Abstract
Wastewater surveillance for SARS-CoV-2 may serve as a useful source of data for public health departments as the virus is shed in the stool of infected individuals. However, for wastewater data to be actionable, wastewater must be collected, concentrated, and analyzed in a timely manner. This manuscript presents modifications on a skimmed milk concentration protocol to reduce processing time, increase the number of samples that can be processed at once, and enable use in resource-limited settings. Wastewater seeded with Human coronavirus OC43 (OC43) was concentrated using a skimmed milk flocculation protocol, and then pellets were directly extracted with the QIAamp Viral RNA Mini kit. This protocol has a higher average effective volume assayed (6.35 mL) than skimmed milk concentration methods, with and without Vertrel XF™, which involve resuspension of the pellets in PBS extraction prior to nucleic acid extraction (1.28 mL, 1.44 mL, respectively). OC43 was selected as a recovery control organism because both it and SARS-CoV-2 are enveloped respiratory viruses that primarily infect humans resulting in respiratory symptoms. The OC43 percent recovery for the direct extraction protocol (3.4%) is comparable to that of skimmed milk concentration with and without Vertrel XF™ extraction (4.0%, 2.6%, respectively). When comparing SARS-CoV-2 detection using McNemar’s chi-square test, the pellet extraction method is not statistically different from skimmed milk concentration, with and without Vertrel XF™ extraction. This suggests that the method performs equally as well as existing methods. Added benefits include reduced time spent per sample and the ability to process more samples at a single time. Direct extraction of skimmed milk pellets is a viable method for quick turnaround of wastewater data for public health interventions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12560-022-09512-5.
Keywords: SARS-CoV-2, Wastewater surveillance, Method development, Environmental surveillance
Introduction
SARS-CoV-2 emerged in late 2019 in Wuhan, China, and within a few months, it was declared as a pandemic by the WHO (World Health Organization, 2020, March 11). Infection with SARS-CoV-2 primarily results in respiratory symptoms but can also cause gastrointestinal disease in a fraction of infected individuals (Jones et al., 2020). Additionally, even if individuals do not show gastrointestinal symptoms, they can shed the virus in their stool for weeks after infection (Wang et al., 2020; Wu et al., 2020; Xiao et al., 2020), which led research groups around the world to begin analyzing wastewater and stool samples for the virus (Ahmed et al., 2020; Medema et al., 2020; Peccia et al., 2020).
Although the infectivity of virus detected in wastewater and stool is still being understood, the presence of the virus in wastewater can serve as a useful source of information for public health departments. Three potential use cases for wastewater surveillance include detecting an introduction of new cases to a water catchment area, monitoring changes in trends of infection, and estimating community prevalence (Water Research Foundation, 2020). Research has shown that an outbreak was averted in a university dorm, in Arizona, USA, because the virus was detected in wastewater before positive clinical tests, giving time for residents to be tested and isolated if positive (Peiser, 2020). Additionally, detection in wastewater has been shown to precede the first positive clinical tests by weeks in Brisbane, Australia (Ahmed et al., 2021) and also frequently matches community infection trends (Gonzalez et al., 2020; Peccia et al., 2020).
However, for wastewater surveillance to be a useful tool for public health officials, results must be available as quickly as possible. Many of the existing concentration methods take hours or are dependent on unreliable supply chains. Here, we present a modified version of the skimmed milk flocculation method (Philo et al., 2021), the skimmed milk pellet extraction protocol. Skimmed milk flocculation was originally developed to concentrate viruses from coastal waters (Calgua et al., 2008) and was previously used by this group to further concentrate the beef extract eluate off of positively charged ViroCap filters (Scientific Methods, Granger, IN, USA) (Falman et al., 2019). However, in all those variations on the method, the skimmed milk pellets were resuspended in PBS with a fraction of the PBS resuspension being used for RNA and/or DNA extraction. In this optimized method, the skimmed milk pellets are resuspended in the lysis buffer of the RNA extraction kit. This allows the entire initially concentrated volume of wastewater to be extracted and results in larger effective volumes assayed.
The skimmed milk flocculation method was chosen for optimization because it performed well for recovery of human coronavirus OC43 (OC43, used to determine method recovery efficiencies) and SARS-CoV-2 detection (Philo et al., 2021). Additionally, it is more feasible in low resource settings because it does not require expensive or hard to obtain supplies, and the entire protocol can be completed in a single day. The method was validated using wastewater collected from three Seattle-area wastewater treatment plants (WWTPs) over fifteen weeks. This method is a viable alternative for public health departments and research groups to conduct timely SARS-CoV-2 wastewater surveillance and successfully detects SARS-CoV-2 in Seattle-area wastewaters.
Methods
Sample Collection and Seeding
Primary wastewater was grab sampled from three Seattle-area wastewater treatment plants (WWTP) from October 2020 through March 2021 (Supplementary Table 5). All wastewater was transported in coolers with ice packs and stored at 4 °C for no longer than 3 days. Grab samples collected from a single WWTP over the course of the day were composited and mixed before being processed. All wastewater samples were seeded with OC43 at a concentration of 3.3 × 104 TCID50/L of wastewater before concentration (OC43 ATCC VR-1558). OC43 viral stocks were produced following previously published protocols (Philo et al., 2021).
Skimmed Milk Concentration
Skimmed milk flocculation with and without Vertrel XF™ (Miller-Stephenson, Inc., Danbury, CT, USA) extraction was carried out as previously published (Philo et al., 2021). Briefly, a 5% skimmed milk solution was added to 0.6 L of seeded wastewater (1% v/v final). The pH of the wastewater with skimmed milk was dropped to 3–4 using 5 M HCl, shaken at 200 rpm for 2 h, and then divided into 50 mL conicals, and centrifuged at 3500 × g for 30 min at 20 °C. The supernatant was then poured off, and the pellets were used for different resuspension and RNA extraction protocols.
Resuspension and RNA Extraction
Resuspension in PBS
The equivalent of 500 mL of wastewater (ten 50 mL pellets) was resuspended in 6.0 mL of sterile PBS (pH 7.4). This resuspension volume was then split into two equal volumes: one for RNA extraction and the other for Vertrel XF™ extraction to separate the viruses from the solids following previously published protocols (Philo et al., 2021). RNA extraction was carried out on both resuspensions using the QIAamp ViralRNA Mini Kit (QIAGEN, Germantown, MD, USA). Each sample was extracted in duplicate with a doubled input volume of 280 μL for a total input extraction volume of 560 μL. Each column was eluted in 60 μL of Buffer AVE, for a total eluate volume of 120 μL. Duplicate extracts were combined and re-aliquoted before being stored at – 20 °C.
Resuspension in Lysis Buffer
Buffer AVL from the QIAamp Viral RNA Mini Kit was prepared with carrier RNA according to the manufacturer’s protocol. Two skimmed milk pellets (100 mL wastewater equivalent) from each WWTP were resuspended in 560 μL each of the Buffer AVL/carrier RNA mixture. This resuspended pellet was then transferred to a PowerBead Pro tube (Cat. No. 19301, QIAGEN, Germantown, MD, USA). The PowerBead pro tubes were then vortexed using a horizontal adaptor at maximum speed for ten minutes. They were then centrifuged at 15,000×g for 1 min to pellet the solids. The supernatant was transferred without disturbing the solids or beads to a sterile 1.5 mL flip cap tube, one tube for each initial pellet. 560 μL of 100% ethanol was then added to the supernatant and mixed by pulse vortexing for 15 s.
Starting with step six of the manufacturer’s instructions, 630 μL was applied to the column and samples were spun at 15,000×g for 5 min. The extraction then followed the manufacturer’s instructions until the final elution step. At this point, 40 μL of Buffer AVE was added to each column and incubated for one minute. After elution, the same 40 μL was applied back onto the column for a second elution. The elution from replicate columns was then mixed and re-aliquoted for immediate molecular processing or storage at – 20 °C, for a total eluate volume of 80 μL. The complete method is available for viewing online at YouTube (https://youtu.be/Xyb8CScM0Ko).
Generation of Synthetic OC43 RNA Control
As OC43 RNA extracts were enumerated using TCID50, an OC43 RNA control (cRNA) was made using published constructed RNA methods for use in RT-qPCR standard curves (Fronhoffs et al., 2002; Vijgen et al., 2005). Briefly, an OC43 DNA amplicon was constructed from extracted RNA from cell culture using the iTaq Universal SYBR Green one-step kit (Cat. No. 172-5151, Bio-Rad Laboratories, Hercules, CA, USA). Reactions contained 0.3 μM of the OC43-FPT7 and OC43-RP primers from Vijgen et al. (2005) (Table 1). Temperatures followed the Bio-Rad mastermix instructions, with an annealing temperature of 60 °C. DNA amplicon concentration was measured using a Nanodrop. The MEGAshortscript T7 transcription kit (Cat. No. AM1354, Invitrogen Corp., Waltham, MA, USA) was used to transcribe 84 ng of amplicon into constructed RNA of 73 bp in three reaction volumes according to the manufacturer’s instructions. DNA was then digested using 3 µl of TURBO dnase, incubated for 15 min at 37 °C, and stopped by adding 15 μl ammonium acetate stop solution and 115 µl of molecular grade water. RNA precipitation was performed on cRNA by mixing in two volumes of 200 proof ethanol and chilling at – 20 °C for 15 min. Chilled cRNA was then spun at 21,000 × g for 15 min at 4 °C. Supernatant was removed, and cRNA pellet was suspended in AE buffer (10 mM Tris–Cl, 0.5 mM EDTA) resulting in 3 × 1013 genome copies per µl as determined by nanodrop. Aliquots of diluted cRNA were stored at – 80 °C.
Table 1.
Sequences and concentrations of each primer and probe used in RT-qPCR
| Target | Name | Sequence (5′–3′) | Final conc. (μM) | References |
|---|---|---|---|---|
| OC43 | OC43-FP | ATGTTAGGCCGATAATTGAGGACTAT | 0.3 | Vijgen et al. (2005) |
| OC43-RP | AATGTAAAGATGGCCGCGTATT | 0.3 | ||
| OC43-TP | FAM-CATACTCTGACGGTCACAAT-NFQ-MGB | 0.2 | ||
| OC43-FPT7 | TAATACGACTCACTATAGGGAGGATGTTAGGCCGATAATTGAGGACTAT | 0.3 | ||
| SARS-CoV-2 N1 | N1-F | GACCCCAAAATCAGCGAAAT | 0.2 | US Centers for Disease Control and Prevention (2020) |
| N1-R | TCTGGTTACTGCCAGTTGAATCTG | 0.2 | ||
| N1-P | FAM-ACCCCGCAT-ZEN-TACGTTTGGTGGACC-3IABkFQ | 0.2 | ||
| SARS-CoV-2 N2 | N2-F | TTACAAACATTGGCCGCAAA | 0.2 | US Centers for Disease Control and Prevention (2020) |
| N2-R | GCGCGACATTCCGAAGAA | 0.2 | ||
| N2-P | HEX-ACAATTTGC-ZEN-CCCCAGCGCTTCAG-3IABkFQ | 0.2 |
RT-qPCR
Reverse-transcription real-time PCR (RT-qPCR) for OC43 and SARS-CoV-2 was carried out on all wastewater RNA extracts using the iTaq Universal Probes One-Step Kit and the BioRad CFX qPCR systems (Bio-Rad Laboratories, Hercules, CA, USA). All samples were run in duplicate, with duplicate tenfold dilutions to control for inhibition. All primers and probes were obtained from IDT (Integrated DNA Technologies, Inc., Coralville, IA, USA). Their sequences and final concentrations are in Table 1. RT-qPCR cycling conditions for both assays were 50 °C for 10 min, 95 °C for three minutes, and 45 cycles of 95 °C for 15 s and 60 °C for 30 s.
The OC43 assay targeting the M protein was carried out as previously published (Philo et al., 2021; Vijgen et al., 2005). Positive control standard curves were generated by making tenfold serial dilutions in nuclease-free water either by using RNA extracts from viral stocks or the above-mentioned synthetic RNA. Negative controls were nuclease-free water, and SARS-CoV-2 was included as an off-template control to ensure there was no cross-reactivity.
RT-qPCR for SARS-CoV-2 was carried out using the US CDC targets for the N1 and N2 genes in a total reaction volume of 15 μL (US Centers for Disease Control & Prevention, 2020). Samples positive for SARS-CoV-2 were identified if either replicate for N1 or N2 had amplification with Cq < 40 in either the non-diluted or 10–1 diluted RT-qPCR reactions. To reduce the time to results, the assay was multiplexed by tagging the N1 probe with FAM and the N2 probe with HEX (Table 1). Positive control standard curves were generated by making tenfold serial dilutions of a plasmid control containing the target genes for SARS-CoV-2 (Integrated DNA Technologies, Inc., Coralville, IA, USA) or RNA extract obtained from BEI (Item No. NR-52285, BEI Resources, Manassas, VA, USA). RNA extract was used because it resulted in more consistent standard curves than the plasmid-positive control. Negative controls were nuclease-free water, and OC43 was included as an off-template control to ensure there was no cross-reactivity.
Recovery Calculations
Fractional recovery was calculated differently depending on the type of positive control used to generate the standard curve. The actual OC43 assayed for each sample is calculated using Eq. 1. Cq is the cycle quotient as determined using the standard curves, and the intercept and slope are the values obtained from the standard curve.
| 1 |
To calculate the theoretical OC43 assayed, the volume adjusted OC43 that we would have detected had 100% of it been recovered was calculated using Eq. 2. Cinoculated is the concentration of OC43 inoculated into the sample and Vinoculated is the effective volume assayed.
| 2 |
Percent recovery was calculated using Eq. 3.
| 3 |
Data Analysis
All RT-qPCR data were analyzed using the Bio-Rad CFX Maestro for Mac program (Bio-Rad Laboratories, Hercules, CA, USA). All data were collated and managed using Microsoft Excel (Microsoft Corp., Redmond, WA, USA) and REDCap electronic data capture tools hosted at the University of Washington, Seattle (Harris et al., 2009, 2019). Statistical comparisons, calculations, and figures were generated using RStudio (2019 RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, URL http://www.rstudio.com/).
To understand method performance over time, control charts were generated for each method using OC43 Cq values from the RT-qPCR assay and paired t tests were conducted. The average value is the OC43 Cq value averaged across treatment plants and time for each method. Upper warning limits (UWL) and lower warning limits (LWL) for each method were calculated by adding or subtracting, respectively, one standard deviation from the mean Cq. Upper confidence limits (UCL) and lower confidence limits (LCL) for each method were calculated by adding or subtracting three times the standard deviation from the mean Cq. These were then plotted against the average Cq value for each sample for a visual representation of variability over time.
To compare SARS-CoV-2 detection between concentration and extraction methods, two-by-two contingency tables were generated comparing the direct extraction method to both skimmed milk methods. Comparisons were made for both undiluted assays and 10–1 diluted assays. A positive sample for SARS-CoV-2 was determined by detection of N1 or N2 target with a representative sigmoidal curve and a Cq of less that 40 in either the undiluted or tenfold diluted sample. McNemar’s Chi-Square test for count data was carried out on the tables using the built in R function to assess differences in SARS-CoV-2 detection between methods. McNemar’s test was chosen because it assesses concordant and discordant pairs of paired data (McNemar, 1947; Smith & Ruxton, 2020).
Results and Discussion
OC43 Surrogate Comparisons
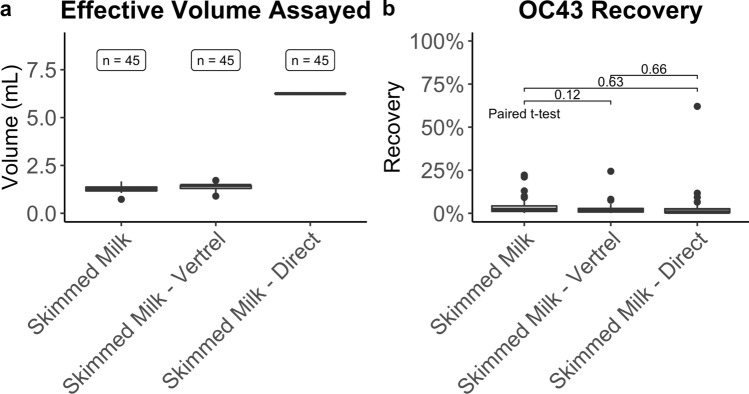
To understand how the three tested methods compared, the effective volume assayed, OC43 percent recovery, and performance over time were all considered. The effective volume assayed was calculated as the proportion of the initial volume of concentrated wastewater that was assayed by RT-qPCR. The mean effective volume assayed for skimmed milk flocculation with and without Vertrel XF™ extraction is 1.28 mL and 1.44 mL, respectively (Table 2A, Fig. 1a). Because the direct pellet extraction method does not rely on variable resuspension volumes of the pellets, the effective volume assayed for each sample is 6.25 mL. This consistency in volume assayed has a substantial benefit because it reduces variability between samples. Variability in SARS-CoV-2 detection cannot be attributed to variable effective volumes assayed because this is a constant number with the direct pellet extraction.
Table 2.
Descriptive statistics for OC43 comparisons
| (A) Effective volume assayed | (B) Percent recovery | |||||
|---|---|---|---|---|---|---|
| Skimmed milk | Skimmed milk—Vertrel | Skimmed milk—direct | Skimmed milk | Skimmed milk—Vertrel | Skimmed milk—direct | |
| Max | 1.67 | 1.72 | 6.25 | 22.16 | 24.38 | 62.06 |
| 75th Quant | 1.37 | 1.46 | 6.25 | 4.29 | 2.81 | 2.62 |
| Median | 1.3 | 1.46 | 6.25 | 2.29 | 1.38 | 0.57 |
| 25th Quant | 1.17 | 1.3 | 6.25 | 1.09 | 0.73 | 0.31 |
| Minimum | 0.73 | 0.9 | 6.25 | 0.12 | 0.13 | 0 |
| Mean | 1.28 | 1.44 | 6.25 | 4.03 | 2.63 | 3.36 |
(A) Effective volume assayed by method. (B) Percent recovery by method
Fig. 1.
Effective volume assayed and percent recovery for each method. a The effective volume assayed is the proportional volume of the original wastewater sample assayed by RT-qPCR. The skimmed milk direct pellet extraction has the highest volume assayed per reaction (6.25 mL). Because it is not dependent on variable resuspension volumes, the effective volume assayed for each sample is the same, unlike skimmed milk flocculation with or without Vertrel XF™ extraction. b The OC43 recovery is the volume adjusted recovery using the RT-qPCR standard curves. The mean recoveries for all three compared methods are comparable
When comparing OC43 percent recoveries, all three methods perform similarly. The average OC43 percent recovery for the direct pellet extraction method is 3.36% compared to 4.03% and 2.63% for skimmed milk flocculation with and without Vertrel XF™ extraction, respectively (Table 2B, Fig. 1b). Paired t tests were run comparing the percent recoveries, and there were no statistically significant differences present in the distributions (Fig. 1b).
These data suggest that there is no loss in performance relative to either of the prior skimmed milk flocculation methods when using this novel extraction method. Although the OC43 recovery values in this study are low, they are similar to those from previous work by the authors (Philo et al., 2021) as well as recoveries obtained for other viruses after wastewater concentration, including < 10% recovery for porcine reproductive and respiratory syndrome virus (PRRSV) (Farkas et al., 2021) and approximately 5% for bovine coronavirus (BCoV) (Gonzalez et al., 2020). Recovery is low for several potential reasons. Concentration and recovery methods focus on viral particles, while detection methods focus on viral RNA. It is not possible to distinguish between virus clusters and individual virus particles with cell culture methods in the way that RT-qPCR differentiates genome copies. Additionally, because OC43, PRRSV, BCoV, and SARS-CoV-2 are enveloped respiratory viruses, they could be less stable in the wastewater matrix and degrade quickly compared to non-enveloped enteric viruses (Bogler et al., 2020; Ye et al., 2016).
Previous work has indicated that larger effective volumes assayed can reduce OC43 recovery due to inhibitors concentrated with the wastewater. The bag-mediated filtration system had an average effective volume assayed of 11.90 mL through RT-qPCR but had an average OC43 percent recovery of 0.69% (Philo et al., 2021). Since the direct pellet extraction method does not have the same relationship, it can be inferred that this method better controls for the presence of inhibitors and recovers more OC43 despite the large volume assayed.
To look at the methods’ performance over time, control charts were developed using the Cq values obtained from the OC43 RT-qPCR assays (Fig. 2). The mean Cq values for all three methods are very similar (Table 3). However, the variability around the mean Cq value for the direct pellet extraction method is substantially larger than for both of the other skimmed milk methods (Table 3; Fig. 2). This is likely due to the method having both the lowest minimum Cq value and the highest maximum Cq value (Table 3). While the large maximum detection value is of concern, the low minimum Cq value suggests that direct pellet extraction can recover more of the seeded OC43 than the other two methods. In addition, there is one direct pellet extraction method sample that has a Cq greater than the UWL level compared to seven with the skimmed milk flocculation method and four with the skimmed milk flocculation with the Vertrel method.
Fig. 2.
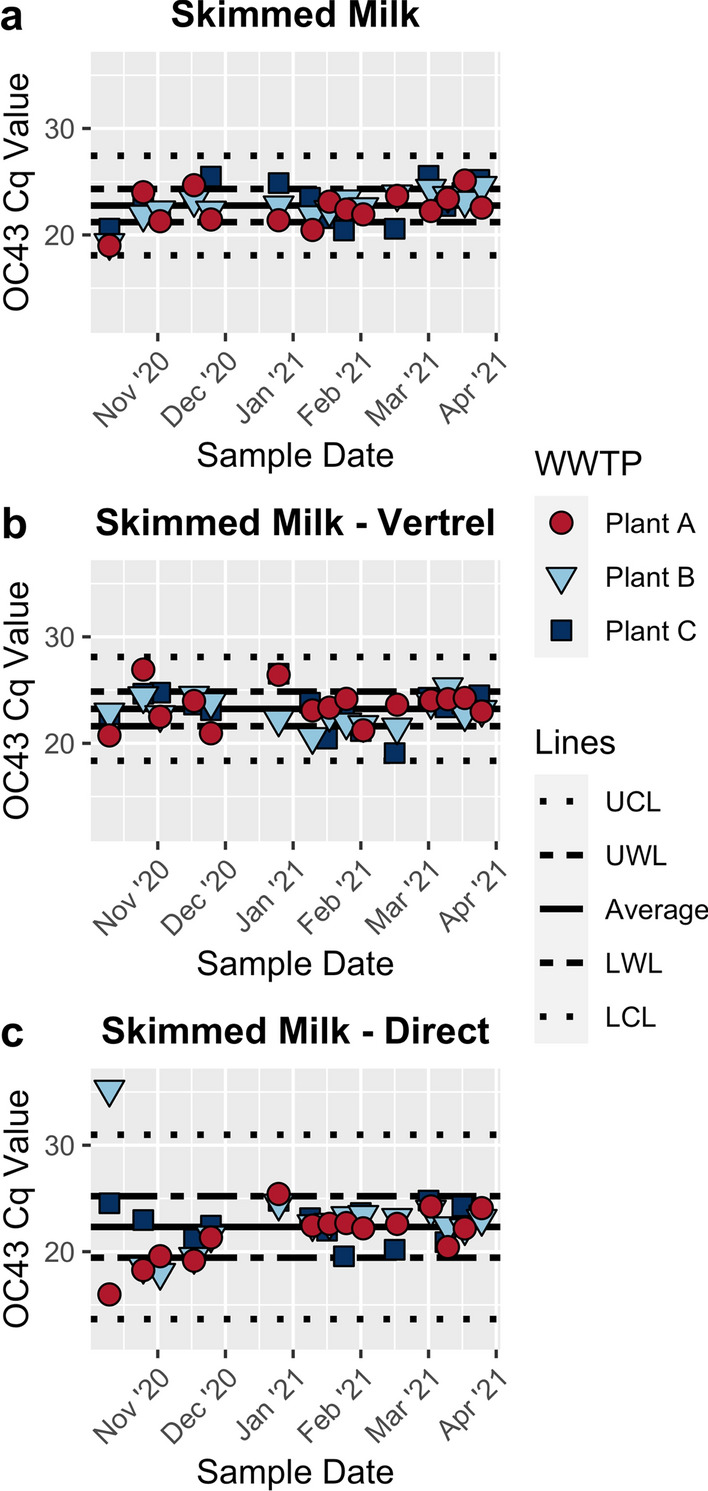
Control charts of the OC43 RT-qPCR Cq value for a skimmed milk flocculation, b skimmed milk flocculation with Vertrel XF™ extraction, and c direct skimmed milk pellet extraction. Average values were calculated by averaging across treatment plant and time. The upper warning limit (UWL) and lower warning limit (LWL) for each method were calculated by adding or subtracting, respectively, the standard deviation from the average Cq. The upper confidence limit (UCL) and lower confidence limit (LCL) were calculated by adding or subtracting, respectively, three times the standard deviation from the average Cq
Table 3.
Descriptive statistics for OC43 RT-qPCR Cq values used to generate control charts
| Skimmed milk | Skimmed milk—Vertrel | Skimmed milk—direct | |
|---|---|---|---|
| Mean | 22.8 | 23.2 | 22.3 |
| Std. dev. | 1.6 | 1.6 | 2.9 |
| Variance | 2.4 | 2.6 | 8.3 |
| Max | 25.6 | 26.9 | 35.2 |
| Min | 19 | 19.1 | 16 |
| Non detect | 0 | 0 | 0 |
OC43 was chosen as the surrogate organism in this study because both OC43 and SARS-CoV-2 are beta-coronaviruses that cause respiratory infections in humans and are spread via droplet and/or aerosol transmission (Greenhalgh et al., 2021; Harrison et al., 2020; Liu et al., 2021). If the purpose of using a surrogate organism is to understand how a method performs over time in the given water matrix, as with this study, how the surrogate behaves in relation to SARS-CoV-2 may not be as important. However, if the purpose is to adjust SARS-CoV-2 values using the recovery of the surrogate, it is crucial that the surrogate behaves similarly to SARS-CoV-2. Adjusting SARS-CoV-2 values using recovery values of an organism without direct comparisons to SARS-CoV-2 adds additional uncertainty to the results.
SARS-CoV-2 Detection
McNemar’s chi-squared tests were run on two-by-two contingency tables assessing detection of SARS-CoV-2 using criteria established in “Data Analysis”. There were no statistically significant differences in detection between the direct pellet extraction method and skimmed milk flocculation with and without Vertrel XF™ extraction in both the non-diluted and 10–1 diluted RT-qPCR reactions (Table 4). Although it is difficult to prove statistical similarity, the inability to reject the null hypothesis that the row and column marginal frequencies are equal (Smith & Ruxton, 2020) indicates that these methods result in similar SARS-CoV-2 detection.
Table 4.
Results of McNemar's chi-squared test for count data analyzing SARS-CoV-2 detection in (A) non-diluted RT-qPCR reactions and (B) 10−1 RT-qPCR reactions (n = 45)
| Method 1 | Method 2 | McNemar’s X2 | Degrees of freedom | p-Value | |
|---|---|---|---|---|---|
| (A) Non-diluted | Skimmed milk—direct | Skimmed milk | 0.23 | 1 | 0.63 |
| Skimmed milk—Vertrel | 0.06 | 1 | 0.80 | ||
| (B) 10−1 dilution | Skimmed milk—direct | Skimmed milk | 1.78 | 1 | 0.18 |
| Skimmed milk—Vertrel | 0.90 | 1 | 0.34 |
There is no statistially significant difference in SARS-CoV-2 detection between the direct pellet extraction method and skimmed milk flocculation with and without Vertrel XF™ extraction in both the (A) non-diluted reactions and (B) 10−1 reactions
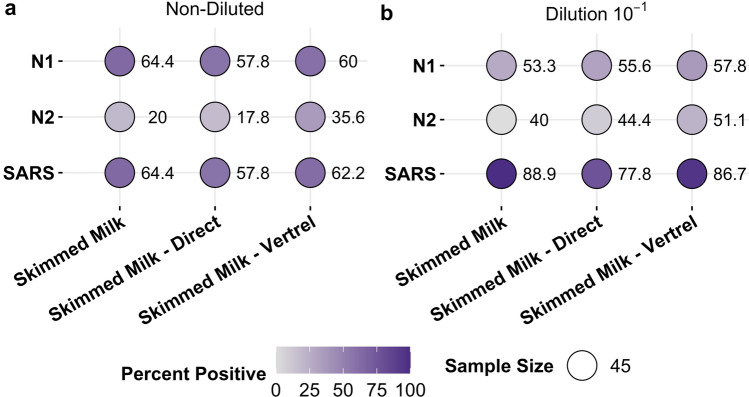
Balloon plot heat maps were also generated to visualize differences in detection between assays and methods (Fig. 3). When looking at the non-diluted RT-qPCR reactions (Fig. 3a), all three methods had similar percent positivity for the individual N1 and N2 reactions and SARS-CoV-2. Because the values for N1 and SARS-CoV2 are the same or very similar (Fig. 3a), it can be inferred that most SARS-CoV-2 detection in the undiluted reactions is due to N1 detection and not N2 detection. Frequency of SARS-CoV-2 detection in the 10–1 reactions (Fig. 3b) is higher for all three methods than in the non-diluted reactions (Fig. 3a). In addition, the proportion of samples positive for N2 is higher in the diluted reactions compared to the non-diluted reactions (Fig. 3). These results together indicate that there are inhibitors from the wastewater present in the RNA extracts, and that N2 is more susceptible to inhibitors than N1. Previous work indicates the limit of detection (LOD) is higher for N2 (100 gene copies) in wastewater than N1 (10 gene copies), likely due to the presence of inhibitors (Philo et al., 2021). Because the Cq values for these samples are at or near the LOD in wastewater (Supplementary Fig. 4), tenfold dilutions could be diluting past the point at which we expect detection in each sample. In addition, other research suggests the N1 primer and probe set is more sensitive than the N2 primer and probe set in nasopharyngeal samples (Vogels et al., 2020). This effect could be amplified in wastewater samples like those collected and processed for this study. To control for inhibitors, all RT-qPCR reactions for these methods should be run using tenfold dilutions in addition to undiluted samples (Loge et al., 2002).
Fig. 3.
Percent positivity for SARS-CoV-2 by assay in either a non-diluted or b 10−1 diluted RT-qPCR reactions. Darker colors indicate more detection. There is higher percent positivity for the N1 assay compared to N2 across all the methods, suggesting N1 contributes more to SARS-CoV-2 detection. SARS-CoV-2 detection is higher in the 10−1 diluted RT-qPCR reactions than the non-diluted reactions, indicating that there are residual effects of inhibitors on the RT-qPCR reactions
This manuscript describes the development and validation of a modification to existing skimmed milk flocculation protocols for surveillance of SARS-CoV-2 in Seattle-area wastewater. Currently, these data are not compared to the clinical case trends in King County, WA. Even though these samples were collected during a large peak in cases over the holiday period (Supplementary Table 5), SARS-CoV-2 was not detected in every sample (Fig. 3). Because the wastewater is well mixed by the time, it reaches the treatment plants, SARS-CoV-2 should have been detectable during the entire sampling period. It is likely that the SARS-CoV-2 signal is diluted beyond the limit of detection before collection. The treatment plants where samples were collected to process tens of millions of gallons of wastewater a day, with up to twice as much processed daily during the winter rainy season. For this reason, in locations that have clear wet and dry seasons, taking flow adjusted composite samples and correcting for flow in the molecular results may better represent local infection trends.
This method was developed because other existing methods require more active time in a laboratory, rely on access to expensive and often difficult to obtain consumable materials, or have limits on daily samples processed. Since the PBS resuspension step is not included in the method described here compared to other uses of skimmed milk flocculation (Calgua et al., 2008; Falman et al., 2019; Philo et al., 2021), samples can be concentrated, extracted, and analyzed for SARS-CoV-2 in a single day. The skimmed milk pellet extraction method requires three to four hours of active time and can process up to 12 samples with a single microcentrifuge, ensuring data can be reported in a timely manner. Finally, methods such as membrane filtration and ultrafiltration require expensive materials and/or advanced laboratory facilities. The direct pellet extraction method can be carried out in more resource-limited settings, making it more feasible in low- and middle-income countries (LMICs).
Despite the success of the skimmed milk direct pellet extraction at detecting SARS-CoV-2, this method can be further optimized. It is unclear where the OC43 RT-qPCR signal is being lost to result in the low mean percent recovery (Table 2B). This suggests that different methods need to be tested to identify one that performs better. In addition, recovery studies should be carried out assessing loss at each step of the method and with different extraction kits. The QIAamp Viral RNA Mini kit was selected to maintain consistency across methods, and because it was suggested by the company in early/mid-2020. However, it is likely that different protocols work better because there are still some effects of inhibitors on RT-qPCR with this extraction kit (Fig. 3). In addition, many other groups are using digital droplet or digital partitioning PCR systems for molecular detection. Optimizing the workflow for these systems will likely increase fractional recovery because they better control for inhibitors than RT-qPCR. Finally, how samples are collected can be better optimized. Understanding how frequently to take samples for a given population, whether to use grab or composite samples, and at what time of day to collect can result in more representative and useful information. Environmental and wastewater surveillance for SARS-CoV-2 will be most useful for public health officials if it provides insight beyond what can be obtained with clinical surveillance. Methods being used must be optimized and performance validated for SARS-CoV-2 in the specific catchment area for this to be true. Although the skimmed milk pellet extraction method could still be improved, it has been shown to be fit for the purpose of detecting SARS-CoV-2 in wastewater.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work could not have been carried out without our collaborators at the wastewater treatment plants who facilitated sample collection. Particularly, the West Point Process Lab, the South Treatment Plant collection team, and the Brightwater Operations staff.
Author Contributions
Conceptualization & methodology: SEP, JSM. Formal analysis and investigation: SEP, AQWO, RS, EKK, and ALK. Writing—original draft preparation: SEP, EKK, and NAZ. Writing—review and editing: SEP, ALK, NAZ, NKB, and JSM. Supervision: NKB and JSM.
Declarations
Conflict of interest
The authors did not receive support from any organization for the submitted work. The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.138764PMID-32387778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Tscharke B, Bertsch PM, Bibby K, Bivins A, Choi P, Clarke L, Dwyer J, Edson J, Nguyen TMH, O'Brien JW, Simpson SL, Sherman P, Thomas KV, Verhagen R, Zaugg JL, Mueller JF. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Science of the Total Environment. 2021 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A, Packman A, Furman A, Gross A, Kushmaro A, Ronen A, Dagot C, Hill C, Vaizel-Ohayon D, Morgenroth E, Bertuzzo E, Wells G, Kiperwas HR, Horn H, Negev I, Zucker I, Bar-Or I, Moran-Gilad J, Balcazar JL, Bibby K, Elimelech M, Weisbrod N, Nir O, Sued O, Gillor O, Alvarez PJ, Crameri S, Arnon S, Walker S, Yaron S, Nguyen TH, Berchenko Y, Hu YX, Ronen Z, Bar-Zeev E. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nature Sustainability. 2020;3(12):981–990. doi: 10.1038/s41893-020-00605-2. [DOI] [Google Scholar]
- Calgua B, Mengewein A, Grunert A, Bofill-Mas S, Clemente-Casares P, Hundesa A, Wyn-Jones AP, López-Pila JM, Girones R. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. Journal of Virological Methods. 2008;153(2):79–83. doi: 10.1016/j.jviromet.2008.08.003PMID-18765255. [DOI] [PubMed] [Google Scholar]
- Falman JC, Fagnant-Sperati CS, Kossik AL, Boyle DS, Meschke JS. Evaluation of secondary concentration methods for poliovirus detection in wastewater. Food and Environmental Virology. 2019;11(1):20–31. doi: 10.1007/s12560-018-09364-yPMID-30612304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K, Hillary LS, Thorpe J, Walker DI, Lowther JA, McDonald JE, Malham SK, Jones DL. Concentration and quantification of SARS-CoV-2 RNA in wastewater using polyethylene glycol-based concentration and qRT-PCR. Methods Protocols. 2021 doi: 10.3390/mps4010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronhoffs S, Totzke G, Stier S, Wernert N, Rothe M, Brüning T, Koch B, Sachinidis A, Vetter H, Ko Y. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Molecular and Cellular Probes. 2002;16(2):99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Curtis K, Bivins A, Bibby K, Weir MH, Yetka K, Thompson H, Keeling D, Mitchell J, Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Research. 2020 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. The Lancet. 2021;397(10285):1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN, REDCap Consortium The REDCap Consortium: Building an International Community of Software Platform Partners. Journal of Biomedical Informatics. 2019;95:03208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010PMID-18929686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends in Immunology. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Baluja MQ, Graham DW, Corbishley A, McDonald JE, Malham SK, Hillary LS, Connor TR, Gaze WH, Moura IB, Wilcox MH, Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Science of the Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.141364PMID-32836117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Liang JQ, Fung TS. Human Coronavirus-229E, -OC43, -NL63, and -HKU1 (Coronaviridae) Encyclopedia of Virology. 2021 doi: 10.1016/B978-0-12-809633-8.21501-X. [DOI] [Google Scholar]
- Loge FJ, Thompson DE, Call DR. PCR detection of specific pathogens in water: A risk-based analysis. Environmental Science & Technology. 2002;36(12):2754–2759. doi: 10.1021/es015777mPMID-12099475. [DOI] [PubMed] [Google Scholar]
- McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12(2):153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environmental Science & Technology Letters. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wang M, Warren JL, Weinberger DM, Arnold W, Omer SB. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-zPMID-32948856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiser, J. (2020). The University of Arizona says it caught a dorm's covid-19 outbreak before it started. Its secret weapon: poop. The Washington post.
- Philo SE, Keim EK, Swanstrom R, Ong AQW, Burnor EA, Kossik AL, Harrison JC, Demeke BA, Zhou NA, Beck NK, Shirai JH, Meschke JS. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Science of The Total Environment. 2021 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MQRP, Ruxton GD. Effective use of the McNemar test. Behavioral Ecology and Sociobiology. 2020;74(11):133. doi: 10.1007/s00265-020-02916-y. [DOI] [Google Scholar]
- US Centers for Disease Control and Prevention. (2020). CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Retrieved 3 March from
- Vijgen L, Keyaerts E, Moës E, Maes P, Duson G, Ranst MV. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. Journal of Clinical Microbiology. 2005;43(11):5452–5456. doi: 10.1128/jcm.43.11.5452-5456.2005PMID-16272469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Catherine Muenker M, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Dela Cruz CS, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nature Microbiology. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Water Research Foundation. (2020). Wastewater Surveillance of the COVID-19 Genetic Signal in Sewersheds. https://www.waterrf.org/sites/default/files/file/2020-06/COVID-19_SummitHandout-v3b.pdf
- World Health Organization. (2020, March 11). WHO Director-General's opening remarks at the media briefing on COVID-19—11 March 2020https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. The Lancet Gastroenterology & Hepatology. 2020;5(5):434–435. doi: 10.1016/s2468-1253(20)30083-2PMID-32199469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833.e1833. doi: 10.1053/j.gastro.2020.02.055PMID-32142773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YY, Ellenberg RM, Graham KE, Wigginton KR. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environmental Science & Technology. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.