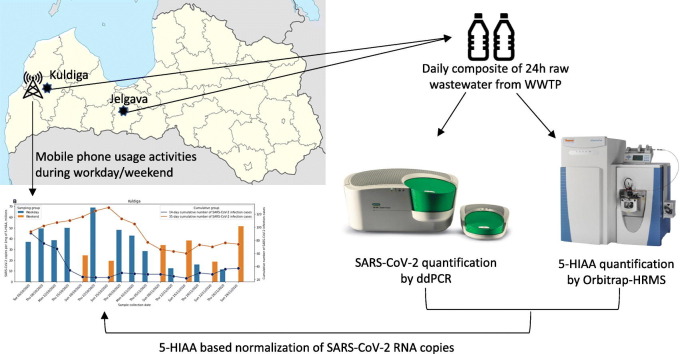
Abstract
Wastewater-based epidemiology (WBE) has regained global importance during the COVID-19 pandemic. The mobility of people and other factors, such as precipitation and irregular inflow of industrial wastewater, are complicating the estimation of the disease prevalence through WBE, which is crucial for proper crisis management. These estimations are particularly challenging in urban areas with moderate or low numbers of inhabitants in situations where movement restrictions are not adopted (as in the case of Latvia) because residents of smaller municipalities tend to be more mobile and less strict in following the rules and measures of disease containment. Thus, population movement can influence the outcome of WBE measurements significantly and may not reflect the actual epidemiological situation in the respective area. Here, we demonstrate that by combining the data of detected SARS-CoV-2 RNA copy number, 5-hydroxyindoleacetic acid (5-HIAA) analyses in wastewater and mobile call detail records it was possible to provide an accurate assessment of the COVID-19 epidemiological situation in towns that are small (COVID-19 28-day cumulative incidence r = 0.609 and 35-day cumulative incidence r = 0.89, p < 0.05) and medium-sized towns (COVID-19 21-day cumulative incidence r = 0.997, 28-day cumulative incidence r = 0.98 and 35-day cumulative incidence r = 0.997, p < 0.05). This is the first study demonstrating WBE for monitoring COVID-19 outbreaks in Latvia. We demonstrate that the application of population size estimation measurements such as total 5-HIAA and call detail record data improve the accuracy of the WBE approach.
Abbreviations: ddPCR, Digital Droplet PCR; WW, Wastewater; WWTP, Wastewater treatment plant; PEG, Polyethylene glycol; WBE, Wastewater based epidemiology; CDR, call detail record; SD, standard deviation
Keywords: SARS-CoV-2, WBE, Wastewater, ddPCR, 5-HIAA, Mobile phone data
Graphical abstract
1. Introduction
The COVID-19 pandemic caused by the SARS-CoV-2 virus reached Latvia in March 2020, with an average of 7.4 new cases daily and a mean 14-day cumulative incidence of 3.1 ± 4.5 until late September 2020. During this period several outbreaks of SARS-CoV-2 were detected and successfully contained due to extensive testing and contact tracing led by the Disease Prevention and Control Centre. However, the incidence increased rapidly starting from October, reaching its maximum on January 10th, 2021, with a 14-day cumulative incidence of 693.9. Since then, the number of COVID-19 cases has remained consistently high (>200 until June 7th, 2021), with only periodical small-scale fluctuations (Centre for Disease Prevention and Control, 2021a), which was associated partially with the spread of emerging SARS-CoV-2 variants. At the moment of manuscript preparation, on December 9th, 2021, 259,215 confirmed infection cases and 4325 deaths have been reported in Latvia (Centre for Disease Prevention and Control, 2021b).
Several studies have analysed the continuous data of the SARS-CoV-2 presence in the wastewater (WW) from different countries: France (Trottier et al., 2020), the USA (Gerrity et al., 2021; Gonzalez et al., 2020), Qatar (Saththasivam et al., 2021), Finland (Hokajärvi et al., 2021), Spain (Randazzo et al., 2020), Germany (Westhaus et al., 2021), the UK (Hillary et al., 2021), Sweden (Saguti et al., 2021) and others; concluding that tracing the viruses that are present in raw WW has proved to be a quick and timely method to track the dynamics of the infection within a community (Ahmed et al., 2020a; Peccia et al., 2020). Nevertheless, an estimation of the approximate number of infected individuals remains a worldwide challenge for several reasons. These reasons include, but are not limited to, diverse viral shedding rates in a population, high dilution of viral RNA and their concentration fluctuations by rain and industrial WWs, the presence of compounds or particles that may either degrade or physically protect the virus and loss of viral RNA during the transit time through the WW network (Hillary et al., 2021; Polo et al., 2020). While some of the above-mentioned reasons might have a low impact on large cities with a high population density for the detection and quantification of WW viral RNA, smaller size populations contributing to a particular WW catchment are affected to a much greater extent.
Common methods for the detection of SARS-CoV-2 RNA in WW are real-time quantitative polymerase chain reaction (RT-qPCR) or droplet digital PCR (ddPCR), which can provide strain-level resolution and quantification (Farkas et al., 2020). At present RT-qPCR is considered conventional, because of its wide use and reliable performance in molecular diagnostics. However, the recent emergence of ddPCR offers greater precision and reproducibility of measurements even at low level concentrations of target nucleic acid (Taylor et al., 2017). Currently, various WW-based SARS-CoV-2 identification strategies have been established and introduced into routine monitoring (Donde et al., 2021; Gonzalez et al., 2020; Kumar et al., 2020; Tiwari et al., 2021; Zhao et al., 2022). However, the detection and estimation of SARS-CoV-2 in the WW encounters problems such as low abundances of the viral RNA and poor sample stability because enveloped ssRNA viruses are more fragile to water treatment processes and environmental conditions than nonenveloped viruses (Ahmed et al., 2022; Corpuz et al., 2020; Kumar et al., 2021; Sbaoui et al., 2021). To ensure a comparison between cities, sewage parameters are normalized by the number of people that are served by the treatment plants. The ideal scenario would be the application of a ubiquitous biomarker that has no other source than human excretion, is stable during the time spent in sewers and is consistent between different populations. Several human faecal presence indicators have been proposed for this role, such as cross-assembly phage (Ai et al., 2021; Crank et al., 2020; Green et al., 2020; Wilder et al., 2021) and pepper mild mottle virus (Ai et al., 2021; D'Aoust et al., 2021; Jafferali et al., 2021). However, these indicators might not be appropriate for the assessment of population size due to the abundant differences in different individuals and cultural differences in cuisine preferences between different populations and no improvement was observed with their application to WW signal correlation in new case numbers (Ai et al., 2021). On the contrary, it has been proposed that an improved estimation of COVID-19 positive cases can be achieved by an appropriate viral concentration choice and the designated biomarker should be correlated with other human viral pathogens (Crank et al., 2020). Apart from biological biomarkers, many chemical population biomarkers have been suggested (caffeine, creatinine, cholesterol, coprostanol, cotinine, cortisol, androstenedione and 5-hydroxyindoleacetic acid (5-HIAA), etc.). Several authors have studied the applicability of these candidate compounds by considering the content in WW, stability in WW, as well as the consistency of inter-day excretion and correlation between excretion and population figures. Most of the studies resulted in the proposals to use ammonium, caffeine, cotinine, and 5-HIAA, which were quantifiable and stable in the WW, thus demonstrating a sufficient correlation with population size (O'Brien et al., 2014; Rico et al., 2017; Thai et al., 2019).
Based on literature research, an HPLC-MS/MS-based assay of neurotransmitter metabolite 5-HIAA has been selected in our study since it is an endogenous compound that is not lifestyle or habit dependent. In addition to biological and chemical biomarkers, the use of mobile data as a real-time data source for population size measurements has become increasingly relevant (Arhipova et al., 2020), and allows an assessment of the trends of regional economic development or to determine the change in population mobility patterns (Chen et al., 2018; Wu et al., 2021). This can be achieved through the use of call detail record (CDR) data, which are collected by mobile network operators and contain information about when, where and how a mobile network user generates voice calls and text messages (Chen et al., 2018). A limited number of studies have estimated population size using CDR and wastewater-based epidemiology (WBE) tools (Thomas et al., 2017), however, further insight into correlation estimation between CDR and WBE data with different population sizes and wastewater treatment plant (WWTP) catchments are needed.
Therefore, the objectives of this study were (i) to demonstrate the applicability of WBE in small and medium-sized municipalities of Latvia and (ii) to test whether the combination of data from biomarkers (5-HIAA) and mobile CDR can increase the accuracy of a relationship assessment between the amount of detected SARS-CoV-2 RNA copies in WW and confirmed COVID-19 cases. To reach the objectives, 24-hour WW composite samples of two municipalities were collected and analysed for the presence and quantity of SARS-CoV-2 viral RNA by ddPCR. To further investigate if the viral data obtained can be associated with the publicly available information on the confirmed COVID-19 cases, additional quantitative measurements for the 5-HIAA in the same WW composite samples were performed using mass-spectrometry based methods. In addition, we evaluated whether 5-HIAA measurements from the WW can be supported by the CDR data.
2. Methods
2.1. Sample collection
Two municipalities of Latvia were selected for continuous monitoring of SARS-CoV-2 in WW: Jelgava (~55,000 inhabitants) and Kuldiga (~11,000 inhabitants). The daily composites of 24-hourly raw WW samples (7.2 l) were collected at the WWTP (after grit removal) using a portable autosampler P6 MINI MAXX (MAXX Mess und Probenahmetechnik GmbH, Rangendingen, Germany), which was operating in a time-dependent mode (meaning 300 ml of WW were automatically collected after every hour, regardless of the WW flow fluctuations at the WWTP). In the design of the sampling procedure, we used the experience of European studies (Gawlik et al., 2021) in which 24 h composite samples were preferred. Collected samples were immediately transferred to the laboratory, stored at 4 °C and further processed within 24 h. Samples at each municipality were collected once or twice per week. Samples from Jelgava WWTP were collected from 17th August 2020 until 30th November 2020, resulting in the acquisition of 16 samples; samples from Kuldiga WWTP started from 7th October 2020 until 29th November 2020, resulting in a collection of 17 samples (Supplementary Table 2). To account for factors that could exert a substantial impact on the stability of SARS-CoV-2 RNA and fluctuations in population size, several physical and chemical measurements were carried out following the sample collection, these included pH, temperature, electrical conductivity and 5-HIAA concentration measurements. To estimate the total volume of WW that flowed through the site during sample collection time we also copied the appropriate water meter reading records. However, because of access restrictions, the meter reading times did not match those of sample collection start and end times, therefore for this study the total volume of municipality WW that flowed into the WWTP during sample collection was estimated through the calculation of average WW volume per hour during the period between two-meter records and multiplied by the amount of time the sample collection was carried out. If the sample collection spanned several meter record time periods, then the calculation was carried out for each period individually and subsequently summed.
2.2. WW treatment and RNA extraction
The WW sample (180 ml, 4 × 45 ml) was centrifuged at 8000 ×g for 30 min at 4 °C to remove larger particles, such as bacterial cells and debris. The supernatant was transferred to new tubes and virus particles were precipitated using polyethylene glycol (PEG) 8000 as described elsewhere (Fuqing et al., 2021). Briefly, 24 g of PEG 8000 (8% w/v, Sigma-Aldrich, St. Louis, MO, USA) and 5.4 g of NaCl (PanReac AppliChem, Darmstadt, Germany) were added to the supernatant. The mixtures were incubated for 2 h at room temperature with gentle agitation. The precipitated virus particles were recovered by centrifugation at 12000 ×g for 10 min (Balke et al., 2018). Total RNA was isolated with Tri reagent (Sigma-Aldrich, cat. No. 93289) according to the manufacturer's instructions and eluted in 30 μl molecular grade water. RNA samples were stored at −80 °C and subjected to RT-qPCR analysis within 24 h after the RNA extraction. RNA concentration was estimated using the Qubit RNA HS Assay kit and Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. To assess the efficiency of the extraction procedure and estimate the viral recovery rate, each sample was supplemented with 5 μl of surrogate - recombinant, replication-defective and GFP gene containing Semliki Forest Virus (SFV) particles (constructed and cultivated at Latvian Biomedical Research and Study Centre). The concentration of the surrogate particles was 2.27 × 107 per 1 ml as estimated by the determination of infectious particle titre. The number of GFP copies per 1 μl of the surrogate was 100,700 as determined by ddPCR. The procedure of copy number determination is described in subsequent sections.
2.3. ddPCR
As ddPCR is superior to qPCR in terms of cDNA detection sensitivity and precision of quantification (Taylor et al., 2017), in this study a ddPCR approach was used for the accurate quantification of SARS-CoV-2 RNA copies within WW samples and assessment of surrogate recovery rate. For this purpose, analyses were carried out on two regions of the SARS-CoV-2 nucleocapsid (N) gene (N1 and N2), a single region of SARS-CoV-2 envelope small membrane protein (E) gene and a single region of the surrogate, the recombinant SFV (rSVF) GFP gene (Supplementary Table 1). All reactions were carried out in a single plex using a One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad, Hercules, CA, USA). For each reaction, negative and positive controls targeting the SARS-CoV-2 N gene were added. The reaction mixture contained 5 μl of Supermix, 2 μl of reverse transcriptase, 1 μl of 300 mM dithiothreitol, appropriate primers (forward and reverse) and probes (Metabion, Planegg, Germany) to a final concentration of 1.21 μM each, 2 μl of extracted WW RNA and RNase free water to a total volume of 22 μl. The following steps included droplet generation with a QX200 Droplet Generator (Bio-Rad), amplification in a T100 Thermal Cycler (Bio-Rad) (under following conditions: Ramp Rate setting 1; 50 °C for 60 min; 95 °C for 60 min; 40 cycles of 94 °C for 30 s and 60 °C for 2 min; 98 °C for 10 min), 4 h equilibration and droplet stabilization at room temperature and positive/negative droplet quantification in a QX200 Droplet Reader (Bio-Rad). Acquired data were analysed using QuantaSoft software (Bio-Rad) and calculated to the number of copies per ml of WW. The average from three SARS-CoV-2 analyses was used in all further viral concentration assessments. The recovery of surrogate SFV was calculated as the percentage of recovered GFP RNA coding copies.
2.4. Measurements of 5-HIAA within WW
All the solvents were of HPLC grade and were purchased from Fluka (Buchs, Switzerland) or Sigma-Aldrich. Deionised water was generated with a Milli-Q water purification system (Millipore, Billerica, MA, USA). All the analytical standards used in this study were obtained from Sigma-Aldrich. Solid-phase extraction was performed on Phenomenex (Torrance, CA, USA) Strata-X cartridges (200 mg/3 ml). Cartridges were conditioned with 3 ml of methanol and 3 ml of deionised water. The samples were filtered through glass fibre filters using a vacuum. After acidification with 100 μl of formic acid, the samples were loaded on columns at the approximate flow rate of 5 ml min−1. Columns were rinsed with 3 ml of 5% methanol solution in water and then dried for 60 min under vacuum, followed by elution with 2 × 3 ml of methanol. The eluates were then evaporated to dryness under a gentle nitrogen stream in a 40 °C water bath. The samples were reconstituted in 200 μl of water/methanol (80/20, v/v) containing the internal standard of salbutamol-d3 at a concentration of 0.05 ng μl−1. Extracts were filtered through Phenomenex PVDF 0.22 μm centrifugal filters. For the analysis of 5-HIAA, final extracts were diluted ten-fold with water/methanol (80/20, v/v).
Instrumental analysis was performed on the UHPLC-Orbitrap-HRMS system (Thermo Fisher Scientific). Chromatographic separation was achieved on a Thermo Hypersil Gold C18 analytical column (100 × 2.1 mm, 1.9 μm) column using mobile phases of 0.01% acetic acid in water (A) and acetonitrile (B) at a flow rate of 0.3 ml min−1. A gradient programme was used: 5% of the mobile phase (B) was used from 0 to 1.0 min, 5% (B) to 95% (B) from 1.0 to 5.0 min, maintained at 95% (B) from 5.0 to 9.0 min, then decreased back to 5% (B) from 9.0 to 10.0 min and finally, the column was re-equilibrated with 5% (B) from 10.0 to 15 min. A 10 μl aliquot of the extract was injected. The column and autosampler were maintained at 30 °C and 15 °C, respectively.
An Orbitrap-HRMS was equipped with a heated electrospray ionisation probe (HESI II) operating in the positive ionisation mode. The 5-HIAA was detected in full-scan mode by measuring two characteristic ions, 192.0655 m/z (quantitative) and 146.0606 m/z (qualitative), using a mass tolerance of 10 ppm. Quantification was performed by external calibration using a five-point calibration curve obtained by spiking pooled WW samples at the concentration range of 2.5 to 50 μg l−1. Multiple fortified WW samples were analysed in each batch for quality control purposes, the obtained recoveries were in the range of 83% to 104%. Since no blank WW samples were available, surface water was used as a blank sample. Limit of quantification (LOQ) was determined by analysing fortified surface water samples. The determined LOQ value was 1 μg l−1. Quantification was performed by external calibration using a five-point calibration curve obtained by spiking pooled WW sample at a concentration range of 2.5 to 50 μg l−1. A linear calibration curve with a correlation coefficient r 2 > 0.99 was obtained.
2.5. Estimation of population size through the use of mobile activity data (CDR)
The unique mobile phone user dataset consisted of the aggregated CDR collected from October 2020 to November 2020 from all base stations in the Kuldiga municipality cellular network (outgoing or incoming calls, SMS). No personal data was collected and used; hence we complied with General Data Protection Regulation and ethical norms. The average activity per hour was calculated for each day and multiplied by the amount of time the sample collection was carried out during that day to acquire comparable datasets (Supplementary Table 2). Consequently, the details of the mobile data applied in the evaluation are lost (daily instead of hourly data is applied), however, the simplified data leads to a statistically correct comparison of datasets.
2.6. Data visualization and statistics
Relative normalization of viral load in WW against population size was performed through the use of population size marker 5-HIAA by dividing the total copies of detected viral RNA with the total amount of 24 h 5-HIAA. Datasets of confirmed SARS-CoV-2 cases for each municipality were acquired from the Centre for Disease and Prevention and Control and can be downloaded from https://data.gov.lv/dati/lv/dataset/covid-19-pa-adm-terit. Cumulative incidence for 14, 21, 28 and 35 days was calculated for each municipality; and the time series obtained were smoothed by locally weighted regression LOWESS (Cleveland, 1979) using the statsmodels (Seabold and Perktold, 2010) package within the Python environment. The fraction of the data points that were considered to fit a regression model was set to 0.3. The same locally weighted regression was performed to the time series measurements of the total detected viral RNA copies and 5-HIAA normalized SARS-CoV-2 RNA copies. In the case of the Kuldiga time series, locally weighted regression with the same parameters was performed separately on the full dataset, workday and weekend dataset. Spearman's rank correlation coefficient with the associated p-value of total viral copies in millions and millions of copies per mg of 5-HIAA versus the cumulative incidence of respective periods was carried out using the scipy.stats package (Virtanen et al., 2020) within the Python environment. Spearman's correlation analysis was also performed to evaluate the association between the total amount of 5-HIAA and the average number of phone calls in Kuldiga. Next, the two-tailed Mann-Whitney test (Mann and Whitney, 1947) was used to compare (i) 5-HIAA measurements between different sampling days of the week, (ii) between total viral copies, 5-HIAA normalized viral copies and cumulative incidence; (iii) normalized viral load measurements between different sampling days of the week within a municipality, and (iv) the average number of phone calls between weekends and weekdays. All figures were created using matplotlib (Hunter, 2007) and seaborn (Waskom, 2021) libraries within the Python environment.
3. Results
3.1. Physicochemical analysis of WW samples
The analysis of parameters that were measured on-site revealed that to some extent they were municipality-specific, but otherwise rather stable over the whole study time (Supplementary Table 2). Thus, the average WW pH value in Jelgava was 7.77 (standard deviation (SD) = 0.14) and the average electrical conductivity was 2015.38 μS/cm (SD = 99.75), while in Kuldiga they were 7.67 (SD = 0.15) and 1215.11 μS/cm (SD = 111.70), respectively. Greater variations were observed in WW temperature, which in Jelgava on average was 15.01 °C (SD = 4.64), whereas in Kuldiga it was 11.78 °C (SD = 1.65), but these variations correlated with the temperature of the surrounding environment and were appropriate for Latvian seasonal changes.
Since it is a well-known fact that industrial WW, as well as rainwater, can significantly affect the concentration of specific analytes through the dilution effect, we decided to work with loads and not the concentration values. In the case of 5-HIAA, this strategy is also supported by “European Neuroendocrine Tumor Society Consensus Guidelines” where 24 h total 5-HIAA measurements are recommended for the diagnostics of neuroendocrine tumours (Oberg et al., 2017) and the normal excretion range of this metabolite is 3 to 15 mg/24 h per person (Lenchner and Santos, 2021). Thus, the multiplication of results from 5-HIAA concentration measurements with the total amount of WW from specific collection events revealed that there is considerable day-to-day variability in the loads of this metabolite (Supplementary Table 2). We observed that on average inhabitants of Jelgava produced 103,080.70 mg of 5-HIAA per day, the SD was 14,489.43 mg, which corresponds to a 14.06% deviation from the average, while the relative range was 52.34% points of the average. Although the cause of these fluctuations has not been investigated in this study, we believe that it might be due to combinatorial effects such as the metabolite degradation rate in WW, metabolite secretion rate and most prominently, population movement. In comparison to Jelgava, the average of the total 5-HIAA load in WW of Kuldiga was 6.02 times lower (17,136.14 mg), which fits well with the differences in population size of both locations, but unlike in the former, here we also observed greater SD 3686.6 mg (21.51%) and greater relative range of the measurements (82.54% points of the average). Due to the fluctuations of total 5-HIAA loads, we divided datasets of both populations according to the day of the week of sample collection start date (Mon, Tue and Thu for Jelgava and Mon, Tue, Thu and Sun for Kuldiga) and where there were enough data points (at least three) all were compared in pairs using two-tailed Mann-Whitney test. As a result, the differences between Mon and Thu datasets from Jelgava were not significant (p = 0.3710) while differences between Thu and Sun datasets from Kuldiga were significant (p = 0.0080). Considering that daytime activities of the general population during weekdays differ from those of weekends, we decided to perform a comparison of weekdays (Mon, Tue, Thu) and weekend data (Sun) using the Kuldiga dataset. Acquired results revealed that both 5-HIAA measurement datasets differed significantly (p = 0.0237). The average of the total 5-HIAA load in WW collected on weekdays was 18,751.69 mg (SD = 2987.32 mg or 15.93%), whereas in WW that was collected on weekends it was 14,174.30 mg (SD = 3061.76 mg or 21.60%), but the relative range of measurements was 54.91% points and 48.31% points of the average, respectively, which in both cases is closer to that of Jelgava. Since the use of 5-HIAA as a population size marker is considered experimental and difference was significant, we decided that an additional verification of this observation using an approach unrelated to WW was necessary.
3.2. Validation of observed differences between weekday and weekend total 5-HIAA measurements by CDR
The use of mobile phones has become prevalent in many European countries and since these devices are always located in the proximity of their owners their territorial density reflects the density of the population within a specific area. Therefore, we speculated that if the observed differences in the total load of 5-HIAA in Kuldiga between weekdays and weekends were true then these should also be reflected in mobile phone usage data, and we performed an evaluation of mobile phone call activities in conjunction with 5-HIAA measurements.
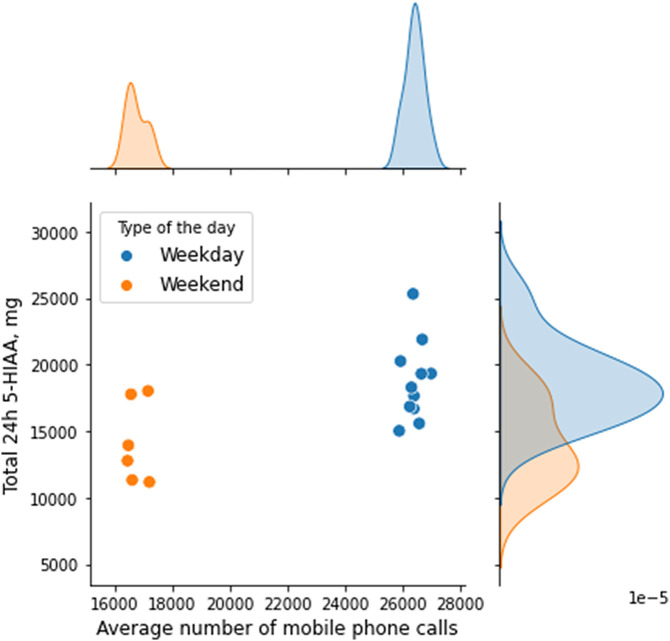
To evaluate if there is a correlation between the loads of 5-HIAA and unique mobile phone usage activity we carried out a Spearman correlation analysis. The results we acquired revealed that there is a moderate (r = 0.5907) yet significant (observed significance p = 0.0125) correlation between the two datasets. The analysis related scatter plot (Fig. 1 ) also uncovered that all values were grouped in two clusters, which correspond to weekdays and weekends. Further comparison of unique mobile phone activities between weekdays and weekends revealed that the two datasets differed significantly (p = 0.0011, two-tailed Mann-Whitney test).
Fig. 1.
The scatter plot of 5-HIAA and unique CDR data in Kuldiga shows the effect of population activity on 5-HIAA measurement data depending on the type of the day (e.g., weekday or weekend).
Since our initial assumption that population size, which contributed to the formation of WW, differed significantly between weekdays and weekends was confirmed, it was decided that all further Kuldiga related data analyses shall be carried out separately for the whole Kuldiga dataset, weekday dataset and weekend dataset.
3.3. Validation of SARS-CoV-2 RNA extraction procedure through the use of a surrogate virus
We used the rSFV as a surrogate to monitor the efficiency and success rate of the extraction procedures. Acquired results revealed that the rSFV recovery ratio in Jelgava on average was 112% (median 100%) and in Kuldiga it was 64% (median 58%). However, it varied greatly from extraction to extraction and displayed no correlation with any of the other measurements that were carried out. Plausible explanations and recommendations are negotiated within the discussion section.
3.4. SARS-CoV-2 RNA copy number correlation with cumulative incidence
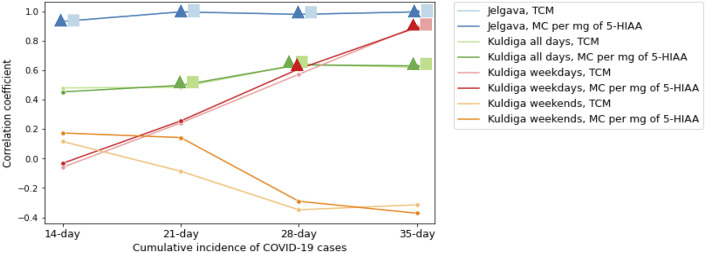
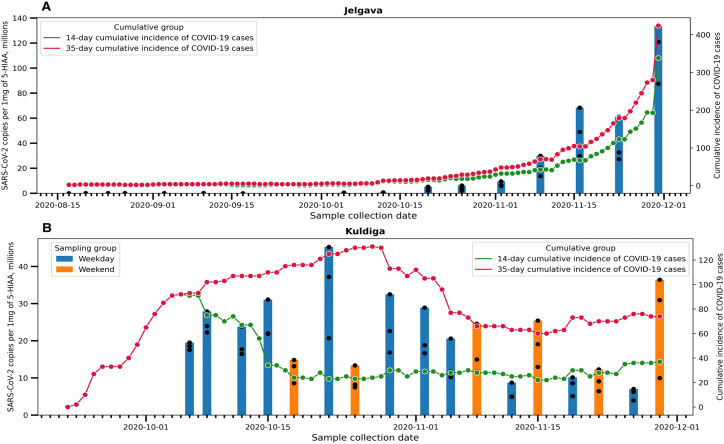
To assess whether the changes in acquired WW viral copy number correlated with the theoretical number of infected patients, we carried out a Spearman correlation analysis. However, since several researchers have reported that feces remain SARS-CoV-2 positive for up to 30 days after the diagnosis (Gupta et al., 2020), the correlation analysis between viral copy number and 21, 28 and 35-day cumulative incidence of COVID-19 cases were also carried out. In addition to assessing whether the normalization against population size marker would provide an improvement to acquired results we also calculated the number of viral RNA copies per 1 mg of 5-HIAA for all samples (Supplementary Table 2) and performed the same correlation analyses. Acquired results revealed that in Jelgava the correlation between both total and 5-HIAA normalized SARS-CoV-2 copy number and the cumulative incidence of disease cases was high regardless of the number of days that was used for calculation (r > 0.932, observed significance p < 0.0001) (Fig. 2, Fig. 3A). In the case of Kuldiga, the correlation between all RNA copy number values and cumulative incidences of cases was weaker, the best were between total RNA copy number and 21-, 28- and 35-day cumulative incidences of COVID-19 cases (r = 0.485, r = 0.64 and r = 0.62, respectively, observed significance p < 0.05). Similar correlations were observed between 5-HIAA normalized copy numbers and 21, 28 and 35-day cumulative incidences of COVID-19 cases (r = 0.498, r = 0.637 and r = 0.629, respectively, observed significance p < 0.05) when evaluating the full Kuldiga dataset. After separating the Kuldiga dataset into the weekday and weekend measurements, a strong correlation between total RNA copy number and 35-day cumulative incidence of SARS-CoV-2 positive tests was observed (r = 0.9, p = 0.0002) for the weekday dataset. Furthermore, a correlation between 5-HIAA normalized copy number and the 28 and 35-day cumulative incidence of COVID-19 cases was observed (r = 0.609 and r = 0.89, with observed significance p < 0.05). By contrast to the weekday dataset, the weekend copy number data displayed a weak and insignificant correlation for all four cumulative incidences with both total RNA copy number and 5-HIAA normalized SARS-CoV-2 RNA copy numbers (Fig. 2, Fig. 3B).
Fig. 2.
Changes in correlation coefficient between detected SARS-CoV-2 RNA copy number and a cumulative incidence of COVID-19 cases depending on the number of days that was used for the calculation of cumulative incidence. In the case of Jelgava, highly similar correlations were observed for all cumulative incidences with the total SARS-CoV-2 RNA copies and 5-HIAA normalized SARS-CoV-2 RNA copies therefore it is difficult to distinguish between TCM and MC values within the figure. Square designation denotes significant correlation (p < 0.05) of a particular cumulative incidence with TCM of SARS-CoV-2 RNA, whereas triangular designation denotes significant correlation of cumulative incidence with the MC of SARS-CoV-2 RNA per mg of 5-HIAA per respective municipality, as indicated by the colouring. Abbreviations: TCM - total copies in millions, MC - millions of copies.
Fig. 3.
5-HIAA normalized quantity of SARS-CoV-2 RNA within the wastewater of two Latvian locations A - Jelgava and B - Kuldiga. Bars represent the quantity of SARS-CoV-2 copies per 1 mg of 5-HIAA whereas line plots represent either 14-day or 35-day cumulative incidence of the officially confirmed COVID-19 cases in the respective municipality.
4. Discussion
WW epidemiology has been used widely to study various aspects of public health. Moreover, the application of WW epidemiology gained worldwide interest during the COVID-19 pandemic. This approach played an important role as an additional tool to analyse trends during disease surveillance, particularly in urban areas. Most of the studies have focussed on large cities where weekly variation in population size is relatively small. The purpose of this study was to evaluate the applicability of SARS-CoV-2 monitoring in the WW for early detection of viral outbreaks and to assess the overall epidemiological situation during an outbreak in small and medium-sized municipalities of Latvia. To achieve this goal the strategy of this study was based on longitudinal WW testing for the presence of SARS-CoV-2 RNA. Considering that this was a pilot study and thus limited in its scope and resources it was decided to perform the surveillance in only two municipalities.
Since Riga is the capital city of Latvia and accommodates approximately 30% of the Latvian population, it was not surprising that the outbreaks of COVID-19 at this location were detected regularly (details of the testing strategy can be found in Supplementary file 3). Although Riga is relevant from an epidemiological point of view, it was not suitable for the early detection of outbreaks. Therefore, we selected a municipality that was relatively “free of SARS-CoV-2”, no more than a two-hour journey from Riga that maintains a strong economic connection to the capital, ensuring daily population movements between the locations. Furthermore, the municipality has an independent economy, signifying that there is a strong necessity for communication between itself and Riga, thus providing a good opportunity for the spread of the virus. Considering all of these factors we selected Jelgava, which is the fourth largest city in the country, a major regional economic centre and residence of ~55,000 inhabitants (approx. 89% are connected to the municipal WW (ISMADE Ltd., 2020)). Additionally, it was decided to closely monitor the epidemiological situation in all COVID-19 free Latvian towns with suitable sewage collection systems and initiate the collection of WW samples in one of them upon the first news of an outbreak. Such an opportunity presented itself at the beginning of October 2020, when an outbreak was detected in Kuldiga, which is the 15th largest town in Latvia and has a population of approximately 11,000 inhabitants (approx. 88% are connected to the municipal WWTP (ISMADE Ltd., 2020)).
An assessment of SARS-CoV-2 RNA presence in collected WW samples was carried out through the ddPCR based quantification of three viral genome regions. The selection of primers and probes for the nucleocapsid (N) gene detection was based on “US CDC” recommendations (National Center for Immunization and Respiratory Diseases and Division of Viral Diseases, 2020) whereas the envelope (E) gene detection was based on World Health Organization (WHO) recommendations (Corman et al., 2020). Acquired data yielded by all three probe/primer sets were comparable, but on average the numbers generated by the N1 set were higher than those generated by the other two sets and those generated by the N2 set were higher than those of the E set. However, this rule was not universal and since observed differences could be the result of a combinatorial effect such as sample-specific RNA degradation, differences in probe/primer set detection efficiencies and others, we decided that rather than focussing on the results of one assay we shall use the average of three. A total number of SARS-CoV-2 RNA copies within the WW from each collection event were calculated to avoid the dilution effect.
Variation in the extraction of the rSVF surrogate, which might potentially affect the SARS-CoV-2 viral RNA extraction efficiency, was observed. According to previous studies (Ahmed et al., 2020c; Fuqing et al., 2021; Kumar et al., 2020; La Rosa et al., 2020, La Rosa et al., 2021; Pulicharla et al., 2021; Sharif et al., 2021; Zhang et al., 2020) and recommendations by the WHO (World Health Organization, 2003), PEG-precipitation methods of viral RNA from WW are known to be an efficient and cost-effective (Farkas et al., 2021; Kumar et al., 2020) approach and can be applicable for SARS-CoV-2 RNA extraction. SARS-CoV-2 remains viable in a wider range of conditions than rSFV which may explain the observed variance in the extraction of the surrogate. Thus, the former can tolerate pH values in a range from 4 to 11 (Chan et al., 2020; Chin et al., 2020), whereas the latter can be inactivated at a pH below 6.0 (Svehag et al., 1966). In addition, WW is an aggressive environment that contains detergents, proteolytic enzymes, RNases and other compounds, and there are significant temperature and pH fluctuations that may affect both viruses differentially (Amoah et al., 2020). Furthermore, the viral particles of SARS-CoV-2 spend a considerably longer period of time in this environment than those of rSFV, and since RNA decay over time is sloped rather than linear, it is possible that at the moment of extraction SARS-CoV-2 RNA is deteriorating at a slower rate in comparison to the surrogate (Ahmed et al., 2020b; Sala-Comorera et al., 2021). Although it is essential to use some kind of surrogate for the plus-minus test to assess whether the extraction was successful, the acquired numbers should not be used for the correction of target virus quantification results unless it has been determined that the stability of both target and surrogate under various conditions is highly similar.
In this study, 5-HIAA measurements were performed to account for fluctuations in the size of the population that contributed to the formation of WW and the choice of this metabolite as a population size marker was based on earlier studies (Chen et al., 2014; Choi et al., 2018; Pandopulos et al., 2020; Thai et al., 2019). Although the excretion of 5-HIAA might be influenced by diet (Burks and Bao, 2016; Mashige et al., 1996), alcohol consumption (Mackus et al., 2021; Voltaire et al., 1992) and neuroendocrine tumours (Ewang-Emukowhate et al., 2019; Zandee et al., 2016), these individualised variations do not seem to influence the overall measurements, as confirmed by the correlation analysis within the present study between 5-HIAA normalized SARS-CoV-2 RNA copy number and cumulative incidence of COVID-19 cases. CDR of the studied region was successfully applied to validate the population further. Thus, we were able to show for the first time that CDR data along with the 5-HIAA measurements can support the disease prevalence studies with an estimation of population size that contributes to the WW. This might be especially important for cities with a population size below 100,000, as this WW could be more heavily influenced by rain drainage water, industrial WW, and population movement, thus introducing the fluctuations of the detected SARS-CoV-2 genetic material and human excreted biomarkers. However, further investigations towards the estimation of the population size by CDR and 5-HIAA in small size cities are necessary.
In this study, we observed a high level of fluctuation within detected SARS-CoV-2 RNA between consecutive sample collections in Kuldiga. We speculated that this might be due to weekly changes in population size, which are characteristic for small-sized municipalities such as Kuldiga when people leave the town during the weekend and return during the weekdays. This hypothesis was supported by 5-HIAA measurements and subsequently also confirmed by CDR data analysis. This, however, led to the conclusion that although individuals with confirmed cases of infection have been put in strict isolation, a considerable proportion of detected SARS-CoV-2 RNA in the WW might arise from asymptomatic individuals, who unknowingly continue to spread the virus. At the same time, recovered individuals might still excrete the virus for a prolonged time. An additional feature of small-sized municipalities is the proportion of people that are travelling during the weekdays from the countryside to their workplace in the town. Therefore, if they are COVID-19 positive, their virus genetic material would be detectable only during the workdays. Furthermore, a proportion of people, who reside in the town could travel abroad during the weekend and introduce an additional bias towards the detection of SARS-CoV-2 RNA in the WW. Another explanation for the observed fluctuations might also be the rural nature of the town, where WW for a significant number of households (in the case of Kuldiga approx. 12%) is collected in designated collection tanks and transportation of accumulated WW from both infected and non-infected households to the WWTP is carried out exclusively on weekdays, which might increase the concentration of viral RNA. Hence, these results suggest that the collection of WW samples from small-sized municipalities, such as Kuldiga, should be performed either on a specific day of the week, preferably during the workdays, or daily to either control the effect of population migration or provide sufficient data to recognize the pattern.
The 14-day cumulative incidence is used for the assessment of the COVID-19 incidence in different countries. The rationale behind the use of this indicator is based on the observation that symptomatic disease lasts approximately 14 days and after this period the viral load of the patient is not sufficient to infect others. However, our data showed that a 14-day cumulative incidence displayed a good correlation with detected SARS-CoV-2 RNA copies only in the WW of Jelgava, where the incidence of COVID-19 cases during the study period were either non-existent or on a steep rise, which might have a significant impact on the outcome. In response to the above-mentioned reasons, we evaluated the cumulative incidence of different durations and concluded that the 35-day cumulative incidence was superior in explaining the variation of SARS-CoV-2 RNA copies in the WW. This correlation was not surprising as studies worldwide have reported that the virus can be excreted via human body fluids up to 40 days from the onset of symptoms (Chen et al., 2020; Gupta et al., 2020; Lo et al., 2020; Xing et al., 2020). Introducing the 35-day cumulative incidence of confirmed COVID-19 cases might provide better resolution in the prediction of the outbreak and possibly aid in developing a model to predict the number of infected individuals in a community based on WW testing.
An additional crucial aspect that should be considered during the conduction of WBE studies is the sample collection. Collection and viral RNA extraction procedures that are presented in this study are efficient yet labour intensive and difficult to apply in massive surveillance programmes. In addition, the number of virus particles in the WW is relatively low, which sets limits to the accuracy of detection methods. Thus, there is a need to improve the sample collection and pre-treatment procedures through a concentration of the samples without a significant increase in labour and costs. Several approaches are proposed including the addition of sorbent materials in the WW (Freda, 2021). However, it would be simpler to concentrate samples through the multi-step membrane ultrafiltration that has been used in surface water analyses (Ferguson et al., 2004; Rusiñol et al., 2020). Moreover, sample collection coupled with WW flow rate measurements and adjusted by this data would improve the overall representativity of the collected sample and increase the accuracy of viral RNA and population size estimation even further.
The use of mobile data as a real-time data source has become increasingly relevant. For instance, it allows an assessment of regional economic development or the change in population mobility patterns. In this study, the analysis of aggregate data from mobile network operators was successfully integrated to confirm 5-HIAA measurement observations and to evaluate whether the highest activity of the population is related to changes in SARS-CoV-2 RNA abundance in WW. However, in our study the correlation between these two datasets was weak most likely due to an insufficient number of measurements and further studies are required to account for various WW collection systems and mobile phone usage related parameters that could influence the correlation.
Many countries have recently adopted WBE as a cost-effective approach for the wide-scale screening of SARS-CoV-2 RNA. Along with the clinical data, it could provide information on potential virus transmission within the community, thus aiding the decision-making process for authorities in the public health sector. However, several issues must be considered when using WBE as an epidemiological tool. First, with WBE, there is a gap of knowledge that prohibits precise estimation of the number of COVID-19 affected individuals. Second, we suggest that for smaller and medium-sized municipalities, the population movement should be considered while evaluating the presence of the SARS-CoV-2 RNA in the WW. Third, there is a necessity to develop and implement time-effective, cost-effective and reliable methods to detect emerging SARS-CoV-2 variants within the population through WBE surveillance.
In summary, this study validates the application of WBE as a reliable tool to investigate the prevalence of infection within a community. We confirmed that WBE based SARS-CoV-2 RNA monitoring could be performed in small and middle-size municipalities in Latvia and acquired viral RNA data correlate with a 35-day cumulative incidence of SARS-CoV-2 infection cases. In addition, results of this study suggest that in small size municipalities due to the high influence of population migration, the collection day of the WW samples should be selected carefully and the continuation of monitoring should be performed only on that particular weekday or carried out daily to facilitate pattern recognition.
5. Conclusion
-
●
This is the first study showing that data from mobile calls could be used effectively to better understand and improve the accuracy of WBE. It not only allows an estimate of the population but also the mobility of people in COVID-19 affected areas.
-
●
It is essential to perform measurements that enable population size control because weekly population movement and/or differences of population daytime activities on different days of the week significantly affect the WBE results in smaller municipalities, thus samples should be collected either daily or weekly on a specific day of the week.
-
●
There is a moderate to strong correlation between 5-HIAA and mobile phone calls, hence both methods could be used to assess population size and movement in WBE studies.
-
●
A 35-day cumulative incidence was superior to that of 14 days in explaining the variation of SARS-CoV-2 RNA copies in WW.
The following are the supplementary data related to this article.
Names and sequences of employed oligonucleotides. All were synthesized by Metabion GmbH (Germany).
Collected raw data of Kuldiga and Jelgava municipality.
Description of the testing strategy and capacity in Latvia.
dMIQE checklist.
CRediT authorship contribution statement
Dita Gudra: Data curation; Formal analysis, Methodology, Visualization, Writing – original draft, Writing-review and edit, Sandis Dejus: Data curation; Formal analysis, Methodology, Project administration, Resources, Validation, Writing – original draft, Writing-review and edit, Vadims Bartkevics: Formal analysis, Methodology, Project administration, Resources, Validation, Writing – original draft, Writing-review and edit, Ance Roga: Methodology, Investigation, Validation, Ineta Kalnina: Methodology, Investigation, Validation, Martins Strods: Investigation, Methodology, Validation, Anton Rayan: Investigation, Methodology, Validation, Kristina Kokina: Methodology, Validation, Project administration, Anna Zajakina: Resources, Validation, Uga Dumpis: Project administration, Funding acquisition, Writing-review and edit, Laura Elina Ikkere: Methodology, Validation, Irina Arhipova: Investigation, Methodology, Validation, Writing-review and edit, Gundars Berzins: Methodology, Validation, Aldis Erglis: Investigation, Methodology, Validation, Juris Binde: Resources, Methodology, Evija Ansonska: Investigation, Methodology, Validation, Aivars Berzins: Methodology, Project administration, Resources, Validation, Writing-review and edit, Funding acquisition, Talis Juhna: Methodology, Project administration, Resources, Validation, Writing-review and edit, Funding acquisition, Davids Fridmanis: Methodology, Project administration, Resources, Validation, Writing – original draft, Writing-review and edit, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work was supported by project No. VPP-COVID-2020/1-0008: “Multidisciplinary approach to monitor, control and confine the COVID-19 and other future epidemics in Latvia”. In addition, we thank Latvian Water and Wastewater Works Association for supporting the sampling and access to wastewater plants.
Editor: Warish Ahmed
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S.L., Bertsch P.M., Bibby K., Bivins A., Blackall L.L., Bofill-Mas S., Bosch A., Brandão J., Choi P.M., Ciesielski M., Donner E., D’Souza N., Farnleitner A.H., Gerrity D., Gonzalez R., Griffith J.F., Gyawali P., Haas C.N., Hamilton K.A., Hapuarachchi H.C., Harwood V.J., Haque R., Jackson G., Khan S.J., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B.A., Lipp E., McLellan S.L., McMinn B., Medema G., Metcalfe S., Meijer W.G., Mueller J.F., Murphy H., Naughton C.C., Noble R.T., Payyappat S., Petterson S., Pitkänen T., Rajal V.B., Reyneke B., Roman F.A., Rose J.B., Rusiñol M., Sadowsky M.J., Sala-Comorera L., Setoh Y.X., Sherchan S.P., Sirikanchana K., Smith W., Steele J.A., Sabburg R., Symonds E.M., Thai P., Thomas K.V., Tynan J., Toze S., Thompson J., Whiteley A.S., Wong J.C.C., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A.G., Shanks O.C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., Ru P., Pan X., Bohrerova Z., Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801:149757. doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah I.D., Kumari S., Bux F. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020;143 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhipova I., Berzins G., Brekis E., Binde J., Opmanis M., Erglis A., Ansonska E. Mobile phone data statistics as a dynamic proxy indicator in assessing regional economic activity and human commuting patterns. Expert. Syst. 2020;37 doi: 10.1111/exsy.12530. [DOI] [Google Scholar]
- Balke I., Reseviča G., Zeltins A. In: Isolation and Characterization of Two Distinct Types of Unmodified Spherical Plant Sobemovirus-like Particles for Diagnostic and Technical Uses BT - Virus-Derived Nanoparticles for Advanced Technologies: Methods and Protocols. Wege C., Lomonossoff G.P., editors. Springer New York; New York, NY: 2018. pp. 19–34. [DOI] [PubMed] [Google Scholar]
- Burks M.L., Bao S. The 24-hour urinary 5-HIAA: a simple test with a common pitfall. AACE Clin. Case Rep. 2016;2:e186–e188. doi: 10.4158/EP15794CR. [DOI] [Google Scholar]
- Centre for Disease Prevention and Control . 2021. COVID-19 Tests, Confirmed Cases and Outcomes. [Google Scholar]
- Centre for Disease Prevention and Control Covid-19 Statistics [WWW Document] 2021. https://www.spkc.gov.lv/lv/covid-19-statistika
- Chan K.-H., Sridhar S., Zhang R.R., Chu H., Fung A.-F., Chan G., Chan J.-W., To K.-W., Hung I.-N., Cheng V.-C., Yuen K.-Y. Factors affecting stability and infectivity of SARS-CoV-2. J. Hosp. Infect. 2020;106:226–231. doi: 10.1016/j.jhin.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kostakis C., Gerber J.P., Tscharke B.J., Irvine R.J., White J.M. Towards finding a population biomarker for wastewater epidemiology studies. Sci. Total Environ. 2014;487:621–628. doi: 10.1016/j.scitotenv.2013.11.075. [DOI] [PubMed] [Google Scholar]
- Chen G., Hoteit S., Viana A.C., Fiore M., Sarraute C. Enriching sparse mobility information in call detail records. Comput. Commun. 2018;122:44–58. doi: 10.1016/j.comcom.2018.03.012. [DOI] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O’Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O’Malley E., Crosbie N.D., Thomas K.V., Mueller J.F. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 2018;105:453–469. doi: 10.1016/j.trac.2018.06.004. [DOI] [Google Scholar]
- Cleveland W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979;74:829–836. doi: 10.1080/01621459.1979.10481038. [DOI] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank K., Li X., North D., Ferraro G.B., Iaconelli M., Mancini P., La Rosa G., Bibby K. CrAssphage abundance and correlation with molecular viral markers in italian wastewater. Water Res. 2020;184 doi: 10.1016/j.watres.2020.116161. [DOI] [PubMed] [Google Scholar]
- D’Aoust P.M., Towhid S.T., Mercier É., Hegazy N., Tian X., Bhatnagar K., Zhang Z., Naughton C.C., MacKenzie A.E., Graber T.E., Delatolla R. COVID-19 wastewater surveillance in rural communities: comparison of lagoon and pumping station samples. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donde O.O., Atoni E., Muia A.W., Yillia P.T. COVID-19 pandemic: water, sanitation and hygiene (WASH) as a critical control measure remains a major challenge in low-income countries. Water Res. 2021;191 doi: 10.1016/j.watres.2020.116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewang-Emukowhate M., Nair D., Caplin M. The role of 5-hydroxyindoleacetic acid in neuroendocrine tumors: the journey so far. Int. J. Endocr. Oncol. 2019;6:IJE17. doi: 10.2217/ije-2019-0001. [DOI] [Google Scholar]
- Farkas K., Hillary L.S., Malham S.K., McDonald J.E., Jones D.L. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr. Opin. Environ. Sci. Health. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Hillary L.S., Thorpe J., Walker D.I., Lowther J.A., McDonald J.E., Malham S.K., Jones D.L. Concentration and quantification of SARS-CoV-2 RNA in wastewater using polyethylene glycol-based concentration and qRT-PCR. Methods Protoc. 2021 doi: 10.3390/mps4010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson C., Kaucner C., Krogh M., Deere D., Warnecke M. Comparison of methods for the concentration of cryptosporidium oocysts and giardia cysts from raw waters. Can. J. Microbiol. 2004;50:675–682. doi: 10.1139/w04-059. [DOI] [PubMed] [Google Scholar]
- Freda K. 2021. The myriad ways sewage surveillance is helping fight COVID around the world [WWW Document] 10 May. [DOI] [PubMed] [Google Scholar]
- Fuqing W., Jianbo Z., Amy X., Xiaoqiong G., Lin L.W., Federica A., Kathryn K., William H., Mariana M., Newsha G., Noriko E., Claire D., Mathilde P., Katya M., D W.A., B E.T., R C.P., Janelle T., J A.E., A G.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2021;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik B., Tavazzi S., Mariani G., Skejo H., Sponar M., Higgins T., Medema G., Wintgens T. 2021. SARS-CoV-2 Surveillance employing Sewage - Towards a Sentinel System. (online),10.2760/909651 (print) [DOI] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H., Wilder M., Collins M., Fenty A., Gentile K., Kmush B.L., Zeng T., Middleton F.A., Larsen D.A. Quantification of SARS-CoV-2 and cross-assembly phage (crAssphage) from wastewater to monitor coronavirus transmission within communities. medRxiv. 2020 doi: 10.1101/2020.05.21.20109181. 10 May. [DOI] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces – a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., McDonald J.E., Malham S.K., Jones D.L. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;117214 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770:145274. doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- ISMADE Ltd . 2020. Notekūdeņu apsaimniekošanas investīciju plāns 2021. – 2027. gadam. Minist. Environ. Prot. Reg. Dev. Repub. Latv.https://www.varam.gov.lv/sites/varam/files/content/notekudeni-21-27-zinojums_precizets_051120.pdf [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Alamin M., Kuroda K., Dhangar K., Hata A., Yamaguchi H., Honda R. Potential discharge, attenuation and exposure risk of SARS-CoV-2 in natural water bodies receiving treated wastewater. Npj cleanWater. 2021;4:8. doi: 10.1038/s41545-021-00098-2. [DOI] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-COV-2 in untreated wastewaters in Italy. medRxiv. 2020 doi: 10.1101/2020.04.25.20079830. 2020.04.25.20079830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since december 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenchner J., Santos C. Hydroxyindoleacetic Acid. Vol. 5. Treasure Isl. StatPearls Publ.; 2021. Biochemistry.https://www.ncbi.nlm.nih.gov/books/NBK551684/ [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X., Tian Y., Sin N.N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackus M., van de Loo A.J.A.E., van den Bogaard W.J.M., Korte-Bouws G.A.H., Garssen J., Verster J.C. The 5HTOL/5HIAA ratio as a biomarker of alcohol hangover. J. Clin. Med. 2021;10:4241. doi: 10.3390/jcm10184241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947;18:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- Mashige F., Matsushima Y., Kanazawa H., Sakuma I., Takai N., Bessho F., Ohkubo A. Acidic catecholamine metabolites and 5-hydroxyindoleacetic acid in urine: the influence of diet. Ann. Clin. Biochem. 1996;33:43–49. doi: 10.1177/000456329603300106. [DOI] [PubMed] [Google Scholar]
- National Center for Immunization and Respiratory Diseases and Division of Viral Diseases, 2020 National Center for Immunization and Respiratory Diseases; Division of Viral Diseases, 2020. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes [WWW Document]. Centers Dis. Control Prev. URL https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- O’Brien J.W., Thai P.K., Eaglesham G., Ort C., Scheidegger A., Carter S., Lai F.Y., Mueller J.F. A model to estimate the population contributing to the wastewater using samples collected on census day. Environ. Sci. Technol. 2014;48:517–525. doi: 10.1021/es403251g. [DOI] [PubMed] [Google Scholar]
- Oberg K., Couvelard A., Delle Fave G., Gross D., Grossman A., Jensen R.T., Pape U.-F., Perren A., Rindi G., Ruszniewski P., Scoazec J.-Y., Welin S., Wiedenmann B., Ferone D. ENETS consensus guidelines for standard of Care in Neuroendocrine Tumours: biochemical markers. Neuroendocrinology. 2017;105:201–211. doi: 10.1159/000472254. [DOI] [PubMed] [Google Scholar]
- Pandopulos A.J., Gerber C., Tscharke B.J., O’Brien J., White J.M., Bade R. A sensitive analytical method for the measurement of neurotransmitter metabolites as potential population biomarkers in wastewater. J. Chromatogr. A. 2020;1612 doi: 10.1016/j.chroma.2019.460623. [DOI] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Omer S.B. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 doi: 10.1101/2020.05.19.20105999. 2020.05.19.20105999. [DOI] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulicharla R., Kaur G., Brar S.K. A year into the COVID-19 pandemic: rethinking of wastewater monitoring as a preemptive approach. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2021.106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico M., Andrés-Costa M.J., Picó Y. Estimating population size in wastewater-based epidemiology. Valencia metropolitan area as a case study. J. Hazard. Mater. 2017;323:156–165. doi: 10.1016/j.jhazmat.2016.05.079. [DOI] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater. Curr. Opin. Environ. Sci. Health. 2020;17:21–28. doi: 10.1016/j.coesh.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., Davidsson F., Dotevall L., Mattsson A., Trybala E., Lagging M., Lindh M., Gisslén M., Brezicka T., Nyström K., Norder H. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021;189 doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Comorera L., Reynolds L.J., Martin N.A., O’Sullivan J.J., Meijer W.G., Fletcher N.F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saththasivam J., El-Malah S.S., Gomez T.A., Jabbar K.A., Remanan R., Krishnankutty A.K., Ogunbiyi O., Rasool K., Ashhab S., Rashkeev S., Bensaad M., Ahmed A.A., Mohamoud Y.A., Malek J.A., Abu Raddad L.J., Jeremijenko A., Abu Halaweh H.A., Lawler J., Mahmoud K.A. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbaoui Y., Bennis F., Chegdani F. SARS-CoV-2 as enteric virus in wastewater: which risk on the environment and human behavior? Microbiol. Insights. 2021;14 doi: 10.1177/1178636121999673. 1178636121999673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold S., Perktold J. 9th Python in Science Conference. 2010. statsmodels: Econometric and statistical modeling with python. [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmed J., Safdar R.M., Rehman L., Mujtaba G., Hussain J., Ali J., Angez M., Alam M.M., Akthar R., Wasif Malik M., Iqbal Baig M.Z., Suleman Rana M., Usman M., Qaisar Ali M., Ahad A., Badar N., Umair M., Tamim S., Ashraf A., Tahir F., Ali N. Detection of SARs-CoV-2 in wastewater using the existing environmental surveillance network: a potential supplementary system for monitoring COVID-19 transmission. PLoS One. 2021;16 doi: 10.1371/journal.pone.0249568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svehag S.E., Leendertsen L., Gorham J.R. Sensitivity of bluetongue virus to lipid solvents, trypsin and pH changes and its serological relationship to arboviruses. J. Hyg. 1966;64:339–345. doi: 10.1017/s0022172400040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.C., Laperriere G., Germain H. Droplet digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7:2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai P.K., O’Brien J.W., Banks A.P.W., Jiang G., Gao J., Choi P.M., Yuan Z., Mueller J.F. Evaluating the in-sewer stability of three potential population biomarkers for application in wastewater-based epidemiology. Sci. Total Environ. 2019;671:248–253. doi: 10.1016/j.scitotenv.2019.03.231. [DOI] [PubMed] [Google Scholar]
- Thomas K.V., Amador A., Baz-Lomba J.A., Reid M. Use of Mobile device data to better estimate dynamic population size for wastewater-based epidemiology. Environ. Sci. Technol. 2017;51:11363–11370. doi: 10.1021/acs.est.7b02538. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Gahlot P., Tyagi V.K., Zhang L., Zhou Y., Kazmi A.A., Kumar M. Surveillance of wastewater for early epidemic prediction (SWEEP): environmental and health security perspectives in the post COVID-19 anthropocene. Environ. Res. 2021;195 doi: 10.1016/j.envres.2021.110831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Heal. 2020;10:100157. doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., van der Walt S.J., Brett M., Wilson J., Millman K.J., Mayorov N., Nelson A.R.J., Jones E., Kern R., Larson E., Carey C.J., Polat İ., Feng Y., Moore E.W., VanderPlas J., Laxalde D., Perktold J., Cimrman R., Henriksen I., Quintero E.A., Harris C.R., Archibald A.M., Ribeiro A.H., Pedregosa F., van Mulbregt P. SciPy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltaire A., Beck O., Borg S. Urinary 5-hydroxytryptophol: a possible marker of recent alcohol consumption. Alcohol. Clin. Exp. Res. 1992;16:281–285. doi: 10.1111/j.1530-0277.1992.tb01377.x. [DOI] [PubMed] [Google Scholar]
- Waskom M.L. Seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2003. Guidelines for Environmental Surveillance of Poliovirus Circulation. [Google Scholar]
- Wu L., Chikaraishi M., Nguyen H.T.A., Fujiwara A. Analysis of post-disaster population movement by using mobile spatial statistics. Int. J. Disaster Risk Reduct. 2021;54 doi: 10.1016/j.ijdrr.2021.102047. [DOI] [Google Scholar]
- Xing Y.-H., Ni W., Wu Q., Li W.-J., Li G.-J., Wang W.-D., Tong J.-N., Song X.-F., Wing-Kin Wong G., Xing Q.-S. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J. Microbiol. Immunol. Infect. 2020;53:473–480. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandee W.T., Kamp K., van Adrichem R.C.S., Feelders R.A., de Herder W.W. Limited value for urinary 5-HIAA excretion as prognostic marker in gastrointestinal neuroendocrine tumours. Eur. J. Endocrinol. 2016;175:361–366. doi: 10.1530/EJE-16-0392. [DOI] [PubMed] [Google Scholar]
- Zhang D., Ling H., Huang X., Li J., Li W., Yi C., Zhang T., Jiang Y., He Y., Deng S., Zhang X., Wang X., Liu Y., Li G., Qu J. Potential spreading risks and disinfection challenges of medical wastewater by the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral RNA in septic tanks of fangcang hospital. Sci. Total Environ. 2020;741 doi: 10.1016/j.scitotenv.2020.140445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Atoni E., Nyaruaba R., Du Y., Zhang H., Donde O., Huang D., Xiao S., Ren N., Ma T., Shu Z., Yuan Z., Tong L., Xia H. Environmental surveillance of SARS-CoV-2 RNA in wastewater systems and related environments in Wuhan: april to may of 2020. J. Environ. Sci. 2022;112:115–120. doi: 10.1016/j.jes.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Names and sequences of employed oligonucleotides. All were synthesized by Metabion GmbH (Germany).
Collected raw data of Kuldiga and Jelgava municipality.
Description of the testing strategy and capacity in Latvia.
dMIQE checklist.