Abstract
Little is known about the relative importance of the four species of Lancefield group G beta-hemolytic streptococci in causing bacteremia and the factors that determine the outcome for patients with group G beta-hemolytic streptococcal bacteremia. From 1997 to 2000, 75 group G beta-hemolytic streptococcal strains were isolated from the blood cultures of 66 patients. Sequencing of the 16S rRNA genes of the group G beta-hemolytic streptococci showed that all 75 isolates were Streptococcus dysgalactiae subspecies equisimilis. The API system (20 STREP) and Vitek system (GPI) successfully identified 65 (98.5%) and 62 (93.9%) isolates, respectively, as S. dysgalactiae subspecies equisimilis with >95% confidence, whereas the ATB Expression system (ID32 STREP) only successfully identified 49 isolates (74.2%) as S. dysgalactiae subspecies equisimilis with >95% confidence. The median age of the patients was 76 years (range, 33 to 99 years). Fifty-six patients (85%) were over 60 years old. All patients had underlying diseases. No source of the bacteremia was identified (primary bacteremia) in 34 patients (52%), whereas 17 (26%) had cellulitis and 8 (12%) had bed sore or wound infections. Fifty-eight patients (88%) had community-acquired group G streptococcal bacteremia. Sixty-two patients (94%) had group G Streptococcus recovered in one blood culture, whereas 4 patients (6%) had it recovered in multiple blood cultures. Fifty-nine patients (89%) had group G Streptococcus as the only bacterium recovered in their blood cultures, whereas in 7 patients other bacteria were recovered concomitantly with the group G Streptococcus in the blood cultures (Staphylococcus aureus in 3, Clostridium perfringens in 2, Citrobacter freundii in 1, and Bacteroides fragilis in 1). Overall, 10 patients (15%) died. Male sex, diagnosis other than cellulitis, hospital-acquired bacteremia, and multiple positive blood cultures were associated with mortality {P < 0.005 (relative risk [RR] = 7.6), P < 0.05 (RR = 3.7), P < 0.005 (RR = 5.6), and P < 0.05 (RR = 5.6), respectively}. Unlike group C beta-hemolytic streptococcal bacteremia, group G beta-hemolytic streptococcal bacteremia is not a zoonotic infection in Hong Kong.
Lancefield groups A, B, C, and G streptococci are the major groups of beta-hemolytic streptococci that cause bacteremia. The major reservoir for group A and B streptococci is humans, whereas most group C beta-hemolytic streptococcal bacteremias in Hong Kong are of animal origin (28). The group G beta-hemolytic streptococci consist of Streptococcus dysgalactiae subspecies equisimilis, S. milleri, S. canis, and S. intestinalis. The reservoirs of S. dysgalactiae subspecies equisimilis and S. milleri are humans, whereas those of S. canis and S. intestinalis are dogs and pigs, respectively. In one study, group G streptococci were shown to be the most common cause of beta-hemolytic streptococcal bacteremia (16). It has been reported that diabetes mellitus (DM), malignancy, cardiovascular disease, bone and joint diseases, and cirrhosis are the major underlying diseases in patients with group G beta-hemolytic streptococcal bacteremia (2, 8, 10, 16, 19). However, little is known about the relative importance of the various species and thus the source of bacteria causing bacteremia, the usefulness of commercial kits in identifying the species, and the factors that determine the outcome for patients with group G beta-hemolytic streptococcal bacteremia.
Although the Vitek system (bioMerieux Vitek, Hazelwood, Mo.), the API system (bioMerieux Vitek), and the ATB Expression system (bioMerieux Vitek) are commonly used for microbial identification of beta-hemolytic streptococci in the clinical laboratory, none of these systems contains all four group G beta-hemolytic streptococci in the database. Since the discovery of PCR and DNA sequencing, comparisons of the gene sequences of bacterial species have shown that the 16S rRNA gene is highly conserved within a species and among species of the same genus and thus can be used as the new “gold standard” for species-level identification of bacteria. Using this new standard, phylogenetic trees based on base differences between species are constructed, and bacteria are classified and reclassified into new genera (13, 14). Recently, we have reported the application of 16S rRNA gene sequencing in the identification of clinical isolates with ambiguous biochemical profiles (20, 21, 24, 25) and of a bacterium that was noncultivable (7). In this study, we used 16S rRNA gene sequencing for the species-level identification of 75 group G beta-hemolytic streptococcal strains isolated from the blood cultures of 66 patients in a 4-year period. The usefulness of the Vitek system (GPI), the API system (20 STREP), and the ATB Expression system (ID32 STREP) in identifying the isolates was compared with this gold standard. The epidemiology, clinical diseases, and outcome for patients that developed group G beta-hemolytic streptococcal bacteremia were also analyzed.
MATERIALS AND METHODS
Patients and microbiological methods.
The 66 patients in this study were hospitalized at the Queen Mary Hospital in Hong Kong during a 4-year period (1997 to 2000). All clinical data were collected prospectively as described in our previous publications (11, 27). Clinical specimens were collected and handled according to standard protocols. The BACTEC 9240 blood culture system (Becton Dickinson, Sparks, Md.) was used. All suspect colonies were identified by standard conventional biochemical methods (12). Lancefield serogrouping was performed using Streptex (Murex Biotech Ltd., Dartford, United Kingdom) according to the manufacturer's instructions. In addition, the Vitek system (GPI; bioMerieux Vitek), API system (20 STREP; bioMerieux Vitek), and ATB Expression system (ID32 STREP; bioMerieux Vitek) were used for the identification of the group G beta-hemolytic streptococci in this study. Multiple isolates obtained from the same patient were counted only once in the calculation of the usefulness of each kit for identifying group G Streptococcus. Antimicrobial susceptibility was tested by the Kirby-Bauer disk diffusion method and results were interpreted according to the NCCLS criteria (3).
Extraction of bacterial DNA for 16S rRNA gene sequencing.
The bacterial DNA extraction method was modified from our previous published protocol (23). Briefly, 80 μl of NaOH (0.05 M) was added to 20 μl of bacterial cells suspended in distilled water and the mixture was incubated at 60°C for 45 min, followed by addition of 6 μl of Tris-HCl (pH 7.0) to achieve a final pH of 8.0. The resultant mixture was diluted 100-fold, and 5 μl of the diluted extract was used for PCR.
PCR, gel electrophoresis, and 16S rRNA gene sequencing.
PCR amplification and DNA sequencing of the 16S rRNA genes were performed according to our previous publications (7, 20, 21, 22, 24, 25). Briefly, DNase I-treated distilled water and PCR master mix (which contains deoxynucleoside triphosphates, PCR buffer, and Taq polymerase) were used in all PCRs by adding 1 U of DNase I (Pharmacia, Uppsala, Sweden) to 40 μl of distilled water or PCR master mix and incubating the mixture at 25°C for 15 min and subsequently at 95°C for 10 min to inactivate the DNase I. The bacterial DNA extracts and controls were amplified with 0.5 μM concentrations of the primers (LPW200, 5′-GAGTTGCGAACGGGTGAG-3′, and LPW205, 5′-CTTGTTACGACTTCACCC-3′) (Gibco BRL, Rockville, Md.). The PCR mixture (50 μl) contained bacterial DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, and 0.01% gelatin), 200 μM concentrations of each deoxynucleoside triphosphate, and 1.0 U of Taq polymerase (Boehringer, Mannheim, Germany). The mixtures were amplified in 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 10 min in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). DNase I-treated distilled water was used as the negative control. A 10-μl aliquot of each amplified product was electrophoresed in a 1.0% (wt/vol) agarose gel, with a molecular size marker (lambda DNA AvaII digest; Boehringer) run in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 1.5 h. The gel was stained with ethidium bromide (0.5 μg/ml) for 15 min, rinsed, and photographed under UV light illumination.
The PCR products were gel purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI 377 automated sequencer according to the manufacturer's instructions (Perkin-Elmer, Foster City, Calif.), using the PCR primers LPW200 and LPW205 and additional primers designed from the sequencing data of the first round of sequencing (LPW99, 5′-TTATTGGGCGTAAAGCGA-3′, and LPW273, 5′-TTGCGGGACTTAACCCAAC-3′). The sequences of the PCR products were compared with known 16S rRNA gene sequences in GenBank by multiple sequence alignment using the CLUSTAL W program (17).
Statistical analysis.
A comparison of characteristics was made between patients who succumbed to and those who survived the group G beta-hemolytic streptococcal bacteremia. A chi-square test was used for categorical variables and Student's t test was used for age analysis. Multivariate logistic analysis was not performed because of the small sample size. A P value of <0.05 was regarded as statistically significant.
RESULTS
16S rRNA gene sequencing.
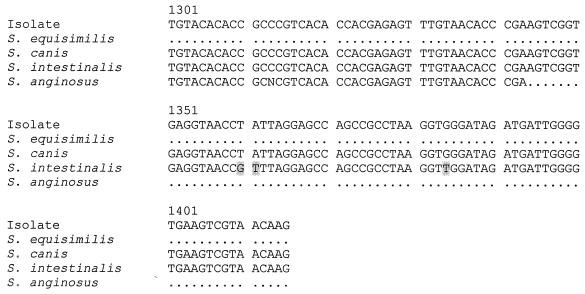
PCR of the 16S rRNA genes of the blood culture group G beta-hemolytic streptococci showed bands at 1,414 bp. Sequencing of the 16S rRNA genes showed that all the isolates obtained from the same patient had the same nucleotide sequence. There were one to four base differences between the isolates and S. dysgalactiae subspecies equisimilis (GenBank accession number AB008926), 79 to 82 base differences between the isolates and S. anginosus (GenBank accession number X58309), 52 to 55 base differences between the isolates and S. canis (GenBank accession number AB002483), and 73 to 76 base differences between the isolates and S. intestinalis (GenBank accession number AB002519), indicating that all 75 isolates were S. dysgalactiae subspecies equisimilis (Fig. 1).
FIG. 1.
DNA sequences of the 16S rRNA genes of the group G beta-hemolytic Streptococcus isolate from the blood culture of patient 1 and those of S. dysgalactiae subspecies equisimilis (GenBank accession number AB008926), S. anginosus (GenBank accession number X58309), S. canis (GenBank accession number AB002483), and S. intestinalis (GenBank accession number AB002519). The shaded bases represent those in the various Streptococcus species that are different from the corresponding ones in the isolate.
Lancefield serogrouping and identification by commercial systems.
All group G beta-hemolytic streptococci (isolated from blood cultures and other clinical specimens) showed agglutination only with the serogroup G antiserum and not with any other antisera. All isolates obtained from the same patient showed the same biochemical profile and hence were counted as one isolate in the calculations. The API system (20 STREP) successfully identified 65 (98.5%) isolates as S. dysgalactiae subspecies equisimilis with >95% confidence. The remaining 1 (1.5%) isolate was identified as S. dysgalactiae subspecies equisimilis with 77% confidence. The Vitek system (GPI) successfully identified 62 (93.9%) isolates as S. dysgalactiae with >95% confidence, 1 (1.5%) isolate as S. agalactiae with 81% confidence, and 3 (4.5%) isolates were “unidentified.” The ATB Expression system (ID32 STREP) only successfully identified 49 (74.2%) isolates as S. dysgalactiae subspecies equisimilis with >95% confidence, 3 (4.5%) isolates as S. dysgalactiae subspecies equisimilis with 83% confidence, 3 (4.5%) isolates as S. dysgalactiae subspecies equisimilis with 65% confidence, 1 (1.5%) isolate as Streptococcus group L with 87% confidence, and 1 (1.5%) isolate as S. agalactiae with >95% confidence (not the isolate that was identified by the Vitek system as S. agalactiae), and 9 (13.6%) isolates were “unidentified.”
Patient characteristics.
The characteristics of the 66 patients with group G streptococcal bacteremia are tabulated and summarized in Tables 1 and 2, respectively. The incidence of group G streptococcal bacteremia was similar throughout the 4-year study period. There were slightly more cases in the late spring and summer months (April to August). The median age was 76 years (range, 33 to 99 years). Fifty-six patients (85%) were over 60 years old. The male/female ratio was 38:28. All patients had underlying diseases. The major underlying conditions included immobilization and/or bed sore in 19 (29%), cerebrovascular accident (CVA) in 18 (27%), malignancy in 17 (26%), hypertension (HT) in 16 (24%), DM in 13 (20%), dementia in 10 (15%), lymphedema in 10 (15%), chronic renal failure (CRF) in 10 (15%), cirrhosis in 5 (8%), and intravenous drug abuse (IVDA) in 5 (8%). No source of the bacteremia was identified (primary bacteremia) in 34 patients (52%), whereas 17 (26%) had cellulitis, 8 (12%) had a bed sore or wound infection, 2 (3%) had infective endocarditis, 2 (3%) had pneumonia, 2 (3%) had an abscess, and 1 (2%) had septic arthritis. Fifty-eight (88%) and 8 (12%) had community- and hospital-acquired group G streptococcal bacteremia, respectively. Sixty-two patients (94%) had group G Streptococcus recovered in 1 blood culture, whereas 4 patients (6%) had it recovered in multiple blood cultures. Fifty-nine patients (89%) had group G Streptococcus as the only bacteria recovered in their blood cultures, whereas in 7 patients other bacteria were recovered concomitantly with the group G Streptococcus in the blood cultures (Staphylococcus aureus in patients 10, 48, and 54; Clostridium perfringens in patients 28 and 56; Citrobacter freundii in patient 21; and Bacteroides fragilis in patient 47). Seven patients with wound infections and two patients with cellulitis had group G Streptococcus recovered from the wound swabs (patients 4, 12, 16, 26, 29, 39, 47, and 56) or aspirate (patient 53). One patient (patient 63) with a pelvic abscess had group G Streptococcus recovered from the abscess pus. The isolates recovered from 63 patients (95.5%) were sensitive to penicillin, cephalothin, erythromycin, clindamycin, and vancomycin. Two isolates (3.0%) were resistant to both erythromycin and clindamycin (patients 38 and 41), and one isolate was resistant to erythromycin but sensitive to clindamycin (patient 39). Overall, 10 patients (15%) died.
TABLE 1.
Characteristics of patients with group G beta-hemolytic streptococcal bacteremia
| Patient no. | Sexa/age (yr) | Underlying conditionb | Diagnosisc | CA or HAd | No. of positive blood cultures | Other bacteria recovered in blood culture | Other clinical specimens with group G Streptococcus isolated | Outcome |
|---|---|---|---|---|---|---|---|---|
| 1 | M/33 | IVDA | IE | CA | 5 | None | None | Died |
| 2 | M/44 | IVDA | Knee septic arthritis | CA | 1 | None | None | Cured |
| 3 | M/78 | CVA | Primary bacteremia | CA | 1 | None | None | Cured |
| 4 | M/76 | DM, CRF | Wound infection | HA | 1 | None | Wound swab | Cured |
| 5 | F/67 | Ca vagina | Cellulitis | CA | 1 | None | None | Cured |
| 6 | F/63 | NPC | Primary bacteremia | CA | 1 | None | None | Cured |
| 7 | F/80 | CRF, dementia | Primary bacteremia | CA | 1 | None | None | Cured |
| 8 | F/87 | Ca cervix | Primary bacteremia | CA | 1 | None | None | Cured |
| 9 | M/87 | DM, CRF | Primary bacteremia | CA | 1 | None | None | Cured |
| 10 | F/92 | Bed bound, bed sore, HT, dementia | Primary bacteremia | CA | 1 | S. aureus | None | Cured |
| 11 | F/69 | HT, Ca breast, lymphedema | Cellulitis | CA | 1 | None | None | Cured |
| 12 | F/87 | Bed bound, HT | Foot gangrene | CA | 1 | None | Wound swab | Cured |
| 13 | M/71 | Ca esophagus | Primary bacteremia | HA | 2 | None | None | Cured |
| 14 | M/78 | CVA, DM, HT | Primary bacteremia | CA | 1 | None | None | Died |
| 15 | M/79 | Ca larynx, Ca esophagus | Primary bacteremia | HA | 1 | None | None | Died |
| 16 | M/60 | IVDA, DM, alcoholic cirrhosis | Wound infection | CA | 1 | None | Wound swab | Died |
| 17 | M/47 | IVDA, cirrhosis | Primary bacteremia | CA | 1 | None | None | Cured |
| 18 | F/76 | CVA, DM, HT | Primary bacteremia | CA | 1 | None | None | Cured |
| 19 | M/80 | CVA, HT, chair bound | Primary bacteremia | CA | 1 | None | None | Cured |
| 20 | M/63 | CVA, HT | Primary bacteremia | CA | 1 | None | None | Died |
| 21 | M/75 | Dementia, CVA, bed bound | Primary bacteremia | CA | 1 | C. freundii | None | Died |
| 22 | M/86 | Dementia | Primary bacteremia | CA | 1 | None | None | Cured |
| 23 | M/80 | CVA, dementia, bed bound | Primary bacteremia | CA | 1 | None | None | Cured |
| 24 | F/71 | Ca breast, lymphedema | Cellulitis | CA | 1 | None | None | Cured |
| 25 | M/83 | Ca lung, bed bound | Primary bacteremia | CA | 1 | None | None | Cured |
| 26 | F/74 | CVA, bed bound | Infected bed sore | CA | 1 | None | Wound swab | Cured |
| 27 | F/64 | Ca breast | Cellulitis | CA | 1 | None | None | Cured |
| 28 | F/76 | CVA, DM, bed bound | Primary bacteremia | CA | 1 | C. perfringens | None | Cured |
| 29 | M/71 | Dementia, DM | Wound infection | HA | 2 | None | Wound swab | Died |
| 30 | F/97 | Blind | Pneumonia | CA | 1 | None | None | Cured |
| 31 | M/80 | Chronic skin problem | Primary bacteremia | HA | 1 | None | None | Died |
| 32 | F/39 | AML | Primary bacteremia | CA | 1 | None | None | Cured |
| 33 | F/81 | CVA | Primary bacteremia | CA | 1 | None | None | Cured |
| 34 | M/82 | DM, Parkinson's disease, bed sore | Infected bed sore | CA | 1 | None | None | Cured |
| 35 | M/91 | CVA, malnutrition | Pneumonia | CA | 1 | None | None | Cured |
| 36 | M/86 | None | Primary bacteremia | CA | 1 | None | None | Died |
| 37 | F/92 | CRF, bed bound | Cellulitis | CA | 1 | None | None | Cured |
| 38 | M/46 | Ca tongue | Cellulitis | CA | 1 | None | None | Cured |
| 39 | F/97 | CRF, chair bound | Cellulitis | CA | 1 | None | Wound swab | Cured |
| 40 | F/76 | DM, CVA, chair bound, Ca breast | Cellulitis | CA | 1 | None | None | Cured |
| 41 | M/45 | Alcoholic cirrhosis | Leg abscess | CA | 1 | None | None | Cured |
| 42 | F/70 | Ca cervix | Primary bacteremia | HA | 1 | None | None | Cured |
| 43 | F/89 | Dementia | Primary bacteremia | CA | 1 | None | None | Cured |
| 44 | M/70 | CVA, HT, CRF, chair bound | Primary bacteremia | CA | 1 | None | None | Cured |
| 45 | M/64 | Lymphedema, recurrent cellulitis | Cellulitis | CA | 1 | None | None | Cured |
| 46 | M/41 | Lymphedema, recurrent cellulitis | Cellulitis | CA | 1 | None | None | Cured |
| 47 | F/99 | CVA, DM, HT, bed bound | Infected bed sore | CA | 1 | B. fragilis | Wound swab | Cured |
| 48 | M/83 | CVA | Cellulitis | CA | 1 | S. aureus | None | Cured |
| 49 | F/73 | CVA, bed bound, DM, HT, gout | Primary bacteremia | CA | 1 | None | None | Cured |
| 50 | M/79 | Ca floor of mouth | Primary bacteremia | CA | 1 | None | None | Cured |
| 51 | M/64 | Lymphedema, recurrent cellulitis | Cellulitis | CA | 1 | None | None | Cured |
| 52 | F/95 | HT, CVA | Cellulitis | CA | 1 | None | None | Cured |
| 53 | M/68 | Varicose vein, recurrent cellulitis | Cellulitis | CA | 1 | None | Wound aspirate | Cured |
| 54 | M/48 | IVDA | IE | CA | 4 | S. aureus | None | Cured |
| 55 | M/81 | Cervical myelopathy, chair bound, HT, CRF | Primary bacteremia | CA | 1 | None | None | Cured |
| 56 | M/72 | DM, peripheral vascular disease, HT, alcoholism | Wound infection | CA | 1 | C. perfringens | Wound swab | Cured |
| 57 | M/82 | HT, gout, CRF | Cellulitis | CA | 1 | None | None | Cured |
| 58 | M/79 | CVA, COPD, bed bound, bed sore | Primary bacteremia | CA | 1 | None | None | Cured |
| 59 | F/82 | HT, dementia | Primary bacteremia | CA | 1 | None | None | Cured |
| 60 | F/64 | Ca maxilla | Cellulitis | CA | 1 | None | None | Cured |
| 61 | F/88 | Bed bound, dementia, HT | Primary bacteremia | CA | 1 | None | None | Cured |
| 62 | M/49 | CRF, renal transplant, cirrhosis | Primary bacteremia | CA | 1 | None | None | Cured |
| 63 | M/80 | Ca rectum, DU, DM | Pelvic abscess | CA | 1 | None | Abscess pus | Cured |
| 64 | F/74 | Dementia, DM, bed sore | Primary bacteremia | HA | 1 | None | None | Cured |
| 65 | F/77 | Ca colon | Cellulitis | CA | 1 | None | None | Cured |
| 66 | M/71 | CRF | Primary bacteremia | HA | 1 | None | None | Died |
M, male; F, female.
IVDA, intravenous drug abuse; CVA, cerebrovascular accident; DM, diabetes mellitus; CRF, chronic renal failure; Ca, carcinoma; NPC, nasopharyngeal carcinoma; HT, hypertension; AML, acute myeloid leukemia; COPD, chronic obstructive pulmonary disease; DU, duodenal ulcer.
IE, infective endocarditis.
CA, community acquired; HA, hospital acquired.
TABLE 2.
Summary of characteristics of patients with group G beta-hemolytic streptococcal bacteremia
| Parameter | No. of patients (%) |
|---|---|
| Year | |
| 1997 | 12 (18) |
| 1998 | 18 (27) |
| 1999 | 17 (26) |
| 2000 | 19 (29) |
| Month | |
| Jan | 4 (6) |
| Feb | 5 (8) |
| Mar | 4 (6) |
| Apr | 9 (14) |
| May | 8 (12) |
| June | 7 (11) |
| July | 4 (6) |
| Aug | 8 (12) |
| Sept | 3 (5) |
| Oct | 5 (8) |
| Nov | 4 (6) |
| Dec | 5 (8) |
| Age [mean ± SEM,a median (range)] | 73 ± 1.9, 76 (33–99) |
| 0–10 | 0 (0) |
| 11–20 | 0 (0) |
| 21–30 | 0 (0) |
| 31–40 | 2 (3) |
| 41–50 | 7 (11) |
| 51–60 | 1 (2) |
| 61–70 | 11 (16) |
| 71–80 | 24 (36) |
| 81–90 | 14 (21) |
| 91–100 | 7 (11) |
| Sex (male:female) | 38:28 |
| Underlying condition | |
| Immobilization and/or bed sore | 19 (29) |
| CVA | 18 (27) |
| Malignancy | 17 (26) |
| HT | 16 (24) |
| DM | 13 (20) |
| Dementia | 10 (15) |
| Lymphedema | 10 (15) |
| CRF | 10 (15) |
| Cirrhosis | 5 (8) |
| IVDA | 5 (8) |
| Diagnosis | |
| Primary bacteremia | 34 (52) |
| Cellulitis | 17 (26) |
| Bed sore/wound infection | 8 (12) |
| Infective endocarditis | 2 (3) |
| Pneumonia | 2 (3) |
| Abscess | 2 (3) |
| Septic arthritis | 1 (2) |
| Community or hospital acquired | |
| Community | 58 (88) |
| Hospital | 8 (12) |
| No. of positive blood cultures | |
| 1 | 62 (94) |
| 2 | 2 (3) |
| 3 | 0 (0) |
| 4 | 1 (2) |
| 5 | 1 (2) |
| Mono- or polymicrobial bacteremia | |
| Monomicrobial | 59 (89) |
| Polymicrobial | 7 (11) |
| Positive culture at other sites | 10 (15) |
| Resistance to antibiotics | |
| Penicillin | 0 (0) |
| Cephalothin | 0 (0) |
| Erythromycin | 3 (5) |
| Clindamycin | 2 (3) |
| Vancomycin | 0 (0) |
| Mortality | 10 (15) |
SEM, standard error of the mean.
The characteristics of patients who succumbed to and those who survived the group G beta-hemolytic streptococcal bacteremia were compared and are summarized in Table 3. Male sex, diagnosis other than cellulitis, hospital-acquired bacteremia, and multiple positive blood cultures were associated with mortality {P < 0.005 (relative risk [RR] = 7.6), P < 0.05 (RR = 3.7), P < 0.005 (RR = 5.6), and P < 0.05 (RR = 5.6), respectively}.
TABLE 3.
Comparison of characteristics of patients who died of and those who survived the group G beta-hemolytic streptococcal bacteremiaa
| Characteristic | No. of patients (%)
|
P value | RR | |
|---|---|---|---|---|
| Died (n = 10) | Survived (n = 56) | |||
| Sex | ||||
| Male | 10 (100) | 28 (50) | <0.005 | 7.6 |
| Female | 0 (0) | 28 (50) | ||
| Age (mean ± SEM) | 69 ± 5.3 | 74 ± 2.0 | NS | |
| Underlying condition | ||||
| Immobilization and/or bed sore | 1 (10) | 18 (33) | NS | |
| CVA | 3 (30) | 15 (27) | NS | |
| Malignancy | 1 (10) | 16 (27) | NS | |
| HT | 2 (20) | 14 (25) | NS | |
| DM | 3 (30) | 10 (18) | NS | |
| Dementia | 2 (20) | 8 (15) | NS | |
| Lymphedema | 0 (0) | 10 (18) | NS | |
| CRF | 0 (0) | 9 (16) | NS | |
| Cirrhosis | 1 (10) | 4 (7) | NS | |
| IVDA | 2 (20) | 3 (5) | NS | |
| Diagnosis | ||||
| Primary bacteremia | 7 (70) | 27 (48) | NS | |
| Cellulitis | 0 (0) | 17 (30) | <0.05 | 3.7 |
| Bed sore or wound infection | 2 (20) | 6 (11) | NS | |
| Infective endocarditis | 1 (10) | 1 (2) | NS | |
| Pneumonia | 0 (0) | 2 (4) | NS | |
| Abscess | 0 (0) | 2 (4) | NS | |
| Septic arthritis | 0 (0) | 1 (2) | NS | |
| Community or hospital acquired | ||||
| Community | 6 (60) | 52 (93) | ||
| Hospital | 4 (40) | 4 (7) | <0.005 | 5.6 |
| No. of positive blood cultures | ||||
| Single | 8 (80) | 54 (96) | ||
| Multiple | 2 (20) | 2 (4) | <0.05 | 5.6 |
| Mono- or polymicrobial bacteremia | ||||
| Monomicrobial | 9 (90) | 50 (89) | NS | |
| Polymicrobial | 1 (10) | 6 (11) | NS | |
| Positive culture at other sites | 2 (20) | 8 (14) | NS | |
| Resistance to antibiotics | ||||
| Penicillin | 0 (0) | 0 (0) | NS | |
| Cephalothin | 0 (0) | 0 (0) | NS | |
| Erythromycin | 0 (0) | 3 (5) | NS | |
| Clindamycin | 0 (0) | 2 (4) | NS | |
| Vancomycin | 0 (0) | 0 (0) | NS | |
Abbreviations: SEM, standard error of the mean; NS, not significant.
DISCUSSION
In this study, we documented that all 75 group G beta-hemolytic streptococci were S. dysgalactiae subspecies equisimilis by using the molecular gold standard of bacterial species-level identification, 16S rRNA gene sequencing. With this gold standard, the relative usefulness of the Vitek system (GPI), the API system (20 STREP), and the ATB Expression system (ID32 STREP) in identifying this species could be compared. The API system (20 STREP) and Vitek system (GPI) were good in identifying S. dysgalactiae subspecies equisimilis, as they were able to confidently identify 99% and 94% of the isolates, respectively. On the other hand, the ATB Expression system (ID32 STREP) could only identify 74% of the isolates correctly, despite this system's containing the largest number of biochemical reactions [32 as opposed to 25 in the API system (20 STREP) and 30 in the Vitek system (GPI)].
The epidemiology of group G beta-hemolytic streptococcal bacteremia in Hong Kong was found to be similar to that reported in previous studies (2, 8, 10, 16, 19). Most patients were old, with a predominance of males. In our series, all patients had underlying diseases, similar to the 74 to 92% rate reported in previous studies (10, 16). The spectrum of underlying conditions was also similar, with immobilization, CVA, HT, and malignancy being the most important conditions (10, 16). Similar to previous studies (i.e., rates of 70 to 75%), most cases of group G beta-hemolytic streptococcal bacteremia were community acquired (88%) (2, 16, 19). The reservoir for S. dysgalactiae subspecies equisimilis is humans, and most infections are probably endogenous. Unlike group C beta-hemolytic streptococcal bacteremia (28), group G beta-hemolytic streptococcal bacteremia is not a zoonotic infection in Hong Kong. Among the 30 patients with documented foci of infection, the skin was the major portal of entry, as cellulitis and bed sore or wound infections made up the majority of the diagnoses. No clinical diagnosis was achieved for 34 patients (52%), which was comparable to the 50% rate reported in a previous study (16).
Cellulitis was the most common diagnosis in those patients who had group G beta-hemolytic streptococcal bacteremia with known sources of infection. In our recent series and in previous reports on cellulitis complicating lymphedema, non-group A beta-hemolytic streptococci (mostly group B and group G streptococci) constituted most of the documented causes of cellulitis (26). Most of the patients had lymphedema because of malignancy with surgery and/or radiotherapy. In the present study, among the 17 patients with group G beta-hemolytic streptococcal bacteremic cellulitis, about half had it as a complication of lymphedema and malignancy, of which carcinoma of the breast was the most common.
The overall mortality rate (15%) in our patients with group G beta-hemolytic streptococcal bacteremia was similar to those reported in previous studies (8 to 17%) (2, 6). In the previous reports, no analysis was made on the risk factors associated with mortality in patients with group G beta-hemolytic streptococcal bacteremia. In this study, we found by univariate analysis that male gender, diagnosis other than cellulitis, hospital-acquired infection, and multiple positive blood cultures were poor prognostic factors for group G beta-hemolytic streptococcal bacteremia. Since there were only 10 patients who died, multivariate analyses for the identification of independent prognostic factors was not performed.
Two S. dysgalactiae subspecies equisimilis isolates were resistant to both erythromycin and clindamycin. Macrolide, lincosamide, and type B streptogramin antibiotics inhibit bacterial growth by binding to the 50S ribosomal subunits of the bacteria and inhibiting protein synthesis. Resistance to these antibiotics is usually due to target modification through the acquisition of erm (erythromycin resistance methylase) genes. These genes encode enzymes that N-6 dimethylate a specific adenine residue of the 23S rRNA of the bacteria, leading to cross-resistance to macrolides, lincosamides, and streptogramin B (MLSB resistance phenotype) (1, 4, 5, 6, 9, 15, 18). We speculate that the cross-resistance to erythromycin and clindamycin in the two S. dysgalactiae subspecies equisimilis isolates was due to an erm gene. Further cloning and characterization of such a gene will delineate whether it belongs to an existing class or a previously undescribed class.
ACKNOWLEDGMENT
This work was partly supported by the Committee of Research and Conference Grants, The University of Hong Kong.
REFERENCES
- 1.Arthur M, Brisson-Noël A, Courvalin P. Origin and evolution of genes specifying resistance to macrolide, lincosamide, and streptogramin antibiotics: data and hypotheses. J Antimicrob Chemother. 1987;20:783–802. doi: 10.1093/jac/20.6.783. [DOI] [PubMed] [Google Scholar]
- 2.Auckenthaler R, Hermans P E, Washington J A., II Group G streptococcal bacteremia: clinical study and review of the literature. Rev Infect Dis. 1983;5:196–204. doi: 10.1093/clinids/5.2.196. [DOI] [PubMed] [Google Scholar]
- 3.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a single disc method. Am J Clin Pathol. 1996;45:493–496. [PubMed] [Google Scholar]
- 4.Brantl S, Kummer C, Behnke D. Complete nucleotide sequence of plasmid pGB3631: a derivative of the Streptococcus agalactiae plasmid pIP501. Gene. 1994;142:155–156. doi: 10.1016/0378-1119(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 5.Ceglowski P, Boitsov A, Chai S, Alonso J C. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 6.Ceglowski P, Alonso J C. Gene organization of the Streptococcus pyogenes plasmid pDB101: sequence analysis of the orf?-orfS region. Gene. 1994;145:33–39. doi: 10.1016/0378-1119(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 7.Cheuk W, Woo P C Y, Yuen K Y, Yu P H, Chan J K C. Intestinal inflammatory pseudotumor with regional lymph node involvement: identification of a new bacterium as the etiologic agent. J Pathol. 2001;192:289–292. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH767>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Finch R G, Aveline A. Group G streptococcal septicaemia: clinical observations and laboratory studies. J Infect. 1984;9:126–133. doi: 10.1016/s0163-4453(84)90922-8. [DOI] [PubMed] [Google Scholar]
- 9.Horinouchi S, Byeon W, Weisblum B A. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983;154:1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C E, Jang T N, Wang F D, Liu C Y. Invasive group G streptococcal infections: a review of 37 cases. Chin Med J. 1995;56:173–178. [PubMed] [Google Scholar]
- 11.Luk W K, Wong S S Y, Yuen K Y, Ho P L, Woo P C Y, Lee R A, Chau P Y. Inpatient emergencies encountered by an infectious disease consultative service. Clin Infect Dis. 1998;26:695–701. doi: 10.1086/514591. [DOI] [PubMed] [Google Scholar]
- 12.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. [Google Scholar]
- 13.Olsen G J, Woese C R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 14.Olsen G J, Overbeek R, Larsen N, Marsh T L, McCaughey M J, Maciukenas M A, Kuan W M, Macke T J, Xing Y, Woese C R. The ribosomal database project. Nucleic Acids Res. 1992;20(Suppl.):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seppälä H, Skurnik M, Soini H, Roberts M C, Huovinen P. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob Agents Chemother. 1998;42:257–262. doi: 10.1128/aac.42.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skogberg K, Simonen H, Renkonen O V, Valtonen V V. Beta-haemolytic group A, B, C and G streptococcal septicaemia: a clinical study. Scand J Infect Dis. 1988;20:119–125. doi: 10.3109/00365548809032427. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Coirralin P. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 1990;18:3660. doi: 10.1093/nar/18.12.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watsky K L, Kollisch N, Densen P. Group G streptococcal bacteremia. Arch Intern Med. 1985;145:58–61. doi: 10.1001/archinte.145.1.58. [DOI] [PubMed] [Google Scholar]
- 20.Woo, P. C. Y., A. S. P. Leung, K. W. Leung, and K. Y. Yuen. Identification of slide-coagulase positive, tube-coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol. Pathol., in press. [DOI] [PMC free article] [PubMed]
- 21.Woo P C Y, Cheung E Y L, Leung K W, Yuen K Y. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species with ambiguous biochemical profile from a renal transplant recipient. Diagn Microbiol Infect Dis. 2001;39:85–93. doi: 10.1016/s0732-8893(01)00206-1. [DOI] [PubMed] [Google Scholar]
- 22.Woo P C Y, Fung A M Y, Wong S S Y, Tsoi H W, Yuen K Y. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J Clin Microbiol. 2001;39:1190–1191. doi: 10.1128/JCM.39.3.1190-1194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo P C Y, Lo C Y, Lo S K, Siau H, Peiris J S M, Wong S S Y, Luk W K, Chan T M, Lim W W, Yuen K Y. Distinct genotypic distributions of cytomegalovirus (CMV) envelope glycoprotein in bone marrow and renal transplant recipients with CMV disease. Clin Diagn Lab Immunol. 1997;4:515–518. doi: 10.1128/cdli.4.5.515-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo P C Y, Tsoi H W, Leung K W, Lum P N L, Leung A S P, Ma C H, Kam K M, Yuen K Y. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S ribosomal RNA sequencing. J Clin Microbiol. 2000;38:3515–3517. doi: 10.1128/jcm.38.9.3515-3517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo P C Y, Leung P K L, Leung K W, Yuen K Y. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol Pathol. 2000;53:211–215. doi: 10.1136/mp.53.4.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo P C Y, Lum P N L, Wong S S Y, Cheng V C C, Yuen K Y. Cellulitis complicating lymphedema. Eur J Clin Microbiol Infect Dis. 2000;19:294–297. doi: 10.1007/s100960050478. [DOI] [PubMed] [Google Scholar]
- 27.Yuen K Y, Woo P C Y, Hui C H, Luk W K, Chen F E, Lie A K W, Liang R. Unique risk factors for bacteraemia in allogeneic bone marrow transplant recipients before and after engraftment. Bone Marrow Transplant. 1998;21:1137–1143. doi: 10.1038/sj.bmt.1701246. [DOI] [PubMed] [Google Scholar]
- 28.Yuen K Y, Seto W H, Choi C H, Ng W, Ho S W, Chau P Y. Streptococcus zooepidemicus (Lancefield group C) septicaemia in Hong Kong. J Infect. 1990;21:241–250. doi: 10.1016/0163-4453(90)93885-v. [DOI] [PubMed] [Google Scholar]