99); (t(44) = -1.73, P = 0.091). The maximum blood THC concentration was significantly elevated in T-allele carriers (M = 56.17, SD = 34.46) compared to C/C individuals (M = 34.04, SD = 28.83); t(44) = -2.17, P = 0.035.
Figure 3.
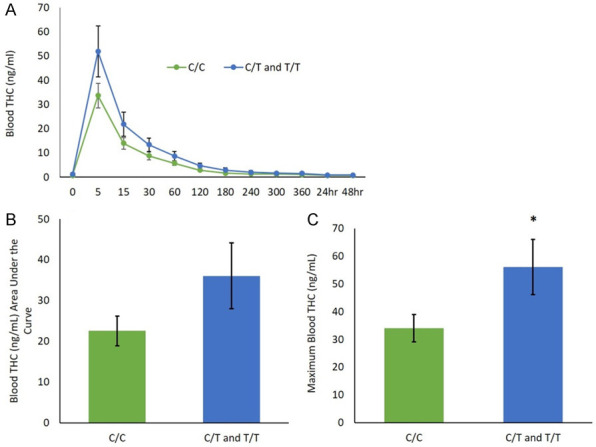
Effect of rs510769 genotype on THC pharmacokinetics. Mean ± SEM of (A) blood THC concentration (ng/ml) over time (minutes or hours) analyzed across genotypes by repeated measures ANOVA, (B) blood THC area under the curve using independent sample t-test, (C) maximum blood THC concentration (ng/ml) using independent sample t-test. C/C genotype compared to C/T and T/T genotypes (C/C: N = 34, C/T and T/T N = 12). *P<0.05.
rs2281617 and rs1799971
No significant associations were found between rs2281617 or rs1799971 and any VAS measures (Table 3).
Table 3.
Maximum VAS scores over time by rs2281617 and rs1799971 genotype groups (Mean (SEM))
| rs1799971 | rs2281617 | |||
|---|---|---|---|---|
|
|
|
|||
| AA (N = 28) | AG + GG (N = 23) | CC (N = 29) | CT + TT (N = 22) | |
| Effect | 74.79 (3.89) | 69.61 (6.67) | 75.17 (4.19) | 68.86 (6.51) |
| High | 70.07 (3.68) | 67.04 (6.65) | 71.24 (4.18) | 65.36 (6.27) |
| Good | 76.75 (3.94) | 68.78 (6.78) | 77.00 (4.35) | 68.09 (6.49) |
| Bad | 28.71 (4.57) | 28.30 (5.79) | 27.03 (4.77) | 30.50 (5.53) |
| Liking | 79.11 (4.35) | 70.74 (6.36) | 79.69 (4.28) | 69.59 (6.49) |
| Rush | 54.18 (5.37) | 52.13 (6.59) | 53.79 (5.46) | 52.55 (6.51) |
| Like Cannabis | 79.78 (5.11) | 71.43 (7.11) | 77.59 (5.38) | 73.95 (6.99) |
Discussion
We provide preliminary evidence that OPRM1 contributes to the variability in subjective responses to smoked cannabis. Out of the three investigated SNPs, the rs510769 T-allele was associated with increased positive responses to cannabis and higher blood THC levels compared to C-allele homozygotes. Genotypes at rs1799971 and rs2281617 had no significant effect on subjective ratings. Variation in OPRM1 has previously been shown to affect responses to alcohol, opioids, and amphetamine [31-33], and may also affect substance dependence liability [25-27]. Our results show an effect of the intronic variant rs510769. T- allele carriers reported significantly higher maximum VAS ratings of “Effect”, “Good”, “Liking”, “Rush”, and “Feels like cannabis”, suggesting increased sensitivity to the drug’s positive effects. This SNP may have a regulatory effect on receptor expression in the brain [37] and has previously been associated with reduced OPRM1 expression in the cerebellum [38]. The C/T and T/T group also had significantly higher maximum levels of blood THC concentrations, indicating that the increased drug effects may be due to pharmacokinetic differences between genotypes. This SNP was previously shown to have the opposite effect on subjective responses to amphetamine, to which T-allele carriers reported reduced euphoria and stimulation [31]. Further research is required to determine how this SNP affects opioid receptor function and expression, THC pharmacokinetics, and the mechanisms by which it affects subjective responses to different drugs.
We found no association between rs1799971 and ratings of subjective effects. This functional polymorphism leads to a change in MOR function, which has unclear effects on intoxication and risk of addiction to different substances. The G allele has been associated with increased alcohol intoxication [39] and found to have no effect on responses to amphetamine [31]. One study that associated the G allele with increased risk of dependence to four pooled substances found no effect on cannabis dependence alone; however, this may have resulted from limited sample size [26]. Due to the small number of G allele homozygotes in our study (N = 2), A/G and G/G genotypes were combined into one group and compared to A allele homozygotes. Interestingly, both G/G individuals in our sample had peak ratings of “Effect” and “Good” over 25% higher than those reported by A/A individuals. Previous studies have associated the G/G genotype with dosage and response to opioid analgesics compared to A/A and A/G genotypes [40-42].
rs2281617 did not affect subjective responses to cannabis. It has been investigated by very few studies which have indicated possible reduced subjective reward in minor allele carriers [31,32]. It is a non-coding SNP with unknown consequences on gene or protein expression. Further research is required to characterize its function and determine its effects on reward processing.
Our findings may have implications for the therapeutic use of naltrexone in cannabis use disorder (CUD). MOR antagonism by naltrexone results in the blunting of rewarding drug effects and a reduction in cravings, thereby reducing drug use and rates of relapse [43]. It is one of the most effective pharmacologic treatments for alcohol use disorder. Polymorphisms in OPRM1 affect subjective alcohol intoxication, the effectiveness of naltrexone in blocking subjective effects of alcohol, and rates of relapse after treatment [43-45]. Our findings may suggest a similar implication of the gene in the treatment of CUD. In addition to affecting therapeutic outcomes, OPRM1 may contribute to the initial development of cannabis dependence. An increased risk of problematic cannabis use has been reported in users with stronger positive reactions to the drug [2-4]. Based on our findings, it is possible that rs510769 T-allele carriers are more susceptible to developing CUD. The association between this SNP and CUD as well as potential prevention strategies require further study.
The results of this study should be interpreted in light of certain limitations. Although demographic characteristics and cannabis use frequency did not vary between genotype groups, our findings should be confirmed in a larger sample, using an adjusted statistical model to control for potential confounding variables. Participant ancestry would be an important variable to control for, as minor allele frequencies for rs1799971 and rs2281617 differ considerably between ethnic groups [46-48]. Importantly, this limitation may have led to false negative results, especially for rs1799971 [48]. In addition, we did not correct for multiple comparisons in our statistical analyses and cannot exclude the possibility of type I error. A larger sample would allow the detection of potential effects with greater statistical power and an adjusted significance level. Despite limitations related to small sample size, our results provide novel, preliminary evidence for the possibility that variation in OPRM1 contributes to differences in subjective responses to smoked cannabis.
Acknowledgements
The CADRI study was supported by a Canadian Institutes of Health Research operating grant (FRN114939). Additional support for the work was provided by the Canada Foundation for Innovation. The authors would like to acknowledge Aurora Cannabis Enterprises Inc. for supplying the active cannabis and the National Institute of Drug Abuse (NIDA) Drug Supply Program for providing the placebo cannabis for the CADRI study. The authors declare no conflict of interest. Dr. Le Foll has obtained funding from Pfizer (GRAND Awards, including salary support) for investigator-initiated projects. Dr. Le Foll has some in-kind donation of cannabis product from Aurora and medication donation from Pfizer and Bioprojet and was provided a coil for TMS study from Brainsway. Dr. Le Foll has obtained industry funding from Canopy (through research grants handled by CAMH or University of Toronto), Bioprojet, ACS and Alkermes. Dr. Le Foll has received in kind donations of nabiximols from GW Pharma for past studies funded by CIHR and NIH. He has been consultant for Shionogi. He is supported by CAMH and a clinician-scientist award from the department of Family and Community Medicine of the University of Toronto and an Addiction Psychiatry Chair from the department of Psychiatry of the University of Toronto. Dr Le Foll also participated in an advisory board meeting for Indivior and got a grant from Indivior for a clinical trial.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Curran HV, Freeman TP, Mokrysz C, Lewis DA, Morgan CJ, Parsons LH. Keep off the grass? Cannabis, cognition and addiction. Nat Rev Neurosci. 2016;17:293–306. doi: 10.1038/nrn.2016.28. [DOI] [PubMed] [Google Scholar]
- 2.Scherrer JF, Grant JD, Duncan AE, Sator CE, Haber JR, Jacob T, Buchloz KK. Subjective effects to cannabis are associated with use, abuse, and dependence after adjusting for genetic and environmental influences. Drug Alcohol Depend. 2009;105:76–82. doi: 10.1016/j.drugalcdep.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeiger JS, Haberstick BC, Corley RP, Ehringer MA, Crowley TJ, Hewitt JK, Hopfer CJ, Stallings MC, Young SE, Rhee SH. Subjective effects to marijuana associated with marijuana use in community and clinical subjects. Drug Alcohol Depend. 2010;109:161–166. doi: 10.1016/j.drugalcdep.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fergusson DM, Horwood LJ, Lynskey MT, Madden PA. Early reactions to cannabis predict later dependence. Arch Gen Psychiatry. 2003;60:1033–1039. doi: 10.1001/archpsyc.60.10.1033. [DOI] [PubMed] [Google Scholar]
- 5.Lyons MJ, Toomey R, Meyer JM, Green AI, Eisen SA, Goldberg J, True WR, Tsuang MT. How do genes influence marijuana use? The role of subjective effects. Addiction. 1997;94:409–417. [PubMed] [Google Scholar]
- 6.Wenzel JM, Cheer JF. Endocannabinoid regulation of reward and reinforcement through interaction with dopamine and endogenous opioid signaling. Neuropsychopharmacology. 2018;43:103–115. doi: 10.1038/npp.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JP, Paredes W, Li J, Smith D, Lowinson J, Gardner EL. Delta 9-tetrahydrocannabinol produces naloxone-blockable enhancement of presynaptic basal dopamine efflux in nucleus accumbens of conscious, freely-moving rats as measured by intracerebral microdialysis. Psychopharmacology (Berl) 1990;102:156–162. doi: 10.1007/BF02245916. [DOI] [PubMed] [Google Scholar]
- 8.Tanda G, Pontieri FE, Di Chiara G. Cannabinoid and heroin activation of mesolimbic dopamine transmission by a common mu1 opioid receptor mechanism. Science. 1997;276:2048–2050. doi: 10.1126/science.276.5321.2048. [DOI] [PubMed] [Google Scholar]
- 9.Salio C, Fischer J, Franzoni MF, Mackie K, Kaneko T, Conrath M. CB1-cannabinoid and mu-opioid receptor co-localization on postsynaptic target in the rat dorsal horn. Neuroreport. 2001;12:3689–3692. doi: 10.1097/00001756-200112040-00017. [DOI] [PubMed] [Google Scholar]
- 10.Pickel VM, Chan J, Kash TL, Rodríguez JJ, MacKie K. Compartment-specific localization of cannabinoid 1 (CB1) and mu-opioid receptors in rat nucleus accumbens. Neuroscience. 2004;127:101–12. doi: 10.1016/j.neuroscience.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Schoffelmeer AN, Hogenboom F, Wardeh G, De Vries TJ. Interactions between CB1 cannabinoid and mu opioid receptors mediating inhibition of neurotransmitter release in rat nucleus accumbens core. Neuropharmacology. 2006;51:773–781. doi: 10.1016/j.neuropharm.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Braida D, Pozzi M, Cavallini R, Sala M. Conditioned place preference induced by the cannabinoid agonist CP 55,940: interaction with the opioid system. Neuroscience. 2001;104:923–926. doi: 10.1016/s0306-4522(01)00210-x. [DOI] [PubMed] [Google Scholar]
- 13.Braida D, Pozzi M, Parolaro D, Sala M. Intracerebral self-administration of the cannabinoid receptor agonist CP 55,940 in the rat: interaction with the opioid system. Eur J Pharmacol. 2001;413:227–234. doi: 10.1016/s0014-2999(01)00766-x. [DOI] [PubMed] [Google Scholar]
- 14.Ghozland S, Matthes HW, Simonin F, Filliol D, Kieffer BL, Maldonado R. Maldonado R. Motivational effects of cannabinoids are mediated by mu-opioid and kappa-opioid receptors. J Neurosci. 2002;22:1146–1154. doi: 10.1523/JNEUROSCI.22-03-01146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solinas M, Goldberg SR. Motivational effects of cannabinoids and opioids on food reinforcement depend on simultaneous activation of cannabinoid and opioid systems. Neuropsychopharmacology. 2005;30:2035–2045. doi: 10.1038/sj.npp.1300720. [DOI] [PubMed] [Google Scholar]
- 16.Williams CM, Kirkham TC. Reversal of delta 9-THC hyperphagia by SR141716 and naloxone but not dexfenfluramine. Pharmacol Biochem Behav. 2002;21:333–340. doi: 10.1016/s0091-3057(01)00694-3. [DOI] [PubMed] [Google Scholar]
- 17.Justinova Z, Tanda G, Munzar P, Goldberg SR. The opioid antagonist naltrexone reduces the reinforcing effects of Delta 9 tetrahydrocannabinol (THC) in squirrel monkeys. Psychopharmacology. 2004;173:186–194. doi: 10.1007/s00213-003-1693-6. [DOI] [PubMed] [Google Scholar]
- 18.Tanda G, Munzar P, Goldberg SR. Self-administration behavior is maintained by the psychoactive ingredient of marijuana in squirrel monkeys. Nat Neurosci. 2000;3:1073–4. doi: 10.1038/80577. [DOI] [PubMed] [Google Scholar]
- 19.Haney M. Opioid antagonism of cannabinoid effects: differences between marijuana smokers and nonmarijuana smokers. Neuropsychopharmacology. 2007;32:1391–1403. doi: 10.1038/sj.npp.1301243. [DOI] [PubMed] [Google Scholar]
- 20.Cooper ZD, Haney M. Opioid antagonism enhances marijuana’s effects in heavy marijuana smokers. Psychopharmacology. 2010;211:141–148. doi: 10.1007/s00213-010-1875-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haney M, Ramesh D, Glass A, Pavlicova M, Bedi G, Cooper ZD. Naltrexone maintenance decreases cannabis self-administration and subjective effects in daily cannabis smokers. Neuropsychopharmacology. 2015;40:2489–2498. doi: 10.1038/npp.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang D, Johnson AD, Papp AC, Sadée W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 23.Bond CK, LaForge S, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters β-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miranda R, Ray L, Justus A, Meyerson LA, Knopik VS, McGeary J, Monti PM. Initial evidence of an association between OPRM1 and adolescent alcohol misuse. Alcohol Clin Exp Res. 2010;34:112–122. doi: 10.1111/j.1530-0277.2009.01073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Türkan H, Karahalil B, Kadıoğlu E, Eren K, Gürol DT, Karakaya AE. The association between the OPRM1 A118G polymorphism and addiction in a Turkish population. Arh Hig Rada Toksikol. 2019;70:97–103. doi: 10.2478/aiht-2019-70-3153. [DOI] [PubMed] [Google Scholar]
- 27.Schwantes-An TH, Zhang J, Chen LS, Hartz SM, Culverhouse RC, Chen X, Coon H, Frank J, Kamens HM, Konte B, Kovanen L, Latvala A, Legrand LN, Maher BS, Melroy WE, Nelson EC, Reid MW, Robinson JD, Shen PH, Yang BZ, Andrews JA, Aveyard P, Beltcheva O, Brown SA, Cannon DS, Cichon S, Corley RP, Dahmen N, Degenhardt L, Foroud T, Gaebel W, Giegling I, Glatt SJ, Grucza RA, Hardin J, Hartmann AM, Heath AC, Herms S, Hodgkinson CA, Hoffmann P, Hops H, Huizinga D, Ising M, Johnson EO, Johnstone E, Kaneva RP, Kendler KS, Kiefer F, Kranzler HR, Krauter KS, Levran O, Lucae S, Lynskey MT, Maier W, Mann K, Martin NG, Mattheisen M, Montgomery GW, Müller-Myhsok B, Murphy MF, Neale MC, Nikolov MA, Nishita D, Nöthen MM, Nurnberger J, Partonen T, Pergadia ML, Reynolds M, Ridinger M, Rose RJ, Rouvinen-Lagerström N, Scherbaum N, Schmäl C, Soyka M, Stallings MC, Steffens M, Treutlein J, Tsuang M, Wall TL, Wodarz N, Yuferov V, Zill P, Bergen AW, Chen J, Cinciripini PM, Edenberg HJ, Ehringer MA, Ferrell RE, Gelernter J, Goldman D, Hewitt JK, Hopfer CJ, Iacono WG, Kaprio J, Kreek MJ, Kremensky IM, Madden PA, McGue M, Munafò MR, Philibert RA, Rietschel M, Roy A, Rujescu D, Saarikoski ST, Swan GE, Todorov AA, Vanyukov MM, Weiss RB, Bierut LJ, Saccone NL. Association of the OPRM1 variant rs1799971 (A118G) with non-specific liability to substance dependence in a collaborative de novo meta-analysis of european-ancestry cohorts. Behav Genet. 2016;46:151–169. doi: 10.1007/s10519-015-9737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergen AW, Kokoszka J, Peterson R, Long JC, Virkkunen M, Linnoila M, Goldman D. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- 29.Glatt SJ, Bousman C, Wang RS, Murthy KK, Rana BK, Lasky-Su JA, Zhu SC, Zhang R, Li J, Zhang B, Li J, Lyons MJ, Faraone SV, Tsuang MT. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franke P, Wang T, Nöthen MM, Knapp M, Neidt H, Albrecht S, Jahnes E, Propping P, Maier W. Nonreplication of association between mu-opioid-receptor gene (OPRM1) A118G polymorphism and substance dependence. Am J Med Genet. 2001;105:114–119. [PubMed] [Google Scholar]
- 31.Dlugos AM, Hamidovic A, Hodgkinson C, Shen PH, Goldman D, Palmer AA, de Wit H. OPRM1 gene variants modulate amphetamine-induced euphoria in humans. Genes Brain Behav. 2011;10:199–209. doi: 10.1111/j.1601-183X.2010.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haghighi A, Melka MG, Bernard M, Abrahamowicz M, Leonard GT, Richer L, Perron M, Veillette S, Xu CJ, Greenwood CM, Dias A, El-Sohemy A, Gaudet D, Paus T, Pausova Z. Opioid receptor mu 1 gene, fat intake and obesity in adolescence. Mol Psychiatry. 2014;19:63–68. doi: 10.1038/mp.2012.179. [DOI] [PubMed] [Google Scholar]
- 33.Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, Adelson M, Kreek MJ. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YT, Tsou HH, Kuo HW, Fang CP, Wang SC, Ho IK, Chang YS, Chen CH, Hsiao CF, Wu HY, Lin KM, Chen A, Tsai-Wu JJ, Liu YL. OPRM1 genetic polymorphisms are associated with the plasma nicotine metabolite cotinine concentration in methadone maintenance patients: a cross sectional study. J Hum Genet. 2013;58:84–90. doi: 10.1038/jhg.2012.139. [DOI] [PubMed] [Google Scholar]
- 35.Brands B, Mann RE, Wickens CM, Sproule B, Stoduto G, Sayer GS, Burston J, Pan JF, Matheson J, Stefan C, George TP, Huestis MA, Rehm J, Le Foll B. Acute and residual effects of smoked cannabis: impact on driving speed and lateral control, heart rate, and self-reported drug effects. Drug Alcohol Depend. 2019;205:107641. doi: 10.1016/j.drugalcdep.2019.107641. [DOI] [PubMed] [Google Scholar]
- 36.Zai CC, Fabbri C, Hosang GM, Zhang RS, Koyama E, de Luca V, Tiwari AK, King N, Strauss J, Jones I, Jones L, Breen G, Farmer AE, McGuffin P, Vincent JB, Kennedy JL, Lewis CM. Genome-wide association study of suicidal behaviour severity in mood disorders. World J Biol Psychiatry. 2021;22:722–731. doi: 10.1080/15622975.2021.1907711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roadmap Epigenetics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28:1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Zhang L, Zhao X, Shen S, Luo X, Zhang Y. Association between MDR1/CYP3A4/OPRM1 gene polymorphisms and the post-caesarean fentanyl analgesic effect on Chinese women. Gene. 2018;661:78–84. doi: 10.1016/j.gene.2018.03.081. [DOI] [PubMed] [Google Scholar]
- 41.Walter C, Lötsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain. 2009;146:270–275. doi: 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Lee SH, Kim JD, Park SA, Oh CS, Kim SH. Effects of μ-opioid receptor gene polymorphism on postoperative nausea and vomiting in patients undergoing general anesthesia with remifentanil: double blinded randomized trial. J Korean Med Sci. 2015;30:651–657. doi: 10.3346/jkms.2015.30.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64:1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- 44.Schacht JP, Randall PK, Latham PK, Voronin KE, Book SW, Myrick H, Anton RF. Predictors of naltrexone response in a randomized trial: reward-related brain activation, OPRM1 genotype, and smoking status. Neuropsychopharmacology. 2017;42:2640–2653. doi: 10.1038/npp.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anton RF. Naltrexone for the management of alcohol dependence. N Engl J Med. 2008;359:715–721. doi: 10.1056/NEJMct0801733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D, Liu L, Xiao Y, Peng Y, Yang C, Wang Z. Ethnic-specific meta-analyses of association between the OPRM1 A118G polymorphism and alcohol dependence among Asians and Caucasians. Drug Alcohol Depend. 2012;123:1–6. doi: 10.1016/j.drugalcdep.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 47.The 1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levran O, Kreek MJ. Population-specific genetic background for the OPRM1 variant rs1799971 (118A>G): implications for genomic medicine and functional analysis. Mol Psychiatry. 2021;26:3169–3177. doi: 10.1038/s41380-020-00902-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.