Abstract
Background
In mammals, modifications to cytosine bases, particularly in cytosine-guanine (CpG) dinucleotide contexts, play a major role in shaping the epigenome. The canonical epigenetic mark is 5-methylcytosine (5mC), but oxidized versions of 5mC, including 5-hydroxymethylcytosine (5hmC), are now known to be important players in epigenomic dynamics. Understanding the functional role of these modifications in gene regulation, normal development, and pathological conditions requires the ability to localize these modifications in genomic DNA. The classical approach for sequencing cytosine modifications has involved differential deamination via the chemical sodium bisulfite; however, bisulfite is destructive, limiting its utility in important biological or clinical settings where detection of low frequency populations is critical. Additionally, bisulfite fails to resolve 5mC from 5hmC.
Scope of review
To summarize how enzymatic rather than chemical approaches can be leveraged to localize and resolve different cytosine modifications in a non-destructive manner.
Major conclusions
Nature offers a suite of enzymes with biological roles in cytosine modification in organisms spanning from bacteriophages to mammals. These enzymatic activities include methylation by DNA methyltransferases, oxidation of 5mC by TET family enzymes, hypermodification of 5hmC by glucosyltransferases, and the generation of transition mutations from cytosine to uracil by DNA deaminases. Here, we describe how insights into the natural reactivities of these DNA-modifying enzymes can be leveraged to convert them into powerful biotechnological tools. Application of these enzymes in sequencing can be accomplished by relying on their natural activity, exploiting their ability to discriminate between cytosine modification states, reacting them with functionalized substrate analogs to introduce chemical handles, or engineering the DNA-modifying enzymes to take on new reactivities. We describe how these enzymatic reactions have been combined and permuted to localize DNA modifications with high specificity and without the destructive limitations posed by chemical methods for epigenetic sequencing.
Keywords: Epigenetics, Bisulfite, 5-Methylcytosine, 5-Hydroxymethylcytosine, DNA cytosine methyltransferases, TET dioxygenases, AID/APOBEC DNA deaminases, Glucosyltransferases
Highlights
-
•
Chemical sequencing methods damage DNA and can confound cytosine modifications.
-
•
DNA modifying enzymes offer non-destructive and selective biotechnological tools.
-
•
DNA deaminases, methyltransferases, oxygenases and glucosyltransferases can be used.
-
•
Permuting enzymes with various activities can reveal distinct cytosine states.
-
•
Engineered enzymes utilizing unnatural co-substrates expand sequencing scope.
1. Introduction — modified cytosine bases
The four chemically distinct bases of DNA—A, C, G, and T—are conserved across phylogeny and constitute the genomic code that can be inherited by future generations. Early in the 20th century, Wheeler and Johnson first synthesized 5-methylcytosine (5mC) and postulated its existence in genomic DNA samples [1]. Presciently called “epicytosine” in studies by Hotchkiss, 5mC was shown to have a distinct chemical identity from its parent base while simultaneously maintaining many of its properties [2].
Several decades later, the ubiquity of 5mC became evident, solidifying its standing as the 5th base of genomic DNA. In organisms from prokaryotes to eukaryotes, a conserved family of DNA methyltransferase enzymes (MTases) has been shown to catalyze the generation of 5mC through a reaction between the unmodified cytosine in DNA and the methyl donor S-adenosyl-l-methionine (SAM). 5mC preserves the capacity for hydrogen bonding with guanine, which is required for successful DNA replication. However, the methyl moiety introduced at the 5-position of cytosine provides a readable chemical handle with the potential to affect DNA-binding proteins and enzymes that often interact within the major groove of DNA, thus implicating 5mC across many diverse processes. In bacterial species, this chemical mark can serve to distinguish self from non-self as part of restriction–modification systems [3]. In eukaryotes, 5mC takes on new functions, serving predominantly as a gene-repressive epigenetic mark with physiological roles in development, imprinting, X-chromosome inactivation, and transposon silencing, as well as pathological roles in oncogenesis [4]. In 5mC, nature has found an opportunity to embellish DNA, thus expanding its information-encoding capacity within each generation without affecting DNA's most important function of facilitating inheritance of information across generations [5].
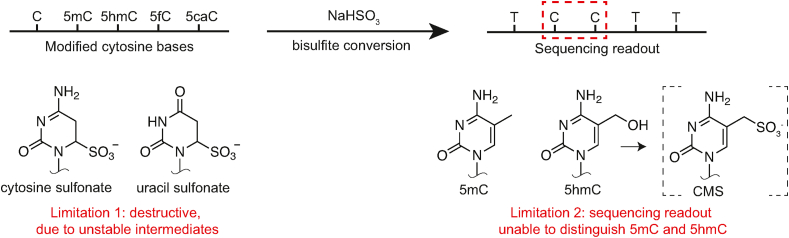
While early approaches such as paper chromatography and restriction digestion provided means for distinguishing 5mC from its parent base [2,6], it was the subsequent application of the chemical sodium bisulfite (NaHSO3) that allowed for the study of methylated cytosines at base resolution (Figure 1). The treatment of genomic DNA with bisulfite under acidic conditions leads to the sulfonation of unmodified cytosines, which promotes their deamination to uracil [7]. By contrast, 5mC does not react efficiently with bisulfite. Following amplification, the unmodified cytosines are read as thymidine in sequencing, while 5mC is still read as cytosine in a quantitative manner at single-base resolution.
Figure 1.
Bisulfite sequencing and its limitations. Bisulfite selectively deaminates various cytosines, which can aid in localizing modifications upon PCR amplification and sequencing. Problematically, sodium bisulfite is both destructive and unable to distinguish between the two most common modifications in mammalian genomes, 5mC and 5hmC.
The last decade has expanded our understanding of the importance of modified cytosines in epigenetics yet further [4,5]. The discovery of the TET family of enzymes [8] demonstrated that 5mC could be oxidized as part of a pathway that promotes the reversion of 5mC back to unmodified cytosine, a pathway known as active DNA demethylation. TET dioxygenases catalyze the stepwise conversion of 5mC to 5-hydroxymethylcytosine (5hmC), 5hmC to 5-formylcytosine (5fC), and 5fC to 5-carboxylcytosine (5caC) (Figure 1) [9,10]. 5hmC is the most prevalent of these modifications, with a presence as high as 10–30% of the level of 5mC in certain contexts such as cerebellar Purkinje cells [11]. Importantly, the field's reliance on bisulfite explains in part why 5hmC was so long overlooked (Figure 1). Unlike 5mC, 5hmC reacts with bisulfite, generating cytosine-5-methylenesulfonate (CMS). However, as CMS base-pairs with G upon amplification, the initial 5hmC base is indistinguishable from 5mC upon sequencing [12].
Despite its centrality to major advances in epigenetics, bisulfite sequencing (BS-Seq) poses at least two major limitations (Figure 1) that have prompted the development of a series of novel sequencing approaches. First, the fact that 5mC and 5hmC are indistinguishable in standard bisulfite sequencing poses a major challenge, as these two most common cytosine modifications appear to have distinct and often antagonistic biological functions. Notably, despite their low prevalence, 5fC and 5caC pose another source of confoundment with bisulfite, as these bases are deaminated in a manner akin to unmodified cytosine [13]. As a second limitation, bisulfite is inherently destructive. Sulfonation of cytosine bases results in unstable intermediates prone to the formation of abasic sites, which can occur at a frequency greater than 1 in every 200 bases after bisulfite conversion [14]. These lesions promote strand scission and can block amplification of DNA, limiting bisulfite's applicability in sparse samples. Despite these limitations, bisulfite-based approaches applied to circulating cell–free DNA have been explored for early cancer detection, highlighting a promise that could be fulfilled by more sensitive and specific techniques [15,16]. As modern epigenetics now aims to develop more robust workflows that can address the central question of how one cell differs from the next, the limitations posed by bisulfite have prompted a renaissance in the sequencing of epigenetic marks, with dedicated efforts aimed at developing alternatives that are less destructive and/or able to distinguish between 5mC and other modifications.
In this review, we highlight recent advances in cytosine-sequencing technologies, emphasizing the utility of either natural or engineered cytosine-modifying enzymes as tools for sequencing. Enzymes have advantages because of their specificity and non-destructive nature, which make them best poised to serve as powerful biotechnological tools in either enrichment-based approaches or more quantitative base-resolution approaches. Notably, while this review focuses on 5-position–modified cytosines, the most prevalent modifications in eukaryotic genomes, approaches to sequencing modified DNA bases more broadly have recently been reviewed [17]. PCR-free methods to directly read DNA modifications, such as those employed in third-generation sequencing technologies, will not be reviewed here [18,19]. Our review aims to introduce several cytosine-modifying enzymes in series, highlighting how permutations of natural or engineered versions of four enzymatic reactions on cytosine bases in DNA—glucosylation, oxidation, deamination, and methylation—have been recently leveraged as a means of distinguishing cytosine's many modification states.
2. Glucosyltransferases
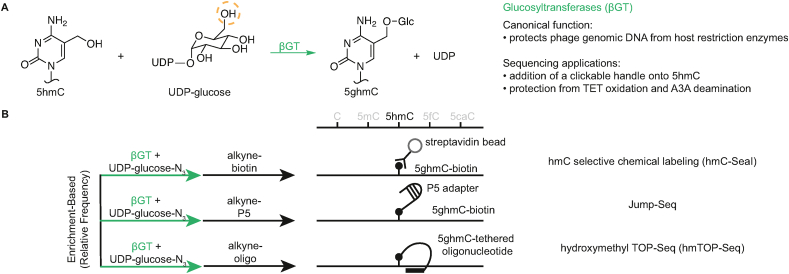
Many phage genomes encode hypermodified DNA bases that predominantly serve as a strategy to evade bacterial immune systems [20], with the T4 phage that infects Escherichia coli being a well-characterized model system. Upon infecting the bacteria, the phage first alters the deoxynucleotide triphosphate (dNTP) pool to contain 5-hydroxymethyl-dCTP in lieu of unmodified dCTP, resulting in the generation of phage genomes exclusively containing 5hmC. These 5hmC bases are then further glucosylated by phage-encoded α- and β-glucosyltransferases that use uridine diphosphate glucose (UDP-glucose) to make either the α- or β-anomer of glucosyl-5hmC (5ghmC) (Figure 2A).
Figure 2.
βGT activity and applications in sequencing. A) Canonical βGT reaction. βGT catalyzes the transfer of glucose from UDP-glucose to the hydroxymethyl group of 5hmC, generating 5ghmC. The dotted orange circle highlights a position on glucose that can be derivatized to transfer chemical groups of interest, such as azides. B) Various applications of the βGT enzyme in sequencing workflows.
Two types of approaches leveraging the phage-derived T4 β-glucosyltransferase (βGT) have been developed, which permit either enrichment-based or near base–resolution detection of 5hmC in genomic samples (Figure 2B). Notably, enrichment-based methods reveal the relative frequency of a modification in a particular genomic location compared to other parts of the genome, but not its absolute abundance. hmC-Seal was the first enzymatic enrichment-based approach for studying 5hmC [21]. In this approach, the native T4-βGT is used, but with an unnatural substrate—a chemically-modified UDP-glucose derivative containing an azide functional group (UDP-6-azide-glucose)—that site-specifically labels all 5hmC bases with the azido-modified glucose. The azido group can then be conjugated to a biotin-containing alkyne using copper-free click chemistry. The canonical biotin-streptavidin interaction is then exploited to enrich for molecules containing 5hmC bases in a manner analogous to an antibody pulldown experiment. These molecules can then be PCR amplified. Subsequent optimizations of this method have obtained information from as few as 1000 cells and have been explored as cancer diagnostic when applied to cell-free circulating DNA [[22], [23], [24], [25]].
A recent derivative technique named Jump-Seq also starts by utilizing T4-βGT to label 5hmC with an azido-modified glucose [26]. However, rather than biotin, the subsequent click chemistry tags the 5hmC-containing DNA with a hairpin oligonucleotide. This hairpin can then prime polymerase extension and, due to the covalent tether, the extended DNA can “jump” onto a 5hmC landing site. The technique can be used to infer near base–resolution information of 5hmC in a cost-effective manner, although it remains unclear how the technique handles multiple modifications in cis. A similar approach called hmTOP-Seq makes use of a tethered oligonucleotide as the template for primed extension and 5hmC localization [27].
3. TET dioxygenases
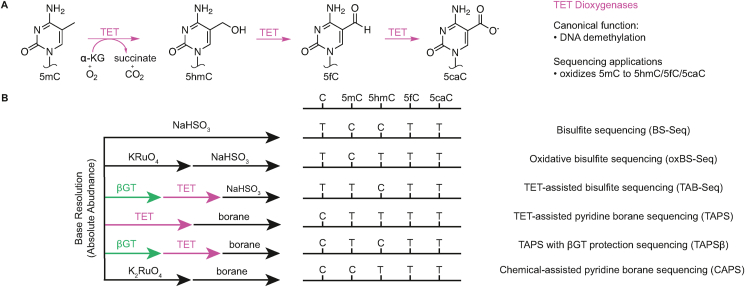
The discovery that bisulfite is unable to distinguish between 5mC and 5hmC motivated efforts to detect these two bases separately with chemical or enzymatic approaches. For chemical approaches, these efforts have relied upon the fact that 5fC and 5caC are both generally susceptible to bisulfite-mediated deamination, although it is important to note that the efficiency of 5fC deamination is not as high as that of unmodified cytosine [13]. One such chemical approach, oxidative bisulfite sequencing (oxBS-Seq), utilizes potassium perruthenate (KRuO4) to selectively oxidize 5hmC to 5fC [28]. Subsequent bisulfite treatment leaves only 5mC resistant to deamination, allowing for 5hmC to be calculated through bioinformatic subtraction between BS-only (5mC + 5hmC as C) and KRuO4 + BS experiments (5mC as C).
In their native role, TET enzymes catalyze the Fe(II)- and α-ketoglutarate–dependent oxidation of 5mC to 5hmC, 5hmC to 5fC, and 5fC to 5caC (Figure 3A). These reactivities have been leveraged in a variety of ways to localize modified cytosine bases (Figure 3B). One early approach used a combination of enzymatic approaches with bisulfite to detect 5hmC. In TET-Assisted Bisulfite Sequencing (TAB-Seq) [29], the activities of TET on 5mC and 5hmC are uncoupled from one another by first quantitatively converting all 5hmC to 5ghmC with UDP-glucose and T4-βGT. These 5ghmC bases are thus protected from TET-mediated oxidation, while 5mC bases are oxidized to 5fC or 5caC. Subsequent bisulfite treatment renders only the original 5hmC bases resistant to deamination. While a single TAB-Seq experiment allows the user to sequence 5hmC as C, comparison with a parallel standard bisulfite sequencing experiment (5mC + 5hmC) lets the user infer 5mC by bioinformatic subtraction. While this approach is convenient, indirect subtraction-based methods increase error, akin to 5hmC detection with oxBS-Seq, and cannot be applied in single cells given the need to process through two independent sequencing pipelines. An added limitation of TET-dependent sequencing approaches is the efficiency of TET enzymes themselves. TET enzymes are required to efficiently convert 5mC to 5caC in these sequencing pipelines, but the enzymes themselves are also prone to self-inactivation given their highly reactive Fe(IV)-oxo intermediates [9] and that the efficiency of oxidation decreases with conversion of 5mC to 5hmC or 5hmC to 5fC [30].
Figure 3.
TET activity and applications in sequencing. A) Canonical TET reaction. TET enzymes iteratively oxidize 5mC to produce the oxidized methylcytosines 5hmC, 5fC, and 5caC. B) Various applications of TET enzymes in sequencing workflows, with traditional bisulfite sequencing and oxBS-Seq for comparison on the top rows.
TET enzymes have also been recently applied in concert with non–bisulfite-mediated chemical schemes for localizing modifications [31]. TET-assisted pyridine borane sequencing (TAPS) starts with TET-catalyzed oxidation of 5mC to 5fC or 5caC. When the genomic DNA is subsequently treated with pyridine borane, 5fC and 5caC are converted to dihydrouracil, a non-aromatic uracil analog which sequences as T. The net result is a direct strategy for sequencing 5mC and 5hmC as T while leaving unmodified C intact. This unique reactivity profile yields both advantages and disadvantages: while efficient alignment and primer design benefit from the predominantly preserved four-base genome (in contrast to the three-base, C-depleted genomes made by bisulfite), it can be difficult to strand-specifically map individual reads, especially when modification prevalence is sparse. A similar borane reduction strategy has also been combined with either T4-βGT (TAPSβ) or potassium ruthenate (CAPS) to sequence 5mC and 5hmC individually, with varying degrees of efficiency [32]. Notably, borane-mediated conversion requires lengthy incubation under acidic conditions but functions by a mechanism that is less destructive than bisulfite deamination, which is inherently dependent on unstable sulfonated intermediates.
4. DNA deaminases
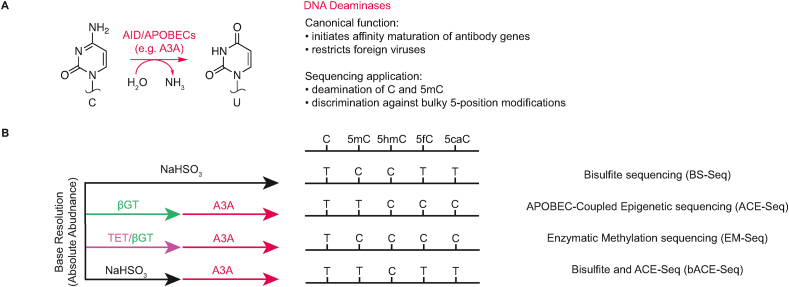
While bisulfite and borane-mediated conversion represent two mechanisms for deaminating cytosines, an enzymatic alternative achieves similar results. The DNA deaminases of the AID/APOBEC family play critical functions in adaptive or innate immunity, initiating antibody maturation and restricting retroviruses replication (Figure 4A) [5,33]. In their canonical roles, AID/APOBECs use a zinc cofactor to activate water molecules for nucleophilic attack on cytosines in single-stranded DNA (ssDNA). Enzymatic deamination by nucleophilic activation thus bypasses the unstable sulfonated intermediate generated by bisulfite-based deamination.
Figure 4.
AID/APOBEC activity and applications in sequencing. A) Canonical AID/APOBEC reaction. AID/APOBECs, including A3A, deaminate either cytosines to generate uracils or 5mCs to generate thymines. B) Various applications of the A3A enzyme in sequencing workflows, with traditional bisulfite sequencing for comparison on the top row.
A series of findings suggesting that DNA deaminases can discriminate between different cytosine modification states revealed new possibilities for their application to sequencing pipelines (Figure 4B). The initial detection of activity on 5mC led to conjectures about possible moonlighting roles for DNA deaminases in epigenetic reprogramming [34]. Subsequent systematic studies revealed that while activity on unmodified C and 5mC can be readily detected, deamination activity against 5hmC is significantly impaired [35]. Based on the analysis of a larger series of natural and unnatural 5-position–modified cytosines, the mechanistic basis for discrimination appeared to be selection against bulky or electronegative substituents [35,36]. This trend was maintained by APOBEC3A (A3A), the most active AID/APOBEC deaminase [37], and extends to discrimination against 5fC and 5caC [38]. Crystal structures have provided a molecular rationale for discrimination against larger 5-position substrates, with an active site residue (Tyr130) positioned to act as a hydrophobic gate adjacent to the C5–C6 face of cytosine in the structure of A3A bound to ssDNA [39].
Grounded in these extensive biochemical and structural studies, A3A has now been used in various approaches for epigenetic sequencing, all linked by their common reliance on discrimination against bulky 5-position–modified cytosine bases. Sequencing using enzymatic DNA deamination was pioneered in APOBEC-Coupled Epigenetic Sequencing (ACE-Seq) [40]. In this strategy, all 5hmCs are first converted to 5ghmC by T4-βGT. The added steric bulk to 5hmC blocks low level deamination, and the remaining unmodified C and 5mC bases can be efficiently deaminated by A3A. ACE-Seq represents the first non-destructive sequencing approach for profiling 5hmC at base resolution and additionally shows a sensitivity and specificity that outpaces bisulfite-based approaches.
A3A has also been combined with both TET enzymes and T4-βGT in a method called Enzymatic Methylation Sequencing (EM-Seq) [41]. In this approach, genomic DNA is oxidized by TET enzymes in the presence of T4-βGT. The 5mC and 5hmC are thus converted to a combination of 5caC and 5ghmC. As these modified bases are resistant to A3A-mediated deamination, subsequent treatment with A3A results in deamination of only unmodified cytosines, providing a readout akin to standard bisulfite. Importantly, this method has been extended to long read platforms, such as PacBio and Nanopore, taking advantage of the non-destructive nature of enzymatic deamination [42].
Enzymatic deamination has also been combined with bisulfite deamination in a manner that exploits the differential reactivity of 5mC and 5hmC. Bisulfite and APOBEC-Coupled Epigenetic Sequencing (bACE-Seq) builds on the fact that although 5hmC does not deaminate, the reaction to form CMS creates a bulky 5-position adduct that makes the modified base resistant to enzymatic deamination (Figure 1). Additional benefit comes from the fact that bisulfite can simultaneously fragment DNA and yield the ssDNA substrate needed for enzymatic deamination [43,44]. In bACE-Seq, after treatment with bisulfite, the DNA can be split into two parallel workflows: one to detect 5mC and 5hmC together (BS-only), and the other to deaminate 5mC by treating it with A3A, leaving only original 5hmC bases reading as C. Thus, the ability for DNA deaminases to discriminate between cytosine modifications has already been exploited to great effect, with a promise of more innovations to come.
5. DNA methyltransferases
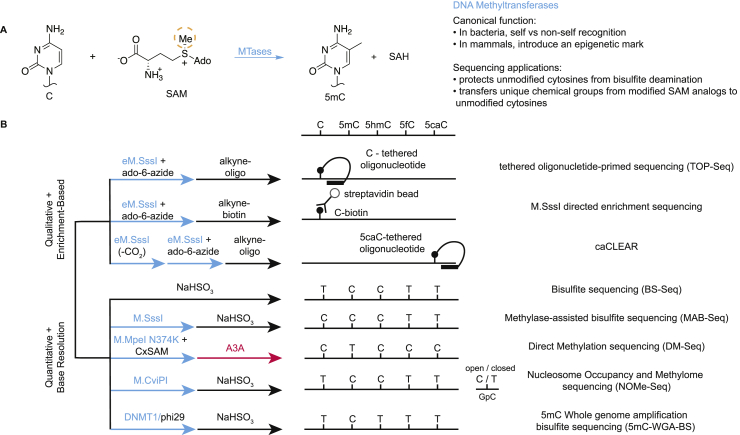
Cytosine DNA Methyltransferases (MTases) function by a conserved mechanism to create 5mC from cytosine and the metabolite SAM (Figure 5A). While cytosine modification occurs predominantly in the CpG context in mammals, there are cytosine MTases across phylogeny which can act in a variety of sequence contexts, and enzymatic sequencing approaches have exploited bacterial, viral, and mammalian MTases [45] (Figure 5B).
Figure 5.
MTase activity and applications in sequencing. A) Canonical MTase reaction. MTases catalyze the addition of a methyl group (from the methyl donor SAM) to cytosine. The dotted orange circle highlights the position on SAM that can be derivatized to transfer chemical groups of interest such as azides. B) Various applications of MTase enzymes in sequencing workflows, with traditional bisulfite sequencing for comparison in the top row of base resolution techniques.
The discovery of bacterial MTases with a preference for the canonical mammalian CpG site provided an initial tool for use in sequencing. M.SssI, derived from a Spiroplasma strain MQ1 [46], is one such CpG-specific MTase. In a strategy termed Methylase-Assisted Bisulfite Sequencing (MAB-Seq) [47,48], wild-type M.SssI is used to convert unmodified CpGs in genomic DNA samples into 5mCpGs. Given that these newly modified CpGs are now protected from deamination, as are the original 5mC and 5hmC, treatment with bisulfite then allows for the quantitative base resolution sequencing of 5fC and 5caC, the two remaining bases susceptible to bisulfite-mediated deamination.
MTases can also be engineered to accept SAM analogs as substrates. As first achieved with the M.HhaI MTase, alteration of the active site via mutagenesis at two conserved polar residues, often a glutamine and asparagine, to alanine allows for the transfer of larger extended alkyl chains from modified SAM analogs [45,46]. Mechanistically, while steric accommodation on the enzyme side is one requirement for analog transfer, a conjugated pi system in the SAM analog that facilitates transfer by increasing the electrophilicity of the transferable moiety serves as a second requirement [49].
This steric engineering strategy has been extended from M.HhaI to M.SssI to create the enzyme-engineered M.SssI (eM.SssI) [50]. In this approach, eM.SssI is used to react unmodified CpGs with a SAM analog containing one of two hex-2-ynyl side chains (Ado-6-Amine or Ado-6-Azide). These derivatized cytosine bases can then be coupled by amine-NHS or azide-DBCO conjugation chemistries in order to tag the modified DNA with biotin. Subsequent streptavidin pulldowns then enrich for fragments of DNA, allowing profiling of the “unmethylome”.
eM.SssI has also been applied in TOP-Seq [51], an earlier iteration of the previously described hmTOP-Seq. In this approach, an oligonucleotide hairpin is appended to unmethylated cytosine instead of 5-hydroxymethylcytosine using a chemical handle introduced via eM.SssI and a SAM analog. Polymerase extension yields fragments enriched for the unmethylated cytosine.
eM.SssI has also been applied to other non-canonical MTase reactions. In the absence of SAM, some MTases have been used to derivatize 5hmC with alkylthio moieties that can be further enriched [52]. It has also been previously shown that MTases can promote the removal of certain 5-position modifications in vitro and in the absence of SAM [53]. In a recently developed method known as caCLEAR, WT M.SssI is first employed to methylate all unmodified CpGs, and 5hmC bases are protected by T4-βGT (reactions not shown for clarity) [54]. Then, subsequent decarboxylation with eM.SssI in the absence of SAM “clears” 5caC residues by converting them to unmodified CpGs. Finally, eM.SssI is used to install Ado-6-Azide on all of the original 5caC residues, leaving the original unmodified cytosines, 5mC, and 5hmC residues unreacted. The azide-labeled 5caC residues can then be clicked to an oligonucleotide hairpin, whereby subsequent polymerase extension can yield fragments enriched for 5caC. Collectively, these results have shown that both WT and rational engineering of the Spiroplasma M.SssI have been useful for studying mammalian cytosine modifications.
In an added extension of MTase reactivity, our group has recently discovered MTases that can be engineered to take on neomorphic carboxymethyltransferase (CxMTase) activity [55]. Building on insights gleaned from the structure of the recently crystallized CpG MTase M.MpeI [56], we found that a single active site point mutation could allow the sparse natural metabolite carboxy-SAM (CxSAM) to be efficiently accepted as a substrate in lieu of SAM. We anticipate that coupling this unique activity to create an A3A-resistant 5-carboxymethylcytosine (5cxmC) base at unmodified CpGs could fit with our existing ACE-Seq workflow and create the first fully enzymatic sequencing workflow to directly sequence 5mC at base resolution.
Interestingly, cytosine-modifying enzymes can be used to simultaneously study DNA modifications and histone occupancy. The Chlorella virus GpC MTase M.CviPI was first leveraged in this regard [57]. In this approach, termed Nucleosome Occupancy and Methylome Sequencing (NOMe-Seq), the GpC MTase first reacts with DNA that is not compacted within inaccessible nucleosomes. Subsequent bisulfite sequencing then reads both regions of open chromatin, marked by non-nucleosomal GpC methylation, as well as native 5mC/5hmC modifications throughout the DNA sample.
In addition to bacterial and viral DNA MTases, mammalian MTases have been harnessed as biotechnological tools for sequencing. Rigorous biochemical characterization of the maintenance MTase DNMT1 has validated its strong preference for methylating opposite a 5mCpG while discriminating against unmodified CpGs and other modifications [58]. This selectivity has recently been leveraged in combination with phi29 polymerase for whole-genome amplification that preserves 5mC marks (5mC-WGA) [59]. Subsequent bisulfite treatment only identifies 5mC marks that have been copied by phi29 and replicated by DNMT1. The creative method is akin to naturally occurring DNA replication and offers a means to overcoming the limitations of destructive bisulfite-based sequencing by increasing the input sample amount. Thus, various aspects of MTase selectivity—sequence specificity, SAM analog preference, and discrimination between hemi-modified and unmodified CpG dyads—all have been exploited, with a promise of more applications to come.
6. Conclusions
In this review, we have highlighted how the very same enzymes that introduce cytosine modifications in natural systems can be leveraged to localize cytosine modifications in biotechnological applications. While classical methodologies have relied on bisulfite-mediated deamination, the discovery of a broader suite of DNA modifications and the need to profile DNA modifications in limited samples have provided the impetus for exploring non-destructive enzymatic approaches to sequencing. As noted above, the natural reactivity and selectivity of various cytosine-modifying enzymes offer a set of useful tools. In vitro reactivity and selectivity can also be viewed as potential challenges for enzyme-based sequencing pipelines, as high efficiency and substrate biases are theoretical limitations. However, structural and biochemical studies can be leveraged to optimize enzymatic features that are desirable for biotechnological applications, and the vast repertoire of homologous enzymes offered by nature offers the opportunity to overcome or complement limitations imposed by any one family member alone. Beyond protein engineering efforts, the ability to exploit non-canonical co-substrate metabolites (e.g., UDP-glucose and SAM analogs) or manipulate enzymes (e.g., eM.SssI or the neomorphic CxMTase) greatly increases the utility and power of enzymatic approaches. We anticipate that the arsenal of useful tools will continue to expand as new DNA-modifying enzymes are discovered, including recent examples such as dsDNA-specific cytidine deaminase enzymes, TET homologs that convert 5mC into 5-glyceryl-mC, and 5-hydroxymethylcytosine carbamoyltransferases [[60], [61], [62]]. Enzymatic approaches have presented and will continue to offer new opportunities for sequencing samples from rare populations at greater resolution than ever before, which will undoubtedly yield insights into fundamental questions of how two cells that share a genome can be distinguished from one another.
Conflict of interest
R.M.K. serves on the Scientific Advisory Board for Cambridge Epigenetix (CEGX) and T.W. is supported by a CEGX Training Fellowship. R.M.K. and T.W. have patents or patent submissions related to epigenetic sequencing methods.
Acknowledgments
The authors thank Emily Schutsky and Walraj Gosal for critical feedback. T.W. is supported by a Cambridge Epigenetix (CEGX) Training Fellowship. This work was supported by the NIH (R01-HG010646 to R.M.K.).
References
- 1.Hesson L.B., Pritchard A.L. 1st ed. Springer; 2019. Clinical epigenetics. [Google Scholar]
- 2.Hotchkiss R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. Journal of Biological Chemistry. 1948;175:315–332. [PubMed] [Google Scholar]
- 3.Wilson G.G., Murray N.E. Restriction and modification systems. Annual Review of Genetics. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 4.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 5.Nabel C.S., Manning S.A., Kohli R.M. The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chemical Biology. 2012;7:20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A.P., Southern E.M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. Journal of Molecular Biology. 1978;118:27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 7.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H., Wu X., Zhang Y. Base-resolution profiling of active DNA demethylation using MAB-seq and caMAB-seq. Nature Protocols. 2016;11:1081–1100. doi: 10.1038/nprot.2016.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka K., Okamoto A. Degradation of DNA by bisulfite treatment. Bioorganic & Medicinal Chemistry Letters. 2007;17:1912–1915. doi: 10.1016/j.bmcl.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Gole J., Gore A., He Q., Lu M., Min J., et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nature Communications. 2020;11:3475-z. doi: 10.1038/s41467-020-17316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., CCGA Consortium Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Annals of Oncology : Official Journal of the European Society for Medical Oncology. 2020;31:745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer A., Liu Z.J., Balasubramanian S. Detection, structure and function of modified DNA bases. Journal of the American Chemical Society. 2019;141:6420. doi: 10.1021/jacs.9b01915. [DOI] [PubMed] [Google Scholar]
- 18.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nature Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 19.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nature Reviews Genetics. 2020;21:597–614. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weigele P., Raleigh E.A. Biosynthesis and function of modified bases in bacteria and their viruses. Chemical Reviews. 2016;116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 21.Song C., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnology. 2010:1–8. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D., Lu X., Shih A.H., Nie J., You Q., Xu M.M., et al. A highly sensitive and robust method for genome-wide 5hmC profiling of rare cell populations. Molecular Cell. 2016;63:711–719. doi: 10.1016/j.molcel.2016.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P., Lin S., Cai M., Zhu Y., Song Y., Sui Y., et al. 5-Hydroxymethylcytosine profiling from genomic and cell-free DNA for colorectal cancers patients. Journal of Cellular and Molecular Medicine. 2019;23:3530–3537. doi: 10.1111/jcmm.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Research. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song C.X., Yin S., Ma L., Wheeler A., Chen Y., Zhang Y., et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Research. 2017;27:1231–1242. doi: 10.1038/cr.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L., Liu Y., Han S., Yang L., Cui X., Gao Y., et al. Jump-seq: genome-wide capture and amplification of 5-hydroxymethylcytosine sites. Journal of the American Chemical Society. 2019;141:8694. doi: 10.1021/jacs.9b02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibas P., NarmontA M., Stasevskij Z., Gordevicius J., Klimasauskas S., KriukienA E., et al. Precise genomic mapping of 5-hydroxymethylcytosine via covalent tether-directed sequencing. PLoS Biology. 2020;18 doi: 10.1371/journal.pbio.3000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 29.Yu M., Hon G.C., Szulwach K.E., Song C.X., Zhang L., Kim A., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu L., Lu J., Cheng J., Rao Q., Li Z., Hou H., et al. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527:118. doi: 10.1038/nature15713. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Siejka-Zielinska P., Velikova G., Bi Y., Yuan F., Tomkova M., et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nature Biotechnology. 2019;37:424–429. doi: 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Hu Z., Cheng J., Siejka-Zielińska P., Chen J., Inoue M., et al. Subtraction-free and bisulfite-free specific sequencing of 5-methylcytosine and its oxidized derivatives at base resolution. Nature Communications. 2021;12:618. doi: 10.1038/s41467-021-20920-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siriwardena S.U., Chen K., Bhagwat A.S. Functions and malfunctions of mammalian DNA-cytosine deaminases. Chemical Reviews. 2016;116:12688–12710. doi: 10.1021/acs.chemrev.6b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. Journal of Biological Chemistry. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 35.Nabel C.S., Jia H., Ye Y., Shen L., Goldschmidt H.L., Stivers J.T., et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nature Chemical Biology. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangam G., Schmitz K.M., Cobb A.J., Petersen-Mahrt S.K. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One. 2012;7:e43279. doi: 10.1371/journal.pone.0043279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito F., Fu Y., Kao S.A., Yang H., Chen X.S. Family-wide comparative analysis of cytidine and methylcytidine deamination by eleven human APOBEC proteins. Journal of Molecular Biology. 2017;429:1787–1799. doi: 10.1016/j.jmb.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schutsky E.K., Nabel C.S., Davis A.K.F., DeNizio J.E., Kohli R.M. APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Research. 2017;45:7655–7665. doi: 10.1093/nar/gkx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi K., Carpenter M.A., Banerjee S., Shaban N.M., Kurahashi K., Salamango D.J., et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nature Structural & Molecular Biology. 2017;24:131–139. doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schutsky E.K., DeNizio J.E., Hu P., Liu M.Y., Nabel C.S., Fabyanic E.B., et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nature Biotechnology. 2018;36:1083–1090. doi: 10.1038/nbt.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaisvila R., Ponnaluri V.K.C., Sun Z., Langhorst B.W., Saleh L., Guan S., et al. EM-seq: detection of DNA methylation at single base resolution from picograms of DNA. bioRxiv. 2020 doi: 10.1101/gr.266551.120. 2019.12.20.884692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z., Vaisvila R., Hussong L.M., Yan B., Baum C., Saleh L., et al. Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Research. 2021;31:291–300. doi: 10.1101/gr.265306.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabyanic E.B., Hu P., Qiu Q., Wang T., Berríos K.N., Flournoy J., et al. 2021. Quantitative single cell 5hmC sequencing reveals non-canonical gene regulation by non-CG hydroxymethylation. [Google Scholar]
- 44.Caldwell B.A., Liu M.Y., Prasasya R.D., Wang T., DeNizio J.E., Leu N.A., et al. Functionally distinct roles for TET-oxidized 5-methylcytosine bases in somatic reprogramming to pluripotency. Molecular Cell. 2021;81:859–869. doi: 10.1016/j.molcel.2020.11.045. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer L.M., Abhiman S., Aravind L. Natural history of eukaryotic DNA methylation systems. Progress in Molecular Biology and Translational Science. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 46.Renbaum P., Abrahamove D., Fainsod A., Wilson G.G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI) Nucleic Acids Research. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H., Wu X., Shen L., Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nature Biotechnology. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neri F., Incarnato D., Krepelova A., Rapelli S., Anselmi F., Parlato C., et al. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Reports. 2015;10:674–683. doi: 10.1016/j.celrep.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Dalhoff C., Lukinavicius G., Klimasauskas S., Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nature Chemical Biology. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 50.Kriukienė E., Labrie V., Khare T., Urbanavičiūtė G., Lapinaitė A., Koncevičius K., et al. DNA unmethylome profiling by covalent capture of CpG sites. Nature Communications. 2013;4:2190. doi: 10.1038/ncomms3190. [DOI] [PubMed] [Google Scholar]
- 51.Staševskij Z., Gibas P., Gordevičius J., Kriukienė E., Klimašauskas S. Tethered oligonucleotide-primed sequencing, TOP-seq: a high-resolution economical approach for DNA epigenome profiling. Molecular Cell. 2017;65:554–564.e6. doi: 10.1016/j.molcel.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liutkeviciute Z., Kriukiene E., Grigaityte I., Masevicius V., Klimasauskas S. Methyltransferase-directed derivatization of 5-hydroxymethylcytosine in DNA. Angewandte Chemie. 2011;50:2090–2093. doi: 10.1002/anie.201007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liutkeviciute Z., Lukinavicius G., Masevicius V., Daujotyte D., Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nature Chemical Biology. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 54.Ličytė J., Gibas P., Skardžiūtė K., Stankevičius V., Rukšėnaitė A., Kriukienė E. A bisulfite-free approach for base-resolution analysis of genomic 5-carboxylcytosine. Cell Reports. 2020;32:108155. doi: 10.1016/j.celrep.2020.108155. [DOI] [PubMed] [Google Scholar]
- 55.Wang T., Kohli R.M. Discovery of an unnatural DNA modification derived from a natural secondary metabolite. Cell Chemical Biology. 2021;28:97–104. doi: 10.1016/j.chembiol.2020.09.006. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wojciechowski Marek, Czapinska Honorata, Bochtler Matthias. CpG underrepresentation and the bacterial CpG-specific DNA methyltransferase M.MpeI. Proceedings of the National Academy of Sciences. 2013;110:105–110. doi: 10.1073/pnas.1207986110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelly T.K., Liu Y., Lay F.D., Liang G., Berman B.P., Jones P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Research. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seiler C.L., Fernandez J., Koerperich Z., Andersen M.P., Kotandeniya D., Nguyen M.E., et al. Maintenance DNA methyltransferase activity in the presence of oxidized forms of 5-methylcytosine: structural basis for ten eleven translocation-mediated DNA demethylation. Biochemistry. 2018;57:6061–6069. doi: 10.1021/acs.biochem.8b00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Cui X., Zhao B.S., Narkhede P., Gao Y., Liu J., et al. DNA 5-methylcytosine-specific amplification and sequencing. Journal of the American Chemical Society. 2020;142:4539. doi: 10.1021/jacs.9b12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue J.H., Chen G.D., Hao F., Chen H., Fang Z., Chen F.F., et al. A vitamin-C-derived DNA modification catalysed by an algal TET homologue. Nature. 2019;569:581–585. doi: 10.1038/s41586-019-1160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mok B.Y., De Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583 doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang W., Lin Y., Johnson W., Dai N., Vaisvila R., Weigele P.R., et al. A Genome-Phenome Association study in native microbiomes identifies a mechanism for cytosine modification in DNA and RNA. bioRxiv. 2021 doi: 10.7554/eLife.70021. 2021.03.23.436658. [DOI] [PMC free article] [PubMed] [Google Scholar]