Summary
Studies suggest that the efficacy of cancer chemotherapy and immunotherapy is influenced by intestinal bacteria. However, the influence of the microbiome on radiation therapy is not as well-understood, and the microbiome comprises more than bacteria. Here, we find that intestinal fungi regulate antitumor immune responses following radiation in mouse models of breast cancer and melanoma and that fungi and bacteria have opposite influences on these responses. Antibiotic mediated depletion or gnotobiotic exclusion of fungi enhances responsiveness to radiation, whereas antibiotic mediated depletion of bacteria reduces responsiveness and is associated with overgrowth of commensal fungi. Further, elevated intratumoral expression of Dectin-1, a primary innate sensor of fungi, is negatively associated with survival in patients with breast cancer and is required for the effects of commensal fungi in mouse models of radiation therapy.
Keywords: Microbiome, Mycobiome, Radiation, Tumor Immunology, Immunotherapy, Macrophages, T cells
eTOC Blurb
Depletion of commensal bacteria leads to expansion of commensal fungi and reduced anti-tumor immunity following irradiation of tumors. Targeting commensal fungi enhanced the radiation induced anti-tumor immune response by reducing macrophage mediated immunosuppression. Thus, Shiao et al demonstrates opposing effects of commensal bacteria and fungi on anti-tumor immunity following radiation.
Graphical Abstract
Introduction
Commensal microorganisms can shape development of the immune system and immune responses to diverse challenges from cancer to autoimmunity (Blander et al., 2017; Levy et al., 2017). Increasingly, recognition that microorganisms in the gut influence both local mucosal immunity and systemic immune responses has led to the exploration of how the microbiota influences immune processes governing the development of cancers (Kostic et al., 2013; Schwabe and Jobin, 2013) and their responses to treatment (Geller et al., 2017; Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2017). For example, murine studies found that the bacterial species Fusobacterium nucleatum promoted more rapid tumor development through a mechanism involving recruitment of inflammatory myeloid cells (Brennan and Garrett, 2019; Kostic et al., 2013). This is consistent with the notion that tumor-associated immune cells can either promote or hinder tumor progression depending on the immune microenvironment (Binnewies et al., 2018; Chen and Mellman, 2017) and that this microenvironment can be regulated in part by the interaction of the immune system with the microbiome (Routy et al., 2018).
In addition to regulating carcinogenesis, recent studies have revealed that intestinal bacterial communities are required for effective antitumor immunity following chemotherapy (Daillere et al., 2016; Geller et al., 2017; Iida et al., 2013; Viaud et al., 2013) and immunotherapy (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2017; Sivan et al., 2015) in both preclinical models and human studies. Depletion of bacteria with antibiotics reduced the efficacy of oxaliplatin (Iida et al., 2013) and cyclophosphamide (Viaud et al., 2013) by reducing treatment-induced myeloid and Th17 responses. Further examination of the relationship between microbiota and the response to chemotherapy revealed a role for Enterococcus hirae and Barnesiella intestinihominis in generating a productive antitumor T cell response following administration of the alkylating agent cyclophosphamide (Daillere et al., 2016). More recent studies have demonstrated that the bacterial microbiota including Akkermansia and Bifidobacterium species are also critical for the response to immunotherapies, particularly the checkpoint inhibitors targeting programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) (Gopalakrishnan et al., 2018; Matson et al., 2018).
Cytotoxic therapies such as chemotherapy and radiation play an integral role in the treatment of virtually all cancers, and it is increasingly recognized that the efficacy of these therapies depends on generating antitumor immune responses in part through induction of immunogenic cell death (Galluzzi et al., 2017). Much effort has been devoted to characterizing the nature of the antitumor immune response following radiation therapy (RT), and multiple studies have identified the importance of CD8+ cytotoxic T cells, CD11b+ myeloid cells, and dendritic cells in mediating the antitumor effects of RT (Deng et al., 2014; Lugade et al., 2005; Shiao et al., 2015b; Stone et al., 1979; Vanpouille-Box et al., 2017), but whether gut bacteria influence this response is not yet known. Further, whether other components of the intestinal microbiota, especially fungi, are important is not yet understood. The fungal microbiome has been recently recognized to regulate inflammatory responses in diverse disease settings including colitis (Iliev et al., 2012; Limon et al., 2019), asthma (Wheeler et al., 2016) and inflammation-associated colon cancer (Malik et al., 2018) at least in part through their effects on dendritic cells and myeloid cells. Thus, we hypothesized that the fungal microbiome may also regulate responses to cancer therapies.
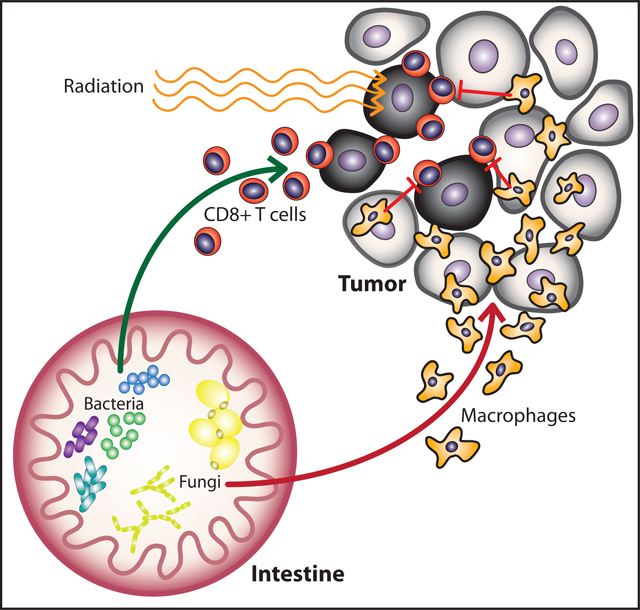
We observe here that bacterial and fungal microbiota differentially regulate tumor responses to RT. Our studies revealed that commensal bacteria are required for efficient antitumor immune responses, while commensal fungi regulate the immunosuppressive microenvironment following treatment. Targeting commensal fungi enhanced the response to RT and, consistent with this observation, we found an association between reduced expression of Dectin-1, a key innate immune receptor for sensing fungi, and survival in breast cancer. Genetic ablation of Dectin-1 abrogated the effect of antifungals on RT indicating that enhanced efficacy observed by targeting commensal fungi with RT depends on a mechanism involving sensing of fungi through the Dectin-1 receptor. These studies indicate that the fungal microbiome regulates immune responses following cytotoxic therapy and that this influence is distinct from the influences of the bacterial microbiome. Together, fungi and bacteria shape radiation-induced therapeutic antitumor immunity.
Results
Depletion of commensal bacteria reduces efficacy of radiation therapy
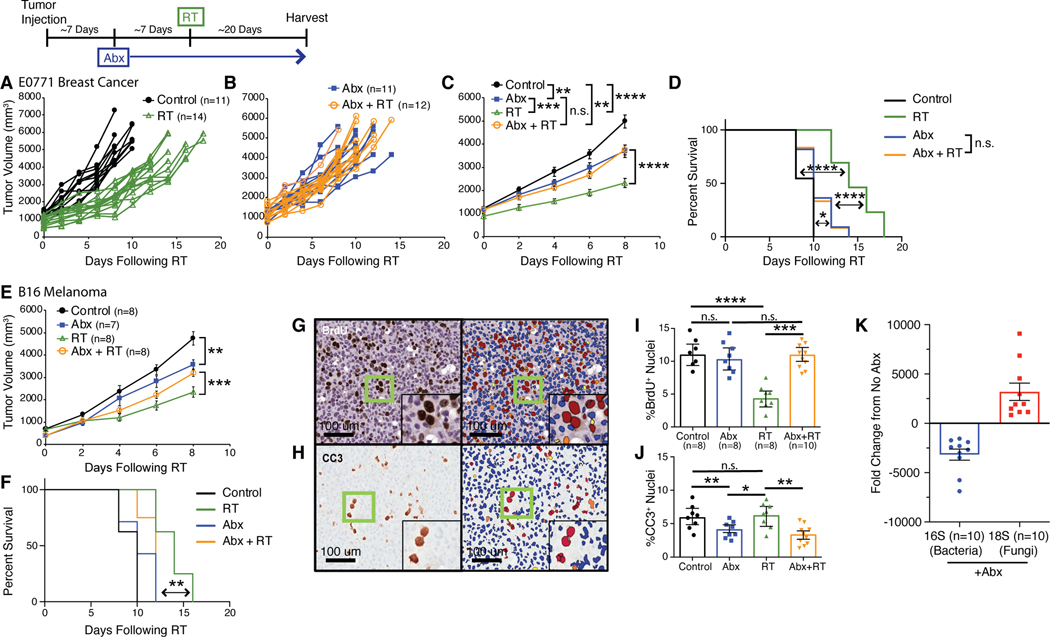
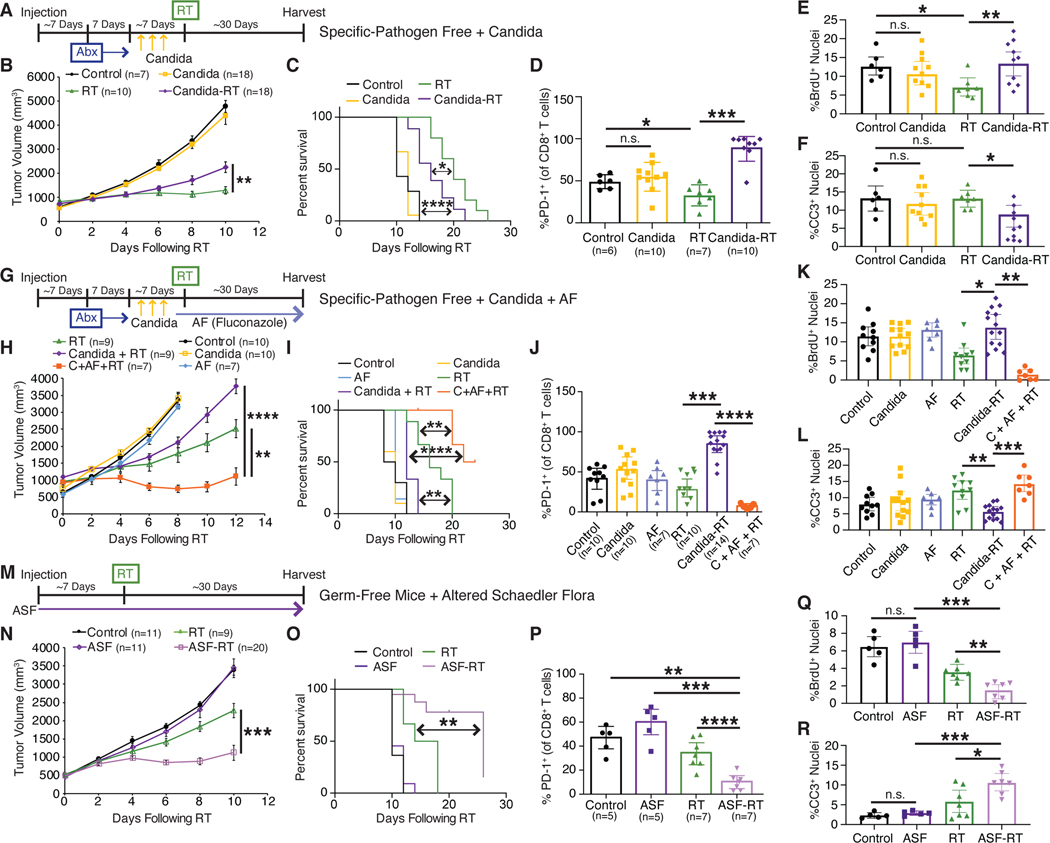
To determine whether commensal bacteria or fungi alter the effect of radiation therapy (RT), we employed an orthotopic syngeneic murine model of breast cancer treatment with focal irradiation (Shiao et al., 2015b). We injected murine breast tumor cells (E0771) into the mammary gland of C57BL/6 mice housed in a specific pathogen-free (SPF) environment. Tumors were then focally irradiated with a single fraction of 16 Gray (Gy) using a high precision small animal irradiator with a beam arrangement designed to avoid delivering radiation to the gut (Figure S1A). This dose was selected empirically from a dose response curve as it reliably slows tumor growth and prolongs survival (Figure S1B). Previous studies have reported that antibiotics decrease the effectiveness of certain chemotherapies such as oxaliplatin (Iida et al., 2013) and cyclophosphamide (Viaud et al., 2013) in murine models of lymphoma, sarcoma and melanoma, and we investigated if this was similarly true for radiation therapy and breast cancer. To test this, we allowed tumors to reach a diameter of >5 mm at which time mice received an antibiotic (Abx) cocktail of ampicillin, imipenem, cilastatin and vancomycin administered continuously until the end of the experiment. Following initiation of Abx treatment, we allowed the tumors to grow to 10 mm in diameter (~5–7 days) at which point they were irradiated. We found that treatment with Abx prior to RT reduced the efficacy of the treatment as determined by the failure of RT to delay tumor growth (Figures 1A-1C) or prolong survival time (Figure 1D). We observed similar decreases in the efficacy of RT in a model of melanoma where mice bearing subcutaneous B16 melanoma tumors receiving Abx showed faster tumor growth (Figure 1E) and decreased survival time (Figure 1F). Other antibiotic combinations similarly decreased the efficacy of RT (Figure S1C-S1F). Interestingly, mice administered Abx alone also exhibited slower tumor growth compared to controls (Figure 1B-1C) which may be a result of alterations in digestion that occur following microbiome depletion leading to metabolic alterations such as reduced serum glucose and short-chain fatty acids that serve to decrease tumor growth (Zarrinpar et al., 2018).
Fig. 1. Bacterial depletion reduces the tumor response to radiation.
Orthotopic E0771 mammary tumors (A-D) were treated with antibiotics (Abx) for one week prior to being treated with localized kV radiation (16 Gy) (RT). Both individual tumors (A, B), mean tumor burden ± SEM (C) and survival (D) for the indicated treatments were assessed every 3 days until endpoint. Antibiotics (Abx) were ampicillin, imipenem, cilastatin and vancomycin. Mean tumor burden/animal ± SEM and survival were also assessed in subcutaneous B16 melanomas (E, F) treated with antibiotics (Abx) for one week prior to being treated with localized kV irradiation (16 Gy). Antibiotics (Abx) were ampicillin, imipenem, cilastatin and vancomycin. E0771 mammary tumors were harvested at one week following RT and stained for bromodeoxyuridine (BrdU) and cleaved caspase 3 (CC3) to assess for proliferation and cell death respectively (G, H, left panels). Slides were scanned using the Aperio slide scanner and analyzed using the ScanScope nuclear algorithm included in the Aperio software package (G, H, right panels) to quantitate the number of BrdU (I) and CC3 (J) positive cells. Significance was determined by two-way ANOVA with post-hoc testing for tumor growth, Log-Rank test for the Kaplan-Meier survival curves and Student’s t-test with Welch’s correction for the immunohistochemistry (IHC). DNA from fecal pellets obtained from Abx treated mice were examined by quantitative PCR for bacterial 16S or fungal 18S ribosomal DNA and the fold change from untreated mice with standard error was calculated (K). Numbers (n) for each experiment are listed on the figure and are pooled data from at least two independent experiments. For all figures, significance is shown as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To better understand how Abx reduced the efficacy of RT, we examined the impact of Abx on tumor cell division and cell death following RT. We stained tumor sections collected from mice that were treated with Abx, RT or both for bromodeoxyuridine (BrdU) which was injected thirty minutes prior to harvesting to assess for the number of dividing cells within in a tumor (Figure 1G). As expected, RT reduced the amount of cell division substantially (Figure 1I). However, treatment with Abx restored the number of proliferating cells to pre-treatment levels. Examination of the number of dying cells using cleaved caspase 3 (CC3) revealed that in addition to increasing the number of proliferating cells, depletion of the bacterial microbiome led to reduced cell death (Figures 1H and 1J). These data support the observation that Abx treatment reduces the overall antitumor efficacy of RT.
Depletion of commensal fungi enhances the efficacy of radiation therapy
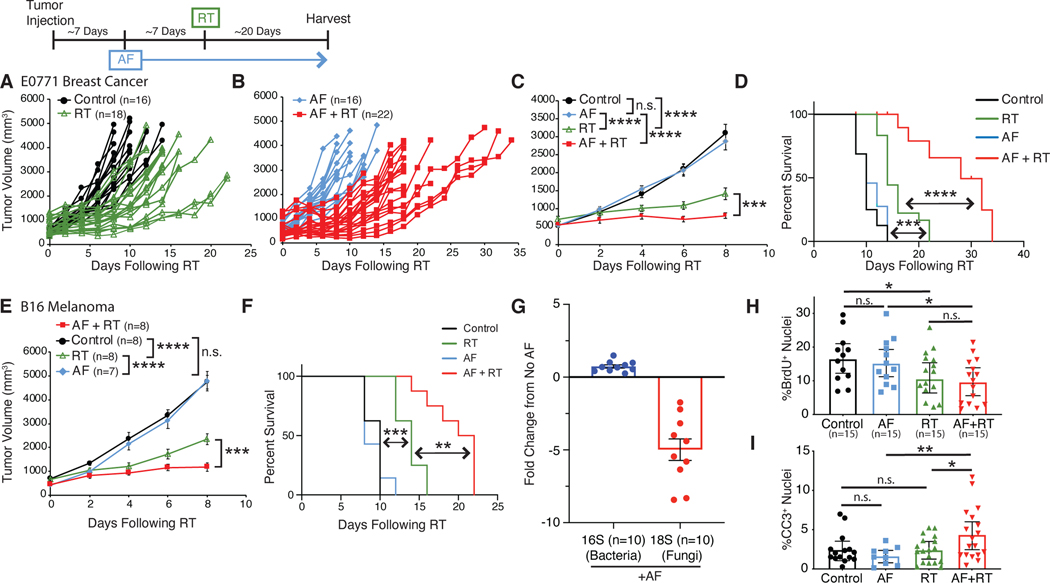
While verifying that our antibiotic treatment effectively depleted intestinal bacteria, we noted that intestinal fungi were, instead, expanded. Quantitative PCR (qPCR) of microbial rDNA revealed that bacteria were depleted over 2000-fold, while at the same time, fungi expanded over 2000-fold (Figure 1K). This led us to wonder whether the effects of antibiotics were due solely to depletion of bacteria or were, alternatively, due to expansion of fungi. To determine whether intestinal fungi impacted the response to RT we treated SPF mice with the antifungal antibiotic (AF) fluconazole (Figures 2A-2D), or 5-fluorocytosine (Figures S2A-D) and found that, in contrast to antibacterial treatment, antifungal treatment enhanced the ability of RT to delay tumor growth and improved survival in both mammary tumors (Figures 2A-D and S2A-D) and melanoma (Figures 2E and 2F). qPCR showed that AF treatment had limited impact on bacterial levels, but reduced commensal fungal levels as expected (Figure 2G).
Fig. 2. Fungal dysbiosis enhances the tumor response to radiation.
Orthotopic E0771 mammary tumors (A-D) were treated with antifungals (AF) for one week prior to being treated with localized kV irradiation (16 Gy). Both individual tumors (A, B), mean tumor burden ± SEM (C) and survival (D) were treated with their indicated treatments and assessed every 3 days until endpoint. The antifungal (AF) was fluconazole with similar results observed using 5-fluorocytosine (Supplemental Figure S2). Subcutaneous B16 melanomas (E, F) were treated with antifungals (AF) for one week prior to being treated with localized kV irradiation (16 Gy). Total tumor burden/animal was assessed every 3 days until endpoint. Mean tumor burden ± SEM (E) and survival (F) with the indicated treatments is shown. DNA from fecal pellets obtained from AF treated mice were examined by quantitative PCR for bacterial 16S and fungal 18S ribosomal DNA and the fold change from untreated mice with standard error was calculated (G). E0771 tumors from mice were harvested at one week following RT and stained for bromodeoxyuridine (BrdU) and cleaved caspase 3 (CC3) to quantitate the number of BrdU (H) and CC3 (I) positive cells to assess for proliferation and cell death respectively. Significance was determined by two-way ANOVA with post-hoc testing for tumor growth, Log-Rank test for the Kaplan-Meier survival curves and Student’s t-test with Welch’s correction for the IHC. Numbers (n) for each experiment are listed on the figure and are pooled data from at least two independent experiments. For all figures, significance is shown as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To evaluate whether the enhanced reduction in tumor growth after RT combined with AF treatment is due to changes in rates of tumor cell division or tumor cell death, we again analyzed tumors from treated mice using immunohistochemistry for BrdU (Figure 2H) or CC3 (Figure 2I) to assess the number of dividing and dying cells respectively. AF-treated mice showed no change in tumor cell proliferation following RT, however AF treatment significantly increased levels of tumor cell death following RT. Together, these findings suggest that commensal bacteria enhance the efficacy of RT, whereas intestinal fungi limit the tumor response to RT.
Commensal fungi suppress antitumor immunity whereas commensal bacteria are required for effective antitumor immunity following RT
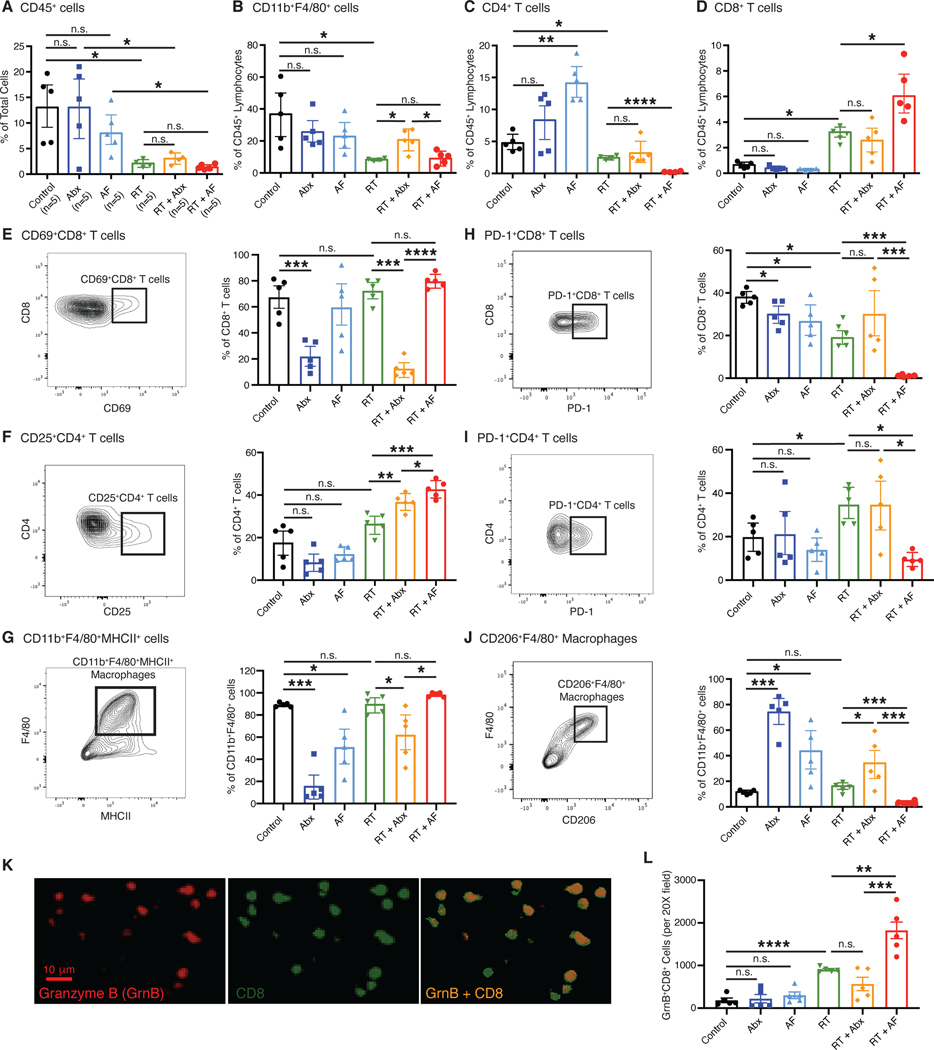
To better understand how commensal bacteria and fungi influence the antitumor immune responses elicited by RT, we digested tumors from Abx- and AF-treated mice one week following RT into single-cell suspensions, isolated CD45+ leukocytes and profiled them using multiparametric flow cytometry (Figure S3A). Examining immune populations as a whole, we found that the frequency and numbers of intratumoral CD45+ leukocytes decreased significantly following RT, and this was not impacted by treatment with Abx or AF (Figures 3A, S3B). Abx treatment was associated with an increase in the frequency and numbers of CD11b+F4/80+ tumor-associated macrophages after RT (Figure 3B, S3C). The percentages and numbers of CD4+ T cells (Figure 3C, S3D) or CD8+ T cells following RT (Figure 3D, S3E) was not altered by Abx treatment. Consistent with the opposing effect of Abx and AF on tumor growth when combined with RT, AF treatment with RT (AF + RT) led to significantly reduced frequency and numbers of CD4+ T cells (Figure 3C, S3D) and increased frequency and numbers of CD8+ T cells (Figure 3D, S3E) compared to RT alone. This is consistent with the observed increase in cell death in the tumors following combined treatment with AF and RT. Importantly, AF with RT treatment did not change the number of activated CD69+CD8+ T cells and CD25+CD4+ T cells (Cibrian and Sanchez-Madrid, 2017) (Figure 3E, S3F, 3F, S3G) nor the number of activated CD11b+F4/80+MHCII+ macrophages (Figure 3G, S3H) when compared to RT alone, all consistent with the idea that AF treatment does not alter the ability of RT to elicit full immune activation. Most notably, we observed that the addition of AF led to a dramatic reduction in PD-1+CD8+ T cells, PD-1+CD4+ T cells and CD206+F4/80+ suppressive macrophages, consistent with a more vigorous cytotoxic T cell response when fungi are depleted (Figures 3H, 3I, 3J, S3I, S3J, S3K). Conversely, Abx treatment led to increased frequency of CD206+F4/80+ macrophages (Figure 3J). The cytotoxic capacity of the CD8+ T cells was also evaluated by looking at the levels of Granzyme B (GzmB) by quantitative immunofluorescence (IF) (Figure 3K) which revealed that treatment with antifungals with RT led to higher numbers of GzmB-expressing CD8+ T cells (Figure 3L). To determine the contribution of CD8+ T cells and macrophages to the effect of combining AF and RT, depletion experiments were undertaken using depleting antibodies to CD8 to look at CD8+ T cells and CCL2 to look at macrophages (Figure S4). These depletion studies revealed that the enhanced efficacy of AF when combined with RT depends on both CD8+ T cells (Figure S4A and B) and macrophages (Figure S4C and D) and the combined effect of AF and RT on tumor growth and survival time were significantly abrogated with depletion of either cell population. Together, we interpret these data to suggest that bacteria are important for the generation of activated T cells following RT, while commensal fungi modulate the immunosuppressive tumor microenvironment through their combined effects on macrophages and T cells.
Fig. 3. Depletion of bacteria, but not fungi, decreases antitumor immunity.
Irradiated E0771 mammary tumors were harvested from mice and dissociated at one week following radiation (RT). CD45+ cells were magnetically isolated and the resulting CD45+ cells stained with fluorescent antibodies as described in Materials and Methods and analyzed by flow cytometry. (A) Total leukocytes (CD45+ cells), (B) CD11b+F4/80+ macrophages, (C) CD4+ T cells and (D) CD8+ T cells were assessed following antibiotic or antifungal treatment. Both activated CD69+CD8+ T cells (E), CD25+CD4+ T cells (F) and CD11b+F4/80+MHCII+ macrophages (G) and immunosuppressive PD-1+CD8+ T cells (H), PD-1+CD4+ T cells (I) and CD11b+F4/80+CD206+ macrophages (J) were examined. Tissue sections were also examined for Granzyme B (GzmB) expressing CD8+ cells (K) and (L). Significance was determined by Student’s t-test with Welch’s correction. Antibiotics (Abx) were ampicillin, imipenem, cilastin and vancomycin. Fluconazole was the antifungal agent (AF) used for these experiments. n=5 per group.
Depletion of commensal bacteria leads to expansion of Saccharomycetes fungi
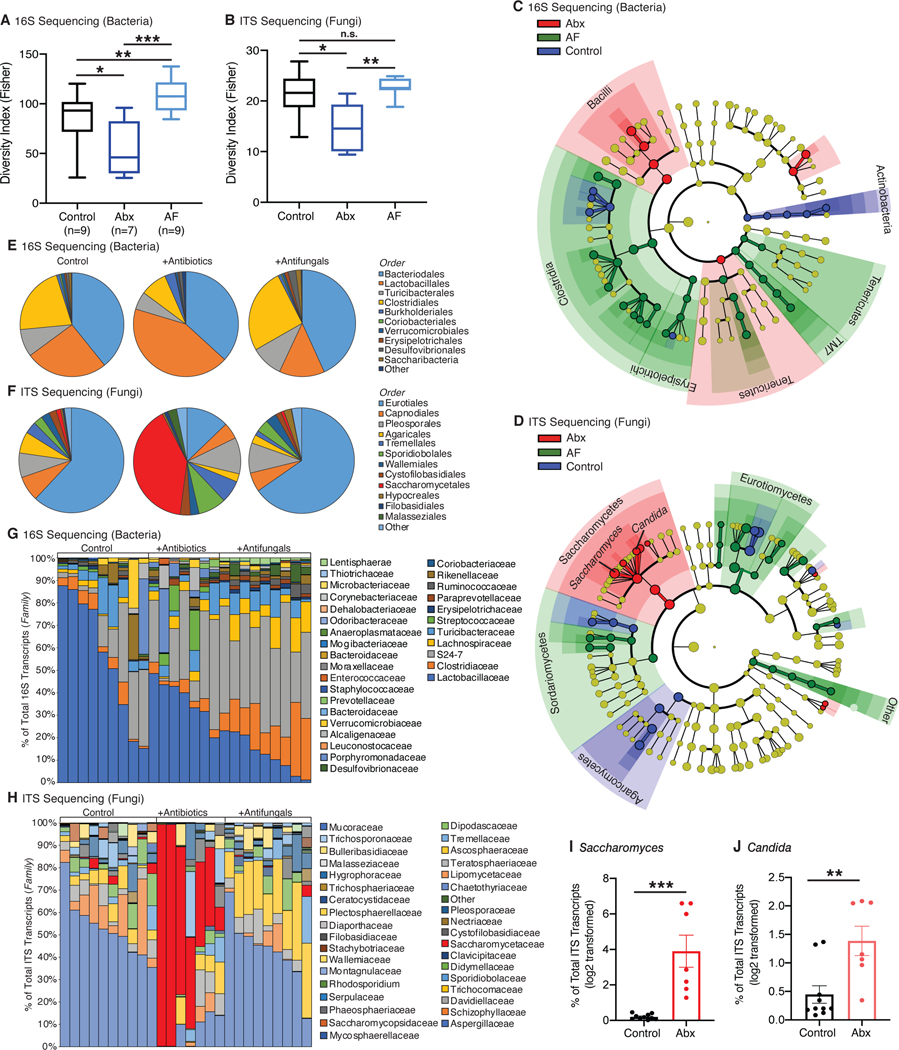
At baseline, SPF mice contain a diverse mixture of both bacteria and fungi (Table S1) and to better understand the nature of the dysbiosis influencing the efficacy of RT in our experiments, we analyzed the changes in the composition of the intestinal bacterial and fungal populations following one week of treatment with either Abx or AF. While Abx treatment led to drastic reduction in the intestinal bacterial burden (Figure 1K), AF treatment had no impact on intestinal bacterial burden (Figure 2G). Examination of the diversity of the samples revealed that Abx decreased the alpha diversity of both bacteria and fungi whereas AF treatment increased bacterial diversity with limited impact on fungal diversity (Figures 4A and 4B). Of the bacteria remaining after Abx treatment, 16S rRNA gene sequencing revealed significant reductions in relative representation across multiple families, although the effect was most pronounced on members of the Clostridiales order (Figures 4C and 4E). Other bacterial orders including Lactobacillales and Burkholderiales were relatively more present (Figures 4E and 4G, Table S2) consistent with reports from other groups (Sivan et al., 2015; Viaud et al., 2013). However, the most striking change following administration of Abx occurred in the intestinal fungal populations, which expanded dramatically (Figure 1K). Fungal ITS-1 gene sequencing revealed a significant overgrowth of Saccharomycetales order following administration of Abx (Figures 4D and 4F) such that they become the dominant order in the majority of cases (Figure 4H, Table S3). Closer examination of the Saccharomycetales order revealed increases in specific genera including Saccharomyces (Figure 4I) and Candida (Figure 4J).
Fig. 4. Antibiotic treatment leads to overgrowth of specific fungi.
DNA from fecal samples was obtained from mice treated with antibiotics (Abx) or antifungals (AF). 16S and ITS sequencing was then performed to identify the types of bacteria (A, C, E, G) and fungal (B, D, F, H) species respectively. Samples were analyzed at the genus level for alpha diversity (Fisher’s alpha index) for both bacterial (A) and fungi (B). Data were analyzed by linear discriminant analysis effect size (LEfSe) method (Segata et al., 2011), and the results displayed as bacterial (C) and fungal (D) cladograms. Microbial prevalence was examined at the order level and pooled for analysis (E, F) or not pooled for visualization of individual samples (G, H). Sequencing data were then examined at the genus level for Saccharomyces (I) and Candida (J). The percentage of ITS transcripts for each genus was log2 transformed and plotted. Antibiotics (Abx) were ampicillin, imipenem, cilastatin and vancomycin. Fluconazole was the antifungal (AF) used for these experiments. Numbers (n) for each experiment are listed on the figure and are pooled data from at least three independent experiments.
To directly investigate the consequences of overgrowth of a Saccharomycetales order on the efficacy of radiation therapy, we evaluated the effects of Candida albicans, the most common of the commensal Saccharomycetales found in humans (Limon et al., 2017) and one that has long been associated with antibiotic use (Seelig, 1966). Using an approach previously described to promote robust C. albicans colonization in mice (Noverr et al., 2004) with limited impact on the bacterial microbiome in the absence of colonization (Erb Downward et al., 2013), we treated tumor-bearing mice with a short course of the antibiotic cefoperazone followed by administration of three doses of C. albicans (Figure 5A). We verified that mice were robustly colonized with C. albicans (Figure S5A), and once tumors reached 10 mm in diameter, they were irradiated with 16 Gy of ionizing RT. We followed tumor growth in these mice post-irradiation and found that, like treatment with antibiotics, overgrowth of C. albicans in the gut led to less delay in tumor growth following RT (Figure 5B) and worse overall survival time (Figure 5C). Interestingly, it did not appear to influence the magnitude of the initial response to tumor growth, but rather led to significantly accelerated tumor regrowth (Figure 5B). Examination of the immune compartment in mice robustly colonized with C. albicans revealed an increased ratio of PD-1+CD8+ T cells indicating a more immunosuppressive tumor microenvironment allowing for increased tumor regrowth (Figure 5D). BrdU and cleaved caspase 3 analysis revealed that Candida colonization led to increased cell division and less cell death following RT consistent with the faster tumor regrowth (Figure 5E and 5F). The deleterious effect of C. albicans colonization was reversed by treatment with fluconazole (Figure 5G) which significantly reduced Candida levels (Figure S5B) and, in so doing, restored tumor responsiveness to RT (Figure 5H and 5I) and improved survival times compared to Candida colonized mice (Figure 5I). Impressively, it also dramatically reduced the number of PD-1+CD8+ T cells (Figure 5J) and the number of dividing cells (Figure 5K) and increased the amount of cell death (Figure 5L). Thus, the results from mice with C. albicans overgrowth parallel the experience with Abx treatment alone where the tumor response to RT is blunted. This suggests that one mechanism by which Abx can influence the antitumor immune response is through its reciprocal effect on the fungal microbiome.
Fig. 5. Fungal-free mice show enhanced efficacy of RT whereas overgrowth of fungus leads to reduced efficacy.
(A) Experimental timeline for SPF mice with orthotopic E0771 mammary tumors treated with cefoperazone (Abx) and then colonized by gavage with Candida. Tumor growth (B) and survival (C) were assessed every three days following radiation (RT). (G) Experimental timeline for antifungal treatment (fluconazole) to suppress colonized C. albicans. Tumor growth (H) and survival (I) were assessed every three days following RT. (M) Experimental timeline for orthotopic E0771 tumors implanted in either normal specific-pathogen free (SPF) mice or germ-free mice colonized with fungal-free altered Schaedler flora (ASF). Tumor growth (N) and survival (O) were assessed every three days following RT. For all three experiments, the tumor infiltrating lymphocyte populations were analyzed in a separate group of mice at one week following RT by flow cytometry for PD-1 expression on cytotoxic CD8+ T cells (D, J, P). Cell proliferation (BrdU) (E, K, Q) and apoptosis (cleaved caspase 3, CC3) (F, L, R) were also assessed by quantitative immunohistochemistry as described in the Methods. Significance was determined by two-way ANOVA with post-hoc testing for tumor growth, Log-Rank test for the Kaplan-Meier survival curves and Student’s t-test with Welch’s correction for the IHC. Numbers (n) for each experiment are listed on the figure and are pooled data from at least three independent experiments. For all figures, significance is shown as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Based on these experiments we hypothesized that the efficacy of RT could be further optimized in a system that contained no intestinal fungi. To examine the effect of fungi in the gut we created a fungal-free gnotobiotic model. To generate this system, germ-free mice were colonized with a defined set of bacteria known as altered Schaedler flora (ASF). Absence of fungi and full colonization by ASF bacteria was verified using PCR of fecal pellets where fungal levels in ASF mice mirrored those of germ-free mice while having bacterial levels near SPF mice (Figure S5C and S5D). Mice were then subsequently maintained in isolators in a gnotobiotic facility. We implanted E0771 breast cancer cells and, when tumors reached approximately 10 mm in diameter, we delivered focal irradiation and followed tumor growth and survival (Figure 5M). Tumors in ASF mice grew similarly to control SPF mice, however, as observed with antifungal treated mice, we found a significant delay in tumor growth following RT for the fungal-free ASF mice (Figure 5N) with a concomitant increase in overall survival time (Figure 5O). Corresponding with the tumor growth and survival, histologic examination of the tumors from these mice showed reduced cell proliferation (Figure 5Q) and increased cell death (Figure 5R). When tumors of these mice were further examined by flow cytometry, a significant reduction in PD-1+CD8+ T cells was again observed consistent with the enhanced response to RT (Figure 5P). Additional studies carried out in germ-free mice monocolonized with Candida showed less delay in tumor growth and reduced survival time following RT compared to similarly irradiated SPF mice (Figure S5E and S5F). Together with the Candida colonization experiment, data from the fungal-free ASF mice and Candida monocolonized germ-free mice indicate that fungi play a significant role in regulating the antitumor immune response elicited by RT. Further, we hypothesize that Abx treatment impacts the efficacy of RT in part through its previously unappreciated effect on the commensal fungal population.
Dectin-1 is expressed in human breast tumors and higher expression is associated with worse survival
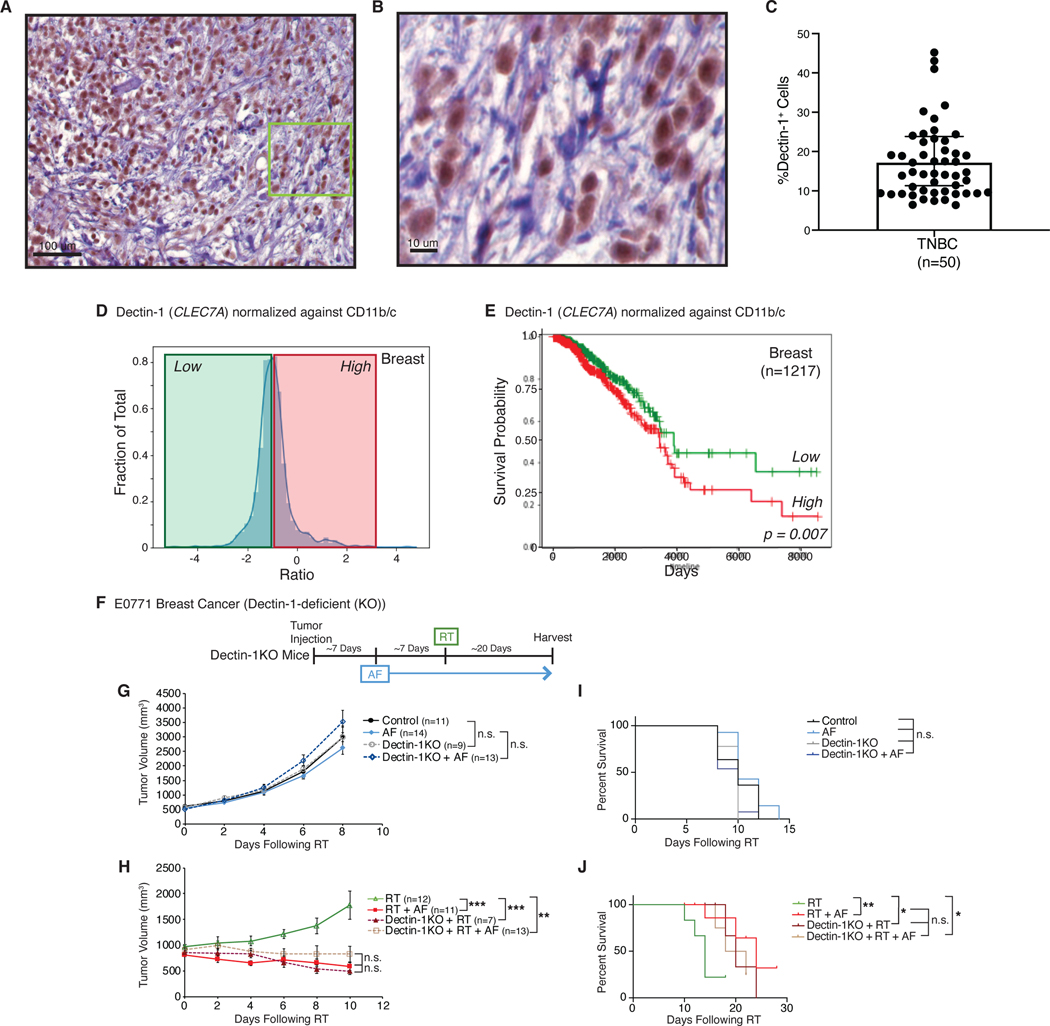
Recognizing the importance of commensal fungi in regulating the response to radiation, we sought to explore the impact of innate immunity to fungi in human cancers. We examined breast tumors for the presence of C-type lectin receptor Dectin-1 (CLEC7A). Dectin-1 specifically recognizes β-glucans found in fungal cell walls and is one of the primary sensors of pathogenic and commensal fungi in humans (Geijtenbeek and Gringhuis, 2009; Wheeler et al., 2017). Mice and humans who have absent or decreased Dectin-1 signaling exhibit increased susceptibility to fungal infections and disseminated fungal disease (Netea et al., 2015). Immunohistochemistry from human breast cancer biopsies showed extensive intratumoral Dectin-1 staining (Figures 6A and 6B), and quantitation of Dectin-1 expression from a cohort of triple-negative breast cancer patients (Table S4) revealed that Dectin-1 is present and expressed in human triple-negative breast tumors (Figure 6C). Previous reports indicated that Dectin-1 expression largely occurs on immune cells such as macrophages and dendritic cells (Brown, 2006; Wheeler et al., 2017). Based on this, we hypothesized that Dectin-1 expression might influence the response to cancer therapy through its ability to sense commensal fungi. We examined the Cancer Genome Atlas (TCGA) for mRNA expression of the C-type lectin receptor Dectin-1 (CLEC7A) in breast tumors and found a wide distribution of Dectin-1 (Figure 6D). Understanding that breast tumors contain significant and variable numbers of phagocytes, we normalized Dectin-1 expression to expression of CD11b (macrophages) and CD11c (DC) and examined whether tumors with macrophages expressing more or less Dectin-1 correlate with survival. Levels that were above the mean (high) predicted for worse survival (Figures 6E). We observed similar findings in the TCGA melanoma cohort looking at Dectin-1 expression normalized to the macrophage marker CSF-1R due to low expression of CD11b (Figure S6A). To test the hypothesis that Dectin-1 sensing of commensal fungi reduces the efficacy of RT, we utilized Dectin-1-deficient mice and found that without Dectin-1 the effect of adding AF to RT was lost (Figure 6H and 6J). Furthermore, Dectin-1-deficiency also enhanced the response to RT similar to administration of AF. Dectin-1-deficiency alone, similar to AF administration by itself, had no impact on tumor growth (Figure 6G and 6I). These data support the notion that increased signals from commensal fungi through the fungal receptor Dectin-1 mediates the response to RT.
Fig. 6. Dectin-1 (CLEC7A) predicts for survival in breast cancer and mediates the effect of fungal dysbiosis on RT.
Human triple-negative breast cancer (TNBC) samples were stained for Dectin-1 (A, B) and the number of positive cells quantitated (C). Expression dataset for all breast cancers from the Cancer Genome Atlas (TCGA) was analyzed for levels of Dectin-1. Dectin-1 levels were then normalized against CD11b and CD11c expression (D) and the elevated normalized Dectin-1 levels divided into high (red line) and low (green line) cohorts. Survival for these cohorts was then compared on Kaplan-Meier plots (E). Orthotopic E0771 mammary tumors were then placed in Dectin-1−/− mice and treated with antifungals (AF) for one week prior to being treated with localized kV irradiation (16 Gy). Tumor growth (G, H) and survival (I, J) were assessed every three days following radiation (RT). Significance was determined by two-way ANOVA with post-hoc testing for tumor growth and Log-Rank test for the Kaplan-Meier survival curves. Numbers (n) for each experiment are listed on the figure and are pooled data from three independent experiments. For all figures, significance is shown as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
The gut microbiota impacts systemic inflammation in multiple disease states largely by influencing the magnitude and character of an immune response (Blander et al., 2017; Limon et al., 2017). Our study highlights the potential for manipulating the gut microflora to enhance the antitumor immune response to radiation therapy. The mechanisms by which the gut microbiota regulates the response to other cancer therapeutic agents including chemotherapy and immunotherapy are only beginning to be elucidated (Daillere et al., 2016; Gopalakrishnan et al., 2018; Iida et al., 2013; Matson et al., 2018; Routy et al., 2017; Sivan et al., 2015; Vetizou et al., 2015; Viaud et al., 2013). Several bacterial subsets including those from the phylum Bacteroidetes, genera Bifidobacterium and Akkermansia have been implicated in regulating antitumor immunity induced by oxaliplatin (Iida et al., 2013), cyclophosphamide (Daillere et al., 2016; Viaud et al., 2013) and PD-1/PD-L1 blockade (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2017; Sivan et al., 2015) although unifying themes for how these bacteria regulate antitumor immunity have not yet emerged. Our work reveals a previously unappreciated role for intestinal fungi in regulating antitumor immunity. Previous studies on the influences of the microbiome on cancer therapy have typically not examined fungal populations, and we postulate that some of the reported influences of altered bacterial microbiota on therapy might be in part attributable to unreported corresponding changes in fungal populations.
Commensal fungi coexist with bacteria in the gut and have been observed to regulate immunity in the intestine (Iliev et al., 2012; Limon et al., 2019), lung (Skalski et al., 2018; Wheeler et al., 2016) and tumors (Aykut et al., 2019). Absence of fungal signaling led to increased inflammation in a model of acute colitis (Iliev et al., 2012) and supplementation with specific fungi including Candida, Malassezia, or Saccharomyces exacerbated disease (Chiaro et al., 2017; Iliev et al., 2012; Limon et al., 2019). Supplementation of gut fungi with Candida, Wallemia, Epicoccum and Aspergillus spp. led to increased allergic airway disease in murine models (Kim et al., 2014; Noverr et al., 2004; Skalski et al., 2018; Wheeler et al., 2016). A recent study in pancreatic ductal adenocarcinoma found that these tumors contain a large amount of the fungi Malassezia spp. and that host sensing of intratumoral fungi via mannose-binding lectin regulates tumor progression (Aykut et al., 2019). These studies all suggest that fungi can regulate systemic immunity and that in certain settings fungi can help shape the nature of an inflammatory response to favor Th2 immunity primarily through sensing of fungal-specific products by pattern recognition receptors including C-type lectin receptors such as Dectin-1, mannose binding receptors and NOD-like receptors (Limon et al., 2017). We noted that the Dectin-1 polymorphism Y238X, which creates a premature stop codon in Dectin-1(Plantinga et al., 2009), is linked to expression levels of Dectin-1 in breast tissue from the Genotype-Tissue Expression (GTEx) Database, suggesting that future targeted studies might find that the polymorphism is associated with increased survival (Figure S6B). By promoting Th2 differentiation, commensal fungi through sensors such as Dectin-1 may inadvertently contribute to cancer progression and therapeutic resistance by promoting a suppressive immune microenvironment most of which is associated with Th2 signals in the tumor (DeNardo et al., 2009; Shiao et al., 2015b). Unlike bacteria, fungi have been shown to induce Th2 immunity in addition to Th1 immunity (Wuthrich et al., 2012), which may underlie the enhanced antitumor cytotoxicity observed following RT when fungi are depleted or absent.
Commensal bacteria can shape antitumor immunity both directly through regulation of immune cells circulating through the gut immune compartment (Gur et al., 2015), but also indirectly through bacterial regulation of metabolites and release of bacterial products (Helmink et al., 2019; Yachida et al., 2019). We hypothesize that fungi behave similarly as we and others have observed that commensal fungi directly regulate intestinal mucosal immunity (Iliev and Cadwell, 2021; Iliev et al., 2012; Limon et al., 2018), but may also release fungal components systemically through the circulation. One recent study has suggested that Malassezia spp. can be found within pancreatic tumors and may promote tumorigenesis (Aykut et al., 2019). However, though bacteria have been described in breast tumors (Nejman et al., 2020), fungi have not yet been identified in tumors outside the gastrointestinal tract. There is evidence that fungal products are present systemically. For example, circulating levels of β-glucan, the main ligand for Dectin-1, can be readily detected in HIV patients and is correlated with some immunosuppressive immunity including regulatory T cell numbers and indolemine-2,3-dioxygenase-1 activity (Mehraj et al., 2020), although the baseline level released from commensal fungi is unclear. Fungi also have complex metabolisms, producing unique metabolites that can be utilized by the host organism, but also may shape the bacterial composition of the microbiome (Keller, 2019). We demonstrate here that alterations in the gut mycobiome can impact distant RT-induced antitumor immune responses by altering levels of intratumoral immunosuppression following RT in a Dectin-1-dependent manner. Future studies examining the fungal metabolome and its impact on systemic immunity and commensal bacteria will help further unravel the complex web of interactions between systemic host immunity and its different commensal communities.
In sum, we introduce here the concept that the response of tumors to radiation can be regulated by the microbiome and that both the bacterial and fungal constituents of the gut microbiota impact antitumor immunity. Our data underscore the importance going forward of fully evaluating the diversity of gut microorganisms in pursuing a deeper understanding of the mechanisms by which gut microbiota influence cancer therapies. The study also suggests a new microflora-based therapeutic target to enhance the efficacy of cancer therapies.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, Stephen L. Shiao MD, PhD (stephen.shiao@cshs.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Sequence reads for 16S and ITS from this study are available from the Sequence Read Archive under the project ID “PRJNA496065”.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human studies
The mRNA expression profiles and corresponding clinical information for breast cancer and melanoma patients were downloaded from the Cancer Genome Atlas (TCGA). After removing patients with incomplete information, 1217 breast and 472 melanoma patients remained for analysis. For breast cancer, we normalized the expression of CLEC7A to the approximate number of macrophages and dendritic cells by dividing the amount of CLEC7A by the sum of the relative expression value of CD11b (ITGAM) and CD11c (ITGAX) found in each tumor. For melanoma, due to the low amount of CD11b in many of the samples, we instead normalized to the CSF1R as an alternative approximation of the number of macrophages. Relative expression of normalized CLEC7A for each patient was then log2 transformed, and values above the mean were included in the high group and values below the mean were included in the low group. The survival of high and low expression groups was then plotted on a Kaplan-Meier curve and significance determined using the Log-Rank test.
To look at the association between a known Dectin-1 polymorphism Y238X, which creates a premature stop codon in Dectin-1, and Dectin-1 expression, the Genotype-Tissue Expression (GTEx) Database (gtexportal.org) was examined for the single-nucleotide polymorphism (SNP) rs16910526 in breast tissue (n=30,971 samples across 34 cohorts). At this SNP loci, the major (normal) and minor (premature stop codon) allele for the SNP were identified and patients separated into three groups: homozygous major allele (AA), heterozygous (AC) and homozygous minor allele (CC). Normalized expression of CLEC7A was then plotted for each group. To obtain normalized gene expression for each tissue, raw RNASeq read counts were normalized by gene length and then normalized to a library size of 1 × 106 transcripts per tissue.
For Dectin-1 expression, 50 formalin-fixed paraffin embedded cases of triple-negative breast cancer were obtained from the Cedars-Sinai Pathology Department with accompanying de-identified patient data. Appropriate blocks containing tumor tissues were selected with the assistance of a breast pathologist (S. Bose). A summary of the patient data characteristics can be found in Table S4. Slides were cut from these blocks onto coated glass microscope slides (VWR International) and were processed as described in the Immunohistochemistry section.
Animal studies
C57BL/6 mice 6–10 weeks of age were used for most experiments and purchased from Jackson Laboratories. To generate tumors, 1 × 107 E0771 murine breast cancer cells, which are syngeneic to C57BL/6, were injected in the mammary gland or 1 × 107 B16 murine melanoma cells were injected subcutaneously on the flank (Crosby et al., 2018; Giavazzi and Decio, 2014). The colony of Altered Schaedler Flora (ASF)-colonized mice was generated by colonizing germ-free mice (C57BL/6, Taconic) with ASF stool (Taconic). Colonization was verified by PCR analysis of the ASF component bacteria, and absence of fungi was assessed by PCR as previously described (Limon et al., 2019). ASF mice were bred and maintained in germ-free microisolator units (CBC). For in vivo experiments, only female mice were used for these experiments. Mice were housed in specific pathogen-free conditions in the Cedars-Sinai animal facility, and all animal experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals on a protocol approved by the institutional animal use and care committee of the Cedars-Sinai Medical Center (IACUC Protocol No. 4841).
For analysis of the experimental murine data, all experiments with comparable treatment protocols and evaluable data were pooled. Mice with insufficient or missing data were censored for the analysis. For tumor growth, all measurements at each time point for a given treatment group were averaged and a standard error was calculated. Comparisons between groups were then done using ANOVA with post-hoc analysis. Survival curves were generated using the IACUC mandated endpoint of when a tumor reached >3500 mm3 (or > 20 mm in any one dimension). Mice who reached experimental endpoint (harvested for analysis), but not survival endpoint were censored from that time point forward. Comparisons between groups were then performed using a Log-Rank test. Standard star notations (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001) were used to designated level of significance for all comparisons.
Fungi
Candida albicans (ATCC 90028) yeasts were grown with shaking overnight at 37°C in Sabouraud dextran broth (SDB). For in vivo Candida colonization experiments, tumor-bearing mice from both the control and Candida-colonized groups approximately 1 week following injection were started on Cefoperazone (0.5 mg/mL) in the water for up to 1 week. Fungi were grown as described above washed 3 times with 0.2M sodium bicarbonate and resuspended at a concentration of 1×109 yeasts/mL of 0.2M sodium bicarbonate. Mice in the Candida-colonized group were dosed by oral gavage with 1×108 yeast cells at the times indicated while control mice received gavage with sodium bicarbonate only. Colonization was verified by qPCR using pan-Candida primers.
Radiation
Radiation was delivered using the X-RAD SmART (PXi, Precision X-Ray). Mice were anesthetized with isoflurane and then placed inside the shielded cabinet. A computed tomography (CT) scan was then performed to determine the location of the tumor. Once the location was determined on the CT the treatment beams were setup on the mice using the accompanying planning software package (SmART Advanced Treatment Planning System, Precision X-Ray). Opposed tangential beams with an energy of 225 kV were typically utilized to treat the tumor and minimize the dose to normal tissue. Mice were then treated with the planned dose of RT.
Antifungal and Antibiotic Treatments
For antifungal experiments, mice were provided with autoclaved water supplemented with 0.5 mg/mL fluconazole (Sigma-Aldrich PHR1160) or 1 mg/mL 5-fluorocytosine (Sigma-Aldrich) for 1–3 weeks prior to radiation treatment as previously published (Wheeler et al., 2016). Antifungal treatment was continued throughout the course of entire experiment. For antibiotics experiments, autoclaved water supplemented with a mixture of 0.25 mg/mL vancomycin, 1 mg/mL imipenem, 1 mg/mL cilastatin and the 1 mg/mL ampicillin was provided to mice unless otherwise indicated. Antibiotic treatment was continued through the course of the entire experiment.
METHOD DETAILS
Immunohistochemistry
Paraffin-embedded tissue sections were fixed in 10% formalin and sectioned onto coated glass microscope slides (VWR International). Slides were then baked at 60°C for 1 hour. To determine proliferation index mice received intraperitoneal injections of bromodeoxyuridine (BrdU; Sigma-Aldrich) dissolved in PBS (50 μg per g of mouse body weight) 30 min prior to sacrifice. 5.0-mm thick paraffin sections were deparaffinized in xylene, rehydrated in graded ethanol, and subjected to antigen retrieval by steam heating in citrate buffer (10x soln) based antigen retrieval solution (Sigma Aldrich). BrdU-positive cells were detected according to manufacturer’s recommendations using the anti-BrdU antibody (Bu20a, 1:100, ThermoFisher). To detect apoptotic cells, cleaved caspase 3 was visualized on paraffin-embedded tissue sections using a polyclonal rabbit anti-mouse cleaved caspase 3, (1:100, Millipore Sigma). Dectin-1 protein was detected using the polyclonal rabbit anti-human Dectin-1 (1:100, abcam). Non-specific binding was blocked using PBS containing 5% donkey serum (Jackson Laboratories). Sections were incubated with the indicated antibody in 0.5X blocking buffer overnight at 4°C. Sections were then washed in PBS and then incubated with biotinylated donkey anti-mouse (BrdU) or donkey anti-rabbit (cleaved caspase 3, Dectin-1) IgG secondary antibodies (1:200, Jackson Laboratories) for 45 min at room temperature. Slides were subsequently incubated with horseradish peroxidase conjugated avidin complex (ABC HRP Kit, Vector Laboratories) for 30 min, followed by incubation with metal-enhanced 3,3 diaminobenzidine (DAB, Thermo Fisher). Sections were counterstained with methyl green, dehydrated, and mounted with Cytoseal 60 (Thermo Fisher). Slides were then scanned on an Aperio AT Turbo (Leica Biosystems) and analyzed using the nuclear algorithm in the ImageScope software package (Leica Biosystems).
Antibody staining and flow cytometry
Single-cell suspensions were prepared from mammary tumors disassociated by manual mincing and enzymatic digestion using the tumor dissociation kit (Miltenyi Biotech) and the gentleMACS dissociator (Miltenyi Biotech). Digestion mixtures were quenched by adding DMEM containing 10% FBS and then filtered through 0.7 μm nylon strainers (Falcon). The resulting single-cell suspension was then incubated with CD45+ magnetic beads (Miltenyi Biotech) and isolated using the positive selection program on an AutoMACS (Miltenyi Biotech). Cells were then incubated for 10 min at 4°C with rat anti-mouse CD16/CD32 mAb (BD Biosciences) at a 1:100 dilution in PBS containing 1.0% of BSA (Sigma) to prevent nonspecific antibody binding. Subsequently, cells were washed twice in PBS/BSA and incubated for 20 min with 100 μl of fluorophore-conjugated anti-mouse antibodies: CD3ε (145–2C11), CD4 (6K1.5), CD8α (53–6.7), CD11b (M1/70), CD45 (30-F11), CD69 (H1.2F3), CD206 (19.2), F4/80 (BM8) and/or MHCII (M5/114.15.2) (all from eBioscience) followed by two washes with PBS/BSA. Zombie UV Fixable Viability Dye (Biolegend) was added (1:10) to discriminate between viable and dead cells. Additionally, to assay for regulatory T cells, cells were incubated for 20 min with 100 μl of fluorophore-conjugated anti-mouse antibodies: CD3ε (145–2C11), CD4 (6K1.5), CD25 (PC61.5). These cells were then fixed for 20 min with the True Nuclear Transcription Factor Set Fixation solution, washed once in PBS/BSA and incubated for 20 min with anti-mouse FoxP3 antibody (FJK-16s) resuspended in the Perm buffer followed by two washes with PBA/BSA. Data acquisition and analysis were performed on a LSRII (BD Biosciences) using the FlowJo version 10 software (Tree Star).
Immune cell isolation and analysis
Immune cells were isolated from tumors using a dual purification strategy with magnetic purification followed by flow cytometric analysis. Single-cell suspensions from tumors were generated as described above. Total leukocytes were isolated using magnetic bead selection for CD45+ cells according to manufacturer’s specifications (Miltenyi Biotec). A portion of unsorted cells was also analyzed to obtain the percentage of CD45+ cells from the total cell population. Magnetically selected cells were incubated for 10 min at 4°C with rat anti-mouse CD16/CD32 mAb (BD Biosciences) at a 1:100 dilution in PBS/BSA then washed twice in PBS/BSA and incubated for 20 min with appropriate fluorescent primary antibodies which included CD45-APC (30-F11), in addition to CD4 (GK1.1), CD3 (145–2C11), CD206 (19.2), CD11b (M1/70) and/or F4/80 (BM8) (all from eBiosciences) at 1:100 dilution depending on the population to be isolated. Stained cells were then analyzed by flow cytometry on an LSRII using FACSDiva software (BD Biosciences). Gating strategies for these populations is provided in Supplementary Figure 4, but has also been described previously (DeNardo et al., 2011; Shiao et al., 2015a).
QUANTIFICATION AND STATISTICAL ANALYSIS
Microbiota analysis
DNA for rtPCR or fungal and bacterial sequencing was isolated from 1–2 fecal pellets following lyticase treatment, bead beating, and processing using QIAmp DNA mini kit (Qiagen) as previously described using a protocol optimized to lyse fungal cell walls for recovery of fungal DNA (Wheeler et al., 2016). DNA extraction from organ tissue was performed by first homogenizing tissue with 0.5 mm steel beads in TissueLyser II (Qiagen) at frequency 30s−1 × 4.5 min then proceeding per DNA extraction protocol used for stool samples. A primer-probe based detection system with iTaq Universal Supermix (BioRad) was used for FungiQuant (fungal 18S) and all species specific fungal rtPCR assays while the 16S, ASF, and pan-Candida rtPCR was performed using a SYBR Green supermix (BioRad) based assay. TaqMan rtPCR assays were run on an Eppendorf Mastercycler realplex2 machine using the following parameters: initial denaturation step of 95°C for 2 min followed by 45 cycles of 95°C x15s then 60°C x1 min. The SYBR green based rtPCR assay was run on an Eppendorf Mastercycler realplex2 machine using the following parameters: initial denaturation step of 95°C for 10 min followed by 40 cycles of 95°C x15s then 60°C x30s then 72°C x32s. The following primer pair and probe sequences were used for rtPCR assays: FungiQuant F: GGR AAA CTC ACC AGG TCC AG FungiQuant R: GSW CTA TCC CCA KCA CGA FungiQuant probe: 5’-(FAM) TGG TGC ATG GCC GTT (3lABkFQ)-3’ (Liu et al., 2012). 16S F: ACT CCT ACG GGA GGC AGC AGT, 16S R: ATT ACC GCG GCT GCT GGC. Pan-Candida F: GCA AGT CAT CAG CTT GCG TT Pan-Candida R: TGC GTT CTT CAT CGA TGC GA (Zhang et al., 2016).
Fungal ITS1 amplicons were generated in 20 μL PCR reactions using 3 μL of each sample with 35 cycles using Invitrogen Platinum SuperFi DNA Polymerase at an annealing temperature of 48°C using the primers ITS1F (CTTGGTCATTTAGAGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGATGC). Amplicons were then used in the second PCR reaction, using Illumina Nextera XT v2 barcoded primers to uniquely index each sample and 2×300 paired end sequencing was performed on the Illumina MiSeq (Illumina), according to manufacturer’s instructions. Raw data processing and run de-multiplexing was performed using on-instrument analytics as per manufacture recommendations. Sequence reads from this study are available from the Sequence Read Archive under the project ID “PRJNA496065”.
16S sequence data were processed and analyzed as previously described including OTU assignment by alignment with the GreenGenes reference database (release of May 2013) at 97% identity (McHardy et al., 2013). For analysis of ITS1 sequence data, raw FASTQ data were filtered to enrich for high quality reads including removing the adapter sequence by cutadapt v1.4.1 (Martin, 2011), truncating reads not having an average quality score of 20 over a 3 base pair sliding window, removing any reads that do not contain the proximal primer sequence or any reads containing a single N (unknown base) by a custom script. Filtered pair-end reads were then merged with overlap into single reads using SeqPrep v1.1 wrapped by QIIME v1.9.1 (Caporaso et al., 2010).
The processed high-quality reads were firstly aligned to previously observed host sequences (including rRNA, olfactory receptor and uncharacterized genes in human and mouse) to deplete potential contamination, then operational taxonomic unit (OTU) were picked by aligning filtered reads to the Targeted Host Fungi (THF) custom fungal ITS database (version 1.6) (Tang et al., 2015), using BLAST v2.2.22 in the QIIME v1.9.1 wrapper with an identity percentage ≥97%.
Statistical analysis
All experiments were conducted with at least triplicate measurements a minimum of two times unless otherwise stated in the text or figure legends. Tumor growth curves were compared using two-way ANOVA to determine differences among groups for the entire experiment with post-hoc testing of individual pair-wise comparisons using Tukey’s multiple comparison test for significance using GraphPad Prism software v7. Survival curves were compared using the Log-Rank test for significance suing GraphPad Prism software v7. Flow and IHC statistics were analyzed for significance using the Student’s t-test with Welch’s correction in GraphPad Prism software v7 or R version 3.4.0.
Supplementary Material
Supplemental Data Table S2. 16S OTU Counts, Related to Figure 4 (.xls)
Supplemental Data Table S3. ITS OTU Counts, Related to Figure 4 (.xls)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD3ε, PerCP-eFluor710, clone 17A2 | eBioscience (ThermoFisher) | Cat#46-0032-82, RRID:AB_1834428 |
| anti-CD4, APC-Fire 750, clone GK1.5 | Biolegend | Cat# 100459, RRID:AB_2572110 |
| anti-CD8, APC, clone 53-6.7 | eBioscience (ThermoFisher) | Cat#17-0081-83, RRID:AB_469336 |
| anti-CD11b, BV421, clone M1/70 | BD Biosciences | Cat#562605, RRID:AB_11152949 |
| anti-CD11c, APC-eFluor780, clone N418 | eBioscience (ThermoFisher) | Cat#47-0114-80, RRID:AB_1548663 |
| anti-CD16/32, purified, clone 93 | Biolegend | Cat# 101320, RRID:AB_1574975 |
| anti-CD25, APC-eFluor780, clone PC61.5 | eBioscience (ThermoFisher) | Cat#47-0251-82, RRID:AB_1272179 |
| anti-CD45, PE-Cy7, clone 30-F11 | eBioscience (ThermoFisher) | Cat#25-0451-82, RRID:AB_2734986 |
| anti-CD69, FITC, clone H1.2F3 | eBioscience (ThermoFisher) | Cat#11-0691-85, RRID:AB_465120 |
| anti-CD206, APC, clone 19.2 | eBioscience (ThermoFisher) | Cat#17-2069-42 RRID:AB_2573182 |
| anti-Dectin-1, clone (rabbit polyclonal) | Abcam | Cat#ab140039 RRID:AB_2877055 |
| anti-F4/80, FITC, clone BM8 | eBioscience (ThermoFisher) | Cat# 14-4801-82, RRID:AB_467558 |
| anti-FoxP3, PE-Cy5, clone FJK-16s | eBioscience (ThermoFisher) | Cat# 15-5773-82, RRID:AB_468806 |
| anti-MHCII, eFluor450, clone M5/114.15.12 | eBioscience (ThermoFisher) | Cat#48-5321-82, RRID:AB_1272204 |
| anti-PD1, PE-Dazzle 594, clone 10F.9G2 | Biolegend | Cat# 124324, RRID:AB_2565639 |
| anti-PD-L1, BV711, clone MIH5 | BD Biosciences | Cat# 563369, RRID: AB_2738163 |
| Anti-BrdU, clone BU20A | eBioscience (ThemoFisher) | Cat# 14-5071-82, RRID:AB_10596495 |
| Anti-Caspase 3 active (cleaved) form, clone (rabbit polyclonal) | Millipore Sigma | Cat# AB3623, RRID:AB_91556 |
| Anti-Granzyme B | Abcam | Cat#ab255598 RRID: AB_2860567 |
| AffiniPure Donkey Anti-Mouse IgG (H+L), biotin-SP | Jackson ImmunoResearch | Cat#715-065-151, RRID:AB_2340785 |
| AffiniPure Donkey Anti-Rabbit IgG (H+L), biotin-SP | Jackson ImmunoResearch | Cat#711-065-152, RRID:AB_2340593 |
| Fungal Strains | ||
| Candida albicans | ATCC | Cat# ATCC 90028 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Zombie UV Fixable Viability Dye | Biolegend | Cat# 423108 |
| True-Nuclear™ Transcription Factor Buffer Set | Biolegend | Cat#424401 |
| Cefoperazone sodium salt | Sigma-Aldrich | Cat#C4292 |
| Metronidazole | Sigma-Aldrich | Cat#M1547 |
| Imipenem-Cilasatin sodium salt (1:1 mixture) | Glentham Life Sciences | Cat#GP8148 |
| Vancomycin hydrochloride | Cayman Chemical | Cat#15327 |
| Streptomycin sulfate | Sigma-Aldrich | Cat#S1277 |
| Ampicillin sodium salt | Sigma-Aldrich | Cat#A0166 |
| 5-fluorocytosine | Sigma-Aldrich | Cat#F7129 |
| Fluconazole | Sigma-Aldrich | Cat#F8929 |
| 5-Bromo-2’-deoxyuridine | Sigma-Aldrich | Cat#B5002 |
| Citrate Buffer, pH 6.0, 10X, Antigen Retriever | Sigma-Aldrich | Cat#C9999 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat#017-000-121, RRID:AB_2337258 |
| Metal Enhanced DAB Substrate Kit | Pierce (ThermoFisher) |
Cat#34065 |
| Vector Methyl Green | Vector Laboratories | Cat#H-3402 |
| Critical Commercial Assays | ||
| Tumor Dissociation Kit, mouse | Miltenyi Biotech | Cat#130-096-730 |
| CD45+ MicroBeads, mouse | Miltenyi Biotech | Cat#130-052-031 |
| QIAamp™ DNA Stool Mini Kit | Qiagen | Cat#51504 |
| DNA Nano LT Kit | Illumina | Cat#20015965 |
| iTaq Universal SYBR Green Supermix | BioRad | Cat#1725122 |
| Vectastain Elite ABC HRP Kit | Vector Laboratories | Cat#PK-6100 |
| Deposited Data | ||
| Sequence reads | Sequence Read Archive | ID: PRJNA496065 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| Mouse: C57BL/6NTac Germ-free-Altered Schaedler Flora (ASF)-colonized | Taconic | Cat# B6 GF |
| Oligonucleotides | ||
| Sequencing: ITS1-F (CTTGGTCATTTAGAGGAAGTAA) | Integrated DNA Technologies | Custom primer |
| Sequencing: ITS2-R (GCTGCGTTCTTCATCGATGC) | Integrated DNA Technologies | Custom primer |
| Sequencing: 16S-F (ACTCCTACGGGAGGCAGCAGT) | Integrated DNA Technologies | Custom primer |
| Sequencing: 16S-R (ATTACCGCGGCTGCTGGC) | Integrated DNA Technologies | Custom primer |
| 18S-F (ATTGGAGGGCAAGTCTGGTG) | Integrated DNA Technologies | Custom primer |
| 18S-R (CCGATCCCTAGTCGGCATAG | Integrated DNA Technologies | Custom primer |
| Pan-Candida ITS1-F (GCAAGTCATCAGCTTGCGTT) | Integrated DNA Technologies | Custom primer |
| Pan-Candida ITS1-R (TGCGTTCTTCATCGATGCGA) | Integrated DNA Technologies | Custom primer |
| FungiQuant-F (GGR AAA CTC ACC AGG TCC AG) | Integrated DNA Technologies | Custom primer |
| FungiQuant-R (GSW CTA TCC CCA KCA CGA) | Integrated DNA Technologies | Custom primer |
| FungiQuant probe: 5’-(FAM) TGG TGC ATG GCC GTT (3lABkFQ)-3’ | Integrated DNA Technologies | Custom primer |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Diva | BD Biosciences | http://www.bdbiosciences.com/us/instruments/clinical/software/flowcytometry-acquisition/bd-facsdivasoftware/m/333333/overview |
| FlowJo v10 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| cutadapt v1.4.1 | cutadapt | https://cutadapt.readthedocs.io/en/stable/guide.html |
| SeqPrep v1.0 | github | https://github.com/jstjohn/SeqPrep |
| QIIME v1.6 | QIIME | http://qiime.org/1.3.0/tutorials/tutorial.html |
| BLAST v2.2.22 | NCBI | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastDocs&DOC_TYPE=Download |
| phyloseq v1.13.3 | github | https://github.com/joey711/phyloseq |
| LEfSe v1.0.7 | Harvard University/ Huttenhower Lab | http://huttenhower.sph.harvard.edu/galaxy/ |
| MaAsLin v0.0.3 | Harvard University/ Huttenhower Lab | http://huttenhower.sph.harvard.edu/galaxy/ |
| SmART Advanced Treatment Planning System | Precision X-Ray | https://pxinc.com/smart-range/ |
| Aperio ImageScope | Leica Biosystems | https://www.leicabiosystems.com/digital-pathology/manage/aperio-imagescope/ |
| Other | ||
| VWR Superfrost Plus Micro Slide | VWR International | Cat#48311-703 |
| Cytoseal Mounting Medium (60) | VWR International | Cat#48212-154 |
| Realplex2 Mastercycler qPCR machine | Eppendorf | N/A |
| LSRII analyzer | BD Biosciences | N/A |
| Mini-Beadbeater-16 | BioSpec | N/A |
| Agilent Bioanalyzer | Agilent Technologies | N/A |
| MiSeq | Illumina | N/A |
| AutoMacs | Miltenyi Biotec | N/A |
| gentleMACS Dissociator | Miltenyi Biotec | N/A |
| Qubit fluorometer | Thermo Fisher Scientific | N/A |
| Aperio AT2 | Leica Biosystems | N/A |
Highlights.
Commensal bacteria support anti-tumor T cell responses following radiation therapy
Depletion of intestinal bacteria leads to expansion of commensal fungal populations
Commensal fungi promote pro-tumor macrophage actions by impeding anti-tumor T cells
Tumor-associated macrophages sense fungi through a Dectin-1 mediated mechanism
Significance.
Recent studies suggest that the bacterial microbiome influences responses to chemotherapy and immunotherapy in animal models and humans. This study explores the previously unappreciated influence of intestinal fungi (the mycobiome) on responses to radiation therapy (RT). In mouse models of breast cancer and melanoma we observe that bacterial and fungal microbiota have opposing effects on radiation-induced antitumor immunity. Response to RT was enhanced in mice depleted of commensal fungi or in gnotobiotic mice lacking fungi, whereas depletion of commensal bacteria reduced the efficacy of RT. In patients, we find evidence indicating that antifungal immunity may be upregulated in cancers that respond poorly to therapy. Thus, the study suggests that modulating the fungal microbiome is important for optimal radiation-induced antitumor immunity.
Acknowledgments
We thank H. Sandler, A. Ribas, J. Weidhaas, B. Knudsen and L. Chung for their support and mentorship. Funding: This study was supported in part by the National Cancer Institute, NIH (CA191139 and CA220000 to S.L.S.), the American Society for Radiation Oncology (S.L.S) and the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (DK093496 to D.M.U.). Further support came from the Janis and William Wetsman Family Chair in Inflammatory Bowel Disease Research (D.M.U.). Data and materials availability: The data presented in this paper are tabulated in the main paper and in the supplementary materials. All sequences generated in this study have been deposited in the National Center for Biotechnology Information, NIH, Short Read Archive (www.ncbi.nlm.nih.gov/Traces/sra, accession no. PRJNA496065).
Inclusion and Diversity
We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Footnotes
Declaration of Interests
S.L.S. and D.M.U. hold a patent for “Targeting Fungi in Combination with Cancer Therapy” (U.S. Patent No. 62/393,546).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, Shadaloey SA, Wu D, Preiss P, Verma N, et al. (2019). The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, Longman RS, Iliev ID, Sonnenberg GF, and Artis D. (2017). Regulation of inflammation by microbiota interactions with the host. Nat Immunol 18, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CA, and Garrett WS (2019). Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol 17, 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD (2006). Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol 6, 33–43. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, and Mellman I. (2017). Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330. [DOI] [PubMed] [Google Scholar]
- Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, et al. (2017). A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med 9, eaaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibrian D, and Sanchez-Madrid F. (2017). CD69: from activation marker to metabolic gatekeeper. European journal of immunology 47, 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby EJ, Wei J, Yang XY, Lei G, Wang T, Liu CX, Agarwal P, Korman AJ, Morse MA, Gouin K, et al. (2018). Complimentary mechanisms of dual checkpoint blockade expand unique T-cell repertoires and activate adaptive anti-tumor immunity in triple-negative breast tumors. Oncoimmunology 7, e1421891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CP, Flament C, Lepage P, Roberti MP, et al. (2016). Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity. [DOI] [PubMed]
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, and Coussens LM (2009). CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer cell 16, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. (2011). Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer discovery 1, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li XD, Mauceri H, Beckett M, Darga T, et al. (2014). STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity 41, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb Downward JR, Falkowski NR, Mason KL, Muraglia R, and Huffnagle GB (2013). Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Scientific reports 3, 2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Buque A, Kepp O, Zitvogel L, and Kroemer G. (2017). Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 17, 97–111. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, and Gringhuis SI (2009). Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 9, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavazzi R, and Decio A. (2014). Syngeneic murine metastasis models: B16 melanoma. Methods in molecular biology 1070, 131–140. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. (2015). Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42, 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, and Wargo JA (2019). The microbiome, cancer, and cancer therapy. Nature medicine 25, 377–388. [DOI] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, and Cadwell K. (2021). Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology 160, 1050–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol 17, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G, and Shibuya A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2). Cell Host Microbe 15, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MAA, Thaiss CA, and Elinav E. (2017). Dysbiosis and the immune system. Nat Rev Immunol 17, 219–232. [DOI] [PubMed] [Google Scholar]
- Limon JJ, Kershaw KM, and Underhill DM (2018). Mucosal immune responses to fungi and the implications for inflammatory bowel disease. Curr Opin Gastroenterol 34, 398–403. [DOI] [PubMed] [Google Scholar]
- Limon JJ, Skalski JH, and Underhill DM (2017). Commensal Fungi in Health and Disease. Cell Host Microbe 22, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, et al. (2019). Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 25, 377–388 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Kachur S, Dwan MG, Abraham AG, Aziz M, Hsueh PR, Huang YT, Busch JD, Lamit LJ, Gehring CA, et al. (2012). FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol 12, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugade AA, Moran JP, Gerber SA, Rose RC, Frelinger JG, and Lord EM (2005). Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. Journal of immunology 174, 7516–7523. [DOI] [PubMed] [Google Scholar]
- Malik A, Sharma D, Malireddi RKS, Guy CS, Chang TC, Olsen SR, Neale G, Vogel P, and Kanneganti TD (2018). SYK-CARD9 Signaling Axis Promotes Gut Fungi-Mediated Inflammasome Activation to Restrict Colitis and Colon Cancer. Immunity 49, 515–530 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17. [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, Graeber TG, Sonnenburg JL, Horvath S, Huttenhower C, et al. (2013). Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehraj V, Ramendra R, Isnard S, Dupuy FP, Ponte R, Chen J, Kema I, Jenabian MA, Costinuik CT, Lebouche B, et al. (2020). Circulating (1-->3)-beta-Dglucan Is Associated With Immune Activation During Human Immunodeficiency Virus Infection. Clin Infect Dis 70, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, and van de Veerdonk FL (2015). Immune defence against Candida fungal infections. Nat Rev Immunol 15, 630–642. [DOI] [PubMed] [Google Scholar]
- Noverr MC, Noggle RM, Toews GB, and Huffnagle GB (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72, 4996–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga TS, van der Velden WJ, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg BJ, Blijlevens NM, and Netea MG (2009). Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis 49, 724–732. [DOI] [PubMed] [Google Scholar]
- Routy B, Gopalakrishnan V, Daillere R, Zitvogel L, Wargo JA, and Kroemer G. (2018). The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 15, 382–396. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2017). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. [DOI] [PubMed]
- Schwabe RF, and Jobin C. (2013). The microbiome and cancer. Nat Rev Cancer 13, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelig MS (1966). Mechanisms by which antibiotics increase the incidence and severity of candidiasis and alter the immunological defenses. Bacteriol Rev 30, 442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C. (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao SL, Ruffell B, DeNardo DG, Faddegon BA, Park CC, and Coussens LM (2015a). TH2-Polarized CD4+ T Cells and Macrophages Limit Efficacy of Radiotherapy. Cancer immunology research 3, 518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao SL, Ruffell B, DeNardo DG, Faddegon BA, Park CC, and Coussens LM (2015b). TH2-Polarized CD4+ T Cells and Macrophages Limit Efficacy of Radiotherapy. Cancer Immunol Res 3, 518–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalski JH, Limon JJ, Sharma P, Gargus MD, Nguyen C, Tang J, Coelho AL, Hogaboam CM, Crother TR, and Underhill DM (2018). Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog 14, e1007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone HB, Peters LJ, and Milas L. (1979). Effect of host immune capability on radiocurability and subsequent transplantability of a murine fibrosarcoma. J Natl Cancer Inst 63, 1229–1235. [PubMed] [Google Scholar]
- Tang J, Iliev ID, Brown J, Underhill DM, and Funari VA (2015). Mycobiome: Approaches to analysis of intestinal fungi. Journal of immunological methods 421, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, Inghirami G, Coleman CN, Formenti SC, and Demaria S. (2017). DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun 8, ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. (2016). Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 19, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler ML, Limon JJ, and Underhill DM (2017). Immunity to Commensal Fungi: Detente and Disease. Annu Rev Pathol 12, 359–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuthrich M, Deepe GS Jr., and Klein B. (2012). Adaptive immunity to fungi. Annu Rev Immunol 30, 115–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature medicine 25, 968–976. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, Saghatelian A, Knight R, and Panda S. (2018). Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun 9, 2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hung GC, Nagamine K, Li B, Tsai S, and Lo SC (2016). Development of Candida-Specific Real-Time PCR Assays for the Detection and Identification of Eight Medically Important Candida Species. Microbiol Insights 9, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data Table S2. 16S OTU Counts, Related to Figure 4 (.xls)
Supplemental Data Table S3. ITS OTU Counts, Related to Figure 4 (.xls)
Data Availability Statement
Sequence reads for 16S and ITS from this study are available from the Sequence Read Archive under the project ID “PRJNA496065”.