ABSTRACT
Bovine alphaherpesvirus 5 causes meningoencephalitis in cattle, belongs to the Herpesviridae family, and can be divided into subtypes a, b, and c. Limited information is available about subtype c. Here, we report the complete genome sequences of two strains, P160/96, and ISO97/45, isolated from cattle in southeast Brazil.
ANNOUNCEMENT
Bovine alphaherpesvirus 5 (BoHV-5) is an important agent of meningoencephalitis in cattle, belonging to the family Herpesviridae, subfamily Alphaherpesvirinae, genus Varicellovirus (1), whose genomes are composed by a single double-stranded DNA molecule with 124.8 to 151.6 kbp (https://talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/herpesviridae/1614/genus-varicellovirus). BoHV-5 is subdivided into subtypes BoHV-5a, BoHV-5b, and BoHV-5c, based on genome restriction endonuclease patterns (2–4). Although BoHV-5 distribution is scarcely known, subtype BoHV-5a seems more widely distributed than BoHV-5b, which has only been detected in Argentina (3–6). Subtype BoHV-5c has only been recovered from a particular region in southeast Brazil (2). There are four complete BoHV-5 genome sequences previously reported, BoHV-5a strain SV507/99 (7) and three BoHV-5b strains (A663, 674/10, and 166/84) (5). Here, two complete genomes of BoHV-5c strains, named P160/96 and ISO97/45, are reported.
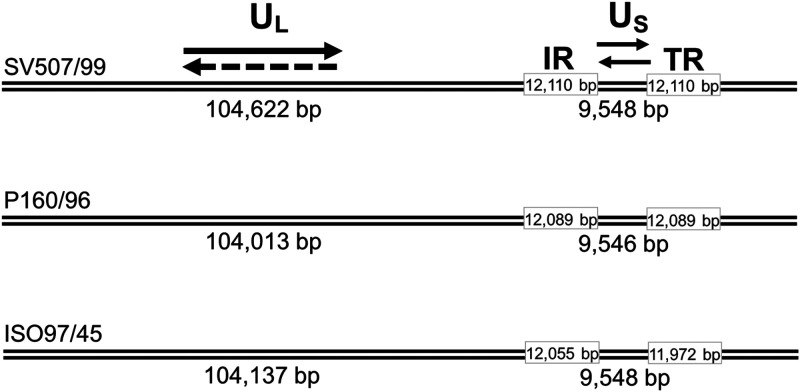
The BoHV-5c strain P160/96 was originally isolated from a case of herpesvirus bovine encephalitis at PESAGRO, in the state of Rio de Janeiro, Brazil. The BoHV-5c strain ISO97/45, also from a case of bovine encephalitis, was recovered from the brain tissues of a calf in 1997. The virus was originally isolated at the Biological Institute of São Paulo, São Paulo, Brazil (8). Both strains had been partially characterized and typed as BoHV-5 by restriction endonuclease and monoclonal antibody analyses (2, 8). For this study, both strains were cultured in Madin-Darby bovine kidney (MDBK) cells (9) and ultracentrifuged (10), and DNA was extracted with phenol-chloroform following standard procedures (11). DNA libraries were prepared with a Nextera kit. High-throughput sequencing was performed in a MiSeq (Illumina) platform, with 500- and 300-cycle kits (version 2) to generate 2 × 250 and 2 × 150 paired-end reads, respectively. The total number of reads mapped to the genomes were 72,887 for P160/96 and 34,096 for ISO97/45. The average read lengths were 250 bp and 150 bp with coverage of 132× and 37×, respectively, of the whole BoHV-5 genome, based on the Lander-Waterman (12) coverage estimate equation. Reads were trimmed using Geneious software (version 9.1) with default settings. Assembly and annotation of the viral genomes were done using template-assisted assembly to the BoHV-5 SV507/99 reference genome (GenBank accession number NC_005261) using a map to reference tools of Geneious version 9.1 with default settings. Both genomes were assembled to full length, including the internal (IR) and terminal repeat (TR) regions (Fig. 1). BoHV-5 genomes showed a classic type D herpesvirus organization (4, 7), with total lengths of 137,741 (P160/96) and 137,712 (ISO97/45) nucleotides (nt), slightly shorter than BoHV-5a SV507/99, which is 138,390 nt long. The difference in genome lengths is shown in Fig. 1. The two BoHV-5 genomes have GC contents of 74.7% (P160/96) and 74.8% (ISO97/45), with 99.4% and 98.9% nucleotide identity to SV507/99, respectively, as determined with the fast Fourier transform (MAFFT) of Geneious version 9.1. The genome sequences of BoHV-5c strains P160/96 and ISO97/45 were also submitted to comparative analyses using MAFFT with default settings, in which each gene was compared individually for identity at the nucleotide level between the different BoHV-5 genomes (Table 1). All bioinformatic tools used here were run with default parameters unless otherwise specified.
FIG 1.
Schematic representation of the genomes reported in this study, highlighting the unique long (UL) and unique short (US) segments and the internal (IR) and terminal (TR) repeat regions. Previously published BoHV-5a strain SV507/99 used as reference (7).
TABLE 1.
Percentages of identity and GC content of nucleotide and amino acid sequences of BoHV-5 strain P160/96 and BoHV-5 strain ISO97/45
| BoHV-5 strain P160/96 |
BoHV-5 strain ISO97/45 |
||||||
|---|---|---|---|---|---|---|---|
| Gene | Predicted product | % nt identitya | % aa identityb | %GC | % nt identitya | % aa identityb | %GC |
| Circ | Myristylated virion protein | 99.9 | 99.6 | 72.8 | 99.7 | 99.6 | 73 |
| UL54 | Regulates and transports RNA | 100 | 100 | 74.1 | 99.7 | 99.8 | 74.1 |
| UL53 | Glycoprotein K | 100 | 100 | 76.9 | 99.6 | 99.7 | 77 |
| UL52 | Component of DNA helicase/primase complex | 100 | 100 | 76.6 | 99.4 | 99.4 | 76.4 |
| UL51 | Palmitoylated protein | 99.9 | 100 | 75.9 | 99.7 | 100 | 76 |
| UL50 | Deoxyuridine triphosphatase | 100 | 100 | 72.2 | 99.2 | 98.8 | 72.4 |
| UL49.5 | Glycoprotein N | 100 | 100 | 68.8 | 100 | 100 | 68.8 |
| UL49 | Tegument protein | 100 | 100 | 77.2 | 99.4 | 99.3 | 77.2 |
| UL48 | Trans-inducing factor | 99.9 | 100 | 72.9 | 99.5 | 99.6 | 73.1 |
| UL47 | Tegument phosphoprotein | 99.9 | 100 | 72.4 | 99.7 | 99.9 | 72.3 |
| UL46 | Tegument protein | 99.7 | 99.7 | 74.8 | 99.5 | 99.7 | 74.6 |
| UL44 | Glycoprotein C | 99.7 | 99.8 | 75.4 | 99.4 | 99.4 | 75.7 |
| UL43 | Virion protein | 100 | 100 | 82 | 99.1 | 100 | 82 |
| UL42 | Processivity factor for DNA polymerase | 99.9 | 100 | 74.5 | 98.6 | 98.8 | 73.8 |
| UL41 | Virion host shutoff factor | 99.9 | 100 | 74 | 99.5 | 99.6 | 73.3 |
| UL40 | Ribonucleotide reductase small subunit | 100 | 100 | 62.8 | 99.9 | 100 | 62.9 |
| UL39 | Ribonucleotide reductase large subunit | 99.9 | 99.9 | 70.6 | 99.8 | 99.6 | 70.4 |
| UL38 | Capsid protein | 99.8 | 99.4 | 75 | 99.3 | 99.2 | 74.4 |
| UL37 | Tegument protein | 99.9 | 99.9 | 79.4 | 99.5 | 99.6 | 79.2 |
| UL36 | Very large tegument protein | 99.9 | 99.8 | 79.8 | 99.7 | 99.5 | 79.8 |
| UL35 | Capsid protein | 100 | 100 | 73.3 | 100 | 100 | 73.3 |
| UL34 | Virion protein | 100 | 100 | 74.8 | 99.9 | 100 | 74.7 |
| UL33 | Capsid packaging protein | 100 | 100 | 71.5 | 99.7 | 100 | 71.2 |
| UL32 | Cleavage and packaging protein | 99.9 | 99.8 | 76.1 | 99.7 | 99.8 | 76.1 |
| UL31 | UL34-associated nuclear protein | 99.6 | 98.9 | 73.6 | 97 | 96.6 | 73.9 |
| UL30 | DNA polymerase, catalytic subunit | 99.7 | 99.5 | 72.2 | 99.6 | 99.7 | 72.5 |
| UL29 | Single-stranded DNA binding protein | 100 | 100 | 73.3 | 99.7 | 99.9 | 73.3 |
| UL28 | Cleavage and packaging protein | 99.1 | 98.7 | 76.2 | 99.8 | 99.4 | 76.5 |
| UL27 | Glycoprotein B | 100 | 100 | 71.6 | 99.8 | 99.8 | 71.4 |
| UL26.5 | Capsid scaffolding protein | 100 | 100 | 79.6 | 99.7 | 99.7 | 79.7 |
| UL26 | Capsid maturation serine protease | 100 | 100 | 78.5 | 99.7 | 99.8 | 78.5 |
| UL25 | DNA packaging virion protein | 99.8 | 99.5 | 77.3 | 99.6 | 99.7 | 77 |
| UL24 | Putative membrane-associated protein | 98.5 | 97.5 | 76.4 | 99.8 | 99.6 | 77.1 |
| UL23 | Thymidine kinase | 99.9 | 100 | 78.6 | 99.7 | 100 | 78.8 |
| UL22 | Glycoprotein H | 99.9 | 99.8 | 76.2 | 99.5 | 99.5 | 76.2 |
| UL21 | Tegument protein | 99.9 | 99.8 | 77.6 | 99.4 | 99.7 | 77.3 |
| UL20 | Virion protein | 98.6 | 97.2 | 76.9 | 99.9 | 100 | 78 |
| UL19 | Major capsid protein | 99.9 | 99.9 | 72.5 | 99.7 | 99.9 | 72.5 |
| UL18 | Capsid protein | 100 | 100 | 75.3 | 99.8 | 100 | 75.4 |
| UL17 | Tegument protein | 100 | 100 | 80.2 | 99.6 | 99.7 | 80.3 |
| UL16 | Virion protein | 99.8 | 99.7 | 77.9 | 99.9 | 100 | 78 |
| UL15 | DNA cleavage, packaging protein | 99.9 | 99.7 | 71.3 | 99.5 | 99.9 | 71.5 |
| UL14 | Minor tegument protein | 100 | 100 | 75.3 | 99.6 | 99.1 | 75.1 |
| UL13 | Virion serine/threonine protein kinase | 100 | 100 | 74.7 | 99.7 | 99.6 | 74.7 |
| UL12 | Alkaline exonuclease | 98.6 | 98.8 | 75.3 | 99.0 | 98.8 | 75.3 |
| UL11 | Myristylated protein | 100 | 100 | 75.2 | 100 | 100 | 75.2 |
| UL10 | Glycoprotein M | 99.9 | 100 | 76 | 99.4 | 99.8 | 75.9 |
| UL9 | Origin-binding protein | 99.9 | 99.9 | 74.5 | 99.6 | 99.9 | 74.5 |
| UL8 | Component of DNA helicase/primase complex | 100 | 100 | 76.7 | 99.7 | 99.7 | 76.6 |
| UL7 | Virion-associated protein | 100 | 100 | 72.7 | 100 | 100 | 72.7 |
| UL6 | Virion protein | 98.3 | 98.3 | 73.4 | 99.5 | 99.9 | 73.8 |
| UL5 | Component of DNA helicase/primase complex | 99.9 | 100 | 66.5 | 97.0 | 98.6 | 68.2 |
| UL4 | Nuclear protein | 100 | 100 | 74.6 | 99.3 | 99.5 | 74.3 |
| UL3.5 | Virion protein | 100 | 100 | 80.1 | 99.6 | 99.3 | 80.1 |
| UL3 | Phosphoprotein | 99.8 | 99.5 | 74.2 | 97.3 | 95 | 74.4 |
| UL2 | Uracil DNA glycosylase | 100 | 100 | 75.5 | 99.8 | 99.3 | 75.3 |
| UL1 | Glycoprotein L | 100 | 100 | 74.4 | 99.8 | 100 | 74.6 |
| UL0.7 | Unknown product | 99.8 | 99.5 | 77.4 | NAc | NA | NA |
| BICP0 | Immediate-early trans-activator protein with zinc finger | 99.9 | 100 | 76.7 | 99.0 | 99 | 76.9 |
| BICP4 | Positive and negative gene regulator | 100 | 100 | 82.2 | 98.3 | 97.3 | 82.2 |
| BICP22 | Transcription factor | 98.3 | 98.1 | 75.3 | 98.4 | 97.8 | 75.1 |
| US1.67 | Virion protein | 100 | 100 | 73.1 | 99.3 | 98.8 | 73 |
| US2 | Tegument protein | 100 | 100 | 72.4 | 99.0 | 98.2 | 72.5 |
| US3 | Virion serine/threonine protein kinase | 99.9 | 100 | 73.9 | 99.8 | 99.5 | 74.1 |
| US4 | Glycoprotein G | 100 | 100 | 68.6 | 99.8 | 99.5 | 68.7 |
| US6 | Glycoprotein D | 100 | 100 | 73.4 | 99.5 | 99.5 | 73.8 |
| US7 | Glycoprotein I | 99.9 | 100 | 76.6 | 99.6 | 99.7 | 76.5 |
| US8 | Glycoprotein E | 99.9 | 100 | 76.3 | 99.1 | 98.8 | 76.2 |
| US9 | Virion protein | 100 | 100 | 76 | 99.8 | 99.3 | 75.8 |
| BICP22 | Transcription factor | 98.3 | 98.1 | 75.3 | 97.8 | 96.8 | 75.2 |
| BICP4 | Positive and negative gene regulator | 100 | 100 | 82.2 | 98.4 | 97.5 | 82 |
nt, nucleotide.
aa, amino acid.
NA, not applicable.
Unlike other bovine herpesviruses, BoHV-5 has a limited geographical distribution; cases have been commonly reported in South American countries, particularly Brazil and Argentina, and sporadically in other continents (2–9, 13), which makes the origin of BoHV-5 and related outbreaks still a mystery. Previously, one BoHV-5a complete genome sequence (7) and three complete sequences of BoHV-5b were reported (5). Regarding BoHV-5c, formerly called BoHV-5 non-a, non-b (2), no other reports on the occurrence of BoHV-5c infections have been made outside a particular region in southeast Brazil, which seems to comprise the state of Rio de Janeiro, northern São Paulo state, and northeastern Minas Gerais, suggesting that adaptive evolution may have played some role in fixing some of the adaptations that, to date, characterize the BoHV-5c subtype.
However, taxonomy, as currently applied to BoHV-1 and BoHV-5, does not reflect the evolutionary history of these viruses since it is not based on full-genome analyses; REA and monoclonal antibody characterization does not entirely express the complexity of genetic alterations (2, 8). Recently, Romera et al. (5) reported the occurrence of naturally generated interspecific recombinants between BoHV-1 and BoHV-5; obviously, such events can influence type or subtype determination, reinforcing the importance of full-genome analyses to allow for more precise classifications. It is expected that the availability of more complete BoHV-5 genomes, such as strains P160/96 and ISO97/45 reported here, will contribute to a better understanding of the genetic evolution of bovine alphaherpesviruses.
Data availability.
The genomes have been deposited in NCBI GenBank and are available under accession numbers KY559403 (BoHV-5 strain P160/96) and KY549446 (BoHV-5 strain ISO97/45). The raw sequencing reads were deposited in the NCBI Sequence Read Archive under BioProject accession numbers PRJNA790921 (SRA experiment number SRX13457950 and SRA run ID SRR17280439) and PRJNA790967 (SRA experiment number SRX13458985 and SRA run ID SRR17281500).
ACKNOWLEDGMENTS
C.T. was in receipt of a master's degree fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). A.C.F., F.R.S., and P.M.R. are research fellows from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Financial support was provided by Fundação de Apoio à Pesquisa do Estado do Rio Grande do Sul (FAPERGS proc. no. 16/2551-0000478-1) and CNPq grant no. 309024/2020-0.
Contributor Information
Paulo M. Roehe, Email: proehe@gmail.com.
Simon Roux, DOE Joint Genome Institute.
REFERENCES
- 1.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Arce RCF, Almeida RS, Silva TC, Franco AC, Spilki F, Roehe PM, Arns CW. 2002. Restriction endonuclease and monoclonal antibody analysis of Brazilian isolates of bovine herpesviruses types 1 and 5. Vet Microbiol 88:315–324. doi: 10.1016/S0378-1135(02)00126-8. [DOI] [PubMed] [Google Scholar]
- 3.Pidone CL, Galosi CM, Echeverria MG, Nosetto EO, Etcheverrigaray ME. 1999. Restriction endonuclease analysis of BHV-1 and BHV-5 strains isolated in Argentina. Zentralbl Veterinarmed B 46:453–456. doi: 10.1046/j.1439-0450.1999.00257.x. [DOI] [PubMed] [Google Scholar]
- 4.Del Médico Zajac MP, Ladelfa MF, Kotsias F, Muylkens B, Thiry J, Thiry E, Romera SA. 2010. Biology of bovine herpesvirus 5. Vet J 184:138–145. doi: 10.1016/j.tvjl.2009.03.035. [DOI] [PubMed] [Google Scholar]
- 5.Romera SA, Perez R, Marandino A, LuciaTau R, Campos F, Roehe PM, Thiry E, Maidana SS. 2022. Whole-genome analysis of natural interspecific recombinant between bovine alphaherpesviruses 1 and 5. Virus Res 309:198656. doi: 10.1016/j.virusres.2021.198656. [DOI] [PubMed] [Google Scholar]
- 6.Maidana SS, Morano CD, Cianfrini D, Campos FS, Roehe PM, Siedler B, De Stefano G, Mauroy A, Thiry E, Romera SA. 2013. Multiplex PCR followed by restriction length polymorphism analysis for the subtyping of bovine herpesvirus 5 isolates. BMC Vet Res 9:111. doi: 10.1186/1746-6148-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delhon G, Moraes MP, Lu Z, Afonso CL, Flores EF, Weiblen R, Kutish GF, Rock DL. 2003. Genome of bovine herpesvirus 5. J Virol 77:10339–10347. doi: 10.1128/JVI.77.19.10339-10347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souza VF, Melo SV, Esteves PA, Schmidt CS, Gonçalves DA, Schaefer R, Silva TC, Almeida RS, Vicentini F, Franco AC, Oliveira EA, Spilki FR, Weiblen R, Flores EF, Lemos RA, Alfieri AA, Pituco EM, Roehe PM. 2002. Monoclonal antibody characterization of bovine herpesviruses types 1 (BHV-1) and 5 (BHV-5). Pesq Vet Bras 22:13–18. doi: 10.1590/S0100-736X2002000100004. [DOI] [Google Scholar]
- 9.Roehe PM, da Silva TC, Nardi NB, Oliveira LG, de Rosa JCA. 1997. Monoclonal antibody differentiation between bovine herpesviruses type 1 and 5. Pesq Vet Bras 17:41–44. doi: 10.1590/S0100-736X1997000100007. [DOI] [Google Scholar]
- 10.de Sales Lima FE, Cibulski SP, Witt AA, Franco AC, Roehe PM. 2015. Genomic characterization of two novel polyomaviruses in Brazilian insectivorous bats. Arch Virol 160:1831–1836. doi: 10.1007/s00705-015-2447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring, NY. [Google Scholar]
- 12.Lander ES, Waterman MS. 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 13.Kumar N, Chander Y, Riyesh T, Khandelwal N, Kumar R, Kumar H, Tripathi BN, Barua S. 2020. Isolation and characterization of bovine herpes virus 5 (BoHV5) from cattle in India. PLoS One 15:e0232093. doi: 10.1371/journal.pone.0232093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genomes have been deposited in NCBI GenBank and are available under accession numbers KY559403 (BoHV-5 strain P160/96) and KY549446 (BoHV-5 strain ISO97/45). The raw sequencing reads were deposited in the NCBI Sequence Read Archive under BioProject accession numbers PRJNA790921 (SRA experiment number SRX13457950 and SRA run ID SRR17280439) and PRJNA790967 (SRA experiment number SRX13458985 and SRA run ID SRR17281500).