Abstract
At least 11 Stx2 variants produced by Shiga toxin-producing Escherichia coli (STEC) isolated from patients and animals have been described. The Stx2 subtyping of STEC isolated from healthy cows positive for stx2 (n = 104) or stx2 and stx1 (n = 63) was investigated. Stx2vh-b, Stx2 (renamed Stx2-EDL933), and Stx2vh-a were the subtypes mostly detected among the bovine isolates (39.5, 39, and 25.5%, respectively). Stx2e was not present, and subtypes included in the Stx2d group (Stx2d-OX3a, Stx2d-O111, and Stx2d-Ount) were found infrequently among the isolates examined (8.5%). A combination of two distinct Stx2 subtypes was observed among 23.5% of the strains. For the first time, a combination of three subtypes (Stx2-EDL933/Stx2vh-b/Stx2d and Stx2vh-a/Stx2vh-b/Stx2d) was detected (3.5% of the isolates). In addition, bovine STEC harboring stx1 and one or two stx2 genes appeared highly cytotoxic toward Vero cells. A new Stx2 subtype (Stx2-NV206), present among 14.5% of the isolates, showed high cytotoxicity for Vero cells. Two amino acid residues (Ser-291 and Glu-297) important for the activation of Stx2 by human intestinal mucus were conserved on the Stx2-NV206 A subunit. The gene encoding Ehx enterohemolysin was prominent among STEC harboring stx2-EDL933 alone (78%) or a combination of stx2-EDL933 and stx2vh-b (85%). In addition, Stx2-EDL933 and/or Stx2vh-b subtypes were highly associated with other putative virulence factors such as Stx1 and EspP extracellular serine protease, but not with EAST1 enterotoxin.
In humans, Shiga toxin (or verocytotoxin)-producing Escherichia coli (STEC) are associated with sporadic cases and outbreaks of watery diarrhea, hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS) (9, 17). Shiga toxins (Stx) are the major pathogenicity factors of STEC and are responsible for the principal manifestations of HC and HUS (9, 17). Genes located in the genome of temperate bacteriophages encode two distinct Stx. The Stx1 and Stx2 toxins possess similar biological activities, including cytotoxicity to Vero and HeLa cells, but are immunologically distinct. Both toxins are composed of an enzymatically active A subunit and a pentameric B subunit (9, 17). The B subunits form a hollow ring and mediate binding to functional receptors. The toxin molecules are then internalized, and the A subunit is able to inhibit the peptide chain elongation during protein synthesis, leading to eukaryotic cell death (9, 17).
Several Stx2 subtypes have been identified on the basis of sequence homology and immunological cross-reactivity. Variations in the Stx2 amino acid sequence have a direct impact on the capacity of a given STEC to cause disease in mice, suggesting that variations may result in Shiga-like toxins with different properties (13). To date, at least 11 different Stx2 subtypes produced by STEC strains from patients with HUS, abdominal cramps, sudden infant death syndrome, or diarrhea and from animals with diarrhea or edema disease have been described (6, 7, 13–16, 18, 22, 26).
Several other virulence factors involved in the pathogenicity of STEC have been described. The intimin encoded by the eae gene located in the chromosomal locus of enterocyte effacement is involved in the intimate attachment of bacteria to enterocytes (23). However, some STEC involved in severe disease do not contain the genetic information encoding intimin. Plasmid-encoded virulence factors are also probably involved in the pathogenicity of STEC (9). Among them, enterohemolysin (Ehx) acts as a pore-forming cytolysin on eukaryotic cells (21), the EspP extracellular serine protease can cleave human coagulation factor V (3), and the EAST1 enterotoxin may contribute to the pathogenesis of watery diarrhea often observed during the early stages of STEC infections (20).
Stx-producing E. coli strains involved in food-borne infections have been studied extensively (9, 17). Human infections are usually a consequence of the consumption of contaminated bovine meat (17). However, although Stx-producing strains are frequently found in the fecal flora of healthy cattle, little is known about the virulence factors of bovine STEC. Recently, a 1-year survey has been undertaken by Pradel et al. to determine the frequency of STEC isolated from the intestinal tract of healthy cows at the city slaughterhouse (19). In this report, the bovine STEC included in Pradel's collection were screened to analyze the frequency of Stx2 subtypes and the putative combination with other virulence genes associated with STEC involved in human infections. The cytotoxity of Stx-producing strains was analyzed according to the Stx2 subtype, and a new Stx2 variant untypeable by the different protocols tested was characterized.
MATERIALS AND METHODS
Bacterial strains.
The 185 STEC strains used in this study are part of a well-characterized bacterial collection obtained during a 1-year prospective study in the same geographic area (19). Bacterial strains isolated from fecal samples from healthy cattle were found to be positive for the presence of stx2 (n = 104), stx1 (n = 18), or stx1 and stx2 (n = 63) by PCR and Southern hybridization (19).
Strains EDL933 (isolated from contaminated meat), Fac9 (isolated from a case of edema disease in swine), and OX3:H11 (isolated from a case of sudden infant death syndrome) were used as positive controls for Stx2, Stx2e, and Stx2d variants, respectively (Table 1). E. coli strain B2F1 isolated from a patient with HUS was used as a positive control for Stx2vh-a and Stx2vh-b variants (Table 1). Strain DH5α was used as a negative control. The reference strains were kindly provided by Eric Oswald (Institut National de la Recherche Agronomique, Toulouse, France), John Fairbrother (GREMIP, Saint-Hyacinthe, Canada), and Francine Grimont (Institut Pasteur, Paris, France).
TABLE 1.
STEC reference strainsa
| Variant | Serotype | Strain | Associated syndrome or origin | Accession no. | Reference |
|---|---|---|---|---|---|
| Stx2 (Stx2-EDL933) | O157:H7 | EDL933 | HUS | Y10775 | 4 |
| Stx2c | O157:H− | E32511 | HUS | M59432 | 22 |
| Stx2-O113 | O113:H21 | 98NK2 | HUS | Not present in data banks | 16 |
| Stx2-O48 | O48:H21 | 94C | HUS | Z37725 | 13 |
| Stx2vh-a | O91:H21 | B2F1 | HUS | Not present in data banks | 6 |
| Stx2vh-b | O91:H21 | B2F1 | HUS | Not present in data banks | 6 |
| Stx2d-OX3a | OX3:H21 | O31 | Suddent infant death | X65949 | 14 |
| Stx2-OX3b | OX3:H21 | O31 | Suddent infant death | L11079 | 15 |
| Stx2d-O111 | O111:H− | PH | HUS | L11078 | 15 |
| Stx2d-Ount | Ount:H21 | EH250 | Abdominal cramps | AF043627 | 18 |
| Stx2e | O139:H1 | 412 | Porcine edema disease | M21534 | 26 |
| Stx2-NV206 | O6:H10 | NV206 | Healthy cow | AF329817 | This study |
The Stx2 subtype produced by the EDL933 reference strain is renamed Stx2-EDL933 in this report. The Stx2-O113, Stx2-O48, Stx2-OX3, and Stx2-O111 subtypes are named according to the O serogroup of the reference STEC strains.
Stx2 subtyping by PCR and RFLP-PCR.
A restriction fragment length polymorphism (RFLP)-PCR system using the VT2c-VT2d primer pair was used to discriminate the genes coding for the Stx2, Stx2vh-a, and Stx2vh-b subtypes (25). Restriction patterns were obtained after digestion of the amplified products by HaeIII, RsaI, and NciI restriction endonucleases (25). The PCR method described by Piérard et al. used the VT2cm-VT2f primer pair to amplify a 256-bp DNA fragment specific to the Stx2d group (Stx2d-Ount, Stx2d-OX3a, and Stx2d-O111) (18). Genotypic detection of the Stx2e variant was performed by amplification of a 230-bp DNA fragment using the VTe-a–VTe-b primer pair described previously (8). Genetic detection of the EAST1 toxin was performed by PCR using primers east1-1a and east1-1b, able to amplify a specific 111-bp DNA fragment (27).
DNA to be amplified was released from whole organisms by boiling. Bacteria were harvested from 1 ml of an overnight Luria-Bertani (LB) broth culture, suspended in 1 ml of sterile water, and incubated at 100°C for 15 min. AmpliTaq DNA polymerase and enzyme buffer were purchased from Appligene-Oncor (Illkirch, France). The PCR procedures were performed on a GeneAmp PCR system 2400 (Perkin-Elmer). DNA extracts from the DH5α recipient strain and appropriate positive controls were included in each PCR run. The PCR products obtained were subjected to electrophoresis on a 1.5% agarose gel and visualized by ethidium bromide staining.
Detection of espP by colony blot hybridization.
A DNA probe amplified using primers espP-A (CACACGGAGCTTATAATATTCTGTCA) and espP-B (AATGTTATCCCATTGACATCATTTGACT) (3) from reference strain EDL933 was used in colony blot hybridization for detection of the espP gene. The probe (1,830 bp) was purified and radiolabeled with [α-32P]dCTP using the random-primed DNA labeling kit (Boehringer-Mannheim, France) according to the manufacturer's instructions. Colony blot hybridization was performed as previously described (19).
Nucleotide sequencing.
The operon coding for the untypeable Stx2 toxin of E. coli NV206 was amplified using two oligonucleotide primers located downstream and upstream of the genes encoding the A and B subunits, respectively (15). The amplified product (approximately 1,500 bp) was purified by using the Prep-A-Gene purification system (Bio-Rad). The nucleotide sequence was determined with double-stranded DNA by the dideoxy chain termination method using a model 373A automatic DNA sequencer (Applied Biosystem Inc.).
Nucleotide sequence accession number.
The nucleotide sequence of the Stx2 toxin of E. coli NV206 (Stx2-NV206) has been submitted to the GenBank database under accession number AF329817.
RESULTS
Subtyping of Stx2 toxins.
To data, at least 11 Stx2 subtypes have been described (Table 1). For better comprehension, the Stx2 subtype produced by reference strain EDL933 is named Stx2-EDL933 in this report, and some of the Stx2 subtypes are named according to the O serogroup of the reference E. coli strains (O113, O48, OX3, and O111) (Table 1). Genotypic methods were used for the Stx2 subtyping of 167 bovine STEC strains positive for stx2 (n = 104) or stx2 and stx1 (n = 63) (Fig. 1). Results are summarized in Table 2.
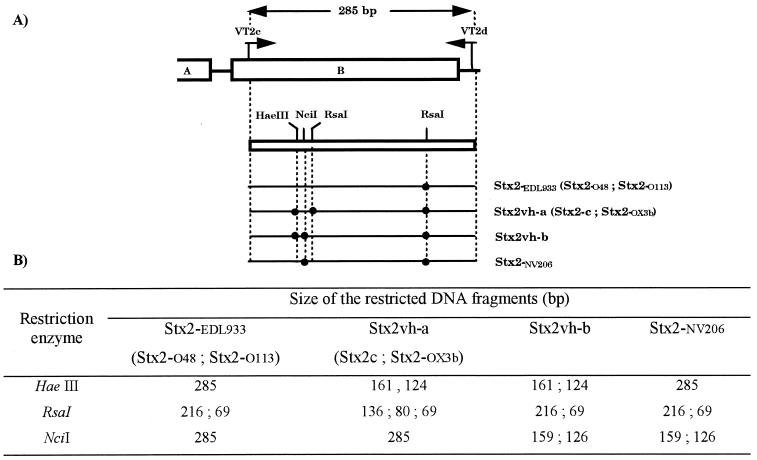
FIG. 1.
Subtyping of Stx2 subtypes by RFLP-PCR. (A) Schematic representation of the restriction patterns obtained with the RFLP-PCR method developed by Tyler et al. (25). (B) Predicted sizes of the amplified products obtained with the VT2c and VT2d oligonucleotide primers and restricted with HaeIII, RsaI, and NciI endonuclease. The method developed to screen the Stx2vh-a subtype should also detect Stx2c and Stx2-OX3b because the nucleotide sequences of primers and sites of the restriction endonucleases used in the RFLP-PCR method were conserved among Stx2vh-a, Stx2c, and Stx2-OX3b genes. Similarly, the protocol used to discriminate Stx2-EDL933 should also detect Stx2-O48. The nucleotide sequence of Stx2-O113 is not available in the databases.
TABLE 2.
Association of Stx2 subtypes with virulence factors
| Stx2 subtype | No. (%) of isolates | No. (%) of bovine STEC isolates positive for:
|
||||
|---|---|---|---|---|---|---|
| stx1 | ehxA | espP | stx1 + ehxA + espP | east1 | ||
| Stx2-EDL933 | 41 (24.5) | 25 (61) | 32 (78) | 31 (75.5) | 21 (51) | 2 (5) |
| Stx2-EDL933 + Stx2vh-a | 1 (0.5) | 0 | 1 | 1 | 0 | 0 |
| Stx2-EDL933 + Stx2vh-b | 20 (12) | 11 (55) | 17 (85) | 14 (70) | 10 (50) | 0 |
| Stx2-EDL933 + Stx2d | 1 (0.5) | 1 | 0 | 0 | 0 | 0 |
| Stx2-EDL933 + Stx2vh-b + Stx2d | 3 (1.8) | 0 | 2 | 3 | 0 | 0 |
| Stx2vh-a | 27 (16) | 8 (30) | 1 (3.5) | 6 (22) | 1 (3.5) | 1 (3.5) |
| Stx2vh-a + Stx2vh-b | 12 (7) | 2 (16.5) | 3 (25) | 7 (58) | 1 (8.5) | 1 (8.5) |
| Stx2vh-a + Stx2vh-b + Stx2d | 3 (1.8) | 0 | 1 | 1 | 0 | 0 |
| Stx2vh-b | 23 (14) | 11 (48) | 12 (52) | 12 (52) | 11 (48) | 1 (4.5) |
| Stx2vh-b + Stx2d | 4 (2.5) | 1 | 0 | 0 | 0 | 0 |
| Stx2d | 2 (1.2) | 0 | 0 | 0 | 0 | 0 |
| Stx2-NV206 | 23 (14) | 3 (13) | 4 (17.5) | 3 (13) | 2 (8.5) | 4 (17.5) |
| Stx2-NV206 + Stx2d | 1 (0.5) | 0 | 1 | 1 | 0 | 0 |
| None | 6 (3.5) | 1 | 2 | 2 | 1 | 2 |
| Total | 167 (100) | 63 (38) | 76 (45.5) | 81 (48.5) | 47 (28) | 11 (6.5) |
The Stx2vh-b, Stx2-EDL933, Stx2vh-a, and Stx2d subtypes were detected (alone or in combination with other Stx2 subtypes) among 39.5, 39, 25.5, and 8.5% of the stx2-positive isolates, respectively. In agreement with previous studies (1, 8), the Stx2e subtype was not detected among the bovine STEC tested. A total of 39 bovine strains (23.5%) were positive for two distinct Stx2 subtypes: associations of Stx2-EDL933/Stx2vh-b, Stx2vh-a/Stx2vh-b, and Stx2vh-b/Stx2d subtypes were observed among 12, 7, and 2.5% of the stx2-positive isolates, respectively. Furthermore, six isolates (3.5%) were positive for three distinct Stx2 subtypes (Stx2-EDL933/Stx2vh-b/Stx2d or Stx2vh-a/Stx2vh-b/Stx2d). Interestingly, Stx2d was more frequently detected when associated with one or two Stx2 subtypes (Table 2).
A total of 24 stx2-positive strains isolated from healthy cattle (14.5%) showed an atypical restriction pattern using the RFLP-PCR method able to discriminate Stx2-EDL933, Stx2vh-a, and Stx2vh-b. A DNA fragment of the expected size (285 bp) was amplified from each of the 24 strains, but a new restriction pattern was observed when HaeIII, NciI, and RsaI restriction endonucleases were used. This new Stx2 subtype is referred to as Stx2-NV206 in Fig. 1 and Tables 1, 2, and 3.
TABLE 3.
Cytotoxicity of STEC strains toward Vero cells according to Stx type and subtypea
| Stx2 subtype | No. (%) of bovine STEC strains
|
|||
|---|---|---|---|---|
|
stx2 positive
|
stx2 and stx1 positive
|
|||
| No. | No. highly cytotoxic | No. | No. highly cytotoxic | |
| Stx2-EDL933 | 16 | 9 (56) | 25 | 18 |
| Stx2-EDL933 + Stx2vh-a | 1 | 1 | 0 | 0 |
| Stx2-EDL933 + Stx2vh-b | 9 | 5 | 11 | 10 |
| Stx2-EDL933 + Stx2d | 0 | 0 | 1 | 1 |
| Stx2-EDL933 + Stx2vh-b + Stx2d | 3 | 2 | 0 | 0 |
| Stx2vh-a | 19 | 8 (42) | 8 | 7 |
| Stx2vh-a + Stx2vh-b | 10 | 8 (80) | 2 | 2 |
| Stx2vh-a + Stx2vh-b + Stx2d | 3 | 1 | 0 | 0 |
| Stx2vh-b | 12 | 4 (33) | 11 | 5 |
| Stx2vh-b + Stx2d | 3 | 2 | 1 | 1 |
| Stx2d | 2 | 0 | 0 | 0 |
| Stx2-NV206 | 20 | 16 (80) | 3 | 3 |
| Stx2-NV206 + Stx2d | 1 | 1 | 0 | 0 |
| None | 5 | 0 | 1 | 0 |
| Total | 104 | 57 (55) | 63 | 47 (74.5) |
| Total no. of strains possessing one Stx2 subtype | 69 | 37 (54) | 47 | 33 (70) |
| Total no. of strains possessing two Stx2 subtypes | 24 | 17 (71) | 15 | 14 (93) |
Culture supernatants of STEC have previously been tested for cytotoxicity on Vero cells (21). Briefly, the bacterial strains were inoculated into brain heart infusion broth and incubated at 37°C overnight. After centrifugation, supernatant filtrates were obtained with a 0.45-μm-pore-size filter. Twofold serial dilutions of bacterial filtrates were done in 96-well microtiter plates, and a total of 100 μl of EMEM buffer containing 105 Vero cells in suspension was added to each well. The culture plates were incubated for 24 h at 37°C in a 5% CO2 atmosphere. The monolayers were washed with PBS (pH 7.2) and stained with a crystal violet solution. The verotoxin titer was expressed as the reciprocal of the highest sample dilution of culture filtrate that caused 50% cell detachment after 24 h of incubation, as judged by the dye intensity and by microscopic observation. On the basis of two independent experiments, the strains were classified into three groups: not cytotoxic for Vero cells (titer ≤2), moderately cytotoxic (titer from 4 to 32), and highly cytotoxic (titer ≥64) (20). The moderate cytotoxicity of STEC was not included in this study because a significant association (P < 0.001) was only found between high cytotoxic activity and the presence of the stx2 gene (20).
In addition, six bovine isolates (3.5%) found to be positive by DNA hybridization using a probe located within the A subunit gene (19) were found to be negative with all the PCR and RFLP-PCR methods used in this study. The six E. coli strains were also negative by PCR for amplification of the operon containing the genes encoding A and B subunits. These results strongly suggested a partial or total deletion of the B subunit-encoding gene that may explain the lack of cytoxicity of these six strains previously demonstrated by Vero cell assays (19).
Sequence of stx2 operon of strain NV206.
Strain NV206 (serotype O6:H10) is one of the 23 bovine E. coli strains which was found to be negative for stx2d and stx2e and untypeable by RFLP-PCR (Table 2). The stx2 operon of E. coli NV206 (stx2-NV206) was amplified, and the nucleotide sequence was determined. The operon contained two open reading frames (ORFs) of 957 and 267 bp, encoding two polypeptides of 319 amino acids (A subunit) and 89 amino acids (B subunit), respectively. Nucleotide sequence comparison of the stx2-NV206 operon with the corresponding ORFs of the eleven Stx2 subtypes described above revealed 94.5 to 99% identity for the A subunit gene and 81.5 to 96% identity for the B subunit gene. At the amino acid level, Stx2-NV206 showed 94 to 99% identity for the A subunit and 87 to 98% identity for the B subunit. Interestingly, the two amino acid residues Ser-291 and Glu-297 of the A subunit of both Stx2vh-a and Stx2vh-b subtypes were conserved at the same position on the Stx2-NV206 A subunit. Melton-Celsa et al. suggest that these two amino acids are involved in the activation of the toxin by mouse or human intestinal mucus (12).
Nucleotide sequence analysis of the B subunit-encoding gene confirmed the stx2-NV206 restriction pattern obtained by RFLP-PCR. A DNA fragment of 285 bp amplified from the B subunit gene was cut into two DNA fragments of 216 and 69 bp by RsaI, two DNA fragments of 159 and 126 bp by NciI, and was not cut by HaeIII (Fig. 1).
Association of different Stx2 subtypes with Stx1 and other virulence factors.
The association of the different stx2 subtypes with the stx1 gene was analyzed among the 63 strains of the collection positive for both stx2 and stx1. Results are summarized in Table 2. The Stx1-encoding gene was prominent among STEC strains that possessed Stx2-EDL933 alone (61%) or both Stx2-EDL933 and Stx2vh-b (55%). The stx1 gene was also detected among 48, 30, 16.5, and 13% of the STEC strains that possessed the genes coding for Stx2vh-b, Stx2vh-a, Stx2vh-a/Stx2vh-b, and Stx2-NV206, respectively. Interestingly, except for the bovine isolates with both Stx2-EDL933 and Stx2vh-b, stx1 was more frequently detected among STEC possessing only one Stx2 subtype.
The presence of the gene encoding the enterohemolysin (ehxA), the extracellular serine protease (espP), and the EAST1 toxin (east1) was analyzed among the 167 bovine isolates positive for stx2 and the 18 isolates positive for stx1 and negative for stx2. The ehxA gene was present among 45.5% of the stx2-positive strains (Table 2) and 14 of the 18 stx1-positive strains (78%) (data not shown). The Ehx-encoding gene was prominent among bovine isolates that possessed both Stx2-EDL933 and Stx2vh-b (85%), Stx2-EDL933 alone (78%), or Stx2vh-b alone (52%) (Table 2). The EspP-encoding gene was detected among 48.5% of the isolates positive for stx2 (Table 2) and 7 of the 18 stx1-positive isolates (39%) (data not shown). A prevalence of espP was observed among bovine strains that possessed Stx2-EDL933 alone (75.5%), a combination of Stx2-EDL933/Stx2vh-b (70%) or Stx2vh-a/Stx2vh-b (58%), and Stx2vh-b alone (52%) (Table 2). The presence of the gene encoding EAST1 was only detected among 6.5% of the stx2-positive isolates (Table 2). In contrast, 9 of the 18 isolates positive for stx1 and negative for stx2 (50%) were east1 positive (data not shown).
A combination of Stx1, Ehx, and EspP was observed among 28% of the stx2-positive strains (Table 2). Interestingly, all 11 stx1-positive strains possessing Stx2vh-b and most of the stx1-positive STEC strains with Stx2-EDL933 subtype alone (21 of 25) or both Stx2-EDL933 and Stx2vh-b (10 of 11) were ehxA and espP positive (Table 2).
Verocytotoxicity of bacterial strains with different Stx2 subtypes.
The culture supernatants of the STEC strains included here have previously been tested for cytotoxicity on Vero cells (19). On the basis of two independent experiments, the strains were classified into three groups: not cytotoxic for Vero cells (titer ≤2), moderately cytotoxic (titer from 4 to 32), and highly cytotoxic (titer ≥64) (19). A significant association between a high cytotoxic activity and the presence of the stx2 gene has been described (P < 0.001) (19). Therefore, only the high cytotoxic activity of STEC was taken into account in the analysis of a putative correlation between the cytotoxicity of the isolates and the Stx2 subtype produced. Strains positive for stx2 (n = 104) and positive for both stx2 and stx1 (n = 63) were analyzed (Table3). In addition, analysis of the cytotoxic potency of the 18 stx1-positive strains was also included in this report.
Of the 104 stx2-positive strains tested, 57 (55%) were found to be highly cytotoxic toward Vero cells (Table 3). Among them, 16 of 20 isolates with Stx2-NV206 (80%), 8 of 10 isolates with both Stx2vh-a and Stx2vh-b (80%), and 9 of 16 isolates with Stx2-EDL933 (56%) were associated with a high cytotoxic activity. In contrast, only 8 of the 19 strains with Stx2vh-a alone (42%) and 4 of the 12 strains with Stx2vh-b alone (33%) were found to be highly cytotoxic toward Vero cells (Table 3). Isolates highly cytotoxic toward Vero cells frequently possessed two distinct Stx2 subtypes (71%). STEC strains with only one Stx2 subtype were found less frequently to have high cytotoxicity (54%). Furthermore, 33 of the 47 isolates with the Stx1 toxin and one Stx2 subtype (70%) and 14 of the 15 isolates with the Stx1 toxin and two distinct Stx2 subtypes (93%) were found to be highly cytotoxic toward Vero cells. In contrast, only 33% of the stx1-positive and stx2-negative E. coli were highly cytotoxic on Vero cells (data not shown).
DISCUSSION
Stx-producing E. coli are mostly transmitted to humans through food contaminated by animal fecal material (9, 17). However, animals do not develop HC or HUS. Furthermore, not all the STEC present in the intestinal tract of cattle are involved in human infections. Because of the lack of suitable animal models that mimic all of the aspects of human diseases caused by STEC, it is difficult to identify the bacterial factors involved. However, it is well documented that STEC strains vary in their capacity to cause serious disease in humans, and this is associated with the type and/or amount of Stx produced (9, 17). Therefore, the type of Stx toxin and/or the Stx2 subtypes produced by STEC isolated from human infections has been extensively studied (6, 22). In contrast, little is known about Stx2 subtype frequency and the combination of virulence factors expressed by STEC from the intestinal tract of healthy cattle.
Considering that bovines are the principal reservoir of STEC potentially pathogenic for humans, a bacterial collection has been constituted from the feces of healthy cows at the city slaughterhouse of Clermont-Ferrand in central France (19). Analysis of the bacterial collection showed that STEC were isolated from 34% of the healthy cows and that 90% of the isolates harbored the Stx2-encoding gene (19). In this report, Stx2 subtyping was undertaken among 167 STEC strains isolated from bovine feces. The Stx2 subtype (renamed Stx2-EDL933 in this study) considered to be the Stx2 prototype for O157:H7 STEC associated with HUS was prominent among the strains isolated from healthy cows. E. coli strains harboring Stx2vh-a- or Stx2vh-b-encoding genes were also frequently detected. Furthermore, the Stx2-EDL933–Stx2vh-b combination was the most frequent among STEC possessing more than one Stx2 subtype. The Stx2d group, including Stx2d-Ount, Stx2d-O111, and Stx2d-OX3a subtypes, was infrequently detected among the bovine isolates included in this report. Recently, a high proportion of strains possessing the Stx2d variant have been isolated from fecal specimens of human asymptomatic carriers (24). In contrast, STEC associated with HUS never showed the Stx2d variants, suggesting that the Stx2d-producing strains might be less pathogenic for humans (14, 15, 18).
Interestingly, a new Stx2 subtype named Stx2-NV206 that is untypeable by Tyler's RFLP-PCR method was detected among 14.5% of the bovine strains studied. In a previous study, the same genotypic method was used to screen STEC isolated from human and meat samples (18). The authors detected 15 isolates (6% of the bacterial collection) possessing an Stx2 toxin referred to as an atypical Stx2vh variant because restriction of amplicons did not correspond to predicted patterns (18). Unfortunately, the restriction pattern of the atypical Stx2 subtype was not described (18), and therefore could not be compared with that of Stx2-NV206.
Among STEC isolated from patients with HUS, both Stx1 and Stx2 toxins are commonly detected, and a few strains produced Stx1 and two distinct Stx2 variants (9, 17). However, Stx2 was 1,000 times more cytotoxic than Stx1 towards human renal endothelial cells, and STEC producing Stx2 are more commonly associated with serious diseases than isolates producing Stx1 or Stx1 and Stx2 (9, 11, 17). In this report, 38% of the STEC were positive for both stx1 and stx2 and 9% of the isolates possessed the genetic information encoding Stx1 and two distinct Stx2 subtypes. Furthermore, a combination of three Stx2 subtypes (Stx2-EDL933/Stx2vh-b/Sx2d or Stx2vh-a/Stx2vh-b/Sx2d) were also detected simultaneously among the bovine E. coli tested. To our knowledge, this is the first report of STEC strains of animal origin that possess a combination of three distinct Stx2 variants. The extent to which multiple stx genes in a given STEC isolate can modulate the level of virulence is unknown. However, it is conceivable that strains possessing a combination of three stx genes are more virulent than those harboring only one or two stx genes, assuming that all three genes are expressed.
Mouse and human intestinal mucus is able to activate Stx2vh-a and Stx2vh-b toxins but not Stx2-EDL933 or Stx2c (12). E. coli B2F1, producing both Stx2vh-a and Stx2vh-b, is highly virulent in an orally infected streptomycin-treated mouse model and becomes more cytotoxic for Vero cells after incubation with mouse or human intestinal mucus (10, 12). Two amino acid residues, Ser and Glu at positions 291 and 297, respectively, of the mature A subunit, are probably involved in activation of the toxin (12). Interestingly, these two amino acid residues, potentially important for activation, were conserved at the same position on the A subunit of Stx2-NV206. Ser-291 and Glu-297 were also conserved on the A subunit of Stx2e, Stx2d-Ount, Stx2d-O111, and Stx2d-OX3a but not among Stx2-EDL933, Stx2-O113, Stx2-O48, or Stx2c. Melton-Celsa et al. suggest that synthesis of an activatable toxin results in an Stx2-producing strain that is more virulent and/or compensates for the absence of additional virulence factors (12).
Although intimin is an essential virulence factor for STEC belonging to serogroups O26, O103, O111, and O157, eae-negative STEC strains belonging to serogroup O91, O104, or O113 have been isolated during cases of HUS and HC (9, 17). Therefore, the pathogenicity for humans of eae-negative STEC from healthy cows could not be excluded. The finding of few eae-positive STEC in the bovine bacterial collection (5%) (19) was in agreement with results obtained from similar European studies (1, 2, 28). Among the 167 STEC included in this study, 12 isolates belonging to the O113:H21 serotype were eae negative (19) and espP positive, and possessed the gene encoding Stx2vh-a and/or Stx2vh-b (results not shown). Recently, Paton et al. described eae-lacking STEC strains belonging to the O113:H21 serotype which were associated with a cluster of cases of HUS (16). Taking into account the fact that STEC are mostly transmitted to humans through food contaminated by animal fecal material, the O113:H21 strains isolated from healthy cattle appeared potentially pathogenic for humans. As suggested for the human STEC strain B2F1, also negative for eae, the presence of Stx2vh-a and/or Stx2vh-b subtypes activatable by human intestinal mucus might compensate for the lack of genetic information coding for attaching and effacing lesion (12).
The ehxA enterohemolysin gene is highly conserved among STEC associated with human diseases (particularly in O157:H7 E. coli strains), suggesting that Ehx is under strong selective pressure and is implicated in STEC survival (1, 17). Furthermore, Gyles et al. demonstrate that ehxA is highly present among STEC belonging to serotypes commonly associated with disease in humans (1, 5). In this report, ehxA was prominent among STEC harboring stx2-EDL933 alone or stx2-EDL933 in association with stx2vh-b (78 and 85% of the strains, respectively), demonstrating that the presence of hlxA was also correlated with the Stx2 subtype. In addition, a close association of genes encoding Stx1, Ehx, and EspP was emphasized among stx2-positive strains harboring the gene encoding Stx2-EDL933 alone, Stx2vh-b alone, or a combination of the two subtypes. In contrast, the EAST1 enterotoxin was infrequently detected and seemed to be more associated with the stx1-positive strains of this collection.
In summary, Stx2 subtyping analysis of STEC isolated from healthy cows emphasized the predominance of Stx2-EDL933 and/or Stx2vh-b toxins and their association with other putative virulence factors such as Stx1, Ehx, and EspP (but not EAST1). For the first time, the combination of three distinct stx2 subtypes was described, and the new Stx2-NV206 subtype was characterized.
REFERENCES
- 1.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco M, Blanco J E, Blanco J, Gonzalez E A, Mora A, Prado C, Fernandez L, Rio M, Ramos J, Alonso M P. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol Infect. 1996;117:251–257. doi: 10.1017/s0950268800001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 4.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gyles C, Johnson R, Gao A, Ziebell K, Pierard D, Aleksic S, Boerlin P. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of shiga-like-toxin-producing Escherichia coli of human and bovine origins. Appl Environ Microbiol. 1998;64:4134–4141. doi: 10.1128/aem.64.11.4134-4141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 7.Jackson M P, Neill R J, O'Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-toxin I and Shigat-toxin II encoded by bacteriphages from Escherichia coli. FEMS Microbiol Lett. 1987;44:109–114. [Google Scholar]
- 8.Johnson W M, Pollard D R, Lior H, Tyler S D, Rozee K R. Differentiation of genes coding for Escherichia coli verotoxin 2 and the verotoxin associated with porcine edema disease (VTe) by the polymerase chain reaction. J Clin Microbiol. 1990;28:2351–2353. doi: 10.1128/jcm.28.10.2351-2353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law D. Virulence factors of Escherichia coli O157 and other Shiga toxin- producing E. coli. J Appl Microbiol. 2000;88:729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 11.Lindgren S W, Melton A R, O'Brien A D. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect Immun. 1993;61:3832–3842. doi: 10.1128/iai.61.9.3832-3842.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louise C B, Obrig T G. Specific interaction of Escherichia coli O157:H7-derived Shiga-like toxin II with human renal endothelial cells. J Infect Dis. 1995;172:1397–1401. doi: 10.1093/infdis/172.5.1397. [DOI] [PubMed] [Google Scholar]
- 13.Melton-Celsa A R, Darnell S C, O'Brien A D. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paton A W, Bourne A J, Manning P A, Paton J C. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect Immun. 1995;63:2450–2458. doi: 10.1128/iai.63.7.2450-2458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paton A W, Paton J C, Heuzenroeder M W, Goldwater P N, Manning P A. Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of sudden infant death syndrome. Microb Pathog. 1992;13:225–236. doi: 10.1016/0882-4010(92)90023-h. [DOI] [PubMed] [Google Scholar]
- 16.Paton A W, Paton J C, Manning P A. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb Pathog. 1993;15:77–82. doi: 10.1006/mpat.1993.1058. [DOI] [PubMed] [Google Scholar]
- 17.Paton A W, Woodrow M C, Doyle R M, Lanser J A, Paton J C. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3357–3361. doi: 10.1128/jcm.37.10.3357-3361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paton J C, Paton A W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierard D, Muyldermans G, Moriau L, Stevens D, Lauwers S. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J Clin Microbiol. 1998;36:3317–3322. doi: 10.1128/jcm.36.11.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradel N, Livrelli V, De Champs C, Palcoux J B, Reynaud A, Scheutz F, Sirot J, Joly B, Forestier C. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J Clin Microbiol. 2000;38:1023–1031. doi: 10.1128/jcm.38.3.1023-1031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savarino S J, Fasano A, Robertson D C, Levine M M. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J Clin Investig. 1991;87:1450–1455. doi: 10.1172/JCI115151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt H, Kernbach C, Karch H. Analysis of the EHEC hly operon and its location in the physical map of the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 1996;142:907–914. doi: 10.1099/00221287-142-4-907. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt C K, McKee M L, O'Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman P, Soni R, Karmali M. Attaching and effacing adherence of Vero cytotoxin-producing Escherichia coli to rabbit intestinal epithelium in vivo. Infect Immun. 1988;56:756–761. doi: 10.1128/iai.56.4.756-761.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephan R, Hoelzle L E. Characterization of shiga toxin type 2 variant B-subunit in Escherichia coli strains from asymptomatic human carriers by PCR-RFLP. Lett Appl Microbiol. 2000;31:139–142. doi: 10.1046/j.1365-2672.2000.00778.x. [DOI] [PubMed] [Google Scholar]
- 26.Tyler S D, Johnson W M, Lior H, Wang G, Rozee K R. Identification of verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1991;29:1339–1343. doi: 10.1128/jcm.29.7.1339-1343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein D L, Jackson M P, Samuel J E, Holmes R K, O'Brien A D. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto T, Echeverria P. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect Immun. 1996;64:1441–1445. doi: 10.1128/iai.64.4.1441-1445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zschock M, Hamann H P, Kloppert B, Wolter W. Shiga-toxin-producing escherichia coli in faeces of healthy dairy cows, sheep and goats: prevalence and virulence properties. Lett Appl Microbiol. 2000;31:203–208. doi: 10.1046/j.1365-2672.2000.00789.x. [DOI] [PubMed] [Google Scholar]