Abstract
With the goal of improving the reproducibility and annotatability of MHC multimer reagent data, we present here the establishment of a new data standard: The Minimal Information about MHC Multimers (MIAMM, miamm.lji.org). Multimers are engineered reagents composed of a ligand and a Major Histocompatibility Complex (MHC), which can be represented in a standardized format using ontology terminology. We provide an online website to host the details of the standard, as well as a validation tool to assist with the adoption of the standard. We hope that this publication will bring increased awareness of MIAMM and drive acceptance, ultimately improving the quality and documentation of multimer data in the scientific literature.
Introduction
Major Histocompatibility Complex (MHC) multimers are engineered reagents with precisely defined compositions that are used to detect antigen-specific T cells [1]. Unfortunately, the compositional precision of these reagents is often obscured by shorthand nomenclatures that are useful in context, but that are an impediment to research reproducibility. Errors and confusion in how data are presented is a well-known problem within the scientific literature [2, 3] In order to improve the description of these popular reagents in the literature and in publicly funded resources such as the Immune Epitope Database [4], we propose minimal nomenclature guidelines for MHC multimers. Standardized nomenclature will ensure that multimer data are presented according to the FAIR principles and is more Findable, Accessible, Interoperable and Reusable [5]. Adherence to such standards has become increasingly endorsed or required across many areas of research [6, 7, 8, 9]. Following the precedent set by the Minimum information about a microarray experiment (MIAME), we present the Minimal Information about MHC Multimers (MIAMM, miamm.lji.org) [10]. As a key provider of such reagents, the National Institutes of Health (NIH) Tetramer Core Facility is driving this effort [11]. Recognized as experts in this field, the NIH Tetramer Core Facility is ideally positioned to develop and endorse such standards. Here, we present the details of MIAMM and additionally, provide a public resource to validate multimer terminology.
These guidelines were established with the expertise of the members of the NIH Tetramer Facility, laboratory users of such reagents, and database curators tasked with describing such reagents in machine-readable and interoperable digital resources. We found that no such existing standards could be identified via the FAIRsharing data standard portal [12]. The only available standards related to the description of MHC molecules are the MHC Restriction Ontology (MRO) [13], which is utilized by this MIAMM effort, as described below, and MaHCO, an Ontology for Major Histocompatibility Complex (MHC) Alleles and Molecules [14], which is no longer maintained. The MIATA (Minimal Information About T cell Assays) project overlaps with our effort, as it prescribes that authors report details about assay procedures including all reagents and materials used, that would allow the accurate repetition of the assay by others (Example reports: Intracellular Cytokine Staining assay, Elispot, Multimer) [15, 16]. We extend the MIATA recommendations to the level of how the details of multimer reagents should be reported. A need for such standards was identified by the NIH Tetramer Core Facility, when viewing publications describing the use of their reagents. Far too often, descriptions of MHC multimer reagents in the published literature are absent, misleading, imprecise, or otherwise incorrect. Likewise, the IEDB, a well-established (National Institute of Allergy and Infectious Diseases) NIAID resource which has manually curated more than 3,860,000 multimer assays from more than 21,600 manuscripts, has encountered similar obstacles when attempting to correctly and consistently represent these data [17]. Therefore, we set out to provide guidelines on how to report MHC multimer reagents in publications, based on three simple rules:
Provide a precise definition of the multimer reagent in the Materials and Methods section following the MIAMM standard. In these guidelines, this will be referred to as the “full name”.
Use reader-friendly abbreviations to refer to multimer reagents elsewhere in the manuscript, including figures, figure legends, and body text.
Within a manuscript, ensure that there is exactly one multimer abbreviation used for each multimer defined in the methods.
Results
Precise definitions of MHC ligand complexes
Most MHC molecules in a multimer contain three distinct subunits and each of these must be precisely defined (or unambiguously assumed). In MHC multimers, the class I heavy chains and the class II α and β chains are always truncated to remove the transmembrane domains, but for the purposes of these guidelines, the details of the truncations should be described in the Material and Methods section and the names of the corresponding MHC alleles from the official nomenclature can be used without introducing ambiguity.
MHC nomenclature
The Immuno Polymorphism Database (IPD, https://www.ebi.ac.uk/ipd/mhc/) is the most authoritative repository of MHC allele names for humans and a number of other important species in immunology research (with the unfortunate notable exception of the mouse) [18]. There are subsections of the IPD devoted to the highly standardized human MHC allele names (IPD-IMGT/HLA) and allele names for other species (IPD-MHC) including dogs, horses, rats, chickens, goats, cows, and a wide array of non-human primates. To provide a unifying source of terms for these vocabularies, the MHC Restriction Ontology (MRO) was developed, which incorporates both human MHC allele names from IMGT/HLA and all other species present in IPD, utilizing their approved nomenclature, and provides consistent additional properties, such as assignments to class I, class II, allele, serotype, and other relevant metadata [13]. MRO also contains standardized MHC nomenclature for species not yet present within the IPD library (e.g. the laboratory mouse), making these terms available for use now, while awaiting future guidelines from IPD, which will be adopted once available. Thus, all MHC terms should be designated using MRO terminology. Importantly for data standards, MRO follows official IPD and IMGT nomenclature standards and thus, stays current with new terminology. MRO is an openly developed and maintained ontology, with regular build cycles and versioning. Input and new term requests from the public are facilitated via a Github issue tracker (https://github.com/IEDB/MRO).
For the full name of an MHC multimer reagent, the full MRO label must be used. For example, an example from the literature, written as “DQB0201” would instead be represented as “HLA-DQB1*02:01”, which has the stable and resolvable ontology identifier of MRO:0000674. It is important to note that MIAMM requires specifying the MHC protein chains included in the tetramer. Thus, for example, HLA-A*02:01 is the appropriate designation, rather than HLA-A*02:01:01 or HLA-A*02:01:02, which identifies alleles that encode for the same protein chain.
Class and Locus-specific nomenclature issues
The class I beta 2-microglobulin (β2m) subunit.
Although alleles of β2m have been discovered in a number of species, it is not regarded as polymorphic and is therefore commonly ignored in nomenclatures [19]. In MRO, the β2m chain is included in the annotations of classical and nonclassical class I MHC complexes, but is not restated in the MHC complex term name, unless there is variability or the species differs between the two chains. In MHC multimer applications, the most notable fact about β2m is that reagents with heavy chains from the mouse (and other species) are commonly made with human β2m, in part because of data suggesting better stability of complexes using human β2m, and in part out of historical practice [20, 21, 22]. Here, we suggest authors describe the source of the β2m in the Materials and Methods section.
The class II α subunit.
While the class II β subunit is always polymorphic and must always be included in the multimer full name, the extent of the polymorphism of the class II α chain varies from locus to locus, and from species to species, and this must be considered in the full name. For example, the HLA-DRA1 locus is essentially monomorphic, and therefore it can be omitted, whereas the SLA-DRA locus is polymorphic and probably should be included. In contrast, there is significant allelic diversity for the HLA-DPA1 and HLA-DQA1 loci, thus it is recommended that these alleles be explicitly included in the full name of the multimer, even when experts may be aware of α and β alleles that are in linkage disequilibrium. For example, while HLA-DQ2.5 is a common shorthand for an isoform associated with autoimmune disease, its full name is referred to as HLA-DQA1*05:01/DQB1*02:01 in MRO. MRO allows users to specify one or both chains for class II molecules, for example, both HLA-DRA*01:01/DRB1*03:01 and HLA-DRB1*03:01 are both valid terms.
Non-natural MHC mutants
Non-natural MHC variants, often introduced to improve or reduce CD4 and CD8 coreceptor binding, are common in the MHC tetramer literature. From one perspective, these are exactly equivalent to discovery of a new allele, but the curators of allele databases generally restrict new entries to naturally occurring alleles. To address this, the MRO includes non-natural MHC molecules that have been engineered. The nomenclature for engineered alleles follows what is commonly used for other gene variants, by first indicating the natural sequence using the IMGT-IPD nomenclature, followed by the wild type amino acid, the position of the mutation within the mature protein chain, and the mutant amino acid. For example, “HLA-A*02:01 A150P” indicates an amino acid sequence that is identical to the wild type A*02:01 sequence apart from a substitution from Alanine to Proline at position 150. The amino acid numbering should be based upon the accepted natural mature protein sequence. Mature protein sequences for many species are currently available from IMGT (http://www.imgt.org) and they are working on making these more accessible. If a researcher engineers a new mutated MHC molecule, it will be added to MRO.
Peptide ligand nomenclature
Peptide definitions must include their complete amino acid sequence using the standard one-letter code established by the Nomenclature Committee of the International Union of Biochemistry (IUPAC-IUB) [23]. Table I contains examples of commonly studied peptidic multimer reagents, together with their standardized names and MRO mappings.
Table I.
Example Peptidic Multimers. Example peptidic multimers demonstrating how common textual names and abbreviations frequently found in publications do not convey precise information. Bold ambiguous terms in the textual names are translated to their precise meanings in the standardized name.
| Example textual name | Example abbreviations | MRO label | MRO ID | MIAMM standardized full name |
|---|---|---|---|---|
| HLA-A2/HIV-1 gag SLYNTVATL | A2/gag, A2/gag.SL9 | HLA-A*02:01 | MRO:0001007 | HLA-A*02:01, SLYNTVATL |
| HLA-A2/NS3 (286–294) | A2/NS3, A2/IIM | HLA-A*02:05 | MRO:0001011 | HLA-A*02:05, IIMDEAHFL |
| YFV VNWEVIIMDEAHFLD DRB3*0101 | YFV DRB3, VNW DRB30101 | HLA-DRB3*01:01 | MRO:0001338 | HLA-DRB3*01:01, VNWEVIIMDEAHFLD |
| TT NYSLDKIIVDYNLQSKITLP DRB3*0101 | TT DRB3, NYS DRB30101 | BoLA-DRB3*001:01 | MRO:0000946 | BoLA-DRB3*001:01, NYSLDKIIVDYNLQSKITLP |
| Ab/LCMVGP DIYKGVYQFKSV | I-Ab/LCMV.GP66 | H2-IAb | MRO:0000972 | H2-IAb, DIYKGVYQFKSV |
| DbNP366–374 | Db/ASN | H2-Db | MRO:0000966 | H2-Db, ASNENMETM |
| DbNP366–374 | Db/ASN | H2-Db/huB2m | MRO:0046252 | H2-Db/huB2m, ASNENMETM |
| DRB1/HA (306–318) | DRB1/HA 306 | HLA-DRB1*01:01 | MRO:0001279 | PKYVKQNTLKLAT |
| DRB1/HA (306–318) | DRB1/HA 306 | HLA-DRA*01:01/DRB1*01:01 | MRO_0001256 | PKYVKQNTLKLAT |
| HLA-DQ2.5/α-I | HLA-DQ2.5/QLQ | HLA-DQA1*05:01/DQB1*02:01 | MRO:0001229 | QLQPFPQPQLPY |
| DQ2.2-glia-α1 | HLA-DQB1*02:01 | MRO:0001237 | NPQAQGSVQPQQLPQF | |
| α-II HLA-DQ2 | α-II DQ2 | HLA-DQA1*05:01/DQB1*02:01 | MRO:0001229 | PQPELPYPQPE |
As peptide sequences are not necessarily unique to a single organism and a single antigen, organisms, antigens, and sequence positions within an antigen should not be included in the full name of a multimer reagent; to do so would often imply more information than is warranted. Nevertheless, we recognize that these attributes aid reader understanding, and we therefore recommend that the study-relevant organism, antigen, and position information be included in reagent abbreviations. Additionally, when D-amino acids or retroinverso peptides are used, this should be stated in the Materials and Methods.
To describe non-standard amino acids and/or amino acid modifications, the Protein Modification Ontology (PSI-MOD) terminology [24] should be used, which is also the convention used by UniProt and the Protein Ontology [25, 26]. PSI-MOD is presented as a standard for protein modification terminology by the Fairsharing resource, and like MRO, is a member of the OBO Foundry [27]. Table II provides all of the commonly used modifications, including those relevant to the NIH tetramer facility and found within the literature curated by the IEDB. Modifications must be described using the modification type followed by the position within the peptide, described using the amino acid abbreviation and the position, such as R5. Table III shows examples of commonly observed peptidic multimers with modifications and their ontology identifiers.
Table II.
Common Modifications. Modifications commonly found in multimer ligands standardized using PSI-MOD terminology.
| PSI-MOD label | Synonym | PSI-MOD ID |
|---|---|---|
| amidated residue | Amidation, AMID | MOD:00674 |
| 2-pyrrolidone-5-carboxylic acid (Glu) | Pyrrolidone carboxylic acid, PYRE, PCA, pyroglutamic acid | MOD:00420 |
| 9-fluorenylmethyloxycarbonyl (Fmoc) | Fluorenylmethyloxycarbonyl, FMOC | MOD:01109 |
| acetylated residue | Acetylation, ACET, AcRes, Acetyl | MOD:02078 |
| biotinylated residue | Biotin, BIOT, BtnRes, Biotinylation | MOD:01885 |
| cysteinylation (disulfide with free L-cysteine) | Cysteinylation, CYSTL, SCysCys | MOD:00765 |
| deamidated and methyl esterified residue | Deamidation followed by a methylation, DEAME | MOD:01369 |
| deamidated residue | Deamidation, DEAM, dNRes, Deamidated | MOD:00400 |
| dehydrated residue | Dehydration, DEHY, Dehydrated | MOD:00704 |
| formylated residue | Formylation, FORM, FoRes, Formyl | MOD:00493 |
| galactosylated residue | Galactosylation, GAL, GalRes | MOD:00728 |
| glucosylated residue | Glucosylation, GLUC, GlcRes, Glycation | MOD:00726 |
| glycosylated residue | Glycosylation, GLYC, GlycoRes | MOD:00693 |
| hydroxylated residue | Hydroxylation, HYL, HyRes | MOD:00677 |
| L-2-aminobutanoic acid (Glu) | L-2-Aminobutyric acid, Abu | MOD:00819 |
| L-citrulline | Citrullination, CITR, Cit, Deamidated | MOD:00219 |
| methylated residue | Methylation, METH, METH, MeRes | MOD:00427 |
| myristoylated residue | Myristoylation, MYRI, MYRI, MyrRes | MOD:00438 |
| mono N-acetylated residue | N-acetylation, NAc, NAcRes, N-Acetyl | MOD:00408 |
| oxidized residue | Oxidation, OX, OxRes | MOD:00675 |
| palmitoylated residue | Palmitoylation|PALM, PALM, PamRes, Palmitoyl | MOD:00440 |
| phosphorylated residue | Phosphorylation, PHOS, PhosRes, Phospho | MOD:00696 |
| reduced residue | Reduction, RED, RedRes | MOD:01472 |
| sulfated residue | Sulfation, SULF, SulfRes, Sulfo | MOD:00695 |
| other | OTH |
Table III.
Examples of Modified Peptidic Multimers. Commonly studied peptide modifications demonstrating how the textual names commonly found in publications do not clearly express which amino acid is modified and in what way.
| Textual name | PSI-MOD label | PSI-MOD ID | MRO label | MRO ID | MIAMM standardized full name |
|---|---|---|---|---|---|
| Aggrecan 200–215 Cit/DR4 | L-citrulline | MOD:00219 | HLA-DRB1*04:01 | MRO:0001290 | HLA-DRB1*04:01, DAGWLADQTVRYPIHT, L-citrylline R11 |
| DQA1*0102/DQB1*06:02/HCRT2–6-NH2 | amidated residue | MOD:00674 | HLA-DQA1*01:02/DQB1*06:02 | MRO:0001205 | HLA-DQA1*01:02/DQB1*06:02, ASGNHAAGILTM, amidated residue M12 |
| f-MIGWII.M3 | formylated residue | MOD:00493 | H2-M3 | MRO:0000999 | H2-M3, MIGWII, formylated residue M1 |
| IAk.beta1AR ac 181–200 | acetylated residue | MOD:00394 | H2-IAk | MRO:0000976 | H2-IAk, TVWAISALVSFLPILMHWWR, acetylated residue T1 |
Non-peptidic ligand nomenclature
Non-peptide definitions must include their Chemical Entities of Biological Interest (ChEBI) name and identifier (https://www.ebi.ac.uk/chebi/) [28]. ChEBI is a database and ontology containing information about chemical entities of biological interest and is also an OBO Foundry member, thus promoting interoperability among different resources. An example of a chemical structure known to have been tested as an multimer reagent is 1-O-(alpha-D-galactosyl)-N-hexacosanoylphytosphingosine, which is identified in ChEBI as CHEBI:466659. The IEDB has been able to accommodate all such structures encountered in the literature via this methodology, demonstrating its utility. Table IV presents some of the most commonly studied non-peptidic multimer ligands with their corresponding ontology identifiers.
Table IV.
Common Non-Peptidic Multimers. Commonly studied non-peptidic multimers and how their components are mapped to ontology identifiers to create a MIAMM standardized name. Quotes are used around nonpeptidic ligands that have a comma in their name.
| Presenting molecule | MRO label | MRO ID | ChEBI label | ChEBI ID | Synonyms | MIAMM standardized full name |
|---|---|---|---|---|---|---|
| CD1a | human CD1a | MRO:0001450 | DDM-838 | CHEBI:62384 | DDM | human CD1a, DDM-838 |
| CD1a | human CD1a | MRO:0001450 | 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine | CHEBI:75034 | Phosphatidylcholine(18:0/18:1) | human CD1a, 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| CD1a | human CD1a | MRO:0001450 | 1-O-oleoyl-sn-glycero-3-phosphocholine | CHEBI:28610 | lysophosphatidylcholine 18:1 | human CD1a, 1-O-oleoyl-sn-glycero-3-phosphocholine |
| CD1b | human CD1b | MRO:0001451 | 6-deoxy-D-glucos-6-yl corynomycolate | CHEBI:74256 | glucose monomycolate (C32) | human CD1b, 6-deoxy-D-glucos-6-yl corynomycolate |
| CD1b | human CD1b | MRO:0001451 | glucose 6-monomycolate | CHEBI:59474 | human CD1b, glucose 6-monomycolate | |
| CD1c | human CD1c | MRO:0001452 | β-D-mannosyl C32-phosphomycoketide | CHEBI:61808 | C32 mannosyl MPM, Mannosyl-1β-phosphomycoketide C32 | human CD1c, β-D-mannosyl C32-phosphomycoketide |
| CD1c | human CD1c | MRO:0001452 | C32 phosphomycoketide | CHEBI:77461 | C32 PM | human CD1c, C32 phosphomycoketide |
| CD1d | human CD1d | MRO:0001453 | 1-O-(α-D-galactosyl)-N-hexacosanoylphytosphingosine | CHEBI:466659 | α-GalCer, KRN7000 | human CD1d, 1-O-(α-D-galactosyl)-N-hexacosanoylphytosphingosine |
| CD1d | human CD1d | MRO:0001453 | 1-O-(6-acetamido-6-deoxy-α-D-galactosyl)-N-[(15Z)-tetracos-15-enoyl]phytosphingosine | CHEBI:63080 | PBS57 | human CD1d, 1-O-(6-acetamido-6-deoxy-α-D-galactosyl)-N-[(15Z)-tetracos-15-enoyl]phytosphingosine |
| CD1d | human CD1d | MRO:0001453 | 1-O-(α-D-galactopyranosyl)-N-tetracosanyl-2-aminononane-1,3,4-triol | CHEBI:495150 | OCH | human CD1d, “1-O-(α-D-galactopyranosyl)-N-tetracosanyl-2-aminononane-1,3,4-triol” |
| CD1d | human CD1d | MRO:0001453 | 1-(3-O-sulfo-β-D-galactosyl)-N-[(15Z)-tetracos-15-enoyl]sphingosine | CHEBI:41539 | sulfatide C24:1 | human CD1d, 1-(3-O-sulfo-β-D-galactosyl)-N-[(15Z)-tetracos-15-enoyl]sphingosine |
| MR1 | human MR1 | MRO:0001457 | 6-formylpterin | CHEBI:70981 | 6FP | human MR1, 6-formylpterin |
| MR1 | human MR1 | MRO:0001457 | 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil | CHEBI:78397 | 5-OE-RU | human MR1, 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil |
| MR1 | human MR1 | MRO:0001457 | 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil | CHEBI:78398 | 5-OP-RU | human MR1, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil |
| MR1 | human MR1 | MRO:0001457 | N2-acetyl-6-formylpterin | CHEBI:82729 | Ac-6-FP | human MR1, N2-acetyl-6-formylpterin |
Validation tool
In order to ensure a common understanding of the guidelines specified above, we developed a publicly accessible, web-based tool that allows a user to either a) parse the full name they generated, and validate that it fulfills the specifications, or b) compose the full name, based on the information added by the user. The tool ensures that the valid nomenclatures for MHC alleles are used based on MRO, valid amino acids are used for a peptide ligand, and valid amino acid modifications are specified using PSI-MOD. The tool interface is shown in Figure 1.
Figure 1. Multimer Validation Tool.
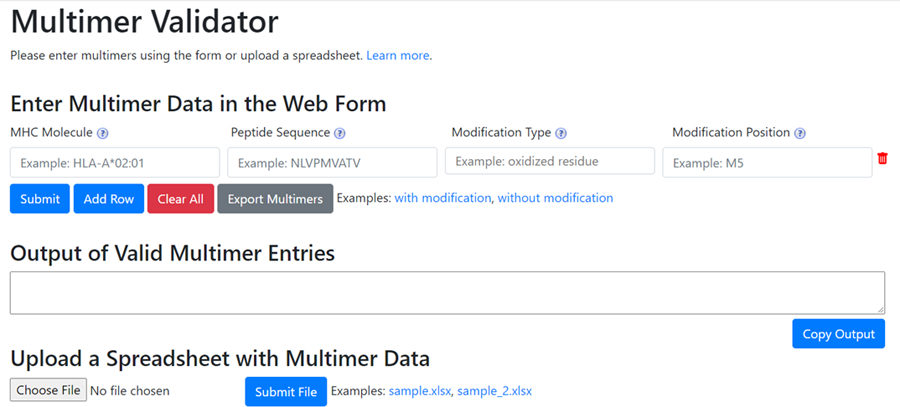
This freely available web tool allows users to validate the terminology used for their multimers at https://multimer.lji.org. Examples are provided and terms may be validated one at a time or uploaded as a set.
This tool accepts manually entered data, as well as provides the ability to validate a set of data via an upload feature. It was tested by research scientists who frequently use MHC multimers in their research as well as biocurators, who commonly encounter this type of data in the literature. Currently, the tool only validates multimers containing peptidic ligands, however, development of a non-peptidic ligand feature is underway. It is freely available at: miamm.lji.org and we welcome the public to use this tool to standardize their MHC multimer terms prior to publication. This website also serves as a portal to inform users of the MIAMM standard and the various ontologies employed by it.
Abbreviations
Once a multimer is precisely defined in the Materials and Methods section, the recommendations for how abbreviations should be constructed are decidedly less prescriptive, and the chosen approach is dependent upon the context and considerations of uniqueness and consistency throughout a manuscript. For abbreviations, there is one absolute rule and one strong suggestion that must be followed:
Abbreviations for multimers must be unique within a manuscript and used consistently.
Abbreviations for multimers should include a reference to both the MHC allele and its ligand.
As an example, in the very first paper describing MHC multimers, we described three tetramers and referred to them as A2-gag, A2-pol, and A2-MP [29]. Within the context of that paper, these abbreviations are unambiguous and meet the criteria of these guidelines.
A variety of peptide name abbreviation systems have been used in the MHC multimer literature. Three examples are:
Use of the antigen name and a position indicator. A common example of this is the I-Ab restricted GP66 epitope from LCMV.
Use of the first three amino acids of the peptide. Common examples are the HLA-B*08:01 restricted epitopes from EBV, RAK and FLR.
Use of the first and last amino acid of the peptide sequence, followed by the length of the peptide. Common examples of this are the Mamu-A*01 restricted epitope from SIV gag referred to as CM9 and the HLA-B*57:01 restricted epitope from HIV-1 gag called IW9.
Any of these approaches (and potentially others) are acceptable as abbreviations for peptides in MHC multimers.
Cautionary notes
While we have embraced the principle of simplicity in these guidelines for multimer abbreviations, we do raise one important cautionary note. The simplest abbreviations are not future proof, and while this is unlikely to be a problem for a manuscript, it may become a problem when labeling reagent aliquots within a laboratory. An abbreviation such as A2-pol may be unique when the reagent is first made, but what happens if additional A2-restricted epitopes are discovered in the HIV pol protein, or if A2-restricted epitopes are discovered in the pol proteins of non-HIV viruses? It is hard to enforce forward-looking nomenclatures in the laboratory, especially for abbreviations, but we encourage it.
Other multimer features
There are numerous other components and formulations of an MHC multimer beyond the typical MHC allele and ligand combinations. For example, class II MHC molecules are often expressed with peptide ligands covalently tethered to the amino terminus of the beta chain [30], sometimes with an added disulfide cross-link [31]. While this current nomenclature proposal does not easily capture these details (suggestions will be collected and discussed on the https://miamm.lji.org/ website), they must be thoroughly described in the Materials and Methods section of the manuscript. Two common additional components, which we will discuss here are (1) fusions to the MHC molecule and (2) the multimerization agent and its linked labels.
Fusions to the MHC molecule.
Features in this category include flexible linkers, leucine zippers, immunoglobulin Fc domains, BirA substrate peptides (or StrepTag equivalents), and affinity tags for purification (e.g. His6 tags). This type of information is critical to be included in the Materials and Methods section of a manuscript, however, they are not required in reagent nomenclatures.
Multimerization agents.
Streptavidin is by far the most common multimerization agent used in MHC multimer technology, but it is not the only one. In addition, it is sometimes combined with additional agents to produce higher-order multimers, such as in the Dextramer technology and the recently developed Spheromer technology [32, 33]. Furthermore, in MHC multimer reagents, the multimerization agent almost always serves a second function – it is the component that is covalently linked to a label for detection.
The multimerization technology should always be described in the Materials and Methods section of a manuscript. Common examples include “tetramer”, “pentamer”, and “dextramer”. Detection labels such as fluorophores must also be included. Conjugation of protein fluorophores such as R-phycoerythrin (PE) and allophycocyanin (APC) results in mixtures of higher order aggregates that vary by vendor, so vendors and catalog numbers for these reagents should be included in Materials and Methods when they are known.
RRID
To extend the standards proposed here and to further contribute to FAIR data, the NIH Tetramer Core Facility is working with the Resource Identification Initiative (RRI) [34] to produce Research Resource Identifiers (RRIDs) for each of our reagents. Furthermore, we and the RRI encourage commercial multimer vendors to do the same (note that we are not imposing any guidelines upon how vendors market their products). We are hopeful that this publication will draw attention to this effort and extend its adoption. We anticipate vendors and other researchers generating multimer reagents will be incentivized to standardize their descriptions and obtain RRIDs because many journals (e.g. Star Methods in Cell press journals) and the NIH increasingly require RRIDs as part of their Rigor and Reproducibility initiatives.
Discussion
Herein, we have introduced guidelines for the Minimal Information about MHC Multimers (MIAMM, miamm.lji.org), as summarized in Table V. The primary goals of these guidelines are two-fold – to enhance the reproducibility and annotatability of the literature using these reagents. We have attempted to provide guidelines that are as least prescriptive as possible, while still adding value. To achieve these goals, we have recommended two basic principles: (1) describe the reagents as precisely as possible in the Materials and Methods section of a manuscript, and (2) use suitable abbreviations elsewhere.
Table V.
Minimal Information about MHC Multimer (MIAM) components and how they are standardized.
| Component | Standardization method | Example | Standardized name |
|---|---|---|---|
| MHC molecule | MHC Restriction Ontology (MRO) | A0201 | HLA-A*02:01 |
| Peptidic ligand | Nomenclature Committee of the International Union of Biochemistry (IUPAC-IUB) | SLYNTVATL | SLYNTVATL |
| Non-peptidic ligand | Chemical Entities of Biological Interest (ChEBI) | α-GalCer | 1-O-(α-D-galactosyl)-N-hexacosanoylphytosphingosine |
| Post translational modification | Protein Modification Ontology (PSI-MOD) | Acetylation | acetylated residue |
| Modification position | Letter followed by position in ligand | the first leucine in SLYNTVATL | L2 |
Adoption of these standards is facilitated by the Multimer Validation Tool, which has been used to standardize the data present in the NIH Tetramer Core Facility. Additionally, it has been used to validate the nearly 18,500 multimer assays in the IEDB, demonstrating the applicability to real life data, as these assays were derived from more than 2,400 separate publications. It has also been shared with stakeholders in both the academic and commercial sectors to facilitate implementation of these standards. We hope that this publication will bring increased awareness of MIAMM and drive acceptance. As a key stakeholder and recognized leader in this field, the NIH Tetramer Core Facility is well-positioned to promote these standards. Both the NIH Tetramer Facility and the IEDB are stable, publicly funded resources well-suited to the maintenance of this project. We hope the application of MIAMM will improve the reproducibility and annotatability of MHC multimer reagent data.
Funding:
JDA, RAW, and DLL acknowledge support from the contract for the NIH Tetramer Facility (75N93020D00005) from the Yerkes National Primate Research Center (P51OD011132), and the Emory Center for AIDS Research (P30AI050409). RV, JAO, AM, and BP acknowledge support from National Institutes of Health grant R24 HG010032. RV, JAO, AM, BP, and AS acknowledge support from National Institutes of Health contract 75N93019C00001
References
- 1.McHeyzer-Williams MG, Altman JD, Davis MM 1996. Enumeration and characterization of memory cells in the TH compartment. Immunol Rev 150: 5–21. [DOI] [PubMed] [Google Scholar]
- 2.Vita R, Vasilevsky N, Bandrowski A, Haendel M, Sette A, Peters B 2016. Reproducibility and Conflicts in Immune Epitope Data. Immunology 147(3): 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, Larocca GM, Haendel MA 2013. On the reproducibility of science: unique identification of research resources in the biomedical literature. PeerJ 1:e148: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B 2019. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47: D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ‘t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, and Mons B 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 15;3:160018: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NIH Data Sharing Policy: http://grants.nih.gov/grants/policy/data_sharing Accessed on July 23,2021.
- 7.Wellcome Trust: https://wellcome.org/sites/default/files/interoperability-standards-oct16.pdf Accessed on July 23,2021.
- 8.Concordat on open research data, Higher Education Funding Council for England (HEFCE), Research Councils UK (RCUK), Universities UK (UUK) and Wellcome Trust: https://www.ukri.org/wp-content/uploads/2020/10/UKRI-020920-ConcordatonOpenResearchData.pdf, Accessed on July 23,2021.
- 9.Boulton G, Campbell P, Collins B, Elias P, Hall W, Laurie G, O’Neill O, Rawlins M, Thorton J, Vallance P, and Walport M 2012. Science as an open enterprise. Alan Turing Institute “Symposium on reproducibility for data-intensive research” report [online]. https://royalsociety.org/-/media/policy/projects/sape/2012-06-20-saoe.pdf:1-105.
- 10.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, and Vingron M 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nature genetics 29.4: 365–371. [DOI] [PubMed] [Google Scholar]
- 11.https://tetramer.yerkes.emory.edu, Accessed on July 23,2021.
- 12.https://fairsharing.org, Accessed on July 23,2021.
- 13.Vita R, Overton JA, Seymour E, Sidney J, Kaufman J, Tallmadge RL, Ellis S, Hammond J, Butcher GW, Sette A, Peters B 2016. An ontology for major histocompatibility restriction. for major histocompatibility restriction. J Biomed Semantics 11;7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca DS, Beisswanger E, Wermter J, Horn PA, Hahn U and Blasczyk R 2009. MaHCO: an ontology of the major histocompatibility complex for immunoinformatic applications and text mining, Bioinformatics (Oxford, England) 25: 2064–2070. [DOI] [PubMed] [Google Scholar]
- 15.Janetzki S, Britten CM, Kalos M, Levitsky HI, Maecker HT, Melief CJM, Old LJ, Romero P, Hoos A, Davis MM 2009. “MIATA”-minimal information about T cell assays. Immunity 16;31(4): 527–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britten CM, Janetzki S, Butterfield LH, Ferrari G, Gouttefangeas C, Huber C, Kalos M, Levitsky HI, Maecker HT, Melief CJM, O’Donnell-Tormey J, Odunsi K, Old LJ, Ottenhoff THM, Ottensmeier C, Pawelec G, Roederer M, Roep BO, Romero P, van der Burg SH, Walter S, Hoos A, Davis MM 2012. T Cell Assays and MIATA: The Essential Minimum for Maximum Impact. Immunity 27;37(1): 1–2. [DOI] [PubMed] [Google Scholar]
- 17.Vita R, Overton JA, Sette A, Peters B 2017. Better living through ontologies at the Immune Epitope Database. Database (Oxford) 1(1): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE 2020. IPD-IMGT/HLA Database. Nucleic Acids Research 48: D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermel E, Robinson PJ, She JX, Lindahl KF 1993. Sequence divergence of B2m alleles of wild Mus musculus and Mus spretus implies positive selection. Immunogenetics 38(2): 106–116. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen LO, Stryhn A, Holter TL, Etzerodt M, Gerwien J, Nissen MH, Thøgersen HC, Buus S 1995. The interaction of beta 2-microglobulin (beta 2m) with mouse class I major histocompatibility antigens and its ability to support peptide binding. A comparison of human and mouse beta 2m. Eur J Immunol 25(6): 1609–1616. [DOI] [PubMed] [Google Scholar]
- 21.Shields MJ, Assefi N, Hodgson W, Kim EJ, Ribaudo RK 1998. Characterization of the interactions between MHC class I subunits: a systematic approach for the engineering of higher affinity variants of beta 2-microglobulin. J Immunol 160(5): 2297–2307. [PubMed] [Google Scholar]
- 22.Altman JD, Davis MM 2016. MHC-Peptide Tetramers to Visualize Antigen-Specific T Cells. Curr Protoc Immunol 115:17.3.1–17.3.44. [DOI] [PubMed] [Google Scholar]
- 23.IUPAC-IUB Commission on Biochemical Nomenclature (CBN), A One-Letter Notation for Amino Acid Sequences. 1968. Arch. Biochem Biophys 125(3),i–v. [Google Scholar]
- 24.Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS 2008. The PSI-MOD community standard for representation of protein modification data. Nat Biotechnol 26(8):864–866. [DOI] [PubMed] [Google Scholar]
- 25.The UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47: D506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natale DA, Arighi CN, Blake JA, Bona J, Chen C, Chen SC, Christie KR, Cowart J, D’Eustachio P, Diehl AD, Drabkin HJ, Duncan WD, Huang H, Ren J, Ross K, Ruttenberg A, Shamovsky V, Smith B, Wang Q, Zhang J, El-Sayed A, Wu CH 2017. Protein Ontology (PRO): enhancing and scaling up the representation of protein entities. Nucleic Acids Res 45(D1): D339–D346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, The OBI Consortium, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S 2007. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature Biotechnology 25, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C 2016. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res 44(D1): D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274(5284): 94–96. [DOI] [PubMed] [Google Scholar]
- 30.Kozono H,, White J, Clements J, Marrack P, Kappler J 1994. Production of soluble MHC class II proteins with covalently bound single peptides. Nature 369(6476): 151–154. [DOI] [PubMed] [Google Scholar]
- 31.Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK 2010. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J Immunol 185(8): 4705–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batard P, Peterson DA, Devêvre E, Guillaume P, Cerottini JC, Rimoldi D, Speiser DE, Winther L, Romero P 2006. Dextramers: new generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8+ T cells. J Immunol Methods 310(1–2): 136–148. [DOI] [PubMed] [Google Scholar]
- 33.Mallajosyula V, Ganjavi C, Chakraborty S, McSween AM, Pavlovitch-Bedzyk AJ, Wilhelmy J, Nau A, Manohar M, Nadeau KC, Davis MM 2021. CD8 + T cells specific for conserved coronavirus epitopes correlate with milder disease in COVID-19 patients. Sci Immunol 6(61):eabg5669; 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, Hof PR, Martone ME, Pols M, Tan SC, Washington N, Zudilova-Seinstra E, Vasilevsky N 2016. The Resource Identification Initiative: A Cultural Shift in Publishing. J Comp Neurol 524(1): 8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]


