Abstract
As one of the most common clinical cardiovascular diseases (CVDs), coronary heart disease (CHD) is the most common cause of death in the world. It has been confirmed that Zhishi Xiebai Guizhi decoction (ZXGD), a classical prescription of the traditional Chinese medicine (TCM), has achieved certain effects in the treatment of CHD; however, the mechanism still remains controversial. In this paper, an integrated approach, including UPLC-UESI-Q Exactive Focus, gene expression profiling, network pharmacology, and experimental validation, was introduced to systematically investigate the mechanism of ZXGD in the treatment of CHD. First, UPLC-UESI-Q Exactive Focus was applied to identify the chemical compounds of ZXGD. Then, the targets of the components for ZXGD were predicted by MedChem Studio software embed in the integrative pharmacology-based research platform of TCM, and the differentially expressed genes (DEGs) of CHD were obtained by gene expression profiling in gene expression omnibus database. The common genes of the above two genes were obtained by Venn analysis as the targets of GXGD in treatment with CHD. Third, the core targets were screened out by protein–protein interaction network analysis, and the kyoto encyclopedia of genes and genomes pathway enrichment analysis was performed by the database for annotation, visualization, and integrated discovery bioinformatics resources. After that, the formula–herb–compound–target–pathway network was constructed to explore the mechanism of ZXGD in the treatment of CHD. Finally, molecular docking and the vitro experiment were carried out to validate some key targets. As a result, a total of 39 compounds, 12 core targets, and 4 pathways contributed to ZXGD for the treatment of CHD. This study preliminarily provided a foundation for the study on the mechanism against CHD for ZXGD and may be a reference for the compatibility mechanism and the extended application of TCM compound prescription.
1. Introduction
Coronary heart disease (CHD), as a high incidence of cardiovascular disease (CVD), is one of the diseases with the highest mortality in the world, and about 8.9 million people die of CHD every year.1 With the characteristics of high mortality, high disability rate, and high recurrence rate, CHD has become one of the major public health problems.2 The pathogenesis of CHD was complicated, mainly related to lipid metabolism disorders, oxidative stress, endothelial cell damage, smooth muscle cell proliferation and migration, and so forth.3 So far, the main clinical treatment drugs for CHD consisted of statins (such as atorvastatin), antiplatelet aggregation drugs (such as aspirin), vasodilators (such as nitroglycerin), and thrombolytic and thrombolytic drugs (such as urokinase).3 Although drugs and surgery have curative effects, the side effects are also inevitable.4−8 For example, statins may cause liver damage and muscle damage, and aspirin can increase the risk of gastrointestinal bleeding. When the area of coronary artery stenosis expands, heart stent surgery must be required. In recent years, traditional Chinese medicine (TCM) has been highlighted for the increasingly important role in the clinical treatment of CHD. With the incidence and mortality of CHD, more and more people use TCM as complementary and alternative treatments.9
The clinical symptoms of CHD are mainly manifested as pectoralgia that belongs to the categories of “chest stuffiness” in TCM. Zhishi Xiebai Guizhi decoction (ZXGD) was first seen in Jin Gui Yao Lue and proposed by Zhongjing Zhang, a famous medical scientist in the Eastern Han Dynasty. This prescription is composed with five herbs, including Aurantii fructus immaturus, Allii macrostemonis bulbus, Trichosanthis fructus, Magnoliae officinalis cortex, and Cinnamomi ramulus and has therapeutic efficacy with chest stuffiness. Modern pharmacological research revealed that A. fructus immaturus has pharmacological effects related to CVD via prolonging myocardial blood perfusion time to reduce myocardial ischemic response.10 A. macrostemonis bulbus and T. fructus are the principal herbs of ZXGD, and modern pharmacology has proved that A. macrostemonis bulbus can significantly reduce the contents of serum total cholesterol, low-density lipoprotein cholesterol, triglyceride level, and atherosclerosis thickness in hyperlipidemic rats. Meanwhile, T. fructus has many functions, such as dilating microvessels, increasing coronary blood flow, increasing hypoxia tolerance, protecting ischemic myocardium, and so on.11 C. ramulus has affected blood vessels by increasing coronary blood flow with different sites of action.12 Some studies have illustrated that M. officinalis cortex could enhance the activity of superoxide dismutase and reduce the damage to mitochondria by inhibiting the lipid peroxidation of the oxygen free-radical production system to alleviate the damage of cardiomyocytes.13 Despite the certain curative effects of herbs, it is still unclear about the mechanism of ZXGD in treating CHD due to the characteristics of multi-components, multi-targets, and multi-pathways of TCM.
As known, TCM formula plays a therapeutic role through multi-component synergistic effects. Therefore, choosing a reasonable and efficient analysis method to quickly identify the chemical composition of TCM formula is the first step to study its mechanism. In recent years, UPLC-UESI-Q Exactive Focus has been recognized as one of the effective methods to identify the components of TCM formula with good resolution, excellent sensitivity, and strong structural characterization capability and has been widely used in the fields of medicine, chemical industry, biology, environmental protection, and so on.14−16 In this study, we used UPLC-UESI-Q Exactive Focus combined with Thermo Scientific Ultimate 3000 system for the rapid qualitative analysis of the chemical composition for ZXGD. Then, the targets of the chemical compositions for ZXGD were determined by a target fishing tool based on molecular similarity embed in the integrative pharmacology-based research platform of TCM (TCMIP). To obtain the genes of disease, with the help of the experimental gene expression data collected in gene expression omnibus (GEO) database, the gene expression profile was introduced to identify the disease-related genes. After that, Venn analysis was performed to obtain the common genes as the targets of GXGD in treatment with CHD, and the core targets were screened out by protein–protein interaction (PPI) network analysis. Afterward, the kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis were performed by the database for annotation, visualization, and integrated discovery (DAVID) bioinformatics resources, and the formula–herb–compound–target–pathway (F–H–C–T–P) network was constructed to explore the mechanism of ZXGD in the treatment of CHD. Ultimately, molecular docking was used to verify the interactions between core targets and compounds, and in vitro experiment was also conducted to validate the potential underlying mechanism of ZXGD in treatment with CHD. This study aimed to provide a comprehensive basis for further research studies on the molecular mechanism of ZXGD in the treatment of CHD.17 The detailed technical strategy of the current study is displayed in Figure 1.
Figure 1.
Flowchart for ZXGD in treatment with CHD.
2. Results
2.1. UPLC-UESI-Q Exactive Focus Analysis of ZXGD
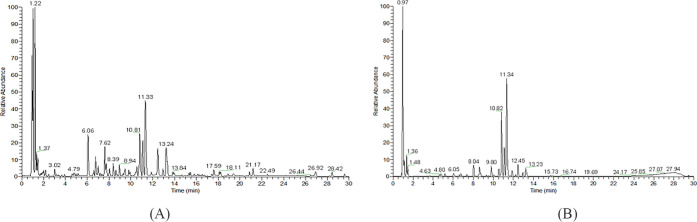
For mapping the chemical profiles of the extracts of ZXGD, all data of mass fragmentation coupled with high-resolution spectrometry provide sufficient information. UPLC-UESI-Q Exactive Focus was used in this study, and the base peak chromatogram of ZXGD in the mode of positive and negative ions is shown in Figure 2A,B. Compounds were identified by determining the elemental compositions of the precursor and product ions. The molecular formula and rational fragmentation patterns and pathways of these compounds were then identified based on the comparison between these data and chemical databases.
Figure 2.
Total ion flow diagram of ZXGD. (A) Positive ion mode and (B) negative ion mode.
Xcalibur 3.2 software was used for peak extraction and peak matching analysis on the collected raw mass spectrometry data. The collected raw data are imported into Compound Discover 3.0 software. The sample name, retention time, molecular formula, mass-to-charge ratio, and data set that was established with the corresponding ion intensity were exported through peak extraction, online and local database search, compound prediction, and other data processing. This method could largely prevent false negative results, thus making the results faster and more accurate. The establishment of our own database was based on an independent database of each herb after checking and summarizing a large number of relevant references. By looking up the literature of ZXGD and other articles about UPLC-UESI-Q Exactive Focus, Xinjuan Liu et al. also used the method of self-built database of each herb material of ZXGD to study the composition of ZXGD.18 It is a general method for the component analysis of TCM by UPLC-MS. Therefore, our analysis method was reliable and feasible.
In this study, a total of 230 compounds were obtained, including 48 ingredients in A. fructus immaturus, 23 ingredients in A. macrostemonis bulbus, 71 ingredients in T. fructus, 86 ingredients in M. officinalis cortex, and 48 ingredients in C. ramulus. These compounds were renumbered as MOL001 to MOL230 (the details are displayed in the Supporting Information). The compounds of A. fructus immaturus are mainly flavonoids, including naringin, hesperidin, poncirin, and so forth. Hesperidin can reduce vascular fragility, lower blood pressure, and can be used as a cardiotonic agent for the adjuvant treatment of CVD.19 Steroid saponins, sulfur-containing compounds, and nitrogen-containing compounds are the main components of A. macrostemonis bulbus.20 Nitrogen-containing compounds, such as adenosine, have pharmacological effects such as dilating coronary blood vessels and have been widely applied in clinical practices. Modern pharmacological studies have revealed that the main component of cinnamon sticks, namely cinnamic aldehyde, has the effects of promoting blood circulation, dilating blood vessels around the center, increasing coronary blood flow, and improving coronary blood circulation.21
2.2. Targets of the Chemical Compositions for ZXGD in the Treatment of CHD
The targets of chemical compositions for ZXGD were obtained by MedChem Studio software, which is a target fishing tool based on molecular similarity embed in the TCMIP. As a result, 2130 targets were considered as candidate targets of ZXGD.
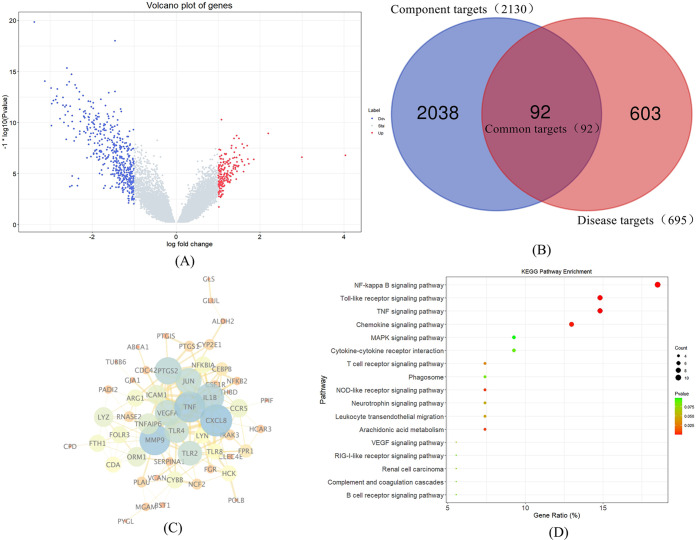
A total of 37,348 genes were annotated by gene expression profiling, and 695 differentially expressed genes (DEGs) between CHD patients and control individuals were screened out with P value < 0.05 and |log FC| > 1, which were considered as CHD-related targets. Among these DEGs, 202 genes were upregulated, and 493 genes were downregulated (Figure 3A).
Figure 3.
(A) Volcano plot of CHD-related genes. Blue represents downregulated genes, red represents upregulated genes, and gray represents genes without significantly different expressions. (B) Venn diagrams of common targets between compound targets and CHD-related targets. Blue area illustrates the target number of ZXGD, red area illustrates the target number of CHD, and purple area illustrates the number of common targets between two data sets. (C) PPI network for ZXGD in treatment with CHD. The nodes represent targets, and the edges represent the interaction between two nodes. The larger the circle, the brighter the color, indicating that it plays a more important role in the network. (D) Bubble plot of KEGG enrichment analysis of ZXGD in treatment with CHD. The gene ratio represented the proportion of targets in 54 core targets, bubble size depended on the target number of pathways, and P value decided the color (red represents a low value, and green represents a high value).
The common targets between the disease-related DEGs and the targets of the compounds were obtained by Venn analysis, and 92 common targets were determined as the potential targets for ZXGD in treatment with CHD (Figure 3B).
2.3. PPI Network Construction and Core Target Determination
The 92 targets of ZXGD were imported into a STRING 11.0 platform for searching the targets that interact with the potential targets, and the confidence level was set as 0.7. After that, the interaction results were imported into Cytoscape 3.7.2 software to draw the PPI network, and 54 nodes and 162 edges were screened out after deleting the nodes and edges that were outside the connection relationship compared with the original network (Figure 3C). In Figure 3C, the nodes represented targets, and the edges marked the interaction between the two nodes. The larger the circle, the brighter the color, indicating that it plays a more important role in the network.
2.4. Results of KEGG Pathway Analysis
Totally, 54 targets were put into the online enrichment analysis website DAVID with species set as “Homo sapiens.” According to the results, 46 KEGG pathways were selected out, and unrelated pathways were obviously filtered out, such as some diseases related to bacterium or virus, “Salmonella infection,” and “Herpes simplex infection.” Finally, the top 17 pathways were regarded as the key pathways for ZXGD in treatment with CHD, including the NF-kappa B signaling pathway, toll-like receptor signaling pathway, and TNF signaling pathway. Moreover, 17 pathways were mapped as a bubble plot with R language (Figure 3D).
2.5. F–H–C–T–P Network Construction of ZXGD in Treatment with CHD
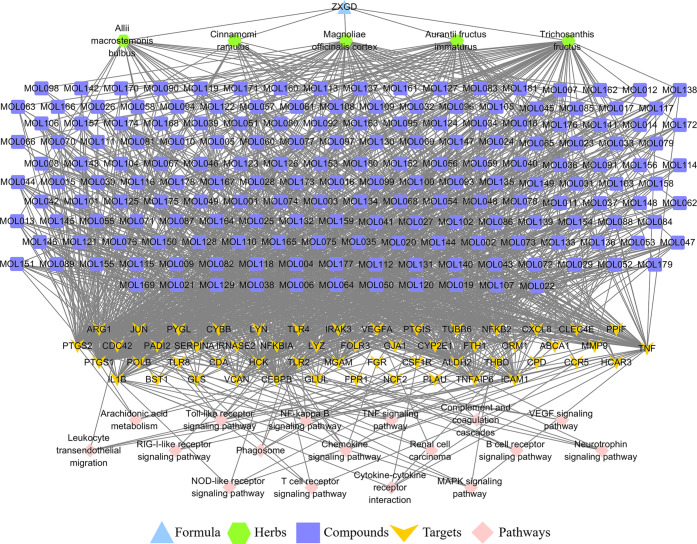
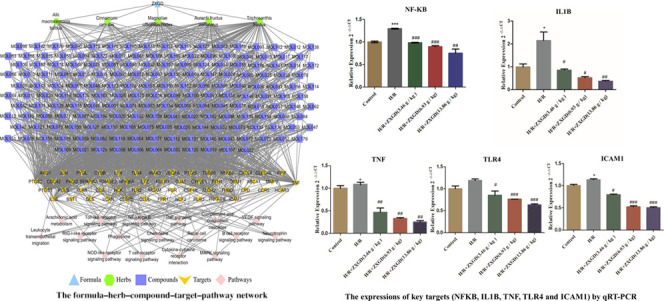
In order to investigate the key compounds, key targets, and key pathways for ZXGD in treatment with CHD, the F–H–C–T–P network was constructed by Cytoscape 3.7.2 (Figure 4). The network consisted of 258 nodes (1 formula, 5 herbs, 181 hub compounds, 54 core targets, and 17 key pathways) and 1408 edges. As shown in Figure 4, the formula ZXGD is shown in sky blue triangle, the herb nodes are shown in green hexagon, the deep blue square represents compound nodes, the targets nodes are shown in orange arrow, and pink diamond represents the pathway nodes. The gray edges stood for the relationship among the herbs, compounds, targets, and pathways. Additionally, centrality is a concept that is applied to network analysis, aiming to spread the significance of a node throughout the whole network, and it is the most direct indicator. Therefore, three topological parameters, namely degree centrality (DC), betweenness centrality (BC), and closeness centrality (CC), were applied by the network analyzer in Cytoscape to reflect the significance of nodes directly. Subsequently, 39 compounds (Table 1), 12 targets (Table 2), and 4 pathways (Table 3) were shifted out, and their topological parameters were greater than the median of DC, CC, and BC.
Figure 4.
F–H–C–T–P network for ZXGD in the treatment of CHD. The formula ZXGD is shown in sky blue triangle, the herb nodes are shown in green hexagon, the deep blue square represents compound nodes, the target nodes are shown in orange arrow, and pink diamond represents the pathway nodes.
Table 1. DC, BC, and CC Values of the Key Compounds for ZXGD in Treatment with CHD.
| no. | compound ID | compound name | DC | BC | CC |
|---|---|---|---|---|---|
| 1 | MOL007 | guanosine | 20 | 0.0339 | 0.4614 |
| 2 | MOL137 | adenosine | 19 | 0.0187 | 0.4631 |
| 3 | MOL020 | thymidine | 16 | 0.0194 | 0.4565 |
| 4 | MOL146 | cytidine | 16 | 0.0117 | 0.4565 |
| 5 | MOL131 | 4-(β-d-glucopyranosyloxy)-3-methoxybenzoic acid | 16 | 0.0057 | 0.4565 |
| 6 | MOL174 | quercetin-3-O-β-d-glucofuranoside | 16 | 0.0054 | 0.4565 |
| 7 | MOL052 | phlorin | 16 | 0.0053 | 0.4549 |
| 8 | MOL054 | prunin | 16 | 0.0050 | 0.4549 |
| 9 | MOL128 | 2,7-dimethyl-2,4-diene deca-α,ω-diacid-8-O-β-d-glu | 15 | 0.0056 | 0.4517 |
| 10 | MOL121 | sinapaldehyde-4-O-β-d-glucopyranoside | 15 | 0.0051 | 0.4565 |
| 11 | MOL057 | rhoifolin | 15 | 0.0042 | 0.4517 |
| 12 | MOL093 | coniferin | 14 | 0.0047 | 0.4517 |
| 13 | MOL113 | manglieside D | 14 | 0.0047 | 0.4517 |
| 14 | MOL123 | syringin | 14 | 0.0047 | 0.4517 |
| 15 | MOL154 | maempferol-3-O-β-d-glucoside-7-O-α-rhamnoside | 13 | 0.0033 | 0.4378 |
| 16 | MOL106 | magnoflorine | 12 | 0.0191 | 0.4423 |
| 17 | MOL006 | glucose | 12 | 0.0038 | 0.4423 |
| 18 | MOL008 | l-arabinose | 12 | 0.0038 | 0.4423 |
| 19 | MOL105 | magnocurarine | 11 | 0.0168 | 0.4363 |
| 20 | MOL152 | glutamic acid | 11 | 0.0166 | 0.4276 |
| 21 | MOL117 | pinoresionol-4-O-β-d-glucopyranoside | 11 | 0.0067 | 0.4439 |
| 22 | MOL122 | syringaresinol-4′-O-β-d-glucopyranoside | 11 | 0.0067 | 0.4439 |
| 23 | MOL001 | acanthoside D | 11 | 0.0063 | 0.4408 |
| 24 | MOL151 | galactonic acid-γ-lactone | 11 | 0.0038 | 0.4454 |
| 25 | MOL003 | chinenoside IV | 11 | 0.0037 | 0.4334 |
| 26 | MOL004 | chinenoside V | 11 | 0.0037 | 0.4334 |
| 27 | MOL173 | quercetin-3-O-α-d-nucleoside | 11 | 0.0033 | 0.4319 |
| 28 | MOL119 | quercitrin | 10 | 0.0039 | 0.4501 |
| 29 | MOL009 | laxogenin | 10 | 0.0036 | 0.4305 |
| 30 | MOL013 | methyl-β-d-fructofuranoside | 10 | 0.0029 | 0.4290 |
| 31 | MOL169 | poriferast-5-ene-3β,4β-diol | 9 | 0.0033 | 0.4349 |
| 32 | MOL045 | marmin | 9 | 0.0032 | 0.4305 |
| 33 | MOL129 | 23,24-dihydrocucurbitacin D | 9 | 0.0031 | 0.4334 |
| 34 | MOL138 | α-spinasterol glucoside | 9 | 0.0029 | 0.4319 |
| 35 | MOL181 | α-spinasterol-3-O-β-d-glucoside | 9 | 0.0029 | 0.4319 |
| 36 | MOL021 | tigogenin | 9 | 0.0029 | 0.4290 |
| 37 | MOL005 | gitogenin | 9 | 0.0028 | 0.4290 |
| 38 | MOL074 | isovanillic acid | 8 | 0.0039 | 0.4485 |
| 39 | MOL095 | eudesobovatol A | 8 | 0.0027 | 0.4290 |
Table 2. DC, BC, and CC Values of the Key Targets for ZXGD in Treatment with CHD.
| no. | gene name | target name | DC | BC | CC |
|---|---|---|---|---|---|
| 1 | PTGS1 | prostaglandin-endoperoxide synthase 1 | 152 | 0.3026 | 0.6076 |
| 2 | PTGS2 | prostaglandin-endoperoxide synthase 2 | 117 | 0.1694 | 0.5130 |
| 3 | NFKB2 | NF-kappa B subunit 2 | 68 | 0.0449 | 0.4220 |
| 4 | ALDH2 | aldehyde dehydrogenase 2 | 66 | 0.0424 | 0.4112 |
| 5 | HCK | hemopoietic cell kinase | 61 | 0.0403 | 0.4165 |
| 6 | TNF | tumor necrosis factor | 59 | 0.0610 | 0.4125 |
| 7 | CEBPB | Ccaat/enhancer-binding protein β | 56 | 0.0183 | 0.3842 |
| 8 | LYZ | lysozyme | 49 | 0.0230 | 0.3948 |
| 9 | PYGL | phosphorylase, glycogen, liver | 48 | 0.0205 | 0.3924 |
| 10 | NFKBIA | NF-kappa B inhibitor α | 43 | 0.0359 | 0.3785 |
| 11 | IL1B | interleukin 1 β | 34 | 0.0173 | 0.3819 |
| 12 | TLR4 | toll-like receptor 4 | 26 | 0.0173 | 0.3625 |
Table 3. DC, BC, and CC Values of the Key Pathways for ZXGD in Treatment with CHD.
| no. | pathway ID | pathway name | targets | DC | BC | CC |
|---|---|---|---|---|---|---|
| 1 | hsa04064 | NF-kappa B signaling pathway | LYN, NFKBIA, CXCL8, PLAU, IL1B, PTGS2, TNF, TLR4, ICAM1, NFKB2 | 10 | 0.0084 | 0.4290 |
| 2 | hsa04668 | TNF signaling pathway | NFKBIA, CEBPB, JUN, IL1B, PTGS2, TNF, MMP9, ICAM1 | 8 | 0.0033 | 0.4276 |
| 3 | hsa04620 | toll-like receptor signaling pathway | NFKBIA, JUN, CXCL8, IL1B, TLR8, TNF, TLR4, TLR2 | 8 | 0.0083 | 0.3604 |
| 4 | hsa04062 | chemokine signaling pathway | LYN, CDC42, NFKBIA, FGR, HCK, CXCL8, CCR5 | 7 | 0.0069 | 0.3422 |
2.6. Molecular Docking between Key Compounds and Targets
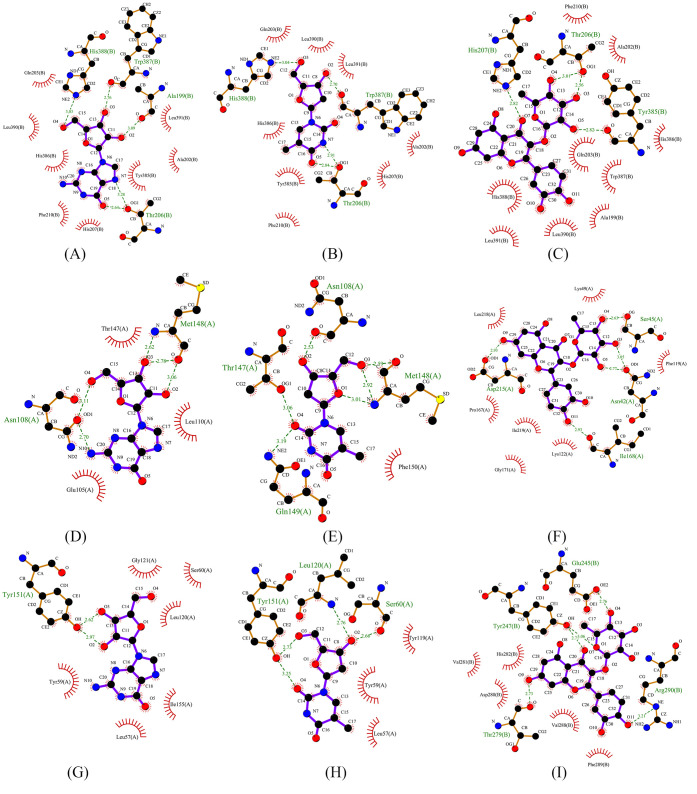
It has been proved that the NF-kappa B signaling pathway is involved in the pathogenesis of CHD and activated to promote the release of inflammatory cytokine, both in animal and clinical experiments.22,23 Considering the significance of the NF-kappa B signaling pathway, the targets enriched in the NF-kappa B signaling pathway and corresponding components were selected to molecular docking to further verify the relative affinity between the compounds and the targets for ZXGD. Five targets enriched in the NF-kappa B signaling pathway and corresponding three compounds were selected for molecular docking, including PTGS2, NFKB2, NFKBIA, TNF, and IL1B and guanosine (MOL007), thymidine (MOL020), and quercitrin (MOL119) (Table 4). As the results showed, the complex between PTGS2 and quercitrin had strong binding abilities, and the binding energy was −8.41 kcal/mol. In addition, the docking results were performed and are shown in Figure 5, demonstrating that the active site of ingredients binds to the targets, which implied the binding mechanism of ZXGD in treatment with CHD.
Table 4. Binding Energy Results between Key Targets and Compounds for ZXGD in Treatment with CHD.
| protein name | gene symbol | PDB ID | ligand name | binding energy (kcal/mol) |
|---|---|---|---|---|
| prostaglandin G/H synthase 2 | PTGS2 | 5F19 | quercitrin | –8.41 |
| prostaglandin G/H synthase 2 | PTGS2 | 5F19 | thymidine | –6.56 |
| prostaglandin G/H synthase 2 | PTGS2 | 5F19 | guanosine | –5.42 |
| NF-kappa-B inhibitor α | NFKBIA | 6Y1J | quercitrin | –5.34 |
| interleukin-1 β | IL1B | 3LTQ | thymidine | –4.39 |
| interleukin-1 β | IL1B | 3LTQ | guanosine | –4.21 |
| nuclear factor NF-kappa B subunit 2 | NFKB2 | 3JV6 | quercitrin | –4.06 |
| tumor necrosis factor | TNF | 5MU8 | thymidine | –3.60 |
| tumor necrosis factor | TNF | 5MU8 | guanosine | –3.33 |
Figure 5.
Interaction between key targets and compounds for ZXGD in treatment with CHD. (A) PTGS2 and guanosine; (B) PTGS2 and thymidine; (C) PTGS2 and quercitrin; (D) IL1B and guanosine; (E) IL1B and thymidine; (F) NFKBIA and quercitrin; (G) TNF and guanosine; (H) TNF and thymidine; and (I) NFKB2 and quercitrin.
2.7. Experimental Validation
2.7.1. Effects of ZXGD on CK, LDH, and MDA Contents and SOD Activity by ELISA
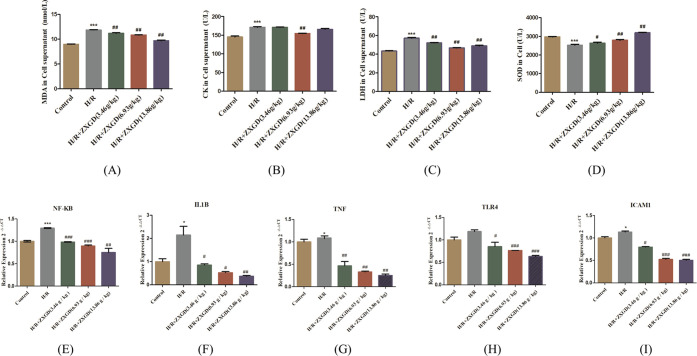
According to the results in vitro experiments, the contents of CK, LDH, and MDA in the model group were 171.73 ± 2.627 U/L, 57.14 ± 1.43 U/L, and 11.87 ± 0.108 nmol/L, respectively, and the contents of these indexes were 145.39 ± 3.764 U/L, 43.32 ± 0.621 U/L, and 8.97 ± 0.073 nmol/L in the control group, respectively. The levels of these indexes in the model group were increased significantly compared with the control group (P < 0.001) (Figure 6A–C). Compared with the model group, the contents of LDH and MDA in all ZXGD-containing serum groups were significantly decreased (P < 0.01), and the contents of CK in the medium dose ZXGD group [6.93 g/(kg·d)] were significantly decreased (P < 0.01). Additionally, the activity of SOD has significantly decreased in the model group [2530.63 ± 74.84 (U/L)], compared with the control group [2975.85 ± 19.375 (U/L)], and the index in the different doses of ZXGD-containing serum groups was significantly increased compared with the model group. All these results showed that establishment of the hypoxia/reoxygenation model (H/R) was successful, and ZXGD-containing serum could alleviate cell injury in H9c2 cells.
Figure 6.
Experimental validation results for ZXGD. (A–D) Contents of CK, LDH, and MDA and SOD activity in the control group, model group, and ZXGD-containing serum groups (3.46, 6.93, and 13.86 g/kg) detected by ELISA; (E–I) mRNA expression of NFKB, IL1B, TNF, TLR4, and ICAM1 in the control group, model group, and ZXGD-containing serum groups (3.46, 6.93, and 13.86 g/kg) detected by qRT-PCR. H/R vs control, *P < 0.05, **P < 0.01, and ***P < 0.001; ZXGD vs H/R, #P < 0.05, ##P < 0.01, and ###P < 0.001.
2.7.2. Verification of Some Key Targets Enriched in the NF-Kappa B Signaling Pathway for ZXGD in Treatment with CHD
According to the results of network pharmacology and molecular docking, the NF-kappa B signaling pathway may play an important role in treatment with CHD for ZXGD. The relative expression of some key targets enriched in the NF-kappa B signaling pathway was detected by quantitative real-time PCR (qRT-PCR) analysis to evaluate the effects of ZXGD, including NFKB, IL1B, TNF, TLR4, and ICAM1. The relative expression of NFKB in the model group was 1.29 ± 0.01; it is increased significantly (P < 0.001) compared with the control group (1.00 ± 0.03), and the levels were significantly decreased in low (P < 0.001), medium (P < 0.001), and high (P < 0.01) ZXGD-containing serum groups. Meanwhile, the consistent results were also appeared on IL1B, TNF, TLR4, and ICAM1 (P < 0.01). All dose groups showed various degrees of decrease in expression. The results suggested that ZXGD can downregulate the expression of NFKB, IL1B, TNF, TLR4, and ICAM1 in vitro.
3. Discussion
TCM plays a significant role in treating CHD,24 with great curative effects on different types of CHD. The mechanism is still not clear due to its characteristics of multi-components, multi-targets, and multi-pathways. In this study, an integrated approach, including UPLC-UESI-Q Exactive Focus, gene expression profiling, and network pharmacology, was introduced to systematically investigate the mechanism of ZXGD in the treatment of CHD. As a result, 39 hub compounds, 12 key targets, and 4 core pathways were closely related to ZXGD in treating CHD.
Obviously, CHD is mainly caused by coronary atherosclerosis that narrows the vascular lumen and eventually leads to myocardial ischemia, hypoxia, and coronary artery spasm, related to inflammation, lipid metabolism alterations, and endothelial injury. The significance of core targets, compounds, and pathways in this study was closely related to the above mechanisms and phenomena.
Among these compounds, guanosine (MOL007) was one of the most important compounds with the highest degree value (DC = 20, BC = 0.0339, and CC = 0.4614). It has been proved that guanosine increased during myocardial ischemia versus extracorporeal circulation to remarkably inhibit platelet aggregation induced by ADP and reduce the risk of thrombosis.25,26 Notably, research studies on a light-induced real-time thrombosis model have concluded that the inhibitory mechanism of guanosine may be triggered by exposing a highly thrombogenic surface to the flowing blood (badimon chamber) or by oxidative stress damage in the mesenteric vessels.25 Experiments demonstrated that guanosine could prevent oxygen/glucose deprivation-induced inflammatory responses by inhibiting nuclear NF-kappa B active subunit into the nucleus.27 Moreover, according to some studies, guanosine may prevent thrombosis and vascular inflammatory injury by reducing the P-selectin surface exposure, and it was a biomarker that favors the progression of atherosclerotic plaque phenotype.25,28
The significance of adenosine (DC = 19, BC = 0.0187, and CC = 0.4631), as one of the main compounds of T. fructus, was mentioned, and adenosine has strong impacts on the cardiovascular system25,29 by attenuating the severity of ischemia and limiting the infarct size due to its coronary vasodilatory action.30,31 It has been proved that exogenous adenosine improves vascular healing via significantly reducing the process of endothelial cell proliferation in the rabbit carotid artery anastomosis model.32 Furthermore, adenosine engages the members of the G-protein-coupled adenosine receptor family to mainly mediate the beneficial adaptive and acute responses within all constituent cells of the heart and vasculature by regulating the heart rate and conduction, coronary vascular tone, cardiac and vascular growth, inflammatory–vascular cell interactions, and cellular stress resistance.33 It was noted that extracellular guanosine also displayed the disposition of extracellular adenosine in rats (in preglomerular vascular endothelial cells and cardiac fibroblasts) and in humans (aortic and coronary artery vascular smooth muscle cells and coronary artery endothelial cells).34 In other words, guanosine and adenosine may promote each other to enhance their roles in ZXGD in treating CHD.
Flavonoids were one of the important compounds in various drugs for the treatment and prophylaxis of cardiovascular disorders. It was proved that, as the significant flavonoid, quercitrin (DC = 10, BC = 0.0039, and CC = 0.4501) contributes to protect against atherosclerosis.35 In the ApoE knockout mice model, a high-fat diet and ingested quercetin for 24 weeks were given, and quercitrin significantly reduced the area of atherosclerotic plaque, alleviated the systemic oxidative stress, and suppressed the aortic p47phox and p67phox expressions.36 Similarly, it was shown that quercetin inhibited oxidant-induced endothelial dysfunction to protect ApoE(-/-) mice against atherosclerosis by largely increasing endothelial nitric oxide synthase activity and heme oxygenase-1 (HO-1) protein expression.36,37 Additionally, a clinical study including 85 patients with CHD has illustrated that quercetin has anti-inflammatory effects38 related to decreasing the transcriptional activities of NFKB.39
As for targets, PTGS2 (DC = 117, BC = 0.1694, and CC = 0.5130) was one of the crucial targets with great topological properties39,40 and was the key target of nonsteroidal anti-inflammatory drugs, including aspirin and ibuprofen. Meanwhile, PTGS2 reduces fatal thrombotic events by inhibiting the platelet activation and aggregation.41 A study containing 66 CHD individuals (including stable angina and unstable angina) has proved that the COX-2 expression of peripheral blood monocytes increased in CHD patients. In particular, the levels of COX-2 protein expression were positively related to the monocyte-platelet aggregate formation rates, and enhanced COX-2 expression was independently associated with CHD risk. This association suggested that COX2 may be the core target of inflammatory regulation in CHD and could result in the development of atherosclerosis.41
The nuclear factor kappa-beta (NFKB) is a transcription factor comprising homo- and/or heterodimers formed from distinct proteins, including REL (cRel), Rel A (p65), RELB, NFKB1 (p50 and its precursor p105), and NFKB 2 (p52 and its precursor p100).42 Notably, NFKB is also the endpoint of a series of signal transduction events and is involved in many biological processes such as inflammation and immunity. Adequate research studies have described the connection of NFKB-related receptor and CVD43,44 because it regulated many of the proinflammatory genes related to atherosclerosis by inhibiting its role in vascular inflammation, proliferation of vascular smooth muscle cells, or foam cell formation.45 NFKB mediates the expression of a variety of proteins to induce the leukocyte adhesion to the vascular wall and infiltration into the subintima that strongly links NFKB with the occurrence of atherosclerosis.46 NFKB2 (DC = 68, BC = 0.0449, and CC = 0.4220) and NFKBIA (DC = 43, BC = 0.0359, and CC = 0.3785) were two crucial proteins that control the primary mechanism of NFKB activation.47 Previous research studies were conducted to analyze the expression of apoptotic genes such as NFKB2 in coronary plaques collected by directional coronary atherectomy, and an overlap between NFKB2 and vascular smooth muscle cells was found.47 Further gene expression analysis exhibited that NFKB2 significantly enhanced expression in acute coronary syndromes without ST elevation (ACS) plaques. What is more, research comprising 402 individuals reported that the expression of NFKBIA was depressed in the blood of patients with CVD compared with controls, thus indicating that NFKBIA may represent novel markers of coronary artery disease susceptibility.48
As the enrichment analysis showed in the bubble plot, perhaps, the NF-kappa B signaling pathway (DC = 10, BC = 0.0084, and CC = 0.4290) is the most significant pathway of ZXGD in the development of CHD and comprises various core targets, such as PTGS2, TNF, IL1B, NFKB2, NFKBIA, and TLR4. The NF-kappa B pathway was involved in many biological processes, including cell immunity, inflammation, and apoptosis, and it played a basic and core role in metabolic inflammation.49 The NF-kappa B signaling pathway occurs by NFKB activation through the classical pathway and noncanonical pathway. In these genes of pathway enrichment analysis, NFKB2 NFKBIA, TNF, and IL1B represented some core targets for the noncanonical pathway. On the other hand, NFKB2 is the key negative regulator that prevents the nuclear translocation of RELB in resting cells.46 Simultaneously, TNF (DC = 59, BC = 0.0610, and CC = 0.4125) is a major inflammatory cytokine involved in activating the NF-kappa B signaling pathway to induce the production of more inflammation mediators and reactive oxygen species.50,51 At the same time, TLR4 involved in the classical pathway depended on IKK-related kinases through phosphorylates NFKBIA at positions S32 and S36.50,51 Based on some research studies, atherosclerotic plaques and neointima were effectively attenuated by regulating the circulatory lipid profile and inhibiting the macrophage ox-LDL uptake via suppressing the LOX-1-NF-kappa B signaling pathway in APOE mice induced by chronic feeding with high lipid diet.52 Similarly, when examining the expression of NFKB-related genes between ruptured and paired stable control segments of endarterectomy plaque specimens, ruptured plaques contained significantly higher gene expression levels of major signaling proteins of the noncanonical pathway.53 These results also provided a link with TNF family in the noncanonical pathway, which further illustrates the association between noncanonical NF-kappa B signaling and adverse cardiovascular events. Interestingly, quercetin (DC = 10, BC = 0.0039, and CC = 0.4501) had anti-inflammatory properties in patients suffering from CHD, indicating a decrease in the transcriptional activity of the nuclear factor of NFKB.38 In other words, quercetin, as a key component of ZXGD, might regulate the expression of NFKB-related receptors through the NF-kappa B signaling pathway for treating CHD.
As mentioned above, the results of bioinformatic analysis illustrated that the NF-kappa B signaling pathway was the most important key pathway. In order to further verify that the NF-kappa B signaling pathway plays an important role in ZXGD in treating CHD, H/R-induced cardiomyocytes cultured in vitro were used to simulate myocardial H/R injury to analyze the CK, LDH, and MDA content, SOD activity, and expression of mRNA of cardiomyocytes cultured in vitro. The protective effects of ZXGD on damaged cardiomyocytes via the upregulation of SOD and downregulation expression of CK, LDH, and MDA were also observed. Inflammatory response is one of the important mechanisms in the occurrence of myocardial ischemia-reperfusion injury. NFKB, IL1B, TNF, TLR4, and ICAM1 as inflammatory markers play important roles in inflammatory signaling by aiding the activation of NFKB. Moreover, almost all studies on ischemia-associated heart injury demonstrated that the inhibition of NF-kappa B activity could attenuate inflammation-associated injury and improve cardiac function.54 In this present study, a model of cardiomyocyte hypoxia/reoxygenation pathology was first constructed. qRT-PCR detection analysis concluded that NFKB, IL1B, TNF, TLR4, and ICAM1 were significantly upregulated in the model group, which further suggested that the cardiomyocyte injury model was highly activated. Drug-containing serum intervention could effectively reduce the expression and secretion of pro-inflammatory factors, including NFKB, IL1B, TNF, TLR4, and ICAM1, notably, the high-dose group had the most significant effects. Different dose groups showed various degrees of decrease in expression, which suggested that ZXGD inhibited the production of inflammatory factors from the aspect of the signaling pathway mechanism.
4. Conclusions
To sum up, an integrated approach, including UPLC-UESI-Q Exactive Focus, gene expression profiling, network pharmacology, molecular docking, and in vitro experimental validation, was introduced to systematically investigate the mechanism of ZXGD in the treatment of CHD. It was found that 39 compounds, 12 targets, and 4 KEGG pathways were closely related to the mechanisms of ZXGD in treatment with CHD. Meanwhile, the significance of key compounds such as quercetin, quercitrin, adenosine, and guanosine in this study has been demonstrated. Furthermore, the NF-kappa B signaling pathway, toll-like receptor signaling pathway, and TNF signaling pathway played a key role by regulating PTGS2, NFKB1A, NFKB2 TNF, and ICAM1. Most importantly, in vitro experiments proved that the ZXGD could improve CHD by downregulating the expression of NFKB, IL1B, TNF, TLR4, and ICAM1 to regulate the NF-kappa B signaling pathway. More experimental verification and research still need to be carried out for ZXGD in treatment with CHD. The results of this work may provide a theoretical basis for further research on the molecular mechanism of ZXGD in the treatment of CHD.
5. Methods and Materials
5.1. Chemical Composition Analysis for ZXGD
5.1.1. Chemicals and Materials
Acetonitrile (HPLC-grade) and methyl alcohol (HPLC-grade) were obtained from Fisher Scientific Company. The MS-grade formic acid was obtained from Sigma-Aldrich (Germany) Trading Co., Ltd. A. fructus immaturus, A. macrostemonis bulbus, T. fructus, M. officinalis cortex, and C. ramulus were all decoction pieces and obtained from Beijing Tongrentang.
5.1.2. UPLC-UESI-Q Exactive Focus Conditions
The UPLC-UESI-Q Exactive Focus conditions were used to analyze the compounds of ZXGD. An instrument, a Thermo Scientific Q exactive focus system coupled with Thermo Scientific Ultimate 3000 system, was used for the chromatographic separation. Separation was performed on an Accucore AQ C18 column (150 mm × 2.1 mm, 2.6 μm), and the column temperature was maintained at 30 °C. The mobile phase consisted of two solvents, namely A (0.1% formic acid in water) and B (methyl alcohol) at a flow rate of 0.3 mL/min. The inlet method used a linear gradient elution of A and B, according to the following program (0–13 min, 95–40% A; 13–27 min, 40–5% A; 27–30 min, 5% A; 30–33 min, 5–95% A; and 33–35 min, 95% A). The injection volume was set at 2 μL. Liquid chromatography HESI-MS analysis was conducted using the positive and negative electrospray modes. The mass spectral scan ranges from 100 to 1500 Da, and the spray voltage was set at 3.5 kV. The other conditions are as follows: capillary temperature, 320 °C; Aux gas heater temperature, 350 °C; sheath gas flow rate, 40 arb; Aux gas flow rate, 10arb; S-lens RF level, 50.0 V; resolution, full MS 70000, dd-MS2 35000; and scanning mode, full MS-dd-MS2.
5.1.3. Preparation of Test Solution
12 g of A. fructus immaturus, 12 g of M. officinalis cortex, 9 g of A. macrostemonis bulbus, 6 g of C. ramulus, and 12 g of T. fructus are weighed out, and all herbs are decoction pieces. Water was added to soak for 20 min and decocted twice, the filtrate was filtered and combined twice, and they were cooled. A proper amount of the above decoction solution was taken, and the solution was fixed with methyl alcohol to a certain solubility. The test product was obtained by filtration through a 0.22 μm microporous membrane.
5.2. Target Prediction for ZXGD in the Treatment of CHD by Network Pharmacology
5.2.1. Target Fishing of the Chemical Compositions for ZXGD
To predict the targets of compounds, the target fishing was performed in terms of MedChem Studio (version 3.0) software that is embed in the TCMIP,55 and it is an efficient similarity tool to identify the similarity between the known drugs and the test compounds. The information of the known drugs and the related targets is originated from DrugBank database.56 The Tanimoto score was used to characterize the degree of similarity between the known drugs and the test compounds in MedChem Studio, and the range of Tanimoto score is in the [0, 1], where “0” indicates the completely different structures between ingredients and known drugs, and “1” represents the same structures of the two components. When the Tanimoto score is higher than 0.6 between the test compounds and the known drugs, it is considered that the targets of the known drugs are that of the test compounds.
5.2.2. Determination of CHD Targets by Gene Expression Profiling
The gene expression profile was carried out by GEO database (www.ncbi.nlm.nih.gov/geo/)57 to identify the related targets of CHD. GEO database is an international public repository for high-throughput microarray and next-generation sequence functional genomic data sets that are submitted by the research community and contain a large number of gene chip sequencing, single cell sequencing, and omics data of clinical, animal, or cell samples. First, chip GSE66360 was selected from GEO database through searching the keywords “coronary heart disease.” The chip contained gene expression data of the circulating endothelial cells in patients with 50 CHD and 49 healthy people and was measured using the HG-133U_PLUS_2 microarray. Second, the whole probe sets of circulating endothelial cells were annotated as gene symbols based on the GPL570 platform. Afterward, differential expressed genes (DEGs) were identified by comparing gene expression between the CHD group and control group with a tool named GEO2R in GEO database. Furthermore, the genes with P value < 0.05 and |log FC| > 1 were considered as the DEGs between CHD patients and healthy individuals. In particular, genes with log FC > 1 were regarded as the upregulated genes, and genes with log FC < −1 were downregulated genes. Consequently, the DEGs of CHD were screened out and diagrammed as a volcano plot with R language.
5.2.3. Common Targets of Component Targets and CHD Targets
The common targets of disease-causing genes and compound targets for ZXGD were determined by the Venn online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), and non-human targets were deleted in this step. As a result, the targets of the candidate compounds were obtained for ZXGD in treating CHD.
5.2.4. Core Targets were Determined by PPI Network Construction for ZXGD in Treating CHD
To screen out the core targets for ZXGD in treating CHD, the common targets were mapped into the online search websites (STRING v11.0, https://string-db.org/),58 which could predict the protein functional associations and PPIs. The protein type was set at “Homo sapiens,” and the core targets would be determined by setting the threshold with the confidence of 0.7. Then, the interactions were introduced to Cytoscape 3.7.2 to construct the PPI network.59
5.2.5. KEGG Analysis on ZXGD in Treating CHD
KEGG pathway enrichment analyses were performed in online enrichment database DAVID (https://DAVID.ncifcrf.gov/tools.jsp)60 to investigate the functions and mechanisms of targets for ZXGD in treating CHD. The KEGG pathways with P value < 0.01 were regarded as the key pathways of ZXGD in treating CHD. Additionally, core pathways were mapped as bubble diagram with R language.
5.2.6. F–H–C–T–P Network Construction of ZXGD in Treating CHD
The data of herbs, compounds, targets, and pathways were leading in the software of Cytoscape 3.7.2 to construct the “F–H–C–T–P” network for ZXGD in the treatment of CHD, and network topological properties were analyzed for each node using the Network analyzer with DC, CC, and BC to identify the hub network of ZXGD in treating CHD.
5.2.7. Molecular Docking of Key Compounds and Targets
Molecular docking was used to predict the directive interaction between core targets and compounds based on the AutoDock Vina.61 The crystal structures of targets were obtained from Protein Data Bank (PDB, http://www.rcsb.org), and the structures of components were obtained from PubChem database. The whole processes comprised the preparation of proteins, determination of docking pockets, and molecular docking with proteins and compounds. Finally, binding energy was calculated using an iterated local search global optimizer. Active interaction site and binding energy score were revealed when the molecule and the protein docked successfully. The smaller the binding energy, the stronger the binding ability between compounds and targets.
5.3. Experimental Validation
5.3.1. Animals and Treatments
12 SD rats (aged eight weeks, weighed 120–160 g, SPF) were provided by FOREVERGEN company; the certificate number was 44822700003542. The rats were housed in the institutional animal facility with standard animal room conditions at 22 °C under a 12 h light/dark cycle with free access to food and water. All animals were randomly divided into the control group (n = 3) and ZXGD group with low, medium, and high concentrations (n = 9).
5.3.2. Herb Materials
A. fructus immaturus, A. macrostemonis bulbus, T., M. officinalis cortex, and C. ramulus of ZXGD were decoction pieces and obtained from Beijing Tongrentang.
5.3.3. Preparation of ZXGD
5.3.4. Preparation of Drug-Containing Serum
12 adult SD rats were randomly divided into four groups. After 2 days of adaptive feeding, ZXGD drug-containing serum of high, medium, and low-dose group and control group was prepared separately. Rats of high-dose group (n = 3), medium-dose group (n = 3), and low-dose group (n = 3) were given ZXGD via intragastric administration with the doses of 13.86 g/(kg·d), 6.93 g/(kg·d), and 3.46 g/(kg·d) for 3 days. For the control group, the rats (n = 3) (1 mL/100 g) were given the saline by intragastric administration once per day for 3 days. After 1 h of the last treatment, abdominal aortic blood was collected under aseptic condition and then centrifuged at 2000 rpm for 15 min to obtain ZXGD drug-containing serum. All serum was filtered through a 0.22 μm filter membrane and inactivated by water bath at 56 °C for 30 min and then stored at −20 °C until all serum could be used for the pharmacological study.
5.3.5. Culture, Passage, and Grouping of H9c2 Cells
H9c2 (ATCC) was grown in the Dulbecco’s modified Eagle’s medium (HyClone, UT, USA) with 10% FBS (Gibco, NY, USA). All the medium were supplemented with 100 units of penicillin per mL and 100 μg of streptomycin per mL (TBD, Tianjin, China).
5.3.6. Establishment of the Hypoxia/Reoxygenation Model (H/R) of H9c2 and Intervention of Drug-Containing Serum
H9c2 cells were divided into five groups, shown as follows: (1) control group: H9c2 cells were kept in a normal incubator; (2) the hypoxia/reoxygenation (H/R) group: H9c2 cells were subjected to 4 h of hypoxia (N2/CO2, 95:5), followed by 2 h of reoxygenation; (3) the H/R + high-dose ZXGD: H9c2 cells were treated with the ZXGD high-dose group containing drug serum and reoxygenated for 2 h; (4) the H/R + medium-dose ZXGD: H9c2 cells subjected to ZXGD medium-dose group containing drug serum and reoxygenated for 2 h; and (5) the H/R + low-dose ZXGD: the treatment of H9c2 cells was the same as mentioned above.
5.3.7. Enzyme-Linked Immunosorbent Assay (ELISA) to Detect the Levels of CK, LDH, and MDA Contents and SOD Activity
The levels of CK, LDH, and MDA contents and SOD activity were determined using commercial ELISA kits (Meimian Industrial Co., Ltd., Jiangsu, China), according to the manufacturer’s instructions. Briefly, 50 μL of the standard and sample was added per well, incubating at 37 °C for 2 h. Next, 100 μL of the HRP-conjugate antibody was added to each well, incubating for 60 min at 37 °C. Before adding 50 μL of stop solution to each well, chromogen solution A (50 μL) and B (50 μL) were added to each well with mixing and incubating for 15 min at 37 °C, respectively. Finally, the absorbance values were measured using a microplate reader (Thermo Fisher, USA) at 450 nm. All experiments were performed independently at least three times.
5.3.8. qRT-PCR to Detect mRNA of NFKB, IL1B, TNF, TLR4, and ICAM1 Expression
The primers were designed and synthesized by Tanzhen Biotechnology. The primer sequences for NFKB, IL1B, TNF, TLR4, and ICAM1 are listed in Table 5. The total RNA for H9c2 cells was extracted by Trizol (BIOTEKE, Beijing, China) and was later employed to determine the RNA concentration. According to the Prime-Script RT reagent kit instruction, the RNA was then reversely transcribed into cDNA. Based on the instruction of the kit, the polymerase chain reaction was initiated at 85 °C for 1 min, followed by 40 cycles of amplification of denaturation at 94 °C for 1 min and annealing at 60 °C for 20 s using a StepOne Plus device (Applied Biosystems). The 2-DDCT method was adopted to analyze the data.
Table 5. Primer Sequences for NFKB, IL1B, TNF, TLR4, and ICAM1 by qRT-PCR.
| gene name | forward (5′–3′) | reverse (5′–3′) |
|---|---|---|
| NFKB | TTTTTGATAACCGTGCCCCC | AGCCAGGTCCCGTGAAATAC |
| IL1B | CAGAACATAAGCCAACAAGTGGT | GCCGTCTTTCATCACACAGG |
| TNF | CTGAACTTCGGGGTGATCGG | GTTTGCTACGACGTGGGCTA |
| TLR4 | CCAGAGCCGTTGGTGTATCT | GAGCATTGTCCTCCCACTCG |
| ICAM1 | CCCACCTCACAGGGTACTT | CAGGTGAGGACCATATAGCACA |
5.3.9. Statistical Analysis
Statistical analysis was performed using SPSS 17.0 software. The measurement data were expressed as mean – standard deviation. The comparison among three or more groups was performed by one-way ANOVA. H/R versus control, *P < 0.05, **P < 0.01, and ***P < 0.001; ZXGD versus H/R, #P < 0.05, ##P < 0.01, and ###P < 0.001. P < 0.05 indicated the statistical difference. P < 0.01 and P < 0.001 indicated the significant difference. The drawing of histogram was performed using Graphad prism 5 software.
Acknowledgments
This research has been financially supported by Innovation Capability Support Program of Shaanxi-Youth Science and Technology Star Plan (Program No. 2021KJXX-55), Shaanxi Administration of Traditional Chinese Medicine Projects (Program No. 2021-ZZ-JC022) and Xi’an Science, Technology Bureau Projects (Program No. 21YXYJ0097), and the Key Research and Development Program of Shaanxi (Program No. S2019-YF-ZDCXL-ZDLSF-0079). Additionally, the corresponding author is thankful to all coauthors who supported in accomplishing this research project.
Glossary
Abbreviations
- ZXGD
Zhishi Xiebai Guizhi decoction
- CAV
cardiovascular diseases
- CHD
coronary heart disease
- TCM
traditional Chinese medicine
- DEGs
differentially expressed genes
- GEO
gene expression omnibus
- PPI
protein–protein interaction
- KEGG
kyoto encyclopedia of genes and genomes
- DC
degree centrality
- BC
betweenness centrality
- CC
closeness centrality
- PTGS2
prostaglandin-endoperoxide synthase 2
- NFKB2
nuclear factor kappa b subunit 2
- NFKBIA
nf-kappa-b inhibitor alpha
- IL1B
interleukin 1 beta
- TLR4
toll-like receptor 4
- ICAM1
intercellular cell adhesion molecule
- TNF
tumor necrosis factor
- CK
creatine kinase
- MDA
malondialdehyde
- qRT-PCR
quantitative real-time polymerase chain reaction
- LDH
lactic dehydrogenase
- SOD
superoxide dismutase
- CCK-8
cell counting kit-8
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c04491.
Detailed information of chemical compounds of herb materials by UPLC-UESI-Q Exactive Focus (PDF)
Author Contributions
∥ Y.L., X.H., and Z.D. have contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Townsend N.; Wilson L.; Bhatnagar P.; Wickramasinghe K.; Rayner M.; Nichols M. Cardiovascular disease in Europe: epidemiological update 2016. Eur. Heart J. 2016, 37, 3232–3245. 10.1093/eurheartj/ehw334. [DOI] [PubMed] [Google Scholar]
- Roffi M.; Patrono C.; Collet J. P.; Mueller C.; Valgimigli M.; Andreotti F.; Bax J. J.; Borger M. A.; Brotons C.; Chew D. P.; Gencer B.; Hasenfuss G.; Kjeldsen K.; Lancellotti P.; Landmesser U.; Mehilli J.; Mukherjee D.; Storey RF.; Windecker S.; 2015 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: Task Force for the Management of Acute Coronary Syndromes in Patients Presenting without Persistent ST-Segment Elevation of the European Society of Cardiology (ESC). G. Ital. Cardiol. 2016, 17, 831–872. [DOI] [PubMed] [Google Scholar]
- Stark K.; Massberg S. Interplay between inflammation and thrombosis in cardiovascular pathology. Nat. Rev. Cardiol. 2021, 18, 666–682. 10.1038/s41569-021-00552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S.; Rashid I. Regression therapy for cardiovascular disease. Eur. Heart J. 2019, 40, 3418–3420. 10.1093/eurheartj/ehz481. [DOI] [PubMed] [Google Scholar]
- Kumbhani D. J.; Cannon C. P.; Beavers C. J.; Bhatt D. L.; Cuker A.; Gluckman T. J.; Marine J. E.; Mehran R.; Messe S. R.; Patel N. S.; Peterson B. E.; Rosenfield K.; Spinler S. A.; Thourani V. H. 2020 ACC Expert Consensus Decision Pathway for Anticoagulant and Antiplatelet Therapy in Patients With Atrial Fibrillation or Venous Thromboembolism Undergoing Percutaneous Coronary Intervention or With Atherosclerotic Cardiovascular Disease: A Report of the American College of Cardiology Solution Set Oversight Committee. J. Am. Coll. Cardiol. 2021, 77, 629–658. 10.1016/j.jacc.2020.09.011. [DOI] [PubMed] [Google Scholar]
- Capodanno D.; Alfonso F.; Levine G. N.; Valgimigli M.; Angiolillo D. J. ACC/AHA Versus ESC Guidelines on Dual Antiplatelet Therapy: JACC Guideline Comparison. J. Am. Coll. Cardiol. 2018, 72, 2915–2931. 10.1016/j.jacc.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Schurgers L. J.; Aebert H.; Vermeer C.; Bültmann B.; Janzen J. Oral anticoagulant treatment: friend or foe in cardiovascular disease?. Blood 2004, 104, 3231–3232. 10.1182/blood-2004-04-1277. [DOI] [PubMed] [Google Scholar]
- Kraakman M. J.; Dragoljevic D.; Kammoun H. L.; Murphy A. J. Is the risk of cardiovascular disease altered with anti-inflammatory therapies? Insights from rheumatoid arthritis. Clin. Transl. Immunol. 2016, 5, e84 10.1038/cti.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X.; Liu Y.; Zhao Z.; Zhao L.; Wang D.; Liu Q. The role of traditional Chinese medicine in the treatment of atherosclerosis through the regulation of macrophage activity. Biomed. Pharmacother. 2019, 118, 109375. 10.1016/j.biopha.2019.109375. [DOI] [PubMed] [Google Scholar]
- Liu X.; Zhou J.; Zhang T.; Chen K.; Xu M.; Wu L.; Liu J.; Huang Y.; Nie B.; Shen X.; Ren P.; Huang X. Meranzin hydrate elicits antidepressant effects and restores reward circuitry. Behav. Brain Res. 2021, 398, 112898. 10.1016/j.bbr.2020.112898. [DOI] [PubMed] [Google Scholar]
- Yu X.; Tang L.; Wu H.; Zhang X.; Luo H.; Guo R.; Xu M.; Yang H.; Fan J.; Wang Z.; Su R. Trichosanthis Fructus: botany, traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 2018, 224, 177–194. 10.1016/j.jep.2018.05.034. [DOI] [PubMed] [Google Scholar]
- Wang S.; Sun L.; Gu L.; Zhang Y.; Zhao S.; Zhao L.-s.; Bi K.-s.; Chen X. The comparative pharmacokinetics of four bioactive ingredients after administration of Ramulus Cinnamomi-Radix Glycyrrhizae herb pair extract, Ramulus Cinnamomi extract and Radix Glycyrrhizae extract. Biomed. Chromatogr. 2016, 30, 1270–1277. 10.1002/bmc.3677. [DOI] [PubMed] [Google Scholar]
- Ho J.; Hong C.-Y. Cardiovascular protection of magnolol: cell-type specificity and dose-related effects. J. Biomed. Sci. 2012, 19, 70. 10.1186/1423-0127-19-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.-L.; Zhu L.; Tang Y.-N.; Kwan H.-Y.; Zhao Z.-Z.; Chen H.-B.; Yi T. Comparative evaluation of chemical profiles of three representative ’snow lotus’ herbs by UPLC-DAD-QTOF-MS combined with principal component and hierarchical cluster analyses. Comp. Study 2017, 9, 1105–1115. 10.1002/dta.2123. [DOI] [PubMed] [Google Scholar]
- Yi T.; Tang Y.; Zhang J.; Zhao Z.; Yang Z.; Chen H. Characterization and determination of six flavonoids in the ethnomedicine “Dragon’s Blood” by UPLC-PAD-MS. Chem. Cent. J. 2012, 6, 116. 10.1186/1752-153x-6-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.; Liang Z.-T.; Yi T.; Ma Y.; Zhao Z.-Z.; Guo B.-L.; Zhang J.-Y.; Chen H.-B. Comparison of chemical profiles between the root and aerial parts from three Bupleurum species based on a UHPLC-QTOF-MS metabolomics approach. Comp. Study 2017, 17, 305. 10.1186/s12906-017-1816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. 10.1016/s1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- Liu X. J.; Jiang M. F.; Yu T.; Wang D. D.; Zhu G. Q.; Gao Q. Based on LC-MS and network pharmacology to explore the study of ginkgo ketone esters in the treatment of new coronavirus pneumonia with cardiovascular disease. Chin. Tradit. Pat. Med. 2020, 42, 2481–2488. [Google Scholar]
- Duan L.; Guo L.; Liu K.; Liu E.-H.; Li P. Characterization and classification of seven citrus herbs by liquid chromatography-quadrupole time-of-flight mass spectrometry and genetic algorithm optimized support vector machines. J. Chromatogr. A. 2014, 1339, 118–127. 10.1016/j.chroma.2014.02.091. [DOI] [PubMed] [Google Scholar]
- Yao Z.-H.; Qin Z.-F.; Dai Y.; Yao X.-S. Phytochemistry and pharmacology of Allii Macrostemonis Bulbus, a traditional Chinese medicine. Chin. J. Nat. Med. 2016, 14, 481–498. 10.1016/s1875-5364(16)30058-9. [DOI] [PubMed] [Google Scholar]
- Zhao H.; Zhang M.; Zhou F.; Cao W.; Bi L.; Xie Y.; Yang Q.; Wang S. Cinnamaldehyde ameliorates LPS-induced cardiac dysfunction via TLR4-NOX4 pathway: The regulation of autophagy and ROS production. J. Mol. Cell. Cardiol. 2016, 101, 11–24. 10.1016/j.yjmcc.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Guo F.; Tang C.; Li Y.; Liu Y.; Lv P.; Wang W.; Mu Y. The interplay of LncRNA ANRIL and miR-181b on the inflammation-relevant coronary artery disease through mediating NF-kappaB signalling pathway. J. Cell. Mol. Med. 2018, 22, 5062–5075. 10.1111/jcmm.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coto E.; Reguero J. R.; Avanzas P.; Pascual I.; Martín M.; Hevia S.; Morís C.; Díaz-Molina B.; Lambert J. L.; Alonso B.; Cuesta-LLavona E.; Díaz-Corte C.; Gómez J. Gene variants in the NF-KB pathway (NFKB1, NFKBIA, NFKBIZ) and risk for early-onset coronary artery disease. Immunol. Lett. 2019, 208, 39–43. 10.1016/j.imlet.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Li H.; Sun K.; Zhao R.; Hu J.; Hao Z.; Wang F.; Lu Y.; Liu F.; Zhang Y. Inflammatory biomarkers of coronary heart disease. Front. Biosci. 2017, 22, 504–515. 10.2741/4498. [DOI] [PubMed] [Google Scholar]
- Fuentes F.; Alarcón M.; Badimon L.; Fuentes M.; Klotz K.-N.; Vilahur G.; Kachler S.; Padró T.; Palomo I.; Fuentes E. Guanosine exerts antiplatelet and antithrombotic properties through an adenosine-related cAMP-PKA signaling. Int. J. Cardiol. 2017, 248, 294–300. 10.1016/j.ijcard.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Valen G.; Öwall A.; Takeshima S.; Goiny M.; Ungerstedt U.; Vaage J. Metabolic changes induced by ischemia and cardioplegia: a study employing cardiac microdialysis in pigs. Eur. J. Cardiothorac. Surg. 2004, 25, 69–75. 10.1016/s1010-7940(03)00672-9. [DOI] [PubMed] [Google Scholar]
- Dal-Cim T.; Ludka F. K.; Martins W. C.; Reginato C.; Parada E.; Egea J.; López M. G.; Tasca C. I. Guanosine controls inflammatory pathways to afford neuroprotection of hippocampal slices under oxygen and glucose deprivation conditions. J. Neurochem. 2013, 126, 437–450. 10.1111/jnc.12324. [DOI] [PubMed] [Google Scholar]
- Woollard K. J.; Lumsden N. G.; Andrews K. L.; Aprico A.; Harris E.; Irvine J. C.; Jefferis A.-m.; Fang L.; Kanellakis P.; Bobik A.; Chin-Dusting J. P. F. Raised soluble P-selectin moderately accelerates atherosclerotic plaque progression. PLoS One 2014, 9, e97422 10.1371/journal.pone.0097422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudry M.; Vairo D.; Marlinge M.; Gaubert M.; Guiol C.; Mottola G.; Gariboldi V.; Deharo P.; Sadrin S.; Maixent J. M.; Fenouillet E.; Ruf J.; Guieu R.; Paganelli F. Adenosine and Its Receptors: An Expected Tool for the Diagnosis and Treatment of Coronary Artery and Ischemic Heart Diseases. Int. J. Mol. Sci. 2020, 21, 5321. 10.3390/ijms21155321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. L.; Linden J. Cloned receptors and cardiovascular responses to adenosine. Cardiovasc. Res. 1993, 27, 62–67. 10.1093/cvr/27.1.62. [DOI] [PubMed] [Google Scholar]
- Su A. I.; Wiltshire T.; Batalov S.; Lapp H.; Ching K. A.; Block D.; Zhang J.; Soden R.; Hayakawa M.; Kreiman G.; Cooke M. P.; Walker J. R.; Hogenesch J. B. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 6062–6067. 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albayrak G.; Silistreli E.; Ergur B.; Kalkan S.; Karabay O.; Erdal A. C.; Acikel U. Inhibitory effect of adenosine on intimal hyperplasia and proliferation of smooth muscle cells in a carotid arterial anastomosis animal model. Vascular 2015, 23, 124–131. 10.1177/1708538114533962. [DOI] [PubMed] [Google Scholar]
- Headrick J. P.; Ashton K. J.; Rose’Meyer R. B.; Peart J. N. Cardiovascular adenosine receptors: expression, actions and interactions. Pharmacol. Ther. 2013, 140, 92–111. 10.1016/j.pharmthera.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Jackson E. K.; Cheng D.; Jackson T. C.; Verrier J. D.; Gillespie D. G. Extracellular guanosine regulates extracellular adenosine levels. Am. J. Physiol.: Cell Physiol. 2013, 304, C406–C421. 10.1152/ajpcell.00212.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumbeckaite S.; Bernatoniene J.; Majiene D.; Jakštas V.; Savickas A.; Toleikis A. The effect of flavonoids on rat heart mitochondrial function. Biomed. Pharmacother. 2006, 60, 245–248. 10.1016/j.biopha.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Xiao L.; Liu L.; Guo X.; Zhang S.; Wang J.; Zhou F.; Liu L.; Tang Y.; Yao P. Quercetin attenuates high fat diet-induced atherosclerosis in apolipoprotein E knockout mice: A critical role of NADPH oxidase. Food Chem. Toxicol. 2017, 105, 22–33. 10.1016/j.fct.2017.03.048. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Ward N. C.; Hodgson J. M.; Puddey I. B.; Wang Y.; Zhang D.; Maghzal G. J.; Stocker R.; Croft K. D. Dietary quercetin attenuates oxidant-induced endothelial dysfunction and atherosclerosis in apolipoprotein E knockout mice fed a high-fat diet: a critical role for heme oxygenase-1. Free Radical Biol. Med. 2013, 65, 908–915. 10.1016/j.freeradbiomed.2013.08.185. [DOI] [PubMed] [Google Scholar]
- Chekalina N.; Burmak Y.; Petrov Y.; Borisova Z.; Manusha Y.; Kazakov Y.; Kaidashev I. Quercetin reduces the transcriptional activity of NF-kB in stable coronary artery disease. Indian Heart J. 2018, 70, 593–597. 10.1016/j.ihj.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Feng J.; Zhang J.; Kang X.; Qian D. Quercetin modulates AMPK/SIRT1/NF-kappaB signaling to inhibit inflammatory/oxidative stress responses in diabetic high fat diet-induced atherosclerosis in the rat carotid artery. Exp. Ther. Med. 2020, 20, 280. 10.3892/etm.2020.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman P.; Kaley G. Cardiac and coronary prostaglandin synthesis and function. N. Engl. J. Med. 1978, 298, 1122–1128. 10.1056/nejm197805182982005. [DOI] [PubMed] [Google Scholar]
- Lucido M. J.; Orlando B. J.; Vecchio A. J.; Malkowski M. G. Crystal Structure of Aspirin-Acetylated Human Cyclooxygenase-2: Insight into the Formation of Products with Reversed Stereochemistry. Biochemistry 2016, 55, 1226–1238. 10.1021/acs.biochem.5b01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q.; Fei X.; Li S.; Xu C.; Tu C.; Jiang L.; Wo M. Predicting significance of COX-2 expression of peripheral blood monocyte in patients with coronary artery disease. Ann. Transl. Med. 2019, 7, 483. 10.21037/atm.2019.08.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzini C.; Cominacini L.; Garbin U.; Fratta Pasini A. M. Endoplasmic Reticulum Stress, NRF2 Signalling and Cardiovascular Diseases in a Nutshell. Curr. Atheroscler. Rep. 2017, 19, 33. 10.1007/s11883-017-0669-7. [DOI] [PubMed] [Google Scholar]
- Coto E.; Reguero J. R.; Avanzas P.; Pascual I.; Martín M.; Hevia S.; Morís C.; Díaz-Molina B.; Lambert J. L.; Alonso B.; et al. Gene variants in the NF-KB pathway (NFKB1, NFKBIA, NFKBIZ) and risk for early-onset coronary artery disease. Immunol. Lett. 2019, 208, 39–43. 10.1016/j.imlet.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Yu X.-H.; Zheng X.-L.; Tang C.-K. Nuclear Factor-kappaB Activation as a Pathological Mechanism of Lipid Metabolism and Atherosclerosis. Adv. Clin. Chem. 2015, 70, 1–30. 10.1016/bs.acc.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Mitchell J. P.; Carmody R. J. NF-kappaB and the Transcriptional Control of Inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. 10.1016/bs.ircmb.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Rossi M. L.; Marziliano N.; Merlini P. A.; Bramucci E.; Canosi U.; Belli G.; Parenti D. Z.; Mannucci P. M.; Ardissino D. Different quantitative apoptotic traits in coronary atherosclerotic plaques from patients with stable angina pectoris and acute coronary syndromes. Circulation 2004, 110, 1767–1773. 10.1161/01.cir.0000142865.04816.89. [DOI] [PubMed] [Google Scholar]
- Özbilüm N.; Arslan S.; Berkan Ö.; Yanartaş M.; Aydemir E. I. The role of NF-kappaB1A promoter polymorphisms on coronary artery disease risk. Basic Clin. Pharmacol. Toxicol. 2013, 113, 187–192. 10.1111/bcpt.12085. [DOI] [PubMed] [Google Scholar]
- Moynagh P. N. The NF-κB pathway. J. Cell Sci. 2005, 118, 4589–4592. 10.1242/jcs.02579. [DOI] [PubMed] [Google Scholar]
- Read M. A.; Whitley M. Z.; Williams A. J.; Collins T. NF-kappa B and I kappa B alpha: an inducible regulatory system in endothelial activation. J. Exp. Med. 1994, 179, 503–512. 10.1084/jem.179.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Singhal V.; Roshan R.; Sharma A.; Rembhotkar G. W.; Ghosh B. Piperine inhibits TNF-alpha induced adhesion of neutrophils to endothelial monolayer through suppression of NF-kappaB and IkappaB kinase activation. Eur. J. Pharmacol. 2007, 575, 177–186. 10.1016/j.ejphar.2007.07.056. [DOI] [PubMed] [Google Scholar]
- Cheng X.-L.; Ding F.; Wang D.-P.; Zhou L.; Cao J.-M. Hexarelin attenuates atherosclerosis via inhibiting LOX-1-NF-κB signaling pathway-mediated macrophage ox-LDL uptake in ApoE-/- mice. Peptides 2019, 121, 170122. 10.1016/j.peptides.2019.170122. [DOI] [PubMed] [Google Scholar]
- Maracle C. X.; Agca R.; Helder B.; Meeuwsen J. A. L.; Niessen H. W. M.; Biessen E. A. L.; de Winther M. P. J.; de Jager S. C. A.; Nurmohamed M. T.; Tas S. W. Noncanonical NF-κB signaling in microvessels of atherosclerotic lesions is associated with inflammation, atheromatous plaque morphology and myocardial infarction. Atherosclerosis 2018, 270, 33–41. 10.1016/j.atherosclerosis.2018.01.032. [DOI] [PubMed] [Google Scholar]
- Potz B. A.; Sabe A. A.; Elmadhun N. Y.; Sabe S. A.; Braun B. J. V.; Clements R. T.; Anny U.; Sellke F. W. Calpain inhibition decreases inflammatory protein expression in vessel walls in a model of chronic myocardial ischemia. Surgery 2017, 161, 1394–1404. 10.1016/j.surg.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.-Y.; Zhang Y.-Q.; Liu Z.-M.; Chen T.; Lv C.-Y.; Tang S.-H.; Zhang X.-B.; Zhang W.; Li Z.-Y.; Zhou R.-R.; Yang H.-J.; Wang X.-J.; Huang L.-Q. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E.; Barrett T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D.; Gable A. L.; Lyon D.; Junge A.; Wyder S.; Huerta-Cepas J.; Simonovic M.; Doncheva N. T.; Morris J. H.; Bork P.; Jensen L. J.; Mering C. v. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl M.; Wiese S.; Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Tan Q.; Kir J.; Liu D.; Bryant D.; Guo Y.; Stephens R.; Baseler M. W.; Lane H. C.; Lempicki R. A. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.