Abstract
The COBAS AMPLICOR CT/NG test for Neisseria gonorrhoeae cross-reacts with certain strains of nonpathogenic Neisseria species. In some strains, the target sequence is identical to that of N. gonorrhoeae, whereas other strains have a small number of mismatches within the regions recognized by the primers or probe used in the COBAS AMPLICOR NG test. These cross-reactive strains are occasionally present in urogenital specimens, causing false-positive results in the COBAS AMPLICOR NG test. Analysis of the data generated in a large multicenter clinical trial showed that 2.9% of the specimens gave signals between A660s of 0.2 and 3.5 but that one-half of these equivocal specimens did not contain N. gonorrhoeae. Most of these equivocal specimens were correctly classified as true positive or true negative by retesting in duplicate and defining a PCR-positive result as two of three results with an A660 of ≥2.0. If specimens had been classified as positive or negative based on a single test result using a cutoff of an A660 of 0.2, specificity would have ranged from 96.2 to 98.9% depending on specimen type, sex, and presence of symptoms. By employing the equivocal zone-retesting algorithm, specificity increased to 98.6 to 99.9% with little effect (0.1 to 4.9% decrease) on sensitivity in most specimen types, enabling the test to achieve a positive predictive value of at least 90% in populations with a prevalence of 4% or higher. In lower-prevalence populations, the test could be used to screen for presumptive infections that would have to be confirmed by an independent test.
The COBAS AMPLICOR CT/NG test provides a powerful diagnostic tool for screening for both chlamydial and gonococcal infections. We and others have observed that the NG portion of the test (COBAS AMPLICOR NG) cross-reacts with some isolates of certain nonpathogenic Neisseria species (4). When the original, recommended cutoff (A660 of 0.2) is used, the COBAS AMPLICOR NG test can produce false-positive results for Neisseria gonorrhoeae, presumably due to the presence of cross-reactive Neisseria species in some urogenital specimens. In one population, approximately 26% of COBAS AMPLICOR NG-positive results were false positives, which corresponded to approximately 3% of the total population (4). In contrast, the same laboratory observed fewer than 1% false-positive results among urogenital specimens from a second population (4).
In this paper, we confirm that the test does cross-react with some isolates of Neisseria subflava (4) and Neisseria cinerea and compare the target sequences in these cross-reactive species with that of N. gonorrhoeae. Using data from a multicenter trial (n = 4,173 patients; prevalence = 13.1%) conducted at six sites in the United States (8), we show how test sensitivity and specificity vary with the cutoff value used. We then assess whether both sensitivity and specificity can be optimized by establishing a large equivocal zone and using an algorithm that involves additional testing of specimens yielding equivocal results. Implementation of the equivocal zone-retesting algorithm identified by this analysis produced good specificity (98.8 to 99.9%) without sacrificing sensitivity in the multicenter trial (8).
MATERIALS AND METHODS
Clinical samples.
The data analyzed here were obtained during a multicenter trial designed to evaluate the performance of the COBAS AMPLICOR NG test (8). Briefly, specimens were collected from consecutive, consenting individuals visiting sexually transmitted disease clinics or family planning centers in six geographical regions. Two endocervical swab specimens from women and two urethral swab specimens from men were collected by standard procedures. The first swab was processed for gonorrhea culture according to each laboratory's standard procedure. Gram-negative diplococci were confirmed as N. gonorrhoeae by glucose utilization profiles or antibody reactivity. The second swab was placed in chlamydial culture transport medium, which was then used for PCR testing (12). Ten to 50 ml of first-catch urine was also collected from both men and women. Four aliquots of each culture transport medium swab and urine specimen were stored at −20°C for retesting and for use in discrepant analysis. NG culture results were available for all patients. PCR results for both swab and urine specimens were available for 4,200 of 4,277 patients. One PCR result was missing for the remaining 77 patients because the sample either was not collected or was inhibitory when tested.
PCR testing.
Each specimen was processed and tested in the COBAS AMPLICOR NG test as previously described (8). For each processed specimen, the Chlamydia trachomatis, N. gonorrhoeae, and internal control (IC) target DNAs were simultaneously amplified in a single reaction that contained two primer pairs, one specific for C. trachomatis and the IC and one specific for N. gonorrhoeae. The resulting amplification products were detected separately by hybridization to magnetic microparticles coated with N. gonorrhoeae-, C. trachomatis-, and IC-specific oligonucleotide probes.
All specimens with N. gonorrhoeae signals above an A660 of 0.2 were retested in duplicate. Where indicated, the results of the repeat testing were used to classify specimens as COBAS AMPLICOR NG positive or negative (see below).
Classification of COBAS AMPLICOR results.
Specimens yielding COBAS AMPLICOR NG signals above an A660 of 0.2 were classified as presumptive COBAS AMPLICOR NG positive. To identify a cutoff that produced the best combination of sensitivity and specificity, these presumptive positive specimens were reclassified as COBAS AMPLICOR NG positive or negative using various cutoff values. Where indicated, the combined results of initial and repeat testing were compared at various cutoff values to determine the COBAS AMPLICOR result. Specimens yielding COBAS AMPLICOR NG signals below an A660 of 0.2 were interpreted as negative, provided that the IC signal was above an A660 of 0.2. If the IC signal was below an A660 of 0.2, another aliquot of the original specimen was processed and tested.
Resolution of presumptive results.
Specimens from culture-negative patients that yielded COBAS AMPLICOR NG signals above an A660 of 0.2 were also tested with a PCR assay for an alternative target DNA sequence located within the N. gonorrhoeae 16S rRNA gene (S. Y. Lu, S. Y. Kao, S. Silver, A. Purohit, M. Longiaru, and T. J. White, Abstr. 91st Gen. Meet. Am. Soc. Microbiol. 1991, abstr. C-115, p. 361, 1991). If the specimen was negative in the 16S rRNA test, the other specimen type (swab or urine) was also tested for 16S rRNA to provide evidence for N. gonorrhoeae infection; this test was performed regardless of whether the second specimen type had originally tested positive for N. gonorrhoeae.
Definition of N. gonorrhoeae-positive and N. gonorrhoeae-negative specimens.
Specimens were classified as positive or negative for N. gonorrhoeae based on a combination of culture, COBAS AMPLICOR NG, and, where necessary, NG 16S rRNA PCR results. For the purpose of this study, a specimen was considered N. gonorrhoeae positive if (i) it was obtained from a patient who was positive by culture or (ii) the initial COBAS AMPLICOR NG test yielded a signal at an A660 of ≥0.2 and either of the two specimen types from the patient tested positive by the NG 16S rRNA PCR assay. Specimens from culture-negative patients were classified as N. gonorrhoeae negative if they either (i) yielded a COBAS AMPLICOR NG signal at an A660 of <0.2 or (ii) yielded an initial COBAS AMPLICOR NG signal at an A660 of ≥0.2 that was not confirmed by a positive NG 16S rRNA PCR result on either the swab or urine specimen.
RESULTS
Cross-reactivity with nonpathogenic neisseriae.
The COBAS AMPLICOR CT/NG test for N. gonorrhoeae gave negative results when multiple isolates of various nongonococcal Neisseria strains were tested at concentrations of 104 cells per amplification reaction (Table 1). In contrast, one of six N. cinerea isolates and two of nine Neisseria subflava subsp. perflava isolates gave strong positive signals (Table 1). In the two cross-reactive N. subflava subsp. perflava isolates, the primer and probe sequences, which are contained within the putative cytosine DNA methyltransferase gene (9), were identical to that found in N. gonorrhoeae; the remainder of the amplified target sequence contained no differences and a 1-bp substitution, respectively. In the cross-reactive N. cinerea isolate, the probe sequence differed from the N. gonorrhoeae sequence by 1 bp; the primer sequences were not determined, and there were six additional base pair substitutions in the remainder of the target sequence. The species identification of these isolates was confirmed by biochemical testing and 16S rRNA gene sequencing.
TABLE 1.
Reactivity of the COBAS AMPLICOR CT/NG test with various species of Neisseria
| Species | n | Strain(s)a | COBAS AMPLICOR NG resultb |
|---|---|---|---|
| N. cinerea | 5 | CDC 10050,c 10051,c 10053, and 10054c; NRL 30003 | Negative |
| 1 | CDC 10052c | Positive | |
| Neisseria dentrificans | 1 | NBL 1998c | Negative |
| Neisseria elongata | 1 | NRL 1558 | Negative |
| Neisseria flavescens | 1 | NRL 1095 | Negative |
| Neisseria gonorrhoeae subsp. kochi | 7 | CDC 10046c; NRL 31291,c 31292,c 31294,c 32895,c and 32896c; RMSCC 2039 | Negative |
| Neisseria lactamica | 1 | ATCC 23970 | Negative |
| Neisseria meningitidis types A, B, X, Y, Z, 29E, and 135 | 7 | ATCC 13077, 13090, 35560, 35561, 35562, 35558, and 43744, respectively | Negative |
| Neisseria mucosa | 5 | ATCC 19695c and 19696; NBL 599c and 2086c; RMSCC 1866 | Negative |
| Neisseria polysaccharea | 5 | ATCC 29256 and 43768; CDC 10047,c 10048,c and 10049c | Negative |
| Neisseria sicca | 1 | ATCC 9266 | Negative |
| Neisseria subflava subsp. flava | 1 | ATCC 14221c | Negative |
| N. subflava subsp. perflava | 7 | ATCC 14799; NRL 30015c; RMSCC 2705,c 2706,c 2707,c 2708,c and 2709c | Negative |
| 2 | RMSCC 2627c and 2628c | Positive | |
| Neisseria subflava subsp. subflava subsp. flava | 3 | ATCC 49275c; NRL 30017; RMSCC 2710c | Negative |
ATCC, American Type Culture Collection; CDC, Centers for Disease Control and Prevention; NBL, Naval Biosciences Laboratory; NRL, Neisseria Reference Laboratory; RMSCC, Roche Molecular Systems Culture Collection.
Negative means that signals were at an A660 of <0.2 and ≥0.2 for the NG and IC probes, respectively. Positive means that the signal was at an A660 of ≥3.5 for the NG probe.
Specimens were tested with the microwell plate format AMPLICOR CT/NG test instead of the COBAS AMPLICOR CT/NG test. Both tests use the same primers and probe.
Clinical trial results.
Specimens were obtained from 1,110 asymptomatic women, 1,155 symptomatic women, 721 asymptomatic men, and 1,291 symptomatic men; the corresponding prevalence of N. gonorrhoeae infection was 5.5, 7.7, 3.3, and 30.0%, respectively. Results for matching swab and urine specimens were available for 4,200 of 4,277 patients to give a total of 8,477 COBAS AMPLICOR NG tests performed. Of these tests, 1,093 were performed on N. gonorrhoeae-positive specimens and 7,384 were performed on N. gonorrhoeae-negative specimens. Nine of the 77 patients with a missing PCR result for one specimen type gave a COBAS AMPLICOR NG-positive result for the other sample (three positive swab and six positive urine specimens); the missing results were due to inhibition in the alternate specimen. All six urine specimens and one of the three swab specimens were confirmed positive by culture. Thus, only two swab specimens may have been affected by the missing urine PCR result. These two samples were classified as N. gonorrhoeae negative (based on the criteria defined in Materials and Methods). These two results would have been classified as false negative had the urine specimen tested NG positive by COBAS AMPLICOR and been confirmed by the 16S rRNA PCR test.
When COBAS AMPLICOR NG results were evaluated using a cutoff of an A660 of 0.2, ≥92.9% sensitivity was observed in all specimen types except female urine (Table 2). In contrast, specificity was relatively low, ranging from 96.2 to 98.9% (Table 2). Sensitivity was very high for all test sites (Table 3). Three test sites obtained an overall specificity of approximately 99%, whereas specificity at the other three sites was approximately 3% lower (Table 3). Presumably, lower specificity was due to the presence of cross-reactive Neisseria species in a small fraction of the specimens. To enhance specificity, we sought a testing algorithm that could distinguish specimens containing cross-reactive Neisseria species from those that contained N. gonorrhoeae.
TABLE 2.
Effect of using an expanded equivocal zone on test sensitivity and specificity for different specimen types
| Sex, symptom | Specimen | % Positive | Sensitivity (%)
|
Specificity (%)
|
||
|---|---|---|---|---|---|---|
| A660 cutoff of 0.2 | Equivocal zone | A660 cutoff of 0.2 | Equivocal zone | |||
| Female, asymptomatic | Endocervical | 5.2 | 98.3 | 98.2 | 98.5 | 99.3 |
| Urine | 5.0 | 83.1 | 72.7 | 97.6 | 99.9 | |
| Female, symptomatic | Endocervical | 7.6 | 96.6 | 95.3 | 98.9 | 99.7 |
| Urine | 7.2 | 76.1 | 65.9 | 98.3 | 99.6 | |
| Male, asymptomatic | Urethral | 3.1 | 95.8 | 90.9 | 96.7 | 99.0 |
| Urine | 1.9 | 92.9 | 78.6 | 98.9 | 99.9 | |
| Male, symptomatic | Urethral | 29.6 | 99.7 | 99.5 | 96.2 | 98.7 |
| Urine | 29.1 | 97.3 | 96.2 | 98.2 | 99.9 | |
TABLE 3.
Effect of using an expanded equivocal zone on test sensitivity and specificity at different sitesa
| Site | n (no. positive)b | Sensitivity (%)
|
Specificity (%)
|
||
|---|---|---|---|---|---|
| A660 cutoff of 0.2 | Equivocal zone | A660 cutoff of 0.2 | Equivocal zone | ||
| Indiana University | 2,588 (485) | 95.9 | 93.5 | 99.1 | 99.9 |
| Thomas Jefferson University Hospital | 1,332 (20) | 95.0 | 85.0 | 98.9 | 99.8 |
| University of California at San Francisco | 1,417 (52) | 98.1 | 94.1 | 99.2 | 99.9 |
| Johns Hopkins University | 1,741 (392) | 93.6 | 92.1 | 95.3 | 98.3 |
| Louisiana State University | 794 (48) | 100.0 | 100.0 | 96.6 | 99.3 |
| University of Texas Medical Branch at Galveston | 605 (96) | 99.0 | 97.9 | 96.3 | 99.8 |
Values represent the combined results for all specimen types at each test site.
Values are the numbers of specimens tested and, in parentheses, the numbers of specimens classified as N. gonorrhoeae positive.
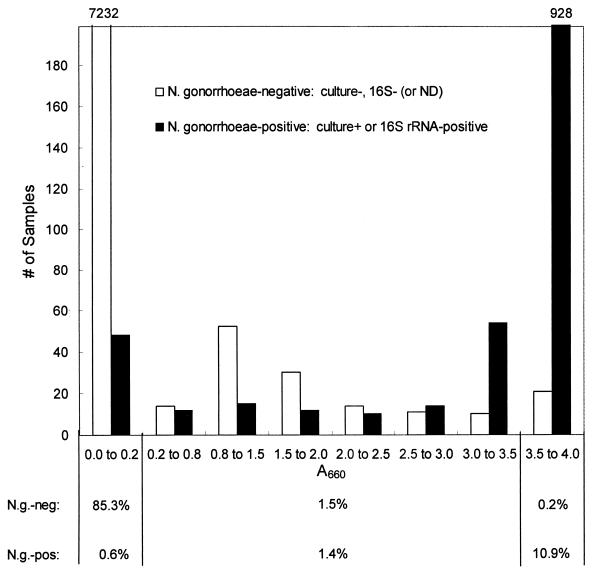
The distribution of COBAS AMPLICOR NG signals suggested that using a higher cutoff could enhance specificity (Fig. 1). Fifty-three percent of the specimens with signals between A660s of 0.2 and 3.5 were N. gonorrhoeae negative. Within this absorbance range, the proportion of N. gonorrhoeae-negative specimens tended to decrease as the signal increased. Similar distributions were obtained when results for each specimen type (urine, urethral swab, and endocervical swab) were analyzed separately (data not shown). Likewise, the distribution of signals was similar for each of the six sites (data not shown). However, the proportion of N. gonorrhoeae-negative specimens with signals at an A660 of ≥0.2 was approximately 3% higher at three sites (Table 3).
FIG. 1.
Histogram showing the distribution of signals produced in the initial COBAS AMPLICOR NG test by swab and urine specimens obtained from symptomatic and asymptomatic men and women. Separate distributions are shown for N. gonorrhoeae-negative specimens and N. gonorrhoeae-positive specimens. Specimens were classified as N. gonorrhoeae negative or N. gonorrhoeae positive as described in Materials and Methods. Within each group, specimens had signals equal to or greater than the lower value and less than the upper value of the range shown on the x axis. The left- and rightmost bars are off scale; the numbers of specimens in these two categories are shown above the bars. The percentages of N. gonorrhoeae-positive and N. gonorrhoeae-negative specimens that give COBAS AMPLICOR NG-negative (A660 < 0.2), -equivocal (A660 ≥ 0.2 and < 3.5), and -positive (A660 ≥ 3.5) results are shown beneath the graph. ND, no data.
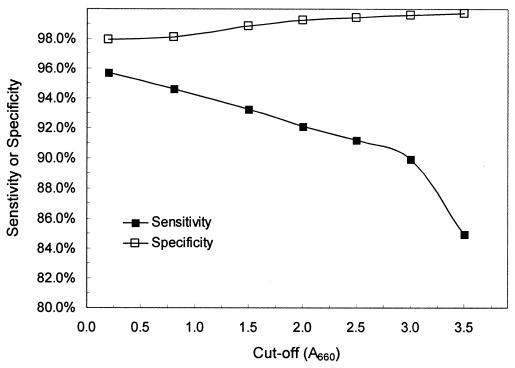
Given this distribution of results, cumulative distribution analysis was performed to determine how sensitivity and specificity varied using different cutoff values (Fig. 2). Maximum sensitivity was achieved with a cutoff of an A660 of 0.2. Sensitivity decreased slowly as the cutoff was increased to an A660 of 3.0 and then decreased more rapidly when the cutoff was further increased to an A660 of 3.5. Specificity increased continuously as the cutoff was increased from an A660 of 0.2 to 3.5. We were unable to identify a cutoff value that produced a combination of greater than 90% sensitivity and 99.5% specificity, although a cutoff of an A660 of 3.0 nearly met these criteria with a sensitivity of 89.9% and a specificity of 99.6%.
FIG. 2.
Cumulative distribution curves for initial test results showing the sensitivity and specificity that would be obtained for various cutoffs. Performance was calculated from the data shown in Fig. 1.
Identification of an equivocal zone and retesting algorithm.
Since cumulative distribution analysis (Fig. 2) did not identify a single cutoff that yielded acceptable performance, we evaluated whether employing an equivocal zone plus additional testing of the original specimen in duplicate could enhance performance. Specimens were classified as COBAS AMPLICOR NG negative if the A660 was below 0.2, positive if the A660 was ≥3.5, and equivocal if the A660 was between 0.2 and 3.5.
For the entire population, 85.9% of specimens were classified as COBAS AMPLICOR NG negative (i.e., A660 < 0.2), of which the overwhelming majority (99.3%) were N. gonorrhoeae negative (Fig. 1). Eleven percent of specimens were classified as COBAS AMPLICOR NG positive (i.e., A660 ≥ 3.5), and the vast majority (98.2%) of these were N. gonorrhoeae positive (Fig. 1). Only 2.9% of specimens were classified as equivocal, with approximately equal numbers being N. gonorrhoeae positive and N. gonorrhoeae negative. Thus, only a small portion of all samples fell into the equivocal zone and had to be adjudicated by additional testing.
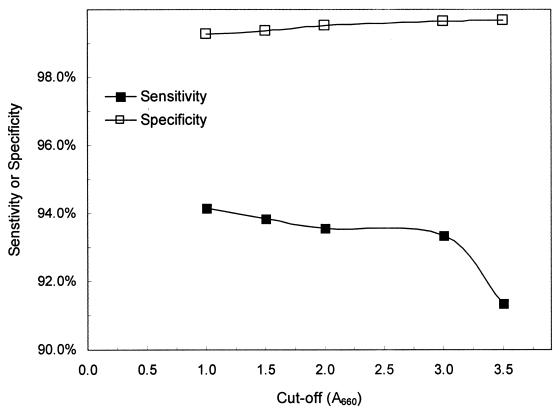
Initially equivocal specimens were interpreted as COBAS AMPLICOR NG positive or COBAS AMPLICOR NG negative if at least two of the three tests (initial plus additional test results) yielded signals above or below the selected cutoff, respectively. Cumulative distribution analysis showed that specificity increased and sensitivity decreased as the cutoff used for repeat testing of these equivocal specimens increased (Fig. 3). Based on these data, we selected a cutoff of an A660 of 2.0 for resolving equivocal results. Use of a cutoff value greater than an A660 of 2.0 would yield somewhat fewer false-positive results, but this would come at the cost of a substantial increase in false-negative results (Fig. 3). For cutoff values less than an A660 of 2.0, the number of equivocal specimens giving false-positive results was too high to achieve acceptable specificity.
FIG. 3.
Cumulative distribution curves for the combination of initial and repeat test results showing the sensitivity and specificity that would be obtained for various cutoffs. All specimens with an initial A660 equal to or greater than 3.5 were interpreted as COBAS AMPLICOR NG positive. The x-axis value represents the cutoff used to interpret the three results for those specimens with initial optical densities of between 0.2 and 3.5. These initially equivocal specimens were interpreted as COBAS AMPLICOR NG positive if at least two of the three results were above the value shown on the x axis.
Performance with the equivocal zone-retesting algorithm.
The equivocal zone-retesting algorithm increased specificity by 0.8 to 2.5% compared to the specificity achieved when each sample type (gender, presence of symptoms, and urine versus swab) was scored as COBAS AMPLICOR NG positive or negative based on a single test result and a cutoff of an A660 of 0.2 (Table 2). The final specificity achieved using this algorithm ranged from 98.6% for urethral swab specimens from symptomatic men to 99.9% for urine specimens from asymptomatic men and women (Table 2). At the three study sites that exhibited 99% specificity for a cutoff of an A660 of 0.2, the equivocal zone-retesting algorithm increased specificity to 99.9% (Table 3). Use of the algorithm increased specificity to similarly high levels at two of the three sites that had exhibited lower specificity using a cutoff of an A660 of 0.2 (Table 3). However, at the remaining site, the specificity increased to only 98.3% (Table 3).
The overall increase in specificity was accompanied by a decrease in sensitivity in all specimen types. The decrease in sensitivity ranged from 0.1 to 1.3% for urethral swab and urine specimens from symptomatic men and for endocervical swab specimens (Table 2). The decrease in sensitivity was limited because a relatively large fraction of specimens with equivocal results gave signals above an A660 of 2.0 in two out of three tests. More substantial decreases in sensitivity were observed for urethral swab and urine specimens from asymptomatic men and for urine specimens from women (Table 2). Most likely, the very low organism load in these specimen types resulted in sample variation, accounting for the failure to consistently obtain strong positive signals.
Five of the six sites exhibited a small (0 to 4%) decrease in sensitivity associated with the use of the equivocal zone-retesting algorithm (Table 3). The more substantial (10%) decrease in sensitivity at the remaining site occurred because this site tested only female specimens. Thus, half of the results from this site were obtained from female urine specimens.
Reduced COBAS AMPLICOR NG signals due to competition from amplification of C. trachomatis DNA in coinfected specimens were not responsible for the decrease in sensitivity. Among 117 N. gonorrhoeae-positive specimens that had an equivocal result in the COBAS AMPLICOR NG, 25 were coinfected with C. trachomatis and 92 were not. Twenty (80%) of the 25 coinfected specimens were interpreted as NG positive using the equivocal zone-retesting algorithm (4 were interpreted as NG negative and 1 was inhibitory when retested). For comparison, 52 (57%) of the 92 specimens infected with NG only were interpreted as NG positive using the equivocal zone-retesting algorithm.
DISCUSSION
The results of this study demonstrate that the specificity of the COBAS AMPLICOR CT/NG test for N. gonorrhoeae is substantially enhanced by employing an equivocal zone and resolving equivocal results by retesting the original specimen. Without an equivocal zone, specificity ranged from 96.2 to 98.9%, which would result in a positive predictive value less than 90% when the prevalence of infection is below 20%. Specificity ranged from 98.6 to 99.9% when the equivocal zone-retesting algorithm was employed, enabling the test to achieve a positive predictive value of at least 90% when performed on urogenital specimens obtained from populations with a prevalence of 4% or higher. The test should not, however, be used to test throat swab specimens, since up to 50% of samples will give false-positive results (J. Weiss and M. Rosenstraus, unpublished results).
Without the equivocal zone, specificity was low (96%) at some sites and high (99%) at others. In another study, a single lab observed a high frequency of false-positive results in samples from one population but a very low frequency in a different population (4). The reasons for this variation are not clear. Our data show that the equivocal zone-retesting algorithm enhances specificity to greater than 99.3% for most populations with a high rate of false-positive results and to 99.9% for populations with a low rate of false positives. This specificity approaches the 99.1 to 100.0% specificity previously reported for the Abbott LCx N. gonorrhoeae test (1–3, 5, 10, 11, 13).
The increase in specificity was accompanied by a slight decrease in sensitivity, but this did not impact the test's diagnostic utility. Sensitivity still exceeded 95% for urethral swab and urine specimens from symptomatic males and for endocervical swab specimens. Sensitivity was more severely compromised for specimens from asymptomatic men and for female urine specimens. Although the sensitivity of asymptomatic male urine testing was somewhat lower (10 to 20%) than that of culture (8), testing noninvasively collected urine specimens may be preferable to collecting and culturing urethral swab samples. The sensitivity decrease for female urine specimens did not impact utility; the test has suboptimal sensitivity for female urine with or without the equivocal zone.
The same equivocal zone-retesting algorithm is used in the microwell plate-based AMPLICOR CT/NG test for N. gonorrhoeae. The COBAS and microwell plate formats exhibited similar performance characteristics when the tests were performed in parallel on the specimens used in the present study (8). With the equivocal zone-retesting algorithm, the specificity of the microwell plate format ranged from 98.4 to 99.7% depending on the specimen type being analyzed (data not shown).
Although a wide equivocal zone is required to maximize specificity, the additional testing burden will be small. Depending on the test population, approximately 1 to 4% of N. gonorrhoeae-negative specimens will give equivocal results, while 20% of N. gonorrhoeae-positive specimens will give equivocal results. Thus, only 2 to 7% of specimens will need to be retested in most populations with an N. gonorrhoeae prevalence ranging from 5 to 20%.
Our observation that false-positive specimens generally give weak signals is consistent with an earlier study, which showed that 14 of 17 specimens with signals between A450s of 0.2 and 0.8 in the microwell plate-based AMPLICOR CT/NG test were false positive (4). These specimens, which presumably contain cross-reactive neisseriae, may yield relatively weak signals for two reasons. First, the cross-reactive neisseriae are likely to be present at a low concentration (6). Second, some, but not all, of the cross-reactive strains contain mismatches in the primer or probe regions, which could reduce the efficiency of amplification or hybridization and result in weak signals when the target is present at a low level. Finally, some of the specimens classified as false positive could actually be N. gonorrhoeae infected. Specimens containing a low concentration of N. gonorrhoeae could have produced some of the initial positive results that were not confirmed by additional testing.
Low organism concentration and probe mismatches also explain how retesting initially equivocal specimens can distinguish most of those containing cross-reactive neisseriae from N. gonorrhoeae-containing specimens. Cross-reactive specimens tend to give negative results when retested because the low target concentration produces aliquot-to-aliquot fluctuation in the amount of target tested. Those specimens that do give reproducible results will often give low signals if the cross-reactive organism has a mismatch in the probe region. Specimens containing substantial amounts of cross-reactive organisms or strains that do not have mismatches in the probe region probably account for the small number of N. gonorrhoeae-negative specimens that give signals above an A660 of 3.5 when initially tested. These specimens, along with the few equivocal, cross-reactive specimens that are not identified by retesting, explain why 100% specificity was not achieved.
Because some cross-reactive isolates contain target sequences identical to those found in N. gonorrhoeae, we hypothesize that these isolates arose by recombination between N. gonorrhoeae and the non-cross-reactive neisseriae. Genetic exchange between neisseriae is a common phenomenon, occurring both in genes coding for housekeeping and in genes coding for pathogen-associated proteins, and may play a role in the generation of new subspecies or species (7). Genetic drift subsequent to the genetic exchange probably accounts for the isolates that have one or more nucleotide differences compared to the N. gonorrhoeae sequence. These differences generally do not result in amino acid changes.
In summary, urogenital specimens that initially give false-positive results with the COBAS AMPLICOR CT/NG test for N. gonorrhoeae can be distinguished from most N. gonorrhoeae-containing urogenital specimens by retesting all samples that give results within an equivocal range. The algorithm used here is the one recommended by the manufacturer in the Food and Drug Administration-cleared version of the COBAS Methods Manual. Specificity ranges from 98.6 to 99.9% when the equivocal zone-retesting algorithm is used, enabling the test to achieve a 90% or higher positive predictive value when performed on urogenital specimens obtained from populations with a prevalence of 4% or higher. Finally, the approach of retesting specimens with relatively low signals may prove useful for enhancing specificity in other N. gonorrhoeae assay systems.
ACKNOWLEDGMENTS
We thank James Williams and Laura Brandenburg for laboratory assistance and the many clinicians at each site for assistance with enrolling patients.
This work was supported by Roche Molecular Systems.
REFERENCES
- 1.Buimer M, Van Doornum G J J, Ching S, Peerbooms P G H, Plier P K, Ram D, Lee H H. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by ligase chain reaction-based assays with clinical specimens from various sites: implications for diagnostic testing and screening. J Clin Microbiol. 1996;34:2395–2400. doi: 10.1128/jcm.34.10.2395-2400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll K C, Aldeen W E, Morrison M, Anderson R, Lee D, Mottice S. Evaluation of the Abbott LCx ligase chain reaction assay for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine and genital swab specimens from a sexually transmitted disease population. J Clin Microbiol. 1998;36:1630–1633. doi: 10.1128/jcm.36.6.1630-1633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ching S, Lee H, Hook III E W, Jacobs M R, Zenilman J. Ligase chain reaction for detection of Neisseria gonorrhoeae in urogenital swabs. J Clin Microbiol. 1995;33:3111–3114. doi: 10.1128/jcm.33.12.3111-3114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell D. Evaluation of AMPLICOR Neisseria gonorrhoeae PCR using cppB nested PCR and 16S rRNA PCR. J Clin Microbiol. 1999;37:386–390. doi: 10.1128/jcm.37.2.386-390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehl S C, Georgakas K, Swain G R, Sedmak G, Gradus S, Singh A, Foldy S. Evaluation of the Abbott LCx assay for detection of Neisseria gonorrhoeae in endocervical swab specimens from females. J Clin Microbiol. 1998;36:3549–3551. doi: 10.1128/jcm.36.12.3549-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knapp J, Rice R J. Neisseria and Branhamella. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 324–340. [Google Scholar]
- 7.Maiden M C J, Malorny B, Achtman M. A global gene pool in the neisseriae. Mol Microbiol. 1996;21:1297–1298. doi: 10.1046/j.1365-2958.1996.981457.x. [DOI] [PubMed] [Google Scholar]
- 8.Martin D H, Cammarata C, Van Der Pol B, Jones R B, Quinn T C, Gaydos C A, Crotchfelt K, Schachter J, Moncada J, Jungkind D, Turner B, Peyton C. Multicenter evaluation of AMPLICOR and automated COBAS AMPLICOR CT/NG tests for Neisseria gonorrhoeae. J Clin Microbiol. 2000;38:3544–3549. doi: 10.1128/jcm.38.10.3544-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyada C G, Born T L. A DNA sequence for the discrimination of Neisseria gonorrhoeae from other Neisseria species. Mol Cell Probes. 1991;5:327–335. doi: 10.1016/s0890-8508(06)80003-4. [DOI] [PubMed] [Google Scholar]
- 10.Smith K R, Ching S, Lee H, Ohhashi Y, Hu H Y, Fisher III H C, Hook E W., III Evaluation of the ligase chain reaction for use with urine for identification of Neisseria gonorrhoeae infections in females attending a sexually transmitted disease clinic. J Clin Microbiol. 1995;33:455–457. doi: 10.1128/jcm.33.2.455-457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stary A, Ching S-F, Teodorowicz L, Lee H. Comparison of ligase chain reaction and culture for detection of Neisseria gonorrhoeae in genital and extragenital specimens. J Clin Microbiol. 1997;33:239–242. doi: 10.1128/jcm.35.1.239-242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Der Pol B, Quinn T C, Gaydos C A, Crotchfelt K, Schachter J, Moncada J, Jungkind D, Martin D H, Turner B, Peyton C, Jones R B. Multicenter evaluation of the AMPLICOR and Automated COBAS AMPLICOR CT/NG tests for detection of Chlamydia trachomatis. J Clin Microbiol. 2000;38:1105–1112. doi: 10.1128/jcm.38.3.1105-1112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu K, Glanton V, Johnson S R, Beck-Sague C, Bhullar V, Candal D H, Pettus K S, Farshy C E, Black C M. Detection of Neisseria gonorrhoeae infection by ligase chain reaction testing of urine among adolescent women with and without Chlamydia trachomatis infection. Sex Transm Dis. 1998;25:533–538. doi: 10.1097/00007435-199811000-00007. [DOI] [PubMed] [Google Scholar]