Abstract
Motivation
Two key steps in the analysis of uncultured viruses recovered from metagenomes are the taxonomic classification of the viral sequences and the identification of putative host(s). Both steps rely mainly on the assignment of viral proteins to orthologs in cultivated viruses. Viral Protein Families (VPFs) can be used for the robust identification of new viral sequences in large metagenomics datasets. Despite the importance of VPF information for viral discovery, VPFs have not yet been explored for determining viral taxonomy and host targets.
Results
In this work, we classified the set of VPFs from the IMG/VR database and developed VPF-Class. VPF-Class is a tool that automates the taxonomic classification and host prediction of viral contigs based on the assignment of their proteins to a set of classified VPFs. Applying VPF-Class on 731K uncultivated virus contigs from the IMG/VR database, we were able to classify 363K contigs at the genus level and predict the host of over 461K contigs. In the RefSeq database, VPF-class reported an accuracy of nearly 100% to classify dsDNA, ssDNA and retroviruses, at the genus level, considering a membership ratio and a confidence score of 0.2. The accuracy in host prediction was 86.4%, also at the genus level, considering a membership ratio of 0.3 and a confidence score of 0.5. And, in the prophages dataset, the accuracy in host prediction was 86% considering a membership ratio of 0.6 and a confidence score of 0.8. Moreover, from the Global Ocean Virome dataset, over 817K viral contigs out of 1 million were classified.
Availability and implementation
The implementation of VPF-Class can be downloaded from https://github.com/biocom-uib/vpf-tools.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Viruses are the most abundant life forms on Earth (Paez-Espino et al., 2016; Suttle, 2007). The number of viral sequences in public databases has increased exponentially due to improvements in computational and experimental methods for detecting viral genomes in metagenome and viral samples. As a result, virus classification has become a new challenge in computational biology. Viruses are extremely diverse in their gene complements, replication mechanisms and even their genetic material (Aiewsakun et al., 2018). Viruses can have DNA or RNA genomes that are double-stranded or single stranded, as reflected in the Baltimore classification system that divided viruses into seven groups (Baltimore, 1971) and the classification from the International Committee on Taxonomy of Viruses (ICTV) which is the main authority for the designation and naming of virus taxa. In addition to genetic classification, viruses may be classified by the host(s) they infect (Mihara et al., 2016) and their means of replication.
Different approaches have been proposed as strategies for viral classification [see the overview presented in (Nooij et al., 2018)]. Most of them provide a clustering of the viral data as a taxonomic classification instead of a taxonomic assignment of each viral sequence. Clustering of the viral data has been performed using either the approach traditionally used in sequence-based analyses of cellular life (Dougan and Quake, 2019; Simmonds and Aiewsakun, 2018), or network-based approaches (Bolduc et al., 2017; Jang et al., 2019; Meier-Kolthoff and Göker, 2017). Other methods are aimed at predicting the viral host. WIsH predicts prokaryotic hosts of phages from phage contig sequences (Clovis et al., 2017). VirHostMatcher predicts hosts under the assumption that virus and host genomes often have similar oligonucleotide frequencies (Ahlgren et al., 2017). Also, a classifier has been developed to distinguish between phages and eukaryotic viruses in Gałan et al. (2019). However, there are no methods that classify a viral genome for both taxonomy and host. Here, we present a new methodology that can classify viral genomes at the family and genus levels according to the Baltimore taxonomy, and can predict each viral host at the domain, family and genus levels.
One of the strategies developed for viral detection and further classification is to consider viral proteins as bait. Viral Protein Families (VPFs) have been extensively used in the identification of new viral sequences in large metagenomic datasets (Paez-Espino et al., 2017b, 2019a; Schulz et al., 2020). Briefly, 14K VPFs were generated from a collection of 167 042 genes from 2353 isolated viruses and retroviruses, clustered using Markov clustering method followed by manual curation to remove families common in plasmid, bacterial and archaeal genomes. An additional set of 11K VPFs were generated from manually curated metagenomic viral contigs larger than 50Kb. Hence, the approach implemented here is based on using the taxonomy and host information from VPFs to further classify viral genomes. A total of 25 281 VPFs from the Integrated Microbial Genome/Virus system (IMG/VR) (Paez-Espino et al., 2017a, 2019b) have been classified and used to infer taxonomy and predict hosts of the viral genomes.
We present VPF-Class, a tool to classify viral genomes with respect to taxonomy and host prediction at multiple taxonomic levels. One of the advantages of our tool is to provide a taxonomic assignment as well as a host prediction of each viral genome instead of a clustering of the viral data. In addition, our tool does not require to download or to select a reference database where the viral data has to be mapped, which avoids any bias on the final classification while makes the tool more user-friendly. We validate the proposed methodology with the results obtained in three different datasets: the NCBI viral sequences, the prophages dataset in Roux et al. (2015) and the Global Ocean Virome (Roux et al., 2016). With the NCBI database, at the genus level, VPF-Class obtained an accuracy of 98% to classify dsDNA, ssDNA and retroviruses, and an accuracy of 86.4% in host prediction. With the prophages dataset, the accuracy in host prediction was 86.6% again at the genus level, whereas with the Global Ocean Virome dataset, taxonomy was assigned to over 817K viral genomes.
2 Materials and methods
In this section, we describe our tool VPF-Class that performs taxonomic assignment and host prediction of viruses.
2.1 The structure of the VPF-Class algorithm
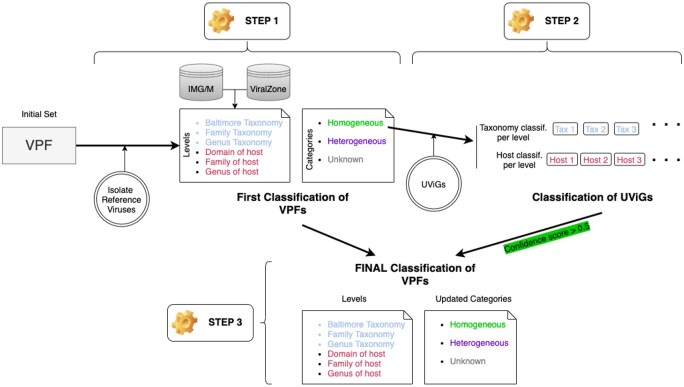
VPF-Class receives as input a set of viral genomes and produces as output a table with the taxonomic classification and host prediction for each genome of the input set. VPF-Class has been implemented in Python and Haskell. The overall graphical representation of the pipeline to classify the VPFs (step 1–3) is shown in Figure 1. The implementation of step 4 as a tool is freely available at https://github.com/biocom-uib/vpf-tools.
Fig. 1.
Graphical representation of the pipeline for viral genome classification (steps from 1 to 3). The first step is the initial classification of the VPFs regarding taxonomy and host infection at three different levels based on the information of the isolate reference viruses retrieved from IMG/M and ViralZone databases. The second step is to classify the uncultured viral genomes (UViGs) based on hits to homogeneous classified VPFs. The third step is the final VPFs classification adding the information from the UViGs classification
The main steps in VPF-Class are:
Taxonomic classification and host prediction of the VPFs
Taxonomic classification and host prediction of viral genomes
Cross validation and second round classification of VPFs
Viral genomes classification and score
2.1.1 Step 1. VPFs classification
The 25 281 viral protein families (VPFs) from the IMG/VR system were classified according to taxonomic classification and host prediction. As a first round of categorization, for every VPF, the hmmsearch tool (Potter et al., 2018) (http://www.ebi.ac.uk/Tools/hmmer) was used with an e-value threshold of 0.001, to obtain hits between VPF proteins and proteins from isolate reference viruses. The information on isolate viruses used to classify the VPFs was retrieved from and contrasted between the ViralZone (Hulo et al., 2011) (https://viralzone.expasy.org/) and IMG/M (Chen et al., 2019) databases.
We considered three levels in taxonomic classification. The first level was the Baltimore classification (Baltimore, 1971) which divides viruses into six groups corresponding to those with double-stranded (ds)DNA genomes, single-stranded (ss)DNA genomes, dsRNA genomes, ss(+)RNA genomes with a sense orientation of genes, ss(-) RNA genomes in antisense orientation and reverse transcribing viruses (RT). The second level was based on refining the previous Baltimore classification to include the family taxonomic level in each of the six groups. The last level was to resolve taxonomy to genus level classification. Host predictions were first made at the domain level and then refined to family and genus levels.
For every feature (taxonomy and host prediction), a VPF was considered to be homogeneous if all its hits were to viruses within the same taxonomic classification. Homogeneous VPFs were classified as category 1. A VPF whose hits were to viruses with no taxonomic information was defined as category 0 whereas a VPF was considered heterogeneous if its hits were to viruses in different classifications, defined as category –1.
Homogeneous, or category 1, VPFs were redefined according to their numbers of hits. For every feature, we considered the number of hits of a VPF as a discrete random variable and analyzed the frequency distribution of number of hits, which turned out to be a χ2 distribution. Next, we defined four subcategories (ranking from 1.1 to 1.4) of category 1 for the Baltimore, family and genus taxonomic classifications. These four subcategories correspond to the quartiles and , respectively. We defined three subcategories (ranking from 1.1 to 1.3) of category 1 for host domain, host family and host genus classification, corresponding to tertiles {7, 3, 0}, {5, 2, 0} and {3, 1, 0}, respectively. For instance, a category 1 VPF with 9 hits in Baltimore classification and 8 hits in host domain classification is classified as a category 1.2 concerning Baltimore classification and a category 1.1 in host domain classification. Table 1 summarizes the different category classification.
Table 1.
Minimal number of required hits per category
| Feature | Level | Cat 1.1 | Cat 1.2 | Cat 1.3 | Cat 1.4 |
|---|---|---|---|---|---|
| Taxonomy | Baltimore | >11 hits | >5 hits | >2 hits | >0 hits |
| Family | >10 hits | >4 hits | >2 hits | >0 hits | |
| Genus | >6 hits | >2 hits | >1 hits | >0 hits | |
| Host | Domain | >7 hits | >3 hits | >0 hits | – |
| Family | >5 hits | >2 hits | >0 hits | – | |
| Genus | >3 hits | >1 hits | >0 hits | – |
Note: Cat denotes the category.
2.1.2 Step 2. UViGs classification
Homogeneous VPFs were used to classify uncultivated virus genomes (UViGs). To classify an UViG, at each of the three levels of classification, we first considered its hits to the category 1 (homogeneous) VPFs. Next, we used the classification assigned to the VPFs to infer the UViG classification. More precisely, we classified an uncultivated virus genome as well as calculated its total score in every level of classification as follows:
Let v be an uncultivated virus genome and let us assume that v has a set of proteins P and a subset H of them has hits to homogeneous VPFs classified under a specific taxonomic level with sequence score of for each . Set where K(p) is the number of Kbase pairs of p. That is, if the number of base pairs of p is 3500 then . We call the normalized sequence score of the hit of p. Let be the homogeneous VPF such that p has the hit. Then, we define the taxonomy classification of v at the specific level, as the set where is the classification at the considered taxonomic level of . Also, we define the membership ratio of every different classification t in T(v) as where
- The score of t is defined as
The total score of v is defined as
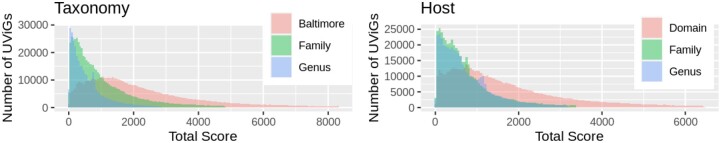
Finally, the confidence score of the taxonomic classification of v at the considered level is defined as the quantile rank of the total score sv in the distribution of all total scores obtained for all UViGs, regarding the considered taxonomic level classification. Figure 2 shows the distribution of all total scores obtained for all UViGs.
Fig. 2.
Score distributions of the UViGs regarding taxonomic classification (left panel) and host prediction (right panel). Scores values are displayed in the x-axis while the y-axis represents the number of UViGs with the corresponding score
Analogously we define the host-prediction score and the host-prediction confidence score, at every level (domain, family and genus).
Example.
Let us assume that a virus v has three proteins that provided hits to homogeneous VPFs: two hits to VPFs classified as Myoviridae and one hit to a VPF classified as Siphoviridae. Suppose that the first hit, h1, has a normalized sequence score of 300 to a category 1.1 Myoviridae VPF. Suppose that the second hit, h2, has a normalized sequence score of 200 to a category 1.2 Myoviridae VPF. And suppose that the third hit, h3, has a normalized sequence score of 240 to a category 1.4 Siphoviridae VPF. Then, the taxonomy classification—at family level—of virus v is the set {Myoviridae, Siphoviridae} under the following scores:
Myoviridae score:
Siphoviridae score:
The total score is the sum of the Myoviridae and Siphoviridae scores, that is . Then, the membership ratio of Myovididae classification is 0.88 () and the membership ratio of Siphoviridae classification is 0.12 (). Thus, the virus is classified as 88% Myoviridae and 12% Siphoviridae. Finally, the total score corresponds to a confidence score of 0.36.
2.1.3 Step 3. Cross validation and second round classification of VPFs
In its third step, VPF-Class compared the results obtained in the previous steps. First, the UViGs classifications providing new information to VPFs were added. Next, a new classification for each VPF was obtained.
Inferring information from the UViGs to the VPFs. UViGs classified with a confidence score above percentile 50 were used to infer new information on VPFs and reclassify them. Namely, all UViGs with a confidence score higher than 0.5, as well as their classification information, were added to the set of isolate viruses. Next, VPFs were reclassified following the same procedure as in Step 1. This implies that some category 0 VPFs were updated to category 1 or category –1 (inferring information from the UViGs proteins), and some category 1 VPFs, where updated to category –1 (when the UViGs and the isolate viruses within a VPF had heterogeneous protein information).
Reclassifying VPFs. We reclassified the homogeneous and category 0 VPFs with hits to UViGs with a confidence score higher than 0.5. Namely, at each level of classification, for proportional reclassification of every VPF we considered the previous hits it had to isolate viruses and its new hits to UViGs to proportionally reclassified it.
More precisely, let F be a VPF either not classified or classified as homogeneous in Step 1 with hits to UViGs classified with a confidence score greater or equal to 0.5. Let us assume that F has hits to a set of proteins Ho of isolate viruses homogeneously classified as t0 under a specific taxonomic level, and hits to proteins He of a set of classified UViGs ending up with a set of different classifications T under the same taxonomic level. Then, the classification of F at this level is with proportion of every t defined by . The score of t is , where
and using v(p) to refer to the virus to which a protein p belongs.
Finally, for every classification tp, a category 1.1, 1.2, 1.3 or 1.4 is assigned to F taking into account the total number of hits such that tp is assigned. We denote it by .
Analogously, we defined the proportion of every predicted host and its score, at every level (domain, family and genus).
Note that we ended up with six and five categories of VPFs regarding the taxonomic and host classification, respectively. The homogenous VPFs that were divided in four and three categories, respectively, the heterogeneous or non-homogeneous VPFs and the unclassified VPFs called category 0.
Example.
Let us assume that a VPF, F, was classified as a category 1.3 Myoviridae providing 4 hits with normalized sequence scores: 300, 350, 400 and 200, respectively, to isolated viruses classified as Myoviridae. Assume also that F has 3 hits to 2 non-homogeneous UViGs: v1 (1 hit) and v2 (2 hits) with normalized sequence scores: 250, 150 and 375, respectively. Finally, assume that the classification of v1 is 30% Myoviridae and 70% Siphoviridae, and that the classification of v2 is 60% Myoviridae and 40% Nimaviridae.
Then, for every family we have the following score:
-
Myoviridae score: for every hit from the isolates, its normalized sequence score is multiplied by 0.5 (Cat. 1.3) and for every hit from the UViGs, its normalized sequence score is multiplied considering the Myoviridae membership ratio. Thus, the normalized sequence score from the hit in v1 is multiplied by 0.3 (30% Myoviridae) and the normalized sequence score from the hits in v2 are multiplied by 0.6 (60% Myoviridae). Then, the score of Myoviridae is:
(300 + 350 + 400 + 200) · 0.5 + 250 · 0:3 + (150 + 375) · 0.6 = 625 + 75 + 315 = 1,015:
-
Siphoviridae score: since the classification of v1 is 30% Myoviridae and 70% Siphoviridae, the normalized sequence score from the hit in v1 is multiplied by 0.7. Thus, the Siphoviridae score is
Nimaviridae score: since v2 is classified as 60% Myoviridae and 40% Nimaviridae, the normalized sequence score from the hits to v2 are multiplied by 0.4. Thus, the Nimaviridae score is
Thus, the sum of all scores is and the proportion of every family is:
Myoviridae:
Siphoviridae:
Nimaviridae:
The result is a reclassification of F as 73% Myoviridae, 12% Siphoviridae and 15% Nimaviridae. In addition, since the number of hits to the Myoviridae family has increased from 4 to 7, the category of the Myoviridae classification is 1.2. The number of hits to the Siphoviridae family is 1 and the number of hits to the Nimaviridae family is 2, so their category is 1.4.
2.1.4 Step 4. Viral genomes classification and score
In its last step, for every set of viral genomes, VPF-Class provides a classification and a confidence score for every classified virus. First, for every viral genome, its protein coding genes are predicted with the Prodigal software (https://github.com/hyattpd/Prodigal). Then, we perform a hmmsearch against the given VPFs to obtain its hits. Next, the viral genomes classification is obtained as described below.
Let v be a viral genome such that v has a set of proteins P and hits to the set of VPFs classified under a specific taxonomic level and let be the set of classifications of for each . Then, v is classified under the specific taxonomic level as T(v) and the membership ratio of every different classification t in T(v) as where
-
The score of t is defined as
The total score of v is defined as
The confidence score for the specific classification of v is, again, the percentile rank of its total score sv in the distribution of all total scores obtained for all UViGs.
Analogously we define the host-prediction score and the host-prediction confidence score, at every level (domain, family and genus).
Example.
Let us assume that v has 5 proteins that provided hits to VPFs classified as shown in Table 2. Then, the virus is classified as Myoviridae, Siphoviridae, Papillomaviridae and Nimaviridae with:
Table 2.
Viral genomes classification–Example
| Hits | Normalized seq. score |
VPF-classification |
|---|---|---|
| h 1 | 300 | 1.1 Myoviridae |
| h 2 | 250 | 1.2 Myoviridae |
| h 3 | 200 | 30% Myoviridae (1.1), 70% Siphoviridae (1.3) |
| h 4 | 350 | 60% Myoviridae (1.3), 25% Siphoviridae (1.4), 15% Papillomaviridae (1.4) |
| h 5 | 275 | 25% Myoviridae(1.4), 75% Nimaviridae (1.1) |
Myoviridae score:
Siphoviridae score:
Papillomaviridae score:
Nimaviridae score:
The total score sv is the sum of the scores, that is
and the membership ratios are 0.69 for Myoviridae (669.686/980.936 = 0.69), 0.09 for Siphoviridae (91.875/980.936 = 0.09), 0.01 for Papillomaviridae (), 0.21 for Nimaviridae (). Thus, the virus is classified as 69% Myoviridae, 9% Siphoviridae, 1% Papillomaviridae and 21% Nimaviridae. Finally, the confidence score for this classification is the quantile rank that the total score (980.936) in the distribution of all scores from the UViGs classification, which corresponds to 0.58.
3 Results and discussion
As a result of VPF-Class, we obtained a taxonomic classification and host prediction for each of the viral protein families and the uncultivated virus genomes available at the IMG/VR database.
3.1 VPFs and UViGs classification
A curated set of 25 281 VPFs and 730 921 uncultivated virus genomes (UViGs) were classified and categorized at different levels regarding taxonomic assignment and host prediction.
Classification of uncultivated virus genomes from IMG/VR . We considered 730 921 UViGs from IMG/VR. For every UViG, we considered the hits of its proteins to the VPFs to define its taxonomic classification and host prediction as well as a confidence score (see Section 2 for the detailed definitions). Table 3 summarizes the results of the UViGs classification and Figure 2 shows the scores distribution.
Table 3.
UViGs classification
| Level | Classified UViGs | Homogeneous | 75% homogeneous |
|---|---|---|---|
| Balt. tax | 713 648 (97, 64%) | 712 129 | 713 528 |
| Fam. tax | 634 397 (86, 79%) | 409 625 | 525 767 |
| Genus tax | 362 962 (49, 65%) | 270 390 | 307 625 |
| Domain host | 633 475 (86, 67%) | 592 920 | 620 938 |
| Fam. host | 489 016 (66, 90%) | 306 747 | 368 612 |
| Genus host | 461 113 (63, 09%) | 289 089 | 344 538 |
In Baltimore classification, 713 648 UViGs had some hit to VPFs classified under the same taxonomy. Among them, 712 129 UViGs were homogeneous, which means that they had genes with hits to VPFs equally classified and 713 528 had a taxonomic class with a proportion greater than 75%. At the family and genus levels, 634 397 and 362 962 UViGs were classified, respectively. Among them, 409 625 and 270 390 were homogeneous. In host prediction, 633 475 UViGs were classified at the host domain level and 489 016 and 461 113 at the family and genus level respectively. Among the host predictions, 592 920, 306 747 and 289 089 were homogeneous at the host domain, family and genus level, respectively.
VPFs classification . In our first-round classification, 16 257 VPFs were classified. Next, to reclassify the VPFs, we considered the new information provided by the set of classified UViGs so that, we transferred information from the UViGs classification to the VPFs that were classified as category 0 or category 1. As a result, in the Baltimore, family and genus level of taxonomy, only 120, 198 and 545 VPFs respectively remained as category 0. In host prediction, only 191, 588 and 579 VPFs remained in category 0 in the domain, family and genus levels. In addition, some VPFs were moved from homogeneous to heterogeneous and others remained homogeneous but increased their category (see the methods section for a detailed description of this reclassification). The final classification is shown in Table 4. For instance, we can observe there that, at the family level of taxonomic classification, close to 4K VPFs were homogeneous and over 18K were heterogeneous which means that we had a total of over 22K classified VPFs at this level. As to the taxonomy distribution of the homogeneous VPFs at the Baltimore classification, 20 698 were dsDNA, 88 were ssDNA and 29 were retrovirus. Among them, we obtained 40 different families and 202 different genera (47 and 286 different families and genera if we include heterogeneous VPFs). The most represented families were Myoviridae (1242), Siphoviridae (860), Herpesviridae (364) and Phycodnaviridae (203). The most represented genera were Chlorovirus (345), Tequatrovirus (213), Alphabaculovirus (151) and Cytomegalovirus (128). Thus, the most represented group is dsDNA while RNA viruses are not represented. This means that our tool presumably will correctly classify dsDNA but it will not classify RNA viruses. Regarding the host infection distribution, 6126 VPFs had Bacteria as a host prediction, 1064 had Eukaryota and 127 had Archaea; these hosts were distributed in 58 different families and 67 different genera. The most represented host families were Enterobacteriaceae (254), Mycobacteriaceae (250) and Hominidae (206). The most represented host genera were Mycobacterium (250), Aeromonas (223), Homo (217), Bacillus (151) and Pseudomonas (124) (see Table Summary in Supplementary Material for a detailed description of all represented families and genera). Finally, we want to stress that every further update of the VPFs classification automatically provides an update of our tool. As for instance the new classification that has been recently accepted in Koonin et al. (2020).
Table 4.
VPFs final classification
| Feature | Level | Homog. | Heterog. | Total |
|---|---|---|---|---|
| 1.1—1.2—1.3—1.4 | ||||
| Taxonomy | Baltimore | 14 264—4042—2122—397 | 4142 | 24 957 |
| Family | 1709—1511—614—81 | 18 124 | 22 039 | |
| Genus | 1710—1425—144—94 | 15 732 | 19 105 | |
| Host | Domain | 4594—1975—748—0 | 16 226 | 23 543 |
| Family | 1109—580—216—0 | 17 877 | 19 782 | |
| Genus | 1427—329—84—0 | 17 216 | 19 056 |
VPF-Class evaluation. In order to evaluate our tool, we performed a series of tests as described in this section.
3.2 Test 1-NCBI database
We considered the viral genome sequences from the NCBI reference sequence database (https://www.ncbi.nlm.nih.gov/refseq/) and used these as input for VPF-Class to evaluate whether the resultant taxonomic classification and the host annotation agreed with the classification from the International Committee on Taxonomy of Viruses (ICTV) and the host annotation from Virus-Host DB (https://www.genome.jp/virushostdb/).
In order to evaluate the results obtained in this test, we took into account the values of coverage (i.e. number of classified viral genomes over the number of viral genomes) and accuracy (i.e. number of correctly classified viral genomes over the number of classified viral genomes) obtained with different confidence score and membership ratio thresholds. Notice that we only considered the annotated sequences in each taxonomic level, therefore the dataset changes for every level.
Taxonomic classification . Table 5 summarizes the results obtained at the genus and family level by VPF-Class. At the family level, we can observe that when the membership ratio and the confidence score ranged from 0 to 0.2, for dsDNA the coverage decreases from 66.5% to 65% while the accuracy increases from 37% to 99%; for ssDNA, the coverage decreases from 49.6% to 48% while the accuracy increases from 94% to 99.9%, and for retroviruses, the coverage is 93% while the accuracy is 100%. Also, at the genus level, the coverage decreases from 56.8% to 55.5% for dsDNA and from 52.5% to 52% for ssDNA, while the accuracy increases from 39% to 98% for dsDNA and from 94% to 98% for ssDNA. Thus, we nearly obtain an accuracy of 100% with a membership ratio and confidence score thresholds of 0.2 without barely decreasing the coverage. For retroviruses, the coverage and the accuracy is 100% at the genus level.
Table 5.
Prediction coverage and accuracy with the NCBI test
| TAXONOMY | ||||
|---|---|---|---|---|
| Thresholds | dsDNA | ssDNA | RT | |
| Family | MR 0, CS0 | 66.5% | 37% | 49.6% | 94% | 93% | 100% |
| MR0.1, CS0.1 | 66% | 98.2% | 49% | 99.8% | 93% | 100% | |
| MR0.2, CS0.2 | 65% | 99% | 48% | 99.9% | 93% | 100% | |
| Genus | MR0, CS0 | 56.8% | 39% | 52.5% | 94% | 100%| 100% |
| MR0.1, CS0.1 | 56.5% | 97% | 52% | 97.5% | 100% |100% | |
| MR0.2, CS0.2 | 55.5% | 98% | 52% | 98% | 100% | 100% | |
| HOST | ||||
|---|---|---|---|---|
| Thresholds | Bacteria | Archaea | Eukaryota | |
| Family | MR0.1, CS0.1 | 53% | 86% | 62% | 82% | 6.5% | 56.6% |
| MR0.2, CS0.2 | 52% | 90.6% | 59% | 84.2% | 5% | 66.6% | |
| MR0.3, CS0.5 | 48.7% | 95.4% | 59% | 86.8% | 3.8% | 78.5% | |
| Genus | MR0.1, CS0.1 | 91.7% | 69.5% | 95.5% | 65.5% | 8% | 64% |
| MR0.2, CS0.2 | 87.3% | 77.6% | 93.3% | 75% | 6.5% | 73.3% | |
| MR0.3, CS0.5 | 78% | 86.7% | 91% | 76.5% | 5% | 93.2% | |
Note: In every entry, the coverage (left) appears separated from the accuracy (right) by a vertical bar. MR and CS denote the membership ratio and confidence score, respectively.
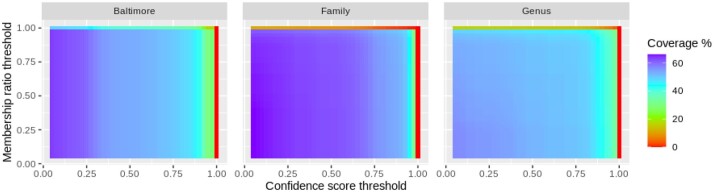
In addition, we also considered the heatmap representation to visualize and analyze the relationship between the number of classified viral genomes (coverage) and the values of the confidence score and membership ratio. In Figure 3 we show the heatmaps obtained in this test for every taxonomic level. The x-axes in the heatmaps are the confidence score thresholds while the y-axes are the membership ratio thresholds. The colors represent the ratio of classified viral sequences with respect to the total number of sequences. They range from the lowest value in red to the highest value in purple. Thus, the color at the point represents the ratio of classified sequences with a confidence score higher or equal to 0.5 that have a taxonomic classification with a membership ratio higher or equal to 0.75. We can observe in this figure how the color vary following the prediction coverage and accuracy numbers presented in Table 5. Also, in this particular test we observe that color changes varies along the x-axes which means that the coverage is sensitive to the confidence threshold, namely, it decreases as the confidence score threshold increases. We refer to the Supplementary Material to visualize all the heatmaps calculated in this test as well as the different tables with the coverage and accuracy information.
Fig. 3.
Heatmaps depicting results obtained with the NCBI test. The x-axes show the confidence score thresholds while the y-axes show the membership ratio thresholds. The colors represent the ratio of classified viral sequences above a confidence score (x value) and a membership ratio (y value) with respect to the total number of sequences
Host prediction . Table 5 summarizes the results obtained at the genus and family level by VPF-Class. If we consider a membership ratio and a confidence score greater or equal to 0.1, the coverage of VPF-Class at the family level is 53% for Bacteria, 62% for Archaea and 6.5% for Eukaryota, while the accuracy is 86%, 82% and 56.6%, respectively. If we consider a membership ratio greater or equal to 0.3 and a confidence score greater or equal to 0.5, then the coverage is nearly the same but the accuracy increases to 95.4%, 86.8% and 78.5%, respectively. At the genus level, again if we consider a membership ratio greater or equal to 0.3 and a confidence score greater or equal to 0.5 (last row), then the coverage is 78% for Bacteria, 91% for Archaea and 5% for Eukaryota, while the accuracy is 86.7%, 76.5% and 93.2%, respectively. The low coverage of Eukaryotic viruses is due to the lack of homogeneous RNA VPFs.
3.3 Test 2-Global ocean virome database
We applied VPF-Class to classify 1 380 523 viral genomes from the Global Ocean Virome (GOV) database (Roux et al., 2016). Most viral contigs, 1 039 064 (75%), hit classified VPFs, so that we were able to provide taxonomic assignments for them as follows:
at the Baltimore level, 1 039 064 viral genomes were classified and half of them had a membership ratio of .
at the family level, 874 652 viral genomes were classified and 27 251 were homogeneous; 437 330 were classified with a membership ratio of and a confidence score of 0.3.
at the genus level, 817 279 viral genomes were classified and 25 441 were homogeneous. Among the viral genomes classified as heterogeneous, 204 517 were classified with a membership ratio greater than or equal to 0.7 and a confidence score of 0.8.
Regarding host prediction, 1 001 386 genomes had a host prediction at the domain level. At the family and genus levels, 843 779 and 834 023 genomes had a host predicted, respectively. Among them, 210 951 and 208 524 had a confidence score above 0.7 (see the Supplementary Material for additional details and the heatmaps calculated in this test).
3.4 Test 3-prophages
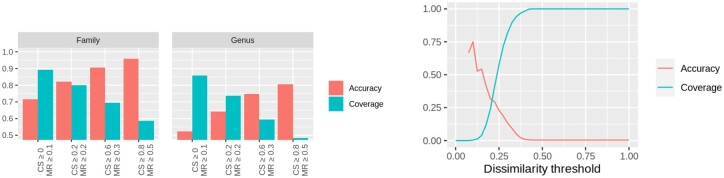
We considered a dataset consisting of 12 498 prophages and their host information (Roux et al., 2015) to further evaluate the host predicted with VPF-Class. At the domain level, 12 444 prophages were classified with a membership ratio above 0.75. If we consider a membership ratio greater or equal to 0.1, the coverage is 89% (and 86%) and the accuracy is 71% (and 52%) at the family (genus) level. The accuracy increases to 96% (and 81%) while the coverage decreases to 59% (and 49%) if we consider a membership ratio greater or equal to 0.5 and a confidence score of 0.8 at the family (genus) level. See left panel of Figure 4 for a better visualization of these results.
Fig. 4.
Coverage and accuracy values obtained by VPF-Class (left panel) and VirHostMatcher (right panel) in host prediction of the prophages test. On the right, we show the coverage and accuracy values (y-axis) obtained by VirHostMatcher with respect to the values of ONF dissimilarity measure (x-axis)
In this test, we obtained that the confidence score and the membership ratio did not correlate with the number of correctly classified viral sequences since almost all sequences were correctly classified (see the Supplementary Material for additional results and to visualize all heatmaps).
3.5 VPF-Class versus other tools
In order to analyze the performance and utility of VPF-Class with respect other implemented tools, we considered vConTACT v.2.0, a recently developed tool for taxonomic classification of viral genomes (Jang et al., 2019) and VirHostMatcher, an oligonucleotide frequency dissimilarity measure for host prediction (Ahlgren et al., 2017).
3.5 vConTACT
The recent published tool vConTACT provides a clustering of an input set of viral genomes together with 2304 classified viral genomes from the NCBI database. The viral sequences within the same cluster are a genus-level group. When the cluster has a classified viral genome (a genome from the NCBI database), its classification may be manually inferred to the cluster’s elements, however, vConTACT does not provide a classification for each individual viral sequence. On the other hand, every viral genome that had some hits to the set of classified VPFs, is individually classified by VPF-Class, and those without hits to the VPFs remain unclassified. Therefore, these tools have differing structures and strategies, as well as output formats. Nevertheless, in order to provide some guidance in the utility of both tools, we considered the three tests run with VPF-Class and analyze the results of both tools. Since vConTACT does not predict the host, we only compared the results on taxonomic classification.
NCBI benchmark. We ran vConTACT on the NCBI dataset previously used to evaluate VPF-Class. Notice that part of the data on the NCBI dataset was used as a training set in both tools and both tools correctly classify over 90% of them. However, in order to compare the results obtained at the genus level of both tools we calculated their agreement in the taxonomy classification. With a membership ratio of 0.25 and a confidence score of 0.75, we obtained an agreement of 95%. Clearly, as the membership ratio increased in VPF-Class, the number of classified viral genomes decreased. Nevertheless, with a membership ratio of 0.75 and a confidence score of 0.75 the agreement was 78%. Therefore, we conclude that, in the NCBI database, the agreement between vConTACT and VPF-Class is very high (), as it was expected.
GOV benchmark. Due to computational restrictions of vConTACT, instead of considering the Global Ocean Virome database (Roux et al., 2016) which has 1 380 523 viral genomes, we had to consider a subset of 14 025 viral genomes of the GOV database also considered in (Jang et al., 2019). At the genus level, we obtained 13 883 viral genomes classified by VPF-Class. Considering a membership ratio of 0.25 and a confidence score lower or equal to 0.75, approximately 7500 were homogeneous (see the corresponding heatmap in the Supplementary Material). And, we obtained 867 clusters (genus groups) from vConTACT. In order to analyze the results and due to the lack of a truth, we calculated the coherence of a cluster (genus group of vConTACT) with the classification with VPF-Class (above a confidence score and a membership ratio threshold) as the number of pairs in the cluster that shared a genus classification by VPF-Class divided by the number of pairs classified by VPF-Class. Next, we calculated the agreement or cluster coherence between these 867 genus groups and the viral genomes classified as homogeneous by VPF-Class. With a membership ratio of 0.25 and a confidence score of 0.5, the cluster coherence was 63%.
UViGs benchmark. Again, due to computational restrictions of vConTACT, we ran vConTACT on a benchmark of 10 171 UViGs from the IMG/VR database. In order to analyze the results obtained with both tools at the genus level, we created a benchmark with 171 UViGs that VPF-Class did not classify, and UViGs classified with different confidence scores. Hence, we randomly selected 1000 UViGs in every decile of the UViGs confidence score distribution obtained with VPF-Class. The classification provided by vConTACT was of 1661 clusters with a mean quality of 0.47; 1628 clusters had a taxon prediction score of 1; and 283 had some classified viral genome from the NCBI database. Thus, we obained a total number of 723 UViGs classified. 184 clusters were homogenous (all classified viral genomes had the same genus) so we obtained a total number of 45 UViGs classified as homogenous. On the other hand, VPF-Class classified 4731 UViGs as homogenous with a membership ratio and a confidence score of 0.5. Among them, 57 were also classified by vConTACT and 31 were equally classified by both tools.
As a summary, we conclude that VPF-Class and vConTACT are transversal tools, and we suggest to use both tools to improve the viral genome classification. On one hand, every viral genome sequence with hits to the set of classified VPFs, is individually classified by VPF-Class. Then, we recommend VPF-Class as a first round of classification. However, those viral genomes without hits to the VPFs remain unclassified by VPF-Class. Then, we recommend vConTACT to obtain a clustering on the unclassified viral data.
3.6 VirHostMatcher
The host prediction tool VirHostMatcher (Ahlgren et al., 2017) is made under the assumption that virus and host genomes often have similar oligonucleotide frequencies. To compare the performance of VirHostMatcher and VPF-Class we run the tool on the dataset of Test 3 which is a dataset consisting of 12 498 prophages and their host information (Roux et al., 2015). Unlike VPF-class, the tool VirHostMatcher requires to introduce the genome sequences of the possible hosts which clearly creates a bias in the host prediction results. As a first attempt, we try to consider all the bacteria from the NCBI dataset as host dataset. However, due to computational restrictions, we considered a random set of 1000 complete bacteria and archaea genomes. We consider the oligonucleotide frequency (ONF) dissimilarity measure defined in Ahlgren et al. (2017). The results obtained in this test, at the genus level, are shown in Figure 4. The right panel of this figure displays the graphical representation of the coverage and accuracy values (y-axis) obtained with respect to the ONF dissimilarity measure (x-axis). We can observe that the coverage decreases when the ONF dissimilarity decreases while the accuracy increases when the ONF dissimilarity decreases. Also, we observe that when the ONF dissimilarity is above 0.4 the coverage rises to one, while the accuracy decreases to zero. When the ONF dissimilarity is around 0.2, intersection of both curves, the coverage and accuracy are both around 30%. On the other hand, the left panel of Figure 4 shows the results obtained by VPF-Class at the family and genus levels. We can observe that, at the genus level, if we consider a membership ratio greater or equal to 0.1, VPF-Class obtained a coverage of 86% and an accuracy of 52%. Also, we can observe there that the accuracy obtained by VPF-Class is 81% and the coverage is 49% with a confidence score and a membership ratio up to 0.8 and 0.5, respectively. Therefore, we conclude that in this test, unlike VirHostMatcher, VPF-Class obtains a very good balance between accuracy and coverage values.
4 Conclusion
In this paper, we proposed a new approach to taxonomic classification and host prediction of viral sequences. Classification was based on orthologous viral proteins from a set of previously classified viral protein families (VPFs) from the IMG/VR database. The characterization of VPFs and uncultivated virus classification was split into two different categories: virus taxonomy and host prediction at different taxonomic levels. For virus taxonomy, we considered a high-rank taxonomic level (Baltimore classification) as well as deeper taxonomy levels (virus family and genus). For host prediction we used the host domain level to separate bacteriophages, archaeal viruses and eukaryotic viruses and the family and genus level assignments of the predicted host. Relying on the VPFs classification, a new methodology to classify metagenome viruses was conceived. As a result, VPF-Class, a tool to predict taxonomy and host of viruses within metagenome samples, has been successfully implemented. Some experiments have been performed in order to validate the proposed methodology. Considering a confidence score and a membership ratio over 0.5, VPF-Class reported 98.9% (resp. 91.3%) of accuracy in the genus taxonomic classification (resp. host prediction) of the RefSeq database. In the host prediction of the prophages dataset (from Roux et al., 2015), VPF-class obtained a right balance between accuracy and coverage values and the accuracy in this test was 77.5%. Also, 817 279 viral genomes were classified at the genus level from the Global Ocean Virome database (Roux et al., 2016).
Acknowledgements
The authors thank S. Roux for his help with generating the vConTACT results from the GOV dataset and Heather Maughan for critical reading of the paper.
Funding
This work was supported by the Ministerio de Ciencia e Innovación (MCI), the Agencia Estatal de investigación (AEI) and the European Regional Development Funds (ERDF); through project PGC2018-096956-B-C43 (FEDER/MICINN/AEI), and by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under contract no. DE-AC02–05CH11231 and made use of resources of the National Energy Research Scientific Computing Center, which is also supported by the DOE Office of Science under contract no. DE-AC02–05CH11231.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Conflict of Interest: none declared.
Contributor Information
Joan Carles Pons, Department of Mathematics and Computer Science, University of the Balearic Islands, Palma 07122, Spain.
David Paez-Espino, Department of Energy Joint Genome Institute, Berkeley, CA 94720, USA.
Gabriel Riera, Department of Mathematics and Computer Science, University of the Balearic Islands, Palma 07122, Spain.
Natalia Ivanova, Department of Energy Joint Genome Institute, Berkeley, CA 94720, USA.
Nikos C Kyrpides, Department of Energy Joint Genome Institute, Berkeley, CA 94720, USA.
Mercè Llabrés, Department of Mathematics and Computer Science, University of the Balearic Islands, Palma 07122, Spain.
References
- *Ahlgren N.A. et al. (2017) Alignment-Free d2 oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res., 45, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun P. et al. (2018) Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: steps towards a unified taxonomy. J. Gen. Virol., 99, 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. (1971) Expression of animal virus genomes. Bacteriol. Rev., 35, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc B. et al. (2017) vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect archaea and bacteria. PeerJ, 5, e3243.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-M.A. et al. (2019) IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res., 47, D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clovis G. et al. (2017) WIsH: who is the host? Predicting prokaryotic hosts from metagenomic phage contigs. Bioinformatics, 33, 3113–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan T., Quake S. (2019) Viral taxonomy derived from evolutionary genome relationships. PLoS One, 14, e0220440.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gałan W. et al. (2019) Host taxon predictor – a tool for predicting taxon of the host of a newly discovered virus. Sci. Rep., 9, e0220440.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo C. et al. (2011) Viralzone: a knowledge resource to understand virus diversity. Nucleic Acids Res., 39, D576–D582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.B. et al. (2019) Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol., 37, 632–639. [DOI] [PubMed] [Google Scholar]
- Koonin E.V. et al. (2020) Global organization and proposed megataxonomy of the virus world. Microbiol. Mol. Biol. Rev., 84, e0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J.,P., Göker M. (2017) VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics, 33, 3396–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara T. et al. (2016) Linking virus genomes with host taxonomy. Viruses, 8, 66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooij S. et al. (2018) Overview of virus metagenomic classification methods and their biological applications. Front. Microbiol., 9, 749.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2016) Uncovering Earth’s virome. Nature, 536, 425–430. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2017a) A database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res., 45, D457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2017b) Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat. Protoc., 12, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2019a) Diversity, evolution, and classification of virophages uncovered through global metagenomics. Microbiome, 7, 157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D. et al. (2019b) IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res., 47, D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S.C. et al. (2018) HMMER web server: 2018 update. Nucleic Acids Res., 46, W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S. et al. ; Tara Oceans Coordinators. (2016) Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature, 537, 689–693. [DOI] [PubMed] [Google Scholar]
- Roux S. et al. (2015) Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. eLife, 4, e08490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F. et al. (2020) Giant virus diversity and host interactions through global metagenomics. Nature, 578, 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Aiewsakun P. (2018) Virus classification – where do you draw the line? Arch. Virol., 163, 2037–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. (2007) Marine viruses – major players in the global ecosysteme. Nat. Rev. Microbiol., 5, 801–812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.
Conflict of Interest: none declared.