Abstract
Background
Mitochondria are cellular organelles responsible for energy production, and dysregulation of the mitochondrial network is associated with many disease states. To fully characterize the mitochondrial network's structure and function, a three-dimensional whole cell mapping technique is required.
Scope of review
This review highlights the use of soft X-ray tomography (SXT) as a relatively high-throughput approach to quantify mitochondrial structure and function under multiple cellular conditions.
Major conclusions
The use of SXT opens the door for mapping cellular rearrangements during critical processes such as insulin secretion, stem cell differentiation, or disease progression. SXT provides unique information such as biochemical compositions or molecular densities of organelles and allows for unbiased, label-free imaging of intact whole cells. Mapping mitochondria in the context of the near-native cellular environment will reveal more information regarding mitochondrial network functions within the cell.
Keywords: Structural biology, Cell mapping, Tomography, Spatial biology, Mitochondria
Abbreviations: Soft X-ray tomography, (SXT); three-dimensional, (3D); mitochondria-associated membranes, (MAMs); stimulated emission depletion, (STED); fluorescence lifetime imaging microscopy, (FLIM); linear absorption coefficient, (LAC); endoplasmic reticulum, (ER); glucagon-like peptide-1 receptor, (GLP-1R); potassium chloride, (KCl)
Highlights
-
•
Soft X-ray tomography (SXT) generates 3D organelle maps of intact cells.
-
•
3D maps reveal the positions of mitochondria and their molecular densities.
-
•
SXT can be used to quantify and compare organelle contacts between conditions.
-
•
SXT is unbiased imaging that identifies the contents of subcellular neighborhoods.
-
•
SXT provides an exciting path for exploring metabolic dysfunction.
1. Introduction
Mitochondria are cellular organelles that produce ATP, contribute to cell stress responses such as apoptosis, and are a hub of biosynthetic processes [1]. The double-membrane structure of mitochondria creates specialized compartments that are critical to its function. The inner membrane has complex folds organized as cristae, which increase the surface area and the capacity for ATP generation. Mitochondria also form a dynamic and interconnected structural network throughout the cell that facilitates communication with other organelles and subcellular microenvironments via calcium signaling. Mitochondrial networks are dynamic because of fluctuations in mitochondria division, fusion, motility, and tethering to other organelles [1]. Mitochondria-associated membranes (MAMs) between the outer membranes of mitochondria and other membranes within the cell allow specialized regions to emerge, such as those important for Ca2+ homeostasis and lipid biosynthesis [2]. Mitochondrial dysfunction is associated with numerous disease states, such as neurodegenerative disorders, metabolic syndromes, cancer, and obesity [1]. While mitochondria have one of the most recognizable subcellular structures, many questions remain about the structural and functional roles that mitochondria play in health and how these are dysregulated in disease.
Advances in modern imaging modalities have allowed researchers to gain increasingly high-resolution images of mitochondrial networks and reveal their functions; for example, confocal and electron microscopy revealed that mitochondria-derived vesicles transport cargo with neighboring organelles such as peroxisomes [3]. The use of stimulated emission depletion (STED) microscopy enabled visualization of the dynamic structure of mitochondria cristae in live cells [4], while light-sheet microscopy was employed to explore the mitochondria interactome in a whole cell [5]. These advancements in imaging technologies, among others, have paved the way for the generation of new hypotheses regarding the function of mitochondria, and they highlight the importance of understanding structure and function relationships. To quantify more complex aspects of mitochondrial network structure, such as associations with other organelles and subcellular neighborhoods during specific cellular processes, an unbiased three-dimensional (3D) cellular mapping approach is required. A comparison of the benefits and limitations for multiple experimental mapping methods is available in Table 1.
Table 1.
A summary of the benefits and limitations of imaging methods used to characterize 3D cellular structure. There are many exemplary case-uses for each method that have contributed significantly to our basic understanding of cell structure and function. For brevity, only a few examples are provided in the above table. Indications for potential variations in experimental setup are indicated in parenthesis in the Approach column.
| Approach | Resolution Range | Benefit | Limitation | Reference Examples |
|---|---|---|---|---|
| Electron Microscopy (scanning, block-face, fluorescence correlative) | 4–8 nm in approximately 1,000 μm3 tissue volume [55] | Can image in the context of tissue, keeping cell-to-cell contacts and neighboring vasculature intact for improved interpretations | Requires laborious serial sectioning which limits the number of cells and conditions that can be investigated. May also require plastic embedding or chemical fixation. | [28,29,56,57] |
| Cryo-electron tomography (thin periphery of cell, thin lamella of interior of cell, single-cell multiple distinct tomogram acquisitions) | 2–5 nm in 180–250 nm thick samples | Provides high resolution details on 3D cellular organization within windows of the cell. Allows for in-situ structural biology. | Require use of thin lamella sections of cells that may not accurately reflect cellular organization due to polarity and uneven distributions of cellular contents | [[58], [59], [60]] |
| Fluorescence Microscopy (confocal, super-resolution) | 70–250 nm in a single cell | Provides live cell dynamics and cellular rearrangements | Requires specific labeling strategy limiting unbiased discovery | [[61], [62], [63], [64]] |
| Soft X-Ray Tomography (cells cryo-fixed on a grid or in solution within capillary. Fluorescence correlative) | Up to 35 nm in 10 μm diameter cell | Produces unbiased and high-throughput 3D reconstructions of intact cells | Requires dissociated tissue for single cell analysis, limited to static timepoints, and limited access to facilities with ideal capabilities | [16,19,27,65] |
This review highlights the advantages of using soft X-ray tomography (SXT) to quantify the 3D organization of mitochondria in intact cells, and it discusses limitations and opportunities for future integration of SXT maps with additional experimental data and computational methods. The combination of multiple types of information and tools will allow us to better understand mitochondrial network structure and function relationships in health and disease.
2. Advantages of using SXT
SXT is an experimental method that is optimally suited to characterize the relationships between cell structures and functions in intact cells in a near-native state because it is an unbiased, mesoscale, quantitative, relatively high-throughput, and correlative approach [6].
2.1. Near-native state
To accurately map cellular structure, it is advantageous to maintain the native structure as much as possible during sample preparation. Unlike other 3D imaging strategies, SXT enables imaging of intact, fully-hydrated cells without the need for chemical fixatives. Instead, cells are cryo-preserved, which provides added protection from radiation damage during data collection [6,7].
SXT datasets are collected using a soft X-ray transmission microscope equipped with a specimen stage that allows for full-rotation or tilted-angle data acquisition [8]. Images are typically collected at 1–2° increments over 180° on a full-rotation stage [6], or ±65° in microscopes with a tilt stage [9,10]. Stacks of projection images are collected to comprehensively capture the cells from different orientations and are used to reconstruct a 3D tomogram of the sample [11,12].
2.2. Unbiased
SXT is a label-free approach that leverages the natural contrast of cellular components rather than stains, probes, or fluorescent protein tags to identify specific cellular structures. This allows for unbiased, discovery-based observations, even when performing focused experiments on a specific organelle distribution.
SXT leverages synchrotron radiation as a light source, where samples are imaged within the “water window” (284–543 eV). Within this energy range, the absorption of X-rays by the sample adheres to the Beer–Lambert law [13]. The absorption of X-rays is a linear and quantitative measure of the biochemical composition of the sample. For example, carbon-rich materials such as lipid droplets or native protein crystals have higher linear absorption coefficient (LAC) values than the cytoplasm, which has higher water content [14,15]. Many organelles can be identified simultaneously in SXT tomograms by using a combination of LAC values and general morphology. For example, based on differences in molecular composition and morphology, the nucleus, nucleolus, mitochondria, lipid droplets, and dense-core insulin secretory vesicles have all been simultaneously identified within single datasets [6,14,16]. In cases where the organelle morphology is ambiguous or is more difficult to observe, correlative imaging can be conducted in parallel to assemble a full organelle atlas. Correlative imaging will help with identifying tubular structures such as the endoplasmic reticulum (ER) and the Golgi, and organelles involved in the recycling process of cellular components, such as lysosomes and endosomes [[17], [18], [19]].
Similar to several advanced fluorescence imaging methods [5,20,21], SXT data provide a near-isotropic resolution which allows for the accurate 3D reconstruction of the whole cell. The advantage of observing the cell using its natural contrast is that it does not require labeling of specific structures. SXT provides information regarding the X-ray absorption of carbon-rich molecules in specific compartments, thus alterations in organelle molecular densities can be identified under many conditions such as stimulation with chemical probes. Leveraging the natural contrast of the cell allows for the unbiased investigation of the subcellular landscape.
2.3. Mesoscale resolution
SXT encompasses the mesoscale range, which includes whole cells (∼10 μm) to objects just larger than molecular machines (50 nm) [6]. Cell thickness spanning from 1 to 15 μm is suitable for SXT imaging, enabling exploration of a large range of cell types. The limiting factor for SXT imaging is the cell thickness, which in general affects the image contrast [22]. Within this scale, the spatial resolution typically ranges from 25 nm to 60 nm and is determined by the numerical aperture of the objective lens. In SXT, the objective is a circular diffracting grating, known as a Fresnel zone plate, where the width of the outermost ring dictates the numerical aperture of the SXT microscope [18,23,24]. Because an increase in spatial resolution leads to smaller field of view [25], images with a size of 15 × 15 μm2 can be easily imaged in a single collection using a 60 nm objective zone plate [26].This provides an optimal range for capturing the mitochondrial network structure and its interactions with neighboring organelles within the entire cell.
2.4. Quantitative
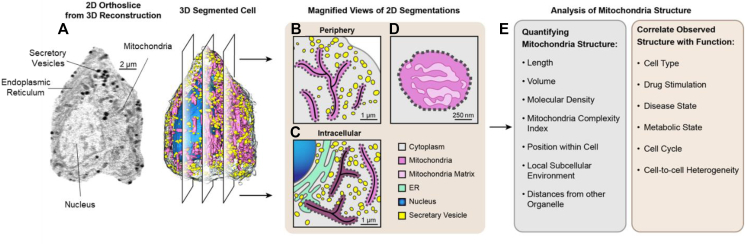
To quantify cell structure, one must first identify and segment each organelle present in the cell and measure and analyze multiple structural and physical parameters of the organelles. Towards this goal, an SXT map provides the numbers, volumes, and positions of organelles within the cell, as well as a map of their associations with other organelles and specific subcellular neighborhoods. SXT also provides a measure of the molecular densities of organelles, which reflect biochemical crowding (Figure 1). The molecular densities of a single organelle type can also be measured to identify subcompartments within an organelle responsible for different functions, such as mitochondria cristae and matrix, allowing for an added level of structural quantification and analysis [6,16,27].
Figure 1.
Process of using SXT to correlate mitochondrial structure and function. Modified from [16]. a) Representative 2D orthoslice and a reconstruction of a β-cell (corresponding 3D map) display the segmented organelles. The segmented cell masks are then used to quantify specific features of the cellular organization such as mitochondrial network structure. b-c) Magnified view of 2D slices of the segmented cell illustrate features that can be extracted and compared between different cellular regions. SXT can reveal slight differences in biochemical compositions of organelles by close analysis of their LAC values. An illustration of mitochondria with different biochemical compositions based on cellular localizations is shown in panel c. The darker pink shade indicates denser mitochondria. d) Illustration of a cross section of a mitochondria with cristae shown in dark pink and the mitochondrial matrix shown in light pink. The degree of mitochondrial complexity or extent of cristae folding can be quantified by segmenting these subcompartments using LAC values. e) List of the specific features that can be quantified from 3D segmentations of a cell and the corresponding cellular conditions that can be compared to correlate structure and function.
2.5. Relatively high throughput
Because thick samples (10–15 μm diameter) can be imaged with SXT, entire cells can be imaged without sectioning. SXT generates a 3D reconstructed volume of an entire cell within 10 min. Other 3D imaging methods require time-consuming serial sectioning and may allow for comparison of only a few cells [28,29] while SXT has been used to compare tens or hundreds of cells, depending on the cell type. For example, the structural organization of 60 INS-1E cells (a rat insulinoma cell line) under different conditions has been compared [16]. The improved scalability of SXT enables the collection of screening datasets comprised of hundreds of cells under multiple cellular conditions to characterize changes in the relationship of structure and function of the cell. The potential for collecting datasets including multiple experimental conditions opens the door for mapping cellular rearrangements during critical cellular processes, such as insulin secretion, stem cell differentiation, or disease progression.
2.6. Correlative
Many organelles are easily recognizable with SXT when considering both their characteristic LAC values and their unique morphologies; one example is the mitochondrial network that is usually identifiable in multiple cells and organisms [16,30,31]. On the other hand, to correlate unknown cellular substructures with organelles and protein co-localizations, correlative cryo-fluorescence microscopy can be used in tandem with SXT [6,19,[32], [33], [34]], providing an unequivocal identification of the subcellular components. For example, the fluorescence signal can be collected first using a high-numerical aperture confocal cryogenic microscope and subsequently aligned and correlated with the 3D tomographic reconstruction of the same sample [6,35]. Alternatively, the correlation of SXT with X-ray fluorescence microscopy allows characterization of the localization of heavy atoms in the subcellular structure and their cellular function. For instance, identifying the subcellular composition of mitochondria would help to quantify the dysfunction of the subcellular compartment with respect to the normal functions of cellular homeostasis [36].
Taken together, these advantages of SXT over other methodologies for cellular imaging allow comparisons to be made of the relationship between structure and function of mitochondria under many diverse cellular conditions, such as drug stimulations, disease states, metabolic states, and cell cycle stages (Fig. 1E).
3. Use of SXT to quantify mitochondrial network structure
Recent studies have utilized SXT to investigate different aspects of mitochondrial structure and function, demonstrating the exciting promise of SXT as a tool for interrogating the relationship between mitochondrial structure and cellular metabolism. Mitochondria have been explored by SXT in the context of healthy single cells [26] and cancer cells [37,38], and after isolation from cells to explore their internal structure [27,39]. For the purpose of this review, we will highlight four unique applications of SXT that have been used to investigate the internal structure of mitochondria [27], evaluate mitochondria-ER contacts [19], explore the interaction between mitochondria and endosome-like vesicles [40], and visualize the mitochondrial network structure during multiple phases of insulin secretion [16].
3.1. Internal structure of mitochondria
Disease states such as cardiomyopathies and neuronal disorders are associated with structure-related dysfunction in mitochondria [41,42]. Polo et al. recently established SXT as a method to characterize mitochondria cristae morphology [27]. Their goal was to establish a quantitative approach for evaluating cristae to more meaningfully compare mitochondria in healthy and diseased cellular states. They used SXT at its highest resolution (35 nm) to investigate the internal organization of mitochondria. By closely examining the LAC values within mitochondria, the authors were able to distinguish between mitochondrial matrix and cristae, providing a new approach for evaluating cristae structure (illustrated in Figure 1D). A mitochondrial complexity index [43] was used to quantify the morphological complexity, including parameters of matrix surface area and volume. This methodology will be useful in future studies for investigating the role of mitochondrial structure in cellular functions both in health and disease.
3.2. Mitochondria and ER interactions
Mitochondrial fission is important to achieve the appropriate distribution of mitochondria needed to accommodate the cell's metabolic needs for ATP [44,45]. The ER is implicated in regulating mitochondrial dynamics, but many questions remain about the molecular mechanisms involved [46]. Elgass et al. employed a sophisticated combination of confocal live-cell imaging, correlative cryogenic fluorescence microscopy and SXT on COS-7 and murine cells to investigate the link between specific mitochondrial dynamics proteins (MiD49 and MiD51) and the ER in mitochondrial fission [19]. The authors established a link between MiD49 and MiD51 to the constriction of mitochondria by ER tubules during fission and characterized the 3D structure of ER-mitochondria contact sites. The authors leveraged LAC values of organelles to assess changes in the concentration of either lipids or proteins in organelles during fission, providing an opportunity to correlate organelle structure and biochemical states. This study elegantly displays the potential of using SXT to map physical associations of organelles and implicates specific proteins in regulating structural rearrangements.
3.3. Mitochondria and endosome-like vesicle contacts
Biolipids such as cholesterol are important for maintaining several biophysical properties of membranes such as fluidity, curvature, permeability, and protein-lipid interactions [40]. Delivery of cholesterol to the mitochondria enables its conversion to sterol metabolites and steroid hormones. To investigate the mechanisms of cholesterol delivery to the mitochondria, SXT and cryo-fluorescence microscopy were used to reveal close contacts of mitochondria to endosome-like organelles in human fibroblasts [40]. The authors used these results to determine the surface fraction of endo-lysosomes in contact with mitochondria, providing clarity on how cholesterol can be delivered to the mitochondria. In a related study, the authors used SXT to observe the transfer of excess cholesterol from endo-lysosomes to the plasma membrane by shedding extracellular vesicles [47]. Taken together, these studies demonstrate the usefulness of correlative cryo-fluorescence microscopy and SXT in mapping cellular rearrangements and quantifying inter-organelle contacts.
3.4. Mitochondrial network structure during insulin secretion
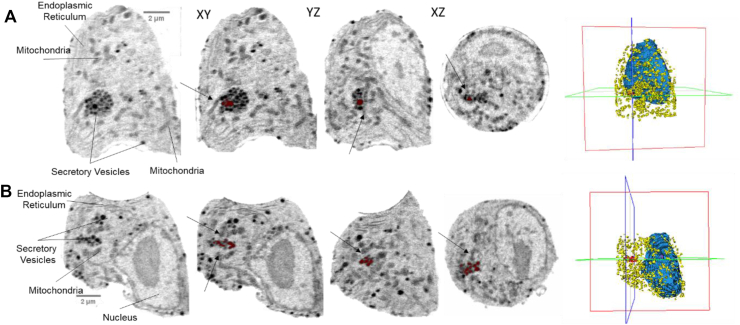
β-Cells are specialized secretory machines that release insulin into the blood stream upon elevations in blood glucose levels. Insulin secretion is mediated by multiple signaling pathways providing an opportunity to investigate how these different pathways initiate cellular rearrangements during the secretion process. To explore rearrangements during cellular processes, SXT was recently used to map 3D rearrangements of a pancreatic β-cell line (INS-1E) during insulin secretion [16]. Imaging was undertaken for multiple experimental conditions to assess cellular rearrangements and how different mechanisms of insulin secretion impact cell structure. The high-throughput nature of SXT allowed examination of multiple cells at different time points, capturing dynamic rearrangements that are evident during the first and second phases of insulin secretion. 3D cellular reconstructions revealed distinct insulin vesicle distribution patterns, reflecting altered vesicle pool sizes as they travel through the secretory pathway. Key findings of this study revealed that glucose stimulation causes rapid changes in insulin vesicle biochemical composition, increased mitochondrial volumes, and increased mitochondria-insulin vesicle contacts (Figure 2). Associations between insulin vesicles and mitochondria had been noted in the literature [48], but without 3D mapping of whole cells under different conditions, it was difficult to determine whether the interaction was relevant for biology. The role of insulin vesicle–mitochondria association remains unclear, but the door has been opened for forming new hypothesis on β-cell function. This study highlights the potential for SXT to be used as a screening tool to investigate how different protein signaling pathways impact mitochondrial structure and interactions with other organelles.
Figure 2.
Mitochondria in close proximity to clusters of insulin vesicles in β-cells stimulated with glucose. Orthoslices from two representative cells (two separate cells shown in A and B, respectively) stimulated with 25 mM glucose for 30 min are displayed. To assist in tracking the cellular orientation between the XY, YZ, and XZ views, a single cluster of secretory vesicles is highlighted in red in each orthoslice. Nearby mitochondria are indicated by black arrows. The position of orthoslices for XY, YZ, and XZ views are indicated in the far-right panel showing the 3D segmentations. The red box indicates the XY view, the blue box indicates the YZ view, and the green box indicates the XZ view. Images with scale bars were generated in Fiji (Image J) and XY, YZ, XZ slices, and 3D orientations were generated in Amira Software (FEI). Modified from [16].
Many of the findings in these studies relied on use of LAC values to identify organelles and observe changes in biochemical compositions, allowing for unbiased mapping of cellular organization. Because mitochondrial networks span much of the volume of the cell, quantifying its structure and relationships with neighboring organelles will be useful in characterizing mitochondrial behavior. Mapping mitochondrial structure under specific cellular conditions or drug stimulations will aid in correlating these structures with metabolic states of the cell.
4. Limitations and future developments
The above examples demonstrate how SXT can be applied to investigate mitochondrial structure and function, and they highlight opportunities for correlating changes of these structures with specific cellular states (Figure 1E). This type of analysis may reveal how disease states alter mitochondrial dynamics and cellular metabolism, providing new avenues for therapeutic design. In this light, significantly more information will surely be extracted from SXT tomograms as the field progresses and develops new analysis methods. Because SXT is a single-cell imaging modality, it can uncover unique structural signatures of cell subtypes, revealing cell-to-cell heterogeneities. Further exploitation of single-cell imaging and omics modalities provides an exciting path for understanding the role of specific cell subtypes in biology. Ideally, one will integrate the structure with biochemical data that reveal details about the biochemical states of subcompartments of the cell. The use of phasor-fluorescence lifetime imaging microscopy (FLIM) can monitor the metabolic state (i.e. distinguish between a more oxidative phosphorylation state and glycolytic state) in living cells before and after stimulation [49]. Future uses of SXT that match experimental conditions performed with complimentary imaging modalities like FLIM will enhance the information that can be extracted from SXT tomograms.
The use of SXT is currently limited by the lack of widespread instrumentation or facilities. Most research universities have core facilities capable of modern live cell imaging and/or electron microscopy imaging, but only a few locations in the world are set up for SXT imaging of whole cells because of the requirement for synchrotron radiation. With this challenge, the field is still developing and exploring how SXT can be applied to address a wide host of biological questions. The recent progress highlighted in this review demonstrates that there are rich opportunities for further application of SXT to characterize mitochondrial structure and function and to analyze cellular metabolism. The development of a laboratory-based SXT microscope would contribute to the continued development of the technique as a tool for structural cell biology [50].
The 3D maps generated by SXT do not characterize all aspects of cell biology, but represent a critical resource for integrating multi-scale data to describe how cellular components fit together and interact in the crowded cellular environment [16,51]. These SXT maps are expected to contribute important data towards assembling an integrative model of an entire cell, opening the door for next-generation structure-based therapeutic design [[52], [53], [54]].
Funding
This work was supported by the Bridge Institute at the USC Michelson Center for Convergent Bioscience, the Burroughs Wellcome Fund Travel Award (KLW). The National Center for X-ray Tomography is supported by NIH NIGMS (P30GM138441) and the DOE's Office of Biological and Environmental Research (DE-AC02-5CH11231).
Author contributions
VL and KLW wrote the manuscript.
Acknowledgments
KLW would like to thank Jitin Singla, Gerry McDermott, Claire Cato and Angela Walker for their comments on this manuscript, and Katya Kadyshevskaya for assistance in constructing the figures. KLW would also like to thank the members of her laboratory, Carolyn Larabell's lab at the National Center for Soft X-ray Tomography (NCXT), and the Pancreatic Beta Cell Consortium for their insightful discussions on the topic of cell mapping.
Conflict of interest
None declared.
References
- 1.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi T., Rizzuto R., Hajnoczky G., Su T.P. MAM: more than just a housekeeper. Trends in Cell Biology. 2009;19(2):81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Current Biology. 2008;18(2):102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 4.Wang C., Taki M., Sato Y., Tamura Y., Yaginuma H., Okada Y., et al. A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proceedings of the National Academy of Sciences of the U S A. 2019;116(32):15817–15822. doi: 10.1073/pnas.1905924116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valm A.M., Cohen S., Legant W.R., Melunis J., Hershberg U., Wait E., et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546(7656):162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekman A.A., Chen J.H., Guo J., McDermott G., Le Gros M.A., Larabell C.A. Mesoscale imaging with cryo-light and X-rays: larger than molecular machines, smaller than a cell. Biologie Cellulaire. 2017;109(1):24–38. doi: 10.1111/boc.201600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss D., Schneider G., Niemann B., Guttmann P., Rudolph D., Schmahl G. Computed tomography of cryogenic biological specimens based on X-ray microscopic images. Ultramicroscopy. 2000;84(3–4):185–197. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
- 8.Cinquin B.P., Do M., McDermott G., Walters A.D., Myllys M., Smith E.A., et al. Putting molecules in their place. Journal of Cellular Biochemistry. 2014;115(2):209–216. doi: 10.1002/jcb.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harkiolaki M., Darrow M.C., Spink M.C., Kosior E., Dent K., Duke E. Cryo-soft X-ray tomography: using soft X-rays to explore the ultrastructure of whole cells. Emerg Top Life Science. 2018;2(1):81–92. doi: 10.1042/ETLS20170086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider G., Guttmann P., Rehbein S., Werner S., Follath R. Cryo X-ray microscope with flat sample geometry for correlative fluorescence and nanoscale tomographic imaging. Journal of Structural Biology. 2012;177(2):212–223. doi: 10.1016/j.jsb.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson D.Y., Epperly L.R., McDermott G., Le Gros M.A., Boudreau R.M., Larabell C.A. Nanoimaging cells using soft X-ray tomography. Methods in Molecular Biology. 2013;950:457–481. doi: 10.1007/978-1-62703-137-0_25. [DOI] [PubMed] [Google Scholar]
- 12.Parkinson D.Y., Knoechel C., Yang C., Larabell C.A., Le Gros M.A. Automatic alignment and reconstruction of images for soft X-ray tomography. Journal of Structural Biology. 2012;177(2):259–266. doi: 10.1016/j.jsb.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer Determination of the aabsorption of red light in colored liquids. Annalen der Physik und Chemis. 1852;86:78–88. [Google Scholar]
- 14.Le Gros M.A., McDermott G., Cinquin B.P., Smith E.A., Do M., Chao W.L., et al. Biological soft X-ray tomography on beamline 2.1 at the advanced light source. Journal of Synchrotron Radiation. 2014;21(Pt 6):1370–1377. doi: 10.1107/S1600577514015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larabell C.A., Le Gros M.A. X-ray tomography generates 3-D reconstructions of the yeast, saccharomyces cerevisiae, at 60-nm resolution. Molecular Biology of the Cell. 2004;15(3):957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White K.L., Singla J., Loconte V., Chen J.H., Ekman A., Sun L., et al. Visualizing subcellular rearrangements in intact beta cells using soft x-ray tomography. Science Advances. 2020;6(50) doi: 10.1126/sciadv.abc8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duke E.M., Razi M., Weston A., Guttmann P., Werner S., Henzler K., et al. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM) Ultramicroscopy. 2014;143:77–87. doi: 10.1016/j.ultramic.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller W.G., Heymann J.B., Nagashima K., Guttmann P., Werner S., Rehbein S., et al. Towards an atlas of mammalian cell ultrastructure by cryo soft X-ray tomography. Journal of Structural Biology. 2012;177(2):179–192. doi: 10.1016/j.jsb.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elgass K.D., Smith E.A., LeGros M.A., Larabell C.A., Ryan M.T. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. Journal of Cell Science. 2015;128(15):2795–2804. doi: 10.1242/jcs.169136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curdt F., Herr S.J., Lutz T., Schmidt R., Engelhardt J., Sahl S.J., et al. isoSTED nanoscopy with intrinsic beam alignment. Optics Express. 2015;23(24):30891–30903. doi: 10.1364/OE.23.030891. [DOI] [PubMed] [Google Scholar]
- 21.Nieves D.J., Baker M.A.B. Pushing the super-resolution limit: recent improvements in microscopy below the diffraction limit. Biochemical Society Transactions. 2021;49(1):431–439. doi: 10.1042/BST20200746. [DOI] [PubMed] [Google Scholar]
- 22.Sakdinawat A., Attwood D. Nanoscale X-ray imaging. Nature Photonics. 2010;4(12):840–848. [Google Scholar]
- 23.Schneider G., Guttmann P., Heim S., Rehbein S., Mueller F., Nagashima K., et al. Three-dimensional cellular ultrastructure resolved by X-ray microscopy. Nature Methods. 2010;7(12):985–987. doi: 10.1038/nmeth.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDermott G., Fox D.M., Epperly L., Wetzler M., Barron A.E., Le Gros M.A., et al. Visualizing and quantifying cell phenotype using soft X-ray tomography. BioEssays. 2012;34(4):320–327. doi: 10.1002/bies.201100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falcone R., Jacobsen C., Kirz J., Marchesini S., Shaprio D., Spence J. New directions in X-ray microscopy. Contemporary Physics. 2011;52(4):293–318. [Google Scholar]
- 26.Weinhardt V., Chen J.H., Ekman A.A., Guo J., Remesh S.G., Hammel M., et al. Switchable resolution in soft x-ray tomography of single cells. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0227601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polo C.C., Fonseca-Alaniz M.H., Chen J.H., Ekman A., McDermott G., Meneau F., et al. Three-dimensional imaging of mitochondrial cristae complexity using cryo-soft X-ray tomography. Scientific Reports. 2020;10(1):21045. doi: 10.1038/s41598-020-78150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noske A.B., Costin A.J., Morgan G.P., Marsh B.J. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. Journal of Structural Biology. 2008;161(3):298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller A., Schmidt D., Xu C.S., Pang S., D'Costa J.V., Kretschmar S., et al. 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse beta cells. The Journal of Cell Biology. 2021;220(2) doi: 10.1083/jcb.202010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth M.S., Gallaher S.D., Westcott D.J., Iwai M., Louie K.B., Mueller M., et al. Regulation of oxygenic photosynthesis during trophic transitions in the green alga chromochloris zofingiensis. The Plant Cell Online. 2019;31(3):579–601. doi: 10.1105/tpc.18.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uchida M., Sun Y., McDermott G., Knoechel C., Le Gros M.A., Parkinson D., et al. Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast. 2011;28(3):227–236. doi: 10.1002/yea.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagen C., Guttmann P., Klupp B., Werner S., Rehbein S., Mettenleiter T.C., et al. Correlative VIS-fluorescence and soft X-ray cryo-microscopy/tomography of adherent cells. Journal of Structural Biology. 2012;177(2):193–201. doi: 10.1016/j.jsb.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith E.A., Cinquin B.P., Do M., McDermott G., Le Gros M.A., Larabell C.A. Correlative cryogenic tomography of cells using light and soft x-rays. Ultramicroscopy. 2014;143:33–40. doi: 10.1016/j.ultramic.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kounatidis I., Stanifer M.L., Phillips M.A., Paul-Gilloteaux P., Heiligenstein X., Wang H., et al. 3D correlative cryo-structured illumination fluorescence and soft X-ray microscopy elucidates reovirus intracellular release pathway. Cell. 2020;182(2):515–530. doi: 10.1016/j.cell.2020.05.051. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagen C., Werner S., Carregal-Romero S., N M A, G K B, Guttmann P., et al. Multimodal nanoparticles as alignment and correlation markers in fluorescence/soft X-ray cryo-microscopy/tomography of nucleoplasmic reticulum and apoptosis in mammalian cells. Ultramicroscopy. 2014;146:46–54. doi: 10.1016/j.ultramic.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapishnikov S., Grolimund D., Schneider G., Pereiro E., McNally J.G., Als-Nielsen J., et al. Unraveling heme detoxification in the malaria parasite by in situ correlative X-ray fluorescence microscopy and soft X-ray tomography. Scientific Reports. 2017;7(1):7610. doi: 10.1038/s41598-017-06650-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscheni C., Malucelli E., Castiglioni S., Procopio A., De Palma C., Sorrentino A., et al. 3D quantitative and ultrastructural analysis of mitochondria in a model of doxorubicin sensitive and resistant human colon carcinoma cells. Cancers. 2019;11(9) doi: 10.3390/cancers11091254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reineck P., Abraham A.N., Poddar A., Shukla R., Abe H., Ohshima T., et al. Multimodal imaging and soft X-ray tomography of fluorescent nanodiamonds in cancer cells. Biotechnology Journal. 2021;16(3) doi: 10.1002/biot.202000289. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y., Kim C., Kwon O.Y., Nam D., Kim S.S., Park J.H., et al. Visualization of a mammalian mitochondrion by coherent X-ray diffractive imaging. Scientific Reports. 2017;7(1):1850. doi: 10.1038/s41598-017-01833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Juhl A.D., Heegaard C.W., Werner S., Schneider G., Krishnan K., Covey D.F., et al. Quantitative imaging of membrane contact sites for sterol transfer between endo-lysosomes and mitochondria in living cells. Scientific Reports. 2021;11(1):8927. doi: 10.1038/s41598-021-87876-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siasos G., Tsigkou V., Kosmopoulos M., Theodosiadis D., Simantiris S., Tagkou N.M., et al. Mitochondria and cardiovascular diseases-from pathophysiology to treatment. Annals of Translational Medicine. 2018;6(12):256. doi: 10.21037/atm.2018.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui M.F., Elwell C., Johnson M.H. Mitochondrial dysfunction in autism spectrum disorders. Autism-Open Access. 2016;6(5) doi: 10.4172/2165-7890.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vincent A.E., White K., Davey T., Philips J., Ogden R.T., Lawless C., et al. Quantitative 3D mapping of the human skeletal muscle mitochondrial network. Cell Reports. 2019;27(1):321. doi: 10.1016/j.celrep.2019.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parone P.A., Da Cruz S., Tondera D., Mattenberger Y., James D.I., Maechler P., et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3(9) doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otera H., Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. Journal of Biochemistry. 2011;149(3):241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- 46.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juhl A.D., Lund F.W., Jensen M.L.V., Szomek M., Heegaard C.W., Guttmann P., et al. Niemann Pick C2 protein enables cholesterol transfer from endo-lysosomes to the plasma membrane for efflux by shedding of extracellular vesicles. Chemistry and Physics of Lipids. 2021;235:105047. doi: 10.1016/j.chemphyslip.2020.105047. [DOI] [PubMed] [Google Scholar]
- 48.Wollheim C.B. Beta-cell mitochondria in the regulation of insulin secretion: a new culprit in type II diabetes. Diabetologia. 2000;43(3):265–277. doi: 10.1007/s001250050044. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z., Gurlo T., Matveyenko A.V., Elashoff D., Wang P., Rosenberger M., et al. Live-cell imaging of glucose-induced metabolic coupling of beta and alpha cell metabolism in health and type 2 diabetes. Commun Biol. 2021;4(1):594. doi: 10.1038/s42003-021-02113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fahy K., Weinhardt V., Vihinen-Ranta M., Fletcher N., Skoko D., Pereiro E., et al. Compact Cell Imaging Device (CoCID) provides insights into the cellular origins of viral infections. Journal of Physics: Photonics. 2021;3(3) [Google Scholar]
- 51.Johnson G.T., Autin L., Al-Alusi M., Goodsell D.S., Sanner M.F., Olson A.J. cellPACK: a virtual mesoscope to model and visualize structural systems biology. Nature Methods. 2015;12(1):85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singla J., McClary K.M., White K.L., Alber F., Sali A., Stevens R.C. Opportunities and challenges in building a spatiotemporal multi-scale model of the human pancreatic beta cell. Cell. 2018;173(1):11–19. doi: 10.1016/j.cell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singla J., White K.L. A community approach to whole-cell modeling. Current Opinion in Structural Biology. 2021;26:33–38. [Google Scholar]
- 54.Sali A. From integrative structural biology to cell biology. Journal of Biological Chemistry. 2021:100743. doi: 10.1016/j.jbc.2021.100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu C.S., Hayworth K.J., Lu Z., Grob P., Hassan A.M., Garcia-Cerdan J.G., et al. Enhanced FIB-SEM systems for large-volume 3D imaging. Elife. 2017;6 doi: 10.7554/eLife.25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao A., McBride E.L., Zhang G., Xu H., Cai T., Notkins A.L., et al. Determination of secretory granule maturation times in pancreatic islet beta-cells by serial block face scanning electron microscopy. Journal of Structural Biology. 2020:107584. doi: 10.1016/j.jsb.2020.107584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffman D.P., Shtengel G., Xu C.S., Campbell K.R., Freeman M., Wang L., et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science. 2020;367(6475) doi: 10.1126/science.aaz5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Cuellar L.K., et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351(6276):969–972. doi: 10.1126/science.aad8857. [DOI] [PubMed] [Google Scholar]
- 59.Zhang X., Carter S.D., Singla J., White K.L., Butler P.C., Stevens R.C., et al. Visualizing insulin vesicle neighborhoods in beta cells by cryo-electron tomography. Science Advances. 2020;6(50) doi: 10.1126/sciadv.abc8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao C.L., Liu Y.T., Sun R., Zhang B., Qi L., Shivakoti S., et al. Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. Journal of Neuroscience. 2018;38(6):1493–1510. doi: 10.1523/JNEUROSCI.1548-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilhelm B.G., Mandad S., Truckenbrodt S., Krohnert K., Schafer C., Rammner B., et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344(6187):1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 62.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., et al. A subcellular map of the human proteome. Science. 2017;356(6340) doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 63.Ounkomol C., Seshamani S., Maleckar M.M., Collman F., Johnson G.R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nature Methods. 2018;15(11):917–920. doi: 10.1038/s41592-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo Y., Li D., Zhang S., Yang Y., Liu J.J., Wang X., et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell. 2018;175(5):1430–1442 e17. doi: 10.1016/j.cell.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 65.Parkinson D.Y., McDermott G., Etkin L.D., Le Gros M.A., Larabell C.A. Quantitative 3-D imaging of eukaryotic cells using soft X-ray tomography. Journal of Structural Biology. 2008;162(3):380–386. doi: 10.1016/j.jsb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]