Summary
Zebrafish regenerate fin rays following amputation through epimorphic regeneration, a process that has been proposed to involve the epithelial-to-mesenchymal transition (EMT). We performed single-cell RNA sequencing (scRNA-seq) to elucidate osteoblastic transcriptional programs during zebrafish caudal fin regeneration. We show that osteoprogenitors are enriched with components associated with EMT and its reverse, mesenchymal-to-epithelial transition (MET), and provide evidence that the EMT markers cdh11 and twist2 are co-expressed in dedifferentiating cells at the amputation stump at 1 dpa, and in differentiating osteoblastic cells in the regenerate, the latter of which are enriched in EMT signatures. We also show that esrp1, a regulator of alternative splicing in epithelial cells that is associated with MET, is expressed in a subset of osteoprogenitors during outgrowth. This study provides a single cell resource for the study of osteoblastic cells during zebrafish fin regeneration, and supports the contribution of MET- and EMT-associated components to this process.
Subject areas: Biological sciences, Developmental biology, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Osteoblasts express EMT/MET signatures during zebrafish fin regeneration
-
•
De/re-differentiating osteoblasts express cdh11, an EMT marker
-
•
A subset of osteoprogenitors express the MET marker esrp1
-
•
Our scRNA-seq data can be explored online
Biological sciences; Developmental biology; Omics; Transcriptomics
Introduction
While humans have limited potential for limb regeneration, some vertebrates can regenerate bony appendages following amputation. Zebrafish can regenerate amputated caudal fins through the formation of a pool of lineage-restricted progenitors called the blastema (Poss et al., 2000). Following fin amputation, mature osteoblasts at the amputation stump dedifferentiate, migrate to the blastema, self-renew, then redifferentiate to form new bone (Knopf et al., 2011; Singh et al., 2012; Sousa et al., 2011; Stewart and Stankunas, 2012). This process begins within the first 24 hours after amputation, with osteoblast redifferentiation beginning by 2 days post amputation (dpa) as detected by the upregulation of the committed osteoblast marker sp7, followed by the expression of the late differentiation marker bglap, which is detectable by 5 to 6 dpa. In addition to dedifferentiation of mature cells, osteoprogenitors also derive from resident mesenchymal precursors within the fin ray joint (Ando et al., 2017). The formation of osteoprogenitors through dedifferentiation in zebrafish contrasts from mammals, where osteoblasts primarily derive from resident precursors. The cell biological processes that enable dedifferentiation and reversible switching between differentiated and self-renewing cell states are poorly understood.
Cell plasticity and reversibility between metastable cell states are hallmarks of the epithelial-to-mesenchymal transition (EMT), and its reverse, mesenchymal-to-epithelial transition (MET). EMT is a developmental program by which cells lose epithelial (E) characteristics (e.g., E-cadherin, desmosomes, cortical actin, and apical–basal polarity) while adopting features of mesenchymal (M) cells (e.g., increased vimentin) (Yang et al., 2020). EMT is often observed during cancer invasion and metastasis, where it results in dedifferentiation and acquisition of cancer stem cell-like properties (Mani et al., 2008). While cancer progression is impaired by complete EMT, cancer cell stemness is enhanced by partial EMT, where cells reversibly switch between metastable cell states with hybrid E/M phenotypes that are epithelial-shifted or mesenchymal-shifted (Saxena et al., 2020). Epimorphic regeneration exhibits a number of biological parallels to the progression of solid tumors. It has been suggested that certain mechanisms enabling regeneration may be co-opted by cancer to promote growth at primary and metastatic sites (Wong and Whited, 2020). In silico evidence of a sequential EMT and MET process occurring during axolotl limb regeneration has been reported (Li et al., 2021); however, we are unaware of other articles reporting the existence of EMT signatures in epimorphic regenerative models. A review by Wong and Whited specifically states that no EMT signatures were identified in two axolotl limb regeneration scRNA-seq studies, indicating that more investigation is needed (Gerber et al., 2018; Leigh et al., 2018; Wong and Whited, 2020).
There is evidence that mammalian osteoblastic cells differentially exhibit characteristics of mesenchymal and epithelial cells, which may be conserved in zebrafish. In mammals, early osteoblastic cells are stromal cell-like in appearance, whereas matrix-secreting osteoblasts are tall or cuboidal and form a continuous layer, bearing a partial resemblance to epithelial cells (Ilvesaro et al., 1999). The primary form of cell-cell adhesion between osteoblasts is through adherens junctions (Cheng et al., 1998; Okazaki et al., 1994), which are typically found in epithelial and endothelial cells and rely on the interaction of cadherins at the cell surface to form an intercellular adhesion complex. Multiple studies have shown that interfering with cadherin function impairs osteoblast differentiation (Stains and Civitelli, 2005). Upon further maturation, active osteoblasts transition into quiescent bone lining cells, appearing as flat and elongated cells on the mineralized surface. Thus, while mesenchymal in origin, mammalian osteoblastic cells differentially exhibit characteristics of mesenchymal and epithelial cells depending on cell state.
During zebrafish fin regeneration, Stewart et al. noted dual mesenchymal and epithelial characteristics in osteoblastic cells, and hypothesized that this was due to EMT (Stewart et al., 2014). Specifically, they observed that blastemal osteoprogenitors expressed the EMT-associated transcription factors (EMT-TFs) twist2 and twist3, exhibited decreased staining of the epithelial marker Ctnna1, and changed from an elongated to a polygonal morphology. However, whether this reflects an EMT-like transition or a function independent of EMT is poorly understood. Only a limited set of EMT markers have been characterized, which may be insufficient to resolve hybrid cells that exhibit both E and M characteristics (Yang et al., 2020). Moreover, multiple markers are often needed to precisely assess the state of a cell undergoing EMT within specific biological contexts (Yang et al., 2020). Finally, many transcription factors and other components critical for EMT also affect other cellular functions. These components may be regulated through programs that involve canonical EMT (Yang et al., 2020). As such, more in-depth molecular characterizations are needed. While single-cell analyses are comprehensive approaches and can provide valuable insight, previous single-cell RNA-seq (scRNA-seq) studies of zebrafish caudal fin regeneration have reported low numbers of osteoblastic cells (Hou et al., 2020), hampering an in-depth analysis.
Here, we performed sci-RNA-seq (single-cell combinatorial indexing RNA sequencing) (Cao et al., 2017) to elucidate osteoblastic transcriptional programs during zebrafish caudal fin regeneration. We speculated that the higher throughput of sci-RNA-seq in concert with differences in sample preparation could help retrieve osteoblastic cells compared to droplet-based scRNA-seq approaches. Specifically, sci-RNA-seq uses combinatorial indexing for transcriptomic profiling, enabling higher throughput compared to droplet-based scRNA-seq where individual cells are isolated within physical compartments (Cao et al., 2017). Moreover, whereas droplet-based scRNA-seq commonly employs tissue dissociation using enzymes and usually heat (Denisenko et al., 2020), sci-RNA-seq commonly employs manual homogenization, which could help retrieve cells easily destroyed by, or not readily liberated by, enzymatic tissue dissociation. Thus, in contrast to existing scRNA-seq resources (Hou et al., 2020), our dataset captures a high number of osteoblastic cells (1,673 nuclei), allowing for their in-depth analysis. We show that osteoprogenitors are enriched with components associated with the epithelial-to-mesenchymal transition (EMT) and its reverse, mesenchymal-to-epithelial transition (MET). In trajectory analyses, osteoblastic cells solely expressed EMT components, or transiently expressed MET components prior to expressing those for EMT. We provide evidence that the EMT markers cdh11 and twist2 (Alimperti and Andreadis, 2015) are co-expressed in dedifferentiating cells at the amputation stump at 1 dpa and in differentiating osteoblastic cells in the regenerate at 3 dpa, and that EMT signatures are enriched in the latter. We also show that esrp1, a regulator of alternative splicing in epithelial cells whose expression is important for MET (Warzecha et al., 2009a), is expressed in a subset of osteoprogenitors during outgrowth. This study provides a single cell resource for the study of osteoblastic cells during zebrafish fin regeneration, and supports the contribution of MET- and EMT-associated components to this process.
Results
Sci-RNA-seq captures rare osteoblastic populations
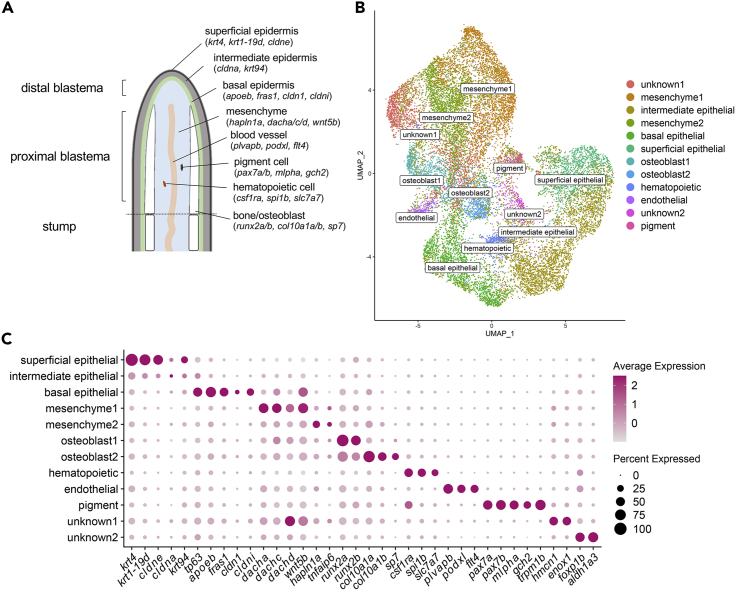
We performed sci-RNA-seq using 225 fins from 3 and 5 dpa, which yielded 19,603 nuclei after filtering for quality control. At these timepoints, a variety of tissues have regenerated (Figure 1A). Unbiased clustering identified 12 different cell populations, of which we defined 10 out of 12 by cell/tissue type using published biomarkers (Figure 1BC, Table S1) (Anbalagan et al., 2018; Avaron et al., 2006; Brown et al., 2009; Carney et al., 2010; Clendenon et al., 2009; Covassin et al., 2006; Gong et al., 2002; Govindan and Iovine, 2014; Hou et al., 2020; Kastenhuber et al., 2013; Knopf et al., 2011; Lee et al., 2009; Li et al., 2009; Marin-Juez et al., 2016; Monnot et al., 1999; Nord et al., 2016; Pagan et al., 2015; Parichy et al., 2000; Pittlik et al., 2008; Rossi et al., 2015; Sheets et al., 2007; Shibata et al., 2018; Sousa et al., 2011; Stewart et al., 2019; Stoick-Cooper et al., 2007; Venkateswaran et al., 2013; Zakrzewska et al., 2010). All populations were observed at both 3 and 5 dpa (see Figure S1A). There was noticeable variability in the proportion of cell types in two different samples from 3 dpa (see Figure S1BC), indicating that the relationship between regeneration timepoint and cell-type proportion could not be reliably inferred. Thus, all timepoints were pooled for final analysis.
Figure 1.
Molecular profiling of fin regeneration reveals distinct cell types including two osteoblast populations
(A) Schematic of zebrafish fin regeneration showing tissues and corresponding gene markers.
(B) UMAP plot visualizing 19,603 cells from sci-RNA-seq analysis of regenerated fins at 3 and 5 dpa. Of the 12 distinct clusters, 10 tissue types could be identified by molecular markers, including two osteoblast populations.
(C) Dotplot visualizing expression of genes demarcating the 12 cell clusters. Circle size represents the percentage of cells expressing the gene, and color indicates average gene expression. See also Figure S1
We identified two populations of osteoblastic cells—osteoblast1 (973 cells) and osteoblast2 (700 cells)—marked by the strong expression of the osteoprogenitor marker runx2a/b (Figure 1C), a transcription factor expressed during early osteoblast differentiation. Mapping of osteoblast1 and osteoblast2 identities to a previous scRNA-seq dataset of fin regeneration (Hou et al., 2020) revealed 16 and 23 osteoblast1 and osteoblast2 cells, respectively (∼40x fewer cells), indicating the difficulty of capturing osteoblastic cells in scRNA-seq studies.
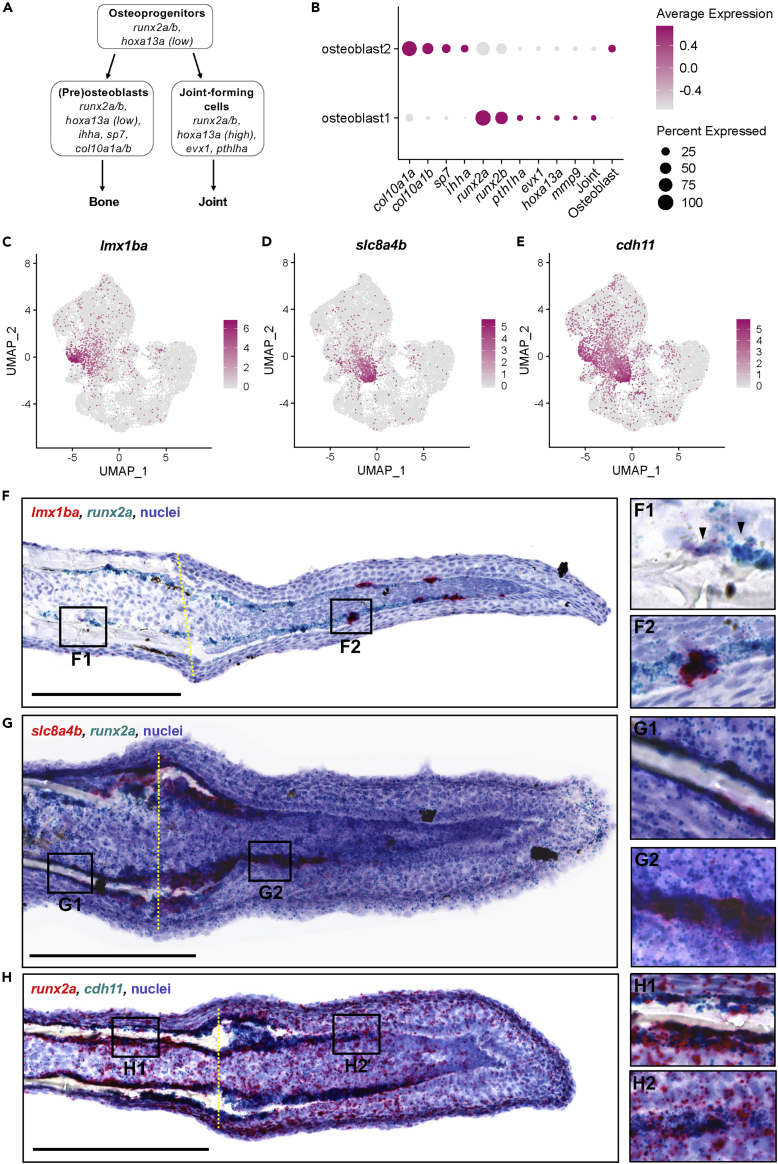
Osteoprogenitors can differentiate into osteoblasts and joint-forming cells (McMillan et al., 2018) (Figure 2A). Cells within the osteoblast1 cluster expressed genes known to be upregulated during osteoprogenitor differentiation into joint cells. This included pthlha, a marker of joint cells (McMillan et al., 2018), and mmp9, a marker of joint-derived resident osteoprogenitors (Ando et al., 2017) (Figure 2B). In contrast, the osteoblast2 cluster expressed genes known to be specifically expressed during osteoblast differentiation, notably col10a1a/b (a marker of pre-osteoblasts), sp7 (a marker of immature osteoblasts), and ihha (McMillan et al., 2018) (Figure 2B). The osteoblast1 and osteoblast2 clusters scored higher for joint cell and osteoblast gene signatures, respectively (Figure 2B). We identified genes with significant differences in expression between the two osteoblast groups (see Table S2). These genes included lmx1ba in the osteoblast1 cluster (Figure 2C), and slc8a4b in the osteoblast2 cluster (Figure 2D). We also identified genes with significantly increased expression in both groups, compared to non-osteoblastic cell types (see Table S3). This included cdh11 (Figure 2E), which was recently identified by scRNA-seq to be a potential marker of osteoblastic cells, but still remains to be experimentally validated (Hou et al., 2020).
Figure 2.
Osteoblast1 and osteoblast2 are likely enriched with cells fated to become joint cells and osteoblasts, respectively
(A) Schematic of osteoprogenitor differentiation into either osteoblasts or joint-forming cells and corresponding gene markers (adapted from McMillan et al., 2018).
(B) Dotplot comparing the expression of joint cell markers and bone markers in the two osteoblast populations. The clusters osteoblast1 and osteoblast2 are enriched with genes associated with joint-forming cells and osteoblasts, respectively.
(C–E) Feature plots for lmx1ba, slc8a4b, and cdh11.
(F-H) Dual RNA in situ hybridization in 3 dpa fin cryosections for the indicated genes. All scale bars are 200 μm. Amputation planes are indicated with a yellow dotted line.
(F) lmx1ba+/runx2a+ cells were present in cells near the native joint (F1 inset, black arrowheads), and in presumptive joint cells in the regenerate (F2 inset).
(G) slc8a4b+/runx2a+ cells were present at the amputation stump, near the regenerating bone, and in cells along the proximal lateral mesenchymal compartment.
(H) cdh11+ cells lined the native bone (H1 inset), the regenerating bone near the amputation stump, and cells along the lateral edges of the mesenchymal compartment of the blastema (H2 inset).
Expression of lmx1ba, slc8a4b, and cdh11 has not previously been characterized during zebrafish fin regeneration. Thus, we performed dual RNA in situ hybridization (ISH) to stain for transcripts for these genes in concert with runx2a. Previous fin regeneration studies have shown that at 3 dpa, osteoprogenitors in the fin blastema are located distal to the amputation stump, extending along the lateral edges of the mesenchymal compartment, just deep to the basal epidermis. We found high expression of runx2a in cells residing in these areas. Although we did not do lineage analysis in our studies, based on their location within the blastema, cells in these areas that were strongly expressing runx2a were most likely to be osteoprogenitors. We also observed lower expression of runx2a in the epidermis and mesenchyme. Our scRNA-seq data indicate that runx2a is expressed in some cells within the epidermis and mesenchyme populations (see Figure 1C). While this was not the focus of this study, our data support the notion of low expression of runx2a in epidermis and mesenchyme during fin rengeneration.
We observed staining for the osteoblast1 marker lmx1ba in distinct, relatively evenly spaced clusters of runx2a+ cells along the lateral mesenchymal compartment of the blastema (Figure 2F2), similar to the staining pattern of the known joint markers pthlha, evx1, and hoxa13a (McMillan et al., 2018). We also detected runx2a+/lmx1ba+ cells in the native fin ray joints (Figure 2F1, black arrowheads). These data support the potential for lmx1ba+ cells in the blastema to be presumptive regenerating joint cells of the fin rays. Mutations in the human ortholog LMX1B have been implicated in focal segmental glomerulosclerosis (a disease of the kidney) and nail-patella syndrome, which is commonly manifested by underdeveloped or irregularly shaped nails, elbows, and knees (Dreyer et al., 1998). Lmx1b-deficient mice exhibit abnormalities in dorsal limb structures attributable to defects in dorsal-ventral patterning (Chen et al., 1998). In mice, Lmx1b appears to be expressed between, rather than within, developing bones (McIntosh et al., 2005) and it has been postulated that Lmx1b could have additional function in anterior-posterior patterning (Dreyer et al., 1998). Further studies of the function of lmx1ba in zebrafish fin regeneration are warranted.
Next, we examined the osteoblast2 marker slc8a4b (Figure 2G). We observed staining for slc8a4b in runx2a+ cells lining the distal end of the amputation stump, around the newly regenerating bone, and in cells along the proximal lateral mesenchymal compartment where new bone was likely to form (Figure 2G2). Staining for slc8a4b was largely absent in runx2a+ cells in the native bone (Figure 2G1). These data support the potential for slc8a4b+ cells to be a population of osteoblast progenitors. slc8a4b was first identified during a screen for homologs of the mammalian genes Slc8a1/2/3 (formerly NCX1/2/3), which are members of the calcium/cation antiporter family of proteins that help regulate intracellular calcium in a variety of cell types. Initially called NCX4, slc8a4b was shown to be a distinct fourth isoform of the NCX family, and orthologs of NCX4/slc8a4b have only been identified in bony fishes (Marshall et al., 2005). In our studies, a tBLASTn of slc8a4b showed highest similarity to mammalian Slc8a1 (XM_006,523,523943.2, E-value: 0, query cover: 96%, identity: 68%). In humans, genetic variants near SLC8A1 have been associated with risk of bone fracture (Kemp et al., 2017). In mice, Slc8a1 is expressed in osteoclasts and plays a role in bone resorption (Albano et al., 2017); however, its function in bone formation requires further study.
Finally, we examined expression of cdh11 (Figure 2H). We observed prominent staining for cdh11 in runx2a+ cells lining the amputation stump, in the lateral mesenchymal compartment (Figure 2H2), and in the native bone (Figure 2H1). CDH11 is highly expressed in osteoblastic cell lines and upregulated during differentiation. Cdh11-deficient mice exhibit hypoplastic synovium (Lee et al., 2007), resistance to inflammatory arthritis (Lee et al., 2007), and osteopenia (Kawaguchi et al., 2001). These studies support the notion that osteoblast1 and osteoblast2 cells are distinct populations enriched for cells fated to become joint cells and osteoblasts, respectively.
Osteoblastic cells exhibit distinct cell states and trajectories
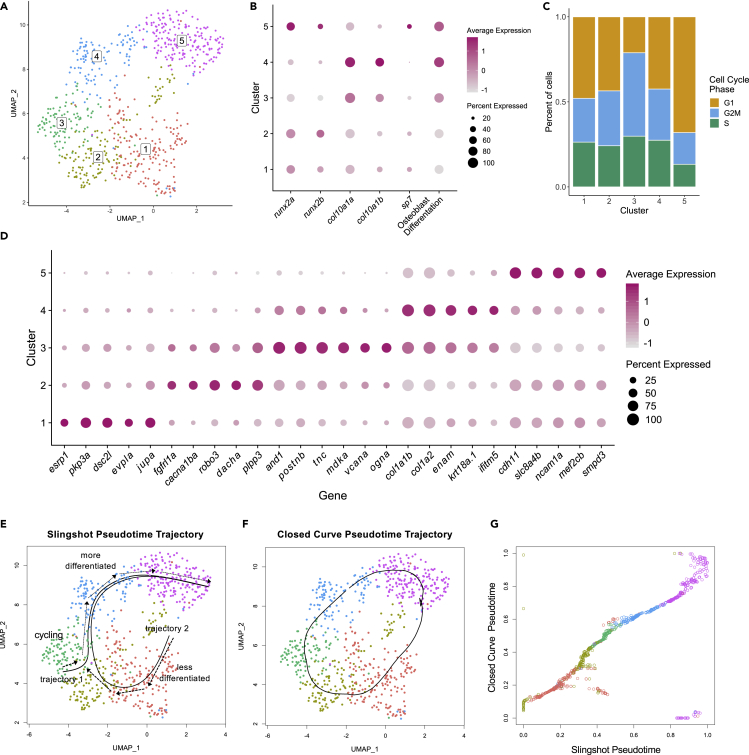
We subset the osteoblast2 cells for further analysis. Unbiased clustering of the osteoblast2 cells revealed five subclusters (Figure 3A). Analysis of markers of osteoblast differentiation (Figure 3B) and cell cycle phase (Figure 3C) revealed distinct cell cycle and differentiation states. Clusters 4 and 5 had the highest osteoblast differentiation scores. Whereas cluster 5 was enriched for sp7, cluster 4 was enriched for col10a1a/b. Moreover, cluster 5 had the lowest percentage of cells in the G2/M and S phases. Cluster 3 had a lower osteoblast differentiation score compared with clusters 4 and 5. Moreover, these cells had relatively high expression of col10a1a/b, and had the highest percentage of cycling cells as indicated by cells in the G2/M and S phases. Cluster 3 also had low runx2a/b expression, which is in agreement with previous studies showing that murine RUNX2 levels regulate cell cycle entry and exit in osteoblasts, with minimal levels of RUNX2 during G2 and mitosis (Galindo et al., 2005; San Martin et al., 2009). Clusters 1 and 2 had the lowest osteoblast differentiation scores, and had higher runx2a/b expression compared with clusters 3 and 4. In regard to statistical analysis, using the Wilcoxon rank-sum test with Bonferroni correction (see Figure S2) we found that for cluster 5, the osteoblast differentiation score was significantly different from clusters 1 and 2; for cluster 4, the osteoblast differentiation score was significantly different from clusters 1, 2, and 3. For cell cycle scores, using a difference of proportions test (prop.test in R) with Bonferroni correction (see Figure S3), we found that the proportion of cycling cells was significantly different between most cluster pairwise comparisons except between clusters 1 and 2, clusters 1 and 4, and clusters 2 and 4. To assess whether specific clusters were enriched with doublets, we used the computeDoubletDensity function from the scDblFinder package in R (Lun et al., 2016). This function generates simulated doublets by randomly pairing cells from the dataset, and then calculates a doublet score for each cell based on its similarity to simulated doublets (see Figure S4). We did not find clear evidence that doublets were enriched in any individual cluster, suggesting that each cluster represented a bona fide subpopulation. The above studies identify diverse osteoblastic subpopulations which differ in cell cycle and differentiation status, three of which (clusters 3, 4, and 5) we could identify based on cell cycle status or known osteoblastic biomarkers.
Figure 3.
Clustering and trajectory analysis of osteoblast 2 population
(A) UMAP plot showing five distinct osteoblastic subclusters.
(B) Dotplot visualizing expression of known genes marking the different stages of osteoblast differentiation and osteoblast differentiation scores.
(C) Percentage of cells in each cell cycle phase for each osteoblast subcluster.
(D) Dotplot showing selected genes found to be differentially expressed within each osteoblastic subcluster.
(E–G) Pseudotemporal ordering of osteoblast 2 cells. (E) Slingshot pseudotime ordering yields two trajectories. Trajectory 1 starts in cluster 3, whereas trajectory 2 starts in cluster 1. (F) Closed curve analysis suggests a similar trajectory to trajectory 2 derived from Slingshot analysis. (G) Comparison of the closed curve trajectory to trajectory 2 derived from Slingshot shows that the orderings from open (Slingshot) and closed curve analyses are highly similar
Next, we performed differential gene expression analysis (Figure 3D, Table S4) and found that osteoblast2 subclusters were uniquely characterized by genes associated with the epithelium or involved in extracellular matrix formation or osteoblast differentiation. Of the 10 most differentially expressed genes in cluster 1, nine were associated with epithelial cells, including genes encoding for components of desmosomes (dsc2l and evpla) and adherens junctions (pkp3a). Among the top marker genes in clusters 3–4 were those encoding for extracellular matrix components. For instance, 10 of the top 20 genes of cluster 3 included markers such as postnb, and1, tnc, vcana, and ogna, while the top two most differentially expressed genes in cluster 4 encoded for type I collagen (col1a1b and col1a2). Cluster 4 was also marked by the expression of ifitm5 which has been implicated in skeletal disease (Farber et al., 2014). The top four markers of cluster 5 included genes whose orthologs are expressed during osteoblast differentiation (cdh11 (Okazaki et al., 1994), mef2c (Stephens et al., 2011), ncam1a (Qin et al., 2019)) and/or are implicated in skeletal disease (smpd3 (Aubin et al., 2005)). A complete analysis of enrichment for pathways and GO terms can be seen in Figure S5.
To assess ordering through cell states, we performed pseudotime and trajectory analysis. In constructing trajectories, the direction of the trajectory may go from more to less differentiated (dedifferentiation), or less to more differentiated (differentiation). At 3–5 dpa, osteoblastic cells are predominantly undergoing differentiation rather than dedifferentiation (Knopf et al., 2011). Thus, we presumed the direction of the trajectory was from less to more differentiated. Because cluster 5 had the highest gene expression for the osteoblast marker sp7, we further presumed that cluster 5 was likely to be the terminal state. Ando et al. previously showed that mmp9 is a marker of joint osteoprogenitors that contribute to fin regeneration (Ando et al., 2017). In separate studies in which we subset all cells (including osteoblast1 and osteoblast2) that expressed either sp7 or runx2a, we did not detect a clear mmp9-enriched cluster which would be needed to construct a trajectory from joint osteoprogenitor cells to differentiated osteoblasts. Thus, joint cells were not included in our trajectory analyses. Unbiased analysis using Slingshot (Street et al., 2018) revealed two trajectories (Figure 3E). Trajectory 1 started in cluster 3 and progressed through clusters 4 and 5. In this trajectory, cycling osteoprogenitors transition into pre-osteoblasts that express col10a1a/b, and then further transition into immature osteoblasts that express sp7. Trajectory 2 started in cluster 1, and then progressed through a trajectory similar to trajectory 1. In both trajectories, extracellular matrix genes such as postnb, and1, tnc, vcanb, and ogna are expressed prior to or concomitantly with genes encoding for type I collagen (col1a1b and col1a2). This is consistent with the synthesis of a transitional matrix rich in tenascin C prior to the synthesis of ECM proteins characteristic of differentiated skeletal tissues like collagen type I (Govindan and Iovine, 2015) during zebrafish fin regeneration and salamander limb regeneration. These studies point to multiple candidate trajectories underlying the identified cell states.
We then evaluated whether the trajectories exhibited characteristics of de- and redifferentiation. We hypothesized that de- and redifferentiation involves a cyclical progression through cell states which would manifest as a closed curve through gene expression space. We therefore sought to examine whether trajectory 2 represented a path through cellular de- and redifferentiation by assessing if the pseudotime orderings in the trajectory were similar to those derived from a closed curve. Algorithms for the analysis of cyclic trajectories are sparse compared to non-cyclic methods (Saelens et al., 2019). Moreover, ANGLE, the highest performing cyclic algorithm benchmarked in Saelens et al., 2019, requires 2D data embeddings. We therefore devised an algorithm for pseudotime inference that enables closed curve trajectory analysis using data embeddings of arbitrary dimension (Figure 3F). Pseudotime orderings derived from the closed curve trajectory were highly similar to trajectory 2 (Figure 3G). In addition, orderings derived using either UMAP or PCA embeddings were comparable to each other (see Figure S6). Thus, pseudotime orderings were similar whether using an open (Slingshot) or closed curve, the latter of which is predicted with a cyclical progression through cellular de- and redifferentiation states.
Osteoblastic cells differentially express MET- and EMT-associated genes
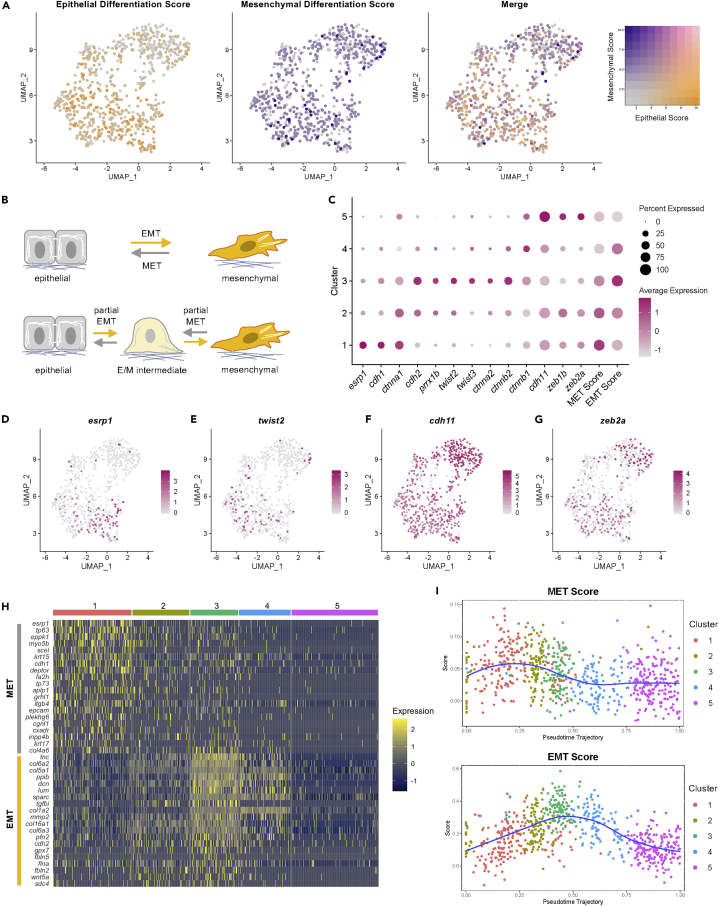
Next, we assessed the mesenchymal and epithelial characteristics of osteoblast2 subclusters. For this, we computed mesenchymal and epithelial differentiation scores (Figure 4A). Whereas mesenchymal differentiation scores were relatively uniform across cell clusters, epithelial differentiation scores were higher in clusters 1–3 compared with clusters 4–5. As a consequence, mesenchymal scores were higher than epithelial scores in clusters 4–5. Hence, osteoblastic cells express genes associated with both mesenchymal and epithelial differentiation, which are differentially expressed depending on cell state. Because expression of epithelial and mesenchymal markers is expected in cells undergoing MET and EMT, respectively, we next investigated whether osteoblast2 subclusters expressed MET and EMT components.
Figure 4.
Osteoblastic cells express EMT- and MET-associated genes
(A) Osteoblastic cells colored by epithelial differentiation score (left), mesenchymal differentiation score (second from left), and merged epithelial and mesenchymal differentiation scores (third from left). Color legend at right.
(B) Partial EMT and MET result in intermediate cellular phenotypes that encompass dual epithelial and mesenchymal phenotypes (bottom).
(C) Dotplot of the expression of canonical EMT and MET markers in the osteoblastic subclusters. Clusters were also scored for the expression of EMT and MET gene signatures.
(D–G) Feature plots for esrp1, twist2, cdh11, and zeb2a. (H) Top differentially expressed MET and EMT genes in the osteoblasts as visualized by heatmap. (I) Pseudotemporal expression of MET (top) and EMT (bottom) gene signatures in the osteoblast 2 cells using trajectory 2
We first assessed whether the top markers for osteoblastic cell clusters included canonical markers for MET and EMT (Alimperti and Andreadis, 2015; Ansieau et al., 2008; Batlle et al., 2000; Bebee et al., 2015; Burguera et al., 2017; Cano et al., 2000; Comijn et al., 2001; Funahashi et al., 1993; Grooteclaes and Frisch, 2000; Jhingory et al., 2010; Kang and Massague, 2004; Kimura et al., 1995; Leptin, 1991; Nieto, 2002; Ocana et al., 2012; Vermeulen et al., 1999; Verschueren et al., 1999; Warzecha et al., 2009b; Zhao et al., 2020). We found that the second most differentially expressed gene in cluster 1 was esrp1. Epithelial splicing regulatory proteins (ESRP) are highly conserved RNA-binding proteins that regulate alternative splicing, and which participate in MET and EMT (Shapiro et al., 2011; Warzecha et al., 2010). There are two paralogs of esrp in vertebrates, ESRP1 and ESRP2. They are required for an epithelial splicing program involving alternative splicing of hundreds of genes, many of which have functions in cell-cell adhesion, cell polarity, and cell migration (Bebee et al., 2015). This includes Fgfr2; Esrp1 and Esrp2 are required for the epithelial isoform of Fgfr2, and knockdown of Esrp1/2 results in the complete switch to the mesenchymal isoform of Fgfr2 (Warzecha et al., 2010). In zebrafish, double knockout of esrp1 and esrp2 leads to multiple developmental abnormalities, including malformed fins (Burguera et al., 2017).
The most differentially expressed gene in cluster 5 was cdh11 (Table S4). CDH11 is expressed during cadherin switching in EMT (Tomita et al., 2000). In this process, expression of CDH1 (epithelial cadherin) decreases in concert with increased expression of CDH11 and CDH2 (mesenchymal cadherins). As such, CDH11 serves as a biomarker of EMT (Zeisberg and Neilson, 2009). In regard to its embryonic functions, CDH11 is predominantly expressed within tissues of mesodermal origin (Hoffmann and Balling, 1995), and is linked to morphogenesis in the head, somite, and limb bud of early mouse embryos (Kimura et al., 1995). In addition to its expression within mesenchymal cells, Cdh11 is also expressed in parts of the neural tube and otic vesicle in early mouse embryos (Kimura et al., 1995). In zebrafish, knockdown of Cdh11 results in otic vesicle (Clendenon et al., 2009) and retinal defects (Clendenon et al., 2012). In chick, CDH11 expression is observed during neural crest cell delamination, which exhibits characteristics of EMT (Manohar et al., 2020).
Next, we tested the hypothesis that EMT- and MET-like transitions underlie transitions between mesenchymal- and epithelial-like cell states in osteoblastic cells. Partial MET and EMT result in intermediate E/M and M/E states which exhibit dual epithelial and mesenchymal characteristics (Figure 4B), necessitating the analysis of multiple markers. We first assessed canonical markers and transcription factors associated with MET and EMT (Figures 4C–4G). Cluster 1 had the highest expression of the epithelial markers esrp1 and cdh1, whereas clusters 3 or 5 had the highest expression of the mesenchymal markers cdh2/11, prrx1b, twist2/3, ctnna2/b1/b2, and zeb1b/2a. The expression of twist2/3 and prrx1b was highest in cluster 3. These cells match the description of twist2+ osteoprogenitors in the blastema identified by Stewart et al. (2014), and which were hypothesized to be undergoing EMT. The expression of zeb1b/zeb2a was highest in cluster 5. Consistent with the notion that ZEB drives a mesenchymal identity partly by repressing ESRP (Jolly et al., 2018), we found strongest expression of the zeb1b/zeb2a paralogs in cluster 5, where there is weakest esrp1 expression. These data indicate that canonical MET and EMT components are expressed in osteoblastic cells.
To assess whether Esrp1/2 was active during fin regeneration, we examined alternative splicing of known ESRP-dependent genes using VAST-TOOLS (Tapial et al., 2017). Because our method of scRNA-seq is biased toward the 3′ end of transcripts, which may miss splicing activity that occurs near the 5′ end, we analyzed bulk RNAseq datasets of fin regeneration from Lee et al. in which paired-end libraries were sequenced (Lee et al., 2020). Analysis detected ESRP1/2-dependent alternative splicing in fgfr2, namely an increase in the epithelial isoform and decrease in the mesenchymal isoform, at 1 dpa (see Figure S7). Within this same dataset, RNAseq was performed on sp7+ cells (Lee et al., 2020). In this dataset, we identified 1144 alternative splicing events (see Table S5).While we did not detect the alternative splicing of fgfr2, of the 1144 alternative splicing events that were found, 110 were identified to be dependent on esrp1/2 in zebrafish (see Table S6) (Burguera et al., 2017). We concluded that esrp1 splicing activity occurs during fin regeneration.
We next assessed whether osteoblastic cells express MET and EMT gene signatures. We constructed an orthologous EMT gene signature based on a hallmark gene set from the Molecular Signatures Database (Liberzon et al., 2015). For MET, we constructed an orthologous signature based on a gene set derived from human prostate and breast cancer cell lines induced to undergo MET via OVOL1/2 expression (Roca et al., 2014). Scoring revealed that cluster 3 had the strongest scoring for the EMT gene signature, whereas cluster 1 had the strongest scoring for the MET gene signature (Figure 4C). In regard to statistical analysis, using the Wilcoxon rank-sum test with Bonferonni correction, we found that the EMT scores were significantly different between nearly all cluster pairwise comparisons except between clusters 2 and 4 (see Figure S8). MET scores were significantly different between most clusters except between clusters 1 and 2, clusters 2 and 3, and clusters 4 and 5. We also identified enrichment of these signatures in mesenchymal cell subclusters (see Figure S9).
Differential gene expression analysis revealed enrichment of MET and EMT genes in distinct osteoblast subclusters (Figures 4H and S10). The top EMT markers in cluster 3 were enriched in genes associated with extracellular matrix, including matrix components (tnc, col6a2, col5a1, col1a2, col16a1, col6a3, fbln5, and fbln2), collagen-binding products (tgbi, lum, and dcn), and matrix remodeling (mmp2). Thus, MET- and EMT-associated gene signatures are expressed in osteoblastic cells during fin regeneration.
Finally, to elucidate cell state transitions, we ordered the osteoblastic cells in pseudotime and scored the cells for MET and EMT gene signatures (Figure 4I). Early in pseudotime, the trajectory passes through cells in clusters 1 and 2 which have high scores for MET and, in addition, low scores for osteoblast differentiation (Figure 3B) and high scores for epithelial differentiation (see Figure S11). The trajectory then progresses through cells in cluster 3. At this point in pseudotime, cells have high EMT scores, but still have low scores for osteoblast differentiation and high scores for epithelial differentiation. Finally, the trajectory progresses through cells in clusters 4 and 5. These cells have high scores for osteoblast differentiation and low scores for epithelial differentiation. Mesenchymal differentiation scores remained relatively constant throughout the trajectory. Collectively, the data suggest that in trajectory 2, as osteoblastic cells progress from less differentiated osteoblasts to more differentiated osteoblasts, they express MET-associated genes prior to expressing EMT-associated genes, and become less enriched in epithelial differentiation components.
Taken together, our studies indicate that osteoblastic cells express canonical MET and EMT components, as well as MET- and EMT-associated gene signatures. Moreover, we have identified multiple candidate trajectories underlying transitions between cell states.
Expression of cdh11, esrp1, and twist2 in situ
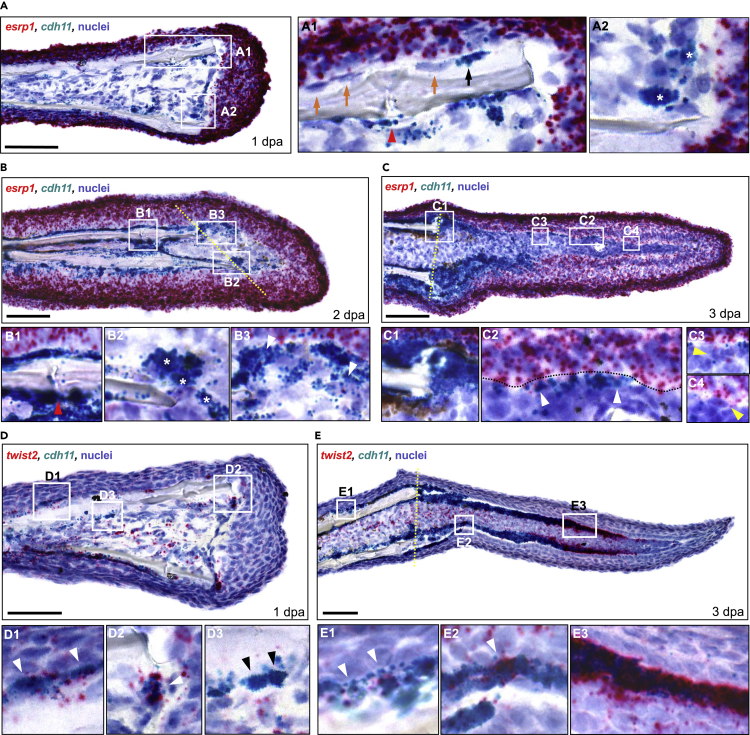
To place our findings into a spatiotemporal context, we performed RNA ISH for cdh11, esrp1, and twist2. In some cases, we also co-stained for runx2a. Prior to performing RNA ISH, we analyzed our sci-RNA-seq data to help assess the relative abundance of different cell populations. Notably, osteoprogenitors that expressed both esrp1 and cdh11 were relatively sparse, as evidenced by the fact that esrp1+/cdh11+ cells comprised 12.7% of all osteoblast2 cells in our sci-RNA-seq analyses. Cells expressing both runx2a/cdh11 and esrp1/twist2 comprised ∼60% and 2% of all osteoblast2 cells, respectively. In general, staining for cdh11 and twist2 was highest in cells within the mesenchymal compartment (Figures 5D and 5E), whereas staining for esrp1 was highest in epithelial cells, with decreasing expression in the deeper epidermis layers (intermediate and basal epidermis) (Figures 5A–5C).
Figure 5.
Osteoprogenitors express esrp1 and cdh11 in a temporal pattern during fin regeneration
(A–C) Dual RNA ISH on cryosections from 1, 2, and 3 dpa fin regenerates using RNAScope. (A) Expression of esrp1 and cdh11 in a 1 dpa fin cryosection. (A1 inset) cdh11 only stained some cells lining the bone (black arrow, positive staining; orange arrows, no staining). cdh11 staining was also observed in cells around the fin ray joints (red arrowhead). (A2 inset) Cells staining for cdh11 were observed along the amputation plane, lining the distal border of the mesenchymal compartment (stars). (B) Expression of esrp1 and cdh11 in a 2 dpa fin cryosection. (B1 inset) cdh11+ cells were prominent along the fin bone, including near the fin joint (red arrowhead). (B2 inset) Multiple cdh11+ cells were observed near the amputation stump (stars). (B3 inset) Arrowhead indicates a cell staining positive for both esrp1 and cdh11 in the forming blastema. (C) Expression of esrp1 and cdh11 in a 3 dpa fin cryosection. (C1 inset) Prominent cdh11 expression is observed at the amputation stump. (C2 inset) Multiple cells showed staining for both cdh11 and esrp1 (arrowheads). (C3, C4 insets) Multiple cells in the lateral mesenchymal compartment were positive for esrp1 but not cdh11 (yellow arrowheads).
(D–E) Dual RNA ISH for twist2 and cdh11 on cryosections from 1 and 3 dpa fin regenerates. twist2+/cdh11- cells were scattered throughout the mesenchyme at both timepoints.
(D) twist2 and cdh11 expression in a 1 dpa fin cryosection. twist2+/cdh11+ cells were detected along the native bone (D1 inset) and the amputation stump (D2 inset) (white arrowheads). (D3 inset) Some cdh11+ cells along the native bone did not express twist2 (black arrowheads).
(E) twist2 and cdh11 expression in a 3 dpa fin cryosection. (E1 inset) twist2+/cdh11+ cells were detected along the native bone (white arrowheads). (E2 inset) Expression of twist2 was less prominent in the proximal lateral mesenchymal compartment than cdh11, although small clusters of cdh11+ cells stained strongly for twist2 (white arrowhead). (E3 inset) Robust twist2 expression was observed in distal cdh11+ cells. All scale bars are 60 μm. Amputation plane is indicated with dashed yellow line.
We first performed dual RNA ISH for cdh11 and esrp1 across several regenerative stages, beginning with fins from 1 dpa (Figure 5A), a timepoint at which the wound epithelium is present but a blastema is not visible. Prior studies by Knopf et al. have shown that at this timepoint, osteoblastic dedifferentiation and migration has initiated (Knopf et al., 2011). Staining for cdh11 was observed in bone lining cells near the amputation stump (Figure 5A1, black arrow), with decreased staining in bone lining cells in more proximal locations (Figure 5A1 orange arrows). We also observed staining for cdh11 in cells near the fin ray joints (Figure 5A1, red arrowhead), a location known to harbor joint-derived resident osteoprogenitors (Ando et al., 2017). Finally, staining for cdh11 was observed in cells aligned with the amputation plane, some of which exhibited a rounded morphology (Figure 5A2, stars). Staining for esrp1 in the mesenchymal compartment was observed occasionally in mesenchymal cells, some of which were cdh11+ (see Figures S12C, S13). Thus, at 1 dpa, staining for cdh11 was present in cells located at sites that harbor dedifferentiated osteoblasts and resident osteoprogenitors, whereas staining for esrp1 was mostly absent from these cells.
At 2 dpa, a forming blastema is visible. At this timepoint, staining for cdh11 was noticeably increased from 1 dpa (Figure 5B). We observed increased staining in cells near the amputation stump (Figure 5B2, stars) as well as in cells near the fin ray joints (Figure 5B1, red arrowhead). Migration of osteoblasts into the blastema has been shown to occur at least between 18 hpa through 2 dpa (Knopf et al., 2011). Thus, it is possible that the broadened expression of cdh11 in the stump from 1 to 2 dpa reflects changes in cell state associated with migration of cells during this period. Within the blastema, staining for cdh11 was present in the lateral mesenchymal compartment (Figure 5B3). We observed staining for esrp1 in several cells within the lateral mesenchymal compartment, and which co-stained with cdh11 (Figure 5B3, white arrowheads). Thus, at 2 dpa, staining for cdh11 was present in cells within the blastema that were likely to be osteoprogenitors, with staining for esrp1 present in some of these cells.
At the 3 dpa timepoint, newly regenerated bone is visible, with more mature osteoblastic cells residing near the amputation stump, and less mature osteoprogenitors located distally (Pfefferli and Jazwinska, 2015). Staining for cdh11 was increased from 2 dpa (Figure 5C) and was prominent in cells lining the amputation stump (Figure 5C1), and in the lateral mesenchymal compartment where new bone was likely to form (Figure 5C2). Staining for cdh11 generally decreased in the distal direction. In regard to esrp1, we observed increased staining in cells within the mesenchymal compartment compared to 2 dpa. Staining for esrp1 was observed in cells in the lateral mesenchymal compartment in both proximal (Figure 5C3) and distal (Figure 5C4) locations. Within these cells, some co-staining of esrp1 and cdh11 was observed (Figure 5C2, white arrowheads). Notably, we did not observe a clear compartment of esrp1+ mesenchymal cells. Rather, esrp1+ cells were relatively sparse, and intercalated among or near cdh11+ cells in both proximal and distal locations within the blastema. Similar findings were observed when performing dual RNA ISH for esrp1 and runx2 (see Figure S12A). Thus, similar to 2 dpa, at 3 dpa, staining for cdh11 was present in cells likely to be osteoprogenitors, with some of these cells staining for esrp1.
We also performed dual RNA ISH for cdh11 and twist2. twist2 is a known biomarker of the mesenchymal lineage and EMT, and plays a role in regulating osteoblast maturation in mammals (Franco et al., 2011). In zebrafish, it has previously been shown to be expressed in regenerating fin at 1 dpa in the stump with whole mount ISH, and in distal runx2+ cells at 3 dpa in longitudinal sections of regenerating fin (Stewart et al., 2014). This suggests that twist2 expression is activated early during fin regeneration when osteoblasts are dedifferentiating (1 dpa), and is expressed in dedifferentiated, immature osteoprogenitors at 3 dpa. To determine if cdh11 marks specific differentiation states in osteoblasts, we performed dual RNA ISH for cdh11 and twist2 at both timepoints. At 1 dpa, staining for twist2 was observed in cells in the mesenchymal compartment (Figure 5D), in elongated cdh11+ bone lining cells along the native bone (Figure 5D1), and in cells at the amputation stump, some of which were cdh11+ (Figure 5D2). Several cdh11+/twist2- cells were also detected along the native bone (Figure 5D3, black arrowheads). At 3 dpa (Figure 5E), we detected cdh11+/twist2+ cells lining the native bone (Figure 5E1), and in the blastema along the lateral edges of the mesenchymal compartment, with the most prominent dual staining at the distal end where less mature osteoprogenitors are thought to reside (Figure 5E3). Small clusters of cdh11+ cells with robust twist2 staining were also found in the proximal blastema near the regenerating bone (Figure 5E2), where maturing osteoprogenitors are likely to reside. cdh11-/twist2+ cells were also observed in scattered cells throughout the mesenchymal compartment of both the blastema and the stump. Our findings suggest that cdh11 is expressed in twist2 dedifferentiating osteoblasts at 1 dpa, dedifferentiated twist2+ osteoprogenitors in the distal regenerate at 3 dpa, as well as more mature, twist2- osteoblastic cells in the proximal regenerate.
Discussion
This study provides a single cell resource for the study of osteoblastic cells during zebrafish fin regeneration, and supports the contribution of MET- and EMT-associated components to this process. Our studies complement previous studies by Stewart et al. who noted dual mesenchymal and epithelial characteristics in osteoblastic cells, and first hypothesized this was due to EMT (Stewart et al., 2014), by demonstrating the expression of MET and EMT signatures in osteoblastic cells. Our studies also extend the identification by Stewart et al. of twist2+ cells in the amputation stump and in the regenerate by showing that these cells co-express the EMT marker cdh11, and that the latter are enriched with EMT signatures. In silico evidence of a sequential EMT and MET process occurring during axolotl limb regeneration was recently reported (Li et al., 2021). In contrast, in their review, Wong and Whited state that no EMT signatures were identified in two axolotl limb regeneration scRNA-seq studies (Gerber et al., 2018; Leigh et al., 2018; Wong and Whited, 2020); however, methods for analysis were not provided. In this context, it is unclear whether the failure to identify EMT signatures during salamander limb regeneration in previous scRNA-seq studies reflects biological differences with zebrafish fin regeneration, or technical differences (e.g., the use of signatures different from those used here). To clarify this, it would be useful to systematically test for the same MET/EMT signatures in different scRNA-seq datasets from different models of appendage regeneration.
In our sci-RNA-seq analyses, we identified two potential trajectories underlying the topology of osteoblastic subclusters, raising questions related to potential reasons for their existence. Whereas trajectory 1 was consistent with osteoblastic differentiation from proliferative cells to form new bone, trajectory 2 appended additional cells to trajectory 1 that were intermediate between sp7-expressing osteoblasts (cluster 5) and proliferative cells (cluster 3). Such intermediate states might reflect cells that are in a dedifferentiating state. While substantial osteoblastic dedifferentiation occurs by 1 dpa (Knopf et al., 2011), the presence of dedifferentiating cells at a later stage of regeneration (3–5 dpa) could be due to several reasons. In lineage tracing studies of zebrafish fin regeneration, it has been observed that the expansion of clones which have migrated into the blastema was temporally variable, with expansion of some clones starting as late as ∼10–14 dpa (Tornini et al., 2016). In this context, it is conceivable that trajectory 2 included dedifferentiating cells which are quiescent or have not yet divided. Alternatively, as we discuss later, it is possible that there is periodic switching between de- and redifferentiation and proliferation, which might help maintain a pool of osteoblastic cells during regeneration.
It is possible that cdh11 has distinct functions in EMT and osteoblast differentiation during fin regeneration. With respect to its potential role in EMT in fin regeneration, our study found evidence that cdh11 is expressed in osteoblastic cells that either scored strongly for the EMT signature or are hypothesized to be undergoing EMT. For example, in the 3 dpa fin regenerate, ISH revealed that cdh11 expression in the distal osteoprogenitors was high, albeit not as high as in more proximal osteoprogenitors (Figure 6, top). These distal osteoprogenitors also stained for twist2 and runx2a and likely corresponded with cells in cluster 3, which scored highest for the EMT signature. In addition, at 1 dpa, cdh11 staining was observed in stump cells that were likely to be dedifferentiating osteoblasts, which have been speculated to be undergoing EMT (Stewart et al., 2014). It is therefore possible that cdh11 has a function in EMT in osteoblasts. Because cdh11 expression was elevated even further in proximal cells compared to distal osteoprogenitors, it is possible that cdh11 may have a role in osteoblast differentiation independent of EMT during fin regeneration. More specifically, at 3 dpa, the strongest cdh11 ISH staining was observed in the proximal osteoprogenitors, which are undergoing osteoblast differentiation and maturation, and staining was comparably reduced in the distal osteoprogenitors, which are less differentiated. From our clustering analysis, the proximal osteoprogenitors likely correspond to cluster 5 cells which not only had the strongest cdh11 expression levels but also scored highest for osteoblastic differentiation, and differentially expressed sp7, a marker of osteoblast maturation. In support of a role for cdh11 in osteoblast differentiation that is independent of a role in EMT, it has been previously shown that Cdh11 is highly expressed in mesenchymal stem cells in mice during osteogenic differentiation (Marie, 2002). Taken together, our study suggests that during fin regeneration, cdh11 could play multiple roles related to EMT and osteoblast differentiation in the regenerate.
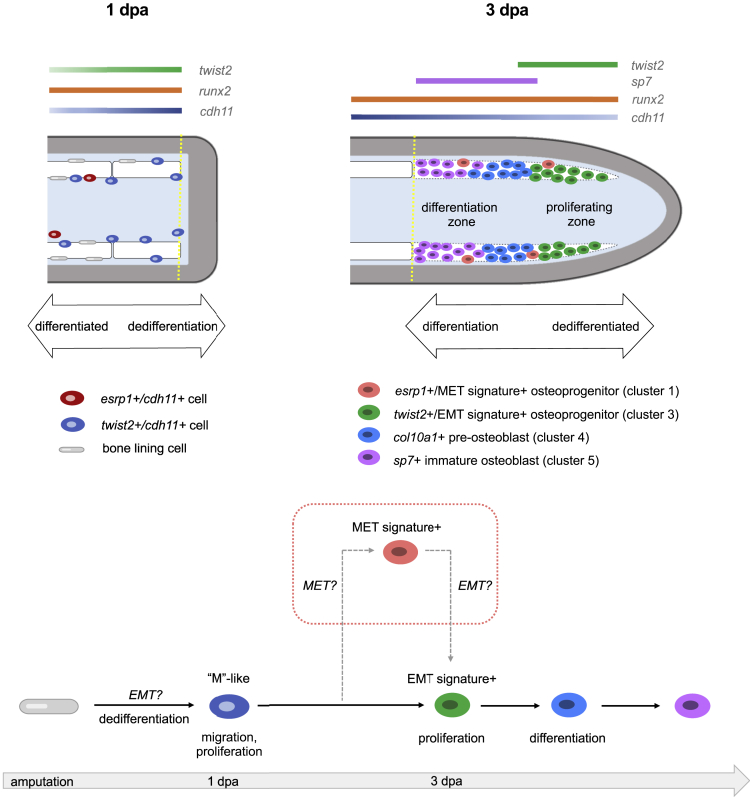
Figure 6.
Schematics showing predicted spatial locations of the different osteoprogenitors from osteoblast2 within the fin (top) and predicted model of osteoblastic de- and redifferentiation during fin regeneration (bottom)
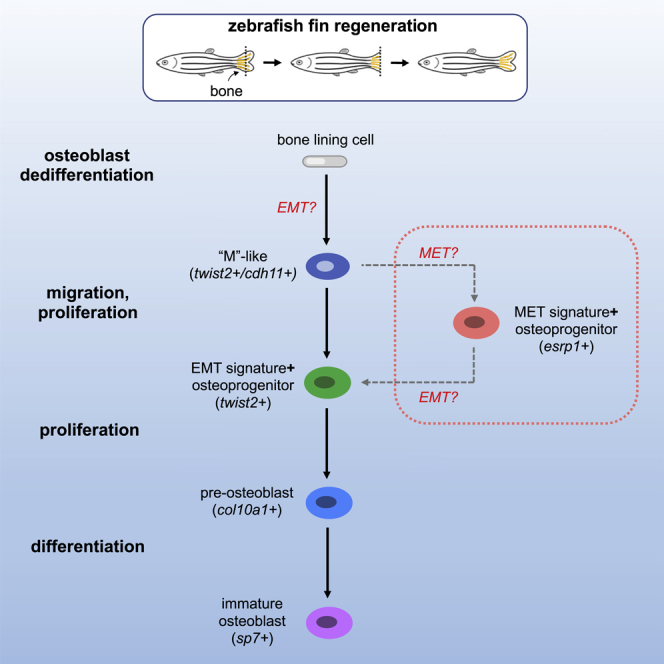
Amputation plane is indicated with a dashed yellow line. (top left) At 1 dpa, dedifferentiated osteoblasts expressing cdh11 and twist2 are observed along the native bone, at fin ray joints, and at the amputation stump. Elongated bone lining cells are still present but are twist2-/cdh11-. esrp1+/cdh11+ cells are occasionally found near the native bone. (top right) At 3 dpa, osteoprogenitors in the blastema are located along the lateral edges of the mesenchymal compartment as indicated by the dashed white region. Early cycling osteoprogenitors expressing a strong EMT gene signature are distally located. These cells also exhibit robust expression of runx2a and twist2, and comparably less cdh11 than in proximal regions of the regenerate. Proximal osteoprogenitors exhibit strong cdh11 expression and have begun maturing into differentiated osteoblasts, first expressing col10a1, then sp7. Non-cycling osteoprogenitors, some of which express a strong MET gene signature, are sporadically located among both the proximal and distal osteoprogenitors. (bottom) Model of osteoblastic de- and redifferentiation during fin regeneration. Bone lining cells dedifferentiate into twist2+/cdh11+ cells, which are mesenchymal-like (“M”-like). These cells then migrate into the blastema and proliferate. A majority of the cells then follow the solid black arrows and progress through redifferentiation and become mature osteoblasts to regenerate bone. Some of the cells follow the dashed gray arrows, first expressing MET components before expressing EMT components, prior to differentiating into mature osteoblasts.
Previous studies have identified spatial domains in the regenerate in which cells differentially express phenotypes linked to MET and EMT, thus it is useful to consider how these domains relate to expression of MET and EMT components in our study. Stewart et al. showed that during zebrafish fin regeneration, osteoblastic cells exhibit a change in shape from more rounded cells in the regenerate to more flattened cells in proximal unamputated tissue (Stewart et al., 2014). Moreover, Mateus et al. showed that mesenchymal cells exhibit a change in shape from more round cells in the distal regenerate to more flat cells in the proximal regenerate (Mateus et al., 2015). In our studies, both cdh11 and twist2 were expressed in osteoblastic cells in the distal regenerate; in the proximal regenerate, cdh11 expression was increased in osteoblastic cells, whereas twist2 was decreased but with intermittent pockets of expression. This suggests that dual expression of cdh11 and twist2 is associated with a more rounded cell shape in distal cells. Future studies that correlate expression of EMT and MET components with cell shape and F-actin localization on a cell-by-cell basis could shed light on the relationship between the expression of these components and cell phenotypes linked to EMT and MET.
Our study identifies relationships between osteoblast differentiation in the regenerate and the expression of genes associated with mesenchymal/epithelial differentiation and MET/EMT. Of the osteoblast2 subclusters, cells with the lowest osteoblast differentiation scores (clusters 1 and 2) had the highest epithelial differentiation and MET scores, cells with intermediate osteoblast differentiation scores (cluster 3) had the highest EMT score and elevated mesenchymal and epithelial differentiation scores, and cells with the highest osteoblast differentiation scores (cluster 4 and 5) exhibited the lowest epithelial differentiation and MET scores. One interpretation of these data is that osteoblasts in a more epithelial-like state are less differentiated. However, this would be opposite to the findings of Stewart et al. in which epithelial characteristics were associated with the differentiated state (Stewart et al., 2014). An alternate interpretation is that osteoblasts in a less differentiated state are in a more hybrid E/M state, whereas more differentiated osteoblasts are in a less hybrid E/M state. This would be similar to what has been shown in cancer where partial EMT can enhance cancer cell stemness, and cells reversibly switch between multiple metastable hybrid E/M states resulting in a spectrum of phenotypes (Saxena et al., 2020). In this context, it is possible that the sporadic appearance of osteoblasts expressing MET signatures among cells that express EMT signatures reflects metastability and sporadic switching between hybrid states that are shifted toward either an epithelial or mesenchymal state, respectively. In addition, there is evidence that cancer cells in an epithelial-shifted hybrid state can exhibit a greater proliferative capacity than more mesenchymal variants (Williams et al., 2019). Accordingly, osteoblasts in an epithelial-shifted hybrid state may be more proliferative, and as fin regeneration takes ∼14 to 21 days to reach completion, osteoblastic cells undergoing periodic switching between epithelial- and mesenchymal-shifted hybrid states could support a replenishing pool of dedifferentiated osteoprogenitors throughout the course of regeneration.
Our study also identifies potential relationships between the processes of EMT, MET, and osteoblast re- and dedifferentiation. Within the osteoblast2 population of the regenerate, the subcluster 3 cells were marked by twist2 expression and high EMT signature scores. These cells also had the highest proportion of cycling cells and a low osteoblast differentiation score. Thus, our data suggest that expression of EMT components is associated with the transition between cell cycling and early osteoblast differentiation and thus it is possible that it facilitates this process. Alternatively, it is possible that the expression of EMT signatures in osteoblasts at this stage is a remnant of an earlier EMT-like event. Previously, Stewart hypothesized that following amputation, osteoblasts dedifferentiate by acquiring a mesenchymal-like cell state through a process similar to EMT. Supporting this notion, at 1 dpa, we observed cdh11 and twist2 co-expression specifically in cells that are likely dedifferentiating osteoblasts. In this context, it is plausible that while osteoblast dedifferentiation in the stump involves cdh11, twist2, and EMT signatures, expression of these components in distal, redifferentiating osteoprogenitors of the regenerate at a later timepoint reflects residual carryover from earlier processes. As for the relationship between MET and re- and dedifferentiation, as we described previously, the cells in subcluster 1, which express esrp1 and MET signatures, may represent osteoblasts that are in a dedifferentiating state at 3 and 5 dpa, timepoints that are later than when a majority of dedifferentiation occurs. Thus, it is conceivable that cells that have already expressed EMT signatures during the initial dedifferentiation may subsequently express MET signatures, followed by another round of EMT, i.e., a process of periodic dedifferentiation, proliferation, and redifferentiation, which could help maintain a pool of osteoblastic progenitor cells during regeneration.
We used our data to develop a model of osteoblastic de- and redifferentiation, which we hypothesize involves two trajectories (Figure 6). The first trajectory is based on trajectory 1 and excludes cluster 1 cells that express MET gene signatures. In this trajectory (solid arrows in Figure 6), following fin amputation, bone lining cells undergo dedifferentiation and express cdh11 and twist2. These cdh11+/twist2+ osteoprogenitors delaminate and migrate into the blastema, a behavior similar to epithelial cells undergoing EMT (denoted by “EMT?” in Figure 6). There, they continue to proliferate and express EMT-associated gene signatures and begin to redifferentiate into osteoblasts. Thus, in this trajectory, osteoblasts acquire a mesenchymal-like (“M”-like) cell state through a process that employs programs similar to EMT, as proposed by Stewart et al. (2014).
The second trajectory is based on trajectory 2 but includes cluster 1 cells that express MET gene signatures (dotted rectangle in Figure 6). This trajectory begins similarly as the first trajectory. However, after initially undergoing EMT, a subset of cdh11+ osteoprogenitors express esrp1 and MET gene signatures. Because esrp1+/cdh11+ osteoprogenitors were relatively sparse, we hypothesize that the “EMT only” trajectory is predominant, whereas the “EMT and MET” trajectory serves as a secondary path to generate specialized cells within the blastema or to help maintain a pool of progenitors. Based on our trajectory analysis, it is conceivable that these cells undergo another round of EMT (denoted by “EMT?” in the dotted rectangle in Figure 6), upon which the cells exhibit an EMT signature and begin to redifferentiate into osteoblasts.
By identifying EMT and MET components expressed during fin regeneration, our studies provide a useful lens to identify pathways underlying de- and redifferentiation. In our studies, paralogs for zeb1/2 and twist (but not snail) were markedly expressed in osteoblastic cells. Moreover, we observed twist2+/cdh11+ cells near the amputation stump at 1 dpa. Thus, osteoblastic dedifferentiation may involve expression of twist, zeb1/2, and cdh11, a combination that has been observed in certain EMT-like processes (Manohar et al., 2020; Stemmler et al., 2019). With respect to the identification of MET signatures in osteoblasts, it has been hypothesized that in cancer, MET may be required for metastasis as mesenchymally-shifted cancer cells often exhibit reduced proliferative capacity. Cancer cells that have undergone a ZEB1/2-induced EMT are particularly associated with decreased proliferation and may as a consequence require MET to proliferate and metastasize (Williams et al., 2019). Further studies are needed to determine whether osteoblastic cells exhibit reversibility between metastable epithelial-shifted and mesenchymal-shifted hybrid states, reminiscent of that associated with increased cancer cell stemness (Saxena et al., 2020).
A growing body of studies suggests that EMT or MET pathways may be harnessed to engineer or enhance dedifferentiation in mammalian cells. For instance, early stages of induced pluripotent stem cell (iPSC) reprogramming from mouse embryonic fibroblasts require MET and transition to an epithelial-like morphology (Li et al., 2010). Esrp1 has been shown to enhance reprogramming of fibroblasts to iPSCs, in part by alternative splicing of the transcription factor Grhl1 (grainyhead-like transcription factor 1) (Cieply et al., 2016). Reprogramming efficiency is also increased by early induction of EMT (Liu et al., 2013; Wang and Unternaehrer, 2019). Moreover, increased differentiation potential of breast epithelial cells similar to mesenchymal stem cells is achieved by overexpression of Snail or Twist (Battula et al., 2010; Wang and Unternaehrer, 2019). Because reprogramming induces much more dramatic changes compared to osteoblastic dedifferentiation during zebrafish fin regeneration, it is not yet clear whether EMT/MET are required for de- and redifferentiation in the latter. Future studies are needed to further elucidate MET pathways and EMT components expressed during zebrafish fin regeneration, and whether such components are necessary and sufficient for osteoblastic de- and redifferentiation in this process.
Limitations of the study
Some limitations of our study should be considered. First, gene expression alone is insufficient to indicate that MET or EMT is occurring (Yang et al., 2020). In our studies, osteoblastic cells expressed several EMT-TFs (including orthologs for ZEB and TWIST, part of a core group of transcription factors that are expressed in all instances of EMT (Yang et al., 2020)), and were enriched with EMT-associated genes. However, we did not assess cell phenotypes that define EMT such as loss of apical–basal polarity, modulation of the cytoskeleton, and cell-cell adhesive properties. Thus, whether the expression of MET- and EMT-associated components in our study are reflective of actual MET and EMT, or are distinct processes that bear resemblance to MET and EMT, should be assessed in future studies. Another limitation of our study is the focus of our sci-RNA-seq analysis on a later window of regeneration (3–5 dpa). While this prevented transcriptomic examination of earlier timepoints, our RNA ISH studies allowed us to connect our single-cell dataset to early osteoprogenitor states. Finally, it should be emphasized that the trajectories constructed in this study are solely based on unbiased inference, and that lineage tracing studies are needed to test their predictions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| RNAscope 2.5HD Duplex Detection Kit | ACD Bio | Cat#322430 |
| RNAscope Dr-runx2a probe | ACD Bio | Cat#409521 |
| RNAscope Dr-cdh11 probe | ACD Bio | Cat#530551 |
| RNAscope Dr-esrp1 probe | ACD Bio | Cat#555081-C2 |
| RNAscope Dr-twist2 probe | ACD Bio | Cat#413131-C2 |
| RNAscope Dr-lmx1ba probe | ACD Bio | Cat#502181-C2 |
| RNAscope Dr-slc8a4b probe | ACD Bio | Cat#1103611-C2 |
| RNAscope 2-plex negative control | ACD Bio | Cat#320751 |
| Deposited data | ||
| zebrafish regenerating caudal fin sci-RNA-seq | This paper | GEO: GSE192498 |
| zebrafish regenerating caudal fin scRNA-seq | Hou Y, Lee HJ, Chen Y, et al. Cellular diversity of the regenerating caudal fin. Sci Adv. 2020;6(33):eaba2084. Published 2020 Aug 12. https://doi.org/10.1126/sciadv.aba2084 | GEO: GSE137971 |
| zebrafish regenerating fin bulk RNA-seq | Lee, H.J., Hou, Y., Chen, Y. et al. Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility.Genome Biol21,52 (2020). https://doi.org/10.1186/s13059-020-1948-0 | GEO: GSE126701 |
| Experimental models: Organisms/strains | ||
| Zebrafish:AB | ZIRC | Cat#ZL1 |
| Software and algorithms | ||
| Seurat v3.1 | Stuart and Butler et al. Comprehensive Integration of Single-Cell Data. Cell (2019) | https://satijalab.org/seurat/articles/install.html |
| VAST-TOOLS v2.5.1 | Tapial, J., Ha, K.C.H., Sterne-Weiler, T., Gohr, A., Braunschweig, U., Hermoso-Pulido, A., Quesnel-Vallières, M., Permanyer, J., Sodaei, R., Marquez, Y., Cozzuto, L., Wang, X., Gómez-Velázquez, M., Rayón, M., Manzanares, M., Ponomarenko, J., Blencowe, B.J., Irimia, M. (2017). An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res, 27(10):1759-1768 | https://github.com/vastgroup/vast-tools |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, W. Joyce Tang (wjtang@uw.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (GEO: GSE192498). This paper also analyzed existing, publicly available data (GEO: GSE137971, GEO: GSE126701). All details associated with the accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Zebrafish husbandry: All studies were performed on an approved protocol in accordance with the University of Washington Institutional Animal Care and Use Committee (IACUC). Zebrafish were reared and maintained at the ISCRM Aquatic Facility under a 14:10 hour light:dark cycle at 28°C and standard conditions. Adult wildtype (AB) zebrafish were used as source material for these studies. Both females and males were used because robust sexual dimorphism in the rate of adult zebrafish caudal fin regeneration has not been observed (Nachtrab et al., 2011).
Method details
Tissue collection
Adult wildtype (AB) zebrafish were anesthetized with 0.02% MS-222 and subjected to 50% amputation of the caudal fin and allowed to recover. At various days post amputation, regenerated fin tissues were collected and either fixed overnight at 4°C in 10% neutral-buffered formalin for cryosectioning, or immediately frozen at −80°C for sequencing. 25 fins were collected at 3 dpa for the pilot sci-RNA-seq study, and 100 fins were collected for each time point for the subsequent sci-RNA-seq study.
Single-cell combinatorial indexing RNA-seq
Detailed methods for single-cell combinatorial indexing RNA-seq (sci-RNA-seq) are described in (Cao et al., 2017, Cao et al., 2019) and are available online (http://atlas.gs.washington.edu/mouse-rna). sci-RNA-seq3 was performed as previously described, using three rounds of barcoding on intact nuclei to identify transcripts with unique barcode combinations to generate single nucleus 3′ RNA-seq data (Cao et al., 2017). Briefly, nuclei were extracted, fixed in 4% paraformaldehyde, and frozen in 100 μl of nuclei wash buffer. Nuclei were then thawed in a 37°C water bath for 5 min, spun down at 500 x g for 5 min, and permeabilized in 400 μl of permeabilization buffer (nuclei wash buffer supplemented with 0.2% Triton X-100) for 3 min. Nuclei were then washed, sonicated for 12 s on low power mode (Diagenode), washed, and resuspended in 100 μl of nuclei wash buffer. The sci-RNA-seq3 scheme of library preparation follows previously published methods for paraformaldehyde-fixed nuclei (Cao et al., 2019)with the following modifications: 2 μl of oligo-dT primers were added to each well with 80,000 nuclei for reverse transcription, the Quick Ligation kit (New England Biolabs) was used in place of T4 ligase, and tagmentation was performed using 1/40thμl per well of i7-loaded TDE1 enzyme, prepared at the University of Washington following published protocols (Hennig et al., 2018). Libraries were sequenced on Illumina NovaSeq S4 flow cells at the University of Washington Northwest Genomics Center core sequencing facility with the following cycles: read 1, 34 cycles; read 2, 100 cycles; index 1, 10 cycles; index 2, 10 cycles. Approximately 23, 42, and 63 million reads were obtained from the 3 dpa pilot, 3 dpa, and 5 dpa samples, respectively. Each sample was sequenced to an average depth of ∼43 million reads, resulting in an average read depth of 6,653 reads/cell. Sequencing reads were processed as previously described, with trimmed reads being mapped to the GRCz11 zebrafish genome using STAR v.2.5.2b and GRCz11.96 (ftp://ftp.ensembl.org/pub/release-96/gtf/danio_rerio/Danio_rerio.GRCz11.96.gtf.gz).
Cryosectioning
Fixed tissues were washed in 1X PBS/0.05%Tween 20 (PBS-T) for 3 × 5 minutes, then dehydrated in increasing graded concentrations of methanol and stored at −20°C for up to 6 months. Tissues were processed for embedding by first rehydrating to PBS-T in increasing graded concentrations, then cryoprotected by incubation in 15% sucrose/1X PBS for 1 h at room temperature, followed by 20% sucrose/1X PBS for 4 h to overnight at 4°C. Embedding was done in O.C.T. compound (Fisher Scientific) in Peel-A-Way embedding molds (Polysciences, Inc., Warrington, PA) and snap frozen in either dry ice or liquid nitrogen and stored at −80°C until ready for sectioning. ∼15μm sections were made on a Leica CM1850 cryostat, collected onto charged slides and allowed to dry for several hours at room temperature before being stored at −20°C.
In situ hybridization with RNAscope
Custom and catalog probes were purchased from the manufacturer (ACD Bio, Newark, CA). The negative control probe was provided by ACD Bio and targeted dapB, a gene found only in bacteria. In situ hybridization was performed using the RNAScope 2.5 HD Duplex Detection kit essentially as per manufacturer instructions, with the following modifications: sections were treated with Target Retrieval solution in a steamer for 8 min at ∼100°C, and permeabilized with Protease Plus for 15 min at 40°C.
Imaging and analysis
Slides were imaged with a 40X objective on an Aperio VERSA 200 slide scanner. All images are representative of at least two different sections. Sections for 1 and 2 dpa analysis were from a single ISH experiment, and sections for 3 dpa analysis were from three separate experiments. Per the manufacturer’s recommendation, more than one spot per ten cells in a field of view at 40X magnification was considered positive staining. We verified this criterion using a negative control (see Figure S12B).
Quantification and statistical analysis
sci-RNA-seq analysis
A pilot dataset from 25 fins collected at 3 dpa was analyzed with Seurat v3.1 using the standard recommended workflow for quality control filtering (200 < nFeatures_RNA <2500) to remove low quality cells or potential doublets, respectively, and data processing. Briefly, the data was normalized by scaling gene content by cell and log normalizing, followed by variable feature selection using the “vst” method. The data was then scaled and principal components were calculated using the identified variable features. Clustering was performed using the Louvain algorithm, followed by non-linear dimensional reduction with UMAP to visualize the data (Butler et al., 2018). Unbiased clustering was performed with the top 15 principal components (PCs) and with clustering resolution set to 0.5. To assign cell types to the resulting clusters, the FindAllMarkers function was used for differential gene expression analysis to identify gene markers for each cluster (logfc.threshold = 0.25, min.pct = 0.2 or 0.25, and/or min.diff.pct = 0.25 as noted in the supplemental tables).
Using the cell type label annotations of our pilot dataset as a reference, we projected the cell type labels onto the main 3 and 5 dpa datasets (n = 100 fins for each time point) using the cell type label transfer method (FindTransfer Anchors, TransferData, AddMetaData functions). The three datasets (3 dpa pilot and larger 3 and 5 dpa datasets) were then combined using the merge function in Seurat. We did not employ Seurat integration workflows (Stuart et al., 2019) since such workflows were designed for situations where substantial differences such as those arising from differences in scRNA-seq technologies or modalities need to be harmonized, which did not apply to our data. Moreover, while we did not observe evidence of batch effects in our data—which would be manifested as cells from each timepoint clustering together—the cell type label transfer workflow accounts for batch differences if present.
The standard quality control filtering and data processing workflow was repeated on the merged datasets (200 < nFeatures_RNA <2500, number of PCs = 15, resolution = 0.5). To subcluster the osteoblast population, we subset the Osteoblast2 cells from the combined dataset and again applied the standard Seurat data processing workflow (number of PCs = 15, resolution = 0.8). Osteoblast scores for gene expression programs were calculated using the AddModuleScore function with the appropriate gene sets and using default parameters. All plots were generated in R with either Seurat, base R graphics, or with the dittoSeq package (Bunis et al., 2020). “Open” trajectory analysis was performed with Slingshot v1.2 (Street et al., 2018) using appropriate data embeddings (either UMAP or the first two PCs from principal component analysis). “Closed” trajectory analysis was performed using the function “principal_curve” (available in base R > 3.0) and a periodic lowess smoother with appropriate data embeddings. Pseudotime values were obtained by computing projections onto the closed curve.
Mapping data onto existing fin regeneration data
Hou et al. (2020), generously shared the Seurat object containing their fin regeneration data. We used the FindTransferAnchors and TransferData functions in Seurat to project the tissue type annotations from our pilot 3 dpa dataset onto the Hou et al. dataset and assessed the efficacy of the mapping by confirming the expression of known marker genes in each tissue type. To determine the number of cells from the Hou et al. dataset which mapped to osteoblast1 and osteoblast2, we counted the number of cells from the Hou et al. dataset which were predicted to belong in the osteoblast1 and osteoblast2 clusters.
Cell scoring, cell cycle analysis, statistical analyses
Gene sets associated with cellular functions such as epithelial differentiation, mesenchymal differentiation, MET and EMT, were obtained from various sources including the Molecular Signatures Database (MSigDB), the Gene Ontology database and the literature, to create “gene signatures” for each cellular function. Using the AddModuleScore function, cells were scored for the expression of each gene signature to calculate the difference between the average expression level of each gene signature and a set of control genes randomly picked as previously described (Tirosh et al., 2016). Zebrafish orthologs for the genes in each gene set were identified if possible. Gene signatures for epithelial differentiation, mesenchymal differentiation, MET and EMT consisted of approximately 100–300 gene orthologs. To test for statistical differences of the scores between each cluster, we first tested for normal distribution using the Shapiro-Wilks test. As the data was not normally distributed, we used the non-parametric Kruskal-Wallis test followed by pairwise comparisons using the Wilcoxon rank-sum test with Bonferroni correction. Cell cycle analysis was performed using the CellCycleScoring function in Seurat, which classifies each cell as being in either G2M, S, or G1 phase based on the expression of G2/M and S phase markers (Tirosh et al., 2016). To test if the proportion of cycling to non-cycling cells was significantly different between clusters, we performed a difference of proportions test followed by a pairwise comparison using Bonferroni correction.
Alternative splicing analysis
The Vertebrate Alternative Splicing and Transcription Tools (VAST-TOOLS) software (Tapial et al., 2017) was used to profile alternative splicing events in bulk RNA-seq data (GSE126701) (Lee et al., 2020). Data from experimental replicates were merged, and differential splicing analysis was performed using the “danRer10” library of alternative splicing profiles in zebrafish as reference (downloaded from the VastDB site, https://vastdb.crg.eu/wiki/Main_Page). All comparisons were performed with the “compare” module and implemented with the following parameters: --min_dPSI 15--min_range 5--p_IR. A custom R script was used to compare the alternative splicing events against those that were previously annotated as esrp-dependent (Burguera et al., 2017).
Additional resources
We have used the ShinyCell package in R (Ouyang et al., 2021) to create a website application that allows users to visually explore our datasets in various plotted formats (UMAP dimensional plots, dot plots, etc), which is available at this URL: https://msblgroup.shinyapps.io/finregeneration/.
Acknowledgments
The authors would like to acknowledge Dr. Ting Wang for generous sharing of scRNA-seq data, as well as members of the MSBL, the UW Regeneration Club, and Dr. Lauren Saunders for helpful discussions. Research reported in this publication was supported by the Brotman Baty Institute Seattle Single Cell Initiative, a Seed Grant from the University of Washington Department of Orthopaedics and Sports Medicine, and the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number AR066061. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, Methodology, and Investigation: W.J.T., C.J.W., and R.Y.K.; Formal Analysis, Data Curation, Writing – Original Draft, and Visualization: W.J.T. and R.Y.K.; Writing – Review & Editing: W.J.T, C.J.W., T.O., C.H.A., and R.Y.K.; Supervision and Funding Acquisition: R.Y.K.
Declaration of interests
The authors declare no competing interests.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103784.
Supplemental information
–S6
References
- Albano G., Dolder S., Siegrist M., Mercier-Zuber A., Auberson M., Stoudmann C., Hofstetter W., Bonny O., Fuster D.G. Increased bone resorption by osteoclast-specific deletion of the sodium/calcium exchanger isoform 1 (NCX1) Pflugers Arch. 2017;469:225–233. doi: 10.1007/s00424-016-1923-5. [DOI] [PubMed] [Google Scholar]
- Alimperti S., Andreadis S.T. CDH2 and CDH11 act as regulators of stem cell fate decisions. Stem Cell Res. 2015;14:270–282. doi: 10.1016/j.scr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan S., Gordon L., Blechman J., Matsuoka R.L., Rajamannar P., Wircer E., Biran J., Reuveny A., Leshkowitz D., Stainier D.Y.R., Levkowitz G. Pituicyte cues regulate the development of permeable neuro-vascular interfaces. Dev.Cell. 2018;47:711–726.e715. doi: 10.1016/j.devcel.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Ando K., Shibata E., Hans S., Brand M., Kawakami A. Osteoblast production by reserved progenitor cells in zebrafish bone regeneration and maintenance. Dev.Cell. 2017;43:643–650.e643. doi: 10.1016/j.devcel.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Ansieau S., Bastid J., Doreau A., Morel A.P., Bouchet B.P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Aubin I., Adams C.P., Opsahl S., Septier D., Bishop C.E., Auge N., Salvayre R., Negre-Salvayre A., Goldberg M., Guenet J.L., Poirier C. A deletion in the gene encoding sphingomyelin phosphodiesterase 3 (Smpd3) results in osteogenesis and dentinogenesis imperfecta in the mouse. Nat. Genet. 2005;37:803–805. doi: 10.1038/ng1603. [DOI] [PubMed] [Google Scholar]
- Avaron F., Hoffman L., Guay D., Akimenko M.A. Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev. Dyn. 2006;235:478–489. doi: 10.1002/dvdy.20619. [DOI] [PubMed] [Google Scholar]
- Batlle E., Sancho E., Franci C., Dominguez D., Monfar M., Baulida J., Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat.Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Battula V.L., Evans K.W., Hollier B.G., Shi Y., Marini F.C., Ayyanan A., Wang R.Y., Brisken C., Guerra R., Andreeff M., Mani S.A. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells. 2010;28:1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., Carstens R.P. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 2015;4 doi: 10.7554/eLife.08954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.M., Fisher S., Iovine M.K. Osteoblast maturation occurs in overlapping proximal-distal compartments during fin regeneration in zebrafish. Dev. Dyn. 2009;238:2922–2928. doi: 10.1002/dvdy.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunis D.G., Andrews J., Fragiadakis G.K., Burt T.D., Sirota M. dittoSeq: universal user-friendly single-cell and bulk RNA sequencing visualization toolkit. Bioinformatics. 2020;36:5535–5536. doi: 10.1093/bioinformatics/btaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguera D., Marquez Y., Racioppi C., Permanyer J., Torres-Mendez A., Esposito R., Albuixech-Crespo B., Fanlo L., D'Agostino Y., Gohr A., et al. Evolutionary recruitment of flexible Esrp-dependent splicing programs into diverse embryonic morphogenetic processes. Nat. Commun. 2017;8:1799. doi: 10.1038/s41467-017-01961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A., Perez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., Nieto M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat.Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney T.J., Feitosa N.M., Sonntag C., Slanchev K., Kluger J., Kiyozumi D., Gebauer J.M., Coffin Talbot J., Kimmel C.B., Sekiguchi K., et al. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. Plos Genet. 2010;6 doi: 10.1371/journal.pgen.1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lun Y., Ovchinnikov D., Kokubo H., Oberg K.C., Pepicelli C.V., Gan L., Lee B., Johnson R.L. Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 1998;19:51–55. doi: 10.1038/ng0598-51. [DOI] [PubMed] [Google Scholar]
- Cheng S.L., Lecanda F., Davidson M.K., Warlow P.M., Zhang S.F., Zhang L., Suzuki S., St John T., Civitelli R. Human osteoblasts express a repertoire of cadherins, which are critical for BMP-2-induced osteogenic differentiation. J. Bone Miner Res. 1998;13:633–644. doi: 10.1359/jbmr.1998.13.4.633. [DOI] [PubMed] [Google Scholar]
- Cieply B., Park J.W., Nakauka-Ddamba A., Bebee T.W., Guo Y., Shang X., Lengner C.J., Xing Y., Carstens R.P. Multiphasic and dynamic changes in alternative splicing during induction of pluripotency are coordinated by numerous RNA-binding proteins. Cell Rep. 2016;15:247–255. doi: 10.1016/j.celrep.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenon S.G., Sarmah S., Shah B., Liu Q., Marrs J.A. Zebrafish cadherin-11 participates in retinal differentiation and retinotectal axon projection during visual system development. Dev. Dyn. 2012;241:442–454. doi: 10.1002/dvdy.23729. [DOI] [PubMed] [Google Scholar]
- Clendenon S.G., Shah B., Miller C.A., Schmeisser G., Walter A., Gattone V.H., 2nd, Barald K.F., Liu Q., Marrs J.A. Cadherin-11 controls otolith assembly: evidence for extracellular cadherin activity. Dev. Dyn. 2009;238:1909–1922. doi: 10.1002/dvdy.22015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol.Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Covassin L.D., Villefranc J.A., Kacergis M.C., Weinstein B.M., Lawson N.D. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc. Natl. Acad. Sci. U S A. 2006;103:6554–6559. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisenko E., Guo B.B., Jones M., Hou R., de Kock L., Lassmann T., Poppe D., Clement O., Simmons R.K., Lister R., Forrest A.R.R. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21:130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer S.D., Zhou G., Baldini A., Winterpacht A., Zabel B., Cole W., Johnson R.L., Lee B. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat. Genet. 1998;19:47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- Farber C.R., Reich A., Barnes A.M., Becerra P., Rauch F., Cabral W.A., Bae A., Quinlan A., Glorieux F.H., Clemens T.L., Marini J.C. A novel IFITM5 mutation in severe atypical osteogenesis imperfecta type VI impairs osteoblast production of pigment epithelium-derived factor. J. Bone Miner Res. 2014;29:1402–1411. doi: 10.1002/jbmr.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco H.L., Casasnovas J., Rodriguez-Medina J.R., Cadilla C.L. Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011;39:1177–1186. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi J., Sekido R., Murai K., Kamachi Y., Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- Galindo M., Pratap J., Young D.W., Hovhannisyan H., Im H.J., Choi J.Y., Lian J.B., Stein J.L., Stein G.S., van Wijnen A.J. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. 2018;362 doi: 10.1126/science.aaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Ju B., Wang X., He J., Wan H., Sudha P.M., Yan T. Green fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Dev. Dyn. 2002;223:204–215. doi: 10.1002/dvdy.10051. [DOI] [PubMed] [Google Scholar]
- Govindan J., Iovine M.K. Hapln1a is required for connexin43-dependent growth and patterning in the regenerating fin skeleton. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0088574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan J., Iovine M.K. Dynamic remodeling of the extra cellular matrix during zebrafish fin regeneration. Gene Expr. Patterns. 2015;19:21–29. doi: 10.1016/j.gep.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Grooteclaes M.L., Frisch S.M. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- Hennig B.P., Velten L., Racke I., Tu C.S., Thoms M., Rybin V., Besir H., Remans K., Steinmetz L.M. Large-scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 (Bethesda) 2018;8:79–89. doi: 10.1534/g3.117.300257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I., Balling R. Cloning and expression analysis of a novel mesodermally expressed cadherin. Dev. Biol. 1995;169:337–346. doi: 10.1006/dbio.1995.1148. [DOI] [PubMed] [Google Scholar]
- Hou Y., Lee H.J., Chen Y., Ge J., Osman F.O.I., McAdow A.R., Mokalled M.H., Johnson S.L., Zhao G., Wang T. Cellular diversity of the regenerating caudal fin. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilvesaro J., Metsikko K., Vaananen K., Tuukkanen J. Polarity of osteoblasts and osteoblast-like UMR-108 cells. J. Bone Miner Res. 1999;14:1338–1344. doi: 10.1359/jbmr.1999.14.8.1338. [DOI] [PubMed] [Google Scholar]
- Jhingory S., Wu C.Y., Taneyhill L.A. Novel insight into the function and regulation of alphaN-catenin by Snail2 during chick neural crest cell migration. Dev. Biol. 2010;344:896–910. doi: 10.1016/j.ydbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M.K., Preca B.T., Tripathi S.C., Jia D., George J.T., Hanash S.M., Brabletz T., Stemmler M.P., Maurer J., Levine H. Interconnected feedback loops among ESRP1, HAS2, and CD44 regulate epithelial-mesenchymal plasticity in cancer. APL Bioeng. 2018;2 doi: 10.1063/1.5024874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Kastenhuber E., Gesemann M., Mickoleit M., Neuhauss S.C. Phylogenetic analysis and expression of zebrafish transient receptor potential melastatin family genes. Dev. Dyn. 2013;242:1236–1249. doi: 10.1002/dvdy.24020. [DOI] [PubMed] [Google Scholar]
- Kawaguchi J., Azuma Y., Hoshi K., Kii I., Takeshita S., Ohta T., Ozawa H., Takeichi M., Chisaka O., Kudo A. Targeted disruption of cadherin-11 leads to a reduction in bone density in calvaria and long bone metaphyses. J. Bone Miner Res. 2001;16:1265–1271. doi: 10.1359/jbmr.2001.16.7.1265. [DOI] [PubMed] [Google Scholar]
- Kemp J.P., Morris J.A., Medina-Gomez C., Forgetta V., Warrington N.M., Youlten S.E., Zheng J., Gregson C.L., Grundberg E., Trajanoska K., et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 2017;49:1468–1475. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Matsunami H., Inoue T., Shimamura K., Uchida N., Ueno T., Miyazaki T., Takeichi M. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev. Biol. 1995;169:347–358. doi: 10.1006/dbio.1995.1149. [DOI] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C.W., Mahatma G., Fisher S., Brand M., Schulte-Merker S., Weidinger G. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev.Cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Lee D.M., Kiener H.P., Agarwal S.K., Noss E.H., Watts G.F., Chisaka O., Takeichi M., Brenner M.B. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007;315:1006–1010. doi: 10.1126/science.1137306. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Hou Y., Chen Y., Dailey Z.Z., Riddihough A., Jang H.S., Wang T., Johnson S.L. Regenerating zebrafish fin epigenome is characterized by stable lineage-specific DNA methylation and dynamic chromatin accessibility. Genome Biol. 2020;21:52. doi: 10.1186/s13059-020-1948-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Hami D., De Val S., Kagermeier-Schenk B., Wills A.A., Black B.L., Weidinger G., Poss K.D. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 2009;331:270–280. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh N.D., Dunlap G.S., Johnson K., Mariano R., Oshiro R., Wong A.Y., Bryant D.M., Miller B.M., Ratner A., Chen A., et al. Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nat. Commun. 2018;9:5153. doi: 10.1038/s41467-018-07604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leptin M. Twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- Li H., Wei X., Zhou L., Zhang W., Wang C., Guo Y., Li D., Chen J., Liu T., Zhang Y., et al. Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein Cell. 2021;12:57–66. doi: 10.1007/s13238-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Felber K., Elks P., Croucher P., Roehl H.H. Tracking gene expression during zebrafish osteoblast differentiation. Dev. Dyn. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sun H., Qi J., Wang L., He S., Liu J., Feng C., Chen C., Li W., Guo Y., et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat.Cell Biol. 2013;15:829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar S., Camacho-Magallanes A., Echeverria C., Jr., Rogers C.D. Cadherin-11 is required for neural crest specification and survival. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.563372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie P.J. Role of N-cadherin in bone formation. J.Cell Physiol. 2002;190:297–305. doi: 10.1002/jcp.10073. [DOI] [PubMed] [Google Scholar]
- Marin-Juez R., Marass M., Gauvrit S., Rossi A., Lai S.L., Materna S.C., Black B.L., Stainier D.Y. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. U S A. 2016;113:11237–11242. doi: 10.1073/pnas.1605431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.R., Fox J.A., Butland S.L., Ouellette B.F., Brinkman F.S., Tibbits G.F. Phylogeny of Na+/Ca2+ exchanger (NCX) genes from genomic data identifies new gene duplications and a new family member in fish species. Physiol. Genomics. 2005;21:161–173. doi: 10.1152/physiolgenomics.00286.2004. [DOI] [PubMed] [Google Scholar]
- Mateus R., Lourenco R., Fang Y., Brito G., Farinho A., Valerio F., Jacinto A. Control of tissue growth by Yap relies on cell density and F-actin in zebrafish fin regeneration. Development. 2015;142:2752–2763. doi: 10.1242/dev.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh I., Dunston J.A., Liu L., Hoover-Fong J.E., Sweeney E. Nail patella syndrome revisited: 50 years after linkage. Ann. Hum. Genet. 2005;69:349–363. doi: 10.1111/j.1529-8817.2005.00191.x. [DOI] [PubMed] [Google Scholar]
- McMillan S.C., Zhang J., Phan H.E., Jeradi S., Probst L., Hammerschmidt M., Akimenko M.A. A regulatory pathway involving retinoic acid and calcineurin demarcates and maintains joint cells and osteoblasts in regenerating fin. Development. 2018;145 doi: 10.1242/dev.161158. [DOI] [PubMed] [Google Scholar]
- Monnot M.J., Babin P.J., Poleo G., Andre M., Laforest L., Ballagny C., Akimenko M.A. Epidermal expression of apolipoprotein E gene during fin and scale development and fin regeneration in zebrafish. Dev. Dyn. 1999;214:207–215. doi: 10.1002/(SICI)1097-0177(199903)214:3<207::AID-AJA4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Nachtrab G., Czerwinski M., Poss K.D. Sexually dimorphic fin regeneration in zebrafish controlled by androgen/GSK3 signaling. Curr. Biol. 2011;21:1912–1917. doi: 10.1016/j.cub.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol.Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nord H., Dennhag N., Muck J., von Hofsten J. Pax7 is required for establishment of the xanthophore lineage in zebrafish embryos. Mol. Biol.Cell. 2016;27:1853–1862. doi: 10.1091/mbc.E15-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocana O.H., Corcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., Barrallo-Gimeno A., Cano A., Nieto M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Okazaki M., Takeshita S., Kawai S., Kikuno R., Tsujimura A., Kudo A., Amann E. Molecular cloning and characterization of OB-cadherin, a new member of cadherin family expressed in osteoblasts. J. Biol. Chem. 1994;269:12092–12098. [PubMed] [Google Scholar]
- Ouyang J.F., Kamaraj U.S., Cao E.Y., Rackham O.J.L. ShinyCell: simple and sharable visualisation of single-cell gene expression data. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab209. [DOI] [PubMed] [Google Scholar]
- Pagan A.J., Yang C.T., Cameron J., Swaim L.E., Ellett F., Lieschke G.J., Ramakrishnan L. Myeloid growth factors promote resistance to mycobacterial infection by curtailing granuloma necrosis through macrophage replenishment. Cell Host Microbe. 2015;18:15–26. doi: 10.1016/j.chom.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy D.M., Ransom D.G., Paw B., Zon L.I., Johnson S.L. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- Pfefferli C., Jazwinska A. The art of fin regeneration in zebrafish. Regeneration (Oxf) 2015;2:72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittlik S., Domingues S., Meyer A., Begemann G. Expression of zebrafish aldh1a3 (raldh3) and absence of aldh1a1 in teleosts. Gene Expr. Patterns. 2008;8:141–147. doi: 10.1016/j.gep.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Poss K.D., Shen J., Nechiporuk A., McMahon G., Thisse B., Thisse C., Keating M.T. Roles for Fgf signaling during zebrafish fin regeneration. Dev. Biol. 2000;222:347–358. doi: 10.1006/dbio.2000.9722. [DOI] [PubMed] [Google Scholar]
- Qin X., Jiang Q., Miyazaki T., Komori T. Runx2 regulates cranial suture closure by inducing hedgehog, Fgf, Wnt and Pthlh signaling pathway gene expressions in suture mesenchymal cells. Hum. Mol. Genet. 2019;28:896–911. doi: 10.1093/hmg/ddy386. [DOI] [PubMed] [Google Scholar]
- Roca H., Pande M., Huo J.S., Hernandez J., Cavalcoli J.D., Pienta K.J., McEachin R.C. A bioinformatics approach reveals novel interactions of the OVOL transcription factors in the regulation of epithelial - mesenchymal cell reprogramming and cancer progression. BMC Syst. Biol. 2014;8:29. doi: 10.1186/1752-0509-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F., Casano A.M., Henke K., Richter K., Peri F. The SLC7A7 transporter identifies microglial precursors prior to entry into the brain. Cell Rep. 2015;11:1008–1017. doi: 10.1016/j.celrep.2015.04.028. [DOI] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- San Martin I.A., Varela N., Gaete M., Villegas K., Osorio M., Tapia J.C., Antonelli M., Mancilla E.E., Pereira B.P., Nathan S.S., et al. Impaired cell cycle regulation of the osteoblast-related heterodimeric transcription factor Runx2-Cbfbeta in osteosarcoma cells. J.Cell Physiol. 2009;221:560–571. doi: 10.1002/jcp.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena K., Jolly M.K., Balamurugan K. Hypoxia, partial EMT and collective migration: emerging culprits in metastasis. Transl Oncol. 2020;13 doi: 10.1016/j.tranon.2020.100845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro I.M., Cheng A.W., Flytzanis N.C., Balsamo M., Condeelis J.S., Oktay M.H., Burge C.B., Gertler F.B. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. Plos Genet. 2011;7 doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets L., Ransom D.G., Mellgren E.M., Johnson S.L., Schnapp B.J. Zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr. Biol. 2007;17:1721–1734. doi: 10.1016/j.cub.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E., Ando K., Murase E., Kawakami A. Heterogeneous fates and dynamic rearrangement of regenerative epidermis-derived cells during zebrafish fin regeneration. Development. 2018;145 doi: 10.1242/dev.162016. [DOI] [PubMed] [Google Scholar]
- Singh S.P., Holdway J.E., Poss K.D. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev.Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Afonso N., Bensimon-Brito A., Fonseca M., Simoes M., Leon J., Roehl H., Cancela M.L., Jacinto A. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- Stains J.P., Civitelli R. Cell-to-cell interactions in bone. Biochem. Biophys. Res. Commun. 2005;328:721–727. doi: 10.1016/j.bbrc.2004.11.078. [DOI] [PubMed] [Google Scholar]
- Stemmler M.P., Eccles R.L., Brabletz S., Brabletz T. Non-redundant functions of EMT transcription factors. Nat.Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- Stephens A.S., Stephens S.R., Hobbs C., Hutmacher D.W., Bacic-Welsh D., Woodruff M.A., Morrison N.A. Myocyte enhancer factor 2c, an osteoblast transcription factor identified by dimethyl sulfoxide (DMSO)-enhanced mineralization. J Biol Chem. 2011;286:30071–30086. doi: 10.1074/jbc.M111.253518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Gomez A.W., Armstrong B.E., Henner A., Stankunas K. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 2014;6:482–498. doi: 10.1016/j.celrep.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 2012;365:339–349. doi: 10.1016/j.ydbio.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Yette G.A., Le Bleu H.K., Henner A.L., Braunstein J.A., Chehab J.W., Harms M.J., Stankunas K. Skeletal Geometry and Niche Transitions Restore Organ Size and Shape during Zebrafish Fin Regeneration. bioRxiv. 2019 doi: 10.1101/606970. [DOI] [Google Scholar]
- Stoick-Cooper C.L., Weidinger G., Riehle K.J., Hubbert C., Major M.B., Fausto N., Moon R.T. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallieres M., Permanyer J., Sodaei R., Marquez Y., et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017;27:1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., van Bokhoven A., van Leenders G.J., Ruijter E.T., Jansen C.F., Bussemakers M.J., Schalken J.A. Cadherin switching in human prostate cancer progression. Cancer Res. 2000;60:3650–3654. [PubMed] [Google Scholar]
- Tornini V.A., Puliafito A., Slota L.A., Thompson J.D., Nachtrab G., Kaushik A.L., Kapsimali M., Primo L., Di Talia S., Poss K.D. Live monitoring of blastemal cell contributions during appendage regeneration. Curr. Biol. 2016;26:2981–2991. doi: 10.1016/j.cub.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran A., Friedman D.B., Walsh A.J., Skala M.C., Sasi S., Rachakonda G., Crooks P.A., Freeman M.L., Sekhar K.R. The novel antiangiogenic VJ115 inhibits the NADH oxidase ENOX1 and cytoskeleton-remodeling proteins. Invest New Drugs. 2013;31:535–544. doi: 10.1007/s10637-012-9884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen S.J., Nollet F., Teugels E., Vennekens K.M., Malfait F., Philippe J., Speleman F., Bracke M.E., van Roy F.M., Mareel M.M. The alphaE-catenin gene (CTNNA1) acts as an invasion-suppressor gene in human colon cancer cells. Oncogene. 1999;18:905–915. doi: 10.1038/sj.onc.1202348. [DOI] [PubMed] [Google Scholar]
- Verschueren K., Remacle J.E., Collart C., Kraft H., Baker B.S., Tylzanowski P., Nelles L., Wuytens G., Su M.T., Bodmer R., et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. J. Biol. Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Wang H., Unternaehrer J.J. Epithelial-mesenchymal transition and cancer stem cells: at the crossroads of differentiation and dedifferentiation. Dev. Dyn. 2019;248:10–20. doi: 10.1002/dvdy.24678. [DOI] [PubMed] [Google Scholar]
- Warzecha C.C., Jiang P., Amirikian K., Dittmar K.A., Lu H., Shen S., Guo W., Xing Y., Carstens R.P. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Sato T.K., Nabet B., Hogenesch J.B., Carstens R.P. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol.Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha C.C., Shen S., Xing Y., Carstens R.P. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009;6:546–562. doi: 10.4161/rna.6.5.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.D., Gao D., Redfern A., Thompson E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.Y., Whited J.L. Parallels between wound healing, epimorphic regeneration and solid tumors. Development. 2020;147 doi: 10.1242/dev.181636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol.Cell Biol. 2020;21:341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska A., Cui C., Stockhammer O.W., Benard E.L., Spaink H.P., Meijer A.H. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116:1–11. doi: 10.1182/blood-2010-01-262873. [DOI] [PubMed] [Google Scholar]
- Zeisberg M., Neilson E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin.Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Louie K.W., Tingle C.F., Sha C., Heisel C.J., Unsworth S.P., Kish P.E., Kahana A. Twist3 is required for dedifferentiation during extraocular muscle regeneration in adult zebrafish. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0231963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
–S6
Data Availability Statement
-
•
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (GEO: GSE192498). This paper also analyzed existing, publicly available data (GEO: GSE137971, GEO: GSE126701). All details associated with the accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.